Abstract
It is well known that metabolism underlies T cell differentiation and functions. The pathways regulating T cell metabolism and function are interconnected, and changes in T cell metabolic activity directly impact the effector functions and fate of T cells. Thus, understanding how metabolic pathways influence immune responses and ultimately affect disease progression is paramount. Epigenetic and posttranslational modification mechanisms have been found to control immune responses and metabolic reprogramming. Sirtuins are NAD+-dependent histone deacetylases that play key roles during cellular responses to a variety of stresses and have recently been reported to have potential roles in immune responses. Therefore, sirtuins are of significant interest as therapeutic targets to treat immune-related diseases and enhance antitumor immunity. This review aims to illustrate the potential roles of sirtuins in different subtypes of T cells during the adaptive immune response.
Subject terms: Acetylation, T cells
Immunity: How to give T cells a boost
Sirtuins, enzymes that regulate how cells respond to stress, regulate T cell metabolism and functions, and therefore blocking or boosting sirtuins influences immune responses. As part of the immune system, some types of T cells attack specific targets; others keep the immune response in check. Imene Hamaidi and Sungjune Kim at H. Lee Moffitt Cancer Center, Tampa, USA, have reviewed how sirtuins affect different subsets of T cells to either promote or suppress immune responses. Boosting sirtuins that increase the function of inflammation-suppressing T cells can improve outcomes for transplant recipients or help treat autoimmune diseases. Conversely, stimulating immune-activating sirtuins can help re-energize exhausted antitumor T cells. Understanding the complex web of sirtuin–T cell interactions may help in developing therapeutic strategies for improving transplant outcomes, and for treating autoimmune diseases and cancer.
Introduction
Overview of the T cell-mediated adaptive immune response
The adaptive immune response elicited by T cells is crucial in mounting specific immune responses against foreign pathogens. T cells develop in the thymus and, upon maturation, are classified by their expression of either CD4 or CD8 receptors. CD4+ and CD8+ T cells exist in several subsets that perform unique functions during an immune response and are characterized by specific surface receptors, cytokine secretion, and expression of lineage-defining transcription factors (Fig. 1)1,2. Depending on the pathogen type and the cytokines generated by antigen-presenting cells (APCs), CD4+ T cells can become activated and differentiate into functionally distinct T helper (Th) 1, Th2, Th17, Th9, or T regulatory (Treg) cells3, while CD8+ T cells differentiate into cytotoxic T lymphocytes (CTLs)4. Th1 cells, which produce interferon-gamma (IFN-γ), interleukin (IL)-2, and tumor necrosis factor-beta (TNF-β), evoke cell-mediated immunity and phagocyte-dependent inflammation and are involved in the elimination of intracellular pathogens5. Th2 cells, which produce IL-4, IL-5, IL-6, IL-9, IL-10, and IL-13, are known to provide immunity against extracellular parasites, including helminths, and play major roles in the induction and persistence of asthma as well as other allergic diseases6. Th9 cells are known to primarily produce IL-9 and facilitate the immune response against helminth parasites7. Th17 cells are characterized by the production of IL-17 and are involved in host protection against microbial infections that are not resolved by Th1 or Th2 immunity, including infections with a subset of extracellular bacteria and some fungi8. Retinoic acid receptor-related orphan nuclear receptor gamma (RORγt) is considered to be one of the master regulators of the development of Th17 cells8. In contrast to the proinflammatory phenotype of Th cells, Treg cells participate in the prevention of uncontrollable inflammatory responses, autoimmune diseases and allergies by suppressing T effector (TEFF) cells and other immune cells9. Treg cells are defined by the expression of the forkhead box P3 (Foxp3) transcription factor10. This cell subset secretes key anti-inflammatory cytokines, including IL-10, tumor growth factor-β (TGF-β), and IL-35. CTLs are CD8+ TEFF cells that participate in cellular immunity, and their role is to directly kill infected or malignant cells through the release of cytotoxic cytokines, including TNF-α and IFN-γ, as well as granules including perforin and granzymes4.
Fig. 1. The metabolic programs of CD4+ T cell subsets.
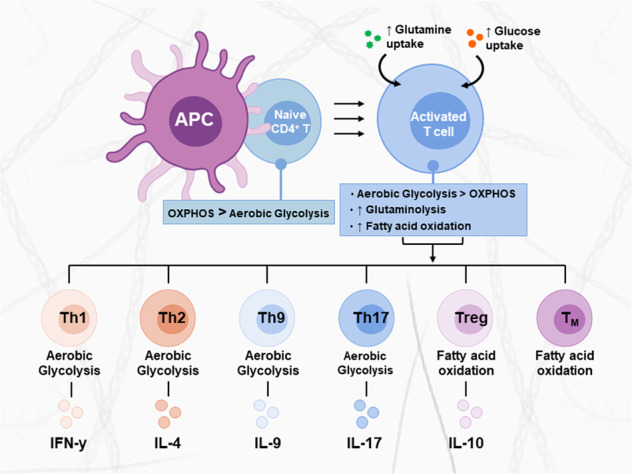
Distinct T cell subsets utilize specific metabolic programs to support their functions. Each functional subset is characterized by specific signaling pathways, transcription factors, metabolic programs, and effector cytokines. Naive T cells are quiescent and rely on OxPhos for their minimal energetic needs. Upon activation, activated T cells switch to aerobic glycolysis and increase glucose and glutamine uptake, which supports cell growth and proliferation. The differentiation of activated T cells into different subsets is due to several metabolic and signaling pathways. Th1, Th2, Th9, and Th17 cells primarily rely on aerobic glycolysis and glutaminolysis; in contrast, Treg and TM cells upregulate fatty acid oxidation.
Metabolic shifts in T cells
Naive T (TN) cells are quiescent and require energy only for survival and circulation. These cells rely predominantly on mitochondrial oxidative phosphorylation (OxPhos) metabolism to generate ATP from glucose or fatty acids (FAs)11. Activation of T cells occurs after stimulation of the T cell antigen receptor (TCR) by a ligand in association with costimulatory signals, which initiates several signaling pathways that promote cell differentiation and growth. This activation is associated with a transition from relatively quiescent oxidative metabolism to intense glycolytic metabolism to support energetic needs for proliferation and cytokine production11–13. In addition to increased glucose catabolism in activated T cells, glutamine uptake and glutaminolysis are also upregulated to supply the tricarboxylic acid (TCA) cycle for ATP generation14,15. The metabolic profile of each specialized T cell subset is optimized to support their unique functions. TEFF cells, including Th1, Th2, and Th17 cells and CTLs, principally rely on aerobic glycolysis and glutaminolysis to promote their rapid growth, proliferation, and effector functions16,17. In contrast, Treg cells rely primarily on FA oxidation (FAO) and glutaminolysis to support their suppressive activity18,19. Following the primary immune response, a portion of CD4+ and CD8+ T cells become memory T (TM) cells that remain ready to rapidly respond to the same antigen. These cells are long-lived, and until reactivation, they exhibit relatively quiescent oxidative metabolism fueled by FAO, similar to that of TN cells (Fig. 1)20,21. Due to competition for glucose and glutamine with cancer cells, tumor-infiltrating T lymphocytes (TILs) are metabolically compromised and functionally exhausted within the tumor microenvironment (TME)22. To partially rescue their effector functions, TILs have been shown to increase FAO metabolism as a way to utilize alternative fuel sources23.
Increasing evidence indicates that immune cell identity, functions, and metabolism are mediated by overlapping signaling pathways that are controlled, at least in part, via epigenetic and posttranslational modification (PTM) mechanisms. Sirtuins are key epigenetic and PTM regulators in T cells. In this review, we will provide relevant insights into the current understanding of T cell-specific immune response regulation by sirtuins and the therapeutic potential of sirtuin modulators in immune-related diseases.
