Abstract
The novel virus “severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)” has been responsible for the worldwide pandemic causing huge devastation and deaths since December 2019. The disease caused by this virus is known as COVID-19. The present study is based on immuno-informatics approach to develop a multi-epitope-loaded peptide vaccine to combat the COVID-19 menace. Here, we have reported the 9-mer CD8 T cell epitopes and 15-mer CD4 T cell epitopes, free from glycosylation sites, belonging to three proteins, viz. surface glycoprotein, membrane glycoprotein and envelope protein of this virus. Immunogenicity, aliphatic amino acid, antigenicity and hydrophilicity scores of the predicted epitopes were estimated. In addition, other physicochemical parameters, namely net charge, Boman index and amino acid contents, were also accounted. Out of all the epitopes, three CD8 T cell epitopes viz. PDPSKPSKR, DPSKPSKRS and QTQTNSPRR and three CD4 T cell epitopes viz. ASYQTQTNSPRRARS, RIGNYKLNTDHSSSS and RYRIGNYKLNTDHSS were found to be efficient targets for raising immunity in human against this virus. With the help of our identified potent epitopes, various pharma industries might initiate efforts to incorporate those epitopes with carrier protein or adjuvant to develop a multi-epitope-loaded peptide vaccine against SARS-CoV-2. The peptide vaccines are usually cost-effective and therefore, could be administered as a preventive measure to combat the spread of this disease. Proper clinical trials must be conducted prior to the use of identified epitopes as vaccine candidates.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00203-022-02845-6.
Keywords: SARS-COV-2, Peptide vaccine, T cell epitopes
Introduction
Humans are common hosts to a few viruses belonging to Coronaviridae family (Hoffmann et al. 2020). In the twenty-first century, three lethal animal-borne coronaviruses, namely MERS-CoV, SARS-CoV and SARS-CoV-2, infected humans by traversing animal–human interface (Walls et al. 2020). Coronaviruses have the largest RNA genome and SARS-CoV-2 under the genus Beta coronavirus is the seventh species of this family (Lai et al. 2020; Wu et al. 2020). The COVID-19 disease, caused by a single-stranded, positive sense virus named SARS-CoV-2, gives rise to troubles in respiratory system, gastrointestinal tract, nervous system, and in liver. Difficulties like fever, dry cough, dyspnea, lymphopenia and fatigue manifest in patients. In extreme cases, viral pneumonia develops and eventually it may result in patient death (Lai et al. 2020; Wu et al. 2020Zhang et al. 2020). Bats are also regarded as the hosts of SARS-CoV-2, as the whole genome study of the virus revealed that it shared 96% similarity with a bat coronavirus (Wu et al. 2020).
The genomic study of SARS-CoV-2 revealed its 79% similarity with SARS-CoV and 50% with MERS-CoV, each virus belongs to Betacoronavirus group. Further resemblance of SARS-CoV-2 with its family members is at the level of structural and non-structural proteins. The structural proteins encoded by SARS-CoV-2 are spike protein (S), nucleocapsid phosphoprotein (N), membrane glycoprotein (M) and envelope protein (E) (Kiyotani et al. 2020). It was reported that SARS-CoV-2 gene expression occurs when it successfully accesses the host cells, where translation of beneficial accessory proteins allows acclimatization of the virus to the host. The two-third segment of coronavirus genome codes for RNA-dependent RNA polymerase (viral polymerase), other transcription enzymes and factors as well as non-structural proteins.
The best preventive measure to combat pathogenic diseases is vaccination (Schlingmann et al. 2018). The emergence of recombinant DNA technology and whole genome sequencing of microbes widened the scope of reverse vaccinology. The computational vaccine design through reverse vaccinology approach analyzes the proteome expressed by the microorganism of interest. It allows prediction of antigens that can act as most efficient vaccine candidates (Sette and Rappuoli 2010). Vaccines can be polyvalent by the use of multiple immunogens from a range of serotypes or use of conserved antigens found in those serotypes (Schlingmann et al. 2018). It has been seen that peptide vaccines are immunodominant B cell and T cell epitopes that can activate cellular immune response (Oyarzún et al. 2013). Synthesis of peptide vaccines is relatively easy, cheaper and the product is stable and non-infectious (Patronov and Doytchinova 2013).
T cell epitopes are exclusively peptides that are either conformational or linear based on their structure and paratope binding. Antigenic peptides complexed with MHC class I or MHC class II molecules are presented to T cells. MHC class I molecules exist on the surface of all nucleated cells, while MHC class II molecules are specifically possessed by specialized antigen presenting cells and lymphocytes. The extracellular peptides of length 8–11 amino acids bind to MHC class I molecules which are recognized by CD8 + T cell receptors. Whereas, 12–25 amino acids’ long peptides are degraded intracellularly and displayed by MHC class II molecules that are identified by CD4 + T cell receptors. Antigen recognition leads CD8 + T cells to become cytotoxic T lymphocytes and CD4 T cells become T helper cells. Cell-mediated immune responses lead to cytokine production and induction of B-cells for antibody maturation. On epitope presentation and recognition, the T cells multiply into effectors that can respond to the same antigen displayed by other cells. Subsequently, memory cells are also generated and adaptive immunity is established (Patronov and Doytchinova 2013; Peters et al. 2020) (Oyarzún et al. 2013) (Sanchez-Trincado et al. 2017).
