Abstract
As evidence has been linking the oral bacterium Fusobacterium nucleatum (F. nucleatum) to colorectal tumorigenesis, we aimed to produce preliminary data on the expression of F. nucleatum in both oral and colorectal body sites in cases diagnosed with colorectal neoplasms (CRN) and CRN-free controls. We conducted a pilot hospital-based case–control study among patients who underwent colonoscopy examination. Saliva samples and biopsies from healthy colon mucosa from CRN cases and CRN-free controls, and from tumors in cases, were collected, as well as data on periodontal condition and potential CRN risk factors. A total of 22 CRN cases and 21 CRN-free controls participated in this study, with a total of 135 biospecimens collected and analyzed by qPCR for detection and quantification of F. nucleatum. The detection rate of F. nucleatum was 95% in saliva samples and 18% in colorectal mucosa specimens. The median (95% CI) salivary F. nucleatum level was 0.35 (0.15–0.82) and 0.12 (0.05–0.65) in case and control groups, respectively, with a Spearman correlation of 0.64 (95% CI 0.2–0.94) between F. nucleatum level in saliva and healthy colorectal mucosa in controls. Our study results support the need for and the feasibility of further studies that aim to investigate the association between oral and colorectal levels of F. nucleatum in CRN cases and controls.
Clinical Relevance: Considering the current evidence linking F. nucleatum to colorectal carcinogenesis, investigating the role of oral F. nucleatum expression in its colorectal enrichment is crucial for colorectal cancer screening and prevention avenues.
Subject terms: Cancer, Microbiology, Gastroenterology
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer and the second cause of death from cancer worldwide, with over 1,900,000 newly diagnosed cases and over 900,000 deaths in 20201. Most CRCs arise from adenomatous polyps, which can eventually degenerate into invasive carcinomas2. The potential for malignant progress depends on the histologic pattern of growth (with villous pattern being an adverse indicator), size, multiplicity of polyps, and high-grade dysplasia status3. A small proportion of CRCs develop under the alternative “serrated pathway”, from serrated polyps frequently located in the proximal colon, and are linked to the Microsatellite Instability phenotype, resulting from a deficiency of the DNA repair system2.
Over the last decade, many studies have reported an enrichment of Fusobacterium nucleatum (F. nucleatum) in colorectal tissues and stools collected from subjects diagnosed with cancerous and even precancerous colorectal lesions4–6. The involvement of F. nucleatum in early colorectal carcinogenesis stages has been suggested by studies that identified the bacterium in colorectal adenomas, with a gradual enrichment of the colon with F. nucleatum in parallel to the adenoma-carcinoma sequence7–9. Moreover, the bacterium was identified with two virulence factors promoting colorectal carcinogenesis. The first factor is FadA, an adhesin that allows F. nucleatum to invade human epithelial cells, activate β-catenin signaling, induce expression of the oncogenic gene, and promote the growth of colorectal tumor cells10. The second factor is a self-transporting protein Fap2, which inhibits the activity of immune cells and thus potentiates the progression of CRC11.
F. nucleatum is also among the dominant species of the oral cavity12,13 and plays an essential role in the formation of dental plaque. It also promotes the colonization and invasion of tooth surfaces by other pathogenic species, which in turn stimulates the recruitment and activation of local immune cells, resulting in destruction of tooth-supporting tissues and progression of periodontitis14,15. F. nucleatum is abundant in salivary samples from patients with gingivitis and chronic periodontitis16.
It has been suggested that gut enrichment with F. nucleatum is sourced intra-individually from the oral cavity17–19, in the presence of periodontal sites, which may explain the association of periodontal disease (PD) with CRC and colorectal adenomas20–23. However, the hypothesis of an intra-individual oral source of gut enrichment with F. nucleatum still needs to be tested, which would require epidemiologic data on paired measures of both oral and gut F. nucleatum levels in subjects diagnosed with colorectal neoplasms (CRN) and in CRN-free controls. To date, few studies have investigated F. nucleatum in saliva from subjects with CRN and CRN-free controls17,18,24–29, only two studies have investigated paired saliva and colorectal specimens in these groups24,28, and none has explored the link between paired oral and colorectal levels of F. nucleatum in subjects with CRN and CRN-free controls (see Table 1 for the studies’ summary).
Table 1.
Summary of studies that investigated Fusobacterium nucleatum in saliva and/or colorectal mucosa or stool in colorectal cancer (CRC) cases and controls (C).
