Abstract
Neuroblastoma (NB) accounts for 15% of cancer-related deaths in childhood despite considerable therapeutic improvements. While several risk factors, including MYCN amplification and alterations in RAS and p53 pathway genes, have been defined in NB, the clinical outcome is very variable and difficult to predict. Since genes of the mechanistic target of rapamycin (mTOR) pathway are upregulated in MYCN-amplified NB, we aimed to define the predictive value of the mTOR substrate-encoding gene eukaryotic translation initiation factor 4E-binding protein 1 (EIF4EBP1) expression in NB patients. Using publicly available data sets, we found that EIF4EBP1 mRNA expression is positively correlated with MYCN expression and elevated in stage 4 and high-risk NB patients. In addition, high EIF4EBP1 mRNA expression is associated with reduced overall and event-free survival in the entire group of NB patients in three cohorts, as well as in stage 4 and high-risk patients. This was confirmed by monitoring the clinical value of 4EBP1 protein expression, which revealed that high levels of 4EBP1 are significantly associated with prognostically unfavorable NB histology. Finally, functional analyses revealed that EIF4EBP1 expression is transcriptionally controlled by MYCN binding to the EIF4EBP1 promoter in NB cells. Our data highlight that EIF4EBP1 is a direct transcriptional target of MYCN whose high expression is associated with poor prognosis in NB patients. Therefore, EIF4EBP1 may serve to better stratify patients with NB.
Subject terms: Tumour biomarkers, Paediatric cancer
Introduction
Neuroblastoma (NB) is a pediatric malignant tumor that develops from progenitor cells of the sympathetic nervous system and the adrenal glands [1, 2]. NB is the most commonly occurring extracranial solid tumor in childhood and the major cause of cancer-related mortality in infants [2]. NB tumors are classified into five stages (1, 2, 3, 4, and 4S) according to tumor size, the presence of metastasis, and the outcome of surgical resection [1]. Noteworthy, stage 4S represents a special form of NB in infants that is associated with a high chance of spontaneous regression despite metastatic spread [1]. Apart from surgical resection, treatment options may include response-adjusted chemotherapy for low to intermediate risk groups or a mix of surgery, high-dose chemotherapy, immunotherapy, and radiation for patients belonging to the high-risk group. The risk level is determined based on the tumor stage combined with age at diagnosis, tumor ploidy, genetic alterations, and tumor histology [1, 3]. However, NB represents a particularly heterogeneous type of cancer, posing challenges to precisely predict therapeutic response and clinical outcome in the individual patient [4, 5]. While some NB tumors may spontaneously regress, high-risk patients have an increased likelihood of relapse and available treatment options for relapsed patients are rarely successful. Indeed, the 5-year overall survival rate for high-risk patients is ranging from 31 to 86%, in contrast to 97–100% for low-risk patients [6]. In addition, success rates of second line treatment in relapsed patients remain poor [5, 7]. Therefore, it is critical to define novel stratification factors for NB patients to better predict individual risk and to facilitate administration of the most appropriate therapeutic option.
NB is rarely familial (1–2%) and only few predisposition genes, such as PHOX2B and ALK, have been reported [4, 8–10]. Genetically, several acquired alterations have been detected in NB and linked to patient outcome. These include gain-of-function mutations in ALK, gain of chromosome arm 17q, loss of chromosome arm 11q, amplification of MYCN [4, 11], and, more recently reported, alterations in genes related to the RAS and p53 pathways [12]. MYCN amplification is found in about 20% of NB and is associated with aggressive tumors, therapy resistance and poor survival [13]. MYCN is a member of the MYC oncogene family and encodes a transcription factor that recognizes a specific DNA element referred to as E-box [14, 15]. This allows MYCN to regulate the transcription of genes involved in cell cycle progression, proliferation, differentiation, and survival [13]. MYCN is a strong driver of NB tumorigenesis, as tissue-specific overexpression of MYCN is sufficient to induce NB tumor development in mouse models [16]. Mechanistically, MYCN is proposed to rewire metabolism to enable NB tumor cells to proliferate, in turn preserving the intracellular redox balance while producing enough energy by inducing a glycolytic switch [17–19]. In particular, MYCN actively augments the transcription of multiple genes whose products are involved in the protein synthesis machinery [18]. Even though MYCN represents a highly attractive therapeutic target in NB, as a transcription factor that lacks hydrophobic pockets which can be targeted by drug-like small molecules, it is still considered as being “undruggable” [20, 21]. Thus, identification of downstream effectors involved in MYCN-driven NB progression is a promising approach to uncover novel targets for molecularly guided therapeutic approaches.
To better delineate the molecular basis of MYCN-amplified NB aggressiveness, several approaches have been undertaken. In particular, RNA-sequencing (RNA-seq) has been used to uncover the set of genes induced in MYCN-amplified compared to MYCN-non-amplified NB [22]. Strikingly, this analysis identified regulators of protein synthesis which are components of the mechanistic target of rapamycin (mTOR) pathway, including the mTOR target eukaryotic initiation factor 4E binding protein 1 (EIF4EBP1). The corresponding protein, 4EBP1, is inhibited through mTOR-mediated phosphorylation when nutrients are available, leading to active mRNA translation initiation [23]. Under nutrient-deprived conditions, when mTOR is inhibited, 4EBP1 gets activated and thus binds to the translation initiation factor eIF4E, in turn blocking cap-dependent mRNA translation initiation [23]. At the cellular level, 4EBP1 is negatively regulating proliferation and mitochondrial activity [24, 25]. The exact role of 4EBP1 in cancer is still debated. 4EBP1 was found to exert a tumor suppressive function in vivo, as 4EBP1 knock-out leads to enhanced tumor formation in mouse models of head and neck squamous cell carcinoma [26], and prostate cancer [27]. In contrast, 4EBP1 was shown to mediate angiogenesis and facilitate tumor growth in a breast cancer model in vivo, highlighting a cancer type-specific function of 4EBP1 [28]. In keeping with that, the clinical relevance of EIF4EBP1 expression depends on the tumor type. EIF4EBP1 was reported to be overexpressed in a number of tumor entities in adults [29], including breast cancer [30], in which EIF4EBP1 is amplified as part of the 8p11–12 amplicon, as well as in ovarian and prostate cancer [31, 32]. In breast and liver cancer, high EIF4EBP1 expression has been associated with poor survival [30, 33]. In contrast, EIF4EBP1 expression was found to be reduced in head and neck cancer, in which low expression is correlated with poor prognosis [26]. In NB, the expression of EIF4EBP1 is deregulated, even though contradictory findings have been reported. While EIF4EBP1 was characterized as a gene upregulated in MYCN-amplified versus MYCN-non-amplified NB tissues and cells [22], another study reported that EIF4EBP1 levels were higher in favorable stages of NB as compared to advanced stage 4 tumors [34]. In addition, Meng et al. showed that EIF4EBP1 is part of a gene signature that predicts poor overall survival [35]. However, it was not investigated whether EIF4EBP1 expression alone can predict NB patient prognosis. Thus, the clinical relevance of EIF4EBP1 expression in NB needs further evaluation. Overexpression of EIF4EBP1 in cancer is mediated by certain transcription factors, such as MYC [36, 37], androgen receptor [38], and the stress regulators ATF4 [39] and HIF-1α [40], which all bind to and thereby modulate the activity of the EIF4EBP1 promoter. More specifically, ChIP-sequencing (ChIP-seq) revealed binding of MYCN to the EIF4EBP1 promoter in NB cells, and MYCN was reported to impact EIF4EBP1 transcription, pointing to EIF4EBP1 as a potential MYCN target gene [41, 42]. However, how MYCN exactly controls the EIF4EBP1 promoter is still poorly understood.
In this study, we analyzed publicly available NB patient data sets and revealed that EIF4EBP1 is overexpressed in NB compared to normal tissues, is significantly co-expressed with MYCN, and is elevated in high-risk relatively to low-risk tumor groups. High EIF4EBP1 levels were found to be significantly linked to poor overall survival in all NB patients, as well as in the more aggressive stage 4 and high-risk groups. In addition, immunohistochemistry staining of NB tissues confirmed the mRNA-based associations and showed that high 4EBP1 protein expression associates with unfavorable histology in NB. Finally, by applying gene reporter assays and by modulating MYCN expression in vitro, we found that MYCN upregulates the EIF4EBP1 promoter activity by binding to three distinct E-boxes.