According to the WHO as on 17 January 2022, the total confirmed cases of COVID-19 were 329,154,731 with 5,559,322 deaths. With an aim to predict the best epitopes of SARS-CoV-2/human/USA/CA-CZB-1053/2020 isolate, it is essential to study the degree of immunogenicity of identified epitopes. In this study, the surface glycoprotein, membrane glycoprotein and envelope protein of the SARS-CoV-2/human/USA/CA-CZB-1053/2020 isolate were retrieved for identification of robust vaccine candidates.
Materials and methodology
Protein sequence extraction
The protein sequences of three proteins viz. surface glycoprotein, membrane glycoprotein and envelope protein of the virus, SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate were selected for the recent study. This set of proteins was extracted from the protein database (http://www.ncbi.nlm.nih.gov) having accession number QJW39952.1, QJW39955.1 and QJW39954.1, respectively.
Prediction of antigenic CD8 and CD4 T cell epitopes
CD8 + T cells (cytotoxic T cells) defend the body against foreign attackers and cancerous cells, while CD4 + T cells (helper T cells) are the cells of immune system that combats against infections. Both CD8 and CD4 T cell epitopes from three proteins of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate were identified by Hopp and Woods method (Hopp and Woods 1981).
Immunogenicity score
The immunogenicity score (the potentiality of T cell to produce antibodies against any antigen) was calculated using AbDesigner algorithm (Pisitkun et al. 2012). The immunogenicity score is directly proportional with the immunogenic response, i.e., with an increase in immunogenicity score the immunogenic response also increases. Different parameters of CD8 and CD4 T cell epitopes viz. immunogenicity, number of aliphatic amino acids, antigenicity and hydrophilicity were enumerated using PERL script program. All the selected epitopes were free from glycosylation site.
Physiochemical properties of T cell epitopes
The physiochemical characterization of T cell epitopes was predicted with the online tool ExPASy-ProtParam (https://web.expasy.org/protparam/). Further, cleavage sites and Boman index value of each epitope were also evaluated with the online tools Peptide cutter (https://web.expasy.org/peptidecutter/) and Antimicrobial Peptide Database (APD3) (http://aps.unmc.edu/AP/prediction/prediction_main.php), respectively. If a protein acquires Boman index greater than 2.48 kcal/mol, then it is more likely to interact with other proteins.
Results
Investigation of immunogenicity
With the use of several bioinformatic tools, five 9-mer CD8 T cell epitopes and five 15-mer CD4 T cell epitopes from three proteins, namely surface glycoprotein, membrane glycoprotein and envelope protein of SARS-CoV-2/human/USA/CA CZB 1055/202 isolate, were identified.
To combat the pandemic of SARS-CoV-2, the immunogenicity scores of the five best 9-mer CD8 T cell epitopes for three proteins viz. surface glycoprotein, membrane glycoprotein and envelope protein were investigated and represented in Table 1a. In surface glycoprotein, the peptide PDPSKPSKR was found with the highest immunogenicity score of 9.20, in membrane glycoprotein two peptides TDHSSSSDN and NTDHSSSSD had the highest immunogenicity score of 8.72 and likewise, in envelope protein, the peptide YSRVKNLNS was found with the highest immunogenicity score of 6.37.
Table 1.
CD8 (a) and CD4 (b) T cell epitopes of SARS CoV2/human/USA/CA CZB 1055/2020 isolate showing number of aliphatic amino acids and immunogenicity (Ig) score (Hopp and Woods 1981)
| Name of proteins | CD8 T- cell epitopes (a) | Aliphatic amino acids | Ig analysis of epitopes | |