| Author -year | Country | No. of CRC cases/C | Exclusion if previous ATB use (period) | Specimen type | Specimen collection time | Collection kits and storage conditions | Bacterial analysis method |
F. nucleatum detection and level quantification outcomes |
|---|---|---|---|---|---|---|---|---|
| Russo et al. 2017 | Italy | 10/10 | Yes (last 3 months) | Unstimulated saliva | 1 day before surgery |
Sterile tube, − 80 °C |
Next generation sequencing & qPCR |
F. nucleatum level: In saliva: No significant difference between CRC cases and C In stool: No significant difference between CRC cases and C Saliva Vs stool: higher abundance in saliva than in stool, in CRC (p < 0.01) and in C (p < 0.002) |
| Stool | ||||||||
|
Fresh tumor mucosa (in CRC cases) |
During surgery | 0.9% NaCl solution, − 80 °C | ||||||
| Guven et al. 2019 | Turkey | 71/77 |
Yes (last 3 months) |
Saliva | Before cancer treatments |
Centrifuge tube, − 20 °C |
qPCR |
F. nucleatum detection: CRC cases: 97.2% Vs C: 96.1%; p > 0.99 F. nucleatum level: (in Log10 copies/ml) CRC cases: 6.89 ± 1.07 Vs C: 6.35 ± 0.78; p = 0.001 |
| Komiya et al. 2019 | Japan |
14/0 (No C) |
Yes (last month) |
Saliva | Before/after colonoscopy | Sterile tubes, anaerobic conditions |
PCR (conventional) |
F. nucleatum detection: Saliva: 100% Tumor mucosa: 57% Saliva and tumor mucosa: 43% (F. nucleatum identical strain in 75% of patients with both saliva and tumor positive to F. nucleatum) |
| Tumor mucosa | During colonoscopy | |||||||
| Kato et al. 2016 | USA | 68 /122 | –- | Oral rinse | –- |
Commercial mouthwash (15% alcohol), − 80 °C |
16SrRNA gene sequencing |
Dominant Phyla: Fusobacteria was not dominant, only 3.7% of all sequences No association between F. nucleatum and CRC |
| Abed et al. 2020 | Israel |
7/0 (No C) |
No: ATB was taken just before surgery |
Saliva | 1 day before surgery, or just after colonoscopy | Sterile tubes, anaerobic conditions | PCR (conventional) |
F. nucleatum detection: Saliva: 100% Tumor mucosa: 100% |
| Tumor mucosa | 45 min after resection | |||||||
|
3/0 (No C) |
Yes (just before surgery) | Saliva | 1 day before surgery, or just after colonoscopy | Sterile tubes, anaerobic conditions | Whole genome sequencing | Great similarity between F. nucleatum strains in saliva and tumor in each subject | ||
| Tumor mucosa | 45 min after resection | |||||||
| Kageyama et al. 2019 | Japan | 24/118 |
Yes (last month) |
Stimulated saliva | Before cancer therapy | Sterile tube, − 80 °C | 16SrRNA gene sequencing |
Differentially abundant OTUs: OTUs corresponding to F. nucleatum were not the most abundant bacteria OTUs in CRC |
| Yang et al. 2019 | USA | 231 / 462 |
Yes (last week) |
Oral rinse | – | Commercial mouthwash, − 80 °C | 16S rRNA gene sequencing |
F. nucleatum detection: CRC cases: 99.6% Vs C: 99.6% (p = 1) |
| Flemer et al. 2017 | Ireland | 99/103 + (32 polyp patients) |
Yes (last month) (ATB during surgery time) |
Oral swabs (45 CRC& 25 C) | – | − 80 °C | 16S rRNA gene amplicon sequencing |
F. nucleatum abundance: Fusobacterium less abundant in oral swabs of CRC cases compared to C |
| Stool | Before colonoscopy | − 80 °C | ||||||
| Colorectal and tumor mucosa | During surgery or colonoscopy | RNA later at 4 °C for 12 h, then at − 20 °C |
No. Number, CRC Colorectal cancer, C Controls, F. nucleatum Fusobacterium nucleatum, ATB Antibiotic, OTU Operational taxonomic unit, – Not reported.
Objectives
This pilot study aimed to generate preliminary data on detection and quantification of F. nucleatum in both saliva and colorectal mucosa in subjects diagnosed with CRN and CRN-free controls. Ultimately, these preliminary data can help in designing a subsequent large epidemiological study investigating F. nucleatum in both oral and colorectal sites concomitantly in subjects diagnosed with CRN and CRN-free controls.
Methodology
We carried out a pilot hospital-based case–control study in the setting of University of Montreal Hospital Center in Montreal, Quebec, Canada. Participants were consecutive patients who underwent colonoscopy in the gastroenterology department between February 2018 and November 2019. Specifically, we identified patients who were scheduled for colonoscopy exam as part of CRC screening, or as a CRC diagnostic test upon recent change in bowel habits, rectal bleeding, unexplained iron deficiency anemia, or a positive Fecal Immunochemical Test. Patients with advanced colorectal adenoma or CRC were also identified among patients scheduled for an endoscopic mucosal resection technique, after they were diagnosed with polyps suspected of being neoplastic, based on a recent medical imaging or colonoscopy. Endoscopic mucosal resection is indicated for resection of the carpet-type adenomatous colonic polyp, and superficial early colorectal cancers that are well and/or moderately differentiated and limited to the mucosa30.
The study inclusion criteria were: (1) aged 40–80 years; (2) resident of Montreal metropolitan area; (3) speaking French and/or English; (4) no prior diagnosis of cancer; (5) no history of hereditary colorectal disease; (6) no history of inflammatory bowel disease; and (7) no history of treatment with antibiotics within the past 3 months.
Patients with histologically confirmed advanced colorectal adenoma or CRC were included in the “case” group of CRN. Advanced colorectal adenoma refers to adenomas with high risk of malignant transformation, which is defined by one or more of the following criteria being met: 3–10 adenomas; high-grade dysplasia; tubulovillous or villous appearance; adenoma > 1 cm in diameter; serrated sessile adenomas31. Patients whose colonoscopy did not result in the diagnosis of CRC, colorectal advanced adenoma, or inflammatory bowel disease were included in the ‘control’ group.
Eligible patients who agreed to participate in the study were invited to complete a multi-item study questionnaire, provide a saliva sample, and provide consent for biopsy collection during colonoscopy examination. The study was approved by the University of Montreal Hospital Centre Research Ethics Committee under the number: 2017-7068, CE 16.375—MJB, and all study participants signed the study consent before undergoing their colonoscopy. Participants were confirmed for eligibility only after colonoscopy examination. Participant status (case or control) was confirmed by histological investigation.