Materials and methods
Databases
The RNA-seq, microarray, and ChIP-seq data were retrieved from ‘R2: Genomics Analysis and Visualization Platform’ (http://r2.amc.nl). Data were visualized with IGV or Affinity Designer. For the MYCN occupancy profile in BE(2)-C cells, the ChIP-seq data by Durbin et al. (GSE94824) were accessed using the human genome GRCh 38/hg 38. For the initial across dataset analysis, “Normal Adrenal gland” dataset from R2 (corresponding to samples taken from multiple data sets [GSE3526, GSE7307, GSE8514] and combined into a single data set) and four publicly available and independent cohorts, namely the Versteeg et al. (GSE16476), Lastowska et al. (GSE13136), Hiyama et al. (GSE16237), and Delattre et al. (GSE14880) datasets were used. The normalization was done automatically by R2 using MAS5.0. The remaining expression, amplification, and survival data consisted of the independent SEQC/ MAQC-III Consortium (GSE49710), Kocak et al. study (GSE45547) and Neuroblastoma Research Consortium [NRC] (GSE85047) cohorts. For the expression analysis of TH-MYCN transgenic NB model, the dataset from Balamuth et al. (GSE17740) was used. For the expression analysis of SH-SY5Y cells treated with all-trans retinoic acid (RA), the dataset from Takeda et al. (GSE9169) was used.
Immunohistochemistry
For immunohistochemistry, deparaffinated tissue sections were pretreated with citrate buffer at 98 °C for 20 min, cooled down to room temperature, and blocked with 2% horse serum, avidin blocking solution, and biotin blocking solution (Avidin/Biotin Blocking Kit, SP-2001, Vector Laboratories, Burlingame, CA, USA) for 10 min each. Staining for 4EBP1 was carried out with monoclonal anti-4EBP1 raised in rabbit (1:200; ab32024, Abcam, Cambridge, UK) for 2 h at 37 °C. Detection was carried out using the Dako REAL detection system, alkaline phosphatase/RED, rabbit/mouse following manufacturer´s instructions (Detection Kit #K5005, Agilent Technologies, Santa Clara, CA, USA). Immunostained tissue sections were counterstained with hematoxylin solution according to Mayer (T865.1, Roth, Karlsruhe, Germany).
Evaluation of immunoreactivity of 4EBP1 was carried out in analogy to scoring of hormone receptor Immune Reactive Score (IRS) ranging from 0–12. The percentage of cells with expression of the given antigen was scored and classified in five grades (grade 0 = 0–19%, grade 1 = 20–39%, grade 2 = 40–59%, grade 3 = 60–79%, and grade 4 = 80 − 100%). In addition, the intensity of marker immunoreactivity was determined (grade 0 = none, grade 1 = low, grade 2 = moderate and grade 3 = strong). The product of these two grades defined the final IRS. IRS 0–6 was considered as “low” staining level while IRS 7–12 was categorized as “high” staining level.
Tissue microarrays (TMAs) were constructed by taking three representative cores (each 1 mm in diameter) from respective blocks exhibiting at least 80% viable tumor tissue. Tumor blocks were retrieved from the archives of the Institutes of Pathology of the LMU Munich or the University Hospital Düsseldorf with IRB approval (study numbers 550-16 UE for LMU Munich and 2018-174 for the University Hospital Düsseldorf). Informed consent was obtained from all patients.
Statistics
All experiments were, if not otherwise stated, independently carried out at least three times. Statistical significance was calculated using Student’s t test or Mann–Whitney U-test in GraphPad Prism 8. For survival analysis, the cohorts were stratified based on relative expression of EIF4EBP1. The median was chosen as expression cutoff to determine high and low EIF4EBP1 level. Statistical significance was determined by the logrank test. Multivariate analysis was performed using the Cox Regression method in SPSS v21 (IBM, Armonk, NY, USA). To calculate significance of the scoring of immunohistochemistry staining, the Chi-square test was used. The data are represented as means ± standard deviation. A p-value of less than 0.05 was considered significant.
Cell culture
Cells were maintained using standard tissue culture procedures in a humidified incubator at 37 °C with 5% CO2 and atmospheric oxygen. NB cell lines IMR-32 and Kelly, and HEK-293-T cells were obtained from American Type Culture Collections (ATCC, Manassas, VA, USA). SHEP-TR-MYCN engineered NB cell lines have been previously described [19]. NB cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA), while HEK-293-T cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Thermo Fisher Scientific). All cell culture media were supplemented with 10% (volume/volume) fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MI, USA) and 1% penicillin/streptomycin (Thermo Fisher Scientific). Cells were treated with 3 µg/ml plasmocin (Invivogen, San Diego, CA, USA) to prevent mycoplasma contamination. To induce MYCN expression, SHEP-TR-MYCN cells were treated with 1 µg/ml doxycycline. All cell lines were routinely confirmed to be mycoplasma-free using Venor®GeM Classic kit (Minerva Biolabs, Berlin, Germany). Cell lines were authenticated by STR-profiling (Genomics and Transcriptomics Laboratory, Heinrich Heine University, Dusseldorf, Germany).
RNA extraction, cDNA synthesis, and quantitative real-time PCR
Total RNA was purified from cells using the RNeasy plus mini kit (QIAgen, Hilden, Germany) according to the manufacturer’s handbook. RNA concentration and purity were assessed by spectrophotometry using the NanoDrop2000 (Thermo Fisher Scientific). Subsequently, each sample was diluted to a concentration of 100 ng/µl in nuclease-free water. For cDNA synthesis, 1 µg RNA was processed in a total reaction volume of 20 µl using the High-Capacity cDNA Reverse Transcription kit (Applied Biosystems, Waltham, MA, USA), following the manufacturer’s protocol. Quantitative real-time reverse transcription (qRT) PCR was performed using SYBR green PCR master mix (Applied Biosystems) and the CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA, USA). Relative expression levels of MYCN and EIF4EBP1 were normalized to internal housekeeping genes GUSB and PPIA. The primer list can be found in supplementary table 1.
Immunoblot analysis of protein expression
Cells were washed with phosphate-buffered saline (PBS) and lysed in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 50 mM Tris-HCl, pH 8, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.1% SDS) supplemented with protease inhibitors (Sigma-Aldrich) and phosphatase inhibitors mix (PhosphoSTOP, Roche, Penzberg, Germany). Cell lysates were centrifuged at 21,000 rpm for 15 min at 4 °C to separate cell debris and DNA from protein lysates. Protein concentration was measured with the BCA protein assay kit (Thermo Fisher Scientific), according to manufacturer’s protocol. Protein lysates were separated by SDS-PAGE and transferred onto a nylon membrane. The membrane was incubated for 1 h in Tris-buffered saline Tween (TBST) (50 mM Tris-Cl, 150 mM NaCl, pH 7.5, 0.1% Tween-20) containing 5% bovine serum albumin (BSA), to prevent non-specific antibody binding, followed by an overnight incubation at 4 °C with the following primary antibodies: 4EBP1 (1:1000; #9644, Cell Signaling Technology, Cambridge, UK), MYCN (1:1,000; #9405, Cell Signaling Technology), GAPDH (1:1000; #2118, Cell Signaling Technology), and β-Actin (1:5,000; #A2228, Sigma-Aldrich). The secondary antibodies IRDye 800CW Goat anti-Rabbit (1:10,000; #926-32211, LI-COR Biosciences, Bad Homburg, Germany) or IRDye 800CW Goat anti-Mouse (1:10,000; #926-32210, LI-COR Biosciences) were incubated at room temperature for 1 h, followed by detection of the fluorescent signal with the Odyssey CLx imager (LI-COR Biosciences).
Plasmid construction
The promoter region of the human EIF4EBP1 gene, spanning from −192 to +1372, was inserted into the SacI and BglII restriction sites of the firefly luciferase expressing pGL4.22 plasmid (Promega, Madison, WI, USA). Each of the three identified MYCN binding site was subsequently mutated alone or in a combination of two sites. Each of the E-box sequence has been mutated to CAAGGC. All cloning was performed by GENEWIZ Germany GmbH (Leipzig, Germany).