|---|---|---|---|---|
| Hydropathy index | Immunogenicity score | |||
| Surface glycoprotein | PDPSKPSKR | 2 | 6.97 | 9.2 |
| DPSKPSKRS | 2 | 6.88 | 9.01 | |
| DYNYKLPDD | 2 | 6.53 | 8.23 | |
| WNSNNLDSK | 2 | 6.34 | 8.12 | |
| QTQTNSPRR | 0 | 7.09 | 8.08 | |
| Membrane glycoprotein | TDHSSSSDN | 0 | 6.46 | 8.72 |
| NTDHSSSSD | 0 | 6.46 | 8.72 | |
| DHSSSSDNI | 1 | 5.88 | 7.59 | |
| GNYKLNTDH | 3 | 6.3 | 7.56 | |
| NYKLNTDHS | 2 | 6.34 | 7.48 | |
| Envelope protein | YSRVKNLNS | 3 | 5.64 | 6.37 |
| KPSFYVYSR | 2 | 5.3 | 5.72 | |
| VYSRVKNLN | 4 | 5.09 | 5.24 | |
| YSFVSEETG | 2 | 4.94 | 4.99 | |
| FYVYSRVKN | 3 | 4.96 | 4.86 | |
| Name of proteins | CD4 T- cell epitopes (b) | Aliphatic amino acids | Ig analysis of epitopes | |
|---|---|---|---|---|
| Hydropathy index | Immunogenicity score | |||
| Surface glycoprotein | NSNNLDSKVGGNYNY | 5 | 5.96 | 7.81 |
| WNSNNLDSKVGGNYN | 5 | 5.93 | 7.71 | |
| ASYQTQTNSPRRARS | 2 | 6.31 | 6.94 | |
| AWNSNNLDSKVGGNY | 6 | 5.58 | 6.92 | |
| HRSYLTPGDSSSGWT | 3 | 5.61 | 6.84 | |
| Membrane glycoprotein | GNYKLNTDHSSSSDN | 3 | 6.26 | 8.14 |
| NYKLNTDHSSSSDNI | 3 | 5.93 | 7.29 | |
| IGNYKLNTDHSSSSD | 4 | 5.73 | 7.05 | |
| RIGNYKLNTDHSSSS | 4 | 5.79 | 6.95 | |
| RYRIGNYKLNTDHSS | 4 | 6.07 | 6.92 | |
| Envelope protein | KPSFYVYSRVKNLNS | 5 | 5.23 | 5.70 |
| VKPSFYVYSRVKNLN | 6 | 4.89 | 5.04 | |
| SLVKPSFYVYSRVKN | 6 | 4.71 | 4.80 | |
| LVKPSFYVYSRVKNL | 7 | 4.41 | 4.23 | |
| YSFVSEETGTLIVNS | 5 | 4.27 | 4.14 | |
The binding efficiency of an epitope is highly enhanced with the presence of aliphatic amino acid sequence on it. Thus, the presence of aliphatic amino acid, namely Gly, Ala, Lys, Ile, Met, Val and Leu, in the desired epitopes can lead to proper interactions with lymphocytes. Here, the CD8 T cell epitopes were analyzed for their total number of aliphatic amino acids (Table 1a). Four epitopes of surface glycoprotein viz. PDPSKPSKR, DPSKPSKRS, DYNYKLPDD and WNSNNLDSK possessed two aliphatic amino acids each, while epitope QTQTNSPRR did not possess any aliphatic amino acid. In membrane glycoprotein, epitope DHSSSSDNI had only one aliphatic amino acid, epitope NYKLNTDHS had two aliphatic amino acids and epitope GNYKLNTDH had three aliphatic amino acids. Further, in envelope protein, the epitopes KPSFYVYSR and YSFVSEETG possessed two aliphatic amino acids; epitopes YSRVKNLNS and FYVYSRVKN had three aliphatic amino acids while epitope VYSRVKNLN possessed four aliphatic amino acids.
The immunogenicity scores of the five best 15-mer CD4 T cell epitopes for three proteins, namely surface glycoprotein, membrane glycoprotein and envelope protein, were evaluated and reported in Table 1b. The epitope NSNNLDSKVGGNYNY of surface glycoprotein, GNYKLNTDHSSSSDN of membrane glycoprotein and KPSFYVYSRVKNLNS of envelope protein possessed the highest immunogenicity score of 7.81, 8.14 and 5.70, respectively.
Aliphatic amino acids in the epitopes of surface glycoprotein varied from 3 (HRSYLTPGDSSSGWT) to 6 (AWNSNNLDSKVGGNY). In membrane glycoprotein, three epitopes (IGNYKLNTDHSSSSD, RIGNYKLNTDHSSSS and RYRIGNYKLNTDHSS) possessed 4 aliphatic amino acids and two epitopes (GNYKLNTDHSSSSDN and NYKLNTDHSSSSDNI) had 3 aliphatic amino acids. Further, in envelope protein, the epitopes KPSFYVYSRVKNLNS and YSFVSEETGTLIVNS had 5 aliphatic amino acids, epitopes VKPSFYVYSRVKNLN and SLVKPSFYVYSRVKN possessed 6 aliphatic amino acids while the epitope LVKPSFYVYSRVKNL had 7 aliphatic amino acids, shown in Table 1b. Epitopes with high immunogenicity scores are usually more immunogenic in nature and thus these peptides could be the potential choice as candidates in vaccine formulation.
