Abstract
The outer membrane of Gram-negative bacteria functions as an impermeable barrier to foreign compounds. Thus, modulating membrane transport can contribute to improving susceptibility to antibiotics and efficiency of bioproduction reactions. In this study, the cellular uptake of hydrophobic and large-scaffold antibiotics and other compounds in Gram-negative bacteria was investigated by modulating the homolog expression of bamB encoding an outer membrane lipoprotein and tolC encoding an outer membrane efflux protein via gene deletion and gene silencing. The potential of deletion mutants for biotechnological applications, such as drug screening and bioproduction, was also demonstrated. Instead of being subjected to gene deletion, wild-type bacterial cells were treated with cell-penetrating peptide conjugates of a peptide nucleic acid (CPP-PNA) against bamB and tolC homologs as antisense agents. Results revealed that the single deletion of bamB and tolC in Escherichia coli increased the uptake of large- and small-scaffold hydrophobic compounds, respectively. A bamB-and-tolC double deletion mutant had a higher uptake efficiency for certain antibiotics and other compounds with high hydrophobicity than each single deletion mutant. The CPP-PNA treated E. coli and Pseudomonas aeruginosa cells showed high sensitivity to various antibiotics. Therefore, these gene deletion and silencing approaches can be utilized in therapeutic and biotechnological fields.
Subject terms: Microbiology, Biotechnology
Introduction
Gram-negative bacteria have inner and outer membranes, and their outer membrane functions as an impermeable barrier to foreign compounds. Compounds > 600 Da in size are not internalized into the cells of Gram-negative bacteria via passive diffusion1,2. Bacteria use various membrane pumps to actively transport compounds across cell membranes. Therefore, the uptake efficiency of foreign compounds is determined by the ratio of influx and efflux through membranes mediated by passive diffusion and active transportation; uptake efficiency has been modified from medical and biotechnological viewpoints3.
As many antibiotics work inside their target cells, shutting off their entry into cells and pumping them out of cells are common antimicrobial resistance mechanisms of bacteria4. Several mutants of Escherichia coli have been isolated to explore ways on how to improve the uptake efficiency of antimicrobial compounds. Mutations in lpx involved in lipid A synthesis decrease the minimum inhibitory concentration (MIC) of antibiotics such as rifampicin, erythromycin, and fusidic acid5. In a comprehensive analysis involving a single-gene deletion collection, a deletion mutant of bamB (encoding the outer membrane lipoprotein), tolC (encoding the outer membrane efflux protein), waaC (participating in lipopolysaccharide biosynthesis), or other cell membrane-related genes has a decreased MIC of various antibiotics6. In many biotechnological applications, bacterial cells are required to uptake foreign compounds and maintain high intracellular concentrations for intracellular reactions. Therefore, the improvement of the substrate uptake rate of microbial cells (e.g., transport engineering) is a key approach for efficient industrial bioproduction3,7. For example, tolC deletion mutants of E. coli have a relatively high uptake of cholesterol derivatives such as vitamin D3 (VD3)8. Although previous studies identified key genes for the improvement of uptake efficiency, the combined effects of these genes have been rarely evaluated and should be further improved.
Various approaches other than gene deletion can increase uptake efficiency; for example, low-molecular-weight sensitizers, along with target compounds, can be used to increase the permeability of cell membranes. One of the most used sensitizers for increasing the membrane permeability of Gram-negative bacteria is polymyxin B nonapeptide, which decreases the MIC of various antibiotics9. Moreover, MAC13243, which inhibits periplasmic chaperone protein LolA activity, sensitizes cells to large-scaffold antibiotics1. Although these sensitizer compounds are generally useful and easy to handle, it is still difficult and laborious to design or find ideal sensitizers having expected effects without causing side effects (e.g., cell toxicity).
In this study, we applied gene deletion and sensitizing approaches to increase the uptake efficiency of E. coli and investigated the influx and efflux properties of several compounds in terms of their hydrophobicity and scaffold size. We selected genes to be targeted according to the following criteria: (1) complete deletion of a chosen gene does not cause severe growth arrest or retardation, (2) the gene is conserved among Gram-negative bacteria, and (3) multiple target genes may have different cellular functions that contrast with one another for investigating the uptake efficiency associated with multiple cellular functions. Based on these criteria, we chose bamB and tolC. The null mutants of both genes are not associated with obvious growth defects8,10, and these genes are well conserved among a broad range of Gram-negative bacteria10–12. BamB is a component of the outer membrane protein assembly complex (β-barrel-assembly machinery [Bam])13,14, whereas TolC is a part of the AcrAB-TolC efflux pump complex that transports various compounds with low specificity15. As they have distinct functions, each deletion could show different effects on membrane permeability; thus, a combined effect is expected. Although the dysfunction of each gene leads to an increase in foreign compound uptake, membrane flux properties and combined effects have not been analyzed in detail. In particular, relationships between flux changes driven by each mutation and the hydrophobicity or scaffold size of foreign compounds are still largely unknown although such information is a key factor to understand and engineer the nature of foreign compound uptake by Gram-negative bacteria. In the sensitizing approach, we used cell-penetrating peptide (CPP) conjugates of peptide nucleic acid (PNA, a nucleic acid analog) as sensitizers to silence gene expression because of their generally low toxicity and easy-to-design nature16. We also investigated the applicability of these approaches in the screening of antibiotic compounds and the bioconversion of value-added compounds. Furthermore, we used the sensitizing approach to Pseudomonas aeruginosa, another Gram-negative pathogenic bacterium, to demonstrate the potential of sensitizers for broad application in Gram-negative bacteria.
Results
Influx and efflux properties of the deletion mutants
bamB (ΔbamB), tolC (ΔtolC), and bamB and tolC (ΔbamBΔtolC) deletion mutants of E. coli were created. N-Phenyl-1-naphthylamine (NPN), a probe compound, was used to investigate the influx and efflux properties of the mutants. It is a small hydrophobic fluorescent dye (molecular weight, 219; 1-octanol/water partition coefficient [log Pow], 4.2 [https://pubchem.ncbi.nlm.nih.gov]) that exhibits high fluorescence in hydrophobic environments, such as lipid bilayers of cell membranes17.
The influx properties of the wild-type strain and the mutants were analyzed by adding NPN to the cultures and monitoring fluorescence over time (Fig. 1a). In this experimental setup, an increase in fluorescence was interpreted as an increase in the influx rate or a decrease in the efflux rate. Fluorescence increased in ΔbamB even at time zero. This result showed that the uptake in ΔbamB occurred too fast to monitor a gradual increase in fluorescence. In ΔbamBΔtolC, fluorescence also immediately increased although it gradually further increased. The fluorescence of ΔtolC at time zero was the same as that of the wild-type strain, and it increased gradually over time. These results suggested that influx was increased by bamB deletion but not by tolC deletion; tolC deletion might cause a decrease in the efflux rate.
Figure 1.
