Abstract
Background
Considerable evidence implicates myeloid-derived suppressor cells (MDSCs) promote tumor progression and drug resistance. Sorafenib is the standard first-line therapy for advanced hepatocellular carcinoma (HCC). Clinical evidence indicates that sorafenib resistance is associated with increased MDSCs, by which MDSCs exerts these effects is obscure. This study aimed to investigate the mechanism of sorafenib resistance mediated by MDSCs.
Methods
A syngeneic mouse-liver cancer cell line BNL was subcutaneously injected to build a tumor-bearing mouse model, and syngeneic MDSCs were adoptive transferred into the tumor-bearing mouse. Tumor tissue was obtained, and transcriptomic analysis of the tumor was carried out on RNAseq data. A coculture system was used to verify the crosstalk between MDSCs and BNL cells.
Results
Adoptive MDSCs transfer into tumor-bearing mice induced an increase of tumor-infiltrating MDSCs, which led to tumor growth and impaired antitumor activity of sorafenib in BNL HCC models. MDSCs transfer contributed to tumor fibrosis and tumor-associated fibroblast (CAF) activation, associated with fibroblast growth factor (FGF1) upregulation. In contrast, MDSC depletion by anti-Ly6G+ reduced fibrosis and increased sorafenib antitumor efficacy. Intriguingly, tumor-infiltrating MDSCs barely expressed FGF1. IL-6 derived from MDSCs increased FGF1 expression in BNL liver cancer cells, and anti-IL-6 attenuated this effect in vitro. MAPK pathway, one of the sorafenib targets, is the downstream signaling of FGF1 and is reactivated by MDSCs-mediated FGF1 upregulation.
Conclusions
Our finding demonstrated that MDSCs led to tumor growth and sorafenib resistance via FGF1 upregulation and subsequent indirect CAF activation. We offered a novel mechanism of MDSCs-driven HCC progression and sorafenib resistance.
Keywords: Myeloid cells, Hepatocellular carcinoma, Sorafenib, Fibroblast cells, Combination therapy
Abbreviations: CAF, tumor-associated fibroblast; Col, collagen formation; CXCR4, C-X-C receptor type 4; ECM, extracellular matrix; FGF1, fibroblast growth factor 1; HCC, hepatocellular carcinoma; H&E, hematoxylin-eosin staining; IF, immunofluorescence; IHC, immunohistochemistry; LAMA1, laminin alpha 1; MAPK, Mitogen-activated protein kinase; MDSC, Myeloid-derived suppressor cells; PDGFR, platelet-derived growth factor receptor; pERK, phosphorylated extracellular regulated protein kinases; SDF, stromal-derived factor 1; TME, tumor microenvironment; THBS 1, thrombospondin-1; VEGFR, vascular endothelial growth factor receptor; VTN, vitronectin; VWF, von willebrand factor
Introduction
Hepatocellular carcinoma (HCC) represents the sixth most commonly diagnosed cancer and the third leading cause of cancer-related mortality [1]. Furthermore, due to lack of effective treatment and frequent resistance, the long-term survival rate has not been significantly improved, further emphasizing the urgent need to deepen our understanding of HCC progression and drug resistance. The tumor microenvironment (TME) comprises immune cells, stromal cells, fibroblasts and endothelial cells, which plays a complex and multidirectional interplay in remodeling characteristics of tumor cells. Therefore, understanding the interaction mechanism between nontumor cells and tumor cells is essential.
Myeloid-derived suppressor cells (MDSCs), a heterogeneous population of immature myeloid cells, induce local and possibly systemic immunosuppression and tumor progression [2]. There are two different types of MDSCs, as identified in studies in both mice and humans: polymorphonuclear MDSC (PMN-MDSC) are phenotypically similar to neutrophils, whereas monocytic MDSC (M-MDSC) are similar to monocytes. The phenotypic characteristics of murine MDSCs have been described CD11b+Ly6G+(PMN-MDSC) and CD11b+Ly6G+(M-MDSC). With potent immunosuppressive activity, both PNM-MDSC and M-MDSC have the ability to support tumor progression, although differences in the phenotypic characteristics. MDSCs correlate with early recurrence and poor prognosis in patients with HCC who underwent curative resection [3], radiotherapy [4], and hepatic arterial infusion chemotherapy [5]. Accompanied by tumor progression, MDSCs were acceleratively expanded, activated and recruited by the molecular from TME [2]. MDSCs drive HCC progression by suppressing CD4+T cells, CD8+ T cells, natural killer cells and the paracrine release of cytokines and chemokines with angiogenesis and recruitment [6]. Little attention, however, has been devoted to analyzing the direct effect of MDSCs on the tumor cells.
Although it offers very moderate survival improvements, Sorafenib is one of the most effective single-drug therapies for advanced HCC [7]. Sorafenib inhibits Raf, as well as the kinase activity of vascular endothelial growth factor receptor (VEGFR) and platelet-derived growth factor receptor (PDGFR) led to antiangiogenic effects. Immunotherapy is promising but depends on combination regimens which has a lower cost-effectiveness [8,9]. Therefore, it is crucial to understand the molecular mechanisms mediating sorafenib resistance to improve clinical outcomes for HCC patients. Here, we investigated the mechanism of MDSC-driven HCC progression and sorafenib resistance.
Methods and materials
Cell line and cell culture
BNL 1ME A.7R.1 (referred as BNL in this study), a balb/c mouse-derived HCC cell lines, were procured through the Shanghai Xin Yu Biotechnology(Shanghai, China) and were routinely maintained. They were tested for mycoplasma contamination by a Mycoplasma Detection Kit. MDSCs was isolated from the peripheral blood of BNL-bearing mice (21 days after inoculation). Then the purity of MDSCs were identified by flow cytometry analysis (FCAS)(>90% purity, defined by CD11b+Gr1+MDSCs). The expression of representative effector molecules of MDSCs Arginase1(Arg1), TGF-β, IL-6 (Fig. S1) and reactive oxygen species(ROS) (Fig. S2) were detected. To effectively simulate the tumor microenvironment, BNL were noncontact cocultured with MDSCs isolated from PBMC of tumor-bearing congenetic mice through Transwell kit (Corning #3450). In particular, BNL cells(2 × 105cells/well) were suspended in 1.5 ml serum-free medium in the top of insert well, and 5 × 104/well MDSCs were seeded in the lower chamber in 2.6 ml complete medium supplemented with 20 ng/ml GM-CSF. After 48 h cultivation, BNL cells were treated with DMSO or sorafenib 5 μM for 24 h.
Animals
Male balb/c mice (6 weeks) were purchased from the Animal Center of Southern Medical University and were maintained under specific pathogen-free conditions. Additional information on methods is available in the supplement.
