Abstract
The cooperative effects of estrogen and oocyte-derived paracrine factors (ODPFs) play critical roles in the normal development of ovarian follicles; however, the mechanism underlying this cooperation has not been well studied. The present study aimed to determine whether ODPFs affect estrogen signaling by regulating the expression of estrogen receptor (ESR) and its coregulators in mouse granulosa cells. Some transcripts encoding ESR coregulators were differentially expressed between cumulus and mural granulosa cells (MGCs). The transcript levels of ESR coregulators, including nuclear receptor corepressor 1 and activator 2, in cumulus cells were significantly suppressed by ODPFs; however, they increased when cumulus cell-oocyte complexes were treated with the transforming growth factor beta receptor I inhibitor, SB431542. Moreover, MGCs exhibited significantly higher ESR2 protein and transcript levels than those in cumulus cells. ODPFs promoted Esr2 expression in cumulus cells but had no effect on that in MGCs. Overall, regulation of the expression of ESR2 and its coregulators in cumulus cells by oocytes seems to be one of the mechanisms underlying estrogen-oocyte cooperation in well-developed antral follicles in mice.
Keywords: Cumulus cells, Estrogen receptor 2, Oocytes
Cumulus cells are a subpopulation of specialized ovarian granulosa cells that support oocyte development. Therefore, normal development and function of cumulus cells are critical for the production of functional oocytes and normal female fertility. The development and function of cumulus cells are governed by multiple intra- and extra-follicular signals, and oocytes play pivotal roles in coordinating these signals [1, 2]. For example, in cumulus cells, oocytes suppress luteinizing hormone (LH) signaling by suppressing LH receptor expression [3], but stimulate epidermal growth factor (EGF) signaling by promoting the expression of EGF receptors in mice [4]. Moreover, oocytes facilitate fibroblast growth factor (FGF) signaling in cumulus cells by suppressing the expression of the FGF receptor antagonist Spry2 [5]. Furthermore, they control the expression of transcriptional regulators, such as transcription factors and their coregulators, in granulosa cells to determine the cumulus cell lineage [6]. This suggests that oocytes regulate the expression of signal receptors and key transcription factors, as well as their coregulators, to coordinate intra- and extra-follicular signals in cumulus cells.
In addition to oocytes, estrogen is important for the normal development of ovarian follicles. Mice deficient in estrogen receptor 2 (ESR2; also known as estrogen receptor beta), a predominant estrogen receptor expressed by granulosa cells, exhibit female subfertility due to the attenuated responsiveness of granulosa cells to gonadotropins [7, 8], reduced estrogen production [9, 10], and ovulation failure, partly due to the impaired development of cumulus cells [9,10,11,12]. Additionally, estrogen participates in natriuretic peptide type C-mediated maintenance of oocyte meiotic arrest by helping to maintain the expression of natriuretic peptide receptor 2 by cumulus cells [13, 14]. Therefore, estrogen signaling is a critical regulator of cumulus cell development and female fertility.
Several studies have shown that oocytes require estrogen signaling to regulate the functions of granulosa cells. This was first shown by Otsuka et al., who reported that signals produced from oocytes are required for the amplification of follicle stimulating hormone (FSH) signaling by estrogen in diethylstilbestrol-primed rat preantral granulosa cells [15]. Furthermore, we have previously shown that estrogen signaling is required for cumulus cells to maintain their competence to undergo cumulus expansion, and that oocytes are required for this estrogen function in mice [16]. The requirement of oocyte signaling for estrogen function seems to be partly due to the ability of oocytes to modulate the effects of estrogen on cumulus cells [17]. However, the mechanism by which oocytes control estrogen signaling in cumulus cells remains largely unknown.
Estrogen signaling in cumulus cells is primarily mediated by the nuclear receptor ESR2. Cytoplasmic ESRs bind to estrogen to form dimers and then translocate into the nucleus, where they act as transcription factors. The transcriptional activity of ESRs is regulated by multiple cofactors, including nuclear receptor coactivators (NCOAs) and corepressors (NCORs). Previously, we have shown that the expression of one of the nuclear receptor coregulators, Nrip1, in cumulus cells is regulated by oocyte-derived paracrine factors (ODPFs) in mice [16]. This suggests that oocytes organize ESR coregulators to regulate estrogen signaling in cumulus cells; however, their effects on the expression of other ESR coregulators in cumulus cells have not been studied.
To understand the mechanisms by which oocytes regulate estrogen signaling in cumulus cells, we examined their effects on the expression of transcripts encoding ESR2 and its coregulators in mouse cumulus cells.
Materials and Methods
Mice
Female BDF1 mice were either purchased from Sankyo Lab Service (Tokyo, Japan) or produced and raised in the research colony of the investigators at the University of Tokyo. All experiments were conducted using three-week-old female BDF1 mice that had been injected with equine chorionic gonadotropin (ASKA Pharmaceutical, Tokyo, Japan) 42–46 h prior to the experiment. All animal protocols were approved by the Institutional Animal Care and Use Committee of the University of Tokyo.
