Abstract
Handedness has low heritability and epigenetic mechanisms have been proposed as an etiological mechanism. To examine this hypothesis, we performed an epigenome-wide association study of left-handedness. In a meta-analysis of 3914 adults of whole-blood DNA methylation, we observed that CpG sites located in proximity of handedness-associated genetic variants were more strongly associated with left-handedness than other CpG sites (P = 0.04), but did not identify any differentially methylated positions. In longitudinal analyses of DNA methylation in peripheral blood and buccal cells from children (N = 1737), we observed moderately stable associations across age (correlation range [0.355–0.578]), but inconsistent across tissues (correlation range [− 0.384 to 0.318]). We conclude that DNA methylation in peripheral tissues captures little of the variance in handedness. Future investigations should consider other more targeted sources of tissue, such as the brain.
Subject terms: Epigenomics, Behavioural genetics
Introduction
Handedness, defined as the preferential use of one hand over the other, is established early in life and represents a highly stable trait that is thought to be accompanied by changes in brain1, corticospinal tract2, peripheral innervation and vascularization of arm skeletal muscles3, arm dynamics4, and possibly the immune system5. Laterality is already observable in very early stages of development: fetuses show coordinated hand movements at 8–12 weeks post-conception with more right than left arm movements in 85% of fetuses6–8. In children and adults, the prevalence of left-handedness is about 10%9. Handedness clearly clusters in families, but its inheritance pattern is not clear and the heritability of handedness is relatively low: approximately 25% in twin studies (with 95% confidence intervals (CI) ranging from 11 to 30%10–12) and 11.9% (95% CI 7.2–17.7) based on autosomal Identity by Descent (IBD) information from closely related individuals in the UK Biobank13. Early genetic hypotheses on the development of hand preference incorporated a component of randomness14,15: depending on which alleles were inherited, a person would be right-handed or have an equal chance of being either left- or right-handed. This randomness has also been referred to as “developmental instability”, or “fluctuating asymmetry”, representing developmental variance unique to the person16. Such randomness could explain monozygotic twin discordance in handedness15 as reported in some twin studies17–19, although very early studies did not confirm zygosity by DNA testing.
Candidate genes associated with handedness and brain and spinal asymmetry include leucine rich repeat transmembrane neuronal 1 (LRRTM1)20, LIM domain only 4 (LMO4)21, neuronal differentiation 6 (NEUROD6)21, proprotein convertase subtilisin/kexin type 6 (PCSK6)22,23 and the androgen receptor gene (AR)24–26. However, genome-wide association studies (GWASs) found no support for these candidate genes27–30, but identified multiple novel loci. Recently, the largest genome-wide association study to date, which included more than 1,5 million right-handed and 194,000 left-handed individuals, found 41 loci associated with left-handedness13. Besides previously described associations of left-handedness with loci that contain microtubule-associated protein 2 (MAP2)29,30 and tubulin beta class 1 (TUBB)29, the list of genome-wide significant associations was expanded with other microtubule formation and regulation genes (TUBB3, NDRG1, TUBB4A, TUBA1B, BUB3 and TTC28)13. Thus, multiple variants were close to genes involved in microtubule functions that form part of the cytoskeleton, and play a role in neurogenesis, axon transport31, and brain asymmetry32. The results of functional analyses suggested involvement of neurogenesis and the central nervous system and brain tissues, including hippocampus and cerebrum, in the etiology of left-handedness. The variance of handedness explained by single nucleotide polymorphisms (SNP heritability) on the liability scale was 3.45% in this meta-analysis13. The estimate in UK Biobank was 5.9%13.
Partly because of the limited success of early genetic association studies, epigenomic studies have been proposed as promising targets to identify mechanisms underlying handedness33–35. Epigenome-wide association studies (EWAS) perform association tests for several hundred thousand of CpGs (cytosine-phosphate-guanine nucleotide base pairing) to identify differentially methylated positions (DMPs) associated with a trait. Approaches that test associations across multiple nearby correlated CpGs to identify differentially methylated regions (DMRs)36, or that combine multiple CpGs into DNA methylation scores37 help improve power by combining the effects of multiple CpG sites and reducing the number of conducted tests. The predictive value of DNA methylation by construction of individual methylation scores has been shown for several outcomes, e.g. body mass index38. Epigenetic variation could be one pathway to connect the hypothesized random component of handedness, and contribute to asymmetrical gene expression in the two brain hemispheres39 and the spinal cord40. The latter was supported by a genome-wide DNA methylation analysis in the right and left part of the fetal spinal cord from six samples obtained between 8 and 12 weeks post conception that detected asymmetrically methylated CpG islands of several genes40.
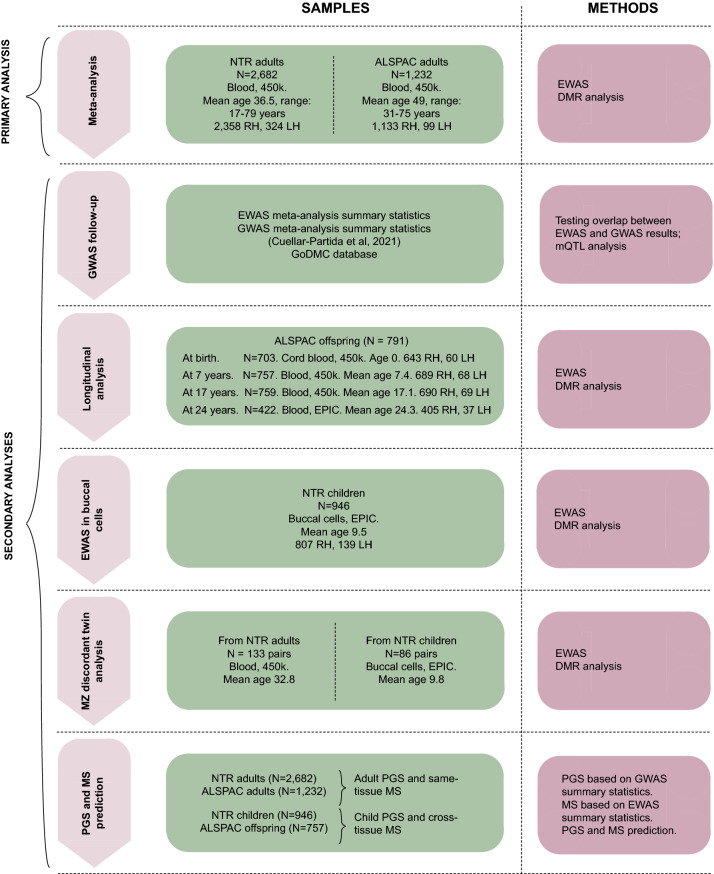
At present, no epigenome-wide association studies of handedness have been performed, and the role of DNA methylation in handedness has only been examined in small candidate-gene studies41,42. Here we analyzed DNA methylation data and left-handedness from two cohorts—the Netherlands Twin Register (NTR) and Avon Longitudinal Study of Parents and Children (ALSPAC). Both cohorts include methylation data in children and adults. In the ALSPAC cohort, adults and children are related (parents and offspring), and in the NTR cohort adults and children come from independent samples. We excluded ambidextrous and mixed-handed persons, and treated handedness as a dichotomous trait (left- or right-handed). First, we performed a meta-analysis of DNA methylation data from the two largest groups with DNA methylation data in adults (total sample size = 3914) to identify differentially methylated positions and differentially methylated regions associated with left-handedness. Next, we performed additional analyses in which we (1) examined if the epigenetic signal for left-handedness was enriched near previously reported GWAS loci13; (2) examined methylation differences between left- and right-handed twins from discordant monozygotic (MZ) twin pairs; (3) characterized the longitudinal and cross-tissue similarity of the genome-wide epigenetic signal associated with left-handedness using data from children; and (4) created methylation scores and estimated their predictive value over and above polygenic scores (Fig. 1).
Figure 1.
Flowchart of epigenome-wide association study of left-handedness. The flowchart summarizes the study design. The primary analyses included EWASs in NTR adults and ALSPAC adults, followed by meta-analysis to identify DNA methylation sites associated with left-handedness. The secondary analyses included: (1) left-handedness GWAS loci follow-up; (2) longitudinal analysis of DNA methylation at four ages in ALSPAC offspring; (3) analysis of buccal cell DNA methylation in NTR children; (4) analysis of DNA methylation differences between left and right-handed co-twins from NTR discordant MZ twin pairs and (5) polygenic and DNA methylation scores prediction. For left-handedness prediction, polygenic scores (PGS) were created based on left-handedness GWAS summary statistics not including NTR/ALSPAC. Methylation scores (MS) were created based on weights from EWASs in NTR adults, ALSPAC adults, NTR children and ALSPAC offspring at 7 years old to estimate the predictive performance. LH, left-handed; RH, right-handed. DMR, differentially methylated region. EWAS, epigenome-wide association study. GWAS, genome-wide association study. GoDMC, Genetics of DNA Methylation Consortium. mQTL, methylation quantitative trait locus. Blood, buccal cells indicate tissue of DNA methylation. 450 k, EPIC indicate the platform for DNA methylation measurement.
Results
Epigenome-wide association meta-analysis of left-handedness
Tables 1, 2 and Supplementary Tables 1–4 display the characteristics of the participants included in the study. The epigenome-wide association study of left-handedness meta-analysed data from adults (N = 3914) with DNA methylation data in peripheral blood (Illumina, 450 k) from NTR (N = 2682, 34% male, mean age at methylation 36.5, standard deviation (SD) 12.7) and ALSPAC (N = 1232, 30% male, mean age at methylation 48.98, SD 5.55). In EWAS discovery cohorts, the prevalence of left-handedness was 12% in NTR, and 8% in ALSPAC. The prevalence of left-handedness as a function of year of birth in NTR is provided in Supplementary Table 2.
Table 1.