Flow cytometric analysis
Single cells were stained by flueorecence-conjugated antibodies (Biolegend) as previously described [10]. Stained cells were analyzed using the FACS Analyzer (FACS Calibur, BD). The results were analyzed with FlowJo10.0 software. Gating strategy for the MDSC subsets are presented in Fig. S3. Additional information on methods is available in the supplement.
Hematoxylin-eosin staining(H&E), Masson's trichrome staining, immunohistochemistry(IHC), immunofluorescence(IF)
HE staining was conducted according to standard protocols. Masson's trichrome staining were performed by kit(Pythonbio, China)according to the instruction. For IHC, the slices were disposed by ElivisionTM plus Polyer HP (Mouse) IHC Kit(MXB, China) and incubated with CD31 (1:2000, abcam ab182981). For immunofluorescence, the tumor tissue slices were incubated with α—SMA(1:500, abcam ab124964) and VEGFA primary antibody (1:500, abcam ab52917) and with Alexa Fluor 488/Dylight-594 conjugated secondary antibody (1:400 Abbkine, Wuhan, China).
RNA-seq
Total RNA was extracted from the frozen tumor tissue using TRIzol® Reagent following the manufacturer's instructions (Invitrogen). Genomic DNA was removed by DNase I (TaKara Jan). RNA quality was examined by 2100 Bioanalyser (Agilent) and quantified using the ND-2000 (NanoDrop Technologies). The methods of library preparation, Illumina Hiseq xten/Nova seq 6000 Sequencing, read and Differential expression and functional enrichment analysis were described in the supplement.
Western blotting and qRT-PCR
The protein of cells and tumor tissue were extracted, quantified, separated, transferred and blocked as described previously [11]. The membranes were incubated with primary antibodies (β-actin, Affinity; GAPDH, Proteintech; FGF1, Ras, ERK, pERK, VEGFA, PDGFB and α-SMA, Abcam) overnight at 4 °C. Protein bands were quantified using ImageJ software with β-actin or GAPDH as the internal control. For qRT-PCR, total RNA was extracted from cells and tumor specimens using trizol reagent (RightGene, Guangzhou, China) following the manufacturer's instructions. The expression of mRNA was measured via qRT-PCR using a SYBR PrimeScript RT-PCR Kit (Takara Bio, Shiga, Japan) in accordance with the manufacturer's instructions. GAPDH was used as an internal control. Primer sequences for various genes are listed in Table 1.
Table 1.
Sequence of primers used in real-time experiments.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| IL-6 | TAGTCCTTCCTACCCCAATTTCC | TTGGTCCTTAGCCACTCCTTC |
| TGFβ | CTCCCGTGGCTTCTAGTGC | GCCTTAGTTTGGACAGGATCTG |
| Arg1 | CTCCAAGCCAAAGTCCTTAGAG | AGGAGCTGTCATTAGGGACATC |
| FGF1 | CCCTGACCGAGAGGTTCAAC | GTCCCTTGTCCCATCCACG |
| PDGF | CATCCGCTCCTTTGATGATCTT | GTGCTCGGGTCATGTTCAAGT |
| GAPDH | AGGTCGGTGTGAACGGATTTG | TGTAGACCATGTAGTTGAGGTCA |
MILLIPLEX® MAP array
Cytokines in the supernatant of cell culture were measured by Mouse High Sensitivity.
Magnetic Bead Panel MILLIPLEX® MAP kit (Merck, Germany) following the manufacturer's instructions.
Statistical analysis
The variances between the groups that are being statistically compared were similar. Quantitative analyses were done using Graphpad prsim 5 (GraphPad Software, Inc. USA). Bubble chart and heatmap was carried out in R software. For experiment in vitro, all data were collected from at least three independent experiments. All data was expressed as mean ± SEM. ANOVA and t-test were used when data accorded with normal distribution and homogeneity of variance. Tukey test was used multiple comparisons. A P-value < 0.05 indicated that the difference was statistically significant.
Results
MDSC promoted tumor growth and attenuated the efficacy of sorafenib in vivo
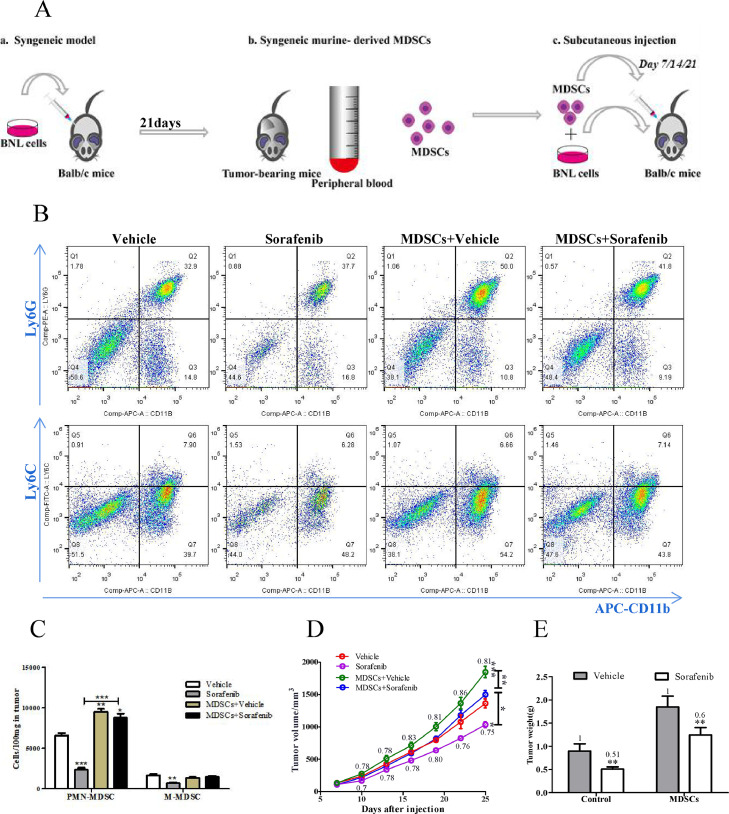
We tested the effect of MDSC in a murine subcutaneous HCC model using BNL HCC cells. Syngeneic MDSCs were generated from peripheral blood of tumor-bearing mice. They were initiated coinjection with BNL cells (1:10) and reinjected them into the tumor at the same amount weekly for three weeks (Fig. 1A). To confirm the mice were successfully transferred MDSC, we examined the number of MDSC in the tumor by Flow cytometric analysis. PMN-MDSC(CD11b+Ly6G+) was significantly increased in tumors of mice with transferred MDSCs, while the number of M-MDSC(CD11b+Ly6C+) was unchanged (Fig. 1B,C,P<0.0001), suggesting that transferred MDSCs promoted PMN-MDSC infiltrating in tumor. In contrast, a reduction of MDSCs was observed in the tumor of mice with sorafenib treatment. Then we analyzed the tumor growth based on tumor volume and weight. As shown in Fig. 1D and E, sorafenib significantly suppressed tumor growth (P = 0.0071) but caused only marginal inhibition in mice transferred MDSCs(P = 0.5097).