Epigenetic mechanisms
In multicellular organisms, all cells contain the same genome; however, gene expression profiles vary from cell to cell, which contribute to differentiation into various cell types within the same organism24. This cell type differentiation is mostly regulated by epigenetic mechanisms, which result in dynamic changes in gene expression patterns without changing the DNA sequence25. Epigenetic regulation of gene expression is known to occur at the DNA, histone, and RNA levels. In this context, DNA methylation, histone methylation, acetylation, ubiquitination, phosphorylation, and microRNA-dependent gene silencing have been well characterized26. Methylation of DNA is mediated by DNA methyltransferases and consists of the addition of a methyl group to cysteine residues within DNA regions that are rich in cysteine-guanine dinucleotides. DNA methylation can act either to inhibit gene transcription if methylation occurs within a promoter region or to promote gene transcription if methylation occurs within the gene body27. Noncoding microRNAs are single-stranded RNA fragments that interact with target messenger (m)RNAs via a perfectly complementary base sequence, resulting in mRNA cleavage and translational repression28. Another important and common epigenetic mechanism that contributes to gene expression regulation is the modification of histones. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are the two classes of enzymes involved in the dynamic regulation of histone acetylation29. Acetylated histones maintain an open and fluid chromatin structure, called euchromatin, promoting gene transcription, while deacetylated histones often form a tightly packed chromatin structure that prevents gene transcription, called heterochromatin30.
Posttranslational modification
PTM is a biochemical mechanism in which amino acid residues from a protein are covalently modified after translation to regulate protein folding, degradation, signaling, localization, stability, enzymatic activity or protein–protein interactions31. There are over 400 known PTM types, including phosphorylation, glycosylation, ubiquitination, nitrosylation, methylation, acetylation, lipidation and proteolysis, which influence almost all aspects of physiological and pathological cell processes32. In contrast to epigenetic mechanisms, PTM allows rapid changes in protein properties in response to cellular needs during acute stress phases. In addition, multisite PTM leads to a vast variety of potential molecular states33. Phosphorylation is the most widely studied PTM and can regulate signaling pathways, the cell cycle, metabolism, the immune response, and cellular growth and differentiation34. Ubiquitin (Ub) is a small and highly conserved protein made of 76 amino acids that can be attached to a lysine substrate through a complex conjugation process requiring a Ub-activating enzyme (E1), Ub-conjugating enzyme (E2) and Ub ligase (E3). Ubiquitination plays important regulatory roles in the life cycle of proteins by leading to the degradation of target substrates35. This process can be reversed by deubiquitinating enzymes (DUBs), which remove conjugated Ub molecules from target substrates36. Protein acetylation has emerged as a key PTM participating in cellular regulation, particularly through the modification of histones, nuclear transcription regulators and metabolic enzymes37. Lysine acetylation is the prevalent modification of metabolic enzymes, and virtually every enzyme in glycolysis, gluconeogenesis, the TCA cycle, the urea cycle, fatty acid metabolism, and glycogen metabolism has been found to be acetylated in human liver tissue38. Protein acetylation status is regulated by a highly dynamic equilibrium between HATs and HDACs39.
Overview on sirtuins
The sirtuin proteins are classified within class III HDACs, which require nicotinamide adenine dinucleotide (NAD+) as a cofactor for their deacetylase activity. Although initially identified as HDACs, later studies revealed that sirtuins can deacetylate a variety of nonhistone proteins40. Moreover, sirtuins show additional enzymatic functions other than deacetylation.
In mammals, seven sirtuins (Sirt1–7) are ubiquitously expressed with distinct subcellular localization and functions (Fig. 2)41–43. Sirt1 is the most studied mammalian sirtuin and is primarily localized in the nucleus. However, under specific conditions, Sirt1 can be transported to the cytoplasm44. Sirt2 is a predominantly cytosolic sirtuin that can migrate to the nucleus during mitosis45. Sirt6 is exclusively localized in the nucleus, whereas Sirt7 is localized in the nucleolus41. All the remaining sirtuins (3–5) are localized in mitochondria46. All sirtuins except Sirt5 have mono-ADP-ribosyl transferase activity, whereas Sirt5 catalyzes the removal of succinyl, malonyl, and glutaryl groups from protein targets47,48.
Fig. 2. The subcellular localization and main functions of the mammalian sirtuins.
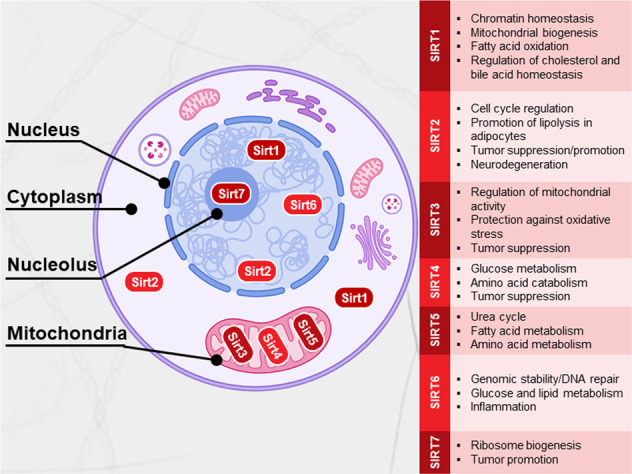
Sirt1 is predominantly located in the nucleus and can also be found in the cytosol. Sirt2 is predominantly localized in the cytosol but can shuttle to the nucleus. Sirt3, Sirt4, and Sirt5 are mitochondrial proteins. Sirt6 and Sirt7 are localized in the nucleus and nucleolus, respectively. The main cellular functions are indicated in the boxes.
The first sirtuin protein was discovered in yeast as silent information regulator 2 (Sir2), whose homolog in mammals is known as Sirt149. Since it was discovered that Sir2 extends yeast lifespan in response to calorie restriction, sirtuins have received considerable attention as anti-aging regulators50. Furthermore, sirtuins have been described to regulate a variety of cell processes, including genome stability and cell metabolism42,43, and are currently gaining extensive interest for their role in mediating several diseases associated with inflammation51, metabolic disorders52, and cancers53 (Fig. 2).
As sirtuins depend on NAD+ (a critical cofactor of metabolism) for their enzymatic activity, any fluctuations in NAD+ levels related to nutrient availability affect their functions54. Hence, sirtuins serve as energy sensors that directly link environmental signals to cellular metabolic homeostasis. Given the link between metabolic reprogramming, nutrient availability and T cell effector function55, sirtuins are speculated to be potential regulators of the immune response, impacting the ability of the immune system to combat foreign pathogens or malignant cells. While each of the sirtuins has been broadly studied and demonstrated to be involved in a number of diseases associated with inflammation56, investigations of the roles of sirtuins in adaptive immunity have been limited to certain aspects of the immune response, and the most intense attention has been given to Sirt1 and Sirt357. This review will discuss the latest advances in the roles of the seven mammalian sirtuins in the T cell adaptive response and include our considerations for targeting sirtuins to manipulate the immune response.
Sirt1
Most studies on Sirt1 have indicated that decreased Sirt1 expression or activity contributes to the enhancement of T cell activation, thereby leading to the occurrence of autoimmune diseases58. The immunoregulatory function of Sirt1 is dependent on the cell types and specific substrates that are targeted within the immune response. In addition, Sirt1-mediated regulation of metabolic processes is critical for optimal immune cell function59.
c-Jun is upregulated after T cell activation to induce IL-2 production, cell proliferation and differentiation60. An early study indicated that Sirt1 inhibits the immune response by blocking c-Jun transcription factor activity, which supports IL-2 production61 and thereby decreases Th1 cell activation. Indeed, enhanced expression and activity of Sirt1 induced by resveratrol treatment impeded CD4+ T cell activation and IFN-γ production, further confirming that Sirt1 negatively impacts Th1 differentiation and IFN-γ secretion62. More recently, in a mouse model of ovarian cancer, Th1 cell differentiation of CD4+ T cells was found to be increased after treatment with artesunate, a promoter of microRNA-142 expression that downregulates Sirt1 expression, again confirming the suppressive role of Sirt1 in Th1 cell differentiation63.
Studies in Sirt1 knockout (Sirt1−/−) mice showed that Sirt1−/− T cells display increased proliferation and IL-2 production, and Sirt1−/− mice are more susceptible to developing autoimmune diseases64. Further studies indicated that IL-2 can reverse T cell anergy by suppressing Sirt1 transcription via cytosolic sequestration of its upstream transcription factor, FoxO3a. The expression of a constitutively active form of FoxO3a blocks IL-2-mediated reversal of T cell tolerance by retaining Sirt1 expression65.