Investigation of antigenicity
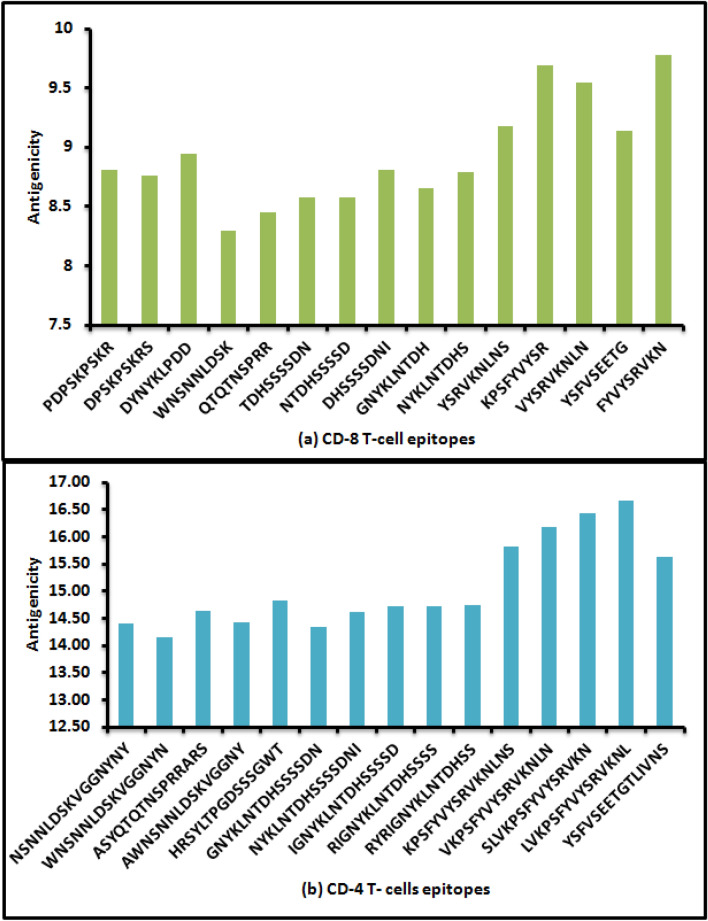
Five CD8 T cell epitopes from each of the three proteins were identified and evaluated for antigenicity (Fig. 1a). The antigenicity score of 15 epitopes varied within the range from 8.29 (WNSNNLDSK) to 9.77 (FYVYSRVKN). Similarly, for CD4 T cell epitopes, the antigenicity score was evaluated and found to vary from 14.14 (WNSNNLDSKVGGNYN) to 16.66 (LVKPSFYVYSRVKNL) shown in Fig. 1b. Epitope with a high antigenicity score normally possesses high antigenic nature and this is considered as a desirable feature of a peptide to be used as vaccine candidate.
Fig. 1.
a Antigenicity of CD8 T cell epitopes of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate, b Antigenicity of CD4 T cell epitopes of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate
Investigation of hydrophilicity
Hydrophilicity score is the average hydrophilicity value of amino acids and the score is helpful in the prediction of protein structure. The hydrophilicity score of each CD8 T cell epitope for three proteins was determined having potential to raise immunogenicity in the host (Hopp and Woods 1981). The hydrophilicity score, the epitope position and total net charge of each CD8 T cell epitope were depicted in Table 1a. Out of five epitopes of surface glycoprotein, the epitope DPSKPSKRS at position 807–815 and net charge 2.00 had the highest hydrophilicity score of 12.90. In case of membrane glycoprotein, two epitopes, namely TDHSSSSDN (position 208–216) and NTDHSSSSD (position 207–215), had the highest hydrophilicity score of 6.50, and net charge -1.75. Likewise, in envelope protein, the epitope YSRVKNLNS possessed the highest hydrophilicity score of 1.40, position 59–67 and net charge 2.00.
Similarly, five CD4 T cell epitopes for each of the three proteins were identified (Hopp and Woods 1981) and their hydrophilicity scores were determined as reported in Table 2b. In surface glycoprotein, epitope ASYQTQTNSPRRARS possessed the highest hydrophilicity score of 6.40, position 671–685 and net charge 3.00. Out of five epitopes of membrane glycoprotein, epitope GNYKLNTDHSSSSDN had the highest hydrophilicity score of 5.80, position 202–216 and net charge − 0.75. In case of envelope protein, hydrophilicity scores of all the identified epitopes were less than 0; thus, these cannot be used as successful vaccine candidates. High hydrophilicity scores of peptides along with their respective locations suggest their hydrophilic sections in the protein molecules and these regions are usually exposed to external surface of the protein molecules. These sections can easily bind with MHC molecules and the complexes formed might exhibit APCs on the outer regions. The T lymphocytes can easily identify those cells and damage them.
Table 2.