Fig. 1.
MDSC promoted tumor growth and attenuated the efficacy of sorafenib in vivo. (A) Schematic diagram of the establishment of MDSCs transfer model. In short, homologous HCC cell line BNL(3 × 106/100 μL) was subcutaneously injected in balb/c mice to build tumor – bearing mice model. 3 weeks later, MDSCs were isolated from the PBMC of tumor bearing mice. Then, MDSCs were mixed with BNL cells and co-injected subcutaneously. MDSCs were injected into the tumor at the indicated dosage weekly for 3 consecutive weeks. (B)Representative flow cytometric plot of PMN-MDSC and M-MDSC. (C)Quantification of the number of fluorescently labelled MDSC from 100mg tumor tissue(n = 3). (D)Tumor volume was measured using a Vernier caliper, V = a × *b2/2, where a is the long diameter and b is the short diameter(n = 5, two - way ANOVA). (E) Tumor weight at experimental end stage(n = 5). (C), (D), (E) MDSCs condition was normalized to the control condition. Data are reported as mean ± SE. Specific n values of biological independent animals. ANOVA for four groups comparisons and Tukey test for multiple comparisons. Asterisks indicate comparison with the control group. *p < 0.05, **p < 0.01, ***p < 0.001.
MDSCs boosted synthesis of extracellular matrix (ECM)
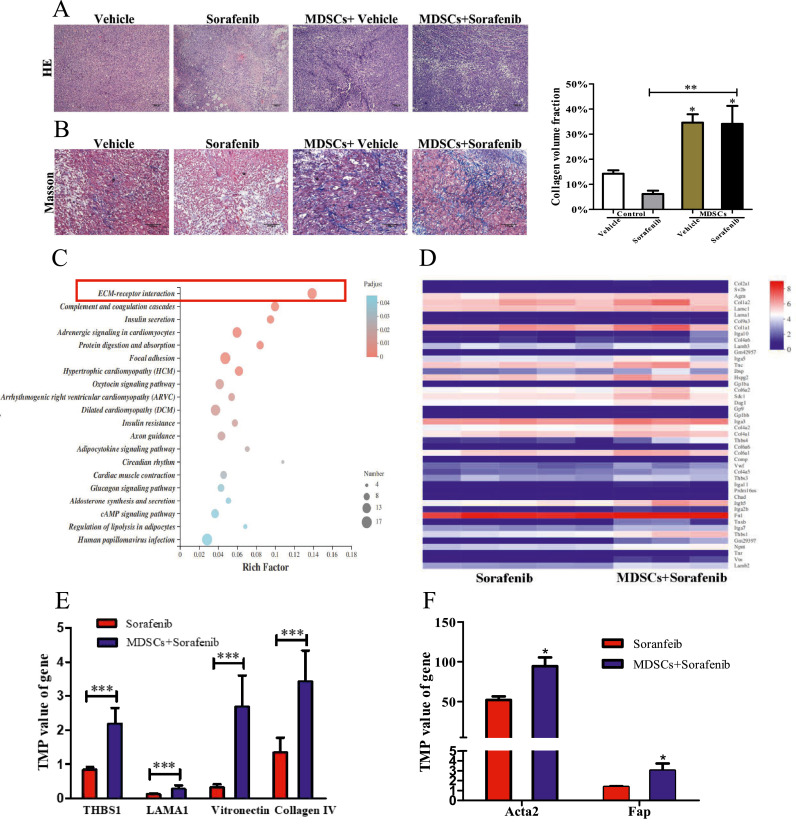
For mechanistic insights, the histopathology of tumor tissue was examined. With H&E staining, we noted a dramatic increase of fibrosis in the tumor from mice with transferred MDSCs (Fig. 2A). In the tumor of mice with transfer MDSCs, we observed a large number of collagen fibers diffused around the tumor cells by Masson staining. Similar phenomena were also observed in the mice transferred MDSCs and treated with sorafenib (Fig. 2B), indicating that MDSCs might induce tumor fibrosis. To further understand this finding, we employed RNA-seq to analyze the gene expression profile of tumors from two major groups’ mice(treated with sorafenib and transferred with MDSCs vs. non-treated by sorafenib). As a result, the Kyoto encyclopedia of genes and genomes enrichment analysis showed significant enrichment for genes of the ECM-receptor interaction (Fig. 2C). And genes upregulated were significantly enriched for signatures related to collagen formation (Col), ECM organization (Col, Thbs, Lam, Vtn, Vwf), and cell motility (ITGA) (Fig. 2D). Genes encoding ECM-related proteins, including thrombospondin-1 (THBS 1), laminin alpha 1 (LAMA1), vitronection and collagen Ⅳ were significantly enriched in tumor transferred MDSCs(Fig. 2E P = 0.0003). CAF is the synthetic machine of ECM [12], the genes of whose defining markers α-SMA(encode by Acta2 gene) and FAP were also enriched in the MDSC+Sorafenib group [12,13]. Overall, these data suggested transferred MDSCs could cause the synthesis of ECM.
Fig. 2.
MDSCs boosted synthesis of extracellular matrix(ECM). (A) Representative images of H&E staining of tumor(Scale bar = 200 μm). (B) Masson staining of tumor tissue, collagen fibers were stained in blue(Scale bar = 100 μm). Collagen volume fraction was analyzed by ImageJ(n = 3). (C) Bubble chart showed kyoto encyclopedia of genes and genomes enrichment analysis for the differential expression of genes between Sorafenib group and MDSCs+Sorafenib group. (D) Heatmap displaying quantification of ECM-receptor interaction gene. (E) The gene expression of THBSI, LAMA1, vitronection and collagen Ⅳ, which encode ECM. (F) The gene expression of Acta2(encode α-SMA protein) and Fap, which are the makers of CAFs. Data are expressed as mean ± SEM of n = 5 or 3 independent animals. t – test for two groups comparisons. *p < 0.05, ***p < 0.001.
