Abstract
SETD5 mutations were identified as the genetic causes of neurodevelopmental disorders. While the whole-body knockout of Setd5 in mice leads to embryonic lethality, the role of SETD5 in adult stem cell remains unexplored. Here, a critical role of Setd5 in hematopoietic stem cells (HSCs) is identified. Specific deletion of Setd5 in hematopoietic system significantly increased the number of immunophenotypic HSCs by promoting HSC proliferation. Setd5-deficient HSCs exhibited impaired long-term self-renewal capacity and multiple-lineage differentiation potentials under transplantation pressure. Transcriptome analysis of Setd5-deficient HSCs revealed a disruption of quiescence state of long-term HSCs, a cause of the exhaustion of functional HSCs. Mechanistically, SETD5 was shown to regulate HSC quiescence by mediating the release of promoter-proximal paused RNA polymerase II (Pol II) on E2F targets in cooperation with HCF-1 and PAF1 complex. Taken together, these findings reveal an essential role of SETD5 in regulating Pol II pausing-mediated maintenance of adult stem cells.
Subject terms: Haematopoietic stem cells, Cell biology
Introduction
Stem cells maintain their self-renewal and differentiation through tight regulation of gene expression patterns at the transcriptional and epigenetic levels. SETD5 is a member of SET domain-containing histone lysine methyltransferases (KMTs) family [1, 2]. Like its orthologs including yeast Set3p and Drosophila UpSET [3, 4], SETD5 has been shown to lack methyltransferase activity in several studies [5–7]. Interestingly, de novo mutations in the Setd5 gene have been identified as the genetic causes of intellectual disability and autism spectrum disorders [8–11]. More recently, SETD5 was identified as a key mediator of pancreatic cancer resistance to MEK1/2 inhibition targeted therapy [7], suggesting that its physiological and pathological roles might be diverse and tissue or cell type specific.
Setd5 is broadly expressed in mouse tissues. Deletion of Setd5 resulted in embryonic lethality at E10.5-11.5 [5], while Setd5 haploinsufficiency led to disrupted developmental gene expression and cognition [6]. Interestingly, in the placenta of Setd5-deficient mice, only maternal-derived mature red blood cells but not embryo-derived nucleated red blood cells were observed. In addition, the number of CD41+ early hematopoietic cells declined in the blood island of Setd5 homozygous knockout mice [5]. These data suggested a potential role of SETD5 in hematopoiesis.
Hematopoietic stem and progenitor cells (HSPCs) maintain hematopoietic homeostasis through well-orchestrated balance between self-renewal and differentiation. The majority of HSCs exist in a quiescent state to prevent HSC exhaustion [12, 13]. Transcriptionally engaged RNA Pol II experiences a temporary stalling after initiation on many metazoan genes and was considered as an important regulatory process in gene transcription [14, 15]. Despite it was reported that Pol II pausing modulates hematopoietic stem cell generation in zebrafish [16], the in vivo effect of Pol II pausing on the maintenance of HSCs in mammals remains to be further elucidated.
Here in this study, we explored the role of Setd5 in hematopoiesis and found that depletion of Setd5 in hematopoietic system significantly increased immune-phenotypically defined HSCs with impaired HSC quiescence at the single cell level. Mechanistic studies revealed a novel transcriptional mechanism of SETD5 in governing HSC quiescence by Pol II pausing-mediated regulation of cell cycle activating genes.
Materials and methods
Animals
Setd5 floxed (Setd5fl/fl) mice were generated by inserting loxp sites flanking exons 3-6, which when deleted, would result in a frame-shift and form a premature stop codon in the reading frame. To induce Mx1-Cre expression, adult mice were intraperitoneally injected (ip) with 10 μg/g polyinosinic-polycytidylic acid (pIpC, InvivoGen) 3 times every 48 h. All primers and antibodies used in this study are listed in Supplemental Table 1. All animal research was approved by the Institutional Animal Care and Use Committee of the State Key Laboratory of Experimental Hematology.
Additional methods
Additional methods are provided in Supplemental Information.
Results
Genetic ablation of Setd5 causes phenotypic HSPC expansion
To investigate the biological roles of Setd5 in hematopoietic system, we generated Vav-Cre;Setd5fl/fl (Setd5CKO) mice and confirmed the deletion efficiency of Setd5 (Figure S1A-E). Setd5CKO mice had slightly higher monocyte count in peripheral blood (PB) and increased spleen (SP) weight but with comparable cellularity than those of the controls (Figure S1F-H). We observed modest decreased frequencies of B and T lymphocytes in the PB and bone marrow (BM) and a significant increased frequency of granulocytes/monocytes in PB of Setd5CKO mice when compared with the control animals (Fig. 1A, Figure S1I).
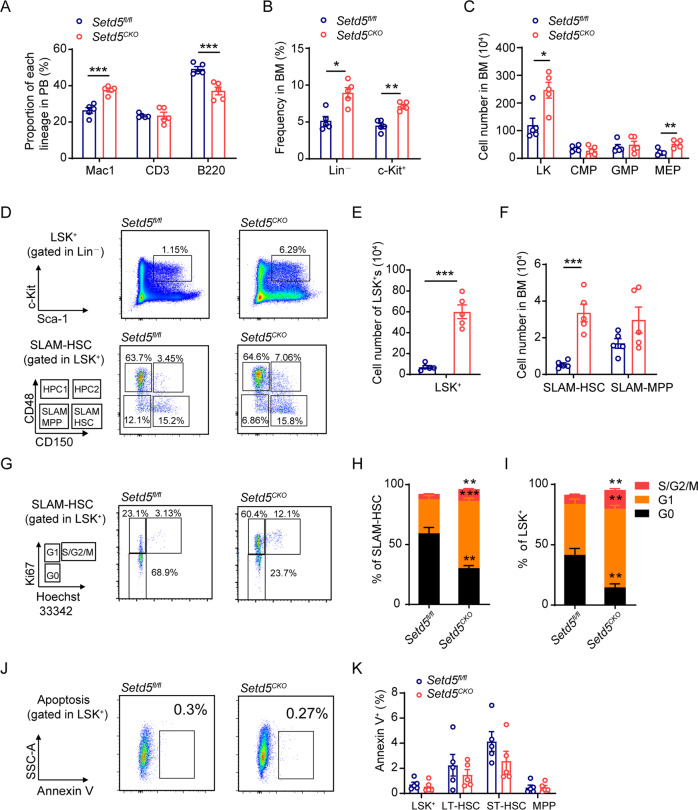
Fig. 1. Setd5 deficiency causes phenotypic HSPC expansion.
A FACS analysis of T, B, and myeloid cells frequency in PB cells; n = 5. B Relative frequency of immature cells (Lin– and c-Kit+) in BM. Lin: lineage cocktail; n = 5. C The absolute cell number of HPC (LK: Lin–c-Kit+Sca-1–), CMP (Lin–c-Kit+Sca1–CD34+CD16/32low), GMP (Lin–c-Kit+Sca1–CD34+CD16/32high) and MEP (Lin–c-Kit+Sca1–CD34–CD16/32low) populations in Setd5fl/fl and Setd5CKO mice; n = 5. D–F FACS analysis of LSK+s (Lin–Sca1+c-Kit+) and SLAM-HSCs (Lin–Sca1+c-Kit+CD150+CD48–) and absolute cell number in BM. SLAM-MPP: Lin–Sca1+c-Kit+CD150–CD48–, HPC1: Lin–Sca1+c-Kit+CD150–CD48+, HPC2: Lin–Sca1+c-Kit+CD150+CD48+; n = 5. G Representative FACS profiles of Ki67 staining of SLAM-HSCs. H, I The frequencies of G0, G1, S/G2/M phases in SLAM-HSCs and LSK+s are shown; n = 4. J, K Apoptosis analysis of HSPCs in Setd5fl/fl and Setd5CKO mice, LT-HSC: Lin–Sca1+c-Kit+CD34–Flt3low, ST-HSC: Lin–Sca1+c-Kit+CD34+Flt3low, MPP: Lin–Sca1+c-Kit+CD34+Flt3+; n = 5. Data are represented as mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001.