Luciferase Reporter Assay
For the promoter reporter assay, HEK-293-T cells were seeded into 12-well plates and co-transfected the following day with 500 ng of the EIF4EBP1 WT or mutant promoter pGL4.22 plasmids, 50 ng of the MYCN overexpressing pcDNA3.1 plasmid or empty pcDNA3.1 plasmid, and 3 ng of the Renilla Luciferase expressing pRL-SV40 plasmid (Promega) for normalization. For transfection, plasmids were incubated with 3 µl CalFectin (SignaGen laboratories, Rockville, MD, USA) in Opti-MEM (Thermo Fisher Scientific) for 20 min before adding the mix dropwise onto the cells. 48 h post-transfection, cells were passively lysed and processed according to the protocol of the Dual-Luciferase® Reporter Assay System (Promega), besides using only half the recommended volume of detection buffers. Firefly and Renilla luciferase activities were sequentially measured using a Tecan Spark plate reader and the ratio of firefly luciferase to Renilla luciferase luminescence was calculated. The experiments were repeated independently for three times.
Results
EIF4EBP1 expression is increased in NB and correlates with MYCN expression
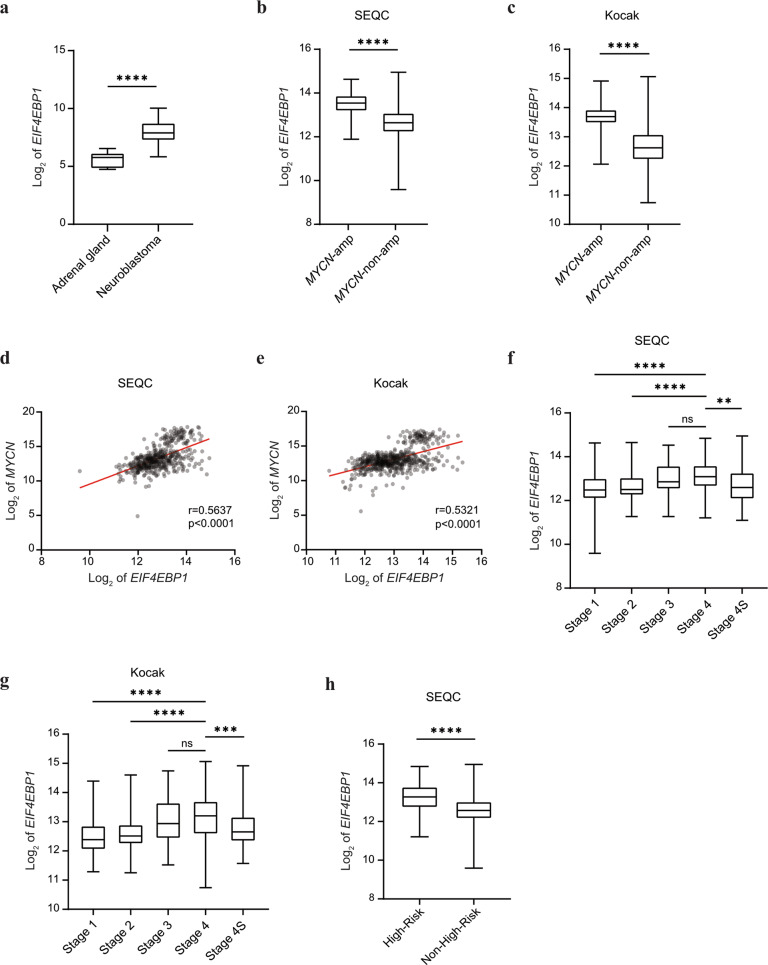
To assess the clinical significance of EIF4EBP1 expression, we first examined EIF4EBP1 mRNA levels in NB tumor tissue samples and normal tissues. We pooled microarray data of four different NB cohorts and retrieved expression data from adrenal tissue used as the corresponding normal tissue (Fig. 1a). This indicated that EIF4EBP1 expression is significantly elevated in NB compared to adrenal gland (p < 0.0001, Fig. 1a). We then determined whether EIF4EBP1 expression is related to the MYCN amplification status. By comparing the level of EIF4EBP1 in MYCN-amplified versus MYCN-non-amplified NB samples, we found that EIF4EBP1 is expressed at higher levels in MYCN-amplified compared to MYCN-non-amplified NB in the SEQC and Kocak cohorts [43, 44] (p < 0.0001, Fig. 1b; p < 0.0001, Fig. 1c). This further supports and extends previous observations made in a limited number of NB samples (n = 20) showing EIF4EBP1 overexpression in MYCN-amplified versus MYCN-non-amplified NB tumors [22]. Since MYCN amplification may result in different levels of MYCN, we next investigated whether expression levels of MYCN and EIF4EBP1 in NB correlate with each other. Our analyses highlighted a significant coexpression between MYCN and EIF4EBP1 in the SEQC (correlation coefficient [r]=0.564, p < 0.0001, Fig. 1d) and Kocak ([r]=0.532, p < 0.0001, Fig. 1e) cohorts. These findings are in line with the reports that EIF4EBP1 is a potential MYCN target gene in NB [41, 42]. We also assessed whether the expression of EIF4EBP1 is determined by NB stages or risk groups, and found that EIF4EBP1 levels are increased according to NB tumor aggressiveness in two cohorts (Fig. 1f, g). In particular, EIF4EBP1 is expressed at higher levels in stage 4 NB tumors as compared to stage 1 and stage 2 tumors (stage 4 versus stage 1, p < 0.0001, Fig. 1f; p < 0.0001 Fig. 1g). Interestingly, samples from stage 4S NB showed significantly lower EIF4EBP1 levels compared to stage 4 tumors (stage 4S versus stage 4, p < 0.01, Fig. 1f; p < 0.001, Fig. 1g). In support of this finding, we observed that in the SEQC cohort EIF4EBP1 expression is higher in high-risk compared to low-risk NB, as based on the Children’s Oncology Group (COG) classification (p < 0.0001, Fig. 1h). Such clinical information was not available in any other publicly available cohorts with mRNA expression data. Taken together, we present evidence that EIF4EBP1 is commonly overexpressed in NB tumors and that EIF4EBP1 level is increased in MYCN-amplified NB and advanced NB stages.
Fig. 1. EIF4EBP1 mRNA expression is associated with MYCN mRNA expression and is increased in more advanced and aggressive NB subsets.
a Expression levels of EIF4EBP1 mRNA in a pool of four different NB cohorts (total n = 203), compared to healthy control tissues (adrenal gland, n = 13). b, c Expression levels of EIF4EBP1 mRNA in MYCN-amplified (n = 92, SEQC [b] and n = 93, Kocak [c]) compared to MYCN-non-amplified (n = 401, SEQC [b] and n = 550, Kocak [c]) NB patients of the SEQC (b) and Kocak (c) cohorts. d, e Expression levels of EIF4EBP1 mRNA plotted against expression levels of MYCN mRNA in SEQC (r = 0.5637, d) and Kocak (r = 0.5321, e) cohorts. f, g Expression levels of EIF4EBP1 mRNA per NB stage in SEQC (f) and Kocak (g) cohorts. h Expression levels of EIF4EBP1 mRNA in high-risk (n = 176) compared to non-high-risk (n = 322) NB in the SEQC cohort. Data were retrieved from the R2: Genomics Analysis and Visualization Platform. Statistics were determined using Mann–Whitney U-test. Exact p-values are presented. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
EIF4EBP1 expression is a factor of poor prognosis in NB
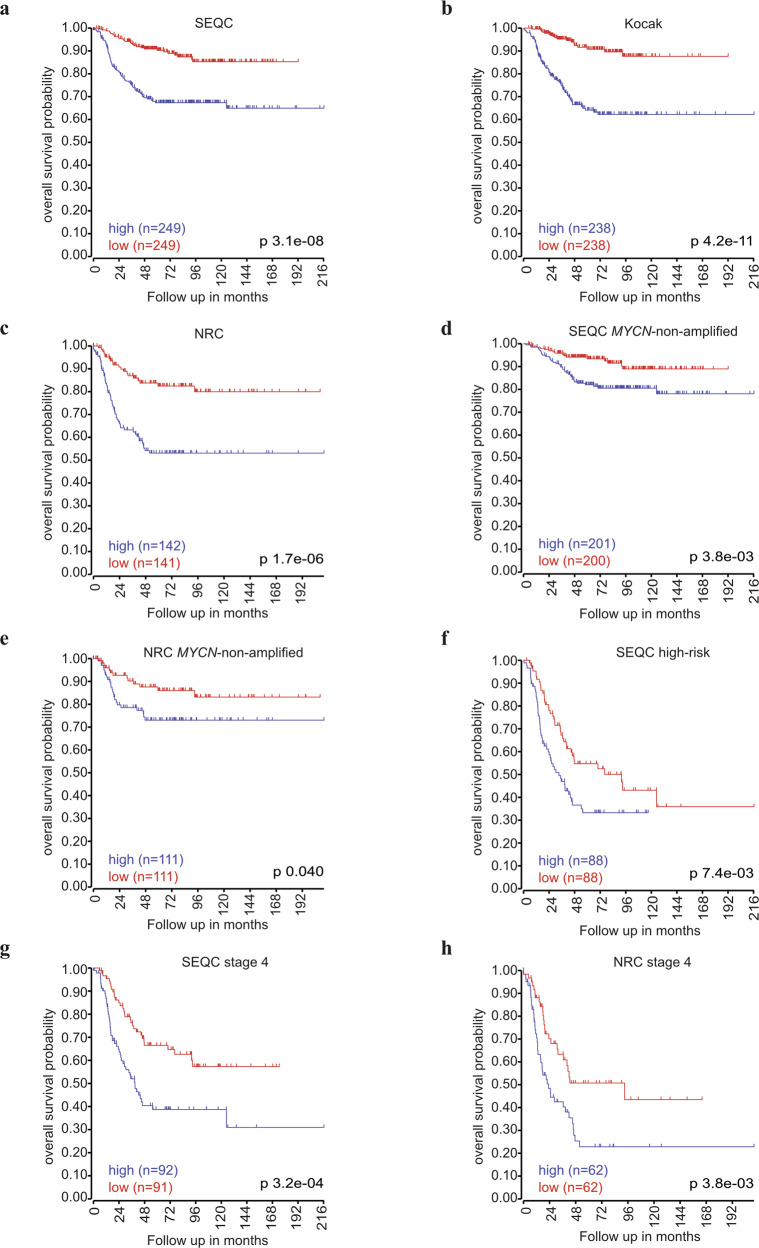
