Abstract
Cutting-edge hyperscanning methods led to a paradigm shift in social neuroscience. It allowed researchers to measure dynamic mutual alignment of neural processes between two or more individuals in naturalistic contexts. The ever-growing interest in hyperscanning research calls for the development of transparent and validated data analysis methods to further advance the field. We have developed and tested a dual electroencephalography (EEG) analysis pipeline, namely DEEP. Following the preprocessing of the data, DEEP allows users to calculate Phase Locking Values (PLVs) and cross-frequency PLVs as indices of inter-brain phase alignment of dyads as well as time-frequency responses and EEG power for each participant. The pipeline also includes scripts to control for spurious correlations. Our goal is to contribute to open and reproducible science practices by making DEEP publicly available together with an example mother-infant EEG hyperscanning dataset.
Keywords: Developmental hyperscanning, Dual EEG analysis, Adult-child interaction, Phase Locking Value, PLV, Cross-frequency PLV, FieldTrip
1. Background
The standard neuroscientific approach studies social information processing from an observer’s viewpoint detached from the social context (Hoehl and Markova, 2018). Experimental designs assessing social processes often consist of paradigms in which participants interact with stimuli presented on a computer screen. Although these experiments have contributed greatly to our understanding of social perception and cognition, they fall short of capturing several crucial aspects of social interaction, such as its bi-directionality and dynamic nature. The second-person neuroscience perspective argues that social cognition in an interactive context may differ fundamentally from situations in which social stimuli are merely passively observed (King-Casas et al., 2005, Schilbach et al., 2013). Interaction partners represent social exchange as a reciprocal process and act in a ‘we-mode’ as compared to acting individually (Gallotti and Frith, 2013). Based on these theoretical accounts, social neuroscience research faces the need to examine interactional phenomena in more naturalistic, ecologically valid contexts. This paradigm shift has paved the way for the emergence of hyperscanning: a technique that allows for the simultaneous recording of neural activity from two or more individuals. Using this new technique, researchers can measure the dynamic mutual alignment of neural processes between interaction partners in naturalistic settings.
As the building blocks of social cognition emerge early on in human development, hyperscanning has gained momentum among developmental researchers in recent years. Hyperscanning studies involving developmental populations have addressed a variety of social exchanges including interactions between infants and adults using EEG (Leong et al., 2017, Leong et al., 2019, Perone et al., 2020, Santamaria et al., 2020, Wass et al., 2018) and functional near-infrared spectroscopy (fNIRS; Nguyen et al., 2020, Nguyen et al., 2021; Piazza et al., 2020); interactions between children and adults using EEG (Bevilacqua et al., 2019), magnetoencephalography (MEG; Hasegawa et al., 2016; Hirata et al., 2014) and fNIRS (Azhari et al., 2019, Hoyniak et al., 2021, Kruppa et al., 2021, Miller et al., 2019, Quiñones‐Camacho et al., 2020, Reindl et al., 2018, Reindl et al., 2021); and interactions between child and adolescent peers using EEG (Dikker et al., 2017, Dikker et al., 2021) and fNIRS (Piazza et al., 2021).
Measures of quantifying the amount of EEG connectivity between individuals (i.e., inter-brain connectivity) are largely similar to methods that have been applied to examine EEG connectivity within individual brains (i.e., intra-brain connectivity). The most popularly used methods include (i) covariance in amplitude or power (i.e., amplitude envelope correlation, power correlation, projected power correlation [PPC]); (ii) phase synchrony, captured by the Phase Locking Value (PLV) (Dumas et al., 2010, Lachaux et al., 1999) or circular correlation (CCorr); and (iii) coherency-based metrics measuring the similarity of both power and phase such as Partial Directed Coherence (PDC) (Leong et al., 2017), wavelet coherence and imaginary coherence (Ayrolles et al., 2021, Burgess, 2013). In this article, we concentrate on phase synchrony. A full discussion of the other available methods is included in the paper by Marriott Haresign and colleagues, in this special issue.