Data collection
Participants were administered a multi-item study-questionnaire that had been used by the research team in a previous population-based case–control study, COLDENT study, investigating the association between PD and sporadic CRC32. The questionnaire included different sections on sociodemographic and medical history information, cigarette smoking, anthropometric measures, non-steroidal anti-inflammatory drugs use, oral health, dietary habits, and total physical activity32–36. A life-course approach was used to document cumulative long-term history regarding cigarette smoking, specific dietary habits, and physical activity.
Thus, collected data enabled the description of study participants regarding sociodemographic characteristics, periodontal health status, as well as potential risk factors of CRN/CRC, namely age, gender, education attainment, income, body mass index, history of type II diabetes, history of CRC in first-degree relatives, history of regular use of non-steroidal anti-inflammatory drugs, lifetime cumulative cigarette smoking, consumption of red meats, processed meats, and total alcoholic drinks since early adulthood, as well as lifetime total physical activity score. Positive history of PD was defined as self-reported PD with bone loss, a previous professional diagnosis or treatment of PD, or history of clinical symptoms and complications of the disease, such as frequent gum bleeding, tooth mobility, or tooth loss because of PD or tooth mobility32.
Collection of biospecimens
In preparation to colonoscopy examination, all participants received the protocol for conventional bowel preparation, which consists of a diet restricted in residue for 2 days, followed by a strict liquid diet and laxatives (Bi-Peglyte® and Dulcolax 5 mg®) in the day before colonoscopy. During colonoscopy, biopsies were taken from healthy mucosa in cases and controls, and from polyps (or tumors) in cases. Given the differences in gut microbial composition between proximal and distal colon sites, biopsies of healthy mucosa were separately collected from ascending and descending colon. Biopsies of polyps were taken from freshly excised polyps before they were sent for histopathology analysis. If a clinical decision was made during colonoscopy to delay a polyp removal and take biopsies for histopathology analysis (when a malignant lesion is suspected), an extra-biopsy was then taken for the present study analysis. All biopsies were collected in physiological solution (Nacl 0.9%), then immediately transferred to empty sterile containers.
Unstimulated saliva was collected from participants the day of colonoscopy, or a few days later (at the time of interview), by spitting in a commercial collection kit for DNA stabilization (DNAGenotek (OMNI gene•ORAL | OM-501 kit®). Participants were warned not to eat, drink, smoke, or chew gum for 30 min before saliva collection. Mucosa and saliva specimens were immediately stored at − 80 °C until analysis.
Bacterial DNA extraction and quantitative polymerase chain reaction (qPCR)
Genomic DNA was isolated from saliva and colon tissue samples using the QIAamp DNA Mini Kit (Cat#51304, Qiagen, USA) and procedures were done according to the manufacturer’s instructions. DNA content was quantified using the Bio-Rad SmartSpec 3000 Spectrophotometer (Bio-Rad, 170–2501, USA). DNA sequences of TaqMan primer and probe used to detect 16S ribosomal RNA gene of F. nucleatum were similar to those described by Mima et al.37: F. nucleatum forward primer, 5′-CAACCATTACTTTAACTCTACCATGTTCA-3′; F. nucleatum reverse primer, 5′-GTTGACTTTACAGAAGGAGATTATGTAAAAATC-3′; F. nucleatum FAM probe, 5′-GTTGACTTTACAGAAGGAGATTA-3′. For human colon tissue, SLCO2A1 was used as endogenous control gene (Hs01114926_m1, FisherThermo Scientific, USA). For human saliva, the MEFE gene (Ba042114926-s1, FisherThermo Scientific, USA) was used as reference gene. A total of 80 ng DNA was used in qPCR reaction and the total reaction volume was 10 ul. Amplification and detection of DNA was performed with the StepOnePlus Real-Time PCR Systems (Applied Biosystems, USA), using the following reaction conditions: 10 min at 95 °C, 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. For quality control, DNA of F. nucleatum strain ATCC 25,586 was used as a positive control. No DNA loading and Diethyl pyrocarbonate (DEPC) treated water were used as negative controls. F. nucleatum positivity was defined as a detectable level of F. nucleatum DNA within 40 PCR cycles, and F. nucleatum negativity was defined as an undetectable level with a proper amplification of human reference gene SLCO2A1. The bacterium level relative quantification is automatically provided by StepOne Plus Realtime PCR Systems (Applied Biosystems, USA) as 2−ΔCq value, with ΔCq = average Cq value of F. nucleatum − average Cq value of total bacteria or of the reference gene.
Statistical analysis
Since this is a pilot study, the statistical analysis performed was purely exploratory, in order to help future studies in study-design decisions, including sample-size calculations.
The distributions of relevant characteristics concerning CRN risk factors in the case and control series were presented with mean and standard deviation, or median and inter-quartile range (when data seemed non-normally distributed) for continuous variables, and percentage for categorical variables. Based on data from qPCR analysis of study specimens, we calculated both frequencies of positive detection of F. nucleatum and medians of 2−ΔCq with their corresponding 95% confidence intervals (CI), in each group (cases and controls) and each specimen type (saliva, colorectal mucosa). Also, coefficients of Spearman correlation between salivary and colorectal F. nucleatum levels, as well as between F. nucleatum levels in heathy mucosa of both the ascending and descending colon were presented with their corresponding 95% CIs. IBM SPSS Statistics version 26 was used for statistical analysis.