Examination of the hematopoietic progenitor cells (HPCs, LK) revealed that the relative frequency of Lin– and c-Kit+ cells was significantly increased in the BM of Setd5CKO mice (Fig. 1B). The cell number of LKs and megakaryocyte–erythroid progenitors (MEP) in the BM was also increased, while common myeloid progenitors (CMP) and granulocyte-monocyte progenitors (GMP) remained unchanged (Fig. 1C, Figure S2A). We also determined the cell number of common lymphoid progenitors (CLPs), which showed no difference between the two groups (Figure S2B-C). Colony forming assay revealed that the frequencies of the functional HPCs remained unchanged with Setd5 deletion (Figure S2D).
We further examined the effect of Setd5 deficiency at HSC levels. CD150 and CD48 [17] were employed to evaluate SLAM-HSCs. LSK+, SLAM-HSC, HPC1 and HPC2 showed a dramatic increase in the cell number by ~5 to ~10 fold in Setd5CKO mice (Fig. 1D-F, S2E). This observation was further corroborated with another set of HSC markers such as CD34 and Flk2 [18, 19], which displayed a dramatic increase in the cell number of LT-HSCs (long-term HSCs), ST-HSCs (short-term HSCs) and MPPs (multipotential progenitors) (Figure S2F-G). Additionally, we observed a decreased expression level of Flk2 and an increase in CD150 and CD48 in Setd5-deficient LSK+ cells (Figure S2H-J).
The increased numbers of HSCs prompted us to further analyze the cell cycle and apoptosis of HSCs. Ki67 staining revealed that significantly higher portions of SLAM-HSC, LSK+ and LK cells underwent active cycling in Setd5CKO than Setd5fl/fl mice (Fig. 1G-I, S2K). Moreover, increased BrdU incorporations in Setd5CKO LSK+ cells and LT-HSCs were observed (LSK+ cells: 42% BrdU+ of Setd5CKO versus 14% of controls, and LT-HSCs: 14% of Setd5CKO versus 5% of controls) (Figure S2L-M), as seen in LK cells. The apoptotic status shows no differences in LT-HSC and LSK+ cells between the two groups (Fig. 1J, K). Together, these observations indicate that deletion of Setd5 increases the number of immune-phenotypical-defined HSCs via promoting cells into S phase.
Setd5 deletion by Vav-Cre impairs repopulation capacity of HSCs
The exit of LT-HSCs from the G0 stage is often associated with HSC exhaustion [12, 20, 21]. In considering this, we further performed competitive serial transplantation experiments by transplanting 1000 sorted-LSK+ cells from Setd5CKO or Setd5fl/fl BM (CD45.2) together with competitor BM cells (CD45.1) into lethally irradiated recipient mice (CD45.1) (Fig. 2A). Setd5CKO derived cells showed a progressive decrease in repopulation capacity than that of Setd5fl/fl in primary and secondary transplantation (Fig. 2B-E). Moreover, Setd5-deficient cells showed an impaired lineage differentiation ability in PB in primary recipient mice (Fig. 2F). Significantly, although Setd5CKO derived mature cells represented only less than 20% of the total bone marrow cells, Setd5CKO derived LSK+s could effectively reconstitute the HSC compartments (LSK+s, SLAM-HSCs, SLAM-MPPs) in the primary recipients but lost the self-renewal ability in secondary transplantation when compared with that of Setd5fl/fl derived LSK+s (Fig. 2G). The tertiary transplantation also showed an even lower engraftment in Setd5CKO recipient PB cells (Figure S3A-D).
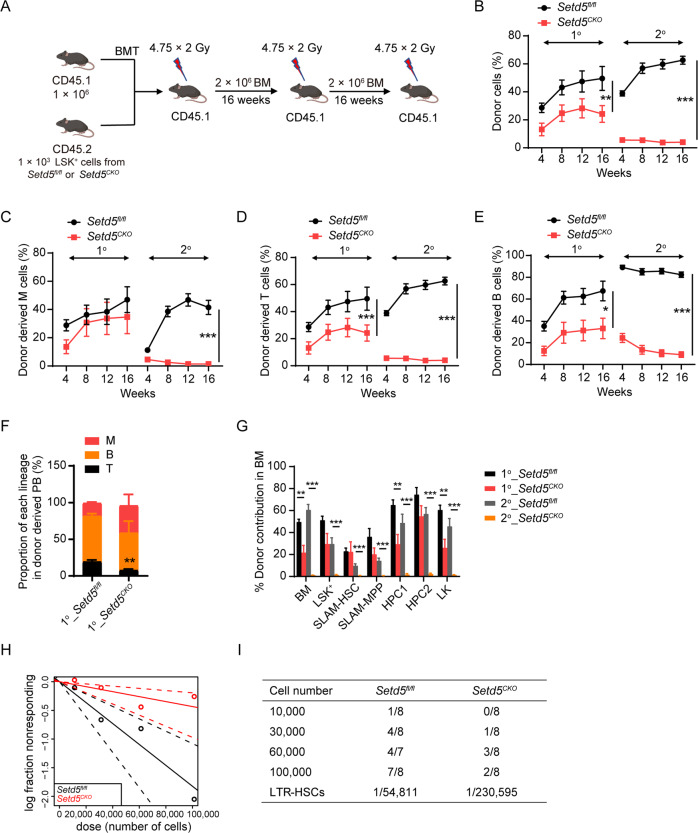
Fig. 2. Setd5 deletion by Vav-Cre impairs repopulation capacity of HSCs.
A–E Serial competitive repopulation assays with Setd5fl/fl and Setd5CKO LSK+ cells. Strategy for serial competitive repopulation assays (A), BMT: BM transplantation. Quantification of donor-derived (CD45.2) cells (B), donor-derived myeloid (M: CD11b+) cells (C), donor-derived T (CD3+) cells (D) and B (B220+) cells (E) in PB at indicated time points in primary (n = 9) and secondary (n = 10) recipients. F Proportion of each lineage in donor derived PB cells; n = 5. G Donor contribution of indicated cell populations in BM cells of primary and secondary recipient mice at indicated times; n = 5. H Poisson statistical analysis from the limiting dilution assay. LTR-HSCs: long-term repopulating (LTR)-HSC. Symbols represent the percentage of negative mice for each dose of cells. Solid lines indicate the best-fit linear model for each dosage. Dotted lines represent 95% confidence intervals. I Frequencies of functional HSCs were calculated according to Poisson statistics. Data are represented as Mean ± SEM; * P < 0.05, ** P < 0.01, *** P < 0.001.
Additionally, we performed the whole BM competitive transplantation assay which showed a similar result with LSK+ competitive transplantation assay (Figure S3E-J), with the chimerism of LSK+s, LT-HSCs and MPPs significantly decreased in Setd5CKO recipients (Figure S3K). Homing assay showed that Setd5CKO BM cells were able to migrate to the BM of recipients as efficiently as the control cells (Figure S3L). In summary, these results indicate that under transplantation pressure, immune-phenotypical-defined Setd5CKO HSCs may have an intrinsic defect to differentiate into committed progenitors and subsequently mature blood cells.
Since HSC subtypes defined by immunophenotype may not accurately represent functional HSCs, we further assessed the absolute number of functional HSCs with limiting dilution assay and observed about 4-fold decrease of HSC number in Setd5CKO mice (Fig. 2H, I). Thus, Vav-Cre mediated Setd5 deletion significantly impaired repopulation capacity of HSCs and decreased number of functional HSCs.