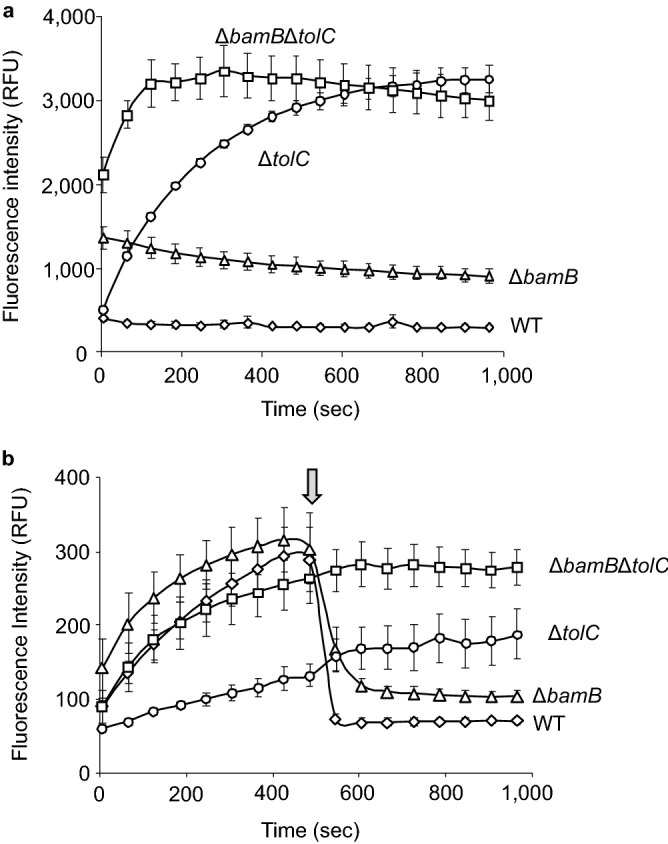
NPN influx and efflux analysis. (a) Result of the influx analysis; fluorescence intensity of NPN is presented as a function of time. The data are presented as the mean ± standard error of mean from triplicate experiments. (b) Efflux analysis. The arrow indicates the time of glucose addition. NPN, N-phenyl-1-naphthylamine; WT, wild-type strain; ΔbamB, outer membrane lipoprotein deletion mutant; ΔtolC, outer membrane efflux protein deletion mutant; ΔbamBΔtolC, bamB and tolC double deletion mutant.
An efflux analysis was performed using NPN and carbonyl cyanide 3-chlorophenylhydrazone (CCCP) that inhibits the proton motive force (PMF) and hence the function of efflux pumps. The cells subjected to CCCP treatment in the absence of glucose, which do not have any PMF driving efflux pumps, were resuspended in buffer with NPN, and fluorescence transition was monitored (Fig. 1b). Because of a lack of PMF, fluorescence intensity was gradually increased solely by the influx in all strains. After the addition of glucose that restored PMF and efflux flow, the fluorescence intensity immediately deceased in ΔbamB and the wild-type strain but not in ΔtolC and ΔbamBΔtolC. This result indicated that tolC deletion affected efflux property. Therefore, bamB and tolC deletion resulted in an increase in influx and a decrease in efflux properties, respectively.
Scaffold size-dependent effect of antibiotics on uptake efficiency in the deletion mutants
The uptake efficiency of foreign compounds in the deletion mutants was further evaluated by determining the MIC of various antibiotics (Table 1). Regarding large-scaffold antibiotics, bamB deletion effectively improved susceptibility: the MICs of vancomycin and actinomycin D decreased by 16- and 8-fold, respectively. Conversely, small-scaffold hydrophobic antibiotics (chloramphenicol, triclosan, and 5-ketoclomazone) were more efficiently taken up by the tolC mutant than those by the other mutants (8-, 64-, and over 4-fold, respectively). An inverse trend of scaffold size dependency of MIC fold change (mutant/wild-type) was observed between ΔbamB and ΔtolC with statistical significance (Fig. 2a). Conversely, no clear correlation was observed in ΔbamBΔtolC. bamB or tolC deletion likely decreased the MIC by improving the inefficient uptake of antibiotics by the wild-type strain due to a low influx or high efflux rate. The uptake of moderate-sized scaffold hydrophobic antibiotics (rifampicin, erythromycin, novobiocin, and fusidic acid) was improved via both deletions (64-, 256-, 256- and 512-fold, respectively). These results suggested that the effect of each deletion on MIC depended on the scaffold size of antibiotics. The hydrophobicity dependency of MIC fold change was commonly observed in the mutants (Fig. 2b), indicating that compounds with high hydrophobicity were more susceptible to influx and efflux changes by bamB or tolC deletion than compounds with low hydrophobicity.
Table 1.
MIC of antibiotics for deletion mutants.
| Antibiotics (molecular weight; log Pow) | Wild-type MIC (μg/mL) | ΔbamB MIC (μg/mL) | ΔtolC MIC (μg/mL) | ΔbamBΔtolC MIC (μg/mL) |
|---|---|---|---|---|
| Vancomycin (1450; − 3.1) | 100 | 6.3 | 100 | 6.3 |
| Actinomycin D (1255; 1.6) | 200 | 25 | 200 | 25 |
| Rifampicin (823; 2.7) | 7.5 | 0.12 | 3.8 | 0.12 |
| Erythromycin (734; 3.06) | 50 | 12.5 | 0.78 | 0.20 |
| Novobiocin (613; 4.1) | 100 | 50 | 1.6 | 0.39 |
| Fusidic acid (517; 6.75) | 400 | 200 | 6.3 | 0.78 |
| Kanamycin (485; − 6.3) | 2.5 | 2.5 | 1.3 | 1.3 |
| Tetracycline (444; − 1.3) | 0.25 | 0.25 | 0.13 | 0.13 |
| Ampicillin (349; 1.35) | 20 | 10 | 10 | 10 |
| Berberine (336; − 1.5) | > 167 | > 167 | 42 | 42 |
| Chloramphenicol (323; 1.14) | 4.3 | 2.1 | 0.54 | 0.54 |
| Triclosan (290; 4.76) | 0.25 | 0.13 | 0.0039 | 0.0019 |
| 5-Ketoclomazone (254; − 0.54) | > 250 | > 250 | 63 | 63 |
| Nalidixic acid (232; 1.59) | 5 | 2.5 | 0.63 | 0.63 |
MIC, minimum inhibitory concentration; ΔbamB, outer membrane lipoprotein deletion mutant; ΔtolC, outer membrane efflux protein deletion mutant; ΔbamBΔtolC, bamB and tolC double deletion mutant. Log Pow values were derived from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/) except for fusidic acid (The European Chemicals Agency, https://echa.europa.eu/registration-dossier/-/registered-dossier/20132/4/8.), berberine18, and 5-ketoclomazone (https://www.hpc-standards.com/msds/681800_Ketoclomazone_Acetonitrile_1_MSDS_EN_HPC-Standards.pdf).
Figure 2.