Ethical approval
The study was approved by the University of Montreal Hospital Centre Research Ethics Committee, and we certify that the study was performed in accordance with the ethical standards as laid down in the 1964 Declaration of Helsinki and its later amendments.
Informed consent
All study participants have provided an informed consent to participate in the study.
Results
A total of 75 potentially eligible participants were solicited, of whom 20 did not meet the study eligibility criteria, and 12 refused to participate. Therefore 43 patients participated to this pilot study, including 22 cases of CRN and 21 CRN-free controls.
All participants, except one case, completed the study questionnaire. Distributions of cases and controls according to sociodemographic and other relevant characteristics are presented in Table 2. In general, case and control groups were similar regarding education attainment (mostly college or university), family history of CRC, and history of diabetes. However, cases were mostly males (81%), slightly older, and less regular users of non-steroidal anti-inflammatory drugs than controls. Also, although the frequency of cigarette smoking was similar in the two groups, the median number of packs-years among smokers in the case group was much higher than for smokers in the control group. Patients in the control group consumed more red and processed meats but fewer alcoholic drinks than cases. Ten participants (7 cases and 3 controls) had a positive history of PD.
Table 2.
Sociodemographic characteristics and potential colorectal neoplasm risk factors in study participants.
| Characteristic | Cases, n = 22 | Controls, n = 21 |
|---|---|---|
|
Age, years Mean (SD) |
63.9 (9.6) | 60.4 (9.1) |
| Gender, n (%) | ||
| Male | 18 (82) | 10 (48) |
| Canadian born | ||
| Yes, n (%) | 16 (73) | 18 (86) |
| Native tongue | ||
| French, n (%) | 15 (68) | 20 (95) |
| Education attainment | ||
| College or university, n (%) | 15 (68) | 15 (71) |
| Living alone | ||
| Yes, n (%) | 5 (23) | 11 (52) |
| BMI, kg ∕m2 | ||
| Mean (SD) | 27.7 (6) | 26.2 (4) |
| Family history of CRC | ||
| Yes, n (%) | 4 (18) | 2 (10) |
| Regular use of NSAIDs | ||
| Yes, n (%) | 3 (14) | 9 (43) |
| Diabetes | ||
| Yes, n (%) | 3 (14) | 3 (14) |
| Periodontal disease | ||
| Yes, n (%) | 7 (32) | 3 (14) |
| Personal income (CAD$ per year) | ||
| Median (IQR) | 35 000 (40 000) | 45 000 (80 000) |
| History of smoking | ||
| Positive, n (%) | 14 (64) | 14 (67) |
| Cigarette smoking, packs-years | ||
| Median (IQR) | 22.5 (33.9) | 14.4 (27) |
| Lifetime average daily total alcoholic drinksa | ||
| Median (IQR) | 1 (1.62) | 0.8 (1.2) |
| Lifetime average weekly servingsb of: | ||
|
Red meats Processed meats Median (IQR) |
2.1 (3.1) 1.5 (2.1) |
3.2 (5.6) 1.8 (3.3) |
|
Lifetime total physical activity score, MET hour/week/year Median (IQR) |
88.7 (95.7) | 70.5 (114) |
BMI Body mass index, NSAID Non-steroidal anti-inflammatory drugs, CAD$ Canadian dollars, MET Metabolic equivalent of task; a: one drink including beer (355 ml bottle or can), wine (180 ml), or liquor (150 ml); b: 1 serving of red meats = 180–240 g, 1 serving of processed meats = 55 g; SD: Standard deviation; IQR: Interquartile range.
All cases had undergone polypectomy or endoscopic mucosal resection during colonoscopy, and one polyp was removed in each patient, except for two patients with two polyps removed during the same colonoscopy, bringing the total CRN specimens to 24. Characteristics of the CRN (location, histological type, and size) are presented in Table 3. Fifteen polyps were located in the proximal colon, and 9 in the distal colon. Upon the histopathology report, 15 polyps were conventional adenomas (namely, of the tubular, tubulovillous, or villous histology type), two were serrated sessile adenomas, and two early-stage CRCs. Besides specimens of CRN, biopsy specimens were collected from healthy mucosa in 17 cases (13 from ascending colon and 14 descending colon) and 21 controls (20 from ascending colon and 21 descending). Saliva samples were collected in all participants. Thus, a total of 135 study biospecimens were analyzed by qPCR for detection and quantification of F. nucleatum levels.
Table 3.
Characteristics of colorectal neoplasms in case group.