Setd5 regulates HSC pool in adult hematopoietic cells and plays an intrinsic role in HSCs
Since Vav-Cre mediated Setd5 deletion occurred at E11.5 [22], and the defect of Setd5CKO HSCs may be due to a long-term accumulated consequence from embryo stage, we generated Mx1-Setd5fl/fl mice to determine the role of Setd5 in adult hematopoiesis and confirmed efficient deletion of Setd5 (Mx1-Setd5fl/fl with pIpC treatment was referred to as Setd5IKO) (Fig. 3A, S4A-B). Similar to Setd5CKO mice, we found an increase in the cell number of LSK+s and SLAM-HSCs in Setd5IKO mice, so as the LT-HSCs, ST-HSCs and MPPs (Fig. 3B, C, S4C). BrdU incorporation assay also revealed that higher percentages of Setd5IKO HSCs and LSK+s were in S-phase than those of the controls (Fig. 3D, E), while the cell number of HPCs remained similar in two groups (Figure S4D). These experiments demonstrated that Setd5 is required for the maintenance of the quiescent state of HSCs in adult hematopoiesis.
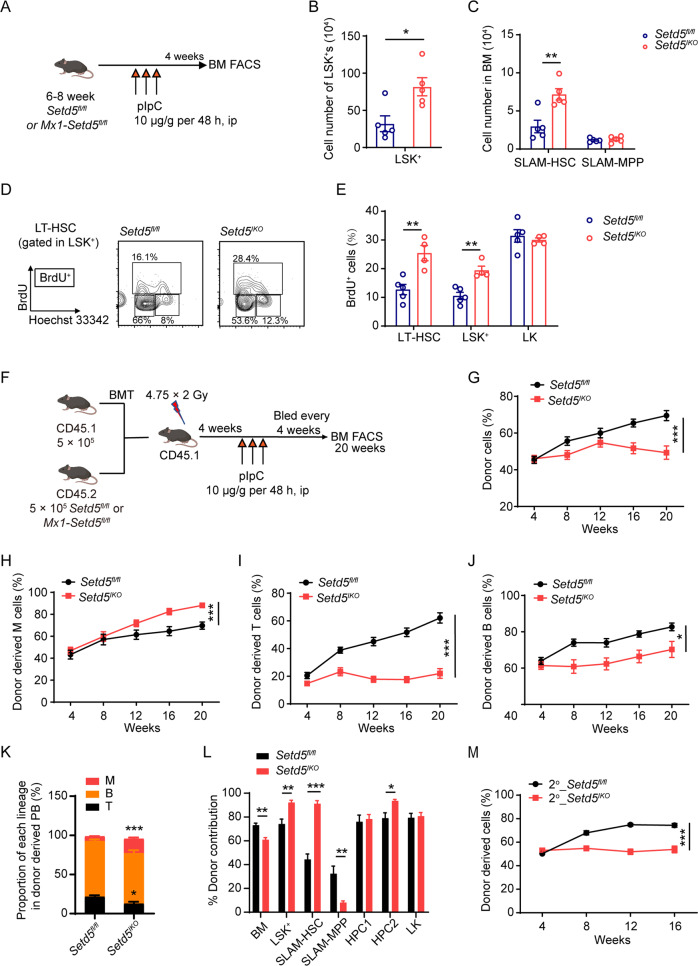
Fig. 3. Setd5 regulates HSC pool in adult hematopoietic cells and plays an intrinsic role in HSCs.
A Schematic diagram of Mx1-Cre mediated Setd5 knockout in adult hematopoiesis. B, C The absolute number of LSK+s and SLAM-HSCs in Setd5fl/fl and Setd5IKO mice; n = 5. D, E BrdU incorporation of LSK+ and HSC populations and representative flow cytometry histogram of LT-HSC populations of BrdU incorporation 24 h after injection; n = 4. F Schematic diagram for transplantation assay with Setd5fl/fl and Setd5IKO BM cells. G–J Quantification of donor-derived (CD45.2) cells in the PB of recipient animals at indicated time points; n = 9. K Proportion of each lineage in donor derived PB cells; n = 5. L Donor contribution of indicated cell populations in BM cells of recipient mice 20 weeks after transplantation; n = 5. M Percentage of donor-derived cells in the PB of Setd5fl/fl and Setd5IKO secondary recipients at the indicated time points; n = 9. Mean ± SEM; * P < 0.05, ** P < 0.01, *** P < 0.001.
To rule out a possible contribution of non-hematopoietic excision of Setd5 by Mx1-Cre to the phenotypes of HSCs, non-competitive BM transplantation (nCBMT) was performed before pIpC injection (Figure S4E). Sixteen weeks after depletion of Setd5, the absolute cell numbers of LSK+, SLAM-HSC, LK, and MEP were significantly increased in Setd5IKO recipients (Figure S4F-H). BrdU incorporations of donor derived LSK+ cells were also increased in Setd5IKO chimeric mice (Figure S4I). These results showed that Setd5 regulates HSC maintenance in a cell-autonomous manner.
To ascertain whether acute deletion of Setd5 in adult mice by pIpC injection would also affect HSC repopulation ability, we performed competitive transplantation with BM cells from Setd5fl/fl and Mx1-Setd5fl/fl before pIpC treatment (Fig. 3F). pIpC was injected after transplantation and Setd5 was efficiently deleted in primary and secondary Setd5IKO recipients (Figure S4J). A progressive decrease in PB chimerism was observed in primary and secondary transplantation. Lymphoid lineage reconstitution was impaired while myeloid lineage reconstitution was increased in Setd5IKO recipients than those of Setd5fl/fl in serial transplantations (Figs. 3G-J and 3M, S4K-M), implying a differentiation bias towards myeloid lineage at the expense of lymphoid lineage of Setd5IKO HSCs (Fig. 3K, S4N). A higher chimerism of LSK+s and SLAM-HSCs was detected in Setd5IKO recipient BM cells (Fig. 3L, S4O). In summary, these results indicated that under transplantation pressure, Setd5 deletion by Mx1-Cre could also affect HSC repopulation ability but enhanced the proliferation of HSCs and promoted myeloid lineage differentiation.
Transcriptome profiling of Setd5-deficient HSCs reveals altered stem cell property and cell cycle signature
To dissect the molecular mechanism of Setd5 in regulating HSC function, we performed bulk RNA-seq (RNA sequencing) of SLAM-HSCs derived from Setd5CKO and control mice, and 443 differentially expressed genes (DEGs) were found (Fig. 4A, Table S2). These DEGs were significantly enriched in cell cycle and Pol II transcription activity (Figure S5A). Additional Gene Set Enrichment Analysis (GSEA) revealed a significantly lower expression of LT-HSC signature genes and a decreased quiescence state, accompanied with a higher expression of intermediate and late progenitor signature genes, as well as an enrichment of cell cycle and E2F targets in Setd5CKO SLAM-HSCs (Fig. 4B). Setd5CKO SLAM-HSCs also exhibited an increased bias towards age-associated patterns accompanied with a decreased lymphoid/myeloid signature (Fig. 4B). Among these DEGs, lymphoid differentiation and pluripotency maintenance related genes, such as Meg3, Flt3 and Foxo1 [23–25] were significantly downregulated, so as p21 and p57. The expression of Vwf, which was involved in specifying myeloid fate [26, 27], was increased in Setd5CKO HSCs (Fig. 4C). We also performed the transcriptome analysis of SLAM-HSCs in Setd5fl/fl and Setd5IKO. 145 upregulated and 185 downregulated genes were found (Figure S5B, Table S3). These DEGs were also enriched in DNA replication and cell cycle (Figure S5C). GSEA also exhibited a decreased LT-HSC and quiescence signature, accompanied with an age-associated bias in Setd5IKO SLAM-HSCs (Figure S5D). Similar expression patterns of Vwf, Foxo1, Gata3, p21 and p57 were also observed in Setd5IKO SLAM-HSCs (Figure S5E). Taken together, the transcriptional alterations triggered by Setd5-deficiency suggested that the absence of Setd5 induced SLAM-HSCs to exit from quiescence phase and led to the exhaustion of functional HSCs.