B-cell lymphoma 2-associated factor 1 (Bclaf1), primarily considered a promoter of cellular apoptosis, has been found to be critical for T cell activation66. Kong et al. reported that Sirt1 suppresses Bclaf1 activity by deacetylating histone lysine residues at the Bclaf1 promoter region, resulting in decreased IL-2 gene transcription. Accordingly, Sirt1−/− T cells displayed higher expression of the Bclaf1 gene and IL-2 production, and specific knockdown of Bclaf1 reversed the increase in IL-2 production and proliferation observed in Sirt1−/− T cells67.
Early studies using ovalbumin-induced asthma models in mice demonstrated that pharmacologic inhibition of Sirt1 reduced allergic reactions compared with mock treatment68,69. Furthermore, Sirt1 inhibition was found to suppress the differentiation of Th2 cells through the B cell lymphoma/leukemia 11B (Bcl11b) transcriptional activator. Bcl11b is essential for Th2 differentiation, and mice lacking Bcl11b in mature T cells have a diminished capacity to mount Th2 responses during helminth infection and allergic asthma70. Sirt1 interacts directly with Bcl11b and is recruited to the promoter template in a Bcl11b-dependent manner to deacetylate histones, leading to transcriptional repression71.
Hypoxia-inducible factor 1-alpha (HIF-1α) activity has consistently been associated with the generation of proinflammatory cytokines and restriction of anti-inflammatory cytokines72. Sirt1 inhibition was found to promote Th9 cell differentiation and IL-9 production via the Sirt1-mTOR-HIF-1α axis73. In the same study, Wang et al. indicated that Sirt1-dependent regulation of glycolysis was critical for directing the differentiation of Th9 cells, highlighting the importance of metabolic reprogramming in controlling T cell fate.
The effects of Sirt1 on Th17 cells are controversial, and more studies are needed to dissect its precise role. A few studies have indicated that the differentiation of Th17 cells is affected by the degree of STAT3 deacetylation. Sirt1 activators such as metformin have been shown to impede Th17 cell differentiation and reduce IL-17A and RORγt expression via deacetylation of the STAT3 transcription factor. STAT3 deacetylation restricts its ability to translocate into the nucleus to induce RORγt transcription74. Another study showed that in vivo activation of Sirt1 using NAD+ supplementation delayed the onset of experimental autoimmune encephalomyelitis (EAE). This protection was hypothesized to be a result of the decrease in Th17-mediated inflammatory responses induced by enhanced Sirt1 expression75. Moreover, treatment with methylene blue, another Sirt1 activator, alleviated Th17 responses and significantly reduced the clinical scores of EAE in mice76. In contrast, other researchers have shown that Sirt1 activation promotes the Th17 phenotype via RORγt deacetylation. Deacetylated RORγt appears to have stronger transcriptional activity than Foxp3, thus strengthening the Th17 proinflammatory phenotype and suggesting that Sirt1 inhibitors may protect against autoimmune diseases77.
Foxp3 is a master regulator of Treg cell development and function and has three lysine acetylation sites (K31, K262, and K267) targeted by Sirt178. Hyperacetylation of Foxp3 prevents its polyubiquitination and proteasomal degradation. Accordingly, Sirt1 deacetylase activity has been found to reduce Foxp3 protein levels, and treatment with Sirt1 inhibitors results in increased functional Treg cells79. In addition to the posttranslational control of Foxp3 by Sirt1, some studies reported that genetic deletion or pharmacologic inhibition of Sirt1 increased the number and suppressive activity of Foxp3+ Treg cells by increasing Foxp3 mRNA levels78,79. In contrast, Sirt1 was reported to promote Treg cell survival by stabilizing the Notch1 intracellular domain proximal to the membrane, given that the Notch receptor is essential for Treg cell survival within caloric-restricted conditions80.
Basic leucine-zipper ATF-like transcription factor (BATF) regulates multiple aspects of the T cell immune response81. Sirt1 was reported to impact CD8+ T cell differentiation and effector functions under the influence of BATF. Indeed, BATF has been shown to transcriptionally inhibit Sirt1 expression, resulting in increased histone acetylation at the T-bet locus. In turn, high levels of T-bet expression promote CD8+ T cell differentiation, and loss of BATF consequently inhibits CD8+ T cell differentiation82.
The metabolic switch toward FAO is a characteristic of TM cell differentiation, and this switch is known to be supported by the transcriptional coactivator PGC-1, which is involved in mitochondrial biogenesis and OxPhos metabolism83. Sirt1-mediated deacetylation has been shown to increase the transcriptional activity of the PGC-1α and PGC-1β cofactors84,85, suggesting a potential role for Sirt1 in promoting TM cell formation. A previous report indicated a decrease in Sirt1 expression levels in terminally differentiated CD8+CD28− TM cells, which concomitantly displayed enhanced glycolytic and cytotoxic capabilities86, suggesting that Sirt1 may restrain glycolytic metabolism in T cells (Fig. 3).
Fig. 3. Mammalian sirtuins and metabolic reprogramming.
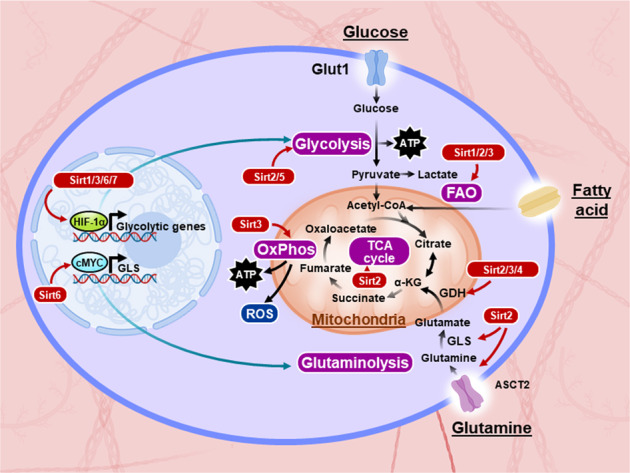
The entry of glucose into the cell is mediated by the glucose transporter Glut1. Glucose is then metabolized to pyruvate, which enters the mitochondrial tricarboxylic acid (TCA) cycle and generates reducing equivalents for ATP production via oxidative phosphorylation (OxPhos). During aerobic glycolysis, pyruvate is fermented into lactate in the cytoplasm despite the availability of oxygen for complete glucose oxidation; this process is called the Warburg effect. Glutamine enters cells using alanine, serine, cysteine-preferring transporter 2 (ASCT2), and it is converted into glutamate by glutaminase (GLS) and into α-ketoglutarate (α-KG) by glutamate dehydrogenase (GDH), which enters the TCA cycle for ATP production via OxPhos. Fatty acid molecules are catabolized into acetyl-CoA via fatty acid oxidation (FAO), and acetyl-CoA enters the TCA cycle for ATP production. Sirtuins are metabolic sensors that modulate a variety of metabolic pathways. Sirt1, Sirt3, and Sirt6 restrain the glycolytic pathway through hypoxia-inducible factor 1-alpha (HIF-1α) inhibition or direct effects. Sirt3 upregulates the OxPhos pathway by enhancing the activity of mitochondrial complexes I, II, and III and dampening reactive oxygen species (ROS) production. Sirt1 is also able to increase FAO by activating PPAR-α and PGC1, while Sirt3 upregulates FAO under caloric restriction conditions. Sirt2 deacetylates and inhibits the activity of many metabolic enzymes involved in glycolysis, glutaminolysis, the TCA cycle and FAO. Sirt3 and Sirt4 activate and inhibit glutaminolysis, respectively, by regulating GDH activity. Sirt6 impacts the glutaminolysis pathway by regulating c-Myc transcriptional activity. Sirt5 increases glycolysis by increasing the activity of the glycolytic enzyme GAPDH. Sirt7 can also repress HIF-1α and, therefore, may inhibit the transcription of glycolytic genes.