CD8 (a) and CD4 (b) T cell epitopes of SARS CoV2/human/USA/CA CZB 1055/2020 isolate showing hydrophilicity score and total net charge (Hopp and Woods 1981)
| Name of proteins | CD8 T- cell epitopes (a) | Epitope position | Hydrophilicity score |
Net charge |
|---|---|---|---|---|
| Surface glycoprotein | PDPSKPSKR | 806–814 | 12.60 | 2.00 |
| DPSKPSKRS | 807–815 | 12.90 | 2.00 | |
| DYNYKLPDD | 419–427 | 5.80 | − 2.00 | |
| WNSNNLDSK | 435–443 | 2.00 | 0.00 | |
| QTQTNSPRR | 674–682 | 6.10 | 2.00 | |
| Membrane glycoprotein | TDHSSSSDN | 208–216 | 6.50 | − 1.75 |
| NTDHSSSSD | 207–215 | 6.50 | − 1.75 | |
| DHSSSSDNI | 209–217 | 5.10 | − 1.75 | |
| GNYKLNTDH | 202–210 | 1.40 | 0.25 | |
| NYKLNTDHS | 203–211 | 1.70 | 0.25 | |
| Envelope protein | YSRVKNLNS | 59–67 | 1.40 | 2.00 |
| KPSFYVYSR | 53–61 | − 2.00 | 2.00 | |
| VYSRVKNLN | 58–66 | − 0.40 | 2.00 | |
| YSFVSEETG | 2–10 | − 0.10 | − 2.00 | |
| FYVYSRVKN | 56–64 | − 3.60 | 2.00 |
| Name of proteins | CD-4 T- cell epitopes (b) | Epitope position | Hydrophilicity score | Net charge |
|---|---|---|---|---|
| Surface glycoprotein | NSNNLDSKVGGNYNY | 436–450 | − 0.30 | 0.00 |
| WNSNNLDSKVGGNYN | 435–449 | − 1.40 | 0.00 | |
| ASYQTQTNSPRRARS | 671–685 | 6.40 | 3.00 | |
| AWNSNNLDSKVGGNY | 434–448 | − 2.10 | 0.00 | |
| HRSYLTPGDSSSGWT | 244–258 | − 1.60 | 0.25 | |
| Membrane glycoprotein | GNYKLNTDHSSSSDN | 202–216 | 5.80 | − 0.75 |
| NYKLNTDHSSSSDNI | 203–217 | 4.00 | − 0.75 | |
| IGNYKLNTDHSSSSD | 201–215 | 3.80 | − 0.75 | |
| RIGNYKLNTDHSSSS | 200–214 | 3.80 | 1.25 | |
| RYRIGNYKLNTDHSS | 198–212 | 3.90 | 2.25 | |
| Envelope protein | KPSFYVYSRVKNLNS | 53–67 | − 1.60 | 3.00 |
| VKPSFYVYSRVKNLN | 52–66 | − 3.40 | 3.00 | |
| SLVKPSFYVYSRVKN | 50–64 | − 3.30 | 3.00 | |
| LVKPSFYVYSRVKNL | 51–65 | − 5.40 | 3.00 | |
| YSFVSEETGTLIVNS | 2–16 | − 5.10 | − 2.00 |
Boman Index (potential protein interaction)
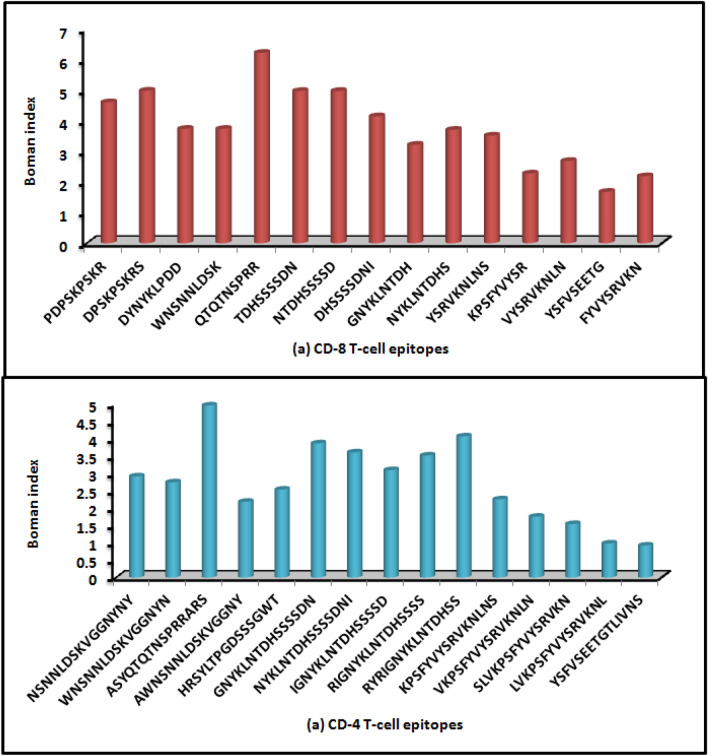
Boman index of each identified CD8 T cell epitope in three proteins of SARS-CoV-2 was estimated and shown in Fig. 2a. A minimum Boman index of 2.48 kcal/mol is considered for efficient binding with the MHC molecule. From this analysis, we could report efficient CD8 T cell epitopes that might be presented to APCs and thereby elicit immune response in the host. Notably, Boman indices of most epitopes were > 2.48 kcal/mol and thus could be used as vaccine candidates. However, a few epitopes, namely KPSFYVYSR, FYVYSRVKN and YSFVSEETG, had Boman index < 2.48 kcal/mol; thus, these epitopes would not be effective as vaccine candidates.
Fig. 2.
a Boman index of CD8 T cell epitopes of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate, b Boman index of CD4 T cell epitopes of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate
Further, the Boman indices of 15 CD4 T cell epitopes across three proteins of SARS-CoV-2 were determined and depicted in Fig. 2b. Here, the epitopes viz. AWNSNNLDSKVGGNY, KPSFYVYSRVKNLNS, VKPSFYVYSRVKNLN, SLVKPSFYVYSRVKN, LVKPSFYVYSRVKNL and YSFVSEETGTLIVNS possessed Boman index < 2.48 kcal/mol and therefore we did not consider them as effective vaccine candidates.