Since we found EIF4EBP1 mRNA levels to be elevated in aggressive NB subsets, we examined whether EIF4EBP1 expression is linked to prognosis in NB patients. Kaplan-Meier estimates univocally showed that high EIF4EBP1 levels (using median expression level as cut off) were significantly associated with reduced overall and event-free survival in three independent cohorts, namely SEQC, Kocak, and NRC cohorts [45] (p = 3.1e-08, Fig. 2a; p = 4.2e-11, Fig. 2b; p = 1.7e-06, Fig. 2c, and supplementary Fig. 1a–c). To test dependence of EIF4EBP1 expression as prognostic factor on established factors of poor prognosis, we performed multivariate analysis to determine the statistical interaction between high EIF4EBP1 expression and MYCN amplification status, tumor stage or age at diagnosis. This indicated that MYCN amplification status, tumor stage and age at diagnosis each influenced the prognostic value of high EIF4EBP1 expression in the SEQC and NRC cohorts (Tables 1 and 2). Therefore, high EIF4EBP1 expression is not an independent factor of poor prognosis in NB. However, we uncovered that EIF4EBP1 expression can predict overall survival in clinically relevant NB subsets, including more advanced and aggressive NB subgroups. Indeed, our analyses highlighted that high EIF4EBP1 expression significantly predicted reduced overall survival in MYCN-non-amplified patients of the SEQC and NRC cohorts (p = 3.8e-03, Fig. 2d; p = 0.04, Fig. 2e), while it was significant for event-free survival only in the SEQC cohort (supplementary Fig. 1d, e). On the other hand, Kaplan–Meier survival estimates in high-risk NB patients (SEQC cohort) revealed that high EIF4EBP1 levels were correlated with poor overall survival (p = 7.4e-03, Fig. 2f), as well as with reduced event-free survival (supplementary Fig. 1f), suggesting that EIF4EBP1 expression can stratify patients within the most aggressive NB subset. We additionally analyzed the prognostic value of EIF4EBP1 expression in stage 4 NB patients. We found high EIF4EBP1 expression to significantly predict decreased overall and event-free survival of stage 4 patients in two independent cohorts (SEQC and NRC cohorts) (p = 3.2e-04, Fig. 2g; p = 3.8e-03, Fig. 2h and supplementary Fig. 1g, h). This highlights that EIF4EBP1 expression robustly stratifies patients within the advanced NB subgroups. Altogether, our analyses support that EIF4EBP1 expression is a factor of poor prognosis in all NB, as well as in high-risk and stage 4 NB.
Fig. 2. EIF4EBP1 mRNA expression correlates with overall survival in NB patients.
a–c Kaplan–Meier survival estimates of overall survival of NB patients stratified by their EIF4EBP1 mRNA expression levels (median cut off) in the SEQC (a), Kocak (b), and NRC (c) cohorts. d–h Kaplan–Meier survival estimates of overall survival of patients with MYCN-non-amplified NB (d, e), high-risk NB (f), or stage 4 NB (g, h) stratified by their EIF4EBP1 mRNA expression levels in the indicated NB cohorts. Significance was determined by log-rank test. Data were obtained from the R2: Genomics Analysis and Visualization Platform.
Table 1.
Multivariate analysis for overall survival of NB patients in the SEQC cohort.
| Variables | HR | 95.0% CI | p value |
|---|---|---|---|
| MYCN amplification | 22.373 | 8.89–56.306 | 0 |
| High EIF4EBP1 mRNA expression | 2.16 | 1.255–3.717 | 0.005 |
| MYCN amplification*high EIF4EBP1 mRNA expression | 0.222 | 0.08–0.614 | 0.004 |
| Stage 4 | 17.618 | 6.694–46.366 | 0 |
| High EIF4EBP1 mRNA expression | 5.457 | 2.026–14.697 | 0.001 |
| Stage 4*high EIF4EBP1 mRNA expression | 0.292 | 0.097–0.879 | 0.029 |
| Age at diagnosis | 33.018 | 7.835–139.139 | 0 |
| High EIF4EBP1 mRNA expression | 12.204 | 2.832–52.598 | 0.001 |
| Age at diagnosis*high EIF4EBP1 mRNA expression | 0.16 | 0.035–0.74 | 0.019 |
Table 2.
Multivariate analysis for overall survival of NB patients in the NRC cohort.
| Variables | HR | 95.0% CI | p value |
|---|---|---|---|
| MYCN amplification | 4.967 | 1.118–22.066 | 0.035 |
| High EIF4EBP1 mRNA expression | 3.031 | 1.543–5.954 | 0.001 |
| MYCN amplification*high EIF4EBP1 mRNA expression | 0.656 | 0.135–3.181 | 0.601 |
| Stage 4 | 15.050 | 4.239–53.432 | 0.018 |
| High EIF4EBP1 mRNA expression | 5.144 | 1.330–19.895 | 0 |
| Stage 4*high EIF4EBP1 mRNA expression | 0.36 | 0.081–1.598 | 0.179 |
| Age at diagnosis | 0.27 | 0.036–2.056 | 0 |
| High EIF4EBP1 mRNA expression | 0.364 | 0.048–2.772 | 0.002 |
| Age at diagnosis*high EIF4EBP1 mRNA expression | 55.427 | 3.258–942.88 | 0.005 |
High 4EBP1 protein expression is associated with prognostically unfavorable histology of NB
To independently confirm the prognostic value of EIF4EBP1/4EBP1 in NB and to determine the biomarker potential of 4EBP1 protein expression in NB, we immunohistochemically analyzed NB TMAs consisting of 69 patient samples. Staining of the TMAs with a 4EBP1-specific antibody revealed a cytoplasmic staining (Fig. 3a), consistent with the expected cellular localization of 4EBP1 [46]. We semi-quantitatively evaluated 4EBP1 staining intensity and correlated 4EBP1 immunoreactivity with the NB histological subtypes according to the International Neuroblastoma Pathology Classification (INPC), which distinguishes patients with favorable or unfavorable histology based on grade of neuroblastic differentiation and mitosis-karyorrhexis index. We found that tumors with unfavorable histology more frequently exhibited a high 4EBP1 staining score (IRS 7–12) as compared to tumors with favorable histology (Fig. 3b), indicating that high 4EBP1 protein expression is associated with more aggressive NB subsets.
Fig. 3. 4EBP1 protein expression is associated with histological subtype of NB.