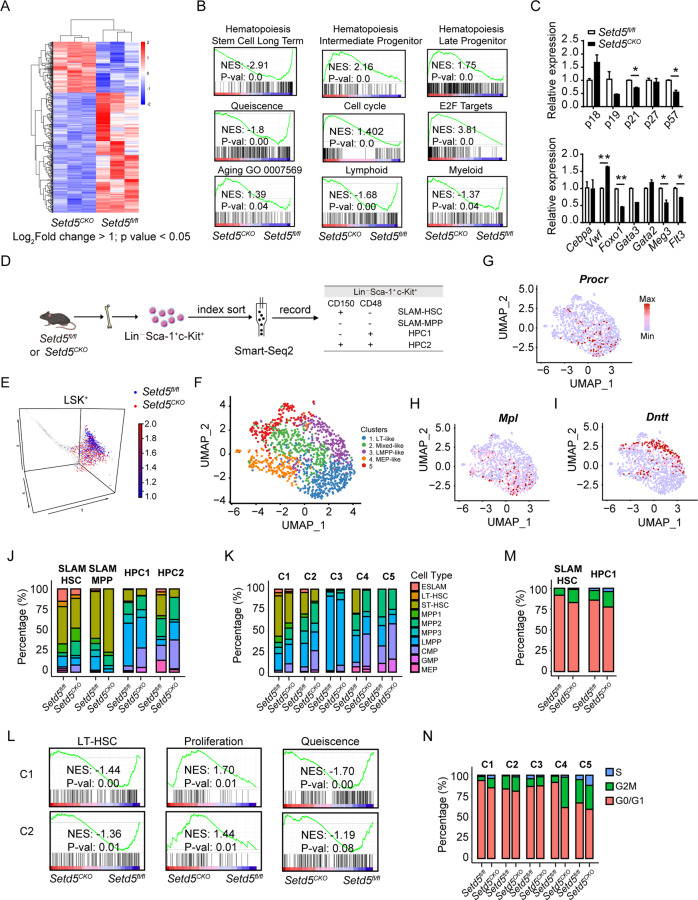
Fig. 4. Transcriptome profiling of Setd5-deficient HSCs reveals altered stem cell property and cell cycle signature.
A Heat map showing differential expression of 443 genes in Setd5fl/fl and Setd5CKO SLAM-HSCs, |Log2foldchange | > 1, p value < 0.05; n = 3. B GSEA analyses for genes affected in the SLAM-HSCs of Setd5fl/fl and Setd5CKO mice. NES, normalized enrichment score. C Relative expression levels of cell cycle and multi-potency genes in HSC cells; n = 3. mRNA levels were normalized to the expression of 18 s. D. Experimental design for single cell sequencing with Smart-seq2, LSK+ cells were indexed sorted into 96-well plates containing lysis buffer. E LSK+ cells were projected onto Nestorowa et al. data [30]. F UMAP visualizations of single cell transcriptomes of identified clusters using Seurat. G–I Diffusion maps of all cells were colored according to the expression levels of selected genes. J, K Histograms showing the compositions of surface markers defined four populations and transcriptome-defined five clusters by ten cell types annotated with Nestorowa et al. data. C1-C5 means cluster 1-5. Ten annotated cells including E-SALM (EPCR+CD48–CD150+), LT-HSC (Lin–Sca1+c-Kit+CD34–Flt3low), ST-HSC (Lin–Sca1+c-Kit+CD34+Flt3lowCD48–CD150–), MPP1 (Lin–Sca1+c-Kit+CD34+Flt3lowCD48–CD150+), MPP2 (Lin–Sca1+c-Kit+CD34+Flt3lowCD48+CD150+), MPP3 (Lin–Sca1+c-Kit+CD34+Flt3lowCD48+CD150–), LMPP (Lin–Sca1+c-Kit+CD34+Flt3+), CMP, GMP and MEP. L GSEA of LT-HSC, proliferation and quiescence signatures comparing between Setd5CKO and Setd5fl/fl in HSC enriched cluster 1 and cluster 2. M, N Proportion of SLAM-HSCs, HPC1s and five clusters identified with Seurat in each of the cell cycle stages between two groups.
To delineate the transcriptional networks of HSCs at the single-cell resolution, we further performed scRNA-seq (single-cell RNA sequencing) of LSK+ cells in Setd5CKO and Setd5fl/fl mice by using Smart-seq2 [28, 29] (Fig. 4D, S5F). We isolated single LSK+ cells stained with CD150 and CD48 and recorded the index-sorting data for each single cell. In total, 1380 cells were processed for scRNA-seq library construction and sequencing, with 1262 cells passed the quality control (see “Supplemental information”). LSK+ cells were projected onto Nestorowa et al. data [30] and matched perfectly with cell types as defined in the paper (Fig. 4E), indicating that our scRNA-seq data is of high quality. Moreover, LSK+s from Setd5CKO mice were largely deviated from the core territory toward more differentiated progenitor state in comparison with their counterparts in Setd5fl/fl mice, suggesting a shift from HSC to HPC at the transcriptional level with Setd5 deletion.
We further grouped cells into 5 clusters based on their transcriptomes with Seurat packages (Fig. 4F, S5G), hereafter termed as LT-like (cluster 1, long-term stem cell-like), Mixed-like (cluster 2, mixed stem cell-like), LMPP-like (cluster 3, lymphoid-primed multipotent progenitor-like) and MEP-like cluster (cluster 4). Nr4a1, Mpl, Egr1, Dntt, Flt3, Icam4 and Vwf were represented as specifically and highly expressed genes in LT-like, Mixed-like, LMPP-like and MEP-like cluster, respectively. The expression levels of individual genes were plotted in the diffusion map (Fig. 4G-I, S5H-J, Table S4). We found a reduced fraction of LT-like and LMPP-like clusters in Setd5CKO LSK+s (Figure S5K). Moreover, by projecting surface markers defined LSK+s to cell types defined in Nestorowa et al. paper including LT-HSCs, ESLAMs, ST-HSCs, MPP1s, MPP2s, MPP3s and LMPPs, we found that the fractions of LT-HSCs and ESLAMs were significantly lower in Setd5CKO SLAM-HSCs than that of the controls (Fig. 4J). We further projected transcriptome-defined 5 clusters to cell types defined by Nestorowa et al., and found that LT-HSCs and ESLAMs were enriched in cluster 1, while LMPPs were enriched in cluster 3 (Figure S5L). We also observed a significant reduced fraction of ESLAMs, LT-HSCs and ST-HSCs in cluster 1 and cluster 2 accompanied with decreased LMPPs in cluster 3 due to Setd5 deletion (Fig. 4K), and the decreased LT-HSC signature in Setd5CKO was also recapitulated by performing GSEA (Fig. 4L). Cluster 3 exhibited a decreased bias towards LMPPs whereas cluster 4 showed bias in erythroid differentiation in Setd5CKO group, accompanied with a decreased myeloid signature in cluster 5 (Figure S5M). These data indicated that both transcriptome- and immunophenotype-defined LT-HSCs lost the long-term stem cell signatures and differentiated toward multipotent progenitors with impaired lymphoid commitment due to Setd5 deficiency.
In addition, the proliferation signature of cluster 1 and 2 was increased, accompanied with a decreased quiescence signature in Setd5CKO group as revealed by GSEA (Fig. 4L). To further characterize the cell cycle changes, we used a recently reported predictor for allocating individual cells to G0/G1, S and G2/M cell cycle categories based on single-cell transcriptomes [31]. We found that the percentages of S and G2/M cell cycle signatures were elevated in Setd5CKO SLAM-HSC and HPC1 cells, as well as in Cluster 1, 2, 4 and Cluster 5 (Fig. 4M-N). Collectively, these data suggest a decreased quiescence state and enhanced proliferation with Setd5 deficiency, that might contribute to the functional lineage biases in Setd5CKO HSCs at the single cell level.