| Cases | Colorectal neoplasms | Paired healthy mucosa: biopsy collection site | ||||
|---|---|---|---|---|---|---|
| Colorectal anatomic site (segment) | Histologic type (subtype) | High grade dysplasia | Sizeǂ (cm) | Ascending colon | Descending colon | |
| 3 | Proximal (Caecum) | SA (sessile serrated) | 2 | X | X | |
| 4 | Proximal (Transverse) | SA (sessile serrated) | 6 | X | X | |
| 5 | Proximal (Ascending) | CA (Tubular) | 4 | X | X | |
| 6 | Proximal (Ascending) | CA (Tubular) | 16 | X | X | |
| 7 | Proximal (Hepatic flexure) | CA (Tubular) | X | 3 | ||
| Proximal (Ascending) | CA (Tubular) | 5 | ||||
| 9 | Distal (Rectum) | CA (Tubular) | 2.5 | X | ||
| 10 | Proximal (Ascending) | CA (Tubulovillous) | X | 5 | X | X |
| 11 | Proximal (Caecum) | CA (Tubulovillous) | 5 | X | X | |
| 12 | Proximal (Caecum) | CA (Tubulovillous) | 4.5 | |||
| 13 | Proximal (Ascending) | CA (Tubulovillous) | X | 3.5 | ||
| 14 | Proximal (Caecum) | CA (Tubulovillous) | 5 | X | X | |
| 15 | Proximal (Hepatic flexure) | CA (Tubulovillous) | 2 | X | ||
| 16 | Distal (Sigmoid) | CA (Tubulovillous) | 2.5 | X | X | |
| 17 | Distal (Rectum) | CA (Tubulovillous) | X | 10 | X | X |
| 18 | Distal (Rectum) | CA (Tubulovillous) | 2.5 | X | ||
| 19 | Distal (Rectum) | CA (Tubulovillous) | 5 | |||
| Distal (Recto-sigmoid) | CA (Tubulovillous) | 2 | ||||
| 21 | Distal (Rectum) | CA (Tubulovillous) | X | 7 | ||
| 22 | Proximal (Transverse) | CA (Villous) | 4 | X | ||
| 23 | Proximal (Ascending) | CA (Villous) | 5 | X | ||
| 24 | Proximal (Caecum) | CA (Villous) | X | 5 | X | |
| 25 | Distal (Rectum) | CRC (High-grade intraepithelial epidermoid neoplasia) | X | X | ||
| 26 | Distal (Sigmoid) |
CRC (moderately differentiated adenocarcinoma developed on a villous adenoma |
X | 7 | X | |
SA Serrated adenoma, CA Conventional adenoma, CRC colorectal cancer, X applicable.
ǂThe largest diameter is reported.
Table 4 shows the F. nucleatum detection rate by biospecimen type (saliva, mucosa), in case and control groups. F. nucleatum was detected in saliva specimens from almost all cases (21/22) and controls (20/21). F. nucleatum levels (measured by qPCR as 2−ΔCq) in saliva ranged from barely detectable (0.000004) to 3.17 and 2.65, in cases and controls respectively. The median (95% CI) of salivary F. nucleatum level was 0.345 (0.15–0.82) and 0.12 (0.05–0.65) in case and control groups respectively (Fig. 1); and 0.4 (0.13–0.53) in participants with positive history of PD vs 0.14 (0.18–0.73) in participants with negative history of PD.
Table 4.
Detection frequency of Fusobacterium nucleatum by specimen type and participant group.
| Specimen type | Case group (n = 22) | Control group (n = 21) | ||
|---|---|---|---|---|
| Total number of specimens | F. nucleatum detected, n | Total number of specimens | F. nucleatum detected, n | |
| Saliva | 22 | 21 | 21 | 20 |
| Healthy mucosa-ascending colon | 13 | 1 | 20 | 9 |
| Healthy mucosa-descending colon | 14 | 1 | 21 | 6 |
| Colorectal neoplasms | 24 | 1 | NA | NA |
F. nucleatum Fusobacterium nucleatum, NA non-applicable.
Figure 1.
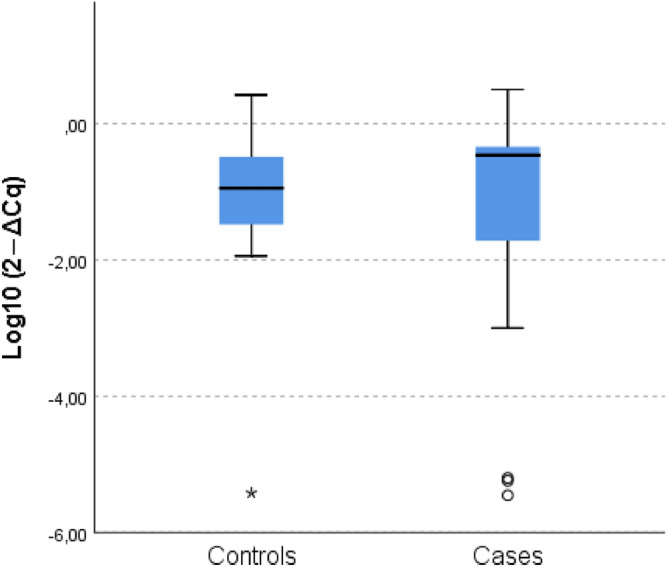
Relative quantification of Fusobacterium nucleatum level in saliva, in case and control groups. Fusobacterium nucleatum level in saliva specimens is measured by qPCR as 2 − ΔCq.
In colorectal mucosa, F. nucleatum was detected in only one case (5%) within both healthy mucosa (from both ascending and descending colon specimens) and polyp, and in 9 controls’ healthy mucosa specimens (ascending and/or descending colon) (43%). The polyp where F. nucleatum was detected was a conventional adenoma, tubular subtype, located in the proximal colon.
The level of F. nucleatum in controls’ healthy mucosa specimens ranged from 0.116 to 2.02 in the ascending colon, and from 0.045 to 1.2 in the descending colon. F. nucleatum level in healthy mucosa from the case detected with F. nucleatum was higher than the maximum observed in controls: 2.574 in the ascending colon, 1.143 in the descending colon, and 1.952 in the polyp.
The Spearman correlation coefficient between F. nucleatum levels in saliva and mucosa samples was 0.64 (95% CI: 0.2–094). This was calculated for controls only, as in cases only one subject had F. nucleatum detected in both saliva and colorectal mucosa specimens. We also explored if there was any correlation between levels of F. nucleatum in heathy mucosa of both ascending and descending colon, and the corresponding Spearman correlation coefficient was calculated as 0.68 (95 CI 0.25–0.96).