MDSCs induced CAF activation by upregulating FGF1 levels
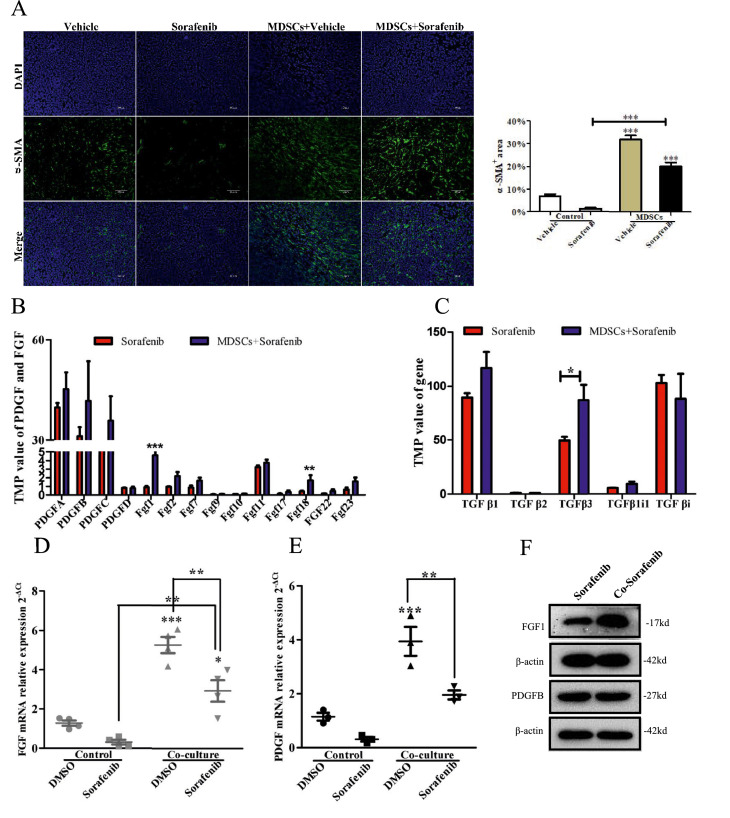
Next, we examined the expression of α-SMA in tumor tissue, and the mice transferred MDSCs expressed increased α-SMA versus controls (Fig. 3A P<0.001). TGFβ, platelet-derived growth factor (PDGF), and FGF are reported the key mediators of CAF activation [13]. We examined the expression of FGF, TGFβ, and PDGF in tumor. The transcript expression level of FGF(FGF1, FGF18) significantly upregulated, whereas PDGF and TGFβ of that did not show an observable difference in mice transferred MDSCs (Fig. 3B,C). This result was further verified in vitro by the coculture of MDSC and BNL (Fig. 3D,E,F). Taken together, we demonstrated that MDSCs might induce CAF activation, which was associated with FGF1 upregulation.
Fig. 3.
MDSCs induced CAF activation by upregulating FGF1 levels. (A) α-SMA immunostaining images of tumor tissues(Scale bar = 100μm). Positive area was analyzed by ImageJ(n = 3 mice). (B), (C) The TMP value of PDGF, FGF1 and TGFβ mRNA in tumor tissue(n = 5 mice). (D) qRT-PCR analyses for FGF1 in the BNL cells(n = 4). (E) qRT-PCR analyses for PDGF in the BNL cells(n = 3). (F)WB for FGF1 and PDGFB in BNL cells(n = 3). Specific n values of biological independent experiments. t – test for two groups comparisons. ANOVA for four groups comparisons and Tukey test for multiple comparisons. Asterisks indicate comparison with the control group. *p < 0.05, **p<0.01***, p < 0.001.
IL-6 was a potential mediator of cross-talk between BNL HCC cells and MDSCs
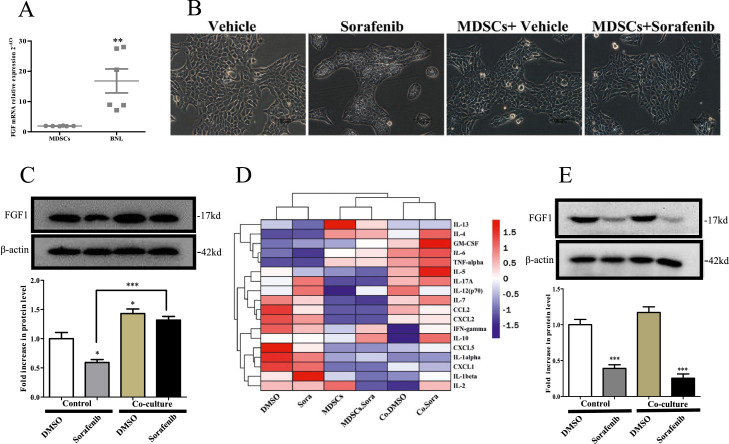
We next questioned whether MDSCs secreted FGF1 and activated CAF based on the findings described above.It was consistent with previous studies, we found that FGF was hardly expressed in MDSC but in BNL (Fig. 4A) [14]. Emerging research suggests that tumor cells are inescapably responsible for CAF activation and proliferation, but the exact mechanism is unclear [15]. We presumed that MDSC-induced FGF1 upregulation of BNL tumor cells drove CAFs activation. To verify this inference, BNL were cultured in the presence (coculture) or absence of MDSC for 48h. As shown in Fig. 4B, the BNL cells cocultured with MDSC showed altered morphology compared to the control. Then, FGF1 expression of BNL cells was analyzed. Sorafenib decreased FGF1 expression, which could be partly reversed by the presence of MDSC (Fig. 4C P = 0.0002). To understand the mediator contributing FGF1 upregulation, we studied cytokines upon coculture system and found IL-6, one cytokine that can be secreted by MDSCs, significantly increased in the coculture system and MDSC supernatant (Fig. 4D). As shown in Fig. 4E, the IL-6 neutralizing antibody (10 ng/mL) blocked cells from MDSC-induced FGF1 upregulation (P<0.0001).
Fig. 4.
IL-6 was a potential mediator of the cross-talk between BNL cells and MDSCs. (A) qRT-PCR analyses for FGF1 in the BNL cells and MDSC(n = 6). (B) Morphology of BNL cell after DMSO or sorafenib treatment following normal culture(control) or co-culture with MDSC(Scale bar = 100μm). (C) WB for FGF1 in BNL cells(n = 3). (D) Heatmap displaying quantification of mouse 18 cytokines in supernatant of culture system by MILLIPLEX® MAP array. (E) When neutralizing Mouse Anti-IL-6 antibody was added to the culture system, the FGF1 expression in BNL cells. Specific n values of biological independent experiments. t – test for two groups comparisons. ANOVA for four groups comparisons and Tukey test for multiple comparisons.Asterisks indicate comparison with the control group. *p < 0.05, **p < 0.01***, p < 0.001.