Isolation and culture of cumulus cell-oocyte complexes (COCs), cumulus cells, mural granulosa cells (MGCs), and oocytes
COCs, MGCs, and fully grown germinal vesicle-stage oocytes were isolated and cultured as described previously [6]. Fresh cumulus cells were obtained by removing oocytes from COCs via repeated pipetting using fine-bore pipettes. Oocytectomized cumulus cells (OOXs) were produced by microsurgically removing oocytes from COCs, as described previously [18]. The culture medium used was bicarbonate buffered MEMα (Life Technologies, Carlsbad, CA, USA) containing 75 μg/ml penicillin G, 50 μg/ml streptomycin sulfate, 3 mg/ml bovine serum albumin, and 10 µM of the phosphodiesterase inhibitor milrinone (all from Sigma-Aldrich, St. Louis, MO, USA). COCs or OOXs were cultured in drops of 30 µl of culture medium in liquid paraffin for 20 h. MGCs were cultured as monolayers in 100 µl of culture medium in each well of a 96-well plate, as described previously [6]. In some experiments, FSH (100 ng/ml; R&D Systems, Minneapolis, MN, USA) or oocytes (2 oocytes/µl) were added to the culture medium. All cultures were maintained at 37ºC in 5% CO2.
Real-time polymerase chain reaction (PCR)
Total RNA was extracted using ReliaPrep RNA Cell Miniprep System (Promega K.K., Tokyo, Japan) and reverse transcribed using RevarTra Ace qPCR RT Master Mix with gDNA Remover (Toyobo, Osaka, Japan), according to the manufacturer’s protocol. Real-time PCR was performed using THUNDERBIRD qPCR Mix (Toyobo) on an ABI Step One Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The PCR primers used are listed in Table 1. The steady-state levels of transcripts were determined via the 2–ΔΔCt method using the housekeeping gene ribosomal protein L19 as an internal control [19]. A single product of the estimated size was identified by agarose gel electrophoresis for each set of primers, and a dissociation curve analysis was performed at the end of the amplification process to confirm the specificity of the PCR products. Each reaction was conducted in duplicate and real-time PCR experiments were repeated at least three times using biologically independent samples.
Table 1. PCR primer sets used in this study.
| Symbol | Accession No. | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|---|
| Slc38a3 | NM_023805 | TATCTTCGCCCCCAACATCTT | TGGGCATGATTCGGAAGTAGA | 107 |
| Lhcgr | NM_013582 | GGATAGAAGCTAATGCCTTTGACAAC | TAAAAGCACCGGGTTCAATGTATAG | 96 |
| Ncor1 | NM_001252313 | CCCATTTCCAGCGTGTTAGT | GGAGCTTCATGTTTGCTTCC | 118 |
| Ncor2 | NM_011424 | TGGGTCTGAAGACCTTACCA | TCAGCTCCTCCTTGGACAGT | 128 |
| Ncoa1 | NM_010881 | GTCACTCCAGCCCACCAC | TATTTGCCCATGGAATCTGA | 125 |
| Ncoa2 | NM_001302702 | TCACTGCATTGGCTCTTCTG | CATTCCTTGCGTTTTCTGGT | 122 |
| Ncoa3 | NM_008679 | GTGGACTCCGAGATCGGTAA | TCGCCTAGTCCACTCATCCT | 94 |
| Ncoa4 | NM_001033988 | TCGCGAGAACTCTGTCTTCA | CCACTCTGTTCCAGGGATGT | 139 |
| Ncoa5 | NM_144892 | CGAGATCATCGAGACCCTGT | TTCCTCCGGCAATAGTCATC | 122 |
| Ncoa6 | NM_019825 | GACCTGTGGCATCTGGAAAT | CAGGGCTCGTCATCCTTATT | 87 |
| Ncoa7 | NM_172495 | AATCCTTTGCCAGTCACACC | CCCATAGACATCAGGAGACCA | 143 |
| Esr2 | NM_207707 | GGGTGATTTCGAAGAGTGGA | CGTGTGAGCATTCAGCATCT | 182 |
| Rpl19 | NM_001159483 | CCGCTGCGGGAAAAAGAAG | CAGCCCATCCTTGATCAGCTT | 103 |
Western blot analysis
Western blot analysis was performed as previously described [20]. Anti-ERβ antibody (sc8974; Santa Cruz Biotechnology, Dallas, TX, USA) and peroxidase-conjugated goat anti-rabbit IgG (AP132P; Millipore, Burlington, MA, USA) were used as the primary and secondary antibodies, respectively. Signals were visualized using the ImmunoStar LD western blotting detection kit (Wako Pure Chemicals, Osaka, Japan) and a C-DiGit Blot Scanner and Image Studio for the C-DiGit system (LI-COR, NE, USA).