Hyperscanning research is at a turning point: the development of transparent and validated data analysis tools is urgently required to ensure the reproducibility of findings. Despite the ever-growing interest in hyperscanning methods, there have been very few attempts to validate analysis techniques across studies (Ayrolles et al., 2021, Burgess, 2013, Nastase et al., 2019). A recent endeavor in this direction was undertaken by Ayrolles and colleagues (2021) who published a data analysis pipeline, namely HyPyP, to perform brain-to-brain connectivity analysis for M/EEG hyperscanning data. HyPyP includes tools for automated data preprocessing and documentation for various inter-brain connectivity measures. Such comprehensive, open-source hyperscanning analysis pipelines are valuable to advance the field. Despite being an important first step, HyPyP does not directly address the challenges of developmental hyperscanning studies, which is what we aim to cover with DEEP. Moreover, while the HyPyP toolbox is integrated with MNE-Python (Gramfort et al., 2013), DEEP offers an alternative solution based on the FieldTrip toolbox operated in MATLAB, which is widely used for neural data analysis.
Hyperscanning methods are also increasingly used alongside naturalistic paradigms in which adults, for example caregivers, are instructed to play with their children as they would do outside of the lab (e.g., Noreika et al., 2020; Wass et al., 2018). Analyzing data acquired using naturalistic paradigms poses several methodological challenges such as shorter recording sessions due to long study preparation times and movement artifacts inherent in naturalistic interactions. The problem of shorter recording sessions and higher rates of data loss due to artifacts (often 25–75%) also means that generally fewer and shorter segments of usable data can be recorded from infants and children (Debnath et al., 2020, Georgieva et al., 2020, Hoehl and Wahl, 2012). This is particularly a problem for dual (or group) EEG studies because the calculation of inter-brain phase alignment measures between two (or more) individuals requires clean data segments from all participants recorded at the same time.
Another important challenge faced by researchers analyzing dual EEG data from developmental populations is that adults and infants inherently have different EEG signal decomposition (Saby and Marshall, 2012). For example, the equivalent of the alpha frequency band, which is widely examined in the 8–12 Hz range in adults, is observed at slower frequencies such as 6–9 Hz in infants (Marshall et al., 2002). We acknowledge the theoretical challenge to explain how perceptual and cognitive processing change as a function of frequency across development, which is beyond the scope of this paper. Here, we address the unique methodological challenge when calculating phase alignment between EEG signals obtained from adult and child participants by introducing the cross-frequency PLV module of DEEP. The cross-frequency PLV method allows for computing phase-alignment across datasets with differing peak frequencies such as adult and infant EEG data. This is an important step forward to establishing transparent and robust data processing approaches and standardized data analysis practices for hyperscanning studies with developmental populations.
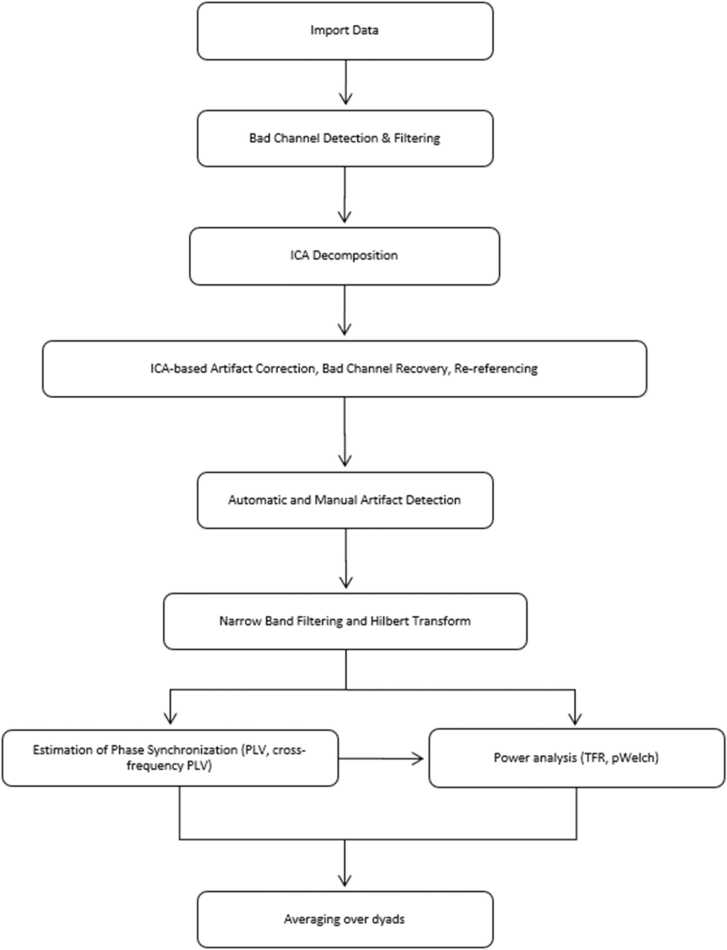
Standardized and automated EEG processing pipelines were developed in recent years to analyze infant EEG data (Debnath et al., 2020, Gabard-Durnam et al., 2018). To our knowledge, however, there have been no attempts to establish standardized and automated preprocessing pipelines to analyze dual EEG data of adults and children or infants. Here, we present and test a dual EEG pipeline, in short DEEP, which aims to address this gap. The pipeline provides users with a step by step command line interface with graphical elements, which we will refer to as graphical user interface (GUI) in the remainder of the paper, to analyze adult-infant/child and adult-adult EEG hyperscanning datasets. As illustrated in Fig. 1, the current version of the pipeline consists of nine data processing steps including: filtering, artifact identification, artifact correction and rejection, interpolation of noisy channels, Hilbert transformation and calculation of inter-brain phase alignment measures such as PLV and cross-frequency PLV.