Characteristics of adult cohorts included in the primary meta-analysis.
| NTR adults | ALSPAC adults | |||
|---|---|---|---|---|
| N = 2682 | N = 1232 | |||
| LH | RH | LH | RH | |
| N | 324 (12%) | 2358 (88%) | 99 (8%) | 1133 (92%) |
| Age at blood sampling | 34.3 (11.2) | 36.8 (12.9) | 49.1 (5.9) | 49.0 (5.5) |
| Sex | ||||
| Males | 119 (37%) | 783 (33%) | 31 (31%) | 333 (29%) |
| Females | 205 (63%) | 1575 (67%) | 68 (69%) | 800 (71%) |
| Multiples | 315 (97.2%) | 2171 (92%) | 0 | 0 |
| BMI | 24.2 (3.7) | 24.2 (3.9) | 25.80 (4.3) | 26.71 (4.7) |
| Smoking (current) | 65 (20%) | 486 (21%) | 37 (37%) | 424 (37%) |
| Cell proportion | Neutrophils | B lymphocytes | ||
| 52.8 (8.7) | 52.4 (9.2) | 10.8 (4.3) | 10.4 (4) | |
| Eosinophils | CD4T | |||
| 3.1 (1.9) | 3.1 (2.3) | 18.4 (7.2) | 18 (6.6) | |
| Monocytes | CD8T | |||
| 8.4 (2.2) | 8.4 (2.4) | 2.2 (3.8) | 1.9 (3.1) | |
| Natural killer cells | ||||
| 21.9 (6.5) | 20.7 (5.7) | |||
| Granulocytes | ||||
| 45.5 (14.7) | 47.9 (12.4) | |||
| Monocytes | ||||
| 7.5 (3.4) | 7.4 (3.5) | |||
NTR Netherlands Twin Register, ALSPAC Avon Longitudinal Study of Parents and Children, LH left-handed, RH right-handed.
Whole blood DNA methylation (Illumina 450 k). Numbers in EWAS basic models are reported.
Values are presented as mean (standard deviation (SD)) or n (%).
Current smokers in ALSPAC were defined as those with cg05575921 methylation below 0.82 (see “Methods”section).
Table 2.
Characteristics of the datasets included in secondary analyses.
| ALSPAC offspring (longitudinal) | NTR children | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| at birth N = 703 | 7 years old N = 757 | 17 years old N = 759 | 24 years old N = 442 | N = 946 | ||||||
| LH | RH | LH | RH | LH | RH | LH | RH | LH | RH | |
| N | 60 (8.5%) | 643 (91.5%) | 68 (9%) | 689 (91%) | 69 (9%) | 690 (91%) | 37 (8.4%) | 405 (91.6%) | 139(15%) | 807(85%) |
| Age at blood sampling | 7.43 (0.1) | 7.4 (0.1) | 16.9 (1.1) | 17.1 (1.0) | 24.3 (0.7) | 24.3 (0.7) | 9.58(1.78) | 9.56(1.86) | ||
| Sex | ||||||||||
| Males | 32 (53.3%) | 302 (47%) | 36 (52.9%) | 332 (48.2%) | 36 (52.2%) | 321 (46.5%) | 16 (43.2%) | 173 (42.7%) | 71 (51%) | 412 (51%) |
| Females | 28 (46.7%) | 341 (53%) | 32 (47.1%) | 357 (51.8%) | 33 (47.8%) | 369 (53.5%) | 21 (56.8%) | 232 (57.3%) | 68 (49%) | 395 (49%) |
| Gestational age | 39.6 (1.5) | 39.6 (1.4) | 39.6 (1.4) | 39.6 (1.6) | 35.51(2.83) | 35.93(2.52) | ||||
| Birth weight | 3567.8 (434.4) | 3477.3 (493.6) | 3583 (445.4) | 3481 (493.3) | 2369 (585.2) | 2407 (533.5) | ||||
| Maternal smoking during pregnancy | 9 (15.2%) | 74 (11.6%) | 10 (14.9%) | 81 (11.8%) | 14(11%) | 56(7%) | ||||
| BMI | 22.5 (3.6) | 22.5 (3.6) | 24.2 (3.6) | 24.4 (4.5) | ||||||
| Smoking (current) | 20 (29.4%) | 161 (23.6%) | 9 (24.3%) | 131 (33%) | ||||||
NTR Netherlands Twin Register (buccal cell DNA methylation); ALSPAC Avon Longitudinal Study of Parents and Children (cord blood and whole blood DNA methylation); LH left-handed; RH right-handed; BMI body mass index.
Numbers in EWAS basic models are reported.
Values are presented as mean (SD) or n (%).
Current smokers in ALSPAC were defined as those with cg05575921 methylation below 0.82 (see “Methods”section).
We tested 409,562 CpGs with adjustment for age, sex, smoking status, body mass index (BMI), measured or estimated cell proportions, and technical covariates. Genome-wide EWAS test statistics from each cohort separately, and from the meta-analysis, showed no inflation (Supplementary Tables 5–11). None of the associations with CpG sites reached epigenome-wide significance (i.e. Bonferroni adjusted P < 0.05 or false discovery rate < 5%) (Supplementary Fig. 1).The CpGs with lowest p-values in meta-analysis (P < 1 × 10–5) are shown in Table 3. Six of eight CpGs were located near transcription start sites on different chromosomes: in the LRRC2 gene on chromosome 3, in the ATP6V1B2 gene on chromosome 8, in the CKAP4, GALNTR6, and UNC1198 genes on chromosome 12, in the C13orf18 gene on chromosome 13, in the MBD2 gene on chromosome 18, and in the NTSR1 gene on chromosome 20. The average difference in DNA methylation between left-handers and right-handers at these CpGs was small (from 0.06 to 0.8%; i.e. 0.0006 to 0.008 on the methylation beta-value scale) with lower methylation level in left-handers at all CpGs except for cg13719901 (LRRC2) (Supplementary Figs. 2 and 3).
Table 3.
Top differentially methylated positions from EWAS meta-analysis of left-handedness.
| CpG | CHR | Positiona | Gene | Location | β | SE | P | FDR | Direction |
|---|---|---|---|---|---|---|---|---|---|
| cg22804475 | 8 | 20,054,597 | ATP6V1B2 | TSS200 | − 0.0007 | 0.0002 | 1.28 × 10–06 | 0.197 | – |
| cg09239756 | 12 | 106,642,360 | CKAP4 | TSS1500 | − 0.0032 | 0.0007 | 1.82 × 10–06 | 0.197 | – |
| cg22541911 | 12 | 51,785,465 | GALNT6 | TSS1500 | − 0.0010 | 0.0002 | 1.90 × 10–06 | 0.197 | – |
| cg13719901 | 3 | 46,608,139 | LRRC2 | 5′UTR; TSS200 | 0.0081 | 0.0017 | 2.60 × 10–06 | 0.197 | + + |
| cg02850812 | 13 | 46,961,666 | C13orf18 | TSS200 | − 0.0014 | 0.0003 | 2.62 × 10–06 | 0.197 | – |
| cg16852837 | 18 | 51,750,955 | MBD2 | 1st Exon; 5′UTR | − 0.0006 | 0.0001 | 3.28 × 10–06 | 0.205 | – |
| cg09893588 | 20 | 61,340,109 | NTSR1 | TSS200 | − 0.0011 | 0.0003 | 9.12 × 10–06 | 0.256 | – |
| cg12402132 | 12 | 121,148,554 | UNC119B | Gene body | − 0.0009 | 0.0002 | 9.54 × 10–06 | 0.256 | – |
β is the regression coefficient for left-handedness in EWAS meta-analysis adjusted model that included NNTR adults = 2663 and NALSPAC adults = 1058.
CpGs with uncorrected P-value < 1.0 × 10–5 are presented.
CHR chromosome; SE standard error; FDR false discovery rate; TSS200 200 base pairs upstream of transcription start site;TSS1500 1500 base pairs upstream of transcription start site; 5′UTR 5′ untranslated region; + +, positive direction of effect in each cohort; –, negative direction of effect in each cohort.
aGenome build Hg19 (build 37). See additional information on CpGs in Supplementary Table 12.
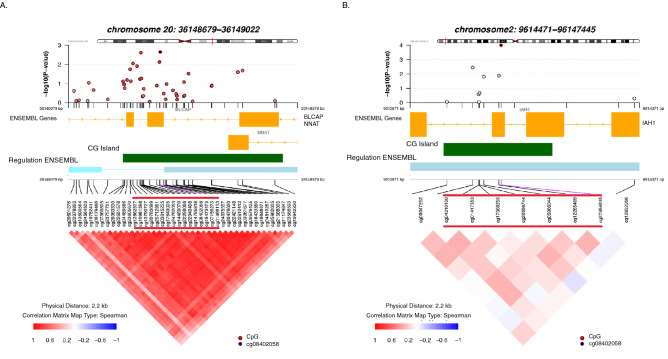
The DMR meta-analysis detected two DMRs associated with left-handedness (Fig. 2, Table 4, Supplementary Table 12). The DMR on chromosome 20 (BLCAP; NNAT; 16 CpGs) had lower DNA methylation in left-handers than in right-handers (P-value adjusted for multiple testing (Padj) = 0.00004). The DMR on chromosome 2 (IAH1; 7 CpGs) also had lower DNA methylation in left-handers (Padj = 0.03). In total, 15 of 16 CpGs in the DMR on chromosome 20 (Supplementary Figs. 4 and 5) and 6 of 7 CpGs in the DMR on chromosome 2 (Supplementary Figs. 6 and 7) were hypomethylated in left-handers. The average absolute DNA methylation difference at these regions between left-handers and right-handers based on meta-analysis regression coefficients for the individual CpGs was 0.4% for the DMR on chromosome 20, and 0.1% for the DMR on chromosome 2. Both DMRs were within CpG islands, and were not detected in the individual cohorts. Some CpG sites within left-handedness-associated DMRs have been previously associated with other traits, these associations are listed in Supplementary Table 13.
Figure 2.
Differentially methylated regions associated with left-handedness in meta-analysis. The figure represents the differentially methylated regions at P-adjusted < 0.05 in meta-analysis of DMR statistics across groups of NTR adults and ALSPAC adults (N = 3712). The top panel of each plot depicts the EWAS P-values for CpGs in the differentially methylated regions (A) at chromosome 20; and (B) at chromosome 2. The x-axis indicates the position in base pair (bp) for the region, while y-axis indicates the strength of association from meta-analysis EWAS with left-handedness (adjusted model). The middle panel shows the genomic coordinates (genome build GRCh37/hg19) and the functional annotation of the region: the ENSEMBL Genes track shows the genes in the genomic region (orange); the CpG Island track shows the location of CpG islands (green); the Regulation ENSEMBL track shows regulatory regions (blue). CpGs from DMR associated with left-handedness are indicated with red lines above the correlation heatmap. More detailed information on the regions is provided in Table 4. The bottom panel shows the Spearman correlation between methylation levels of CpGs in the window. The figure was computed using the R package coMET. See additional information on CpGs from the regions in Supplementary Table 12.