Statistical analysis
Statistical analyses were performed using a standard t-test in Microsoft Excel (Microsoft, Redmond, WA, USA). Statistical significance was set at p < 0.05. Values are presented as the mean ± standard error of the mean.
Results
Differential expression of transcripts encoding NCORs and NCOAs between cumulus cells and MGCs in vivo
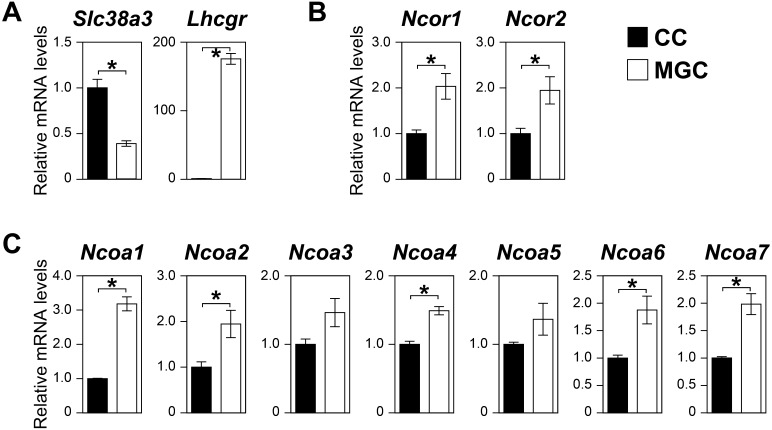
Because of the difference in the distance from oocytes, transcripts regulated by ODPFs tend to be differentially expressed between cumulus cells and MGCs [21]. Therefore, we first compared the expression levels of transcripts encoding NCORs (Ncor1 and Ncor2) and NCOAs (Ncoa1-7) between the two cell types. Efficient isolation of each cell type was confirmed by assessing the levels of known cumulus cell- and MGC-enriched transcripts, namely solute carrier family 38 member 3 (Slc38a3; encodes an amino acid transporter) and LH/choriogonadotropin receptor (Lhcgr; encodes the LH receptor) (Fig. 1A) [3, 22]. The expression levels of Ncor1, Ncor2, Ncoa1, Ncoa2, Ncoa4, Ncoa6, and Ncoa7 were significantly lower in cumulus cells than those in MGCs (Fig. 1B, C), suggesting that ODPFs suppress the expression of these transcripts in nearby cumulus cells in vivo.
Fig. 1.
Differential expression of nuclear receptor coregulators between cumulus cells and MGCs. Levels of transcripts encoding (A) SLC38a3 (cumulus cell-enriched transcript) and LHCGR (MGC-enriched transcript), (B) NCORs, and (C) NCOAs were examined using real-time PCR. Asterisks denote a significant difference (P < 0.05).
Effects of oocytes on the expression of transcripts encoding NCORs and NCOAs in cumulus cells in vitro
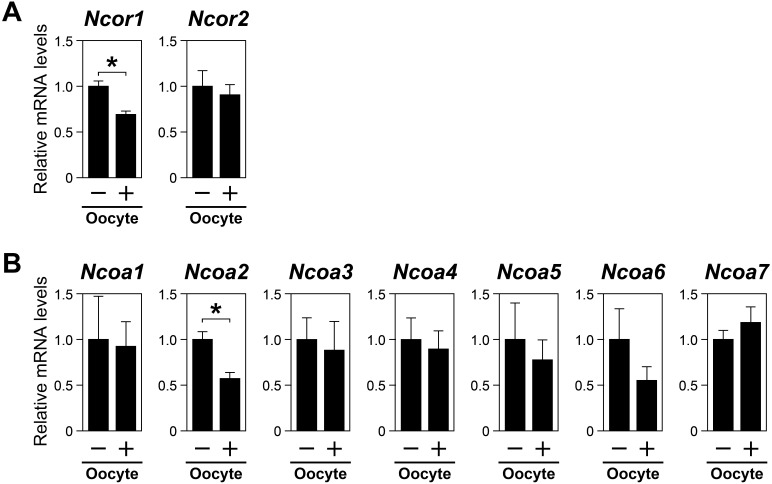
To directly assess the effects of oocytes on the expression levels of transcripts encoding NCORs and NCOAs in cumulus cells in vitro, oocytes were microsurgically removed from COCs, and the resulting oocytectomized cumulus cell complexes (OOXs) were cultured with or without oocytes (2 oocytes/µl). The relative expression levels of transcripts in cumulus cells were determined after 20 h of culture. As shown in Fig. 2, the expression levels of Ncor1 (Fig. 2A) and Ncoa2 (Fig. 2B) transcripts were significantly downregulated in OOXs cocultured with oocytes as compared with those in OOXs cultured without oocytes. This suggested that steady-state levels of these transcripts in cumulus cells were suppressed by oocytes in vitro.
Fig. 2.
Effect of oocytes on the expression levels of nuclear receptor coregulators in cumulus cells. The isolated cumulus cell complexes (OOXs) were cocultured with oocytes (2 oocytes/µl), and the levels of transcripts encoding (A) NCORs and (B) NCOAs were determined using real-time PCR. Asterisks denote a significant difference (P < 0.05).