Sirt2
Until recently, our knowledge on the role of Sirt2 within the immune system was limited to its anti-inflammatory function via negative regulation of the NF-κB p65 subunit87. Accordingly, Sirt2-deficient mice were found to develop severe forms of dextran sodium sulfate (DSS)-induced colitis via polarization of bone marrow-derived macrophages88. Furthermore, that study observed a greater proportion of activated CD4+CD69+ T cells in the mesenteric lymph nodes of Sirt2−/− mice in response to DSS-induced colitis, suggesting a role for Sirt2 in limiting CD4+ TEFF cell functions88.
A recent publication uncovered a new function of Sirt2 as a master regulator of T cell metabolism and functions. Hamaidi et al. found increased proliferation of CD4+ and CD8+ Sirt2−/− T cells upon TCR activation and activated CD8+ T cells displayed increased IFN-γ production and granzyme B expression, leading to superior cytotoxic activity. This hyperreactive phenotype of Sirt2−/− T cells endowed mice with superior antitumor immunity upon tumor challenge in vivo89.
Mechanistically, this hyperactivation and enhanced function of Sirt2−/− T cells is linked to Sirt2 regulation of multiple metabolic pathways that play crucial roles in T cell effector functions. Sirt2 deacetylates and negatively impacts the activity of key metabolic enzymes of the glycolysis, TCA cycle, FAO and glutaminolysis pathways. Accordingly, Sirt2 deletion during T cell activation was followed by hyperacetylation of multiple metabolic enzymes and amplification of their activities, leading to increased aerobic glycolysis, OxPhos, FAO, and glutaminolysis (Fig. 3)89.
Sirt2 was found upregulated in TM cell stages. TM cells rely on FAO metabolism, and Sirt2 negatively impacts FA catabolism; thus, Sirt2 deletion promoted CD4+ and CD8+ TM cell formation ex vivo, which was associated with superior cell survival and decreased apoptosis. Consistently, increased accumulation of TM cells within the TME and the secondary lymphoid organs of Sirt2−/− mice was observed upon tumor challenge, whereas no qualitative or quantitative phenotypic differences were observed between unchallenged WT and Sirt2−/− control mice89.
That study investigated the role of Sirt2 during T cell activation and maturation and within the antitumor immune response89. However, the role of Sirt2 in different CD4+ T cell subsets with distinct metabolic programs, including Treg, Th1, Th2, Th9 and Th17 cell subsets, requires further investigation.
Sirt3
Sirt3 localizes mainly to the mitochondrial matrix and plays an important role in regulating elements of mitochondrial metabolism, including the TCA cycle, urea cycle, FAO, and reactive oxygen species (ROS) detoxification (Fig. 3)90. Sirt3 promotes energy generation; thus, its expression is higher in metabolically active tissues.
A recent report indicated that T cells from Sirt3-deficient (Sirt3−/−) donor mice caused reduced graft-versus-host disease (GVHD) severity in comparison to T cells from control donor mice, suggesting that Sirt3 targeting can improve transplant recipient outcome. In that study, the protective effect of allogeneic Sirt3−/− T cells was related to reduced T cell proliferation and CXCR3 expression, with no significant impact on cytokine secretion or cytotoxic functions91.
Furthermore, Sirt3 deficiency in mouse models had no impact on immune responses against bacterial and fungal infections92, suggesting that Sirt3 may play a limited role in TEFF cell functions.
Inhibition of OxPhos impairs Treg cell function, and given the key role of Sirt3 in OxPhos, it is expected that Sirt3 promotes the suppressive activity of Treg cells. In fact, Treg cells from Sirt3−/− mice exhibited impaired suppressive functions, as demonstrated in in vitro suppression assay and an in vivo allograft model, and HDAC9 deletion was found to increase Treg suppressive activity by increasing Sirt3 expression93.
Sirt3 is also involved in CD8+ T cell function. Sirt3−/− CD8+ T cells exhibited reduced ROS production and CXCR3 expression upon activation. Moreover, T cells from Sirt3−/− donor mice were able to reduce GVHD within the gastrointestinal tract, which is probably due to decreased CXCR3-dependent CD8+ T cell trafficking to the site93.
Sirt4
Sirt4 is a mitochondrial sirtuin with ADP-ribosylation activity. No deacetylase activity of Sirt4 has yet been identified. By ADP-ribosylating glutamate dehydrogenase (GDH), Sirt4 represses its enzymatic activity, which limits the metabolism of glutamine into glutamate to generate ATP (Fig. 3)94. It is thus conceivable that T cell metabolic activity and effector functions can be enhanced by Sirt4 inhibition via increasing glutaminolysis, another ATP-generating pathway used by activated T cells. Furthermore, given the role of glutaminolysis in Th17 cell differentiation95 and Treg cell development18,19, we would also expect a proinflammatory phenotype with Sirt4 deficiency. Sirt4 was found to physiologically break immune tolerance and to resolve acute inflammation in a model of sepsis by coordinately reprogramming the metabolism and bioenergetics of human monocytes96.
Recently, in a mouse model of neuroinflammation following traumatic spinal cord injury, Sirt4 expression was found to be upregulated in infiltrating Treg cells in the spinal cord parenchyma97. Interestingly, Sirt4 overexpression in splenic naive Treg cells was found to alleviate the expression of Foxp3, IL-10, and TGF-β and to weaken the inhibitory activity of Treg cells. Additionally, Sirt4 overexpression blocked in vitro inducible Treg cell generation from conventional T cells97. Consistently, Sirt4 knockdown increased the anti-inflammatory activity of infiltrating Treg cells in the parenchyma of injured spinal cords97. The authors concluded that Sirt4 inhibits the anti-neuroinflammatory activity of Treg cells by blocking AMPK signaling given that an AMPK agonist restored the expression of Foxp3 and IL-10 in Treg cells97. However, this result could also be related to Sirt4’s impact on glutamine metabolism and consequently on Treg cell development.
Sirt5
Sirt5 is another mitochondrial sirtuin that displays a weak deacetylase activity. However, Sirt5 is unique in executing novel enzymatic activities involving lysine desuccinylation, demalonylation, and deglutarylation98. Given the emerging role of protein succinylation in the immune response99, future studies focusing on the role of Sirt5 in T cell function are needed.
An anti-inflammatory protective role of Sirt5 was reported in a DSS-induced colitis mouse model. Sirt5 was found to suppress IL-1β production and proinflammatory responses in macrophages by regulating pyruvate kinase M2 (PKM2) succinylation. Sirt5-dependent succinylation promotes PKM2 entry into the nucleus, where the PKM2-HIF1α complex is formed at the promoter of the IL-1β gene to stimulate its transcription100. Paradoxically, proinflammatory activity of Sirt5 was described during the acute and immunosuppressive phases of sepsis. The induction and persistence of a hypoinflammatory and immunosuppressive state in severe sepsis are commonly associated with increased risks of secondary infections and mortality. Sirt5 was found to rescue the innate inflammatory response of endotoxin-tolerant macrophages by promoting acetylation of NF-κB p65. Mechanistically, Sirt5 competes with Sirt2 to interact with NF-κB p65 and block its deacetylation by Sirt2, which consequently leads to increased acetylation of p65 and activation of the NF-κB pathway with its downstream cytokines101.
Although little work has been done to study Sirt5 in the adaptive immune response, a recent study has demonstrated a pivotal role of Sirt5 in regulating T cell activation and differentiation102. Indeed, Sirt5 deficiency was found to promote mouse naive T cell activation and increased IFN-γ production upon TCR ligation and to further influence T cell differentiation. Sirt5 deletion enhanced Th1 cell and CTL differentiation and decreased CD4+ Treg cell differentiation, whereas no qualitative or quantitative phenotypic differences were observed in the peripheral lymphoid organs of WT and Sirt5−/− mice under steady state conditions102. More importantly, even though Sirt5−/− mice were found to be highly susceptible to DSS-induced colitis100, Sirt5−/− mice were resistant to colorectal tumorigenesis following experimentally induced colitis, and this resistance was related to increased IFN-γ production in the colon tissue by immune cells102. While these data indicate the importance of Sirt5 in T cell activation and antitumor functions, the molecular mechanisms of Sirt5 activity need to be further elucidated.