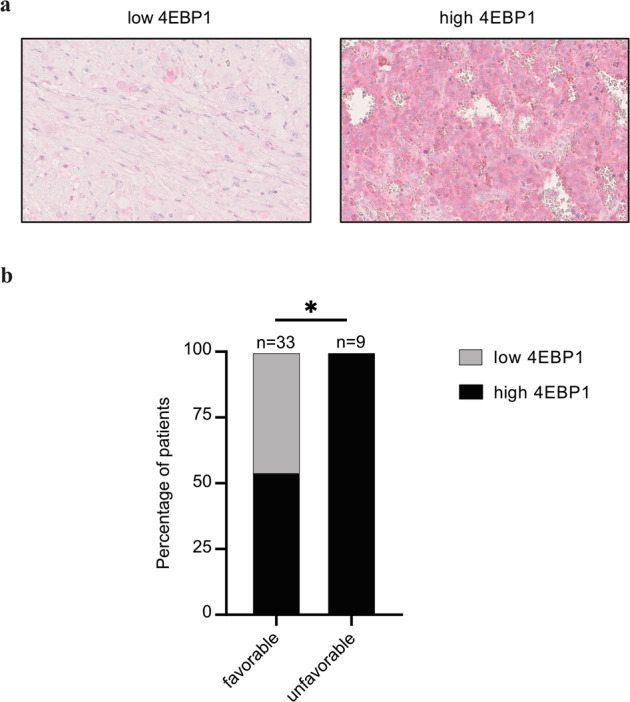
a Representative images at 40X magnification of low (left panel) and high (right panel) 4EBP1 immunohistochemical staining levels of selected NB samples represented on the NB TMAs. b Distribution of NB cases showing low (IRS 0–6) versus high (IRS 7–12) 4EBP1 protein expression in prognostically favorable versus unfavorable histological subtypes according to International Neuroblastoma Pathology Classification (INPC). Fisher’s exact test was used to calculate significance. *P < 0.05.
EIF4EBP1 promoter activity and transcription is controlled by MYCN
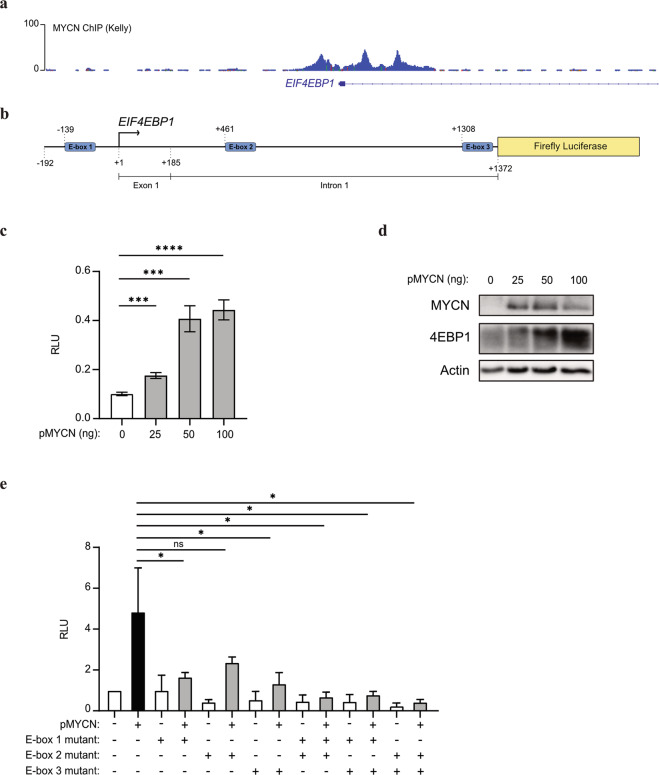
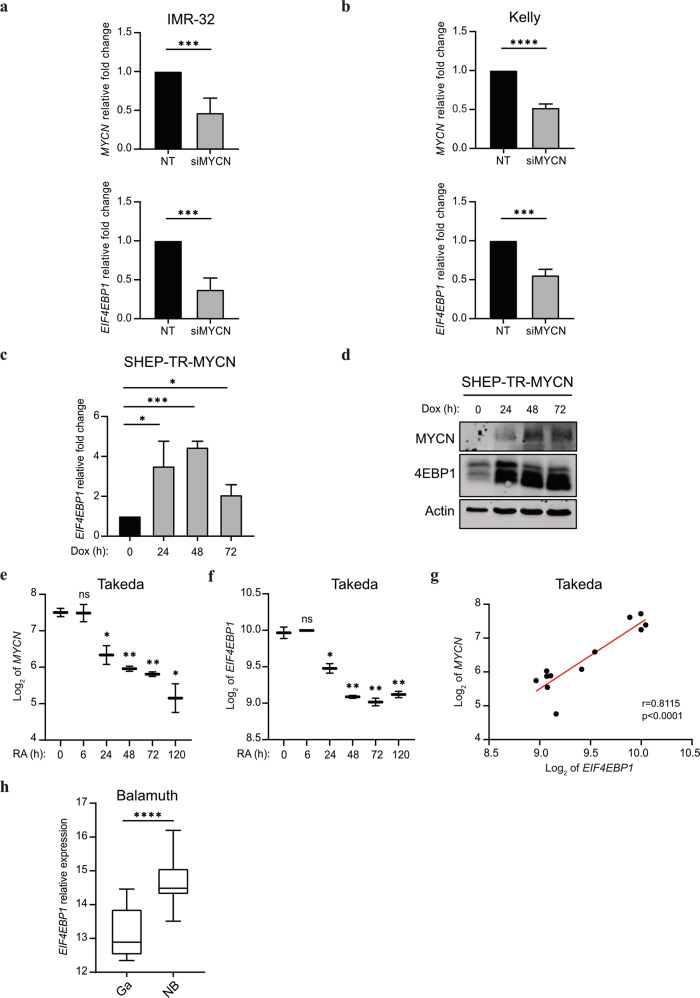
To delineate how elevated EIF4EBP1 expression is mechanistically connected to MYCN amplification and overexpression in NB, we investigated the transcriptional regulation of EIF4EBP1 by MYCN. A previous report detected the presence of MYCN on EIF4EBP1 promoter by ChIP-seq in BE(2)-C, a MYCN-amplified NB cell line [41, 42]. We validated and further extended this finding by analyzing ChIP-seq data available from an additional MYCN-amplified NB cell line, Kelly. This revealed that MYCN binds the endogenous EIF4EBP1 promoter region (which encompasses exon 1 and a part of intron 1) at three distinct positions, indicating three potential MYCN binding sites (Fig. 4a). In silico analysis of the promoter region sequence confirmed the presence of structural E-boxes at the three occupied locations (Fig. 4b). To evaluate the impact of MYCN on the regulation of EIF4EBP1 promoter activity, we designed a luciferase-based gene reporter assay by cloning the EIF4EBP1 promoter region (−192 to +1372) in front of a firefly luciferase gene (Fig. 4b). The activity of the wildtype EIF4EBP1 promoter was dose-dependently increased upon forced expression of MYCN in HEK-293-T cells (Fig. 4c), which was accompanied by an upregulation of endogenous 4EBP1 protein level (Fig. 4d). To investigate which E-boxes are necessary for the transcriptional activation of the EIF4EBP1 promoter by MYCN, either a single or a combination of two of the three potential binding sites were mutated. Mutation of either of the three binding sites alone was sufficient to significantly reduce MYCN-induced promoter activity (Fig. 4e). Any combinations of two mutated binding sites further reduced promoter activity driven by MYCN overexpression (Fig. 4e), suggesting that two binding sites, without a specific preference of one over another, are needed for full induction of EIF4EBP1 promoter activity by MYCN. We next intended to confirm whether MYCN directly regulates EIF4EBP1 transcription in NB cell lines. To do so, we chose two MYCN-amplified NB cell lines, IMR-32 and Kelly, in which we knocked down MYCN expression by siRNA and examined the impact on EIF4EBP1 mRNA levels by qPCR. The depletion of MYCN caused a significant reduction of EIF4EBP1 transcript levels in both cell lines (Fig. 5a, b). To further support these observations, we assessed the impact of forced MYCN expression on EIF4EBP1 transcript and protein levels by using SHEP-TR-MYCN cells, which are MYCN-non-amplified NB cells engineered to express exogenous MYCN with a tetracycline inducible system [19]. Doxycycline treatment markedly increased EIF4EBP1 mRNA level over time (Fig. 5c), in parallel with progressive upregulation of MYCN expression (Fig. 5d). This was accompanied by a net increase in the 4EBP1 protein level (Fig. 5d), supporting that MYCN positively controls EIF4EBP1 mRNA and protein expression in NB cells. To determine whether MYCN regulation of EIF4EBP1 has relevance during NB differentiation, we analyzed expression data of MYCN-non-amplified SH-SY5Y cells treated with RA. This indicated that both MYCN and EIF4EBP1 expression are decreased over time upon treatment, and that levels of both genes are correlated during NB differentiation (Fig. 5e–g). Finally, analyses of expression data from a transgenic mouse model of MYCN-driven NB (TH-MYCN; [47]) revealed that EIF4EBP1 expression is upregulated in NB tumors as compared to the corresponding normal tissue, i.e. the ganglia (Fig. 5h). Taken together, our data provide further evidence that EIF4EBP1 is a transcriptional target of MYCN, potentially providing a mechanistic basis for the observed overexpression of EIF4EBP1 in MYCN-amplified NB patients.