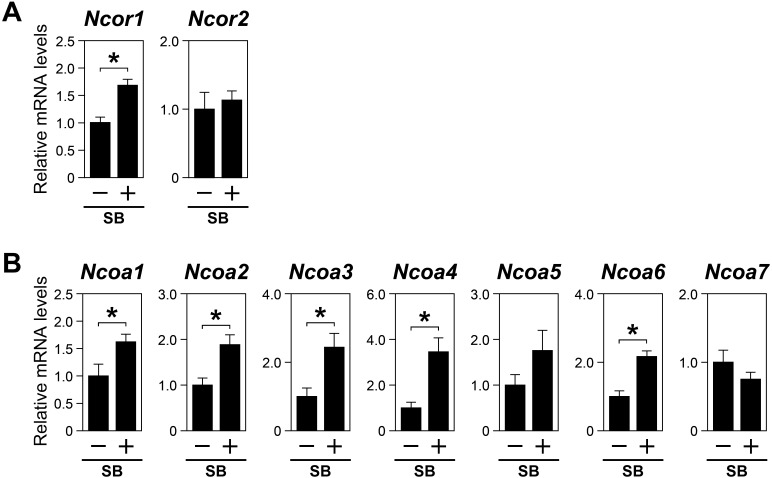
Growth differentiation factor 9 (GDF9) is an ODPF that plays a critical role in cumulus cell development. The GDF9 signal in cumulus cells is mediated by transforming growth factor beta receptor I (TGFBR1), also known as activin receptor-like kinase 5. To determine whether oocyte-derived GDF9 is required for the proper regulation of coregulators in cumulus cells, COCs were treated with the TGFBR1 inhibitor SB431542, and the expression levels of these transcripts were determined. Ncor1 and Ncoa2 expression levels were significantly higher in COCs treated with SB431542 than in those without the treatment with SB431542 (Fig. 3A, B), suggesting that ALK5-mediated ODPF signaling, including GDF9, suppressed the expression of these transcripts in vitro. In addition, while oocyte coculture had no significant effects on the levels of Ncoa1, Ncoa3, Ncoa4, and Ncoa6 (Fig. 2B), SB431542 treatment significantly promoted the expression of these transcripts (Fig. 3B). This indicated that GDF9 may suppress the expression of these transcripts in cumulus cells, but its suppressive effects may be masked by other ODPFs, which may promote the expression of these transcripts.
Fig. 3.
Effects of SB431542 on the expression levels of nuclear receptor coregulators in cumulus cells. COCs were treated with SB431542 (SB) (10 µM), and the levels of transcripts encoding (A) NCORs and (B) NCOAs were determined using real-time PCR. Asterisks denote a significant difference (P < 0.05).
Effects of oocytes on ESR2 expression in cumulus cells and MGCs
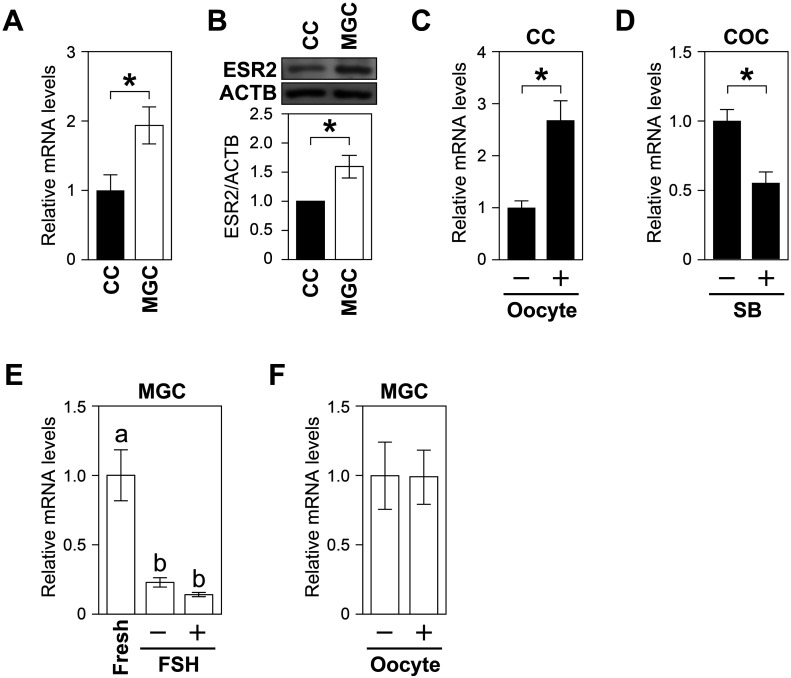
To assess the effects of oocytes on the expression of ESR2 in cumulus cells, we first compared the mRNA and protein levels of ESR2 between cumulus cells and MGCs (Fig. 4A, B), and found that both transcript and protein expression levels were significantly higher in MGCs than those in cumulus cells. These high Esr2 levels in MGCs compared to those in cumulus cells suggest that oocytes suppress the expression of Esr2 in cumulus cells in vivo; however, Esr2 expression in OOXs was significantly promoted by coculture with oocytes (Fig. 4C). Moreover, Esr2 expression in COCs significantly decreased after the treatment with SB431542 (Fig. 4D). These results suggested that Esr2 expression in cumulus cells depends on the oocyte stimulation.