Table 4.
Significant differentially methylated regions associated with left-handedness in meta-analysis and secondary analysis.
| Cohort | Chromosome | Start | End | n CpGs | Genes | Effect size | SE | P | Padjust |
|---|---|---|---|---|---|---|---|---|---|
| Meta-analysis | |||||||||
| NTR and ALSPAC adults (blood DNA methylation) | chr20 | 36,148,679 | 36,149,022 | 16 | BLCAP, NNAT | − 0.153 | 0.024 | 9.80 × 10–11 | 4.31 × 10–05 |
| chr2 | 9,614,471 | 9,614,744 | 7 | IAH1 | − 0.102 | 0.019 | 7.33 × 10–08 | 0.03 | |
| Secondary analysis | |||||||||
| NTR children at 9 years (buccal cell DNA methylation) | chr8 | 145,024,929 | 145,025,064 | 4 | PLEC1 | − 0.056 | 0.008 | 1.07 × 10–11 | 9.14 × 10–06 |
| chr22 | 36,011,405 | 36,011,843 | 2 | MB | − 0.119 | 0.022 | 4.09 × 10–08 | 0.035 | |
| chr9 | 111,696,674 | 111,697,545 | 10 | EELP1, ABITRAM | − 0.134 | 0.024 | 4.59 × 10–08 | 0.039 | |
| chr12 | 899,323 | 899,559 | 2 | WNK1 | − 0.117 | 0.021 | 4.69 × 10–08 | 0.040 | |
Effect size is a weighted sum of the EWAS effects for each CpG site in the DMR (i.e. methylation differences between LH and RH) where the weights account for dependence between CpG sites and uncertainty in the EWAS effects (see “Methods” section). Nmeta-analysis = 3721, NNTR children = 866.
NTR Netherlands Twin Register. ALSPAC Avon Longitudinal Study of Parents and Children. SE standard error; Padjust P-value multiplied by the total number of tests performed; the number of tests is equal to the number of regions for which DMR statistics are calculated.
GWAS follow-up
We tested the overlap of our EWAS meta-analysis results with findings from the most recent GWAS meta-analysis of handedness13. CpGs located within 1 Mb window of SNPs associated with left-handedness (at P < 5 × 10–8) were on average more strongly associated with left-handedness in the EWAS meta-analysis than the other tested CpGs (β = 0.027, P = 0.04). The effect was weaker when less stringent GWAS p-value cut-offs were applied (i.e. SNPs with P < 1 × 10–6, and SNPs with P < 1 × 10–5). Importantly, in a control analysis substituting genetic loci with loci associated with type 2 diabetes43, we did not observe a statistically significant overlap (β = 0.005, P = 0.265) (see Supplementary Table 14, Supplementary Fig. 8).
A look-up of left-handedness associated SNPs13 in the methylation quantitative trait locus (mQTL) database by the Genetics of DNA Methylation Consortium (GoDMC)44 showed that 95% of left-handedness associated SNPs associated with methylation levels of nearby (cis; 86%) or distant (trans; 14%) CpGs (254 unique CpGs). We repeated the GWAS enrichment analysis with these CpGs driven by mQTLs removed, and obtained similar results, i.e. CpGs near GWAS loci (but not driven by significant mQTLs) were still more strongly associated with left-handedness compared to other genome-wide CpGs (β = 0.027, P = 0.027). None of the CpGs driven by mQTL was located in significant DMRs from our EWAS meta-analysis, or among the top 100 CpGs (by p-value) from our EWAS meta-analysis, which illustrates that our top EWAS findings are not driven by mQTL effects of the top GWAS loci.
Longitudinal analysis
While handedness is a stable trait, DNA methylation can vary over age45. We analyzed DNA methylation in ALSPAC offspring measured in cord blood at birth, and in peripheral blood at 7, 17, and 24 years old (N = 791, Table 2 and Supplementary Table 3) to examine the association between DNA methylation and left-handedness throughout childhood and adolescence. No associations survived adjustment for multiple tests at any time point (Bonferroni adjusted P < 0.05; Supplementary Fig. 9e–l, Supplementary Tables 19–26). The correlations of effects for the top 100 CpG by P-value between time points were moderate to strong (mean correlation = 0.414; correlation range from r = 0.355; P = 0.0002 to r = 0.578, P = 1.2 × 10–10), except for a weak correlation between top effects at 17 years and 24 years (rALSPAC17–ALSPAC24 = 0.079; P = 0.435) (Supplementary Fig. 10). There were no overlapping CpGs amongst the top 100 CpGs between analyses at different time points (Supplementary Fig. 11). Correlations between top CpG effects between ALSPAC adults (mothers and fathers) and offspring at birth were strong negative (rALSPACadults–ALSPACatbirth = − 0.68; P = 7.2 × 10–15) (Supplementary Fig. 12), and between ALSPAC adults and offspring at 7, 17, 24 years were weak (r from − 0.006 to 0.141, P > 0.0003) (Supplementary Fig. 10).
DNA methylation in buccal cells
In NTR, buccal DNA methylation data (measured with the EPIC array at 787,711 CpG sites) were available in children (N = 946, mean age 9.5, SD = 1.85). The EWAS did not detect associations of DMPs with left-handedness (Supplementary Fig. 9m, n, Supplementary Table 27–28). The effects for top 100 CpG in EWAS of handedness in buccal cells had weak correlations with effects in blood in the meta-analysis (from r = 0.086, P = 0.39 to r = 0.179, P = 0.07), NTR adults (from r = 0.193, P = 0.05 to r = 0.268, P = 0.007), ALSPAC adults (from r = − 0.008, P = 0.94 to r = − 0.04, P = 0.95), and ALSPAC offspring at different ages (from r = − 0.384, P = 7.9 × 10–05 to r = 0.312; P = 0.002). Four DMRs in buccal cell DNA associated with left-handedness: DNA methylation was lower in left-handers at a DMR on chromosome 8 (4 CpGs; Padj = 9.14 × 10–06, average difference in DNA methylation in left-handers and right-handers 0.07%), a DMR on chromosome 9 (10 CpGs; Padj = 0.039, average difference 0.3%), a DMR on chromosome 12 (2 CpGs; Padj = 0.04, average difference 0.98%), and a DMR on chromosome 22 (2 CpGs; Padj = 0.035, average difference 1.1%) (Table 4, Supplementary Fig. 13). These DMRs did not overlap with DMRs detected in the analyses of blood methylation data. Sixteen of 18 CpGs from these regions had a lower methylation level in left-handed children than in right-handed (Supplementary Table 29).
Sensitivity analyses
We reported the DNA methylation and left-handedness association study with adjustment for prenatal and postnatal factors that influence DNA methylation as shown in previous studies: BMI38 and smoking46 in adults, and gestational age, birth weight and prenatal maternal smoking in children47,48. We examined if the EWAS results for handedness differ without taking these factors into account. Across all analyses, the correlations between the effects for the top 100 CpGs were strong between the models with and without adjustment for these factors (r ranged from 0.99 to 1), and overlaps of the top 100 CpGs were substantial (32–87 CpGs). Adjustment for the factors increased the number of DMRs associated with left-handedness in meta-analysis (1 DMR without adjustment and 2 DMRs with adjustment), and in EWAS in children (2 DMRs without adjustment, and 4 with adjustment in buccal cells in NTR).
Discordant MZ twins
In NTR, the DNA methylation datasets included 1039 monozygotic (MZ) adult twins with blood samples (from the meta-analysis in NTR adults) and 794 MZ children with buccal samples (from NTR children in secondary analysis) from complete twin pairs with handedness data. We found that 21% of the MZ twin adult pairs (N = 133 pairs) and 24% of the MZ twin child pairs (N = 86 pairs) were discordant for handedness. Characteristics of MZ discordant twins are presented in Supplementary Table 4. In both groups, we performed an MZ discordant within-pair EWAS analysis, comparing left- and right-handed twins. Within-pair analyses of DNA methylation of left- and right-handed twins did not identify DMPs or DMRs in blood or buccal samples at Bonferroni or FDR threshold (Supplementary Fig. 9a–d, Supplementary Tables 15–18). We compared the methylation results obtained in discordant MZ twins to the EWAS meta-analysis results for the top 100 CpGs ranked on ascending P-value from each analysis. To avoid sample overlap, we repeated the EWAS meta-analysis after exclusion of the MZ discordant twin pairs. Correlations between the meta-analysis top 100 effects and mean methylation differences from the within-pair analysis were weak in adults (rMZ disc adults blood–Meta-analysis = 0.189, P = 0.06; rMeta-analysis–MZ disc adults blood = 0.188, P = 0.06, α = 0.0003) and children (rMZ disc children buccal–Meta-analysis = 0.134, P = 0.18; rMeta-analysis–MZ disc children buccal = 0.252, P = 0.01) (Supplementary Fig. 10). There were few overlapping CpGs among the top 100 CpGs from the within-pair analyses and other analyses (Supplementary Fig. 11).
Handedness methylation scores
To examine if variation in handedness can be predicted by DNA methylation levels across multiple CpGs, methylation scores (MS) were created. These were based on EWAS summary statistics in NTR to predict into ALSPAC, and on ALSPAC summary statistics to predict into NTR given the following p-value thresholds to include CpGs: P < 1 × 10–1, P < 1 × 10–3, P < 1 × 10–5. To estimate the variance explained by MS above genetic variants, polygenic scores (PGS) were created based on the summary statistics from the handedness GWAS of Cuellar-Partida et al.13 with exclusion of NTR, ALSPAC and 23andMe cohorts (NGWAS = 196,419). Since four scores were tested (3 methylation scores, one polygenic score), we applied Bonferroni correction for four tests (α = 0.05/4 = 0.0125) (Supplementary Table 30, Supplementary Fig. 14). The results are summarized in Supplementary Table 31. MS did not predict left-handedness in NTR and ALSPAC adults, or in children at 7–9 years old and did not explain variance over and above the variance explained by the PGS in the combined model (R2MS from -0.17 to 1.28%, R2PGS from 0.002 to 0.46%). The largest amount of explained variance was in ALSPAC at 7 years old for the MS based on CpGs at P < 1 × 10–5 (R2MS = 1.28%, P = 0.1, NCpGs = 7).