Fig. 4. EIF4EBP1 promoter activity is regulated by MYCN.
a ChIP peaks of MYCN in the EIF4EBP1 promoter region in Kelly NB cell line. b Scheme of the EIF4EBP1 promoter reporter highlighting the three E-boxes corresponding to MYCN binding sites. c HEK-293-T cells were transfected with the wildtype EIF4EBP1 promoter firefly luciferase construct and with the indicated amounts of MYCN expressing plasmid (pMYCN). A Renilla Luciferase vector was used as an internal control. d MYCN and 4EBP1 protein expression were monitored in cell lysates from c by immunoblot analyses using the indicated antibodies. e HEK-293-T were transfected with wildtype or different E-box mutants EIF4EBP1 promoter firefly luciferase constructs with or without a MYCN expressing plasmid (pMYCN). A Renilla Luciferase vector was used as an internal control. Statistics were determined using Student’s t test or Mann–Whitney U-test. Exact p-values are presented. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Fig. 5. EIF4EBP1 expression is regulated by MYCN in NB.
a, b Relative MYCN and EIF4EBP1 mRNA levels upon siRNA-mediated knockdown of MYCN in the MYCN-amplified IMR-32 (a) and Kelly (b) cell lines, as measured by qRT-PCR. c, d SHEP-TR-MYCN cells were treated with doxycycline (1 µg/ml) for the indicated times; EIF4EBP1 mRNA levels were determined by qRT-PCR (c) and levels of MYCN and 4EBP1 proteins were monitored by immunoblot using the indicated antibodies (d). The different 4EBP1 bands correspond to different phosphorylated forms of 4EBP1. e, f Expression levels of MYCN (e) or EIF4EBP1 (f) mRNA in SH-SY5Y cells treated with RA for the indicated times (Takeda’s dataset, n = 2 for each time point; [60]). Statistics were calculated for each time point compared to the control 0 h time point. g Expression levels of EIF4EBP1 mRNA plotted against expression levels of MYCN mRNA in SH-SY5Y cells treated with RA (Takeda’s dataset, n = 2 for each time point; [60]). h Relative EIF4EBP1 mRNA expression in healthy control tissues (ganglia, n = 9) and NB tumors (n = 26) of a TH-MYCN transgenic mouse model of NB (Balamuth’s dataset; [47]). Data were retrieved from the R2: Genomics Analysis and Visualization Platform. Statistics were determined using Student’s t test or Mann–Whitney U-test. Exact p-values are presented. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Discussion
MYCN-amplification is accountable for aggressive NB subsets as it has been associated with increased risk of relapse and reduced overall survival of patients [13]. Since MYCN is considered “undruggable”, there is a demand for identifying targetable downstream effectors of MYCN [20, 21]. In addition, since NB is a clinically heterogenous disease, ranging from spontaneous regression to progression despite aggressive therapies, novel markers that improve patient risk stratification and hence allow for optimal treatment allocation are warranted [4, 48, 49]. Here, we report that EIF4EBP1 expression levels are significantly elevated in NB compared to corresponding non-tumor tissues and positively correlate with both MYCN expression and MYCN amplification status in at least two independent NB patient cohorts. Furthermore, using three independent NB cohorts, we report that high EIF4EBP1 expression is a strong predictor of poor overall and event-free survival across all NB patients. This was not independent of MYCN amplification status, tumor stage or age at diagnosis, which can be explained in part by the regulation of EIF4EBP1 promoter by MYCN which we characterized. However, EIF4EBP1 expression can predict prognosis within distinct patient groups like the MYCN-non-amplified patients subset, for which few biomarkers have been identified. Moreover, we observed that high EIF4EBP1 expression was associated with poor prognosis in the group of patients with aggressive stage 4 NB. Of note, less than a third of stage 4 patients carry a MYCN amplification. Thus, it may be worth considering that, in addition to MYCN amplification status, levels of EIF4EBP1 expression could help identifying patients carrying clinically more aggressive tumors within the stage 4 NB patients group. EIF4EBP1 expression was also linked to worse outcome among high-risk NB patients. Given that MYCN amplification is not able of predicting outcome within high-risk NB patients [50], it appears that EIF4EBP1 expression has a prognostic power beyond MYCN amplification in this patient subset. Thus, EIF4EBP1 expression may represent a promising biomarker for prognostic stratification of high-risk NB patients, in addition to the recently reported genetic alterations in the RAS and p53 pathways [12]. This is further supported by the association we observed between high 4EBP1 protein expression and unfavorable NB histological subtype. Together, our findings highlight a previously underappreciated prognostic factor, i.e., EIF4EBP1/4EBP1, which may help refining risk stratification of NB patients, including MYCN-non-amplified, stage 4 and high-risk patients, and could potentially assist in tailoring more personalized treatment options. Beyond NB, EIF4EBP1 expression was reported to be a factor of poor prognosis in breast and liver cancers [30, 33], as well as in all TCGA tumor types combined [29]. While our data indicate that EIF4EBP1 expression has prognostic power in pediatric cancer, together this supports that EIF4EBP1 expression represents a factor of poor prognosis in a large number of different tumor types.
Our study also extends previous knowledge by providing further experimental evidence to explain the association between EIF4EBP1 and MYCN expression in NB and the overexpression of EIF4EBP1 in MYCN-amplified NB. Our data revealed that MYCN induces transcription of EIF4EBP1 by regulating its promoter through multiple binding sites, which was originally suggested by detection of MYCN binding to the EIF4EBP1 promoter by ChIP analysis [41, 42]. However, whether MYCN could transcriptionally regulate the EIF4EBP1 promoter was still elusive. We demonstrate that MYCN activates the EIF4EBP1 promoter through binding at three distinct E-boxes, which in turn leads to transcriptional increase of EIF4EBP1 even with low to medium MYCN expression, suggesting a threshold for MYCN level. Together with the previous ChIP analysis, this supports that EIF4EBP1 is a direct target gene of MYCN in NB cells. These findings are in line with previous studies reporting that MYC controls EIF4EBP1 by binding its endogenous promoter in colorectal and prostate cancer cells [36, 37], as demonstrated by ChIP, highlighting a general regulation of EIF4EBP1 by MYC family members in cancer cells.
Expression levels of EIF4EBP1 appear not only elevated in MYCN-amplified versus MYCN-non-amplified NB but are also upregulated in MYCN-non-amplified tumors relative to control tissue. It might be speculated that in MYCN-non-amplified NB, EIF4EBP1 expression may be regulated by transcription factors other than MYCN. In particular, ATF4, which is critical for the metabolic response of NB cells to glutamine starvation [51, 52], has been shown to control EIF4EBP1 promoter and transcription in pancreatic beta cells [39]. This transcription factor is highly expressed in NB, and in particular in advanced stage 4 [52]. In addition, another transcription factor that is commonly overexpressed in NB is OCT4 [53]. Of note, this transcription factor has been identified by ChIP-seq to bind the promoter region of EIF4EBP1 in human embryonic stem cells [54, 55], thus OCT4 may also activate EIF4EBP1 transcription in NB cells. Together, these data suggest potential mechanisms underlying the MYCN independent regulation of EIF4EBP1 expression in MYCN-non-amplified NB patients.
Given the prognostic significance of EIF4EBP1/4EBP1 in NB, it is possible that 4EBP1 confers advantages to NB tumor growth or tumor cell survival. As evidenced by the presence of necrotic areas flanked by HIF-1α positive staining [56], NB experience metabolic stress, corresponding to nutrient deprivation and hypoxia, as a consequence of abnormal and immature vascularization [57, 58]. One important mechanism for cancer cells to adapt to metabolic stress is through reprogramming of mRNA translation [59]. As a major regulator of mRNA translation, 4EBP1 may aid NB cells to cope with hypoxia and nutrient deprivation. This is supported by the report that 4EBP1 promotes survival of breast tumors under hypoxia by stimulating the synthesis of pro-angiogenic factors, like HIF-1α and VEGF, to facilitate tumor angiogenesis in vivo [28]. In addition, the control of mRNA translation was shown to be critical to prevent the deleterious effects of MYCN and MYC overexpression, as we and others previously reported [37]. In fact, 4EBP1, by reducing overall protein synthesis, was reported to prevent cell death induced upon MYC overexpression, likely by blunting accumulation of misfolded proteins and proteotoxic ER stress [37]. It is possible that in a similar manner 4EBP1 contributes to inhibit cell death induced by MYCN overexpression in MYCN-amplified NB.