Ly6G+ myeloid cells depletion reduced fibrosis and enhanced sorafenib efficacy
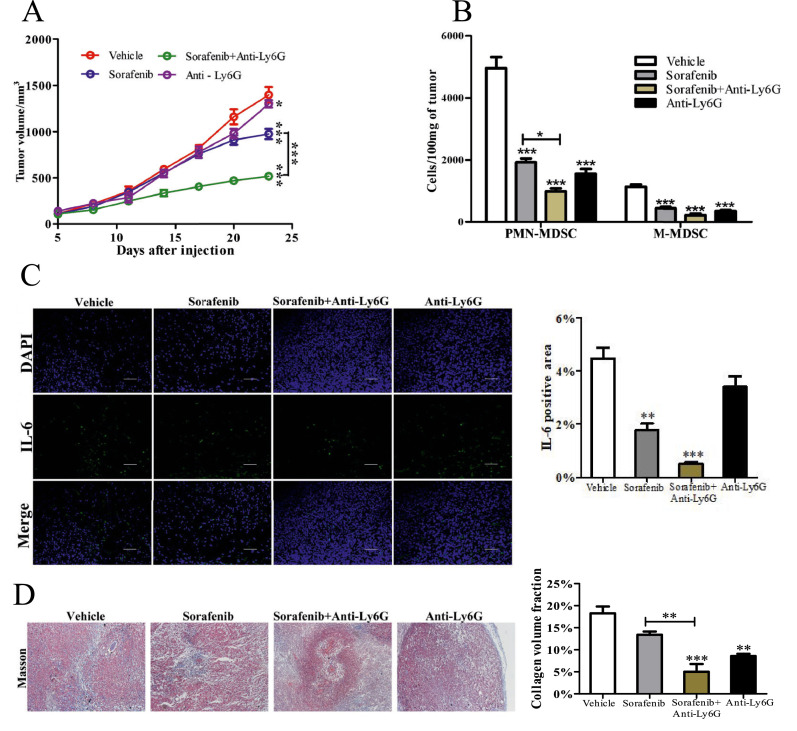
To confirm whether MDSCs was responsible for tumor fibrosis, neutralizing antibody(Anti-Ly6G, a invivo antibody) was injected into mice to decrease (PMN-MDSC)s. The addition of anti-Ly6G mAb significantly improved the ability of sorafenib to suppress tumor growth (P<0.0001), while anti-Ly6G mAb alone did not present significant suppression for tumor (Fig. 5A P>0.05). Anti-Ly6G did not significantly decrease IL-6 level in the tumor(Fig. 5C), although with an MDSC depletion(Fig. 5B P<0.0001). The reason is that a variety of cells in the tumor microenvironment can secrete IL-6, yielding conflicting results. Moreover, Ly6G+ myeloid cells depletion reduced collagenous fiber(Fig. 5D P<0.01) and showed a correlation with the number of MDSCs.
Fig. 5.
Ly6G+ myeloid cells depletion enhanced sorafenib efficacy and reduced fibrosis. (A) Tumor volume was measured using a Vernier caliper, V = a × b2/2, where a is the long diameter and b is the short diameter( n = 5, two – way ANOVA). (B) Number of MDSC were measured using flow cytometry(n = 3). (C) IL-6 immunostaining images of tumor tissues (Scale bar = 100 μm.). Positive area was analyzed by ImageJ(n = 3). (D) Masson staining of tumor tissue(Scale bar = 200 μm).Collagen volume fraction was analyzed by ImageJ(n = 3). Specific n values of biological independent animals. ANOVA for four groups comparisons and Tukey test for multiple comparisons. Asterisks indicate comparison with the control group. *p < 0.05, **p < 0.01, ***p < 0.001.
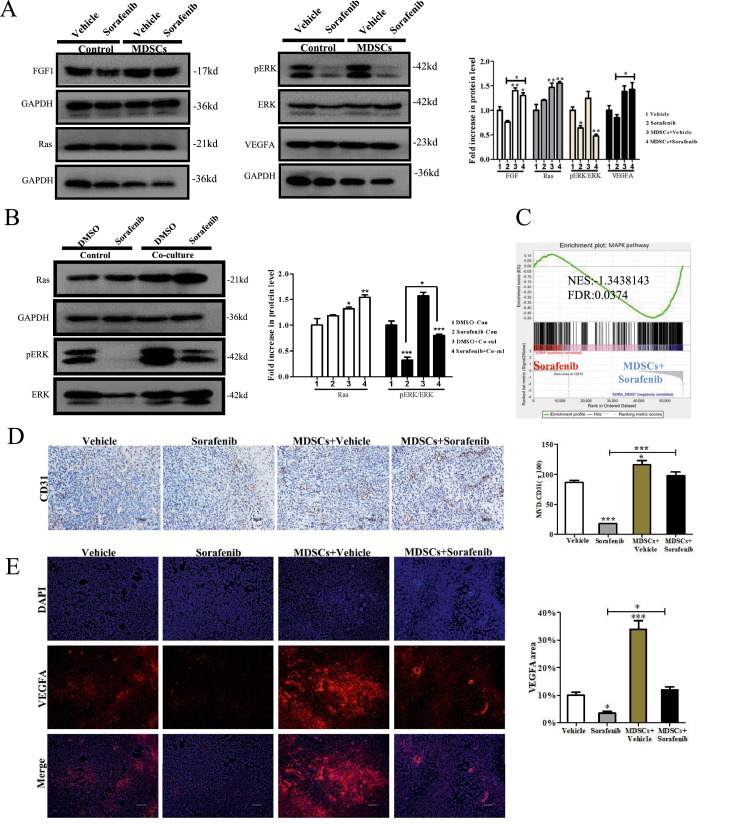
MAPK signaling re-activated led to angiogenesis and resistance to sorafenib
Mitogen-activated protein kinase (MAPK) might exert a central role in these findings, for the reason that which is the downstream of FGF1R activation as well as one of the targets of sorafenib [16,17]. Thus, we detected the expression of major molecules (FGF1, Ras, ERK and VEGFA) of the MAPK pathway. We showed sorafenib drastically suppressed the expression of phosphorylated extracellular regulated protein kinases (pERK) in vivo and vitro (Fig. 6A,B), and MDSCs impaired this effect in vitro (Fig. 6B). A similar result was shown by gene set enrichment analysis(GSEA)(Fig. 6C FDR = 0.0374). CD31, an endothelial cell marker, revealed that microvessel density decreased in tumor tissues of the sorafenib group and significantly increased in that derived from mice with the adoptive transfer MDSCs, whether or not treated with sorafenib (Fig. 6D P<0.0001). VEGFA staining showed a similar result (Fig. 6E P<0.0001). These results suggested MDSC re-actived MAPK pathway which mediated angiogenesis and sorafenib resistance.
Fig. 6.