Discussion
We have performed an epigenome-wide association study of left-handedness, including left- and right-handed individuals from two population-based cohorts from the Netherlands and the UK. In the meta-analysis, combining the NTR and ALSPAC adult cohorts, two DMRs associated with left-handedness. The first DMR (genomic location: chr20q11.23, 36,148,679:36,149,022) is located within the 5’UTR of the BLCAP apoptosis inducing factor (BLCAP) gene and nearby the transcription start site (TSS1500) of the neuronatin (NNAT) gene. BLCAP encodes a protein that reduces cell growth by stimulating apoptosis. NNAT is located within intron of BLCAP and is involved in brain development and nervous system structure maturation and maintenance. BLCAP and NNAT are imprinted in the brain49. The second intron of NNAT regulates the expression of BLCAP transcripts acting as an imprint control region regulating also allele-specific DNA methylation and histone modifications50. Around 50% of CpGs in a region covering the NNAT CpG islands are methylated in brain and other tissues, suggestive of differential allele-specific methylation51. The imprinted DMR for these genes [chr20:36,139,941-36,159,19052] overlaps with the left-handedness DMR that we identified (chr20:36,148,679-36,149,022). A potential connection of genomic imprinting with handedness was previously suggested in a study of another imprinted gene LRRTM141. CpGs from the handedness-associated region at chromosome 20 previously associated with myalgic encephalomyelitis and chronic fatigue syndrome, preterm birth, obesity, metabolic parameters, and arm fat mass (DXA scan measurement).
The second DMR (genomic location: chr2p25.1, 9,614,471:9,614,744) is located within the isoamyl acetate hydrolyzing esterase 1 (IAH1) gene. The IAH1 gene encodes an acyl esterase and is associated with neonatal inflammatory skin and bowel disease, and a disease with an inborn error of leucine metabolism (3 methylglutaconic aciduria type 1). CpGs from the region previously associated with gestational age, bone mineral density, metabolic parameters, and schizophrenia. Some of these traits have been reported to be associated with handedness in epidemiological studies, e.g. BMI53 and gestational age54, for which we adjusted in our analyses. Previous analysis of the genetic correlations between left-handedness and 1349 complex traits using LD-score regression did not reveal any genetic correlations at FDR < 5%, but suggestive positive correlations were observed with neurological and psychiatric traits, including schizophrenia13.
Even though no DMPs were identified after correction for multiple testing, and effect sizes of top CpGs were small (mean differences between left- and right-handed individuals smaller than 1%), the high-ranking CpGs are of potential interest. The second-ranking CpG cg09239756 (genomic location: chr12, 106,642,360) is located near the cytoskeleton associated protein 4 (CKAP4) gene. This gene mediates the anchoring of the endoplasmic reticulum to microtubules. Microtubules are an important cytoskeleton component that play a role in neuronal morphogenesis and migration, and axon transport31. Microtubules have been widely discussed in association with handedness15,29 and brain anatomical asymmetry32, and genes involved in microtubule pathways were enriched in the GWAS of handedness13. Moreover, in our enrichment analysis, we found that CpGs located within a 1 Mb window from SNPs associated with left-handedness in the GWAS meta-analysis by Cuellar-Partida et al.13 were more strongly associated with left-handedness in our meta-analysis compared to CpGs outside of this window. Larger EWAS meta-analysis or replication in additional independent cohorts is necessary to establish the robustness of the top DMPs.
Hand movements together with other lateralized movements and molecular signs of lateralization are observed at very early stages of human development in the uterus6–8. Therefore, DNA methylation differences associated with hand preference are expected to emerge early in development. While DNA methylation at some CpGs in the genome changes throughout the lifespan45, the DNA methylation signal associated with left-handedness was moderately consistent from birth throughout the lifespan: DMP effects correlated in ALSPAC offspring from birth to 24 years old, although genome-wide significance for DMPs was not reached. Consistency in DNA methylation signal associated with left-handedness at different time-points may indicate that the pattern for left-handedness is conserved through the lifespan.
Several DMRs were detected in buccal cells in children around 9 years old (genomic locations: chromosomes 8, 22, 9, and 12) after correction for multiple testing. Annotation of these regions implicate the following protein coding genes: the plectin gene (PLEC1), the myoglobin gene (MB) gene, the elongator complex protein 1 gene (ELP1), the actin binding transcription modulator gene (ABITRAM), and the WNK lysine deficient protein kinase 1 gene (WNK1). The CpGs in these regions are mostly hypomethylated in left-handed individuals. The genes encode for proteins that participate in cytoskeleton functions, chromatin organization, development of neurons, and metabolism. CpGs from DMRs in buccal cells previously associated with other phenotypes: CpGs from the DMR on chromosomes 8 with myalgic encephalomyelitis, chronic fatigue syndrome, multiple sclerosis, and gestational age; CpGs from the DMR on chromosome 9 with bone mineral density, tissue mass of the arm (DXA scans measurement), and multiple metabolic parameters. Interestingly, some of these phenotypes also associated with CpGs from our meta-analysis in blood DNA methylation.
A difference in handedness preference in MZ twin pairs has always fascinated parents of twins and twins themselves: how can children with almost identical genes differ for such a prominent trait? Handedness discordance in identical twins was described a long time ago17,18, and the percentage of MZ discordant twins were reported as 20% of 3486 MZ twins in East Flanders19, and 19% of 1724 MZ twins from a London twin study28. We observed that 21% of adult MZ twins and 24% of young MZ twins were discordant for handedness in our study, but we did not detect DNA methylation differences among them in blood or buccal cells. This null finding could mean that handedness discordance is not associated with methylation differences in the tissues that we studied (but might be present in other tissues), or that our analysis was underpowered to detect methylation differences associated with handedness discordance. Our discordant MZ twin analysis may be underpowered to detect small DNA methylation differences55, as it included only 133 MZ discordant adult twin pairs and 86 child twin pairs. Different, not mutually exclusive, hypotheses have been proposed for handedness discordance of MZ twins, including large unique environmental effects, that are deduced based on the low heritability of around 25% as estimated in the meta-analysis by Medland et al.10. McManus16 emphasizes that such unique environmental factors may represent what in biology is referred to as noise, randomness or fluctuating asymmetry. McManus quotes Mitchell56 saying that such processes are “caused not by any factors outside the organism, but by inherent variation in the processes of development”. These reflect developmental variance unique to the person. In a discussion and review of the human and animal studies literature Molenaar et al.57 called these processes ‘the third source of individual differences’, i.e. a third source besides genetic and environmental influences on individual differences and discuss how deterministic growth process may give rise to highly variable results.
There is a growing interest to improve the prediction of traits with use of other omics data than SNPs, like DNA methylation37 and by non-genetic early life factors (e.g. earlier factors associated with left-handedness including birth weight, being multiple, month of birth, breastfeeding etc.58). Given the low heritability of handedness (~ 25%10,11), it is expected that non-genetic factors play a role. Although together early life factors had minimal predictive value58, they may inspire the search for DNA methylation signatures, as DNA methylation signatures later in life were found for birthweight47. Single CpGs did not individually reach statistical significance in our EWAS, but combining information across multiple CpGs into an overall methylation score can be a more powerful approach to capture variation in handedness. We calculated methylation scores as weighted sums of the individual’s methylation loci beta values of a pre-selected number of CpG sites. However, the predictive value of polygenic and methylation scores for handedness was low, which likely reflects that current GWAS and EWAS analyses for handedness are still underpowered.