Discussion
In this pilot study, we generated preliminary data on detection and quantification of F. nucleatum in both saliva and colorectal mucosa from patients diagnosed with CRN and CRN-free controls.
We were able to collect and analyze a total of 135 biospecimens including saliva samples and healthy-colon mucosa biopsies from most cases and controls, and CRN biopsies from all cases.
F. nucleatum in saliva from CRN cases and CRN-free controls
Analysis with qPCR showed high detection rates of F. nucleatum in saliva from both case and control groups, consistent with previous studies25,29. F. nucleatum is in fact a commensal bacterium of the oral cavity, which explains its more common detection in saliva regardless of disease status. We also found a higher median level of F. nucleatum in saliva from the case group than from controls. A recent study in Turkey with a large number of participants (71 CRC cases and 77 controls)25, and also using qPCR for microbial saliva analysis, showed a higher mean amount of F. nucleatum in CRC group than in control group (6.89 Log10 copies / ml in the case group vs 6.35 in the control group, p = 0.001). However, two other studies that applied 16SrRNA gene sequencing found similar salivary levels of F. nucleatum relative abundance in cases and controls. The first study was conducted in the USA27, and saliva was collected by oral rinse with a commercial mouthwash, among 68 CRC cases and 122 controls. The second was conducted in Japan26, and included unstimulated saliva from 24 CRC cases (and other cancers of the digestive tract) and 118 controls. In both studies, Fusobacterium was not the dominant bacterium.
F. nucleatum in colorectal mucosa from CRN cases and CRN-free controls
We found a low global detection rate of F. nucleatum in colorectal mucosa specimens in the controls, and it was even lower in the cases. At first, this finding might appear contradictory to previous reports finding F. nucleatum to be associated with CRN7–9. However, when considering the histologic type and the location of CRN in the patients in our study, our results can be seen to be consistent with those previous findings. Mima et al.38 analyzed 1,102 colorectal tumors with qPCR, in 13% of which F. nucleatum was detected. When analyzing by colorectal tumor site, F. nucleatum detection was 15% and 9% in proximal and distal-rectal sites, respectively. Also, in a previous study by Yu et al.39, where F. nucleatum was investigated in 280 CRNs and 20 healthy mucosa specimens from independent controls using FISH technique, that was further validated in 20 samples by F. nucleatum-specific PCR primers, F. nucleatum was prevalent in proximal serrated sessile adenomas, but rare in conventional adenomas39. According to that study, the frequency of F. nucleatum positivity (defined as > 5 visualized probes per field) was 29% in proximal conventional adenomas, 24% in distal conventional adenomas, 79% in serrated sessile adenomas, 90% in proximal CRCs, 42% in distal CRCs, and 20% in healthy mucosa from independent controls. High abundance of invasive F. nucleatum (defined as ˃ 20 visualized probes per field) was present in 5.3% of proximal conventional adenomas, 2.4% of distal conventional adenomas, 49% of serrated sessile adenomas, 71% of proximal CRCs, 38% of distal CRCs, and none of the 20 healthy mucosa samples. We point out that the CRNs sampled in our study included 12 proximal conventional adenomas, 6 distal conventional adenomas, 2 serrated sessile adenomas and 2 distal CRCs, and that F. nucleatum was detected in a proximal conventional adenoma.
In this pilot study, we noticed that F. nucleatum was usually either detected in both subject’s proximal and distal colon sites (ascending and descending colon healthy mucosa specimens), or not detected at all in both colon sites. On the other hand, we noticed that F. nucleatum level in the ascending colon moderately correlated with level in the descending colon. This could probably be because some subjects naturally harbor F. nucleatum in their gut microbiome, whereas others do not, which can also explain the detection of F. nucleatum in healthy mucosa of some controls in many previous studies. We can also think F. nucleatum may be associated to the intestinal disorders that led patients in the control group to undergo colonoscopy, and that it may be particularly involved in the serrated neoplasia pathway (where sessile serrated adenomas are precursors to tumors with sporadic microsatellite instability), and less in the conventional adenoma-carcinoma sequence, as suggested by Yu et al.39.
Comparison of F. nucleatum in saliva and in colorectal mucosa within CRN cases and CRN-free controls
Detection rate of F. nucleatum in colorectal mucosa was much lower than in saliva, and few subjects had F. nucleatum detected in both sites. We found a moderate correlation between F. nucleatum level in saliva and healthy proximal colorectal mucosa in controls, but we could not explore this correlation in cases as F. nucleatum was detected in colorectal mucosa specimens of only one CRN case. The only data that could serve as comparison to our finding came from two previous studies that investigated F. nucleatum in a few samples of saliva and colorectal tumors in the same CRC cases, without control group and without bacterium quantification, as only conventional PCR (non-quantitative) was used18,28. F. nucleatum was detected less commonly in mucosa samples than in saliva in the first study (in 8/14 tumors and 14/14 saliva)18, and in all specimens in the second one (in 10 tumors and 10 saliva)28.