Remarkably, studies on a large panel of preclinical mouse models of sepsis showed that Sirt5 deficiency has no impact on antimicrobial host immune defenses103, suggesting that Sirt5 plays a limited role in TEFF cell functions. On the other hand, these observations support the assumption that therapies directed against Sirt5 do not impair antibacterial host defenses.
Sirt6
Sirt6 is a chromatin-associated sirtuin implicated in numerous biological functions, including transcriptional repression, glucose homeostasis, DNA repair, telomeric function, cellular differentiation, mitosis, and meiosis104. Although Sirt6 has mono-ADP-ribosylase activity, the most robust activity of Sirt6 is histone deacetylation. Sirt6 deacetylates histone H3 lysine 9 (H3K9), H3K18, and H3K56 to induce transcriptional repression. Sirt6 can also remove the long-chain fatty acyl group from lysine105.
Recent studies have revealed that Sirt6 possesses anti-inflammatory properties. Sirt6 represses proinflammatory gene transcription by affecting chromatin structure rather than by directly deacetylating NF-κB, while Sirt1 and Sirt2 directly deacetylate p65 and inhibit its transcriptional activity87,106. Although Sirt6 physically associates with the p65 subunit, it modulates its transcriptional activity by deacetylating H3K9 on the promoter of selected NF-κB target genes107. Sirt6 was also reported to interact with JUN and deacetylate H3K9 at the promoter of proinflammatory genes whose expression involves JUN. The same study also indicated that Sirt6-null mice develop chronic liver inflammation attributable to Sirt6 deficiency in immune cells and macrophages, and these cells express increased levels of MCP1, IL-6, and TNF108. While Sirt6 appears to exert anti-inflammatory action when acting at the transcriptional level, scattered evidence seems to suggest that Sirt6 induces proinflammatory effects by modulating distinct intracellular signaling factors, such as Ca2+ homeostasis109 and mRNA translation110. Indeed, Sirt6 has been shown to promote the expression of cytokines and chemokines, including IL-6, TNF, CXCL2, and IFN-γ, and to positively regulate TNF secretion by increasing mRNA translational efficiency in several immune cells110–112.
Little is known about the role of Sirt6 in the adaptive immune response. HIF-1α is a transcriptional regulator of glycolysis and is known to regulate the production of several cytokines113 and to enhance Th17 cell differentiation while concomitantly inhibiting Treg cell function114,115. Furthermore, Myc is a global regulator of the immune response and plays a crucial role in glutamine metabolism15. Glycolysis and glutaminolysis are critical metabolic pathways on which TEFF cells depend to meet their energy needs. Dynamic switching between these metabolic pathways needs to be tightly regulated to achieve optimal function within immune cells.
Sirt6 appears to strongly oppose glycolysis by inhibiting the transcription of many glycolytic genes116. Sirt6 has been shown to physically interact with HIF-1α and to corepress its transcriptional activity by deacetylating histone H3K9 at the promoters of several glycolytic genes117. Furthermore, Sirt6 also regulates glutaminase expression, thus modulating the glutaminolysis pathway. Sirt6 interacts with Myc and reduces its transcriptional activity by deacetylating histones at the promoter region of Myc target genes118. Collectively, these observations suggest that Sirt6 potentially has an immunomodulatory effect on T cell metabolism and functions by regulating the HIF-1α and Myc pathways (Fig. 3).
Sirt7
Sirt7 is the last and least studied member of sirtuins. Sirt7 is a vital regulator of ribosome biogenesis, and it is enriched in nucleoli, where it facilitates RNA polymerase I-dependent transcription of ribosomal (r)RNA genes119. Sirt7 expression is linked to cell proliferation and oncogenic activity, connecting Sirt7-dependent regulation of ribosome biogenesis with cell cycle progression, metabolic homeostasis, stress resistance, aging and tumorigenesis120–122.
Recent studies suggest that Sirt7 is also involved in inflammation. Vakhrusheva et al. showed that Sirt7 deletion predisposes mice to heart hypertrophy and cardiac inflammation. These authors observed an increased infiltration of immune cells, with higher levels of proinflammatory cytokine production123. Information regarding the possible involvement of Sirt7 in the adaptive immune response is currently very limited.
Concluding remarks and future perspectives
The well-documented capacity of sirtuins to promote the resolution of the inflammatory response has led many research groups to focus on developing therapeutic strategies to target their enzymatic activity in vivo. However, to achieve the desired clinical outcomes, it is paramount to decide whether to stimulate or block sirtuin activity and how to modulate a specific sirtuin within a specific T cell subset. All these challenges are worthy of further study due to the ubiquity of sirtuins. In addition, these proteins have intricate and partially unknown functions within an organism, reflecting the immense challenge of targeting any of them.
For example, enhancing Treg cell suppressive activity with Sirt3 agonists to attenuate inflammation represents a potential strategy to treat autoimmunity. Similarly, the use of Sirt1 activators to suppress inflammation could help treat autoimmune diseases. Metformin, classically prescribed for the management of type 2 diabetes, appears to have a stimulatory effect on sirtuins. Sirt1 activation by metformin has been shown to decrease inflammation by decreasing Th17 cell differentiation74. Resveratrol, a compound with sirtuin-activating effects, is currently of intense interest within both scientific and lay communities for its anti-inflammatory properties124. Resveratrol stimulates Sirt1 activity, which decreases c-Jun acetylation, consequently restraining T cell activation62. Resveratrol has been shown to improve outcomes in several experimental autoimmunity models125. For instance, administration of resveratrol protected mice against experimental rheumatoid arthritis by inhibiting Th17 cell differentiation126. Furthermore, resveratrol was proven to have both preventative and therapeutic effects by reverting the advanced stages of insulitis in a nonobese diabetic (NOD) mouse model due to reduced Th17 cells127. Moreover, resveratrol induced protective effects against high-fat diet-induced obesity in mice in part by increasing total Treg cells128.
Despite the obvious progress in the organ transplantation field, the commonly required immunosuppressive therapies are known to ultimately induce cancer and quite often infections. Presently, sirtuins are being thoroughly studied for their role in promoting organ transplant tolerance. Treg and Th17 cells drive transplant tolerance, and in relation to these T cell subsets, Sirt1 has emerged as an eligible target for clinical interventions in transplantation. Indeed, Sirt1 deletion improved Treg cell suppressive activity in vitro and in vivo, and mice with specific Sirt1 deletion in Foxp3+ Treg cells exhibited prolonged survival after cardiac allografts concomitant with increased Treg cell infiltration. In the same study, similar results were obtained when using Sirt1 inhibitors in vivo129. Later, comparable findings were reported after kidney transplantation from BALB/c mice into C57BL/6 recipient mice treated with EX-527, a Sirt1 inhibitor130.
Recently, the effect of sirtinol, a Sirt1 inhibitor, on heart allograft survival was evaluated, and it was found that sirtinol-treated recipients exhibited increased numbers of Foxp3 Treg cells and reduced numbers of Th17 cells in addition to decreased expression of the IL-17A and RORγt transcription factor131. Therefore, Sirt1 inhibition might affect the Th17/Treg cell ratio in favor of Treg cells. In contrast, Sirt1 activators such as metformin were found to decrease IL-17A and RORγt expression in mouse models as described earlier74. Certainly, more studies are required to define the appropriate action of Sirt1 targeting for allograft retention.
To date, the research community has focused on promoting the anti-inflammatory activity of sirtuins to treat a multitude of autoimmune diseases and other chronic diseases associated with inflammation. However, we must extend our interest toward blocking the activity of sirtuins to promote the effector activity of T cells in the context of antitumor immunity. Indeed, enhancing tumor rejection by boosting tumor-reactive T cell activity would be tremendously effective in fighting cancer. Recent reports demonstrating the immunotherapeutic potential of Sirt2 and Sirt5 blockade in T cells to reject tumor challenge89 and to protect against colorectal tumorigenesis in murine models, highlight the expected effectiveness of such strategy102. In addition, sirtuins have been shown to play an oncogenic role in many tumor types132, and sirtuin inhibitor drugs have been shown to have tumor-suppressive activity on cancer cells, suggesting an intriguing possibility that in certain cancer patients, targeting sirtuins may suppress tumor proliferation while simultaneously boosting antitumor immunity. Alternatively, manipulation of sirtuins could be achieved by gene editing during TIL expansion or chimeric antigen receptor (CAR) T cell generation ex vivo, thus selectively targeting T cells in the context of immune cell therapy133,134.