Investigation of amino acid contents
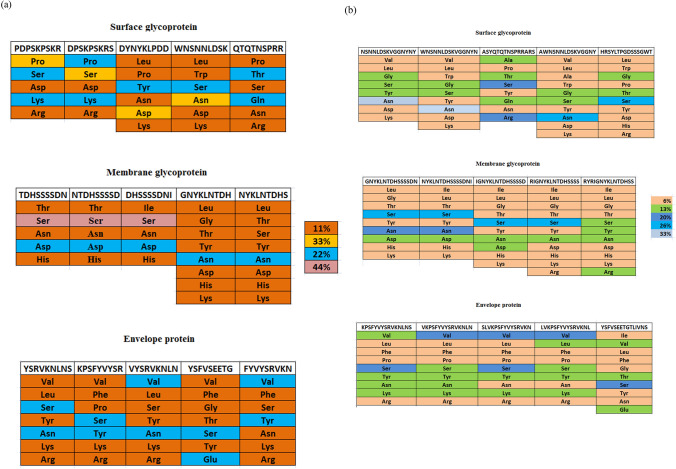
The amino acid content of CD8 T cell epitopes of SARS-CoV-2 was analyzed and represented in Fig. 3a. Distributions of amino acids were found to be different across different epitopes. Interestingly, a few amino acids viz. Pro, Ser, Asp and Asn in surface glycoprotein comprised 33% of the total composition, while Ser amino acid in membrane glycoprotein consisted of 44% of the total peptide composition.
Fig. 3.
a Composition of amino acids of each CD8 T cell epitope of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate, b Composition of amino acids of each CD4 T- cell epitope of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate
Likewise, the amino acid composition for CD4 T cell epitopes for SARS-CoV-2 proteins was analyzed and shown in Fig. 3b. The epitopes exhibited different patterns of amino acid compositions. Most CD4 T cell epitopes possessed amino acid to the tune of 6% in terms of total composition, while a few other amino acids were found to vary from 13 to 33% contents.
Moreover, the identified CD8 T cell and CD4 T cell epitopes, if used as vaccine candidates, against SARS-CoV-2 might be amenable to cleavage by several enzymes on entering the host cell, so the results of their cleavage sites are depicted in supplementary files (S1, S2).
Discussion
The outbreak of SARS-CoV-2 virus has drastically affected our globe. Not only the mortality rate that is of concern, however people who still sustain are highly affected in terms of both physical and mental health. Till date, several scholars have contributed with various paths and techniques to tackle with this pandemic situation, in addition to which different countries have also developed vaccines to combat COVID disease. However, as SARS-CoV-2 is an RNA virus, it is found to mutate itself at a very faster rate. That, in turn, leads to hurdle the effectiveness of any existing vaccine or drug. Designing of peptide vaccine using reverse vaccinology approach is less time-consuming and easy to generate. Therefore, peptide vaccine generation for any new strain of virus is easier, effective and less time-taking as compared to any other resistive method.
In current investigation, the efficient peptide sequences for potential use as vaccine candidates were identified in three proteins viz. surface glycoprotein, membrane glycoprotein and envelope protein of SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate. All the peptides identified as epitopes were evaluated using several bioinformatics tools based on the immunogenicity, antigenicity, hydrophilicity, Boman index and amino acid compositions. A few peptides were discarded in the study for not fulfilling the criteria for vaccine formulation. While, a few epitopes were reported to fulfill the selected criteria and thus we could conclude that the following epitopes of 9-mer peptides: PDPSKPSKR, DPSKPSKRS, QTQTNSPRR, and 15-mer peptides ASYQTQTNSPRRARS, RIGNYKLNTDHSSSS and RYRIGNYKLNTDHSS might generate effective immune response in the host against SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate. As the identified candidate peptide vaccines are both CD4 and CD8 cells, immunity developed through them will be of better response. These potential epitopes might be considered in the development of a novel peptide vaccine for fighting the COVID situation through prophylactic measure.
Alkhurma hemorrhagic fever virus is mostly prevalent in human beings of sub-tropical and tropical regions. This virus of Flaviviridae family was considered as a major public health concern. A study reported a total of 25 T cell (MHC-I = 17, MHC-II = 8) and 5 B cell epitopes from the envelope protein of Alkhurma hemorrhagic fever virus that could be efficiently used as epitope-based peptide vaccine against this virus (Ul-Rahman and Shabbir 2019).
Foot-and-mouth disease in swine is a contagious viral infection caused by foot-and-mouth disease virus (FMDV). Wang et al., designed a few T helper sites as peptide-based vaccine to combat their effects in swine. The G–H loop VP1 site showed cross-reactivity to virus by inclusion of cyclic constraints and adjoining sequences. On clinical trial, it was seen that 20/21 pigs got successfully immunized from infection in Taiwan using peptide dosage as low as 12.5 μg (Wang et al. 2002).