Fig. 4.
Expression and regulation of ESR2 in granulosa cells. (A) mRNA and (B) protein expression levels of ESR2 in the two cell types were compared via real-time PCR and western blotting, respectively. (C) Effects of oocytes on Esr2 expression in cumulus cells. (D) Effects of SB431542 (SB) on Esr2 expression in COCs. (E) Effects of FSH (10 ng/ml) on Esr2 expression in MGCs. (F) Effects of oocytes (2 oocytes/µl) on Esr2 expression in MGCs. Asterisks or different letters (a and b) denote significant differences (P < 0.05).
It is well accepted that FSH plays a critical role in MGC development [23]. As Esr2 expression was significantly higher in MGCs than that in cumulus cells, we hypothesized that FSH may promote Esr2 expression in MGCs. Therefore, we examined the effect of FSH on the expression of Esr2 in MGCs. MGCs were cultured as monolayers with or without FSH, and the levels of Esr2 transcripts were examined. Esr2 expression significantly decreased with culture, and this reduction was not prevented by FSH supplementation (Fig. 4E). Moreover, oocyte coculture had no significant effect on Esr2 levels in MGCs (Fig. 4F). Therefore, it appears that Esr2 expression in MGCs is independent of control by FSH or oocytes, but requires some factors present within the follicles.
Discussion
Although evidence has shown that oocytes play a critical role in the modulation of estrogen signaling in cumulus cells, the underlying mechanism remains unknown. Herein, we showed that transcripts encoding NCORs and NCOAs were differentially expressed between cumulus cells and MGCs, and that some of these transcripts were regulated by oocyte-derived signals. Moreover, the mRNA and protein levels of ESR2 in cumulus cells increased after coculture with oocytes. These results suggest that mouse oocytes regulate estrogen signaling by modulating the expression of both ESR2 and its coregulators in cumulus cells. In this study, we did not examine the effects of other intrafollicular factors, such as FSH, on gene expression in cumulus cells. However, interactions among interfollicular factors are critical for the regulation of granulosa cell development, and therefore need to be assessed in future studies.
The requirement of estrogen signals in the development of ovarian follicles and female fertility is well documented in the phenotypes of estrogen receptor-knockout mice. Female mice deficient in estrogen receptor 1 (Esr1), also known as estrogen receptor alpha, are infertile because of their anovulation phenotype [24], whereas female Esr2 knockout mice are subfertile [11]. The Esr2-deficient ovary contains reduced numbers of large antral follicles and corpora lutea compared to wild-type mice, while atresia of large antral follicles is increased [9]. Moreover, Esr2-deficient MGCs exhibit an attenuated response to FSH-induced differentiation, and cumulus cells of Esr2-deficient mice fail to undergo cumulus expansion [7, 10, 25]. These deficiencies in granulosa cell development in Esr2 knockout mice probably account, at least in part, for the reduced fertility in the mutant mice. Therefore, although both ESR1 and ESR2 are indispensable for female fertility, ESR2 seems to be a critical mediator of estrogen signaling in granulosa cells. The results presented here suggest that the expression of ESR2 and its coregulators in cumulus cells is regulated by oocytes. This may provide useful insights into the mechanism by which oocytes regulate the development and function of granulosa cells as well as female fertility.
In the present study, Ncor1 and Ncoa2 were differentially expressed in cumulus cells and MGCs in vivo, and their expression in cumulus cells was suppressed by oocytes in vitro. Therefore, it is likely that the differential expression of these coregulators observed in vivo can be attributed to oocyte control. Since NCOR1 is a corepressor of nuclear receptors, whereas NCOA2 is a coactivator, it is possible that by repressing both Ncor1and Ncoa2 expression, oocytes may consequently affect the quality of estrogen signaling without affecting its intensity.
NOCR1 was originally identified as a coregulator of thyroid hormone and retinoic acid receptor [26] and is now reported to be a corepressor of various other nuclear receptors, including peroxisome proliferator-activated receptor, liver X receptor, and estrogen-related receptor alpha [27,28,29]. Although few studies have assessed the involvement of NCOR1 in ESR2 signaling, recruitment of a corepressor complex containing NCOR1 to the ESR1 promoter region by ESR2 has been reported in human breast cancer cell lines [30]. Transcripts encoding NCOR1 have been reported to be expressed in rat ovaries and bovine corpus luteum [31, 32]. Moreover, increased NCOR1 expression in granulosa cells has been implicated in the pathogenesis of polycystic ovary syndrome (PCOS) and clinical pregnancy failure in humans [33]. Interestingly, some variants of ODPF-encoding genes, such as GDF9 and bone morphogenetic protein 15, have also been reported to be associated with PCOS [34, 35]. Therefore, it may be surmised that dysregulation of oocyte suppression of NCOR1 expression in cumulus cells may partly account for the pathogenesis of PCOS, however, this requires further research.