In conclusion, concerning the objectives of the pilot study, our study findings provide potentially useful preliminary data on expression of F. nucleatum in both oral and colorectal body sites in patients diagnosed with CRN and CRN-free controls. Further studies that aim to assess the association between oral and colorectal levels of F. nucleatum in CRN cases are still needed and can draw from our methods and results in making study-design decisions, including the inclusion/exclusion criteria, selection of study participants, data collection instruments and analysis, and sample size calculation. They also should pay attention to both the histologic type and site of CRNs to be included, and optimally focus on proximal location and adenoma of sessile type. To quantify F. nucleatum in colorectal healthy mucosa, there may no longer be a need for collection and analysis of two different specimens from both proximal and distal colon sites, as one specimen (preferably from the proximal colon) can be sufficiently informative, especially given that some patients may not consent to provide biopsies from healthy mucosa even if they agree to provide specimens of their tumor, which they know will be excised anyway. Finally, studies should generally plan a quantitative microbial analysis of F. nucleatum in saliva specimens: non-quantitative techniques (such as conventional PCR, for example) only assess the presence of the bacterium.
Our preliminary results encourage future research to investigate oral and colorectal enrichment in F. nucleatum, in patients with precancerous lesions as well as cancerous lesions at different stages of colorectal malignant transformation, to overcome the difficulty of conducting prospective research on the causal role of the oral bacterium F. nucleatum in colorectal carcinogenesis. If the hypothesis of an intra-individual oral origin of the colorectal enrichment in F. nucleatum is confirmed, this may have potential impact on colorectal cancer prevention, diagnosis, and treatment. Thus, many studies are investigating the potential of candidate fecal bacteria as biomarkers for early detection of adenomatous polyps and colon cancer40,41. Under the same perspective, saliva can be a promising non-invasive screening tool for colorectal adenoma and cancer. Also, and more importantly, investigating the association between the oral and colorectal levels of F. nucleatum in colorectal neoplasms patients can advance the understanding of the mechanism(s) underlying the connection between periodontal disease and colorectal cancer, which may involve the translocation of periodontal pathogens to the gut and the release of their pro-oncogen and pro-inflammatory virulence products. Periodontal disease is suggested as a risk factor for periodontal disease42, but the mechanisms of the association have yet to be elucidated. Thus, a subsequent, larger epidemiological study on this topic is highly recommended.
Acknowledgements
We thank all study participants, as well as Dr Sacha Sidani and Dr Simon Bouchard who helped in participant identification and biopsy collection. We also thank the Network for Oral and Bone Health Research for its financial support to publication costs.
Abbreviations
- CRC
Colorectal cancer
- CRN
Colorectal neoplasm
- F. nucleatum
Fusobacterium nucleatum
- PCR
Polymerase chain reaction
- PD
Periodontal disease
- qPCR
Quantitative polymerase chain reaction
Author contributions
A.I.J. contributed to conception, design, data acquisition, and interpretation, and drafted and critically revised the manuscript. I.K. and E.E. contributed to conception, design and interpretation, and critically revised the manuscript. D.V.R. and M.B. contributed to conception and biopsy collection, and critically revised the manuscript. Y.L. and S.D.T. contributed to conception, performed microbial analysis, and critically revised the manuscript. All authors gave their final approval and agreed to be accountable for all aspects of the work.
Funding
Idrissi Janati, A was supported by a doctoral scholarship from the Fonds de Recherche du Québec-Santé (FRQ-S).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sung H, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Keum N, Giovannucci E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019;16(12):713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 3.Pecori S, et al. Intestinal Polyps and Polyposis: From Genetics to Treatment and Follow-up. Springer; 2009. Pathological Features of Sporadic Colonic Adenoma; pp. 19–47. [Google Scholar]
- 4.Flanagan L, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur. J. Clin. Microbiol. Infect. Dis. 2014;33(8):1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 5.Yu YN, et al. Berberine may rescue Fusobacterium nucleatum-induced colorectal tumorigenesis by modulating the tumor microenvironment. Oncotarget. 2015;6(31):32013–32026. doi: 10.18632/oncotarget.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SH, et al. Quantification of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut. 2017;66(8):1441–1448. doi: 10.1136/gutjnl-2016-312766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rezasoltani S, et al. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018;124:244–249. doi: 10.1016/j.micpath.2018.08.035. [DOI] [PubMed] [Google Scholar]
- 8.McCoy AN, et al. Fusobacterium Is associated with colorectal adenomas. PLoS ONE. 2013;8(1):8. doi: 10.1371/journal.pone.0053653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye XC, et al. Fusobacterium nucleatum subspecies animalis influences proinflammatory cytokine expression and monocyte activation in human colorectal tumors. Cancer Prev. Res. 2017;10(7):398–409. doi: 10.1158/1940-6207.CAPR-16-0178. [DOI] [PubMed] [Google Scholar]
- 10.Rubinstein MR, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gur C, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–355. doi: 10.1016/j.immuni.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapatral V, et al. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 2002;184(7):2005–2018. doi: 10.1128/JB.184.7.2005-2018.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han YW. Fusobacterium nucleatum interaction with host cells. In: Kolenbrander PE, editor. Oral Microbial Communities: Genomic Inquiry and Interspecies Communication. Springer; 2011. pp. 221–223. [Google Scholar]