Correlation between MIC fold change and scaffold size or hydrophobicity. The MIC fold change (each mutant/the wild type strain) and (a) scaffold size or (b) hydrophobicity of antimicrobial compounds are plotted for each mutant. The dotted line represents a regression line. The correlation coefficient (r) and p value by Pearson’s correlation analysis are indicated within each panel; p value in bold means significant at p = 0.05.
bamB and tolC deletions elicited combined effects on the uptake of some antibiotics (erythromycin, novobiocin, fusidic acid, and triclosan; Table 1). Therefore, interactions between bamB and tolC deletions were investigated by using a fractional inhibitory concentration index (FICI), which is used to evaluate interactions (synergism, no interaction, or antagonism) between two inhibitory agents (Supplementary Table S1). The results showed that two deletions worked synergistically in moderate-sized scaffold antibiotics showing a combined effect. Moreover, triclosan, a small-scaffold antibiotic with a combined effect, also had an FICI of 0.502, which is close to the criterion for synergism (FICI ≤ 0.5).
Uptake efficiency of CPP-PNAs by the deletion mutants
Antisense CPP-PNAs are considered new anti-infective or antimicrobial drugs. PNA is a charge-neutral oligonucleotide analog that possesses good hybridization properties and resistance to degradation by nucleases and proteases19. The uptake efficiency of PNA alone by bacteria is usually poor; thus, CPPs such as KFFKFFKFFK are frequently conjugated to PNAs19. Therefore, we evaluated whether the uptake efficiency of KFF-acpP, a CPP-PNA, also increased in the deletion mutants. acpP is essential for growth, and KFF-acpP has bactericidal effects19. Subinhibitory KFF-acpP concentrations (0.3 μM) caused a slight growth delay in the single deletion mutants (ΔbamB and ΔtolC) and a relatively strong growth delay in the double deletion mutant (ΔbamBΔtolC). However, it did not affect the growth curve of the wild-type strain, indicating that KFF-acpP was taken up more efficiently in the mutants than in the wild-type strain (Fig. 3). The differential effect of KFF-acpP on growth between the mutants and the wild-type strain was also supported by the decrease in MIC in the mutants (Supplementary Table S2).
Figure 3.
Growth of the wild-type and mutant strains in the presence of KFF-acpP. Cell growth in the presence or absence of CPP-PNAs is indicated. The CPP-PNAs used were KFF-NC (□) and KFF-acpP (○). In a control experiment, distilled water was added instead of CPP-PNAs (△). The final CPP-PNA concentration was 0.3 μM. Data are presented as the mean ± standard error of the mean from triplicate experiments. KFF-NC, negative control; KFF-acpP, CPP-PNA targeting acyl carrier protein.
Proof-of-concept usages: detection of antibiotic compounds at low concentrations
In screening novel antibiotics from natural samples, antibiotic concentrations in samples are generally low20. Therefore, the presence of antibiotic compounds in such samples is often difficult to be detected; consequently, sensitized strains are preferable as indicator strains. On this basis, we tested the sensitivity of the present mutants to the extracts of environmental soil samples as follows: potential antibiotic-containing samples were prepared by inoculating a soil sample with a liquid medium and extracting the culture with ethyl acetate; drops of the extracts were placed onto agar plates containing the wild-type or mutant cells. We found halos (zones of inhibition) only in the plates with ΔbamBΔtolC cells (Fig. 4a). Therefore, ΔbamBΔtolC was more sensitive to antibacterial compounds than the wild-type strain and could be used for detecting active compounds at low concentrations.
Figure 4.
Detection of antibiotic activity in the culture samples of natural soil and gene expression titration assay. (a) Aliquots of the extract from soil culture with R2A (R2A sample) and M9 (M9 sample) media and control (DMSO) were spotted on the lawn of indicator strains on an agar plate as indicated by a diagram. After incubation, antibiotic activity was detected as the inhibition zone formed on the plate. The arrow indicates a halo. (b) 5-Ketoclomazone was spotted on the lawn of indicator strains on an agar plate at concentrations indicated by a diagram. (c) Control experiment was performed with triclosan and fusidic acid as test antibiotic compounds. ΔbamB, outer membrane lipoprotein deletion mutant; ΔtolC, outer membrane efflux protein deletion mutant; ΔbamBΔtolC, bamB and tolC double deletion mutant; dxs AS, expression of dxs antisense RNA.
Proof-of-concept usages: gene expression titration assay (GETA)
A GETA can also be used to increase the sensitivity of cells to antibiotics at low concentrations21. In GETA, an antisense agent is used to partially silence the expression of a specific mRNA, whose translation product is a target for a certain antibiotic; consequently, host cells become sensitized because antibiotics can elicit their effects at low concentrations.
5-Ketoclomazone was used as a test antibiotic here because it showed a low uptake by the wild-type strain (Table 1). dxs encoding 1-deoxy-d-xylulose 5-phosphate synthase, the putative cellular target molecule of 5-ketoclomazone22, was silenced by the endogenous expression of a short antisense RNA against dxs by using an expression vector23. The cells became more susceptible to 5-ketoclomazone when the antisense RNA against dxs was expressed (Fig. 4b), and this effect was greater in ΔtolC and ΔbamBΔtolC than in the wild-type strain and ΔbamB even at low 5-ketoclomazone concentrations. In the negative control experiment involving antibiotics unrelated to Dxs (triclosan and fusidic acid), dxs silencing did not change the sizes of the halo (Fig. 4c). This result indicated that dxs silencing specifically increased the susceptibility to 5-ketoclomazone. Therefore, target gene silencing conferred the higher susceptibility of tolC deletion mutants to 5-ketoclomazone than that of the wild-type strain, and the present bamB/tolC mutants could be useful high-sensitive host strains for GETA.
Proof-of-concept usages: conversion of inactive VD3 to active hydroxylated VD3 in the mutants
VD3 is a hydrophobic vitamin that contains a steroid skeleton24. Several forms of VD3 exist; cholecalciferol (molecular weight, 385; log Pow, 7.5 [https://pubchem.ncbi.nlm.nih.gov]) is an inactive form of VD3, and its hydroxylated forms 25-hydroxycholecarciferol (calcifediol) and 1,25-dihydroxycholecalciferol (calcitriol) are active forms of VD3 (showing a hormonal activity) and valuable as pharmaceuticals24. An enzymatic conversion of inactive to active VD3 in microbes is desirable because an enzymatic hydroxylation approach is more cost-effective than chemical synthesis25. However, a low VD3 uptake rate in E. coli retards enzymatic conversion8. Therefore, cells expressing VD3 hydroxylase (vdh), ferredoxin (aciB), and ferredoxin reductase (aciC) genes were prepared to improve the conversion of VD3. When the wild-type strain was used as an expression host, no VD3 conversion occurred, whereas in ΔtolC and ΔbamBΔtolC, an effective conversion (1.4 and 4.6 μM, respectively) was detected (Fig. 5). Therefore, the conversion rate of inactive VD3 was improved by tolC deletion and further enhanced by double deletion with bamB.
Figure 5.