SETD5 regulates the expression of E2F targets associated with HCF-1
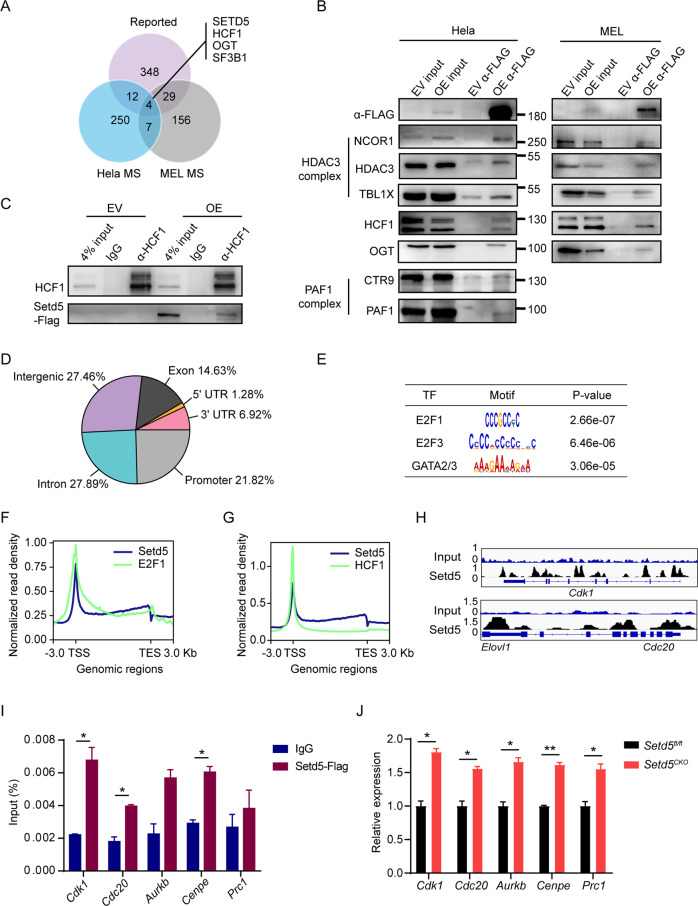
Although SETD5 has a SET domain, its enzyme activity was and may still be in question [6, 7, 32]. By immunoblot analysis, no obvious changes in majority of histone methylation were observed (Figure S6A), suggesting that SETD5 might lack histone methyltransferase activity and exert its function via interaction with other proteins. To explore the potential partners of SETD5, we performed co-immunoprecipitation (Co-IP) assay coupled with quantitative mass spectrometry (MS) using SETD5-FLAG stably expressed Hela or MEL (a murine erythroleukemia cell) cell lines. Overlapping of our Hela and MEL SETD5-IP/MS data with previously reported SETD5 associated proteins [5–7], revealed four proteins including SETD5, HCF-1 (host cell factor C1), OGT (O-linked N-acetylglucosamine (GlcNAc) transferase) and SF3B1 (splicing factor 3b subunit 1) (Fig. 5A). Additional components of HDAC3 complex (including HDAC3, NCOR1 and TBL1X), PAF1 (RNA Pol II–associated factor 1) complex (including PAF1 and CTR9) and OGT that have been previously identified [5–7] were also found in our IP/MS experiments (Fig. 5A, B, Table S5-6). We further confirmed that HCF-1 could be immunoprecipitated by SETD5 both in MEL and Hela cells in Co-IP assays and their interaction was further verified with reciprocal co-immunoprecipitation with HCF-1 antibody in MEL cells (Fig. 5B, C).
Fig. 5. SETD5 regulates the expression of E2F targets associated with HCF-1.
A Venn diagrams showing the overlap between Hela/MEL MS-identified proteins and reported SETD5 interactors. B Anti-FLAG immunoprecipitation of lysates from Hela and MEL cells transfected with SETD5-FLAG (OE) and empty FLAG vector (EV). Input: cell extract before immunoprecipitation, FLAG-IP: immunoprecipitation elute with 2×loading buffer. C Reciprocal immunoprecipitation with antibodies against HCF-1 in MEL cells transfected with SETD5-FLAG and empty FLAG vector. D Pie chart showing the distribution of ChIP-seq peaks for SETD5 with respect to promoter (define as a 3.0-kb proximal region centered on the gene start sites, including TSS), 5’UTR (5’-untranslated region), 3’UTR (3’-untranslated region), exon, intron and intergenic regions. E MEME motif analysis of SETD5 peaks, TF: transcription factor. F–G Density plots of SETD5 (blue) and E2F1/HCF-1 (green) normalized ChIP-seq signals, TES: Transcription End Site. H IGV tracks of target genes of SETD5-FLAG, and input DNA in MEL cells. The coverage data are represented as normalized reads of bins per million (BPM). I ChIP-qPCR of the binding of SETD5-FLAG to the promoters of indicated genes, Prc1 is a negative control which was not found in FLAG-SETD5’s ChIP-seq data. J Relative expression levels of target genes in control and Setd5CKO LSK+ cells measured by real-time PCR; n = 3. mRNA levels were normalized to the expression of 18 s. Mean ± SEM; * P < 0.05, ** P < 0.01, *** P < 0.001.
HCF-1 was reported to interact with both repressive and activating E2Fs, and to regulate the expression of its downstream targets including cell cycle entry [33–35]. The function of HCF-1 in cell cycle regulation prompted us to further examine the potential roles of SETD5 in regulating E2F targets via its association with HCF-1. To identify the potential targets of SETD5 unbiasedly in the blood cells, we performed chromatin immunoprecipitation followed by next-generation sequencing (ChIP-seq) using SETD5-FLAG stably expressed MEL cells. We found that the genomic distribution of SETD5 was highly enriched on promoter region (defined as a 3.0-kb region centered by the transcription start sites (TSS)) and gene bodies (intron and exon) (Fig. 5D). Motif enrichment analysis of SETD5 peaks revealed an enrichment of E2F1, E2F3, and GATA2/3 (Fig. 5E). Further analysis revealed a similar distribution along genomic regions between SETD5 and E2F1/4 as well as HCF-1 (Fig. 5F-G, S6B), suggesting that SETD5 binds the E2F targets with HCF-1. Among E2F1 targets, 66.5% (4794/7204) were enriched with SETD5 bindings and 34.8% (4794/13774) of SETD5 targets were E2F1 targets. However, only 3.88% (186/4794) SETD5/E2F targets showed differential expression (Figure S6C). These 186 SETD5/E2F targeted DEGs were enriched with cell-cycle process and cell division (Figure S6D). The binding of SETD5 at the promoter region of several E2F target genes, including Cdk1, Cdc20, Aurkb, and Cenpe, were observed (Fig. 5H, S6E) and further confirmed with ChIP-qPCR (Fig. 5I). Moreover, we found that the expression of these genes was significantly upregulated in Setd5CKO LSK+ cells (Fig. 5J). Taken together, our results demonstrated that SETD5 could be localized to E2F-responsive promoters in association with HCF-1 to regulate gene transcription.
Considering that the interaction between SETD5 and HDAC3/NCOR complex was observed in previous studies [5, 6, 36] as well as ours, and higher H3 pan-Kac (pan-acetylation of H3 lysine) and H3K9ac were observed in Setd5−/− mouse embryonic stem cells (mESC) [5], we further examined the histone acetylation levels in Setd5−/− hematopoietic stem/progenitor cells. Immunoblot analysis of Setd5fl/fl control and Setd5CKO c-Kit+ cells showed similar histone acetylation levels of H3K9, H3K27, H3K14, H4K5, H4K8, and H4K12 in both groups (Figure S6F), suggesting that the disrupted interaction of SETD5 and HDAC3 in hematopoietic cells may not affect histone acetylation levels.