Hepatitis C virus that belongs to family Flaviviridae, is responsible for liver inflammation and it is estimated that about 58 million people have chronic hepatitis C virus infection, with nearly 1.5 million cases each year. This virus is transmitted primarily via the fecal–oral route from infected person. Yutani et al. (2007) developed personalized peptide vaccines with cytotoxic T lymphocytes (CTLs) in HLA-A24+ patients who failed to respond to interferon-based therapy to which patients responded well in Phase 1 clinical study (Yutani et al. 2007).
Peptide vaccine has also opened an area for treatment of cancer patients by cloning of genes coding for cancer antigens. In a study, gp100 melanoma-associated antigen was used to identify immunodominant peptides as a synthetic peptide to increase the binding with HLA-A2 molecules so as to treat metastatic cancer patients successfully. On clinical trial, it was seen that 91% patients were successfully immunized and holds considerable promise for development of novel cancer immunotherapies (Rosenberg et al. 1998).
Lassa virus (LASV), assigned to Arenaviridae virus family, is mainly responsible for fatal hemorrhagic fever disease known as lassa fever disease (LASV). The virus is transmitted to humans by Mastomys natalensis, a rodent and it had infected a large number of people in western Africa leading to 5000–10,000 deaths per annum. A common potential B cell and T cell epitope SSNLYKGVY that would induce immunity was identified as an efficient vaccine candidate against LASV (Hossain et al. 2018).
Aichi virus 1 (AiV-1) of Picornaviridae family causes gastroenteritis in humans and it is hazardous for children in the developing countries. From a study on this virus, three B cell epitopes, i.e., PPLPTP, LPPLPTP and PLPPDT and one T cell epitope FSIPYTSPL (binds to both MHC-I and MHC-II) were found to be efficient targets for boosting immunity against this virus (Hassan et al. 2019).
Chikungunya virus (CHIKV), transmitted by Aedes mosquitoes, causes an infectious viral disease i.e., Chikungunya infection. Based on in silico analysis, the T cell and B cell epitopes, capable of inducing immune response, were identified in the proteins of the viral strains. The potential T cell epitopes identified were KKKPGRRERMCMKIE, AEEEREAEL and DAEKEAEEEREAELT, likewise the potential B cell epitopes identified in this virus were SSKYDLECAQ and QVLKAKNIGL (Kori et al. 2015).
Merkel cell polyomavirus (MCPYV) is associated with Merkel cell carcinoma (Klionsky et al.), a type of deadly skin malignancy. MCPYV, a member of Polyomaviridae family, under the genus Orthopolyomavirus mostly affects the light-complexioned, older and immunosuppressed individuals (diagnosed with AIDS). A study predicted three B cell epitopes viz. QEKTVYP, QEKTVY and KTVYPK and T cell epitopes viz. LLVKGGVEV, SLFSNLMPK and LQMWEAISV (binding to MHC-I alleles) and ISSLINVHY, IELYLNPRM and NSLFSNLM (binding to MHC II alleles) as efficient components for peptide vaccine against MCPYV (Awad-Elkareem et al. 2017).
Conclusion
From the current study using an in silico approach, we identified a few potential CD8 T cell and CD4 T cell epitopes in three proteins (viz. surface glycoprotein, membrane glycoprotein and envelope protein) for use as components in peptide vaccine formulation against SARS-CoV-2/human/USA/CA CZB 1055/2020 isolate. To design an effective vaccine, primary criterion is to identify the epitopes with high immunogenicity score, as it would induce higher immune response in the host. Out of 15 epitopes identified as CD8 T cell epitopes, only three 9-mer peptides, namely PDPSKPSKR, DPSKPSKRS and QTQTNSPRR, of surface glycoprotein were found to have high scores for immunogenicity, antigenicity, hydrophilicity, Boman index and hydropathy and we concluded that these epitopes have great potential as vaccine candidates. In membrane glycoprotein, the epitopes, i.e., GNYKLNTDH and NYKLNTDHS possessed high immunogenicity, antigenicity, Boman index and hydropathy index; however, their hydrophilicity scores were low, so we did not consider them as potential vaccine candidate. Likewise, in envelope protein, the epitope YSRVKNLNS possessed high values of immunogenicity, antigenicity and hydropathy index; however, their hydrophilicity and Boman index were low and therefore we excluded them as vaccine candidates. Further, the epitopes possessing net negative charge were discarded from our analysis.
In the case of 15-mer CD4 T cell epitopes, we observed that the epitopes ASYQTQTNSPRRARS of surface glycoprotein; RIGNYKLNTDHSSSS and RYRIGNYKLNTDHSS of membrane glycoprotein possessed high scores of immunogenicity, antigenicity, hydrophilicity, Boman index and hydropathy index and hence we identified them as effective vaccine candidates. While, in the case of envelope protein, the hydrophilicity scores of all the five epitopes were very low and thus none of them could qualify as potential component of peptide vaccine.