In contrast to NCOR1, the expression and function of NCOA2 in mammalian ovaries has not been well studied. NCOA2 is a member of the steroid receptor coactivator (SRC) family, which is known as a coactivator for many members of the nuclear receptor superfamily, including those required for reproductive biology [36]. Although no direct evidence for the recruitment of NCOA2 in ESR2-dependent transcription has been reported, colocalization of ESR2 and NCOA2 has been reported in human placental syncytiotrophoblast cells [37]. Loss of the Ncoa2 gene results in subfertile phenotypes in both sexes of mice; Ncoa2-deficient males are subfertile due to defects in spermatogenesis and age-dependent testicular degeneration, whereas females exhibit subfertility mainly due to placental hypoplasia [38]. In addition, Ncoa2-deficient mice exhibit higher energy expenditure and are resistant to diet-induced obesity [39] owing to elevated glycolysis and fatty acid degradation in the liver [40]. Therefore, it is well accepted that the SRC proteins, including NCOA2, are important regulators of cellular metabolic activities [41]. Interestingly, our previous studies have identified several metabolic pathways that are promoted by ODPFs, including glycolysis, cholesterol biosynthesis, and amino acid uptake by cumulus cells [22, 42,43,44]. Accordingly, the present study showed that ODPFs suppressed Ncoa2 expression in cumulus cells. Therefore, it is possible that ODPFs regulate metabolic activities in cumulus cells by regulating Ncoa2 expression. Further analysis is warranted to assess this possibility.
In addition to Ncor1 and Ncoa2, the present analyses identified several other nuclear receptor coregulators that were also differentially expressed between cumulus cells and MGCs. Nevertheless, the mechanism by which the differential expression of these coregulators is maintained remains to be determined. The differential expression of these coregulators likely comprises differential intracellular signaling of nuclear receptors between the two granulosa cell populations, which may account for the differences in the cellular response to extracellular signals of these two cell types.
In summary, the present study provides valuable insights into the mechanism by which oocytes control estrogen signaling in cumulus cells. In addition, considering that the follicular estrogen production is also influenced by oocytes [45, 46], the interaction between oocytes and estrogen signaling seems to exist in multiple layers of signal regulation, including ligand (estrogen) production, receptor expression, and receptor coregulator expression.
Conflicts of interests
The authors declare that there are no conflicts of interests.
Acknowledgments
This work was supported by a Grant-in-Aid for Exploratory Research from the Japan Society for the Promotion of Science (Nos. 20H03124 and 21K19183 to KS).
References
- 1.Emori C, Sugiura K. Role of oocyte-derived paracrine factors in follicular development. Anim Sci J 2014; 85: 627–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sugiura K, Konuma R, Kano K, Naito K. Role of oocyte-derived factors in ovarian follicular development and ovulation. J Mamm Ova Res 2011; 28: 8–17. [Google Scholar]
- 3.Eppig JJ, Wigglesworth K, Pendola F, Hirao Y. Murine oocytes suppress expression of luteinizing hormone receptor messenger ribonucleic acid by granulosa cells. Biol Reprod 1997; 56: 976–984. [DOI] [PubMed] [Google Scholar]
- 4.Su YQ, Sugiura K, Li Q, Wigglesworth K, Matzuk MM, Eppig JJ. Mouse oocytes enable LH-induced maturation of the cumulus-oocyte complex via promoting EGF receptor-dependent signaling. Mol Endocrinol 2010; 24: 1230–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sugiura K, Su YQ, Li Q, Wigglesworth K, Matzuk MM, Eppig JJ. Fibroblast growth factors and epidermal growth factor cooperate with oocyte-derived members of the TGFbeta superfamily to regulate Spry2 mRNA levels in mouse cumulus cells. Biol Reprod 2009; 81: 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emori C, Ito H, Fujii W, Naito K, Sugiura K. Oocytes suppress FOXL2 expression in cumulus cells in mice. Biol Reprod 2020; 103: 85–93. [DOI] [PubMed] [Google Scholar]