SETD5 regulates the promoter-proximal paused Pol II release on E2F targets
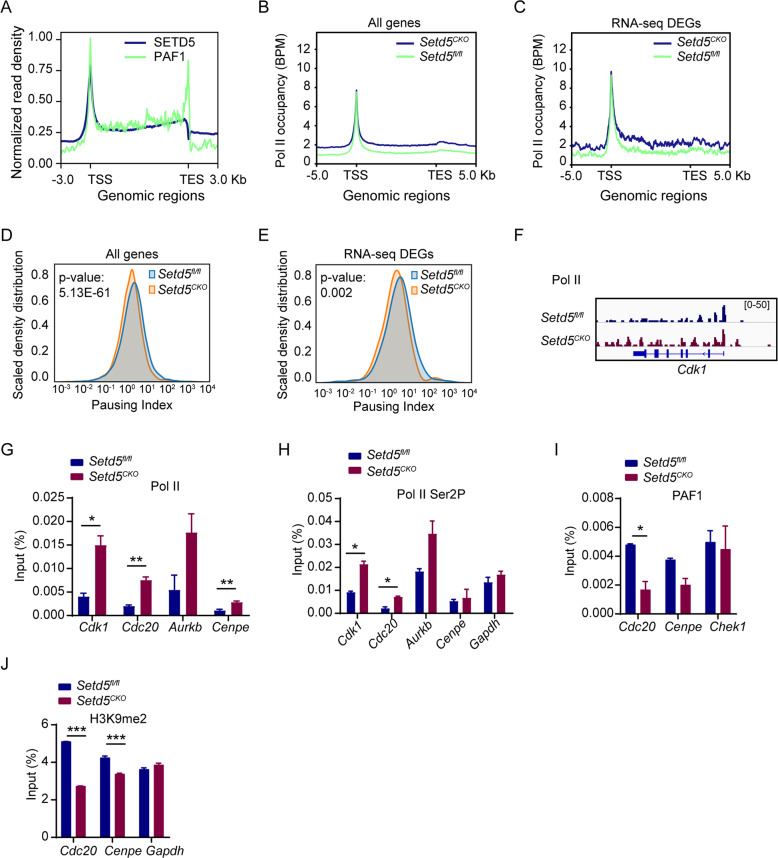
We further investigated the mechanism of SETD5 in regulating the expression of E2F targets since HCF-1 was reported to recruit MLL and Set-1 H3K4 HMT complexes to E2F responsive promoters to regulate gene expression [35, 37]. However, no obvious change among the genomic distribution of H3K4me3 was observed due to Setd5 deficiency (Figure S6G). Interestingly, HCF-1 was recently found to associate with multiple transcription initiation and elongation complexes to regulate Pol II pausing [38]. Besides, PAF1, a critical regulator of Pol II pausing [39, 40], was immunoprecipitated with SETD5 (Fig. 5B), and showed a similar distribution pattern with SETD5 (Fig. 6A). We thus postulated that SETD5 modulates the release of promoter-proximal paused Pol II. By analyzing the genome-wide distribution of Pol II in control and Setd5CKO c-Kit+ cells by ChIP-Seq, we found an increase in Pol II occupancy on all genes and DEGs upon SETD5 deficiency (Fig. 6B, C and S6H-I). We further calculated pausing index (the ratio between promoter-proximal and gene body Pol II density) to measure the release of Pol II from promoters. Deficiency of SETD5 led to a significant decrease of pausing index on average (Fig. 6D), as well as on the DEGs (Fig. 6E), which represented a transcription elongation state. Pausing index was decreased in 5622 genes (59.5%) and increased in 3828 genes (40.5%) (Fig. S6J). We observed significantly increased Pol II occupancy at Cdk1 locus (Fig. 6F). Using ChIP-qPCR, we confirmed the increased Pol II and Pol II Ser2P (phosphorylated Ser2) occupancy at Cdk1, Cdc20, Aurkb and Cenpe in Setd5CKO cells in comparison with the controls (Fig. 6G, H). These data suggested that SETD5 deficiency may promote the release of paused Pol II into productive elongation. Although the total protein level of PAF1 was not significantly changed (Fig. S6K), decreased occupancies of PAF1 at Cdc20 and Cenpe in Setd5CKO c-Kit+ cells were observed (Fig. 6I), suggesting that SETD5 remodeled Pol II pausing and elongation state on E2F targets to regulate their expression, most likely via its interaction with the PAF1 complex.
Fig. 6. SETD5 regulates the promoter-proximal paused Pol II release on E2F targets.
A Density plots of SETD5 (blue) and PAF1 (green) normalized ChIP-seq signals. B, C Occupancy of total Pol II on all genes and DEGs (Fold change>1.5 and p adj value<0.05), ChIP normalization was implemented by integrating Spike-in Chromatin. D, E Density plot of all genes and DEGs with reduced average Pol II pausing index in Setd5CKO. F IGV tracks comparing occupancy of Pol II within Cdk1 locus. G, H ChIP-qPCR of Pol II and Pol II Ser2P at the promoters of indicated genes in c-Kit+ cells from control or Setd5CKO mice; n = 2. I ChIP-qPCR analysis of PAF1 at the promoters of indicated genes in c-Kit+ cells; n = 2. J The H3K9 dimethylation levels at Cdc20 and Cenpe promoters; Gapdh as an internal control; n = 2. Mean ± SEM; *P < 0.05, **P < 0.01, *** P < 0.001.
In consistent with a previously reported role of SETD5 to affect H3K9me2 levels via association with HDACs-G9a co-suppressor complex [7], we also observed a mild decrease of H3K9me2 occupancy at E2F targets (Fig. 6J). Recently, a robust correlation between histone acetylation and transcription elongation had been reported, and HDAC inhibition was shown to stimulate the release of promoter-proximal paused Pol II [41, 42]. The interaction between SETD5 and HDAC3/NCOR complex (Fig. 5B) prompted us to further examine whether the loss of SETD5 would alter H3 pan-Kac, H3K9ac, and H3K27ac around the promoter and 5’UTR of Cdk1, Cdc20, Aurkb, and Cenpe. However, no significant differences were found (Fig. S7A-Q), suggesting that the effect of SETD5 on the regulation of Pol II promoter pausing is probably independent of SETD5-HDAC3/NCOR complexes in normal hematopoiesis. We also examined the genomic distribution of H3K36me3 and found no change between two groups (Fig. S7R).
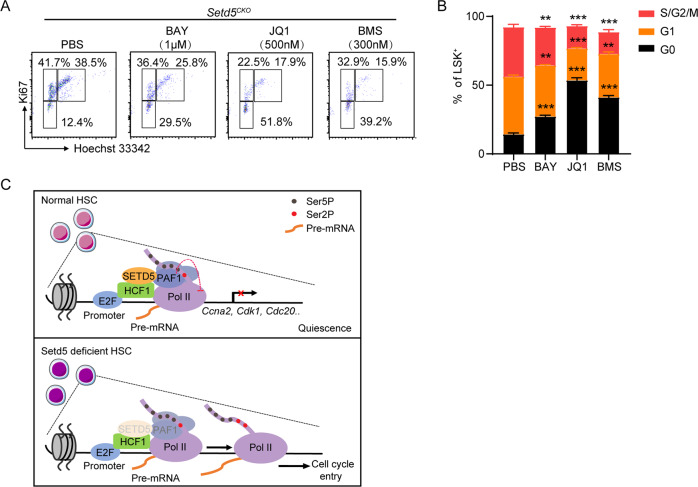
To further determine whether increased cell-cycle transition was mainly caused by the increased Pol II elongation at E2F targets, we employed two inhibitors BAY 1143572 (p-TEFb/CDK9 inhibitor) and JQ1 (Brd4 inhibitor) to inhibit the transition of paused Pol II to elongation and analyzed the cell cycle of LSK+ cells, with CDK inhibitor BMS-387032 as a positive control. The c-Kit+ cells from Setd5CKO were treated in an in vitro culture system with individual inhibitors for 24 h. Significantly decreased S phases were observed in LSK+ cells in all drug-treated groups when compared with PBS treated group (Fig. 7A, B). Taken together, these data indicated that increased transition of paused Pol II to elongation mediated by Setd5 depletion could be a main cause for the exit of quiescence stage of Setd5CKO HSC cells.
Fig. 7. Setd5CKO deficiencies could be partially rescued by super elongation complex-related inhibitors.