Fig. 1.
Nine data processing steps of DEEP.
We focus here on applying phase synchronization methods as they are flexible (i.e., they can be computed over time or trials), easy to implement, and they are grounded within a large body of literature using them to look at both intra- and inter-brain phase synchrony (Liu et al., 2018). We aim to keep up with the continual development of new and existing methods for computing EEG connectivity that can be applied to dual EEG data and integrate these into the current toolbox.
In addition to dual EEG data, DEEP allows users to analyze individual EEG data using time-frequency responses and/or Welch's method. Unlike other hyperscanning pipelines, DEEP allows users to run control analyses with surrogate data to account for differences in the level of phase alignment that would be observed in the data by chance (Nguyen et al., 2020, Reindl et al., 2018; see also Burgess, 2013).
Following the overview of the pipeline, we will test the pipeline using a dataset recorded from mothers and their 8-month-old infants using dual EEG, including resting state and free play conditions. By making a ready-to-use, open-access toolbox and a complete mother-infant dual EEG dataset publicly available, we aim to facilitate open science practices in developmental research.
The scripts used in the current version of the pipeline are available at https://github.com/dmatthes1982/MPI_CBS-DEEP including code descriptions. The anonymized data used in this study is publicly available. In Supplementary Materials, among other documentation, we include a tutorial on how to operate the pipeline with snapshots of the GUI so that users can easily navigate the analysis workflow (see Supplementary Materials S1).
2. Overview of the pipeline
Below, we will describe each processing step in the pipeline in detail. We also provide a document in Supplementary Materials S3, in which we list the parameters and settings used for the preprocessing of the dataset in this paper. Please note that listed passband values were used in cross-frequency PLV calculations. For PLV estimations, we used infant passband values to process both the infant and the mother data.
2.1. Step-1: Data import
In the current version of the pipeline, only data recorded with Brain Vision Recorder software (Brain Products GmbH, Germany) can be analyzed. For every new session, first, the data needs to be imported. The raw data consist of an * .eeg file including the EEG data, a * .vhdr header file including information about the recorded electrodes and the sampling rate, and a * .vmrk file including the trigger information. The files should be named as specified in the pipeline (e.g., Studyname_Dyadnumber). In Supplementary Materials S1, we include a code to convert data recorded by other EEG systems (e.g., BioSemi) to use the pipeline.
The user can import all of the channels or exclude certain channels from the processing by manually typing the channel names. The reason behind this implementation is that channels in the periphery are known to be particularly affected by muscle and movement artifacts rendering them potentially unsuitable for analysis in many adult-infant hyperscanning studies (Georgieva et al., 2020, Wass et al., 2018). The electroocular (EOG) electrodes are omitted from exclusion in order to avoid accidental removal of these electrodes because they are necessary for Independent Component Analysis (ICA; for more information, see steps 3 and 4).