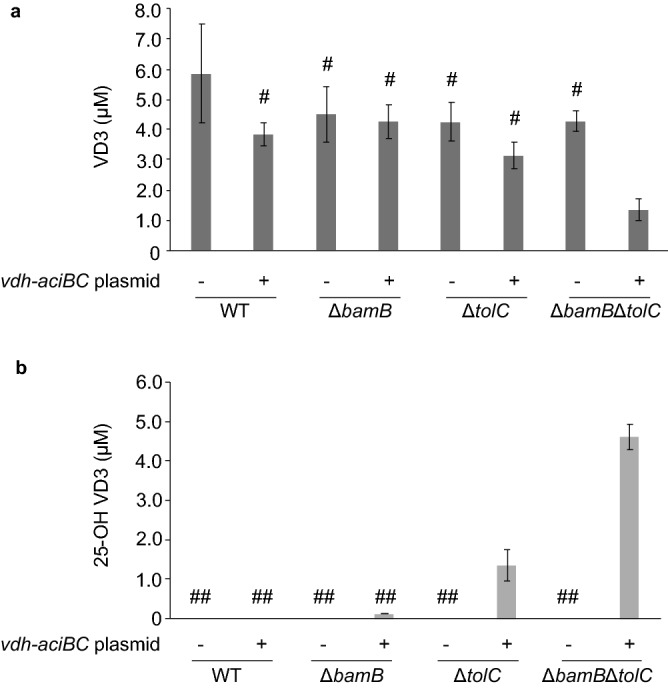
VD3 conversion efficiency of the mutants. Inactive VD3 was converted to active VD3 by using cells transformed with pHN1387 (empty vector control; vdh-aciBC plasmid−) or pHN4136 (vdh-aciBC expression plasmid; vdh-aciBC plasmid+). The host cells used are indicated at the bottom of the graph. The quantification result of inactive VD3 that remained after the conversion reaction from triplicate experiments is presented as the mean ± standard error of the mean. Data were statistically analyzed using ANOVA, and significant differences were evaluated with Tukey’s multiple comparison test. One-way ANOVA showed a significant difference at F = 2.7 (p = 0.046). The bars marked with “#” correspond with the groups that did not differ significantly from each other at adjusted p < 0.05 in the post hoc test. The unmarked bars correspond with the group that significantly differed from the other groups. (b) The quantification result of active VD3 produced after the conversion reaction is shown as in (a). VD3, vitamin D3; ΔbamB, outer membrane lipoprotein deletion mutant; ΔtolC, outer membrane efflux protein deletion mutant; ΔbamBΔtolC, bamB and tolC double deletion mutant. One-way ANOVA showed a significant difference at F = 82 (p = 2.5 × 10–11). “##” is marked as in (a).
Sensitizing wild-type E. coli with anti-bamB and tolC CPP-PNAs
The aforementioned results depended on gene recombination techniques; thus, the generated mutant strains cannot be readily used in pharmaceutical and food industries because of restrictions and regulations for the use of genetically modified organisms. Instead of using gene recombinant mutants, we tested whether CPP-PNAs targeting bamB and tolC could function as efficient sensitizers for increasing the uptake of several antibiotics. Before CPP-PNAs as sensitizers were evaluated, their effect on the growth of the wild-type strain in the absence of antibiotics was assessed. Low CPP-PNA concentrations marginally affected growth, but it caused a slight growth delay at high concentrations, indicating a dose-dependent negative effect of CPP-PNAs on E. coli growth (Supplementary Fig. S1). The wild-type strain was then treated with antibiotics in the presence of the following CPP-PNAs: KFF-NC, KFF-bamB, and KFF-tolC (Table 2). Unexpectedly, the treatment with KFF-NC decreased the MIC of certain antibiotics, namely, actinomycin D, novobiocin, and fusidic acid (compared with the results in Table 1); this result indicated that the KFFKFFKFFK peptide fragment affected the uptake efficiency of some antibiotics. Nevertheless, KFF-bamB and KFF-tolC more effectively decreased the MICs than KFF-NC did. For example, KFF-bamB decreased the MICs of actinomycin D, novobiocin, and fusidic acid by 2-, 16-, and 4-fold, respectively; KFF-tolC decreased the MIC of 5-ketoclomazone by 2-fold. Furthermore, the combination of KFF-bamB and KFF-tolC decreased the MIC of the tested antibiotics largely than the single use of each CPP-PNA except for the MIC of vancomycin, which was not decreased by any CPP-PNAs. The interaction between KFF-bamB and KFF-tolC was evaluated by using FICI (Supplementary Table S3), and two CPP-PNAs were found to work synergistically (FICI ≤ 0.5) for actinomycin D and fusidic acid. These results indicated that KFF-bamB and KFF-tolC are effective sensitizers for E. coli against some antibiotic compounds.
Table 2.
MIC of antibiotics for E. coli and P. aeruginosa in the presence of CPP-PNAs.
| E. coli | |||||
|---|---|---|---|---|---|
| Antibiotics | KFF-NC MIC (μg/mL) | KFF-bamB MIC (μg/mL) | KFF-tolC MIC (μg/mL) | KFF-bamB KFF-tolC MIC (μg/mL)a | |
| Vancomycin | 100 | > 200 | 100 | 100 | |
| Actinomycin D | 50 | 25 | 50 | < 6.3 | |
| Novobiocin | 12.5 | 0.8 | 12.5 | < 0.4 | |
| Fusidic acid | 50 | 12.5 | 50 | < 0.4 | |
| 5-Ketoclomazone | > 250 | > 250 | 125 | 62.5 | |
| P. aeruginosa | |||||
|---|---|---|---|---|---|
| Antibiotics | Control (no CPP-PNA) MIC (μg/mL) | RXR-NC MIC (μg/mL) | RXR-bamB MIC (μg/mL) | RXR-oprM MIC (μg/mL) | RXR-bamB RXR-oprM MIC (μg/mL)b |
| Vancomycin | 1024 | 1024 | 256 | 512 | 256 |
| Erythromycin | 128 | 128 | 32 | 64 | < 16 |
| Carbenicillin | 64 | 64 | 8 | 32 | < 4 |
CPP-PNAs were added at the concentration of 5 and 3 μM for E. coli and P. aeruginosa, respectively.
aIn the presence of both CPP-PNAs, equimolar KFF-bamB and KFF-tolC were mixed.
bIn the presence of both CPP-PNAs, equimolar RXR-bamB and RXR-oprM were mixed.
Sensitizing Pseudomonas aeruginosa with anti-bamB and oprM CPP-PNAs
To explore the applicability of CPP-PNA approach, we tested the effect of CPP-PNAs on antibiotic susceptibility in a notorious opportunistic pathogen, P. aeruginosa. We additionally created CPP-PNAs that targeted bamB and oprM, a tolC homolog in the P. aeruginosa genome and evaluated the potential of these CPP-PNAs as sensitizers in the same manner as E. coli. CPP-PNAs negatively affected the growth of P. aeruginosa PAO1 in the absence of antibiotics; although the same trend was observed as in E. coli, the effect on the growth of P. aeruginosa PAO1 was stronger than that on the growth of E. coli (Supplementary Fig. S2). Similar to E. coli, P. aeruginosa PAO1 treated with the CPP-PNAs targeting bamB and oprM was sensitized by certain antibiotics (Table 2). For all antibiotics tested, the highest MIC decrease (4-, > 8-, and > 16-fold decrease for vancomycin, erythromycin, and carbenicillin, respectively) was observed when both CPP-PNAs were simultaneously added although a synergistic interaction between RXR-bamB and RXR-oprM was not detected (Supplementary Table S3). Therefore, CPP-PNAs targeting bamB and tolC homologs function as efficient sensitizers for improving the uptake of various antibiotics by Gram-negative bacteria.