Many reports indicate that exhausted tumor-infiltrating lymphocytes display dysregulated metabolism within the metabolically restricted tumor microenvironment22,23,135,136. Modulation of sirtuins to enhance the metabolic activity of T cells could provide a viable strategy to restore exhausted tumor-reactive T cells.
Acknowledgements
This work was supported in part by 1R37CA248298-01A1 and 5K08CA194273-05.
Author contributions
I.H. wrote the manuscript, and S.K. revised the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nicholson LB. The immune system. Essays Biochem. 2016;60:275–301. doi: 10.1042/EBC20160017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaplin DD. 1. Overview of the immune response. J. Allergy Clin. Immunol. 2003;111:S442–S459. doi: 10.1067/mai.2003.125. [DOI] [PubMed] [Google Scholar]
- 3.Luckheeram RV, Zhou R, Verma AD, Xia B. CD4(+)T cells: differentiation and functions. Clin. Dev. Immunol. 2012;2012:925135. doi: 10.1155/2012/925135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Russell JH, Ley TJ. Lymphocyte-mediated cytotoxicity. Annu. Rev. Immunol. 2002;20:323–370. doi: 10.1146/annurev.immunol.20.100201.131730. [DOI] [PubMed] [Google Scholar]
- 5.Spellberg B, Edwards JE., Jr. Type 1/Type 2 immunity in infectious diseases. Clin. Infect. Dis. 2001;32:76–102. doi: 10.1086/317537. [DOI] [PubMed] [Google Scholar]
- 6.Del Prete G. Human Th1 and Th2 lymphocytes: their role in the pathophysiology of atopy. Allergy. 1992;47:450–455. doi: 10.1111/j.1398-9995.1992.tb00662.x. [DOI] [PubMed] [Google Scholar]
- 7.Neurath MF, Kaplan MH. Th9 cells in immunity and immunopathological diseases. Semin. Immunopathol. 2017;39:1–4. doi: 10.1007/s00281-016-0611-z. [DOI] [PubMed] [Google Scholar]
- 8.Annunziato F, et al. Phenotypic and functional features of human Th17 cells. J. Exp. Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rudensky AY. Regulatory T cells and Foxp3. Immunol. Rev. 2011;241:260–268. doi: 10.1111/j.1600-065X.2011.01018.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang CH, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho PC, et al. Phosphoenolpyruvate is a metabolic checkpoint of anti-tumor T cell responses. Cell. 2015;162:1217–1228. doi: 10.1016/j.cell.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 14.Carr EL, et al. Glutamine uptake and metabolism are coordinately regulated by ERK/MAPK during T lymphocyte activation. J. Immunol. 2010;185:1037–1044. doi: 10.4049/jimmunol.0903586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang R, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity. 2011;35:871–882. doi: 10.1016/j.immuni.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342:1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahl DR, Byersdorfer CA, Ferrara JL, Opipari AW, Jr., Glick GD. Distinct metabolic programs in activated T cells: opportunities for selective immunomodulation. Immunol. Rev. 2012;249:104–115. doi: 10.1111/j.1600-065X.2012.01148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klysz D, et al. Glutamine-dependent alpha-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci. Signal. 2015;8:ra97. doi: 10.1126/scisignal.aab2610. [DOI] [PubMed] [Google Scholar]
- 19.Pacella I, et al. Fatty acid metabolism complements glycolysis in the selective regulatory T cell expansion during tumor growth. Proc. Natl Acad. Sci. Usa. 2018;115:E6546–E6555. doi: 10.1073/pnas.1720113115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raud B, McGuire PJ, Jones RG, Sparwasser T, Berod L. Fatty acid metabolism in CD8(+) T cell memory: Challenging current concepts. Immunol. Rev. 2018;283:213–231. doi: 10.1111/imr.12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raud B, et al. Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 2018;28:504–515 e507. doi: 10.1016/j.cmet.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CH, et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, et al. Enhancing CD8(+) T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell. 2017;32:377–391 e379. doi: 10.1016/j.ccell.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Romanowska J, Joshi A. From genotype to phenotype: through chromatin. Genes (Basel) 2019;10:76. doi: 10.3390/genes10020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chisolm DA, Weinmann AS. Connections between metabolism and epigenetics in programming cellular differentiation. Annu. Rev. Immunol. 2018;36:221–246. doi: 10.1146/annurev-immunol-042617-053127. [DOI] [PubMed] [Google Scholar]
- 26.Margueron R, Reinberg D. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 2010;11:285–296. doi: 10.1038/nrg2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, et al. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review) Oncol. Rep. 2017;37:3–9. doi: 10.3892/or.2016.5236. [DOI] [PubMed] [Google Scholar]
- 29.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 30.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteom. 2013;12:3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aebersold R, et al. How many human proteoforms are there? Nat. Chem. Biol. 2018;14:206–214. doi: 10.1038/nchembio.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prabakaran S, Lippens G, Steen H, Gunawardena J. Post-translational modification: nature’s escape from genetic imprisonment and the basis for dynamic information encoding. Wiley Interdiscip. Rev. Syst. Biol. Med. 2012;4:565–583. doi: 10.1002/wsbm.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bodenmiller B, et al. Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 2010;3:rs4. doi: 10.1126/scisignal.2001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner SA, et al. Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell Proteom. 2012;11:1578–1585. doi: 10.1074/mcp.M112.017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Wang F. Post-translational modifications of deubiquitinating enzymes: expanding the ubiquitin code. Front. Pharmacol. 2021;12:685011. doi: 10.3389/fphar.2021.685011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 38.Zhao S, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.North BJ, Verdin E. Sirtuins: Sir2-related NAD-dependent protein deacetylases. Genome Biol. 2004;5:224. doi: 10.1186/gb-2004-5-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol. Biol. Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guarente, L. Franklin H. Epstein Lecture: Sirtuins, aging, and medicine. N. Engl. J. Med.364, 2235–2244 (2011). [DOI] [PubMed]
- 43.Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat. Rev. Mol. cell Biol. 2012;13:225–238. doi: 10.1038/nrm3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 45.Vaquero A, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Verdin E, Hirschey MD, Finley LW, Haigis MC. Sirtuin regulation of mitochondria: energy production, apoptosis, and signaling. Trends Biochem. Sci. 2010;35:669–675. doi: 10.1016/j.tibs.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hawse WF, Wolberger C. Structure-based mechanism of ADP-ribosylation by sirtuins. J. Biol. Chem. 2009;284:33654–33661. doi: 10.1074/jbc.M109.024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haigis MC, Guarente LP. Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 51.Vachharajani VT, et al. Sirtuins link inflammation and metabolism. J. Immunol. Res. 2016;2016:8167273. doi: 10.1155/2016/8167273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444:868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 53.Sebastian C, Mostoslavsky R. The role of mammalian sirtuins in cancer metabolism. Semin. Cell Dev. Biol. 2015;43:33–42. doi: 10.1016/j.semcdb.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 54.Anderson KA, Madsen AS, Olsen CA, Hirschey MD. Metabolic control by sirtuins and other enzymes that sense NAD(+), NADH, or their ratio. Biochim. Biophys. Acta Bioenerg. 2017;1858:991–998. doi: 10.1016/j.bbabio.2017.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newsholme P. Cellular and metabolic mechanisms of nutrient actions in immune function. Nutr. Diabetes. 2021;11:22. doi: 10.1038/s41387-021-00162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mendes KL, Lelis DF, Santos SHS. Nuclear sirtuins and inflammatory signaling pathways. Cytokine Growth Factor Rev. 2017;38:98–105. doi: 10.1016/j.cytogfr.2017.11.001. [DOI] [PubMed] [Google Scholar]
