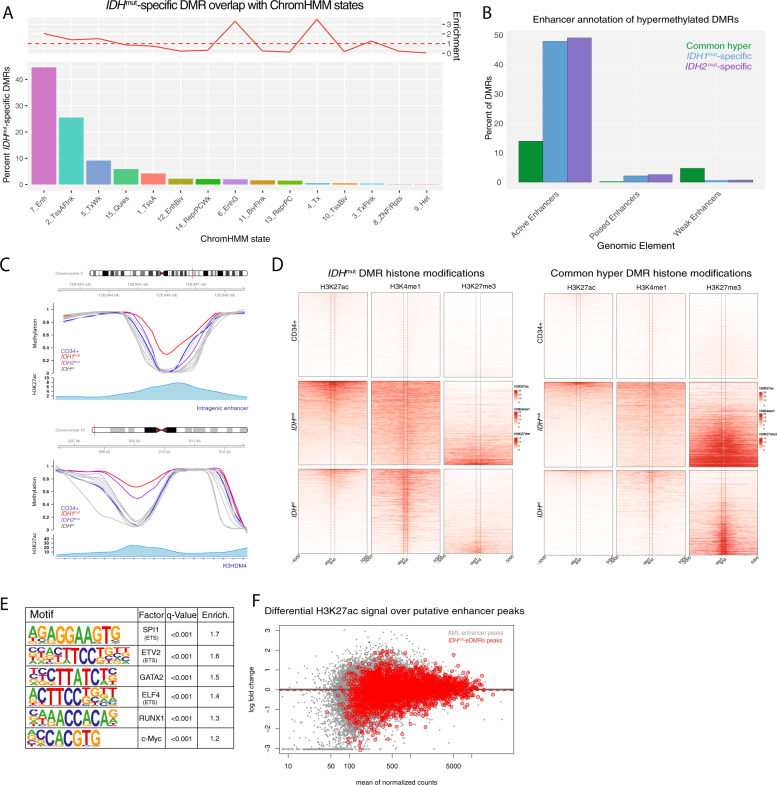
Fig. 5. IDHmut-specific DMRs are enriched for putative enhancers.
A Distribution of ChromHMM chromatin states from CD34+ cells represented in IDHmut-specific DMRs. Enrichment of chromatin states within IDHmut-specific DMRs is shown with respect to the frequency of states overlapping regions of common CpG island hypermethylation. B Enhancer-based annotation of common hypermethylated regions, IDH1mut, and IDH2mut DMRs, where DMRs overlapping an H3K27ac peak alone or in combination with H3K4me1 were called active enhancers, those overlapping a H3K27ac peak in combination with H3K27me3 were called poised enhancers, and those overlapping a H3K4me1 alone were called weak enhancers. C Examples of intragenic and genic enhancer regions exhibiting IDH1mut, IDH2mut, or IDH1/2mut hypermethylation compared with CD34+ cells and other AML subtypes. D Heatmap of enhancer histone modifications and heterochromatin modifications over IDHmut-specific DMRs (left) and generic hypermethylation (right) in CD34+ cells (N = 4 H3K27ac, N = 7 H3K3me1, and N = 7 H3K27me3), IDHmut AML (n = 3), and IDHwt AML samples (N = 9 H3K27ac, N = 10 H3K3me1, and N = 24 H3K27me3). E HOMER motif enrichment analysis of IDH1/2mut-specific DMRs with respect to a background set of generically hypermethylated regions. F Differential active enhancer signal (H3K27ac) for all AML-associated putative enhancers (black points) compared with putative enhancers intersecting an IDH1/2mut-specific DMR (red points).