Discussion
In E. coli and Salmonella enterica, the loss of BamB function leads to reduced levels of most major outer membrane proteins11. The resulting perturbation in outer membrane composition likely increases the outer membrane permeability. In this study, the increase in permeability mediated by bamB deletion was particularly effective in improving the uptake of large-scaffold compounds (LSCs; Table 1). Conversely, tolC deletion remarkably affected permeability for improving the uptake of moderate-sized to small-scaffold compounds (SSCs; Table 1). The present result of tolC deletion is consistent with a previous finding, which demonstrated that tolC deletion is ineffective in improving the uptake of large-scaffold antibiotics26; however, information about the effect of bamB deletion on permeability in terms of scaffold size is lacking. The observed changes in the scaffold size-dependent uptake efficiency in the mutants could be attributed to the continuous influx and efflux of SSCs and the blocking influx of LSCs by the outer membrane. The increased membrane permeability by bamB deletion results in an increased influx of SSCs and LSCs, but cellular SSCs could be excreted by the efflux function attributed by membrane pumps such as TolC. Therefore, as long as the efflux function is working, the cellular concentration of SSCs can be maintained at certain levels. This finding was supported by constant concentrations of NPN in the wild-type strain and ΔbamB (Fig. 1a). In addition, a previous study indicated that the loss of BamB function exceptionally leads to an increase in TolC abundance in the outer membrane11,27, thereby contributing to a slight change in MICs for small-scaffold antibiotics in ΔbamB. However, an increase in the influx of LSCs is not compensated by the efflux function and results in the accumulation of LSCs; for this reason, ΔbamB was mainly sensitized to large-scaffold antibiotics. As for ΔtolC, the deletion caused efflux dysfunction; consequently, SSCs accumulated, whereas the influx of LSCs were not affected. As a result, sensitivity to small-scaffold antibiotics increased. The MICs of moderate-sized scaffold antibiotics decreased in ΔbamB and ΔtolC (Table 1), indicating that moderate-sized scaffold compounds could have intermediate traffic properties compared with those of SSCs and LSCs.
Combined effects of bamB and tolC deletions were detected, and they worked synergistically in a few moderate-sized scaffold antibiotics (Supplementary Table S1). Intriguingly, these antibiotics have relatively higher hydrophobicity than the other compounds tested here, indicating that hydrophobicity is another key parameter of the influx and efflux properties of foreign compounds in bamB and/or tolC deletions. Normally, as the outer membrane of Gram-negative bacteria can work as an effective barrier for hydrophobic compounds28, disturbance in the outer membrane affected the traffic of hydrophobic compounds irrespective of scaffold size.
The number of antibiotics that are newly developed and approved for clinical use has decreased consistently29, and the Infectious Diseases Society of America has stated that the development of new antibiotics against Gram-negative bacteria showing multi-drug resistance (GNMDR) is insufficient30. In addition, the effects of large-scaffold or hydrophobic antibiotics on GNMDR strains have not been studied extensively1. Therefore, an antisense-based sensitizing approach shows potential for inhibiting the growth of GNMDR strains of Pseudomonas, Acinetobacter, Enterobacteriaceae, and other bacteria. In contrast to conventional low-molecular-weight sensitizer compounds whose screening and development are labor intensive, CPP-PNA sensitizers can be easily designed and synthesized according to peptide and nucleotide sequence information. Furthermore, peptide and nucleotide sequences of CPP-PNAs can be changed easily if sensitizer-resistant mutants emerge. Previous studies demonstrated that other gene silencing agents, such as peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs) that target AcrAB-TolC components, can sensitize E. coli and P. aeruginosa to some antibiotics31,32. Otoupal et al.33 implemented CPP-PNAs targeting various genes, including tolC, to potentiate antibiotic efficacy in E. coli. Previous and present results indicated the potential of CPP-PNAs as E. coli and P. aeruginosa sensitizers for the development of future therapeutic use of many antibiotics at low concentrations34,35. For the first time, we demonstrated not only the potential of bamB as a target of antisense oligonucleotide-based techniques but also the combined effect of bamB-targeting CPP-PNAs with those targeting tolC homologs. The combined effects of CPP-PNAs targeting distinct cellular processes could contribute to the reduction in the dose of sensitizers and design of sensitizer cocktails to combat various pathogens with different antimicrobial resistance mechanisms. In the present study, CPP-PNAs worked additively in many cases (Table 2), indicating their potential for combined use. However, CPP-PNAs were ineffective in some cases; for example, 5 μM CPP-PNAs efficiently worked with vancomycin for P. aeruginosa but not for E. coli (Table 2). This difference could be attributed to variations in the effective concentrations of CPP-PNAs among species, targeting genes, and antibiotics. This dose dependency can explain the discrepancy between the results of MIC tests obtained by gene deletion (complete loss of function; Table 1) and silencing (transcriptional downregulation; Table 2). The trend of scaffold-size dependency showed that ΔbamB and ΔtolC increased the permeability of LSCs and SSCs, respectively, but this trend was reversed for some compounds in CPP-PNAs (e.g., novobiocin and fusidic acid). Likewise, although the occurrence of synergism between CPP-PNAs targeting bamB and tolC homologs was detected in only some antibiotics in E. coli (Supplementary Table S3), precise information on the effective concentration must be obtained to evaluate the interactions between CPP-PNAs with distinct target genes. Therefore, determination of the effective concentration of CPP-PNAs for each species and for each antibiotic would be a key step for using CPP-PNAs as sensitizers.
In addition to therapeutic applications, many biotechnological fields, such as drug screening and bioproduction processes, can benefit from the proposed approach that modulates uptake efficiency. Sensitized strains can serve as a strong tool for drug screening by detecting low concentrations of bactericidal compounds in natural samples (Fig. 4). Similar gene silencing techniques have also been investigated for antibiotic dereplication during screening and antibiotic adjuvant screening36,37. GETA-based antibiotic screening not only increases sensitivity but also allows for target-oriented screening and investigations into novel antibiotics targeting cellular processes which were not targeted previoursly (Fig. 4)21,23,38. GETA involving the mutants described in this study may contribute to the discovery of antibiotics with distinct modes of action from known antimicrobials. As the present mutants showed a higher uptake efficiency for an antisense agent (Fig. 3), they could be used as host cells to perform GETAs with higher sensitivity than the wild-type strain. The results could also be used as a basis for improving the uptake efficiency of bacteria for industrial applications. Active VD3 for industrial use is currently produced from cholesterol via chemical synthesis, which involves over 20 reaction steps and has a yield of < 1%39. With this ineffective production, active VD3 becomes costly. The typical price of active VD3 sold as a general-use reagent is > 100,000-fold higher than that of inactive VD324. Therefore, our study (Fig. 5) may contribute to the development of an alternative cost-effective production of active VD3.