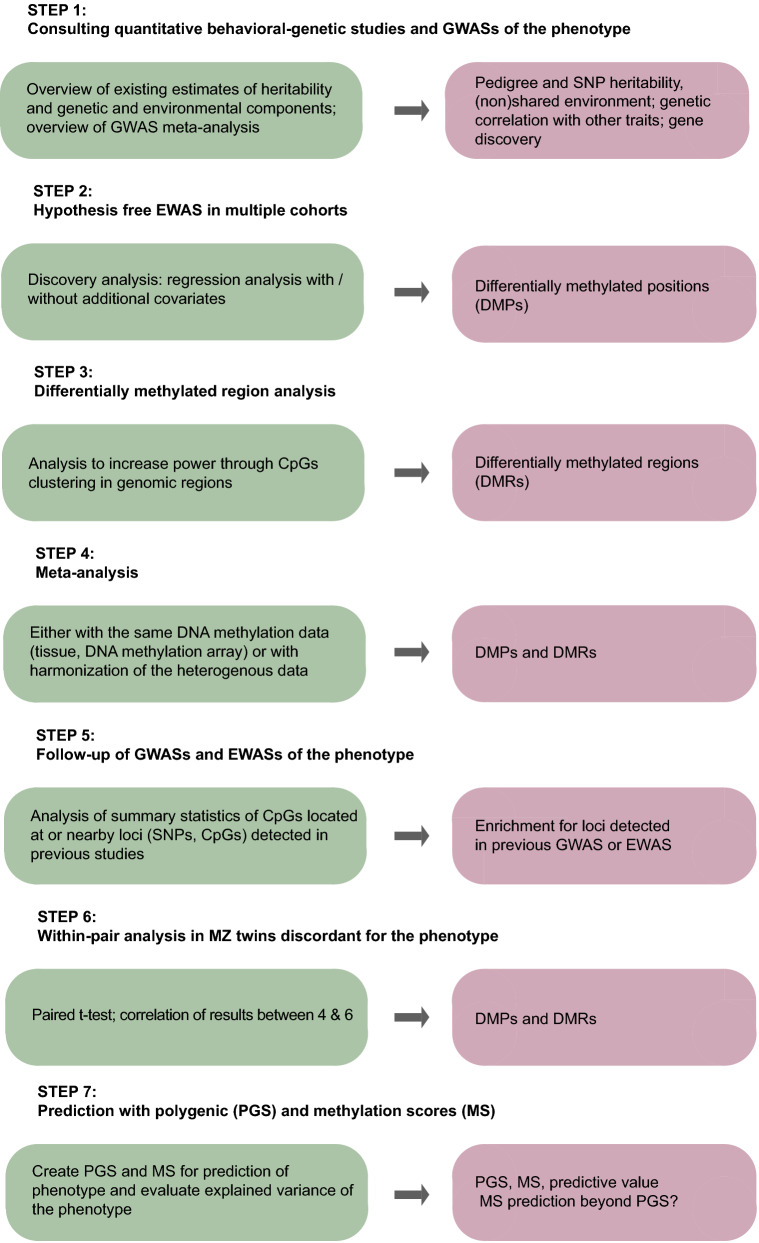
Our multi-cohort epigenome-wide association study can be summarized in several key steps presented in Fig. 3. We examined DNA methylation data in different tissues (whole blood, cord blood, buccal cells) and ages (from birth to adulthood). The limitations of the study are related to handedness measurements, available tissues, differences in platforms used for DNA methylation (Illumina 450 k, EPIC), and study power. There were slight differences in the assessment of left-handedness between NTR and ALSPAC. Power might increase by investigating a refinement of the handedness phenotype (e.g. hand skill measurement rather than self-report of the preferred hand), analysis of DNA methylation in more relevant tissues, and an increase in sample size. The difference in left-handedness rates among children born before and after 1960 may be due to a move away from being forced to use the right hand prior to 196059. We accounted for this trend by including age (which correlates almost perfectly with birth year in these samples) as a covariate in the analyses, however, it should be noted that the forced use of the right hand in older generations may render the phenotype definition of handedness less precise. Our meta-analysis was based on whole blood methylation data, where the methylation level represents the overall level of DNA obtained from millions of white blood cells. We observed methylation differences between left- and right-handed individuals of up to 0.8% at top-DMPs. This small effect size could reflect that a methylation difference is present in only a sub-set of cells in an individual, or a sub-set of individuals in the population, or a combination. The biological implications of these findings remain to be established and our top-DMPs remain to be replicated in additional cohorts or larger meta-analysis. The primary tissues of interest for handedness are brain2,29, spinal cord40, and arm muscle tissues4, and the timing when these tissues are collected could also play a role, but the collections of these tissues are not widely available in population cohorts for obvious reasons. Although 4000 is considered a decent sample size for EWAS and smaller sample sizes have allowed for the successful detection of many loci where DNA methylation robustly associates with traits such as body mass index38, the effect sizes for handedness were unknown a priori and 4000 may still be too small to detect methylation differences associated with left-handedness. Similarly, sample sizes of EWASs of behavioral and psychiatric traits are now increasing beyond 10,00060. For GWAS of handedness that applied similar phenotyping as the current study, the increase of the sample size from about 400,000 to 1.7 million increased the number of associated loci from a handful to over forty13, and similar increases in the number of detected loci may occur when EWAS sample sizes for left-handedness increase. Finally, our study focused on DNA methylation, but other epigenetic processes could play a role in handedness such as histone modifications, post-translational regulation by miRNAs and X-chromosome inactivation that remain to be explored.
Figure 3.
Seven-step approach to DNA methylation signature discovery, incorporating twin design. The figure represents the methodology of DNA methylation signature discovery study of a phenotype in multiple steps. It integrates behavioral-genetic and SNP-based methods (step 1) to estimate heritability, epigenome-wide study methods (steps 3–4) for association analyses, follow-up of results using summary statistics from previous EWASs and GWASs (step 5), the discordant twin design (step 6), and methods integrating polygenic and DNA methylation data (step 7) in enrichment analysis. Specific methods for each step are presented on the left and outcomes on the right. GWAS, genome-wide association study. EWAS, epigenome-wide association study. SNP, single nucleotide polymorphisms. CpG, cytosine-phosphate-guanine. PGS, polygenic scores. MS, methylation scores. DMPs, differentially methylated positions. DMRs, differentially methylated regions.
We reported an EWAS of left-handedness in two large population-based cohorts with data from children and adults, and examined performance of methylation scores and polygenic scores. Despite the plausible rationale of multiple genetic and non-genetic factors that may act via epigenetic pathways to influence the development of handedness, we did not uncover support for the hypothesis that DNA methylation in peripheral tissues captures much if any of the variation in handedness. We propose that future studies consider other tissues, such as related to central nervous system.
Methods
Overview
The primary epigenome-wide association study (EWAS) of left-handedness was performed in two cohorts with DNA methylation data in whole blood (Illumina, 450 k): NTR adults61 (N = 2682 individuals including twins, mean age at methylation 36.5, SD 12.7), and ALSPAC adults62,63 (N = 1232, mean age at methylation 48.98, SD 5.55). EWAS analyses were performed in each dataset separately, and summary statistics were combined in the meta-analysis (N = 3914) testing 409,563 CpGs. As this is a meta-analysis of existing DNA methylation datasets, no analyses were done to pre-determine sample sizes, but the sample size is larger compared to previously published DNA methylation studies of handedness41,42. We tested whether the EWAS signal was enriched in nearby loci detected in the previous GWAS on handedness13. Secondary analyses were performed in different tissues: in cord blood and peripheral blood in ALSPAC offspring64, i.e. the children of ALSPAC participants that contributed to the primary EWAS (N = 791 with DNA methylation data at birth, at 7, and 17 years old (Illumina 450 k chip), and/or at 24 years old (Illumina EPIC array)), and in buccal cells from an independent group of children from the NTR65,66 (N = 946 twins, mean age 9.5, SD 1.85, Illumina EPIC array). The number with DNA methylation and covariate data in ALSPAC differed at different time points from 442 to 759. We carried out within-pair twin analysis in NTR MZ twins discordant for handedness (Nadults = 133 twin pairs, Nchildren = 86 twin pairs). We performed EWAS analyses in each dataset and examined correlations between the regression coefficients of top CpGs (ranked on ascending p-value) from each EWAS analysis. Finally, we created and tested polygenic and DNA methylation scores for left-handedness. The study method and design are presented in Figs. 1 and 3. Detailed cohort information is provided in Appendix 1.
Handedness
NTR
Information on hand preference for adults and children was collected by surveys and in small subgroups from laboratory-based projects. Parental reports on children were collected at 5 years and included seven items for different activities, from which the item “What hand does child use for drawing?” was selected. The four answer categories were left-handed, right-handed, both hands and do not know. Multiple adult surveys included the question: “Are you right-handed or left-handed?” (4 surveys) or “Are you predominantly left-handed or right-handed?” (3 surveys). The three answer categories were left-handed, right-handed, and both. For a small number of adult self-reports at younger ages (14, 16 or 18 years) or parental assessment at age 5 were also available.
ALSPAC
Adults (mothers and fathers) were asked which hand they used to write, draw, throw, hold a racket or bat, brush their teeth, cut with a knife, hammer a nail, strike a match, rub out a mark, deal from a pack of cards or thread a needle (11 questions). Child handedness was assessed at 42 months by questionnaire in which the mother was asked which hand the child used to draw, throw a ball, color, hold a toothbrush, cut with a knife, and hit things (6 questions). Responses were scored − 1, 0 or 1 for left, either or right, respectively. Handedness was coded as 1 for left-handed or 0 for right-handed in both cohorts.
DNA methylation and genotyping
DNA methylation was measured with the Infinium HumanMethylation450 BeadChip Kit which measures more than 450,000 methylation sites (primary analysis in adults in NTR and ALSPAC and secondary analysis in ALSPAC offspring at birth, 7 and 17 years old), or the Infinium MethylationEPIC BeadChip which measures more than 850,000 methylation sites (secondary analysis in ALSPAC offspring at 24 years old and NTR children). Genotyping for polygenic risk scores was done on multiple platforms with imputation of the target data using reference haplotypes from 1000 genomes reference panel. Cohort-specific details on biosample collection, DNA methylation profiling, quality control, cell-type proportions measurements and genotyping are described in Appendix 1.
Intergroup differences
We tested if there were differences in characteristics that were included in EWAS models (such as age at biological sample collection, sex, body mass index (BMI), smoking status at blood collection for adults, and gestational age, maternal smoking during pregnancy, birth weight for children, cell proportions/percentages in buccal swabs and in blood samples) between left- and right-handed individuals by generalized estimating equations (GEE) to accommodate the relatedness among the twins in NTR, and by standard logistic regression in ALSPAC. The R package ‘gee’ was used with the following specifications: binomial (for ordinal data) link function, 100 iterations, and the ‘exchangeable’ option to account for the correlation structure within families and within persons. Right- and lefthanded MZ discordant twins were compared with paired t-test for the traits that were not identical in twins (birth weight, BMI, smoking, cell percentages). All statistical tests here and below were two-tailed.
Epigenome-wide association analyses
Primary analyses
The association between DNA methylation levels and left-handedness was tested for each site under a linear model (ALSPAC) or generalized estimating equation (GEE) model accounting for relatedness of twins (NTR). DNA methylation beta-values were the dependent variable and were typically normally distributed. The following predictors were included in the basic model: handedness (coded as 0 = right-handed and 1 = left-handed), sex, age at blood sampling, percentage of blood cells for blood samples, and technical covariates in NTR and ALSPAC (see Appendix 1). An adjusted model was fitted to account for BMI and smoking status at blood draw in both NTR and ALSPAC adult cohorts, because BMI and smoking have large effects on DNA methylation in adults38,46. The primary results reported in the paper are based on the fully adjusted model. The models are described in Appendix 2. Throughout the text, we refer to regression coefficients from the EWAS, which represent the methylation difference between left-handed and right-handed individuals on the methylation beta-value scale. A positive regression coefficient (β) means a higher methylation level in left-handed individuals. The value of an individual on the methylation scale is commonly also symbolized as beta (β) and ranges from 0 to 1, where 0 represents a methylation level of 0% and 1 represents a methylation level of 100%.
Secondary analyses
The same basic models were fitted to the data from ALSPAC and NTR children. For DNA methylation in buccal cells, cell proportions (epithelial cells, natural killer cells) for buccal samples were included instead of percentage of blood cells. As several characteristics, such as gestational age and birthweight, affect DNA methylation48,67, we included these in the adjusted model in children (see Appendix 2).