MAPK signaling re-activated led to angiogenesis and resistance to sorafenib. (A) WB for MAPK pathway in tumor tissue(n = 5). (B) WB for MAPK pathway in BNL cells(n = 3). (C) GSEA of tumor tissue involved in MAPKpathways(n = 5). The normalized enrichment scores (NES) and tests of statistical significance (FDR) are shown. (D) Representative images(Scale bar = 200 μm) of tumor sections stained IHC with CD31 for sprouting neomicrovessels. Positive area was analyzed by ImageJ(n = 3) (E)VEGFA immunostaining images of tumor tissues(Scale bar = 100 μm). Positive area was analyzed by ImageJ(n = 3). Specific n values of biological independent experiments. ANOVA for four groups comparisons and Tukey test for multiple comparisons. Asterisks indicate comparison with the control group. **p < 0.01, ***p < 0.001.
Discussion
Growing evidence implicating MDSCs mediated the progression, poor prognosis, and therapeutic resistance of HCC, which are mainly linked to its immunosuppressive function [18]. Erewhile, several studies suggeste that MDSCs directly impacte tumor cells' characteristics [19,20] while underlying mechanisms are still unknown. Our study indicated that MDSCs promoted the FGF1 expression in BNL liver cancer cells and IL-6 is a mediator.
In a mouse xenograft model, we observed increased tumor growth and angiogenesis accompanied by tumor fibrosis and improved synthesis of ECM in tumors derived from the mice transferred MDSCs. With the depletion of MDSCs by anti-Ly6G, sorafenib significantly inhibited tumor growth, accompanied by reduced tumor fibrosis and angiogenesis. Several studies provided evidence that MDSC reduction positively correlated with sorafenib response and prognosis [21], [22], [23]. Targeting Ly6G improves sorafenib efficacy [24,25]. However, models of HCC have not yet conclusively clarified the involvement of MDSC for tumor fibrosis. It is reported that MDSCs regulate fibrosis in the lungs, liver, and kidneys and help repair the central nervous system in non-neoplastic inflamed organs [26]. In HCC mice models, MDSCs accumulated in fibrotic livers, which significantly correlated with reduced tumor-infiltrating lymphocytes and increased tumorigenicity [25]. Chen's research found that sorafenib caused tumor hypoxia and an increase in MDSCs, which promoted tumor fibrosis via the stromal-derived factor 1 (SDF) alpha/C-X-C receptor type 4 (CXCR4) axis [27]. We found sorafenib reduced the number of MDSCs, the different results of which might be attributable to the subcutaneous HCC model [24]. HIF, SDF, and CXCR4 were consistently elevated in the tumors of mice transferred MDSCs in this study.
FGF1, PDGF, and TGFβ pathways are the primary mechanisms contributing to CAF activation [13]. In this study, FGF significantly differed between groups. FGF1 activation plays a positive role in the expansion of CAFs through transcriptional repression of TP53 and escapes from p53-dependent stroma cell senescence [28] and section with induction of several key CAF effector genes(MMP1, HGF) [29]. However, no study answers whether MDSCs directly act on fibroblasts. Our finding showed that MDSC barely expresses FGF1, as described in another study. As previously speculated, the stimulation of MDSC-induced fibrosis of tumor tissue may be an indirect effect [14]. In TME, tumor cells and macrophages can mediate MDSC-induce fibrosis. Tumor cells are the central member of tumor tissue and can be effect by MDSC. A study in head and neck squamous cell carcinoma shows that tumor cell-secreted FGF1 increases mitochondrial oxidative phosphorylation and promotes lactate consumption of CAFs [30]. We carried out a series of experiments to support the conjecture. In several disease models, MDSC differentiated into macrophage-like phenotypes. Nevertheless, references to the role of macrophages on tumor fibrosis are contradictory [31]. More research is needed to understand this complex question. All in all, our findings and previous research showed that MDSCs triggered tumor fibrosis, facilitating enhanced growth, angiogenesis, and resistance to sorafenib.
Cytokines are essential mediators linking inflammation and cancer and exert vast immunoregulatory actions to tumor cells [32]. To deeper understand the interaction between MDSCs and BNL, we analyzed the most common mouse 18 cytokines. Four cytokines (GM-CSF, IL-4, TNF-alpha, and IL-6) secreted more in coculture than in monoculture. Among them, IL-6 was shown to have the most dramatic and specific increase(more than 3-fold) in secretion. Anti-IL6 neutralized the effect of MDSC. Our data showed that MDSC-secreted IL-6 was essential for FGF1 upregulation of BNL cells. IL-6 is a prototypical cytokine that exerts a pro-tumorigenic role in inflammation-associated cancers [33]. Serum IL-6 concentrations were significantly higher in patients with HCC than healthy donors [34]. IL-6/IL-6 receptor promotes epithelial-mesenchymal transition progression, cancer stemness of triple-negative breast cancer [35]. Anti-IL-6/IL-6 receptor monoclonal antibodies developed for tumor-targeted therapy have demonstrated promising results in preclinical studies and clinical trials [36]. We provided the first evidence that MDSCs secreted IL-6, a potential mediator of tumor progression and resistance to sorafenib in HCC. Notably, we only analyzed the most common mouse 18 cytokines and did not verify other cytokines secreted by MDSCs. In addition, we found IL-1, CXCL2, CCL2, CXCL5 were decreased in the coculture system compared with tumor cells in mono-culture, which might result from the negative regulation of GM-CSF [37].
Our findings are presented with a limitation that only a single cell line was carried out to investigate the crosstalk among MDSC, HCC, and CAFs. Further study will be performed in multiple cell lines. Nevertheless, a recent study supports the validity of our findings. This study showed that FGF1 upregulated in CAF of HCC patients at all stages and high levels of tumor-associated neutrophils infiltrated in the tumor, which indicated a poor prognosis among patients in middle and late stage. Considering nomenclature of immunosuppressive neutrophils, multiple populations of neutrophils with immunosuppressive features might be PMN-MDSCs. In addition, another clinical and preclinical study demonstrated that tumors with sorafenib-acquired resistance were enriched with FGF signaling cascades and FGF blockade delayed tumor growth and improved survival in sorafenib-resistant tumors, although variations of MDSCs was not mentioned.
In conclusion, we demonstrated that MDSCs facilitated CAF activation led to tumor growth, angiogenesis, and sorafenib resistance by inducing FGF1 upregulation. Our findings offer some insights into the crosstalk among MDSC, HCC and CAFs.
CRediT authorship contribution statement
Xue Deng: Writing – original draft, Methodology, Formal analysis, Data curation, Writing – review & editing. Xueyan Li: Writing – review & editing. Xuan Guo: Methodology, Writing – review & editing. Yantong Lu: Methodology, Writing – review & editing. Yingjie Xie: Formal analysis, Data curation, Writing – review & editing. Xuhui Huang: Funding acquisition, Writing – review & editing. Juze Lin: Funding acquisition, Writing – review & editing. Wei Tan: Funding acquisition, Writing – review & editing. Changjun Wang: Conceptualization, Writing – review & editing, Funding acquisition.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Acknowledgments
We thank Ying Cao for her technical contributions in the analysis of RNA-seq data.