- 57.Warren JL, MacIver NJ. Regulation of adaptive immune cells by sirtuins. Front. Endocrinol. (Lausanne) 2019;10:466. doi: 10.3389/fendo.2019.00466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Qiu Y, Zhou X, Liu Y, Tan S, Li Y. The role of sirtuin-1 in immune response and systemic lupus erythematosus. Front. Immunol. 2021;12:632383. doi: 10.3389/fimmu.2021.632383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen X, et al. Intercellular interplay between Sirt1 signalling and cell metabolism in immune cell biology. Immunology. 2015;145:455–467. doi: 10.1111/imm.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Atsaves V, Leventaki V, Rassidakis GZ, Claret FX. AP-1 Transcription factors as regulators of immune responses in cancer. Cancers (Basel) 2019;11:1037. doi: 10.3390/cancers11071037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J, et al. The type III histone deacetylase Sirt1 is essential for maintenance of T cell tolerance in mice. J. Clin. Invest. 2009;119:3048–3058. doi: 10.1172/JCI38902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou T, et al. Resveratrol Inhibits CD4+ T cell activation by enhancing the expression and activity of Sirt1. PLoS ONE. 2013;8:e75139. doi: 10.1371/journal.pone.0075139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen X, Zhang XL, Zhang GH, Gao YF. Artesunate promotes Th1 differentiation from CD4+ T cells to enhance cell apoptosis in ovarian cancer via miR-142. Braz. J. Med. Biol. Res. 2019;52:e7992. doi: 10.1590/1414-431X20197992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sequeira J, et al. sirt1-null mice develop an autoimmune-like condition. Exp. Cell Res. 2008;314:3069–3074. doi: 10.1016/j.yexcr.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 65.Gao B, Kong Q, Kemp K, Zhao YS, Fang D. Analysis of sirtuin 1 expression reveals a molecular explanation of IL-2-mediated reversal of T-cell tolerance. Proc. Natl Acad. Sci. USA. 2012;109:899–904. doi: 10.1073/pnas.1118462109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McPherson JP, et al. Essential role for Bclaf1 in lung development and immune system function. Cell Death Differ. 2009;16:331–339. doi: 10.1038/cdd.2008.167. [DOI] [PubMed] [Google Scholar]
- 67.Kong S, et al. The type III histone deacetylase Sirt1 protein suppresses p300-mediated histone H3 lysine 56 acetylation at Bclaf1 promoter to inhibit T cell activation. J. Biol. Chem. 2011;286:16967–16975. doi: 10.1074/jbc.M111.218206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim SR, et al. Involvement of sirtuin 1 in airway inflammation and hyperresponsiveness of allergic airway disease. J. Allergy Clin. Immunol. 2010;125:449–460.e414. doi: 10.1016/j.jaci.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 69.Legutko A, et al. Sirtuin 1 promotes Th2 responses and airway allergy by repressing peroxisome proliferator-activated receptor-gamma activity in dendritic cells. J. Immunol. 2011;187:4517–4529. doi: 10.4049/jimmunol.1101493. [DOI] [PubMed] [Google Scholar]
- 70.Lorentsen KJ, et al. Bcl11b is essential for licensing Th2 differentiation during helminth infection and allergic asthma. Nat. Commun. 2018;9:1679. doi: 10.1038/s41467-018-04111-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Senawong T, et al. Involvement of the histone deacetylase SIRT1 in chicken ovalbumin upstream promoter transcription factor (COUP-TF)-interacting protein 2-mediated transcriptional repression. J. Biol. Chem. 2003;278:43041–43050. doi: 10.1074/jbc.M307477200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Imtiyaz HZ, Simon MC. Hypoxia-inducible factors as essential regulators of inflammation. Curr. Top. Microbiol. Immunol. 2010;345:105–120. doi: 10.1007/82_2010_74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y, et al. Histone deacetylase SIRT1 negatively regulates the differentiation of interleukin-9-producing CD4(+) T cells. Immunity. 2016;44:1337–1349. doi: 10.1016/j.immuni.2016.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Limagne E, et al. Sirtuin-1 activation controls tumor growth by impeding Th17 differentiation via STAT3 deacetylation. Cell Rep. 2017;19:746–759. doi: 10.1016/j.celrep.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 75.Wang J, et al. Treatment with NAD(+) inhibited experimental autoimmune encephalomyelitis by activating AMPK/SIRT1 signaling pathway and modulating Th1/Th17 immune responses in mice. Int. Immunopharmacol. 2016;39:287–294. doi: 10.1016/j.intimp.2016.07.036. [DOI] [PubMed] [Google Scholar]
- 76.Wang J, et al. Methylene blue alleviates experimental autoimmune encephalomyelitis by modulating AMPK/SIRT1 signaling pathway and Th17/Treg immune response. J. Neuroimmunol. 2016;299:45–52. doi: 10.1016/j.jneuroim.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 77.Lim HW, et al. SIRT1 deacetylates RORgammat and enhances Th17 cell generation. J. Exp. Med. 2015;212:973. doi: 10.1084/jem.2013237805062015c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwon HS, et al. Three novel acetylation sites in the Foxp3 transcription factor regulate the suppressive activity of regulatory T cells. J. Immunol. 2012;188:2712–2721. doi: 10.4049/jimmunol.1100903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Loosdregt J, et al. Regulation of Treg functionality by acetylation-mediated Foxp3 protein stabilization. Blood. 2010;115:965–974. doi: 10.1182/blood-2009-02-207118. [DOI] [PubMed] [Google Scholar]
- 80.Marcel N, Perumalsamy LR, Shukla SK, Sarin A. The lysine deacetylase Sirtuin 1 modulates the localization and function of the Notch1 receptor in regulatory T cells. Sci. Signal. 2017;10:eaah4679. doi: 10.1126/scisignal.aah4679. [DOI] [PubMed] [Google Scholar]
- 81.Betz BC, et al. Batf coordinates multiple aspects of B and T cell function required for normal antibody responses. J. Exp. Med. 2010;207:933–942. doi: 10.1084/jem.20091548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kuroda S, et al. Basic leucine zipper transcription factor, ATF-like (BATF) regulates epigenetically and energetically effector CD8 T-cell differentiation via Sirt1 expression. Proc. Natl Acad. Sci. USA. 2011;108:14885–14889. doi: 10.1073/pnas.1105133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol. Cell. Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Canto C, Auwerx J. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol. 2009;20:98–105. doi: 10.1097/MOL.0b013e328328d0a4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelly TJ, Lerin C, Haas W, Gygi SP, Puigserver P. GCN5-mediated transcriptional control of the metabolic coactivator PGC-1beta through lysine acetylation. J. Biol. Chem. 2009;284:19945–19952. doi: 10.1074/jbc.M109.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jeng MY, et al. Metabolic reprogramming of human CD8(+) memory T cells through loss of SIRT1. J. Exp. Med. 2018;215:51–62. doi: 10.1084/jem.20161066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J. Cell. Sci. 2010;123:4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 88.Lo Sasso G, et al. SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. PLoS One. 2014;9:e103573. doi: 10.1371/journal.pone.0103573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamaidi I, et al. Sirt2 Inhibition enhances metabolic fitness and effector functions of tumor-reactive T cells. Cell Metab. 2020;32:420–436.e12. doi: 10.1016/j.cmet.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang J, et al. Mitochondrial Sirtuin 3: new emerging biological function and therapeutic target. Theranostics. 2020;10:8315–8342. doi: 10.7150/thno.45922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Toubai T, et al. Mitochondrial deacetylase SIRT3 plays an important role in donor T cell responses after experimental allogeneic hematopoietic transplantation. J. Immunol. 2018;201:3443–3455. doi: 10.4049/jimmunol.1800148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ciarlo E, et al. Sirtuin 3 deficiency does not alter host defenses against bacterial and fungal infections. Sci. Rep. 2017;7:3853. doi: 10.1038/s41598-017-04263-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Beier UH, et al. Essential role of mitochondrial energy metabolism in Foxp3(+) T-regulatory cell function and allograft survival. Faseb. J. 2015;29:2315–2326. doi: 10.1096/fj.14-268409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 95.Johnson MO, et al. Distinct regulation of Th17 and Th1 cell differentiation by glutaminase-dependent metabolism. Cell. 2018;175:1780–1795 e1719. doi: 10.1016/j.cell.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Tao J, Zhang J, Ling Y, McCall CE, Liu TF. Mitochondrial Sirtuin 4 resolves immune tolerance in monocytes by rebalancing glycolysis and glucose oxidation homeostasis. Front. Immunol. 2018;9:419. doi: 10.3389/fimmu.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lin W, Chen W, Liu W, Xu Z, Zhang L. Sirtuin4 suppresses the anti-neuroinflammatory activity of infiltrating regulatory T cells in the traumatically injured spinal cord. Immunology. 2019;158:362–374. doi: 10.1111/imm.13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang X, Liu B, Zhu W, Luo J. SIRT5, functions in cellular metabolism with a multiple enzymatic activities. Sci. China Life Sci. 2015;58:912–914. doi: 10.1007/s11427-015-4902-8. [DOI] [PubMed] [Google Scholar]