In the within-pair analysis of discordant MZ twins, paired t-tests were employed to test for methylation differences between the left-handed and the right-handed twins. Paired t-tests were performed in R on residual methylation levels, which were obtained by adjusting the DNA methylation β-values for sample plate, array row, cell proportions in buccal samples in children and sample plate, array row, and percentages of blood cells in adults. Additional covariates, birth weight in children and BMI and smoking status in adults, were added in adjusted model. Age, sex, maternal smoking, and gestational age were not included because these variables are identical in MZ twins.
To account for multiple testing, we considered Bonferroni correction and a False Discovery Rate (FDR) of 5%. The Bonferroni corrected p-value threshold was calculated by dividing 0.05 by the number of genome-wide CpGs tested, and false discovery rate (FDR) q-values were computed with the R package ‘qvalue’ with default settings. The Bayesian inflation factor (λ) was calculated with the R package Bacon68 (see Supplementary Table 5).
Meta-analysis
A meta-analysis was performed in METAL69 based on estimates (regression coefficients) and standard errors from the EWAS of handedness performed with GEE in NTR and linear regression in ALSPAC. NTR and ALSPAC adult cohorts were combined. In total, 409,563 CpG sites present in both cohorts were tested with statistical significance evaluated after Bonferroni correction and at an FDR q-value < 0.05.
Comparison of top CpGs from different analyses
To compare top CpGs from different analyses, we repeated the NTR EWAS analyses in adults in children and meta-analysis with discordant MZ twin pairs removed to avoid sample overlap. We selected methylation sites that overlapped in 13 analyses with adjusted model (meta-analysis, meta-analysis without discordant MZ twins, EWAS NTR adults, EWAS NTR adults without discordant MZ twins, EWAS ALSPAC adults, EWASs ALSPAC at birth, 7, 17, 24 years, EWAS NTR children, EWAS NTR children without discordant twins, and within-pair analyses of discordant MZ twin adults and children) that resulted in 379,924 methylation sites. We calculated Pearson correlations for effect estimates of the top 100 CpGs ranked by p-value from one analysis with the effect estimates of the same CpGs in other analyses. Statistical significance of correlations was assessed after Bonferroni correction for the number of correlations tested: α = 0.05/(13 × 13 − 13) = 0.0003.
Differentially methylated regions
We used the R dmrff library36 for R to identify regions where CpG sites showed evidence for association with handedness. Dmrff identifies DMRs by meta-analysing EWAS summary statistics from CpG sites in each region while adjusting for dependencies between the sites and uncertainty in the EWAS effects (https://github.com/perishky/dmrff). In this study, dmrff was applied separately in the NTR and ALSPAC cohorts, and then used to identify DMRs in common between the cohorts by meta-analysis. As previously described36, DMR meta-analysis preceded by first identifying candidate DMRs using the EWAS meta-analysis summary statistics, calculating DMR statistics for these candidates in each cohort separately, and then meta-analysing the DMR statistics across the two cohorts. The DMR effect size is a weighted sum of the EWAS effects for each CpG site (i.e. methylation differences between LH and RH). All dmrff p-values were adjusted for multiple tests (Bonferroni adjustment) by multiplying them by the total number of DMRs considered. We report significant regions (Padj < 0.05) including at least two CpG sites within a 500 bp window observed to be nominally associated with handedness by EWAS (P < 0.05). The average absolute DNA methylation difference in the region between left-handers and right-handers is calculated as the sum of absolute regression coefficients of each CpG in the region divided by the number of CpGs. We plotted the DMRs with the coMET R Bioconductor package70 to graphically display additional information on physical location of CpGs, correlation between sites, statistical significance, and functional annotation (annotation tracks included genes Ensembl, CpG islands (UCSC), regulation Ensembl).
GWAS follow-up
GWAS follow-up analyses were performed to examine whether CpGs within a 1 Mb window of loci detected by the GWAS for left-handedness13, on average, showed a stronger association with left-handedness than other genome-wide methylation sites (Infinium HumanMethylation450 BeadChip). We obtained a SNP list based on the GWAS meta-analysis without NTR, ALSPAC, and 23andMe by Cuellar-Partida et al.13 (196,419 individuals, NSNPs = 13,550,404), from which we selected all SNPs with a P-value < 1.0 × 10–08, < 1.0 × 10–06, and < 1.0 × 10–05, and determined the distance of each Illumina 450 k methylation site to each SNP. To test whether methylation sites near GWAS loci were more strongly associated with left-handedness, meta-analysis EWAS test statistics were regressed on a variable indicating if the CpG is located within a 1 Mb window from SNPs associated with handedness (1 = yes, 0 = no):
where |Zscore| represents the absolute Zscore for a CpG from the EWAS meta-analysis of handedness; βcategory x represents the estimate for category x, i.e. the change in the EWAS test statistic associated with a one-unit change in category x (e.g. being within 1 Mb of SNPs associated with left-handedness). For each enrichment test, bootstrap standard errors were computed with 2000 bootstraps with the R-package “simpleboot”. Statistical significance was assessed at α = 0.05. As control analysis, the same follow-up was performed using GWAS summary statistics on a trait that is unrelated to handedness type 2 diabetes in UK Biobank cohort (N = 244,890) 43. GWAS summary statistics were downloaded from GWASAtlas (https://atlas.ctglab.nl/traitDB/3686; 41204_E11_logistic.EUR.sumstats.MACfilt.txt; accessed on February 1 2021).
We looked up CpG sites associated with handedness-associated SNPs with a P-value < 1.0 × 10–08 (based on the GWAS meta-analysis by Cuellar-Partida et al.13 without 23andMe, NTR and ALSPAC, resulting in a list of 420 SNPs) using the mQTL database maintained by the Genetics of DNA Methylation Consortium (GoDMC, N = 27,750 European samples; http://mqtldb.godmc.org.uk/about)44. We then checked if associations with handedness were observed for these sites in our EWAS meta-analysis and DMR meta-analysis.
EWAS follow-up
To examine previously reported associations for epigenome-wide significant DMRs associated with left-handedness in our study, we looked up CpGs from the regions in the EWAS Atlas71 (https://bigd.big.ac.cn/ewas/tools; accessed on August 1 2020) and EWAS catalogue72 (http://www.ewascatalog.org; access on November 1 2020).
Polygenic and methylation scores
Polygenic scores (PGS) for handedness were calculated based on the GWAS meta-analysis without 23andMe by Cuellar-Partida et al.13. To avoid overlap between the discovery and target samples, summary statistics without NTR and ALSPAC were requested (196,419 individuals, NSNPs = 13,550,404). The linkage disequilibrium (LD) weighted betas were calculated using a LD pruning window of 250 KB, with the fraction of causal SNPs set at 0.50 by LDpred73. We randomly selected 2500 2nd degree unrelated individuals from each cohort as a reference population to calculate the LD patterns. The resulting betas were used to calculate the PGSs in each dataset using the PLINK 1.9 software. All PGSs were standardized (mean of 0 and standard deviation of 1). Methylation scores (MS) were calculated in NTR based on EWAS summary statistics obtained from ALSPAC, and vice versa, as previously done to create methylation scores for BMI and height74. We calculated same-tissue same-age DNA-methylation scores based on methylation data from NTR adults (blood) and ALSPAC parents (blood), and cross-tissue DNA-methylation scores based on data from NTR and ALSPAC offspring, with DNA methylation measured in buccal cells, and blood, respectively (see Fig. 1). For each individual, a weighted score sum was calculated for left-handedness by multiplying the methylation value at a given CpG by the effect size of the CpG (β), and then summing these values over all CpGs: DNA methylation score (i) = β1*CpG1i + β2*CpG2i··· + βn*CpGni, where CpGn is the methylation level at CpG site n in participant i, and βn is the regression coefficient at CpGn taken from summary statistics of the EWAS analysis. All methylation scores were standardized (mean of 0 and standard deviation of 1). We used weights from summary statistics of EWASs in four cohorts: NTR adults, ALSPAC adults, NTR children, ALSPAC offspring at 7 years old. Subsets of CpGs to be included in methylation scores were selected based on P-value < 1 × 10–1, < 1 × 10–3, and < 1 × 10–5. We analysed the predictive value of the left-handedness polygenic scores and methylation scores in NTR and ALSPAC adult and child cohorts from our EWAS study. To quantify the variance explained by the PGS and MS, we used the approach proposed by Lee et al.75, where coefficients of determination (R2) for binary responses are calculated on the liability scale. The equations of all models are provided in Appendix 2. Statistical significance was assessed following Bonferroni correction for the number of scores tested (PGS and 3 MSs). This resulted in α = 0.05/4 = 0.0125, nominal significance at 0.05.
Ethics statement
All methods were performed in accordance with the Declaration of Helsinki. For NTR, the study was approved by the Central Ethics Committee on Research Involving Human Subjects of the VU University Medical Centre, Amsterdam, an Institutional Review Board certified by the U.S. Office of Human Research Protections (IRB Number IRB00002991 under Federal-wide Assurance FWA00017598; IRB/institute codes, NTR 03-180). All subjects provided written informed consent. For children, written informed consent was given by their parents. For ALSPAC, ethical approval for the study was obtained from the ALSPAC Ethics and Law Committee and the Local Research Ethics Committees. All subjects provided written informed consent. For children, written informed consent was given by their mothers.
Supplementary Information
Acknowledgements
GRANT SUPPORT: Amsterdam Public Health Institute Methodology travel grant VVO; KNAW Academy Professor Award (PAH/6635) to DIB. JvD is supported by NWO Large Scale infrastructures, X-omics (184.034.019). The UK Medical Research Council and Wellcome (Grant Ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grantacknowledgements.pdf). This research was specifically funded by the BBSRC (grant numbers BBI025751/1 and BB/I025263/1). M.S., C.R. and D.C. are funded by the MRC (Grant Numbers MC_UU_00011/5 and MC_UU_00011/1). GWAS data was generated by Sample Logistics and Genotyping Facilities at Wellcome Sanger Institute and LabCorp (Laboratory Corporation of America) using support from 23andMe. The NTR received funding from the Netherlands Organization for Scientific Research (NWO): Biobanking and Biomolecular Research Infrastructure (BBMRI–NL, 184.021.007; 184.033.111) and Netherlands Twin Registry Repository NWO 480-15-001/674. This work was also supported by “Aggression in Children: Unraveling gene-environment interplay to inform Treatment and InterventiON strategies” project (ACTION). ACTION received funding from the European Union Seventh Framework Program (FP7/2007-2013) under Grant Agreement No. 602768. SEM is supported by NHMRC Investigator Grant APP1172917. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript we are extremely grateful to all the families who took part in this study, the midwives for their help in recruiting them, and the whole NTR and ALSPAC teams, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses.