In summary, the findings reported here indicate that EIF4EBP1 is a direct target gene of MYCN in NB, explaining the observed high expression of EIF4EBP1 in NB, and that EIF4EBP1 mRNA and protein expression have prognostic values in NB patients, especially for stratifying high-risk NB patients.
Supplementary information
Acknowledgements
We would like to thank Dr. Bastian Malzkorn (Institute of Neuropathology, Heinrich Heine University Düsseldorf) for helpful discussions. GL was supported by funding from the Elterninitiative Düsseldorf e.V., the Research Commission of the Medical Faculty of Heinrich Heine University, the Deutsche Forschungsgemeinschaft (Grant LE 3751/2-1), and the German Cancer Aid (Grant 70112624). The laboratory of TGPG is supported by the Barbara und Wilfried Mohr Foundation. BR is supported by the Israel Science Foundation (grant No. 1436/19).
Author contributions
Conception and design: KV and GL. Provision of study material and patients: IE and TK. Financial and administrative support: GR. Data analysis and interpretation: KV, TGPG, AS, and GL. Critical review and discussion: BR, MR, AS, GR, and GL. Experimental support: KV, KS, CF, AK, DP, LH, and MFO. Manuscript writing: KV, GR, and GL. Final approval of the manuscript: all authors.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Competing interests
TK received honoraria for Consulting/Advisory by Amgen, AstraZeneca, BMS, Merck KGaA, MSD, Novartis, Pfizer, Roche, for Research Funding by Merck KGaA and Roche; for talks by Merck KGaA, AstraZeneca. The other authors declare no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41420-022-00963-0.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.van Arendonk KJ, Chung DH. Neuroblastoma: tumor biology and its implications for staging and treatment. Children (Basel) 2019;6. 10.3390/children6010012 [DOI] [PMC free article] [PubMed]
- 3.Tolbert VP, Matthay KK. Neuroblastoma: clinical and biological approach to risk stratification and treatment. Cell Tissue Res. 2018;372:195–209. doi: 10.1007/s00441-018-2821-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maris JM. The biologic basis for neuroblastoma heterogeneity and risk stratification. Curr Opin Pediatrics. 2005;17:7–13. doi: 10.1097/01.mop.0000150631.60571.89. [DOI] [PubMed] [Google Scholar]
- 5.London WB, Castel V, Monclair T, Ambros PF, Pearson ADJ, Cohn SL, et al. Clinical and biologic features predictive of survival after relapse of neuroblastoma: a report from the International Neuroblastoma Risk Group project. J Clin Oncol. 2011;29:3286–92. doi: 10.1200/JCO.2010.34.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto NR, Applebaum MA, Volchenboum SL, Matthay KK, London WB, Ambros PF, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;33:3008–17. doi: 10.1200/JCO.2014.59.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simon T, Berthold F, Borkhardt A, Kremens B, Carolis B, de, Hero B. Treatment and outcomes of patients with relapsed, high-risk neuroblastoma: results of German trials. Pediatr Blood Cancer. 2011;56:578–83. doi: 10.1002/pbc.22693. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–4. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 9.Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, Pontual L, de, Combaret V, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–70. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 11.Amelio I, Bertolo R, Bove P, Candi E, Chiocchi M, Cipriani C, et al. Cancer predictive studies. Biol Direct. 2020;15:18.. doi: 10.1186/s13062-020-00274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ackermann S, Cartolano M, Hero B, Welte A, Kahlert Y, Roderwieser A, et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science. 2018;362:1165–70. doi: 10.1126/science.aat6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang M, Weiss WA. Neuroblastoma and MYCN. Cold Spring Harb Perspect Med. 2013;3:a014415. doi: 10.1101/cshperspect.a014415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zeid R, Lawlor MA, Poon E, Reyes JM, Fulciniti M, Lopez MA, et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat Genet. 2018;50:515–23. doi: 10.1038/s41588-018-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu R, Shi P, Wang Z, Yuan C, Cui H. Molecular mechanisms of MYCN dysregulation in cancers. Front Oncol. 2020;10:625332. doi: 10.3389/fonc.2020.625332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliynyk G, Ruiz-Pérez MV, Sainero-Alcolado L, Dzieran J, Zirath H, Gallart-Ayala H, et al. MYCN-enhanced oxidative and glycolytic metabolism reveals vulnerabilities for targeting neuroblastoma. iScience. 2019;21:188–204. doi: 10.1016/j.isci.2019.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boon K, Caron HN, van Asperen R, Valentijn L, Hermus MC, van Sluis P, et al. N-myc enhances the expression of a large set of genes functioning in ribosome biogenesis and protein synthesis. EMBO J. 2001;20:1383–93. doi: 10.1093/emboj/20.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tjaden B, Baum K, Marquardt V, Simon M, Trajkovic-Arsic M, Kouril T, et al. N-Myc-induced metabolic rewiring creates novel therapeutic vulnerabilities in neuroblastoma. Sci Rep. 2020;10:7157. doi: 10.1038/s41598-020-64040-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bell E, Chen L, Liu T, Marshall GM, Lunec J, Tweddle DA. MYCN oncoprotein targets and their therapeutic potential. Cancer Lett. 2010;293:144–57. doi: 10.1016/j.canlet.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 21.Wolpaw AJ, Bayliss R, Büchel G, Dang CV, Eilers M, Gustafson WC, et al. Drugging the “undruggable” MYCN oncogenic transcription factor: overcoming previous obstacles to impact childhood cancers. Cancer Res. 2021;81:1627–32. doi: 10.1158/0008-5472.CAN-20-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schramm A, Köster J, Marschall T, Martin M, Schwermer M, Fielitz K, et al. Next-generation RNA sequencing reveals differential expression of MYCN target genes and suggests the mTOR pathway as a promising therapy target in MYCN-amplified neuroblastoma. Int. J. Cancer. 2013;132:E106–15. doi: 10.1002/ijc.27787. [DOI] [PubMed] [Google Scholar]
- 23.Musa J, Orth MF, Dallmayer M, Baldauf M, Pardo C, Rotblat B, et al. Eukaryotic initiation factor 4E-binding protein 1 (4E-BP1): a master regulator of mRNA translation involved in tumorigenesis. Oncogene. 2016;35:4675–88. doi: 10.1038/onc.2015.515. [DOI] [PubMed] [Google Scholar]
- 24.Morita M, Gravel S-P, Chénard V, Sikström K, Zheng L, Alain T, et al. mTORC1 controls mitochondrial activity and biogenesis through 4E-BP-dependent translational regulation. Cell Metab. 2013;18:698–711. doi: 10.1016/j.cmet.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Dowling RJO, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–6. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Z, Feng X, Molinolo AA, Martin D, Vitale-Cross L, Nohata N, et al. 4E-BP1 is a tumor suppressor protein reactivated by mTOR inhibition in head and neck cancer. Cancer Res. 2019;79:1438–50. doi: 10.1158/0008-5472.CAN-18-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding M, van der Kwast TH, Vellanki RN, Foltz WD, McKee TD, Sonenberg N, et al. The mTOR targets 4E-BP1/2 restrain tumor growth and promote hypoxia tolerance in PTEN-driven prostate cancer. Mol Cancer Res. 2018;16:682–95. doi: 10.1158/1541-7786.MCR-17-0696. [DOI] [PubMed] [Google Scholar]
- 28.Braunstein S, Karpisheva K, Pola C, Goldberg J, Hochman T, Yee H, et al. A hypoxia-controlled cap-dependent to cap-independent translation switch in breast cancer. Mol Cell. 2007;28:501–12. doi: 10.1016/j.molcel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Wu S & Wagner G. Deep computational analysis of human cancer and non-cancer tissues details dysregulation of eIF4F components and their interactions in human cancers. bioRxiv 2020. 10.1101/2020.10.12.336263