From these industrial viewpoints, it is advantageous that the growth of cells was almost unaffected after gene deletion(s) in E. coli (Fig. 2) because this property is important to facilitate cost-effective industrial biotechnological applications. However, the CPP-PNA treatment at high concentrations negatively affected the growth of E. coli (Supplementary Fig. S1), and the CPP-PNA treatment retarded the growth of P. aeruginosa (Supplementary Fig. S2). Therefore, in the future use of CPP-PNA approach in bioproduction, this undesirable and unexpected growth-inhibiting activity should be analyzed and reduced. However, from a therapeutic viewpoint, the observed growth defect by CPP-PNAs is interesting because it can be applied to inhibit the growth of pathogens33. Several studies have demonstrated the potential of silencing agents targeting growth essential genes of pathogens as an alternative to conventional antibiotics19,40,41. The effect of growth inhibition observed in this study was milder than that of other bactericidal agents and likely more durable against the generation of resistance. Reducing pathogen growth might indirectly contribute to pathogen eradication by the immune system.
In conclusion, we implemented two approaches, namely, gene deletion and gene silencing, to improve the cellular uptake of foreign compounds with various scaffold sizes. Thus, E. coli and P. aeruginosa became sensitized to various antimicrobial compounds, and an industrially important compound in E. coli could be converted. The two approaches could be useful for therapeutics and many biotechnology fields. They might be further utilized for constructing sensitive cell-based biosensors to detect environmental compounds at low concentrations (e.g., endocrine disruptor detection) or for conducting efficient bioremediation (e.g., pollutant degradation). Further analysis on the detailed mechanisms of the improved uptake (e.g., direct microscopic observation) would contribute to future applications of these two approaches in medical and biotechnological fields.
Methods
General genetic manipulations
Escherichia coli MG1655 (wild-type strain; National BioResource Project, Mishima, Japan) and its derivative strains were used in the study. All strains were cultured in Luria–Bertani broth (LB; 10 g/L Difco Bacto tryptone, 5 g/L Difco yeast extract, 10 g/L NaCl) in the presence and absence of appropriate antibiotics at 37 °C with shaking unless otherwise stated. Mueller–Hinton broth (MHB) was purchased from BD Biosciences (Detroit, MI, USA). The composition of the M9AFC medium was as follows: Na2HPO4·12H2O (17 g/L), KH2PO4 (3 g/L), NaCl (0.5 g/L), NH4Cl (1 g/L), MgSO4·7H2O (0.49 g/L), CaCl2·2H2O (0.015 g/L), FeSO4·7H2O (0.0083 g/L), thiamine-HCl (0.01 g/L), glucose (1 g/L), 5-amino levulinic acid (0.08 g/L), and casamino acid (10 g/L). Plasmids were constructed as described in “Supplementary Methods S1”, and genes were deleted in accordance with previously described methods42. CPP-PNAs used in this study are shown in Table 3. All CPP-PNAs were synthesized by Panagene Inc. (Daejeon, South Korea), and a stock solution (200 μM) was prepared using distilled water.
Table 3.
List of CPP-PNAs used in this study.
| CPP-PNA | Target mRNA | Sequencec | References |
|---|---|---|---|
| KFF-acpP | E. coli acpP | KFFKFFKFFK-O_linker-ctcatactct | 19 |
| KFF-NC | Nonea | KFFKFFKFFK-O_linker-atactaacag | This study |
| KFF-bamB | E. coli bamB | KFFKFFKFFK-O_linker-catcgggtcc | This study |
| KFF-tolC | E. coli tolC | KFFKFFKFFK-O_linker-tgcattcctt | This study |
| RXR-NC | Noneb | RXRRXRRXRRXRXB-ctacttatgcd | This study |
| RXR-bamB | P. aeruginosa bamB | RXRRXRRXRRXRXB-catatcattg | This study |
| RXR-oprM | P. aeruginosa oprM | RXRRXRRXRRXRXB-tcaggcctct | This study |
CPP-PNA, cell-penetrating peptide conjugate of peptide nucleic acid; acpP, a gene for acyl carrier protein AcpP; bamB, a gene for outer membrane lipoprotein BamB; tolC, a gene for outer membrane efflux protein TolC; oprM, a gene for outer membrane protein OprM.
aNo match with E. coli MG1655 genome as antisense.
bNo match with P. aeruginosa PAO1 genome as antisense.
cUppercase and lowercase letters indicate peptide and PNA sequences, respectively, and PNA sequences are shown from the N-terminus to the C-terminus. The O_linker portion is composed of polyethylene glycol.
d X and B indicate 6-aminohexanoic acid and beta-alanine residues, respectively.
Influx and efflux experiments using NPN
The influx and efflux rates of NPN were measured as described previously43. The detailed protocol is described in “Supplementary Methods S1”.
Monitoring of E. coli and P. aeruginosa growth and measurement of MIC
Microbial cultures were prepared by diluting an overnight preculture in MHB and cultured in a well of a 96-well clear microtiter plate (Coaster, Cambridge, CA, USA; Product No. 3997). All antibiotics were tested in 2-fold serial dilutions, and the total culture volume was adjusted to 150 μL. Other conditions for MIC determination were applied as described by Muheim1. Water was used as the solvent for preparing stock solutions of ampicillin and kanamycin. NaOH solution (10 mM), ethanol (50%), absolute ethanol, and dimethyl sulfoxide were used to prepare nalidixic acid, tetracycline, chloramphenicol, and other antibiotics, respectively. The turbidity of the culture at 600 nm was measured using a Multiskan Sky microplate reader (Thermo Fisher Scientific, Foster City, CA, USA) every 1 h.
Pseudomonas aeruginosa strain PAO1 was used in this study. MIC was measured as described earlier except for the total culture volume (100 μL), incubation (Thermomixer, Eppendorf, Germany), and bacterial growth determination (visual observation). Water was used as the solvent for preparing stocks of vancomycin and carbenicillin, and ethanol (100%) was used to prepare erythromycin. RXR-bamB and RXR-oprM (Table 3) were synthesized44,45 and used in a similar manner to experiments involving E. coli. The effect of CPP-PNAs on the growth of PAO1 was assessed by culturing the strain in MHB containing CPP-PNAs (3 μM) at 37 °C with shaking; turbidity was measured using a HiTS microplate reader (Scinics Co., Tokyo, Japan). All MIC tests were repeated at least two times.
VD3 conversion experiment
A VD3 conversion experiment and inactive and active VD3 quantification were performed as described previously8,46. In brief, wild-type and mutant strains were transformed with pHN4136 to express vdh-aciBC or with pHN1387, a control empty vector (“Supplementary Methods S1”). The transformant was then cultured in the M9AFC medium. Because VD3 is insoluble in water, partially methylated beta-cyclodextrin (2 g/L) and inactive VD3 solution (0.1 mM; stock solution dissolved in dimethyl sulfoxide) were added to the reaction buffer.