A c-Kit+ BM cells from Setd5fl/fl and Setd5CKO mice were treated with BAY-1143572 1 μM, JQ1 500 μM, and BMS-387032 300 nM for 24 h before cell cycle analysis. B The frequencies of G0, G1, S/G2/M LSK+s in drug-treated c-Kit+ cells for 24 h from Setd5fl/fl and Setd5CKO mice were shown; n = 4. C A model for the role of SETD5 in modulating hematopoietic stem cell homeostasis. In normal HSCs, SETD5 occupies at E2F-responsive promoters in association with HCF-1, maintains a paused Pol II state with PAF1 complex, leading to transcriptional silencing of E2F targets and a quiescent state of HSCs. Once Setd5 is depleted, PAF1 occupancy is weakened and triggers Pol II elongation to active transcription of E2F target genes and promote G1 to S phase transition.
Discussion
In this study, we investigated the roles of Setd5 in HSCs during hematopoiesis. We found that deletion of Setd5 led to an enhanced proliferation and accumulation of HSCs both in Vav-Cre and Mx1-Cre;Setd5fl/fl (abbreviated as Setd5CKO and Setd5IKO respectively) murine models. However, this was accompanied by a strikingly decreased number of functional HSCs and a reduced competitive reconstitution ability with long-term depletion of Setd5 in Setd5CKO mice. When Setd5 was deleted in Setd5CKO, more HSCs exited from quiescence and entered into G1 phase, and consequently, cell cycle perturbed Setd5CKO HSCs displayed impaired self-renewal and multi-lineage differentiation abilities. Interestingly, while more Setd5IKO HSCs were also in proliferative stages, these cells exhibited enhanced proliferation and myelopoiesis in serial transplantations. These different results probably are a differential time and developmental stage effect at which Setd5 is depleted in Vav-Cre or Mx1-Cre mediated Setd5 excision.
Moreover, both bulk and single-cell RNA-seq revealed that transcriptome- and immunophenotype-defined LT-HSCs showed reduced long-term stem cell and quiescence signatures upon Setd5 deletion, accompanied by impaired lymphoid commitment and more cell cycle entrance. These results exemplified a key role of Setd5 in the balance between proliferation and differentiation of HSCs. Mechanistically, although HCF-1 was on a list of potential SETD5 associated proteins identified with previous IP-MS assay [5], the biological function of the interaction between HCF-1 and SETD5 was not explored. Here, we verified the interaction between HCF-1 and SETD5 and found the enrichment of SETD5 at the E2F targets. More recently, HCF-1 was reported to promote transcription elongation of viral Immediate Early genes via the interaction with transcriptional elongation factors including PAF1 in response to herpes virus infection [38]. Our observation suggests that the loss of Pol II pausing of Setd5 deficient HSCs may be caused by the disrupted association of HCF-1 and transcriptional elongation factors including but not limited to PAF1. Previous study reported the interaction between PAF1 and SETD5 and found that depletion of Setd5 could promote Pol II pausing in mESCs [6]. PAF1 has been identified as a regulator of release of promoter-proximal paused RNA Pol II or promoter-proximal pausing by RNA Pol II [39, 40]. Our results reveal that SETD5 restricts Pol II release from pausing to elongation associated with PAF1 at cell cycle related genes to maintain HSC quiescence. These results suggest that the role of Setd5 in the regulation of paused Pol II release is tissue specific.
Although higher bulk H3 pan-Kac and H3K9ac and increased H3K9ac levels at target genes were observed in mESCs and murine PDAC (pancreatic ductal adenocarcinoma) cells upon Setd5 deficiency respectively [5, 36]. Deliu E et al. reported that higher histone acetylation observed in Setd5+/− hippocampal samples and ESCs was independent of HDAC3 activity [6]. In our study, deletion of Setd5 in hematopoiesis has no effect on bulk levels of histone acetylation and site-specific histone acetylation, suggesting that the effect of SETD5 on histone acetylation may be tissue or cell type specific.
Although SETD5 was reported to have an intrinsic methyltransferase activity on H3K36 [32], several other groups had shown that SETD5 lacks histone lysine methyltransferase activity [5, 6, 36]. In our study, no obvious differences were observed of H3K36me3 on both protein level and ChIP-seq signal upon SETD5 deletion, strongly indicating that SETD5 may indeed lack methyltransferase activity on H3K36. The genomic distribution of SETD5 in our study was slightly different with the ChIP-seq data by Sessa et al. Neuron 2019 [32], which may be due to different cell types and antibodies used in the experiments.
Interestingly, SETD5 mutations were reported to cause congenital abnormalities, and multiple congenital anomalies were also observed in two boys with mutation in HCF-1 [43]. PAF1 complex was also reported to has mutations associated with autism [44, 45], suggesting a model of potential interplay among HCF-1, PAF1 and SETD5 proposed in our study may be applicable in the onset/development of certain mental diseases.
In summary, our study provided a new insight into the molecular mechanism of Setd5 in regulating HSC quiescence by Pol II pausing-mediated differential expression of cell cycle genes via HCF-1-SETD5-PAF1-Pol II axis (Fig. 7C). We also revealed that regulation of Pol II pausing and release plays an essential role in HSC maintenance.
Supplementary information
Acknowledgements
We would like to thank Profs. Hideo Ema and Deqing Hu for their insightful discussion, Ms. Zining Yang for her assistance with the quality control of scRNA-seq libraries, Dr. Xiaowei Xie for her helpful discussion with scRNA-seq analysis and Dr. Yangpeng Chen for his help in breeding F1 generation of Setd5 floxed mice. This work was supported by funds from the Ministry of Science and Technology of China (2018YFA0107801 to W. Yuan and 2020YFE0203000 to Y. Chu); the Chinese Academy of Medical Sciences Innovation Fund for Medical Sciences, CIFMS (2021-1-I2M-040 and 2017-I2M-3-015 to W. Yuan); the National Natural Science Foundation of China (82170117 and 81770155 to Y. Chu; 81629001 and 81970149 to W. Yuan).
Author contributions
Y. Chu and W. Yuan conceived the project, supervised the research, and revised the paper. M. Li and Y. Chu designed all of the experiments, performed the experiments, analyzed the data, and wrote the manuscript. C. Qiu performed the computational data analysis. Y. Bian, D. Shi, B. Wang, and Q. Ma helped with the in vivo experiments and flow cytometry analysis. X. Wang, J. Shi, L. Zhang, P. Zhu, Y. Ma, and T. Cheng contributed to the reagents, data analysis, and paper discussion.
Data availability
All sequence data generated in this study have been deposited in the Genome Sequence Archive [46] in National Genomics Data Center [47], China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number CRA003816 that is publicly accessible at https://bigd.big.ac.cn/gsa.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yajing Chu, Email: chuyajing@ihcams.ac.cn.
Weiping Yuan, Email: wpyuan@ihcams.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41375-021-01481-1.