Ethics statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Southern Medical University (Guangzhou, China).
Funding statement
This study was supported by the following foundation projects: The National Natural Science Foundation of China (No. 81774261), Science and Technology Program of Guangzhou (No. 202102080232)
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100788.
Appendix. Supplementary materials
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Gabrilovich D.I. Myeloid-derived suppressor cells. Cancer Immunol Res. 2017;5:3–8. doi: 10.1158/2326-6066.CIR-16-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao X.H., Tian L., Wu J., Ma X.L., Zhang C.Y., Zhou Y., Sun Y.F., Hu B., Qiu S.J., Zhou J., Fan J., Guo W., Yang X.R. Circulating CD14(+) HLA-DR(-/low) myeloid-derived suppressor cells predicted early recurrence of hepatocellular carcinoma after surgery. Hepatol Res. 2017;47:1061–1071. doi: 10.1111/hepr.12831. [DOI] [PubMed] [Google Scholar]
- 4.Wang D., An G., Xie S., Yao Y., Feng G. The clinical and prognostic significance of CD14(+)HLA-DR(-/low) myeloid-derived suppressor cells in hepatocellular carcinoma patients receiving radiotherapy. Tumour Biol. 2016;37:10427–10433. doi: 10.1007/s13277-016-4916-2. [DOI] [PubMed] [Google Scholar]
- 5.Mizukoshi E., Yamashita T., Arai K., Terashima T., Kitahara M., Nakagawa H., Iida N., Fushimi K., Kaneko S. Myeloid-derived suppressor cells correlate with patient outcomes in hepatic arterial infusion chemotherapy for hepatocellular carcinoma. Cancer Immunol Immunother. 2016;65:715–725. doi: 10.1007/s00262-016-1837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C., Rong D., Zhang B., Zheng W., Wang X., Chen Z., Tang W. Current perspectives on the immunosuppressive tumor microenvironment in hepatocellular carcinoma: challenges and opportunities. Mol Cancer. 2019;18:130. doi: 10.1186/s12943-019-1047-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llovet J.M., Kelley R.K., Villanueva A., Singal A.G., Pikarsky E., Roayaie S., Lencioni R., Koike K., Zucman-Rossi J., Finn R.S. Hepatocellular carcinoma. Nat Rev Dis Prim. 2021;7:6. doi: 10.1038/s41572-020-00240-3. [DOI] [PubMed] [Google Scholar]
- 8.Verma V., Sprave T., Haque W., Simone C.N., Chang J.Y., Welsh J.W., Thomas C.J. A systematic review of the cost and cost-effectiveness studies of immune checkpoint inhibitors. J Immunother Cancer. 2018;6:128. doi: 10.1186/s40425-018-0442-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green A.K. Challenges in assessing the cost-effectiveness of cancer immunotherapy. Jama Netw Open. 2021;4 doi: 10.1001/jamanetworkopen.2020.34020. [DOI] [PubMed] [Google Scholar]
- 10.Xie Y., Zhang Y., Wei X., Zhou C., Huang Y., Zhu X., Chen Y., Wen H., Huang X., Lin J., Wang Z., Ren Y., Fan B., Deng X., Tan W., Wang C. Jianpi Huayu decoction attenuates the immunosuppressive status of H(22) hepatocellular carcinoma-bearing mice: by targeting myeloid-derived suppressor cells. Front Pharmacol. 2020;11:16. doi: 10.3389/fphar.2020.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Wen H., Zhou C., Su Q., Lin Y., Xie Y., Huang Y., Qiu Q., Lin J., Huang X., Tan W., Min C., Wang C. TNF-α derived from M2 tumor-associated macrophages promotes epithelial-mesenchymal transition and cancer stemness through the Wnt/β-catenin pathway in SMMC-7721 hepatocellular carcinoma cells. Exp Cell Res. 2019;378:41–50. doi: 10.1016/j.yexcr.2019.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Zeltz C., Primac I., Erusappan P., Alam J., Noel A., Gullberg D. Cancer-associated fibroblasts in desmoplastic tumors: emerging role of integrins. Semin Cancer Biol. 2020;62:166–181. doi: 10.1016/j.semcancer.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16:582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 14.Im J.H., Buzzelli J.N., Jones K., Franchini F., Gordon-Weeks A., Markelc B., Chen J., Kim J., Cao Y., Muschel R.J. FGF2 alters macrophage polarization, tumour immunity and growth and can be targeted during radiotherapy. Nat Commun. 2020;11:4064. doi: 10.1038/s41467-020-17914-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biffi G., Tuveson D.A. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101:147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caruso S., Calatayud A.L., Pilet J., La Bella T., Rekik S., Imbeaud S., Letouzé E., Meunier L., Bayard Q., Rohr-Udilova N., Péneau C., Grasl-Kraupp B., de Koning L., Ouine B., Bioulac-Sage P., Couchy G., Calderaro J., Nault J.C., Zucman-Rossi J., Rebouissou S. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology. 2019;157:760–776. doi: 10.1053/j.gastro.2019.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T.F., Galle P.R., Seitz J.F., Borbath I., Häussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 18.Vetsika E.K., Koukos A., Kotsakis A. Myeloid-derived suppressor cells: major figures that shape the immunosuppressive and angiogenic network in cancer. Cells. 2019;8 doi: 10.3390/cells8121647. Basel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ai L., Mu S., Sun C., Fan F., Yan H., Qin Y., Cui G., Wang Y., Guo T., Mei H., Wang H., Hu Y. Myeloid-derived suppressor cells endow stem-like qualities to multiple myeloma cells by inducing piRNA-823 expression and DNMT3B activation. Mol Cancer. 2019;18:88. doi: 10.1186/s12943-019-1011-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Veirman K., Menu E., Maes K., De Beule N., De Smedt E., Maes A., Vlummens P., Fostier K., Kassambara A., Moreaux J., Van Ginderachter J.A., De Bruyne E., Vanderkerken K., Van Valckenborgh E. Myeloid-derived suppressor cells induce multiple myeloma cell survival by activating the AMPK pathway. Cancer Lett. 2019;442:233–241. doi: 10.1016/j.canlet.