2.2. Step-2: Detection of channels to be interpolated and filtering data
In the second step, the user can select channels that will be interpolated later. To help the user to decide which channels should be interpolated, the noisiest channels are first identified. All available data of each participant are concatenated and the total power of each channel is calculated from 3 Hz onwards using the “DEEP_estNoisyChan” function. Based on a common definition of outliers, if values for a particular channel are above 1.5 * IQR + Q3 or below Q1 - 1.5 * IQR, the channel is considered noisy. Users should keep in mind that these graphs only highlight the channels that are noisier as compared to the other channels. We recommend users to double check their decision on which channels to interpolate by running the preprocessing steps 1–5 without any intervention on the channels and inform their decision using artifact values at step-5. In its current version, the pipeline requires some level of experience with EEG processing to make a decision on which channels to interpolate.
First, the adult data is examined. In the GUI, two figures and a window appear. The first figure depicts the total power of all channels including the bad channels that are marked in red. Using the FieldTrip data browser function (i.e., ft_databrowser), the second figure plots raw data for each channel across time. The user can scroll through the unfiltered data and zoom in and out to inspect the data. The figures together help the user to decide on the channels to interpolate at later steps.
In the “select bad channels'' window, the user can choose the channels to interpolate at step 4. If the user selects neighboring channels for interpolation, the pipeline will continue to operate. However, bad channels will be excluded from the repairment of a likewise bad neighbor. If one channel has no good neighboring channels left, the values of this channel will be set to zero. If a bad channel has only one good neighbor, the interpolated channel will be a duplicate of the neighboring good channel (for more information, see step-4). The GUI will display a warning message if a channel that will be used for re-referencing (e.g., TP10 for the linked mastoids method) is selected for interpolation.
Once bad channels for the adult data are selected, the same process starts for the infant data. Afterwards, the pipeline filters the data using a 1–48 Hz bandpass filter suppressing electrical line noise. To process the current dataset, we have used the default parameters and settings of DEEP, which can be found in DEEP_main_2 script (see https://github.com/dmatthes1982/MPI_CBS-DEEP/blob/master/DEEP_main_2.m). Users can change the settings in the code. In Supplementary Materials S1, we present a schematic of the customized channel configuration used in the current study.
2.3. Step-3: Cleaning prior to ICA
To plan the processing time, the user can either continue with the ICA decomposition step for all of the dyads (i.e., default) or select certain dyads. In this step, the preprocessed EEG data segments are again concatenated to continuous data. Next, transient artifacts in all channels are detected and rejected applying the criterion that within a 200 ms segment with a 50% overlap (i.e., 100 ms): the difference between the maximum and the minimum voltage cannot be more than 200 μV. This ensures that large artifacts will not influence the ICA process. The remaining clean segments are concatenated to a continuous data stream followed by the ICA decomposition using the default method “runica” (for more information on specifications, see https://www.fieldtriptoolbox.org/reference/ft_componentanalysis/). This step only involves the ICA decomposition. The selection of the ICA components takes place in the next step.
2.4. Step-4: Selection of ICA components, interpolation of bad channels and re-referencing the data
2.4.1. Selection of ICA components
At this step, if EOG data are available, the pipeline first detects the eye movement artifacts for subsequent correction based on the criterion that the detected eye movement components show a minimum of 80% correlation with the horizontal and vertical eye electrodes (i.e., calculation of the horizontal vertical EOG signal components [EOGH, EOGV] using channels F9, F10 and V1, V2 in the current layout, respectively). This default threshold can be reduced by the user manually. Using the ft_icabrowser function (https://www.fieldtriptoolbox.org/reference/ft_icabrowser/), the GUI displays a window in which the topography of the detected components over the entire scalp are shown and highlighted with a “Reject” label. Additionally, the power spectrum density graphs as a function of frequency are illustrated together with graphs that show the variance of power over time to help the user better evaluate the detected components.
If eye electrodes are missing, no correlations can be found, thus, the pipeline does not generate automatic suggestions. In that case, the users can inspect the ICA components and mark certain components as eye artifacts manually. Whereas the ICA algorithms work well with adult data, careful visual inspection of the components might be needed for verifying eye movement components in the infant data. In that case, users are expected to have some experience with identifying eye movement artifacts. Following the verification of the eye movement components, which are done separately for the data of infants and adults, eye movement artifacts are corrected.