Detection of antibiotic compounds at low concentrations
A black soil sample was collected from Fukuzumi Ogawa Park (Sapporo, Japan), and 1.5 g of soil was cultured in 20 mL of M9 (17 g/L Na2HPO4·12H2O, 3 g/L KH2PO4, 0.5 g/L NaCl, 1 g/L NH4Cl, 0.49 g/L MgSO4·7H2O, 0.015 g/L CaCl2·2H2O, 0.0083 g/L FeSO4·7H2O, 0.01 g/L thiamine-HCl, and 10 g/L glucose) or R2A medium (Becton Dickinson and Company, Detroit, MI). The sample was left at 25 °C for 7 days, and 1 mL of the soil suspension was extracted using 1 mL of ethyl acetate. The resulting supernatant was added to a new 50 mL conical tube. This supernatant was dried at room temperature overnight, and the dried sample was dissolved in 33 μL of dimethyl sulfoxide. A halo formation assay was performed on an agar plate22.
Gene expression titration assay
5-Ketoclomazone was used as a test antibiotic compound because its uptake by the wild-type E. coli strain was low (Table 1). The cellular target molecule of 5-ketoclomazone has been considered to be 1-deoxy-d-xylulose 5-phosphate synthase (Dxs), which catalyzes the first reaction step of the methyl-d-erythritol 4-phosphate pathway47,48; however, it remains unclear whether Dxs is the actual target, especially in vivo49. As an antisense agent, a short antisense RNA against dxs was expressed endogenously using an expression vector22. In a negative control experiment, triclosan and fusidic acid, which are antibiotics unrelated to Dxs, were used. The wild-type and mutant strains were transformed with pHN4165 to express an antisense RNA against dxs mRNA or with pHN1257, a control empty vector (“Supplementary Methods S1”). An overnight preculture of the transformant was prepared, and 50 μL of the culture was mixed with 11 mL of autoclaved soft LB agar (1.2% agarose and 11 μL of 1 M isopropyl β-d-1-thiogalactopyranoside) and cooled to approximately 40 °C. After solidification, the antibiotic solutions (2.5 μL) were spotted on the plates at appropriate concentrations.
Supplementary Information
Acknowledgements
We would like to thank Mr. Johnathan Adriel, Mr. Davin Saviro Wijaya, Mr. Andrew Steven Liyanto, Ms. Masako Sato, Ms. Ayane Osanai, and Ms. Miyako Hata for their assistance. We would also like to express our gratitude to our research group members for their help and valuable discussions.
Author contributions
K.Y., N.Y., and N.N. designed and performed the experiments. Y.Y. and N.N. designed the plasmids. K.Y., S.A., Y.Y., and N.N. analyzed the data and wrote the manuscript. K.I. performed statistical analysis and revised the manuscript. All authors reviewed the manuscript. N.N. provided funding.
Data availability
No datasets were generated or analyzed in this study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-09635-6.
References
- 1.Muheim C, et al. Increasing the permeability of Escherichia coli using MAC13243. Sci. Rep. 2017;7:17629. doi: 10.1038/s41598-017-17772-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van den Berg B, Prathyusha Bhamidimarri S, Dahyabhai Prajapati J, Kleinekathöfer U, Winterhalter M. Outer-membrane translocation of bulky small molecules by passive diffusion. Proc. Natl Acad. Sci. USA. 2015;112:E2991–E2999. doi: 10.1073/pnas.1424835112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van der Hoek SA, Borodina I. Transporter engineering in microbial cell factories: The ins, the outs, and the in-betweens. Curr. Opin. Biotechnol. 2020;66:186–194. doi: 10.1016/j.copbio.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016 doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vuorio R, Vaara M. The lipid A biosynthesis mutation lpxA2 of Escherichia coli results in drastic antibiotic supersusceptibility. Antimicrob. Agents Chemother. 1992;36:826–829. doi: 10.1128/AAC.36.4.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu A, et al. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: Generating an antibiotic bar code. Antimicrob. Agents Chemother. 2010;54:1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pérez-García F, Wendisch VF. Transport and metabolic engineering of the cell factory Corynebacterium glutamicum. FEMS Microbiol. Lett. 2018;365:fny166. doi: 10.1093/femsle/fny166. [DOI] [PubMed] [Google Scholar]
- 8.Fujii T, Fujii Y, Machida K, Ochiai A, Ito M. Efficient biotransformations using Escherichia coli with tolC acrAB mutations expressing cytochrome P450 genes. Biosci. Biotechnol. Biochem. 2009;73:805–810. doi: 10.1271/bbb.80627. [DOI] [PubMed] [Google Scholar]
- 9.Vaara M. Polymyxin derivatives that sensitize Gram-negative bacteria to other antibiotics. Molecules. 2019;24:249. doi: 10.3390/molecules24020249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim KH, Aulakh S, Paetzel M. The bacterial outer membrane β-barrel assembly machinery. Protein Sci. 2012;21:751–768. doi: 10.1002/pro.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malinverni JC, Silhavy TJ. Assembly of outer membrane β-barrel proteins: The Bam complex. EcoSal Plus. 2011;4:1. doi: 10.1128/ecosalplus.4.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen C. Channel-tunnels: Outer membrane components of type I secretion systems and multidrug efflux pumps of Gram-negative bacteria. Rev. Physiol. Biochem. Pharmacol. 2003;147:122–165. doi: 10.1007/s10254-003-0008-y. [DOI] [PubMed] [Google Scholar]
- 13.Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β-barrel assembly machinery complex. J. Mol. Biol. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 14.Ricci DP, Silhavy TJ. The Bam machine: A molecular cooper. Biochim. Biophys. Acta. 2012;1818:1067–1084. doi: 10.1016/j.bbamem.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du D, et al. Structure of the AcrAB–TolC multidrug efflux pump. Nature. 2014;509:512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sharma C, Awasthi SK. Versatility of peptide nucleic acids (PNAs): Role in chemical biology, drug discovery, and origins of life. Chem. Biol. Drug Des. 2017;89:16–37. doi: 10.1111/cbdd.12833. [DOI] [PubMed] [Google Scholar]
- 17.Loh B, Grant C, Hancock RE. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984;26:546–551. doi: 10.1128/AAC.26.4.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battu SK, et al. Physicochemical characterization of berberine chloride: A perspective in the development of a solution dosage form for oral delivery. AAPS PharmSciTech. 2010;11:1466–1475. doi: 10.1208/s12249-010-9520-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Good L, Awasthi SK, Dryselius R, Larsson O, Nielsen PE. Bactericidal antisense effects of peptide-PNA conjugates. Nat. Biotechnol. 2001;19:360–364. doi: 10.1038/86753. [DOI] [PubMed] [Google Scholar]