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto A., Sarker D., Reebye V., Jarvis S., Sodergren M.H., Kossenkov A., Sanseviero E., Raulf N., Vasara J., Andrikakou P., Meyer T., Huang K.W., Plummer R., Chee C.E., Spalding D., Pai M., Khan S., Pinato D.J., Sharma R., Basu B., Palmer D., Ma Y.T., Evans J., Habib R., Martirosyan A., Elasri N., Reynaud A., Rossi J.J., Cobbold M., Habib N.A., Gabrilovich D.I. Upregulation of C/EBPα inhibits suppressive activity of myeloid cells and potentiates antitumor response in mice and patients with cancer. Clin Cancer Res. 2021;27:5961–5978. doi: 10.1158/1078-0432.CCR-21-0986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho V., Lim T.S., Lee J., Steinberg J., Szmyd R., Tham M., Yaligar J., Kaldis P., Abastado J.P., Chew V. TLR3 agonist and Sorafenib combinatorial therapy promotes immune activation and controls hepatocellular carcinoma progression. Oncotarget. 2015;6:27252–27266. doi: 10.18632/oncotarget.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizukoshi E., Yamashita T., Arai K., Terashima T., Kitahara M., Nakagawa H., Iida N., Fushimi K., Kaneko S. Myeloid-derived suppressor cells correlate with patient outcomes in hepatic arterial infusion chemotherapy for hepatocellular carcinoma. Cancer Immunol Immunother. 2016;65:715–725. doi: 10.1007/s00262-016-1837-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang C.J., Yang Y.H., Chiu C.J., Lu L.C., Liao C.C., Liang C.W., Hsu C.H., Cheng A.L. Targeting tumor-infiltrating Ly6G(+) myeloid cells improves sorafenib efficacy in mouse orthotopic hepatocellular carcinoma. Int J Cancer. 2018;142:1878–1889. doi: 10.1002/ijc.31216. [DOI] [PubMed] [Google Scholar]
- 25.Liu M., Zhou J., Liu X., Feng Y., Yang W., Wu F., Cheung O.K., Sun H., Zeng X., Tang W., Mok M., Wong J., Yeung P.C., Lai P., Chen Z., Jin H., Chen J., Chan S.L., Chan A., To K.F., Sung J., Chen M., Cheng A.S. Targeting monocyte-intrinsic enhancer reprogramming improves immunotherapy efficacy in hepatocellular carcinoma. Gut. 2020;69:365–379. doi: 10.1136/gutjnl-2018-317257. [DOI] [PubMed] [Google Scholar]
- 26.Sendo S., Saegusa J., Morinobu A. Myeloid-derived suppressor cells in non-neoplastic inflamed organs. Inflamm Regen. 2018;38:19. doi: 10.1186/s41232-018-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y., Huang Y., Reiberger T., Duyverman A.M., Huang P., Samuel R., Hiddingh L., Roberge S., Koppel C., Lauwers G.Y., Zhu A.X., Jain R.K., Duda D.G. Differential effects of sorafenib on liver versus tumor fibrosis mediated by stromal-derived factor 1 alpha/C-X-C receptor type 4 axis and myeloid differentiation antigen-positive myeloid cell infiltration in mice. Hepatology. 2014;59:1435–1447. doi: 10.1002/hep.26790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Procopio M.G., Laszlo C., Al L.D., Kim D.E., Bordignon P., Jo S.H., Goruppi S., Menietti E., Ostano P., Ala U., Provero P., Hoetzenecker W., Neel V., Kilarski W.W., Swartz M.A., Brisken C., Lefort K., Dotto G.P. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat Cell Biol. 2015;17:1193–1204. doi: 10.1038/ncb3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bordignon P., Bottoni G., Xu X., Popescu A.S., Truan Z., Guenova E., Kofler L., Jafari P., Ostano P., Röcken M., Neel V., Dotto G.P. Dualism of FGF and TGF-β signaling in heterogeneous cancer-associated fibroblast activation with ETV1 as a critical determinant. Cell Rep. 2019;28:2358–2372. doi: 10.1016/j.celrep.2019.07.092. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar D., New J., Vishwakarma V., Joshi R., Enders J., Lin F., Dasari S., Gutierrez W.R., Leef G., Ponnurangam S., Chavan H., Ganaden L., Thornton M.M., Dai H., Tawfik O., Straub J., Shnayder Y., Kakarala K., Tsue T.T., Girod D.A., Van Houten B., Anant S., Krishnamurthy P., Thomas S.M. Cancer-associated fibroblasts drive glycolysis in a targetable signaling loop implicated in head and neck squamous cell carcinoma progression. Cancer Res. 2018;78:3769–3782. doi: 10.1158/0008-5472.CAN-17-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weston C.J., Zimmermann H.W., Adams D.H. The role of myeloid-derived cells in the progression of liver disease. Front Immunol. 2019;10:893. doi: 10.3389/fimmu.2019.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spangler J.B., Moraga I., Mendoza J.L., Garcia K.C. Insights into cytokine-receptor interactions from cytokine engineering. Annu Rev Immunol. 2015;33:139–167. doi: 10.1146/annurev-immunol-032713-120211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mauer J., Denson J.L., Brüning J.C. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36:92–101. doi: 10.1016/j.it.2014.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Lai S.C., Su Y.T., Chi C.C., Kuo Y.C., Lee K.F., Wu Y.C., Lan P.C., Yang M.H., Chang T.S., Huang Y.H. DNMT3b/OCT4 expression confers sorafenib resistance and poor prognosis of hepatocellular carcinoma through IL-6/STAT3 regulation. J Exp Clin Cancer Res. 2019;38:474. doi: 10.1186/s13046-019-1442-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weng Y.S., Tseng H.Y., Chen Y.A., Shen P.C., Al H.A., Chen L.M., Tung Y.C., Hsu H.L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol Cancer. 2019;18:42. doi: 10.1186/s12943-019-0988-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao X., Huang J., Zhong H., Shen N., Faggioni R., Fung M., Yao Y. Targeting interleukin-6 in inflammatory autoimmune diseases and cancers. Pharmacol Ther. 2014;141:125–139. doi: 10.1016/j.pharmthera.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Kumar V., Donthireddy L., Marvel D., Condamine T., Wang F., Lavilla-Alonso S., Hashimoto A., Vonteddu P., Behera R., Goins M.A., Mulligan C., Nam B., Hockstein N., Denstman F., Shakamuri S., Speicher D.W., Weeraratna A.T., Chao T., Vonderheide R.H., Languino L.R., Ordentlich P., Liu Q., Xu X., Lo A., Puré E., Zhang C., Loboda A., Sepulveda M.A., Snyder L.A., Gabrilovich D.I. Cancer-associated fibroblasts neutralize the anti-tumor effect of CSF1 receptor blockade by inducing PMN-MDSC infiltration of tumors. Cancer Cell. 2017;32:654–668. doi: 10.1016/j.ccell.2017.10.005. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.