2.4.2. Interpolation of bad channels
Next, the noisy channels that are selected at step-2 are interpolated by using the default FieldTrip channel repair method, namely the weighted neighbor approach. The weighted neighbor approach replaces signals from bad channels with the average signal of all neighboring channels. This method inherently requires good signal from surrounding channels, thus, it does not work optimally if several noisy channels need to be interpolated that are located close to each other (for more information, see https://www.fieldtriptoolbox.org/reference/ft_channelrepair/). DEEP, by default, uses an interpolation quota by only allowing 10% of the total number of channels to be interpolated. The deactivation of this function is possible but not recommended, as it is common across developmental EEG studies to interpolate a maximum of 10% of the overall number of channels to obtain meaningful results (Debnath et al., 2020).
2.4.3. Re-referencing the data
In developmental EEG studies, researchers commonly use vertex/Cz or the left or right mastoid channels as the recording reference (Hoehl and Wahl, 2012). In the recording of the current dataset, we referenced all electrodes to the left mastoid (i.e., TP9) online. However, in principle, the data can be re-referenced offline to any other electrode (e.g., right mastoid) or the average of a group of electrodes (e.g., common average re-referencing). Depending on the recorded number of channels and data quality, the pipeline allows the user to choose from the following re-referencing methods: 1) common average re-reference; 2) linked mastoids re-reference; 3) robust average re-reference. The linked mastoids offline re-referencing method uses the average of two electrodes at left and right mastoids (i.e., TP9 and TP10, respectively), whereas the common average offline re-reference method takes into account all of the electrodes.
Here, we added a third offline re-referencing method, namely robust average re-referencing, which was used in the preprocessing of the example dataset. The robust average re-referencing method is similar to the common average re-referencing approach with one main difference. Instead of taking the average of all electrodes, the robust average re-referencing method uses a group of clean channels (as identified in step-2) and calculates the average of these channels to become the reference channel for re-referencing (Bigdely-Shamlo et al., 2015). We think this method is particularly useful when recording EEG data from developmental populations using naturalistic paradigms, since motion contamination results in fewer channels with sufficient data quality compared to EEG recordings from adults. DEEP uses all electrodes that were not marked as bad at step-2 in robust average re-referencing. In other words, all electrodes that are not chosen for interpolation are the ones that are included in robust average re-referencing. In Supplementary Materials S3, we provide the list of interpolated channels with which users can infer which channels were used in robust average re-referencing for each participant.
2.5. Step-5: Automatic and manual artifact detection
We acknowledge the fact that developmental labs vary in their approach to artifact detection and removal. Accordingly, in the fifth step, DEEP allows the user to choose between one of four different algorithms for performing an automatic artifact detection to clean the data further. The four options are as follows: (1) minimum-maximum threshold: the algorithm checks whether the minimum or the maximum value within the segment exceeds a default value of + −75μV, which was used for processing the data presented in this paper. The user can also adjust this default threshold value by selecting a value from a range of 50–200 μV. If the voltage values exceed the upper and lower thresholds, the segment will be marked as an artifact. This approach was successfully applied in infant EEG studies before (e.g., Wahl et al., 2012). (2) Range threshold within 200 ms, sliding window: in a sliding 200 ms window, the algorithm checks whether the difference between the minimum and maximum value within the segment exceeds a certain threshold. If it is true, the segment will be marked as an artifact. This criterion was also used in infant EEG studies (e.g., Michel, 2017). (3) Standard deviation threshold within 200 ms, sliding window: in a 200 ms sliding window, the algorithm checks whether the standard deviation is larger than a certain threshold. If it is true, the segment will be marked as an artifact. This criterion was used for infant data in previous studies (e.g., Hoehl et al., 2008). (4) Multiple of median absolute deviation, sliding window. Thresholds and channels can be chosen for adults and infants separately.