- 7.Deroo BJ, Rodriguez KF, Couse JF, Hamilton KJ, Collins JB, Grissom SF, Korach KS. Estrogen receptor beta is required for optimal cAMP production in mouse granulosa cells. Mol Endocrinol 2009; 23: 955–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez KF, Couse JF, Jayes FL, Hamilton KJ, Burns KA, Taniguchi F, Korach KS. Insufficient luteinizing hormone-induced intracellular signaling disrupts ovulation in preovulatory follicles lacking estrogen receptor-beta. Endocrinology 2010; 151: 2826–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emmen JM, Couse JF, Elmore SA, Yates MM, Kissling GE, Korach KS. In vitro growth and ovulation of follicles from ovaries of estrogen receptor (ER)alpha and ERbeta null mice indicate a role for ERbeta in follicular maturation. Endocrinology 2005; 146: 2817–2826. [DOI] [PubMed] [Google Scholar]
- 10.Couse JF, Yates MM, Deroo BJ, Korach KS. Estrogen receptor-beta is critical to granulosa cell differentiation and the ovulatory response to gonadotropins. Endocrinology 2005; 146: 3247–3262. [DOI] [PubMed] [Google Scholar]
- 11.Krege JH, Hodgin JB, Couse JF, Enmark E, Warner M, Mahler JF, Sar M, Korach KS, Gustafsson JA, Smithies O. Generation and reproductive phenotypes of mice lacking estrogen receptor beta. Proc Natl Acad Sci USA 1998; 95: 15677–15682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng G, Weihua Z, Mäkinen S, Mäkelä S, Saji S, Warner M, Gustafsson JA, Hovatta O. A role for the androgen receptor in follicular atresia of estrogen receptor beta knockout mouse ovary. Biol Reprod 2002; 66: 77–84. [DOI] [PubMed] [Google Scholar]
- 13.Zhang M, Su YQ, Sugiura K, Xia G, Eppig JJ. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science 2010; 330: 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang M, Su YQ, Sugiura K, Wigglesworth K, Xia G, Eppig JJ. Estradiol promotes and maintains cumulus cell expression of natriuretic peptide receptor 2 (NPR2) and meiotic arrest in mouse oocytes in vitro. Endocrinology 2011; 152: 4377–4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otsuka F, Moore RK, Wang X, Sharma S, Miyoshi T, Shimasaki S. Essential role of the oocyte in estrogen amplification of follicle-stimulating hormone signaling in granulosa cells. Endocrinology 2005; 146: 3362–3367. [DOI] [PubMed] [Google Scholar]
- 16.Sugiura K, Su YQ, Li Q, Wigglesworth K, Matzuk MM, Eppig JJ. Estrogen promotes the development of mouse cumulus cells in coordination with oocyte-derived GDF9 and BMP15. Mol Endocrinol 2010; 24: 2303–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emori C, Wigglesworth K, Fujii W, Naito K, Eppig JJ, Sugiura K. Cooperative effects of 17β-estradiol and oocyte-derived paracrine factors on the transcriptome of mouse cumulus cells. Endocrinology 2013; 154: 4859–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buccione R, Vanderhyden BC, Caron PJ, Eppig JJ. FSH-induced expansion of the mouse cumulus oophorus in vitro is dependent upon a specific factor(s) secreted by the oocyte. Dev Biol 1990; 138: 16–25. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 20.Sumitomo J, Emori C, Matsuno Y, Ueno M, Kawasaki K, Endo TA, Shiroguchi K, Fujii W, Naito K, Sugiura K. Mouse oocytes suppress miR-322-5p expression in ovarian granulosa cells. J Reprod Dev 2016; 62: 393–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiura K, Eppig JJ. Society for Reproductive Biology Founders’ Lecture 2005. Control of metabolic cooperativity between oocytes and their companion granulosa cells by mouse oocytes. Reprod Fertil Dev 2005; 17: 667–674. [DOI] [PubMed] [Google Scholar]
- 22.Eppig JJ, Pendola FL, Wigglesworth K, Pendola JK. Mouse oocytes regulate metabolic cooperativity between granulosa cells and oocytes: amino acid transport. Biol Reprod 2005; 73: 351–357. [DOI] [PubMed] [Google Scholar]
- 23.Diaz FJ, Wigglesworth K, Eppig JJ. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J Cell Sci 2007; 120: 1330–1340. [DOI] [PubMed] [Google Scholar]
- 24.Schomberg DW, Couse JF, Mukherjee A, Lubahn DB, Sar M, Mayo KE, Korach KS. Targeted disruption of the estrogen receptor-alpha gene in female mice: characterization of ovarian responses and phenotype in the adult. Endocrinology 1999; 140: 2733–2744. [DOI] [PubMed] [Google Scholar]
- 25.Dupont S, Krust A, Gansmuller A, Dierich A, Chambon P, Mark M. Effect of single and compound knockouts of estrogen receptors alpha (ERalpha) and beta (ERbeta) on mouse reproductive phenotypes. Development 2000; 127: 4277–4291. [DOI] [PubMed] [Google Scholar]
- 26.Hörlein AJ, Näär AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Söderström M, Glass CK, et al. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 1995; 377: 397–404. [DOI] [PubMed] [Google Scholar]