References
- 1.Herz HM, Garruss A, Shilatifard A. SET for life: biochemical activities and biological functions of SET domain-containing proteins. Trends Biochem Sci. 2013;38:621–39. doi: 10.1016/j.tibs.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schuettengruber B, Martinez AM, Iovino N, Cavalli G. Trithorax group proteins: switching genes on and keeping them active. Nat Rev Mol Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 3.Rincon-Arano H, Halow J, Delrow JJ, Parkhurst SM, Groudine M. UpSET recruits HDAC complexes and restricts chromatin accessibility and acetylation at promoter regions. Cell. 2012;151:1214–28. doi: 10.1016/j.cell.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, et al. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osipovich AB, Gangula R, Vianna PG, Magnuson MA. Setd5 is essential for mammalian development and the co-transcriptional regulation of histone acetylation. Development. 2016;143:4595–607. doi: 10.1242/dev.141465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deliu E, Arecco N, Morandell J, Dotter CP, Contreras X, Girardot C, et al. Haploinsufficiency of the intellectual disability gene SETD5 disturbs developmental gene expression and cognition. Nat Neurosci. 2018;21:1717–27. doi: 10.1038/s41593-018-0266-2. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z, Hausmann S, Lyu R, Li TM, Lofgren SM, Flores NM, et al. SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell. 2020. [DOI] [PMC free article] [PubMed]
- 8.Grozeva D, Carss K, Spasic-Boskovic O, Parker MJ, Archer H, Firth HV, et al. De novo loss-of-function mutations in SETD5, encoding a methyltransferase in a 3p25 microdeletion syndrome critical region, cause intellectual disability. Am J Hum Genet. 2014;94:618–24. doi: 10.1016/j.ajhg.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuechler A, Zink AM, Wieland T, Ludecke HJ, Cremer K, Salviati L, et al. Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur J Hum Genet. 2015;23:753–60. doi: 10.1038/ejhg.2014.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauch A, Wieczorek D, Graf E, Wieland T, Endele S, Schwarzmayr T, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 11.Fernandes IR, Cruz ACP, Ferrasa A, Phan D, Herai RH, Muotri AR. Genetic variations on SETD5 underlying autistic conditions. Dev Neurobiol. 2018;78:500–18. doi: 10.1002/dneu.22584. [DOI] [PubMed] [Google Scholar]
- 12.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135:1118–29. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 13.Bryder D, Rossi DJ, Weissman IL. Hematopoietic stem cells: the paradigmatic tissue-specific stem cell. Am J Pathol. 2006;169:338–46. doi: 10.2353/ajpath.2006.060312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilmour DS. Promoter proximal pausing on genes in metazoans. Chromosoma. 2009;118:1–10. doi: 10.1007/s00412-008-0182-4. [DOI] [PubMed] [Google Scholar]
- 15.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Q, Liu X, Zhou T, Cook J, Nguyen K, Bai X. RNA polymerase II pausing modulates hematopoietic stem cell emergence in zebrafish. Blood. 2016;128:1701–10. doi: 10.1182/blood-2016-02-697847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–21. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 18.Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Yang L, Bryder D, Adolfsson J, Nygren J, Månsson R, Sigvardsson M, et al. Identification of Lin(-)Sca1(+)kit(+)CD34(+)Flt3- short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105:2717–23. doi: 10.1182/blood-2004-06-2159. [DOI] [PubMed] [Google Scholar]
- 20.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–28. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 21.Passegué E, Wagers AJ, Giuriato S, Anderson WC, Weissman IL. Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J Exp Med. 2005;202:1599–611. doi: 10.1084/jem.20050967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bustelo XR, Rubin SD, Suen KL, Carrasco D, Barbacid M. Developmental expression of the vav protooncogene. Cell Growth Differ. 1993;4:297–308. [PubMed] [Google Scholar]
- 23.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–39. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Sitnicka E, Bryder D, Theilgaard-Mönch K, Buza-Vidas N, Adolfsson J, Jacobsen SE. Key role of flt3 ligand in regulation of the common lymphoid progenitor but not in maintenance of the hematopoietic stem cell pool. Immunity. 2002;17:463–72. doi: 10.1016/s1074-7613(02)00419-3. [DOI] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, et al. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–81. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanjuan-Pla A, Macaulay IC, Jensen CT, Woll PS, Luis TC, Mead A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502:232–6. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 27.Beerman I, Bhattacharya D, Zandi S, Sigvardsson M, Weissman IL, Bryder D, et al. Functionally distinct hematopoietic stem cells modulate hematopoietic lineage potential during aging by a mechanism of clonal expansion. Proc Natl Acad Sci USA. 2010;107:5465–70. doi: 10.1073/pnas.1000834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Picelli S, Faridani OR, Björklund AK, Winberg G, Sagasser S, Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–81. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 29.Li L, Dong J, Yan L, Yong J, Liu X, Hu Y, et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:858–73. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Nestorowa S, Hamey FK, Pijuan Sala B, Diamanti E, Shepherd M, Laurenti E, et al. A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood. 2016;128:e20–31. doi: 10.1182/blood-2016-05-716480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lun AT, McCarthy DJ, Marioni JC. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sessa A, Fagnocchi L, Mastrototaro G, Massimino L, Zaghi M, Indrigo M, et al. SETD5 regulates chromatin methylation state and preserves global transcriptional fidelity during brain development and neuronal wiring. Neuron. 2019;104:271–89. doi: 10.1016/j.neuron.2019.07.013. [DOI] [PubMed] [Google Scholar]
- 33.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deplus R, Delatte B, Schwinn MK, Defrance M, Mendez J, Murphy N, et al. TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 2013;32:645–55. doi: 10.1038/emboj.2012.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tyagi S, Chabes AL, Wysocka J, Herr W. E2F activation of S phase promoters via association with HCF-1 and the MLL family of histone H3K4 methyltransferases. Mol Cell. 2007;27:107–19. doi: 10.1016/j.molcel.2007.05.030. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Hausmann S, Lyu R, Li TM, Lofgren SM, Flores NM, et al. SETD5-coordinated chromatin reprogramming regulates adaptive resistance to targeted pancreatic cancer therapy. Cancer Cell. 2020;37:834–49. doi: 10.1016/j.ccell.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou P, Wang Z, Yuan X, Zhou C, Liu L, Wan X, et al. Mixed lineage leukemia 5 (MLL5) protein regulates cell cycle progression and E2F1-responsive gene expression via association with host cell factor-1 (HCF-1) J Biol Chem. 2013;288:17532–43. doi: 10.1074/jbc.M112.439729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alfonso-Dunn R, Turner AW, Jean Beltran PM, Arbuckle JH, Budayeva HG, Cristea IM, et al. Transcriptional elongation of HSV immediate early genes by the super elongation complex drives lytic infection and reactivation from latency. Cell Host Microbe. 2017;21:507–17. doi: 10.1016/j.chom.2017.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen FX, Woodfin AR, Gardini A, Rickels RA, Marshall SA, Smith ER, et al. PAF1, a molecular regulator of promoter-proximal pausing by RNA polymerase II. Cell. 2015;162:1003–15. doi: 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu M, Yang W, Ni T, Tang Z, Nakadai T, Zhu J, et al. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science. 2015;350:1383–6. doi: 10.1126/science.aad2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stasevich TJ, Hayashi-Takanaka Y, Sato Y, Maehara K, Ohkawa Y, Sakata-Sogawa K, et al. Regulation of RNA polymerase II activation by histone acetylation in single living cells. Nature. 2014;516:272–5. doi: 10.1038/nature13714. [DOI] [PubMed] [Google Scholar]
- 42.Vaid R, Wen J, Mannervik M. Release of promoter-proximal paused Pol II in response to histone deacetylase inhibition. Nucleic Acids Res. 2020;48:4877–90. doi: 10.1093/nar/gkaa234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gérard M, Morin G, Bourillon A, Colson C, Mathieu S, Rabier D, et al. Multiple congenital anomalies in two boys with mutation in HCFC1 and cobalamin disorder. Eur J Med Genet. 2015;58:148–53. doi: 10.1016/j.ejmg.2014.12.015. [DOI] [PubMed] [Google Scholar]
- 44.Wang T, Hoekzema K, Vecchio D, Wu H, Sulovari A, Coe BP, et al. Large-scale targeted sequencing identifies risk genes for neurodevelopmental disorders. Nat Commun. 2020;11:4932. doi: 10.1038/s41467-020-18723-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Y, Song F, Zhu J, Zhang S, Yang Y, Chen T, et al. GSA: genome sequence archive. Genomics Proteom Bioinf. 2017;15:14–18. doi: 10.1016/j.gpb.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Database Resources of the National Genomics Data Center. China National Center for bioinformation in 2021. Nucleic Acids Res. 2021;49:D18–d28. doi: 10.1093/nar/gkac1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data generated in this study have been deposited in the Genome Sequence Archive [46] in National Genomics Data Center [47], China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number CRA003816 that is publicly accessible at https://bigd.big.ac.cn/gsa.