- 99.Tannahill GM, et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang F, et al. SIRT5 Desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1beta production and to prevent DSS-induced colitis in mice. Cell Rep. 2017;19:2331–2344. doi: 10.1016/j.celrep.2017.05.065. [DOI] [PubMed] [Google Scholar]
- 101.Qin K, et al. NAD(+) dependent deacetylase Sirtuin 5 rescues the innate inflammatory response of endotoxin tolerant macrophages by promoting acetylation of p65. J. Autoimmun. 2017;81:120–129. doi: 10.1016/j.jaut.2017.04.006. [DOI] [PubMed] [Google Scholar]
- 102.Wang K, et al. SIRT5 Contributes to colorectal cancer growth by regulating T cell activity. J. Immunol. Res. 2020;2020:3792409. doi: 10.1155/2020/3792409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Heinonen T, et al. Sirtuin 5 deficiency does not compromise innate immune responses to bacterial infections. Front. Immunol. 2018;9:2675. doi: 10.3389/fimmu.2018.02675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang AR, Ferrer CM, Mostoslavsky R. SIRT6, a mammalian deacylase with multitasking abilities. Physiol. Rev. 2020;100:145–169. doi: 10.1152/physrev.00030.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jiang H, et al. SIRT6 regulates TNF-alpha secretion through hydrolysis of long-chain fatty acyl lysine. Nature. 2013;496:110–113. doi: 10.1038/nature12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawahara TL, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62–74. doi: 10.1016/j.cell.2008.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xiao C, et al. Progression of chronic liver inflammation and fibrosis driven by activation of c-JUN signaling in Sirt6 mutant mice. J. Biol. Chem. 2012;287:41903–41913. doi: 10.1074/jbc.M112.415182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bauer I, et al. The NAD+-dependent histone deacetylase SIRT6 promotes cytokine production and migration in pancreatic cancer cells by regulating Ca2+ responses. J. Biol. Chem. 2012;287:40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Van Gool F, et al. Intracellular NAD levels regulate tumor necrosis factor protein synthesis in a sirtuin-dependent manner. Nat. Med. 2009;15:206–210. doi: 10.1038/nm.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bruzzone S, et al. Catastrophic NAD+ depletion in activated T lymphocytes through Nampt inhibition reduces demyelination and disability in EAE. PLoS ONE. 2009;4:e7897. doi: 10.1371/journal.pone.0007897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Montecucco F, et al. Inhibition of nicotinamide phosphoribosyltransferase reduces neutrophil-mediated injury in myocardial infarction. Antioxid. Redox Signal. 2013;18:630–641. doi: 10.1089/ars.2011.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krzywinska E, Stockmann C. Hypoxia, metabolism and immune cell function. Biomedicines. 2018;6:56. doi: 10.3390/biomedicines6020056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dang EV, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi LZ, et al. HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J. Exp. Med. 2011;208:1367–1376. doi: 10.1084/jem.20110278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim HS, et al. Hepatic-specific disruption of SIRT6 in mice results in fatty liver formation due to enhanced glycolysis and triglyceride synthesis. Cell Metab. 2010;12:224–236. doi: 10.1016/j.cmet.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhong L, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sebastian C, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen S, et al. Repression of RNA polymerase I upon stress is caused by inhibition of RNA-dependent deacetylation of PAF53 by SIRT7. Mol. Cell. 2013;52:303–313. doi: 10.1016/j.molcel.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 120.Kiran S, Anwar T, Kiran M, Ramakrishna G. Sirtuin 7 in cell proliferation, stress and disease: rise of the seventh Sirtuin! Cell Signal. 2015;27:673–682. doi: 10.1016/j.cellsig.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 121.Paredes S, Villanova L, Chua KF. Molecular pathways: emerging roles of mammalian Sirtuin SIRT7 in cancer. Clin. Cancer Res. 2014;20:1741–1746. doi: 10.1158/1078-0432.CCR-13-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Blank MF, Grummt I. The seven faces of SIRT7. Transcription. 2017;8:67–74. doi: 10.1080/21541264.2016.1276658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Vakhrusheva O, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ. Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 124.Gao X, Xu YX, Janakiraman N, Chapman RA, Gautam SC. Immunomodulatory activity of resveratrol: suppression of lymphocyte proliferation, development of cell-mediated cytotoxicity, and cytokine production. Biochem. Pharmacol. 2001;62:1299–1308. doi: 10.1016/s0006-2952(01)00775-4. [DOI] [PubMed] [Google Scholar]
- 125.Gianchecchi E, Fierabracci A. Insights on the effects of resveratrol and some of its derivatives in cancer and autoimmunity: a molecule with a dual activity. Antioxidants (Basel) 2020;9:91. doi: 10.3390/antiox9020091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xuzhu G, et al. Resveratrol modulates murine collagen-induced arthritis by inhibiting Th17 and B-cell function. Ann. Rheum. Dis. 2012;71:129–135. doi: 10.1136/ard.2011.149831. [DOI] [PubMed] [Google Scholar]
- 127.Lee SM, et al. Prevention and treatment of diabetes with resveratrol in a non-obese mouse model of type 1 diabetes. Diabetologia. 2011;54:1136–1146. doi: 10.1007/s00125-011-2064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang B, et al. Regulatory effects of resveratrol on glucose metabolism and T-lymphocyte subsets in the development of high-fat diet-induced obesity in C57BL/6 mice. Food Funct. 2014;5:1452–1463. doi: 10.1039/c3fo60714c. [DOI] [PubMed] [Google Scholar]
- 129.Beier UH, et al. Sirtuin-1 targeting promotes Foxp3+ T-regulatory cell function and prolongs allograft survival. Mol. Cell Biol. 2011;31:1022–1029. doi: 10.1128/MCB.01206-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Levine MH, et al. Targeting Sirtuin-1 prolongs murine renal allograft survival and function. Kidney Int. 2016;89:1016–1026. doi: 10.1016/j.kint.2015.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ye Q, et al. Sirtinol regulates the balance of Th17/Treg to prevent allograft rejection. Cell Biosci. 2017;7:55. doi: 10.1186/s13578-017-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chalkiadaki A, Guarente L. The multifaceted functions of sirtuins in cancer. Nat. Rev. Cancer. 2015;15:608–624. doi: 10.1038/nrc3985. [DOI] [PubMed] [Google Scholar]
- 133.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lim WA, June CH. The principles of engineering immune cells to treat cancer. Cell. 2017;168:724–740. doi: 10.1016/j.cell.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shevchenko I, Bazhin AV. Metabolic checkpoints: novel avenues for immunotherapy of cancer. Front. Immunol. 2018;9:1816. doi: 10.3389/fimmu.2018.01816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gemta LF, et al. Impaired enolase 1 glycolytic activity restrains effector functions of tumor-infiltrating CD8(+) T cells. Sci. Immunol. 2019;4:eaap9520. doi: 10.1126/sciimmunol.aap9520. [DOI] [PMC free article] [PubMed] [Google Scholar]