- 30.Karlsson E, Pérez-Tenorio G, Amin R, Bostner J, Skoog L, Fornander T, et al. The mTOR effectors 4EBP1 and S6K2 are frequently coexpressed, and associated with a poor prognosis and endocrine resistance in breast cancer: a retrospective study including patients from the randomised Stockholm tamoxifen trials. Breast Cancer Res. 2013;15:R96. doi: 10.1186/bcr3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kremer CL, Klein RR, Mendelson J, Browne W, Samadzedeh LK, Vanpatten K, et al. Expression of mTOR signaling pathway markers in prostate cancer progression. Prostate. 2006;66:1203–12. doi: 10.1002/pros.20410. [DOI] [PubMed] [Google Scholar]
- 32.Lee M, Kim EJ, Jeon MJ. MicroRNAs 125a and 125b inhibit ovarian cancer cells through post-transcriptional inactivation of EIF4EBP1. Oncotarget. 2016;7:8726–42. doi: 10.18632/oncotarget.6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cha Y-L, Li P-D, Yuan L-J, Zhang M-Y, Zhang Y-J, Rao H-L, et al. EIF4EBP1 overexpression is associated with poor survival and disease progression in patients with hepatocellular carcinoma. PLoS ONE. 2015;10:e0117493. doi: 10.1371/journal.pone.0117493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fransson S, Abel F, Kogner P, Martinsson T, Ejeskär K. Stage-dependent expression of PI3K/Akt‑pathway genes in neuroblastoma. Int J Oncol. 2013;42:609–16. doi: 10.3892/ijo.2012.1732. [DOI] [PubMed] [Google Scholar]
- 35.Meng X, Li H, Fang E, Feng J, Zhao X. Comparison of stage 4 and stage 4s neuroblastoma identifies autophagy-related gene and LncRNA signatures associated with prognosis. Front Oncol. 2020;10:1411. doi: 10.3389/fonc.2020.01411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balakumaran BS, Porrello A, Hsu DS, Glover W, Foye A, Leung JY, et al. MYC activity mitigates response to rapamycin in prostate cancer through eukaryotic initiation factor 4E-binding protein 1-mediated inhibition of autophagy. Cancer Res. 2009;69:7803–10. doi: 10.1158/0008-5472.CAN-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tameire F, Verginadis II, Leli NM, Polte C, Conn CS, Ojha R, et al. ATF4 couples MYC-dependent translational activity to bioenergetic demands during tumour progression. Nat Cell Biol. 2019;21:889–99. doi: 10.1038/s41556-019-0347-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Horn JL, Banda K, Goodman AZ, Lim Y, Jana S et al. The androgen receptor regulates a druggable translational regulon in advanced prostate cancer. Sci Transl Med. 2019;11. 10.1126/scitranslmed.aaw4993 [DOI] [PMC free article] [PubMed]
- 39.Yamaguchi S, Ishihara H, Yamada T, Tamura A, Usui M, Tominaga R, et al. ATF4-mediated induction of 4E-BP1 contributes to pancreatic beta cell survival under endoplasmic reticulum stress. Cell Metab. 2008;7:269–76. doi: 10.1016/j.cmet.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 40.Azar R, Lasfargues C, Bousquet C, Pyronnet S. Contribution of HIF-1α in 4E-BP1 gene expression. Mol. Cancer Res. 2013;11:54–61. doi: 10.1158/1541-7786.MCR-12-0095. [DOI] [PubMed] [Google Scholar]
- 41.Cheung CHY, Hsu C-L, Tsuei C-Y, Kuo T-T, Huang C-T, Hsu W-M, et al. Combinatorial targeting of MTHFD2 and PAICS in purine synthesis as a novel therapeutic strategy. Cell Death Dis. 2019;10:786. doi: 10.1038/s41419-019-2033-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hsu C-L, Chang H-Y, Chang J-Y, Hsu W-M, Huang H-C, Juan H-F. Unveiling MYCN regulatory networks in neuroblastoma via integrative analysis of heterogeneous genomics data. Oncotarget. 2016;7:36293–310. doi: 10.18632/oncotarget.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SEQC/MAQC consortium. SEQC/MAQC consortium: a comprehensive assessment of RNA-seq accuracy, reproducibility and information. Nat Biotechnol. 2014;32:903–14. doi: 10.1038/nbt.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kocak H, Ackermann S, Hero B, Kahlert Y, Oberthuer A, Juraeva D, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rajbhandari P, Lopez G, Capdevila C, Salvatori B, Yu J, Rodriguez-Barrueco R, et al. Cross-cohort analysis identifies a TEAD4-MYCN positive feedback loop as the core regulatory element of high-risk neuroblastoma. Cancer Discov. 2018;8:582–99. doi: 10.1158/2159-8290.CD-16-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Armengol G, Rojo F, Castellví J, Iglesias C, Cuatrecasas M, Pons B, et al. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 2007;67:7551–5. doi: 10.1158/0008-5472.CAN-07-0881. [DOI] [PubMed] [Google Scholar]
- 47.Balamuth NJ, Wood A, Wang Q, Jagannathan J, Mayes P, Zhang Z, et al. Serial transcriptome analysis and cross-species integration identifies centromere-associated protein E as a novel neuroblastoma target. Cancer Res. 2010;70:2749–58. doi: 10.1158/0008-5472.CAN-09-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rugolo F, Bazan NG, Calandria J, Jun B, Raschellà G, Melino G, et al. The expression of ELOVL4, repressed by MYCN, defines neuroblastoma patients with good outcome. Oncogene. 2021;40:5741–51. doi: 10.1038/s41388-021-01959-3. [DOI] [PubMed] [Google Scholar]
- 49.Pieraccioli M, Nicolai S, Pitolli C, Agostini M, Antonov A, Malewicz M, et al. ZNF281 inhibits neuronal differentiation and is a prognostic marker for neuroblastoma. Proc. Natl Acad. Sci. USA. 2018;115:7356–61. doi: 10.1073/pnas.1801435115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee JW, Son MH, Cho HW, Ma YE, Yoo KH, Sung KW, et al. Clinical significance of MYCN amplification in patients with high-risk neuroblastoma. Pediatr Blood Cancer. 2018;65:e27257. doi: 10.1002/pbc.27257. [DOI] [PubMed] [Google Scholar]
- 51.Qing G, Li B, Vu A, Skuli N, Walton ZE, Liu X, et al. ATF4 regulates MYC-mediated neuroblastoma cell death upon glutamine deprivation. Cancer Cell. 2012;22:631–44. doi: 10.1016/j.ccr.2012.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ren P, Yue M, Xiao D, Xiu R, Gan L, Liu H, et al. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J Pathol. 2015;235:90–100. doi: 10.1002/path.4429. [DOI] [PubMed] [Google Scholar]
- 53.Yang S, Zheng J, Ma Y, Zhu H, Xu T, Dong K, et al. Oct4 and Sox2 are overexpressed in human neuroblastoma and inhibited by chemotherapy. Oncol Rep. 2012;28:186–92. doi: 10.3892/or.2012.1765. [DOI] [PubMed] [Google Scholar]
- 54.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell. 2013;153:1149–63. doi: 10.1016/j.cell.2013.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Čančer M, Hutter S, Holmberg KO, Rosén G, Sundström A, Tailor J, et al. Humanized stem cell models of pediatric medulloblastoma reveal an Oct4/mTOR axis that promotes malignancy. Cell Stem Cell. 2019;25:855–870.e11. doi: 10.1016/j.stem.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Påhlman S, Mohlin S. Hypoxia and hypoxia-inducible factors in neuroblastoma. Cell Tissue Res. 2018;372:269–75. doi: 10.1007/s00441-017-2701-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schaaf MB, Garg AD, Agostinis P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis. 2018;9:115. doi: 10.1038/s41419-017-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lugano R, Ramachandran M, Dimberg A. Tumor angiogenesis: causes, consequences, challenges and opportunities. Cell Mol Life Sci. 2020;77:1745–70. doi: 10.1007/s00018-019-03351-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leprivier G, Rotblat B, Khan D, Jan E, Sorensen PH. Stress-mediated translational control in cancer cells. Biochim. Biophys. Acta. 2015;1849:845–60. doi: 10.1016/j.bbagrm.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 60.Nishida Y, Adati N, Ozawa R, Maeda A, Sakaki Y, Takeda T. Identification and classification of genes regulated by phosphatidylinositol 3-kinase- and TRKB-mediated signalling pathways during neuronal differentiation in two subtypes of the human neuroblastoma cell line SH-SY5Y. BMC Res Notes. 2008;1:95. doi: 10.1186/1756-0500-1-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.