- 20.Singh SB, Phillips JW, Wang J. Highly sensitive target-based whole-cell antibacterial discovery strategy by antisense RNA silencing. Curr. Opin. Drug Discov. Devel. 2007;10:160–166. [PubMed] [Google Scholar]
- 21.Moir DT, Opperman TJ, Butler MM, Bowlin TL. New classes of antibiotics. Curr. Opin. Pharmacol. 2012;12:535–544. doi: 10.1016/j.coph.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 22.Matsue Y, et al. The herbicide ketoclomazone inhibits 1-deoxy-D-xylulose 5-phosphate synthase in the 2-C-methyl-d-erythritol 4-phosphate pathway and shows antibacterial activity against Haemophilus influenzae. J. Antibiot. (Tokyo) 2010;63:583–588. doi: 10.1038/ja.2010.100. [DOI] [PubMed] [Google Scholar]
- 23.Nakashima N, Tamura T. Conditional gene silencing of multiple genes with antisense RNAs and generation of a mutator strain of Escherichia coli. Nucleic Acids Res. 2009;37:e103. doi: 10.1093/nar/gkp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yasutake Y, Tamura T. Efficient production of active form of vitamin D3 by microbial conversion. Synthesiology. 2011;4:222–229. doi: 10.5571/synth.4.222. [DOI] [Google Scholar]
- 25.Kang DJ, Im JH, Kang JH, Kim KH. Bioconversion of vitamin D3 to calcifediol by using resting cells of Pseudonocardia sp. Biotechnol. Lett. 2015;37:1895–1904. doi: 10.1007/s10529-015-1862-9. [DOI] [PubMed] [Google Scholar]
- 26.Krishnamoorthy G, et al. Breaking the permeability barrier of Escherichia coli by controlled hyperporination of the outer membrane. Antimicrob. Agents Chemother. 2016;60:7372–7381. doi: 10.1128/AAC.01882-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Charlson ES, Werner JN, Misra R. Differential effects of yfgL mutation on Escherichia coli outer membrane proteins and lipopolysaccharide. J. Bacteriol. 2006;188:7186–7194. doi: 10.1128/JB.00571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassetti M, Merelli M, Temperoni C, Astilean A. New antibiotics for bad bugs: Where are we? Ann. Clin. Microbiol. Antimicrob. 2013;12:22. doi: 10.1186/1476-0711-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lushniak BD. Antibiotic resistance: A public health crisis. Public Health Rep. 2014;129:314–316. doi: 10.1177/003335491412900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sturge CR, et al. AcrAB-TolC inhibition by peptide-conjugated Phosphorodiamidate morpholino oligomers restores antibiotic activity in vitro and in vivo. ACS Infect. Dis. 2019;5:1446–1455. doi: 10.1021/acsinfecdis.9b00123. [DOI] [PubMed] [Google Scholar]
- 32.Ayhan DH, et al. Sequence-specific targeting of bacterial resistance genes increases antibiotic efficacy. PLOS Biol. 2016;14:e1002552. doi: 10.1371/journal.pbio.1002552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otoupal PB, et al. Potentiating antibiotic efficacy via perturbation of non-essential gene expression. Commun. Biol. 2021;4:1267. doi: 10.1038/s42003-021-02783-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gadelii, A., Hassan, K.-O. & Hakansson, A. P. Sensitizing agents to restore antibiotic resistance. In Capelo-Martínez, JL. & Igrejas, G. (eds.) Antibiotic Drug Resistance 429–452 (Wiley, 2019).
- 35.Wright GD. Antibiotic adjuvants: Rescuing antibiotics from resistance. Trends Microbiol. 2016;24:862–871. doi: 10.1016/j.tim.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 36.Cox G, et al. A common platform for antibiotic dereplication and adjuvant discovery. Cell Chem. Biol. 2017;24:98–109. doi: 10.1016/j.chembiol.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 37.King AM, et al. Aspergillomarasmine A overcomes metallo-β-lactamase antibiotic resistance. Nature. 2014;510:503–506. doi: 10.1038/nature13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patil SD, Sharma R, Srivastava S, Navani NK, Pathania R. Downregulation of yidC in Escherichia coli by antisense RNA expression results in sensitization to antibacterial essential oils eugenol and carvacrol. PLoS ONE. 2013;8:e57370. doi: 10.1371/journal.pone.0057370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu G-D, Okamura WH. Synthesis of vitamin D (calciferol) Chem. Rev. 1995;95:1877–1952. doi: 10.1021/cr00038a007. [DOI] [Google Scholar]
- 40.Geller BL, Deere J, Tilley L, Iversen PL. Antisense phosphorodiamidate morpholino oligomer inhibits viability of Escherichia coli in pure culture and in mouse peritonitis. J. Antimicrob. Chemother. 2005;55:983–988. doi: 10.1093/jac/dki129. [DOI] [PubMed] [Google Scholar]
- 41.Greenberg DE, et al. Antisense phosphorodiamidate morpholino oligomers targeted to an essential gene inhibit Burkholderia cepacia complex. J. Infect. Dis. 2010;201:1822–1830. doi: 10.1086/652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakashima N, Miyazaki K. Bacterial cellular engineering by genome editing and gene silencing. Int. J. Mol. Sci. 2014;15:2773–2793. doi: 10.3390/ijms15022773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Misra R, Morrison KD, Cho HJ, Khuu T. Importance of real-time assays to distinguish multidrug efflux pump-inhibiting and outer membrane-destabilizing activities in Escherichia coli. J. Bacteriol. 2015;197:2479–2488. doi: 10.1128/JB.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang H, et al. oprM as a new target for reversion of multidrug resistance in Pseudomonas aeruginosa by antisense phosphorothioate oligodeoxynucleotides. FEMS Immunol. Med. Microbiol. 2010;60:275–282. doi: 10.1111/j.1574-695X.2010.00742.x. [DOI] [PubMed] [Google Scholar]
- 45.Ghosal A, Nielsen PE. Potent antibacterial antisense peptide-peptide nucleic acid conjugates against Pseudomonas aeruginosa. Nucleic Acids Ther. 2012;22:323–334. doi: 10.1089/nat.2012.0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasutake Y, Nishioka T, Imoto N, Tamura T. A single mutation at the ferredoxin Binding Site of P450 Vdh enables efficient biocatalytic production of 25-hydroxyvitamin D(3) J. Chem. Biol. 2013;14:2284–2291. doi: 10.1002/cbic.201300386. [DOI] [PubMed] [Google Scholar]
- 47.Ferhatoglu Y, Barrett M. Studies of clomazone mode of action. Pestic. Biochem. Physiol. 2006;85:7–14. doi: 10.1016/j.pestbp.2005.10.002. [DOI] [Google Scholar]
- 48.Mueller C, Schwender J, Zeidler J, Lichtenthaler HK. Properties and inhibition of the first two enzymes of the non-mevalonate pathway of isoprenoid biosynthesis. Biochem. Soc. Trans. 2000;28:792–793. doi: 10.1042/bst0280792. [DOI] [PubMed] [Google Scholar]
- 49.Han M, et al. Enzyme inhibitor studies reveal complex control of methyl-d-erythritol 174-phosphate (MEP) pathway enzyme expression in Catharanthus roseus. PLoS ONE. 2013;8:e62467. doi: 10.1371/journal.pone.0062467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed in this study.