- 14.Dumitrescu AL, Ohara M. Etiology and Pathogenesis Of Periodontal Disease. Berlin: Springer; 2010. Periodontal microbiology; pp. 39–73. [Google Scholar]
- 15.Signat B, Roques R, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr. Issues Mol. Biol. 2011;13(2):25–36. [PubMed] [Google Scholar]
- 16.Saygun I, et al. Salivary infectious agents and periodontal disease status. J. Periodontal Res. 2011;46(2):235–239. doi: 10.1111/j.1600-0765.2010.01335.x. [DOI] [PubMed] [Google Scholar]
- 17.Abed J, et al. Colon cancer-associated Fusobacterium nucleatum may originate from the oral cavity and reach colon tumors via the circulatory system. Front. Cell Infect. Microbiol. 2020;10(400):10. doi: 10.3389/fcimb.2020.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komiya Y, et al. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut. 2019;68(7):1335–1337. doi: 10.1136/gutjnl-2018-316661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmidt, T.S. et al. 2019. Extensive transmission of microbes along the gastrointestinal tract. Elife 8. [DOI] [PMC free article] [PubMed]
- 20.Xuan K, et al. Is periodontal disease associated with increased risk of colorectal cancer? A meta-analysis. Int. J. Dent. Hyg. 2020;2:2. doi: 10.1111/idh.12483. [DOI] [PubMed] [Google Scholar]
- 21.Lee D, et al. Association between oral health and colorectal adenoma in a screening population. Medicine (Baltimore) 2018;97(37):e12244. doi: 10.1097/MD.0000000000012244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo CH, et al. Periodontal disease, tooth loss, and risk of serrated polyps and conventional adenomas. Cancer Prev. Res. (Phila). 2020;13(8):699–706. doi: 10.1158/1940-6207.CAPR-20-0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim GW, et al. Periodontitis is associated with an increased risk for proximal colorectal neoplasms. Sci. Rep. 2019;9(1):7528. doi: 10.1038/s41598-019-44014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flemer B, et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guven DC, et al. Analysis of Fusobacterium nucleatum and Streptococcus gallolyticus in saliva of colorectal cancer patients. Biomark. Med. 2019;13(9):725–735. doi: 10.2217/bmm-2019-0020. [DOI] [PubMed] [Google Scholar]
- 26.Kageyama S, et al. Characteristics of the salivary microbiota in patients with various digestive tract cancers. Front. Microbiol. 2019;10:1780. doi: 10.3389/fmicb.2019.01780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kato I, et al. Oral microbiome and history of smoking and colorectal cancer. J. Epidemiol. Res. 2016;2(2):92–101. doi: 10.5430/jer.v2n2p92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Russo E, et al. Preliminary comparison of oral and intestinal human microbiota in patients with colorectal cancer: a pilot study. Front. Microbiol. 2017;8:2699. doi: 10.3389/fmicb.2017.02699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, et al. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer. 2019;144(10):2381–2389. doi: 10.1002/ijc.31941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mannath J, Ragunath K. Endoscopic mucosal resection: Who and how? Therap. Adv. Gastroenterol. 2011;4(5):275–282. doi: 10.1177/1756283X10388683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Imperiale TF, et al. Multitarget stool DNA testing for colorectal-cancer screening. N. Engl. J. Med. 2014;370(14):1287–1297. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 32.Idrissi Janati A, Karp I, Latulippe J-F, Charlebois P, Emami E. Periodontal disease as a risk factor for sporadic colorectal cancer: Results from COLDENT study. Cancer Causes Control. 2022;33(3):463–472. doi: 10.1007/s10552-021-01541-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedenreich CM, Courneya KS, Bryant HE. The lifetime total physical activity questionnaire: Development and reliability. Med. Sci. Sports Exerc. 1998;30(2):266–274. doi: 10.1097/00005768-199802000-00015. [DOI] [PubMed] [Google Scholar]
- 34.Willett WC, Reynolds RD, Cottrell-Hoehner S, Sampson L, Browne ML. Validation of a semi-quantitative food frequency questionnaire: Comparison with a 1-year diet record. J. Am. Diet. Assoc. 1987;87(1):43–47. doi: 10.1016/S0002-8223(21)03057-1. [DOI] [PubMed] [Google Scholar]
- 35.Cade J, Thompson R, Burley V, Warm D. Development, validation and utilisation of food-frequency questionnaires: A review. Public Health Nutr. 2002;5(4):567–587. doi: 10.1079/PHN2001318. [DOI] [PubMed] [Google Scholar]
- 36.Siemiatycki J, Krewski D, Franco E, Kaiserman M. Associations between cigarette smoking and each of 21 types of cancer: A multi-site case-control study. Int. J. Epidemiol. 1995;24(3):504–514. doi: 10.1093/ije/24.3.504. [DOI] [PubMed] [Google Scholar]
- 37.Mima K, et al. Fusobacterium nucleatum and T cells in colorectal carcinoma. JAMA Oncol. 2015;1(5):653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mima K, et al. Fusobacterium nucleatum in colorectal carcinoma tissue according to tumor location. Clin. Transl. Gastroenterol. 2016;7(11):e200. doi: 10.1038/ctg.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, et al. Invasive Fusobacterium nucleatum may play a role in the carcinogenesis of proximal colon cancer through the serrated neoplasia pathway. Int. J. Cancer. 2016;139(6):1318–1326. doi: 10.1002/ijc.30168. [DOI] [PubMed] [Google Scholar]
- 40.Rezasoltani S, et al. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J Microbiol. Methods. 2018;155:82–88. doi: 10.1016/j.mimet.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Suehiro Y, et al. Highly sensitive stool DNA testing of Fusobacterium nucleatum as a marker for detection of colorectal tumours in a Japanese population. Ann. Clin. Biochem. 2017;54(1):86–91. doi: 10.1177/0004563216643970. [DOI] [PubMed] [Google Scholar]
- 42.Xuan K, et al. Is periodontal disease associated with increased risk of colorectal cancer? A meta-analysis. Int. J. Dent. Hyg. 2021;19(1):50–61. doi: 10.1111/idh.12483. [DOI] [PubMed] [Google Scholar]