- 27.Perissi V, Jepsen K, Glass CK, Rosenfeld MG. Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 2010; 11: 109–123. [DOI] [PubMed] [Google Scholar]
- 28.Mottis A, Mouchiroud L, Auwerx J. Emerging roles of the corepressors NCoR1 and SMRT in homeostasis. Genes Dev 2013; 27: 819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fan W, Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Curr Opin Cell Biol 2015; 33: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartella V, Rizza P, Barone I, Zito D, Giordano F, Giordano C, Catalano S, Mauro L, Sisci D, Panno ML, Fuqua SA, Andò S. Estrogen receptor beta binds Sp1 and recruits a corepressor complex to the estrogen receptor alpha gene promoter. Breast Cancer Res Treat 2012; 134: 569–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velez LM, Heber MF, Ferreira SR, Abruzzese GA, Reynoso RM, Motta AB. Effect of hyperandrogenism on ovarian function. Reproduction 2015; 149: 577–585. [DOI] [PubMed] [Google Scholar]
- 32.Rekawiecki R, Kowalik MK, Kotwica J. The expression of progesterone receptor coregulators mRNA and protein in corpus luteum and endometrium of cows during the estrous cycle. Anim Reprod Sci 2017; 183: 102–109. [DOI] [PubMed] [Google Scholar]
- 33.Qu F, Wang FF, Yin R, Ding GL, El-Prince M, Gao Q, Shi BW, Pan HH, Huang YT, Jin M, Leung PC, Sheng JZ, Huang HF. A molecular mechanism underlying ovarian dysfunction of polycystic ovary syndrome: hyperandrogenism induces epigenetic alterations in the granulosa cells. J Mol Med (Berl) 2012; 90: 911–923. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, Zhou S, Wang J, Liu J, Ni F, Yan J, Mu Y, Cao Y, Ma X. Identification of novel missense mutations of GDF9 in Chinese women with polycystic ovary syndrome. Reprod Biomed Online 2010; 21: 344–348. [DOI] [PubMed] [Google Scholar]
- 35.Persani L, Rossetti R, Di Pasquale E, Cacciatore C, Fabre S. The fundamental role of bone morphogenetic protein 15 in ovarian function and its involvement in female fertility disorders. Hum Reprod Update 2014; 20: 869–883. [DOI] [PubMed] [Google Scholar]
- 36.Szwarc MM, Kommagani R, Lessey BA, Lydon JP. The p160/steroid receptor coactivator family: potent arbiters of uterine physiology and dysfunction. Biol Reprod 2014; 91: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SC, Park MN, Lee YJ, Joo JK, An BS. Interaction of steroid receptor coactivators and estrogen receptors in the human placenta. J Mol Endocrinol 2016; 56: 239–247. [DOI] [PubMed] [Google Scholar]
- 38.Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol 2002; 22: 5923–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Picard F, Géhin M, Annicotte J, Rocchi S, Champy MF, O’Malley BW, Chambon P, Auwerx J. SRC-1 and TIF2 control energy balance between white and brown adipose tissues. Cell 2002; 111: 931–941. [DOI] [PubMed] [Google Scholar]
- 40.Jeong JW, Kwak I, Lee KY, White LD, Wang XP, Brunicardi FC, O’Malley BW, DeMayo FJ. The genomic analysis of the impact of steroid receptor coactivators ablation on hepatic metabolism. Mol Endocrinol 2006; 20: 1138–1152. [DOI] [PubMed] [Google Scholar]
- 41.Stashi E, York B, O’Malley BW. Steroid receptor coactivators: servants and masters for control of systems metabolism. Trends Endocrinol Metab 2014; 25: 337–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sugiura K, Pendola FL, Eppig JJ. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol 2005; 279: 20–30. [DOI] [PubMed] [Google Scholar]
- 43.Sugiura K, Su YQ, Diaz FJ, Pangas SA, Sharma S, Wigglesworth K, O’Brien MJ, Matzuk MM, Shimasaki S, Eppig JJ. Oocyte-derived BMP15 and FGFs cooperate to promote glycolysis in cumulus cells. Development 2007; 134: 2593–2603. [DOI] [PubMed] [Google Scholar]
- 44.Su YQ, Sugiura K, Wigglesworth K, O’Brien MJ, Affourtit JP, Pangas SA, Matzuk MM, Eppig JJ. Oocyte regulation of metabolic cooperativity between mouse cumulus cells and oocytes: BMP15 and GDF9 control cholesterol biosynthesis in cumulus cells. Development 2008; 135: 111–121. [DOI] [PubMed] [Google Scholar]
- 45.Vanderhyden BC, Cohen JN, Morley P. Mouse oocytes regulate granulosa cell steroidogenesis. Endocrinology 1993; 133: 423–426. [DOI] [PubMed] [Google Scholar]
- 46.Vitt UA, Hayashi M, Klein C, Hsueh AJ. Growth differentiation factor-9 stimulates proliferation but suppresses the follicle-stimulating hormone-induced differentiation of cultured granulosa cells from small antral and preovulatory rat follicles. Biol Reprod 2000; 62: 370–377. [DOI] [PubMed] [Google Scholar]