Author contributions
J.V.D., D.I.B., C.R., M.S, and V.V.O made the study design. V.V.O, M.S., J.V.D wrote the main manuscript text. V.V.O, M.S., G.C.-P. performed meta-analysis. V.V.O, M.S., R.P., J.J.H, and C.D. performed secondary analyses. V.V.O. prepared Figs. 1, 2 and 3. V.V.O. and M.S. prepared all supplementary figures. F.H., CvB, L.L., G.W., and E.d.G. collected data. E.E., J.B., and BIOS Consortium performed laboratory analyses. All the authors reviewed the manuscript.
Data availability
The HumanMethylation450 BeadChip data from the NTR are available as part of the Biobank-based Integrative Omics Studies (BIOS) Consortium in the European Genome-phenome Archive (EGA), under the accession code EGAD00010000887 (https://ega-archive.org/datasets/EGAD00010000887). The Infinium MethylationEPIC from NTR are available from the Netherlands Twin Register on reasonable request (https://tweelingenregister.vu.nl/information_for_researchers/working-with-ntr-data). DNA methylation data from ALSPAC are available at ALSPAC and can be provided on request. The study website contains details of all the data that is available through a fully searchable data dictionary and variable search tool (http://www.bristol.ac.uk/alspac/researchers/our-data). The code used to perform the primary and secondary analyses is available at https://github.com/MRCIEU/handedness-ewas. The pipeline for the DNA methylation array analysis developed by the Biobank-based Integrative Omics Study (BIOS) consortium are available here: https://molepi.github.io/DNAmArray_workflow/. EWAS summary statistics for the top 100 CpGs are given in Supplemental Tables 6–11 and 15–29. The full EWAS summary statistics from the meta-analysis with basic and adjusted model are provided in Supplemental Tables 32 and 33. The full summary statistics for all other analyses are available upon request from the corresponding author.
Competing interests
GC-P contributed to this study while employed at The University of Queensland. He is now an employee of 23andMe Inc., and he may hold stock or stock options. All other authors report no conflicts of interest.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Veronika V. Odintsova and Matthew Suderman.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Veronika V. Odintsova, Email: v.v.odintsova@vu.nl
Jenny van Dongen, Email: j.van.dongen@vu.nl.
BIOS Consortium:
Bastiaan T. Heijmans, Peter A. C. ’t Hoen, Joyce van Meurs, Aaron Isaacs, Rick Jansen, Lude Franke, Dorret I. Boomsma, René Pool, Jenny van Dongen, Jouke J. Hottenga, Marleen M. J. van Greevenbroek, Coen D. A. Stehouwer, Carla J. H. van der Kallen, Casper G. Schalkwijk, Cisca Wijmenga, Lude Franke, Sasha Zhernakova, Ettje F. Tigchelaar, P. Eline Slagboom, Marian Beekman, Joris Deelen, Diana van Heemst, Jan H. Veldink, Leonard H. Van den Berg, Cornelia M. van Duijn, Bert A. Hofman, Aaron Isaacs, André G. Uitterlinden, Joyce van Meurs, P. Mila Jhamai, Michael Verbiest, H. Eka D. Suchiman, Marijn Verkerk, Ruud van der Breggen, Jeroen van Rooij, Nico Lakenberg, Hailiang Mei, Maarten van Iterson, Michiel van Galen, Jan Bot, Dasha V. Zhernakova, Rick Jansen, Peter van ’t Hof, Patrick Deelen, Irene Nooren, Peter A. C. ’t Hoen, Bastiaan T. Heijmans, Matthijs Moed, Lude Franke, Martijn Vermaat, Dasha V. Zhernakova, René Luijk, Marc Jan Bonder, Maarten van Iterson, Patrick Deelen, Freerk van Dijk, Michiel van Galen, Wibowo Arindrarto, Szymon M. Kielbasa, Morris A. Swertz, Erik. W. van Zwet, Rick Jansen, Peter-Bram ’t Hoen, and Bastiaan T. Heijmans
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-08998-0.
References
- 1.Willems, R. M. et al. On the other hand: including left-handers in cognitive neuroscience and neurogenetics. Nature reviews. Neuroscience vol. 15 193–201 (Nature Publishing Group, 2014). [DOI] [PubMed]
- 2.De Gennaro L, et al. Handedness is mainly associated with an asymmetry of corticospinal excitability and not of transcallosal inhibition. Clin. Neurophysiol. 2004;115(6):1305–1312. doi: 10.1016/j.clinph.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Sarkar A, Dutta S, Bal K, Biswas J. Handedness may be related to variations in palmar arterial arches in humans. Singapore Med. J. 2012;53:409–412. [PubMed] [Google Scholar]
- 4.Diederichsen LP, et al. The effect of handedness on electromyographic activity of human shoulder muscles during movement. Int. Soc. Electrophysiol. Kinesiol. 2007;17:410–419. doi: 10.1016/j.jelekin.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 5.Stoyanov Z, Decheva L, Pashalieva I, Nikolova P. Brain asymmetry, immunity, handedness. Cent. Eur. J. Med. 2012;7(1):1–8. [Google Scholar]
- 6.Hepper PG. The developmental origins of laterality: Fetal handedness. Dev. Psychobiol. 2013;55:588–595. doi: 10.1002/dev.21119. [DOI] [PubMed] [Google Scholar]
- 7.de Vries JIP, et al. Fetal handedness and head position preference: A developmental study. Dev. Psychobiol. 2001;39:171–178. doi: 10.1002/dev.1042. [DOI] [PubMed] [Google Scholar]
- 8.Parma V, Brasselet R, Zoia S, Bulgheroni M, Castiello U. The origin of human handedness and its role in pre-birth motor control. Sci. Rep. 2017;7:16804. doi: 10.1038/s41598-017-16827-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadatou-Pastou M, et al. Human handedness: A meta-analysis. Psychol. Bull. 2020;146:481–524. doi: 10.1037/bul0000229. [DOI] [PubMed] [Google Scholar]
- 10.Medland SE, Duffy DL, Wright MJ, Geffen GM, Martin NG. Handedness in twins: Joint analysis of data from 35 samples. Twin Res. Hum. Genet. 2006;9:46–53. doi: 10.1375/183242706776402885. [DOI] [PubMed] [Google Scholar]
- 11.Medland SE, et al. Genetic influences on handedness: Data from 25,732 Australian and Dutch twin families. Neuropsychologia. 2009;47:330–337. doi: 10.1016/j.neuropsychologia.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vuoksimaa E, Koskenvuo M, Rose RJ, Kaprio J. Origins of handedness: A nationwide study of 30,161 adults. Neuropsychologia. 2009;47:1294–1301. doi: 10.1016/j.neuropsychologia.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuellar-Partida G, et al. Genome-wide association study identifies 48 common genetic variants associated with handedness. Nat. Hum. Behav. 2021;5:59–70. doi: 10.1038/s41562-020-00956-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Annett, M. Left, right, hand, and brain : the right shift theory. (L. Erlbaum Associates, 1985).
- 15.McManus C. Half a century of handedness research: Myths, truths; fictions, facts; backwards, but mostly forwards. Brain Neurosci. Adv. 2019;3:239821281882051. doi: 10.1177/2398212818820513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McManus C. Is any but a tiny fraction of handedness variance likely to be due to the external environment? Laterality. 2021;26(3):310–314. doi: 10.1080/1357650X.2021.1892126. [DOI] [PubMed] [Google Scholar]
- 17.Rife DC. Handedness, with special reference to twins. Genetics. 1940;25:178–186. doi: 10.1093/genetics/25.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal, N. L. Twin Mythconceptions : False beliefs, fables, and facts about twins. (2017).
- 19.Derom C, Thiery E, Vlietinck R, Loos R, Derom R. Handedness in twins according to zygosity and chorion type: A preliminary report. Behav. Genet. 1996;26(4):407–408. doi: 10.1007/BF02359484. [DOI] [PubMed] [Google Scholar]
- 20.Francks C, et al. LRRTM1 on chromosome 2p12 is a maternally suppressed gene that is associated paternally with handedness and schizophrenia. Mol. Psychiatry. 2007;12(1057):1129–1139. doi: 10.1038/sj.mp.4002053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun T, et al. Early asymmetry of gene transcription in embryonic human left and right cerebral cortex. Science. 2005;308:1794–1798. doi: 10.1126/science.1110324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arning L, et al. PCSK6 VNTR polymorphism is associated with degree of handedness but not direction of handedness. PLoS ONE. 2013;8:e67251. doi: 10.1371/journal.pone.0067251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brandler WM, et al. Common variants in left/right asymmetry genes and pathways are associated with relative hand skill. PLoS Genet. 2013;9:e1003751. doi: 10.1371/journal.pgen.1003751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arning L, et al. Handedness and the X chromosome: The role of androgen receptor CAG-repeat length. Sci. Rep. 2015;5:8325. doi: 10.1038/srep08325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hampson E, Sankar JS. Hand preference in humans is associated with testosterone levels and androgen receptor gene polymorphism. Neuropsychologia. 2012;50(8):2018–2025. doi: 10.1016/j.neuropsychologia.2012.04.027. [DOI] [PubMed] [Google Scholar]
- 26.Medland SE, et al. Opposite effects of androgen receptor CAG repeat length on increased risk of left-handedness in males and females. Behav. Genet. 2005;35:735–744. doi: 10.1007/s10519-005-6187-3. [DOI] [PubMed] [Google Scholar]
- 27.Eriksson N, et al. Web-based, participant-driven studies yield novel genetic associations for common traits. PLoS Genet. 2010;6:e1000993. doi: 10.1371/journal.pgen.1000993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armour JAL, Davison A, McManus IC. Genome-wide association study of handedness excludes simple genetic models. Heredity. 2014;112(3):221–225. doi: 10.1038/hdy.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiberg A, et al. Handedness, language areas and neuropsychiatric diseases: Insights from brain imaging and genetics. Brain A J. Neurol. 2019;142:2938–2947. doi: 10.1093/brain/awz257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Kovel CGF, Francks C. The molecular genetics of hand preference revisited. Sci. Rep. 2019;9:5986. doi: 10.1038/s41598-019-42515-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conde C, Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 2009;10:319–332. doi: 10.1038/nrn2631. [DOI] [PubMed] [Google Scholar]
- 32.Sha Z, et al. The genetic architecture of structural left-right asymmetry of the human brain. Nat. Hum. Behav. 2021;5(9):1226–1239. doi: 10.1038/s41562-021-01069-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klar AJS. An epigenetic hypothesis for human brain laterality, handedness, and psychosis development. Cold Spring Harb. Symp. Quant. Biol. 2004;69:499–506. doi: 10.1101/sqb.2004.69.499. [DOI] [PubMed] [Google Scholar]
- 34.Crow TJ. A theory of the origin of cerebral asymmetry: Epigenetic variation superimposed on a fixed right-shift. Laterality. 2010;15:289–303. doi: 10.1080/13576500902734900. [DOI] [PubMed] [Google Scholar]
- 35.Schmitz J, Metz GAS, Güntürkün O, Ocklenburg S. Beyond the genome-Towards an epigenetic understanding of handedness ontogenesis. Prog. Neurobiol. 2017;159:69–89. doi: 10.1016/j.pneurobio.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Suderman, M. et al. dmrff: Identifying differentially methylated regions efficiently with power and control. bioRxiv 508556 (2018) 10.1101/508556.