Pharma industries might use these identified CD8 T cell epitopes viz. PDPSKPSKR, DPSKPSKRS and QTQTNSPRR and CD4 T cell epitopes viz. ASYQTQTNSPRRARS, RIGNYKLNTDHSSSS and RYRIGNYKLNTDHSS for developing a multi-epitope-loaded peptide vaccine by combining in various proportions with a carrier protein or strong adjuvant. These epitopes could be administered as a prophylactic vaccine, following proper clinical trials to confirm their efficacy and safety in host. This might prevent the devastating spread of the virus by boosting immunity in human.
In view of recent outbreak of SARS-CoV-2, an endless struggle for survival is continuing worldwide. It is, therefore, imperative to construct peptide vaccine comprising different sets of epitopes in various combinations. Immunity against SARS-CoV-2 could be induced in human through the large-scale use of peptide vaccine as a prophylactic measure in human populations across the globe.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to Assam University, Silchar, Assam, India for providing necessary facilities to carry out this research work. Further, the research work is dedicated to those great souls who died of global pandemic SARS-CoV-2 due to lack of proper medication and medical facilities. In addition, this research is also dedicated to all those great warriors who are in the job to combat the devastation of this virus.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by SC, BD, DN and DM. The first draft of the manuscript was written by all authors and later modified as per corresponding author’s comments on previous versions of the manuscript. All authors read and approved the final manuscript. All authors in this manuscript share equal authorship.
Funding
Not applicable.
Declarations
Conflict of interest
Authors declare no conflict of interests in the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Awad-Elkareem M, Osman S, Mohamed H, Hassan H, Abu-haraz A. Prediction and conservancy analysis of multiepitope based peptide vaccine against merkel cell polyomavirus: an immunoinformatics approach. Immunome Res. 2017;13:2. doi: 10.4172/1745-7580.1000134. [DOI] [Google Scholar]
- Hassan AA, et al. Multi epitope vaccine prediction against aichi virus using immunoinformatic approach. BioRxiv. 2019 doi: 10.1101/795427. [DOI] [Google Scholar]
- Hoffmann M, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020 doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc Natl Acad Sci. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MU, et al. Design of peptide-based epitope vaccine and further binding site scrutiny led to groundswell in drug discovery against Lassa virus. 3 Biotech. 2018;8:81. doi: 10.1007/s13205-018-1106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyotani K, Toyoshima Y, Nemoto K, Nakamura Y. Bioinformatic prediction of potential T cell epitopes for SARS-Cov-2. J Hum Genet. 2020 doi: 10.1038/s10038-020-0771-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kori P, Sajjan SS, Madagi SB. In silico prediction of epitopes for Chikungunya viral strains. J Pharm Investig. 2015;45:579–591. doi: 10.1007/s40005-015-0205-0. [DOI] [Google Scholar]
- Lai C-C, Shih T-P, Ko W-C, Tang H-J, Hsueh P-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020 doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyarzún P, Ellis JJ, Bodén M, Kobe B. PREDIVAC: CD4+ T-cell epitope prediction for vaccine design that covers 95% of HLA class II DR protein diversity. BMC Bioinformatics. 2013;14:52. doi: 10.1186/1471-2105-14-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patronov A, Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters B, Nielsen M, Sette A. T cell epitope predictions. Annu Rev Immunol. 2020;38:123–145. doi: 10.1146/annurev-immunol-082119-124838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T, Hoffert JD, Saeed F, Knepper MA. NHLBI-AbDesigner: an online tool for design of peptide-directed antibodies. Am J Physiol Cell Physiol. 2012;302:C154–C164. doi: 10.1152/ajpcell.00325.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg SA, et al. Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat Med. 1998;4:321–327. doi: 10.1038/nm0398-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Trincado JL, Gomez-Perosanz M, Reche PA. Fundamentals and methods for T-and B-cell epitope prediction. J Immunol Res. 2017 doi: 10.1155/2017/2680160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlingmann B, Castiglia KR, Stobart CC, Moore ML. Polyvalent vaccines: High-maintenance heroes. PLoS Pathog. 2018 doi: 10.1371/journal.ppat.1006904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette A, Rappuoli R. Reverse vaccinology: developing vaccines in the era of genomics. Immunity. 2010;33:530–541. doi: 10.1016/j.immuni.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ul-Rahman A, Shabbir MAB. In silico analysis for development of epitopes-based peptide vaccine against Alkhurma hemorrhagic fever virus. J Biomol Struct Dyn. 2019 doi: 10.1080/07391102.2019.1651673. [DOI] [PubMed] [Google Scholar]
- Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, et al. Effective synthetic peptide vaccine for foot-and-mouth disease in swine. Vaccine. 2002;20:2603–2610. doi: 10.1016/S0264-410X(02)00148-2. [DOI] [PubMed] [Google Scholar]
- Wu D, Wu T, Liu Q, Yang Z. The SARS-CoV-2 outbreak: what we know. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yutani S, et al. Phase I clinical study of a personalized peptide vaccination for patients infected with hepatitis C virus (HCV) 1b who failed to respond to interferon-based therapy. Vaccine. 2007;25:7429–7435. doi: 10.1016/j.vaccine.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang J-J, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan China. Allergy. 2020 doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.