- 37.Hüls A, Czamara D. Methodological challenges in constructing DNA methylation risk scores. Epigenetics. 2020;15:1–11. doi: 10.1080/15592294.2019.1644879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahl S, et al. Epigenome-wide association study of body mass index, and the adverse outcomes of adiposity. Nature. 2017;541:81–86. doi: 10.1038/nature20784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmitz J, Güntürkün O, Ocklenburg S. Building an asymmetrical brain: The molecular perspective. Front. Psychol. 2019;10:982. doi: 10.3389/fpsyg.2019.00982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ocklenburg S, et al. Epigenetic regulation of lateralized fetal spinal gene expression underlies hemispheric asymmetries. Elife. 2017;6:e22784. doi: 10.7554/eLife.22784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leach EL, Prefontaine G, Hurd PL, Crespi BJ. The imprinted gene LRRTM1 mediates schizotypy and handedness in a nonclinical population. J. Hum. Genet. 2014;59:332–336. doi: 10.1038/jhg.2014.30. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz J, Kumsta R, Moser D, Güntürkün O, Ocklenburg S. DNA methylation in candidate genes for handedness predicts handedness direction. Laterality. 2018;23:441–461. doi: 10.1080/1357650X.2017.1377726. [DOI] [PubMed] [Google Scholar]
- 43.Watanabe K, et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 2019;51:1339–1348. doi: 10.1038/s41588-019-0481-0. [DOI] [PubMed] [Google Scholar]
- 44.Min JL, et al. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021;53(9):1311–1321. doi: 10.1038/s41588-021-00923-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mulder RH, et al. Epigenome-wide change and variation in DNA methylation in childhood: Trajectories from birth to late adolescence. Hum. Mol. Genet. 2021;30(1):119–134. doi: 10.1093/hmg/ddaa280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joehanes R, et al. Epigenetic signatures of cigarette smoking. Circ. Cardiovasc. Genet. 2016;9:436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Küpers LK, et al. Meta-analysis of epigenome-wide association studies in neonates reveals widespread differential DNA methylation associated with birthweight. Nat. Commun. 2019;10(1):1893. doi: 10.1038/s41467-019-09671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Joubert BR, et al. DNA methylation in newborns and maternal smoking in pregnancy: Genome-wide consortium meta-analysis. Am. J. Hum. Genet. 2016;98:680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schulz R, et al. Transcript- and tissue-specific imprinting of a tumour suppressor gene. Hum. Mol. Genet. 2009;18:118–127. doi: 10.1093/hmg/ddn322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thamban T, et al. The putative Neuronatin imprint control region is an enhancer that also regulates the Blcap gene. Epigenomics. 2019;11:251–266. doi: 10.2217/epi-2018-0060. [DOI] [PubMed] [Google Scholar]
- 51.Evans HK, Weidman JR, Cowley DO, Jirtle RL. Comparative phylogenetic analysis of blcap/nnat reveals eutherian-specific imprinted gene. Mol. Biol. Evol. 2005;22:1740–1748. doi: 10.1093/molbev/msi165. [DOI] [PubMed] [Google Scholar]
- 52.Court F, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Propper R, Struble C, Brunyé T. Handedness and physical measures II: Objectively measured height and weight. Int. J. Sch. Cognit. Psychol. 2015;2:1000127. [Google Scholar]
- 54.Domellöf E, Johansson A-M, Rönnqvist L. Handedness in preterm born children: A systematic review and a meta-analysis. Neuropsychologia. 2011;49:2299–2310. doi: 10.1016/j.neuropsychologia.2011.04.033. [DOI] [PubMed] [Google Scholar]
- 55.Tsai P-C, Bell JT. Power and sample size estimation for epigenome-wide association scans to detect differential DNA methylation. Int. J. Epidemiol. 2015;44:1429–1441. doi: 10.1093/ije/dyv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell, K. J. Innate. (Princeton University Press, 2018).
- 57.Molenaar PC, Boomsma DI, Dolan CV. A third source of developmental differences. Behav. Genet. 1993;23:519–524. doi: 10.1007/BF01068142. [DOI] [PubMed] [Google Scholar]
- 58.de Kovel CGF, Carrión-Castillo A, Francks C. A large-scale population study of early life factors influencing left-handedness. Sci. Rep. 2019;9:584. doi: 10.1038/s41598-018-37423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Porac C, Friesen IC. Hand preference side and its relation to hand preference switch history among old and oldest-old adults. Dev. Neuropsychol. 2000;17:225–239. doi: 10.1207/S15326942DN1702_05. [DOI] [PubMed] [Google Scholar]
- 60.van Dongen J, et al. DNA methylation signatures of aggression and closely related constructs: A meta-analysis of epigenome-wide studies across the lifespan. Mol. Psychiatry. 2021;26(6):2148–2162. doi: 10.1038/s41380-020-00987-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Willemsen G, et al. The Netherlands twin register biobank: A resource for genetic epidemiological studies. Twin Res. Hum. Genet. 2010;13:231–245. doi: 10.1375/twin.13.3.231. [DOI] [PubMed] [Google Scholar]
- 62.Boyd A, et al. Cohort profile: The ‘children of the 90s’—The index offspring of the avon longitudinal study of parents and children. Int. J. Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fraser A, et al. Cohort profile: The avon longitudinal study of parents and children: ALSPAC mothers cohort. Int. J. Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Northstone K, et al. The avon longitudinal study of parents and children (ALSPAC): An update on the enrolled sample of index children in 2019. Wellcome Open Res. 2019;4:51. doi: 10.12688/wellcomeopenres.15132.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boomsma DI. Aggression in children: Unravelling the interplay of genes and environment through (epi)genetics and metabolomics. J. Pediatr. Neonatal Individ. Med. 2015;4:e040251–e040251. [Google Scholar]
- 66.Bartels M, et al. Childhood aggression and the co-occurrence of behavioural and emotional problems: results across ages 3–16 years from multiple raters in six cohorts in the EU-ACTION project. Eur. Child Adolesc. Psychiatry. 2018;27:1105–1121. doi: 10.1007/s00787-018-1169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Küpers LK, et al. DNA methylation mediates the effect of maternal smoking during pregnancy on birthweight of the offspring. Int. J. Epidemiol. 2015;44:1224–1237. doi: 10.1093/ije/dyv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Iterson M, van Zwet EW, Heijmans BT. Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genom. Biol. 2017;18:19. doi: 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Martin TC, Yet I, Tsai P-C, Bell JT. coMET: Visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinform. 2015;16:131. doi: 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li M, et al. EWAS Atlas: A curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019;47:D983–D988. doi: 10.1093/nar/gky1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Battram T, et al. The EWAS Catalog: A database of epigenome-wide association studies [version 1; peer review: awaiting peer review] Wellcome Open Res. 2022;7:41. doi: 10.12688/wellcomeopenres.17598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vilhjálmsson BJ, et al. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am. J. Hum. Genet. 2015;97:576–592. doi: 10.1016/j.ajhg.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shah S, et al. Improving phenotypic prediction by combining genetic and epigenetic associations. Am. J. Hum. Genet. 2015;97:75–85. doi: 10.1016/j.ajhg.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SH, Goddard ME, Wray NR, Visscher PM. A better coefficient of determination for genetic profile analysis. Genet. Epidemiol. 2012;36:214–224. doi: 10.1002/gepi.21614. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HumanMethylation450 BeadChip data from the NTR are available as part of the Biobank-based Integrative Omics Studies (BIOS) Consortium in the European Genome-phenome Archive (EGA), under the accession code EGAD00010000887 (https://ega-archive.org/datasets/EGAD00010000887). The Infinium MethylationEPIC from NTR are available from the Netherlands Twin Register on reasonable request (https://tweelingenregister.vu.nl/information_for_researchers/working-with-ntr-data). DNA methylation data from ALSPAC are available at ALSPAC and can be provided on request. The study website contains details of all the data that is available through a fully searchable data dictionary and variable search tool (http://www.bristol.ac.uk/alspac/researchers/our-data). The code used to perform the primary and secondary analyses is available at https://github.com/MRCIEU/handedness-ewas. The pipeline for the DNA methylation array analysis developed by the Biobank-based Integrative Omics Study (BIOS) consortium are available here: https://molepi.github.io/DNAmArray_workflow/. EWAS summary statistics for the top 100 CpGs are given in Supplemental Tables 6–11 and 15–29. The full EWAS summary statistics from the meta-analysis with basic and adjusted model are provided in Supplemental Tables 32 and 33. The full summary statistics for all other analyses are available upon request from the corresponding author.