At this step, the pipeline generates two graphs that illustrate the artifact maps for each condition. Each artifact map illustrates the artifacts in each channel over time. On the left side of the graph, the user can also view the number of artifacts for each channel in numeric form. Artifact free segments are depicted in green whereas the segments which exceed the artifact threshold are marked in red (see Supplementary Materials S1). In addition to automatic artifact detection, artifacts can be selected manually in the data browser window. After the completion of step-5, the user can select further processing options such as processing individual EEG data (i.e., step-6 followed by step-8) or running inter-brain phase alignment analyses (i.e., step-6 followed by step-7). It should be noted that the artifacts are identified at this step; however, the removal of the artifacts takes place at step-7 or step-8 depending on the analysis of interest.
2.6. Step-6: Narrow-band filtering and Hilbert transformation
The GUI asks the user to choose either the same default passband settings to be used in subsequent processing steps or separate passbands for the adult and the infant data. The default settings include the following passbands for the adult data: 1) theta [4–7 Hz] 2) alpha [8–12 Hz] 3) beta [13–30 Hz] 4) gamma [31–48 Hz]. Because of the differences in frequency bands between adult and infant data, the following settings are used for the infant data: 1) theta [3–5 Hz] 2) alpha [6–9 Hz] and 3) beta [13–30 Hz] 4) gamma [31–48 Hz]. In this paper, we examine theta and alpha frequencies, as they are most commonly used in developmental studies (e.g., Leong et al., 2017; Wass et al., 2018). We would like to note that the pipeline also allows the users to specify passband ranges manually and analyze frequency ranges such as beta and gamma. At this step, the pipeline also estimates the Hilbert phase in each of the passbands.
2.7. Step-7: Calculation of phase locking values (PLVs) and cross-frequency PLVs
PLV is the main metric used to estimate phase-locking between two signals. It measures the extent to which phase angles are similar between two signals over time or trials. PLV is calculated as follows:
| (1) |
Where N is the number of observations, is the phase on observation k, at time t, in channel and at channel. If the phase angles from the two signals fluctuate over time with a consistent difference, this will lead to PLVs close to 1. If the phase angles fluctuate over time with little consistency between each of the two signals, PLVs will be close to 0. Phase locking measures connectivity between signals with a zero lag. It is worth noting that, as phase synchrony (or phase locking) is a measure of the consistency of phase angles between two signals, where these two cycles are in relation to each other is less important than how they co-vary.
Cross-frequency phase entrainment or cross-frequency PLV shares the same underlying assumption with PLV. Cross-frequency phase entrainment or PLV m:n is calculated similarly to PLV as follows:
| (2) |
N is the number of trials and is calculated as follows:
| (3) |
Where n and m are the center frequencies of the two signals, which should be integer values satisfying the equation , and , is the phase angle at channel , at time t, on trial k, and channel . Similar to PLV, cross-frequency PLV can be applied over trials or in a time window within a trial (see Marriott Haresign et al. paper, in this special issue for more information).
DEEP applies the following steps to calculate PLVs or cross-frequency PLVs. The pipeline first segments all of the data into epochs of 1 or 5 s duration. The epoch durations can be adjusted by the user. The pipeline then rejects all of the segments which contain artifacts that were defined at step-5 (i.e., automatic and manual artifact detection). For each good epoch, it estimates one PLV or cross-frequency PLV. Following this, all estimated PLVs or cross-frequency PLVs are averaged for each condition for all of the passbands. In the current dataset, we will examine PLVs and cross-frequency PLVs at theta and alpha frequency bands. Here, we segmented the data into 1 s epochs both for theta and alpha frequencies (cf. Leong et al., 2017), as we obtained cleaner data with shorter epoch durations.
2.8. Step-8: Time-frequency responses and power spectral densities using Welch’s method
DEEP also allows users to analyze individual EEG data. In this step, users can calculate either time-frequency responses (TFRs) or power spectral densities (PSDs), separately for each participant (e.g., for infants and adults). TFRs are calculated using the FieldTrip function ft_freqanalysis (for more information on default settings, see https://www.fieldtriptoolbox.org/reference/ft_freqanalysis/). As default, the response is calculated with wavelet time-frequency transformation using Morlet wavelets, applying a Hanning window in the frequency range of 2–50 Hz in steps of 1 Hz and a time stamp of 500 ms on which the analysis is centered. These settings can manually be changed in the functions DEEP_main_8.m and in the sub-function DEEP_timeFreqanalysis.m.
At a next step, the power spectrum can be analyzed with the FieldTrip function ft_freqanalysis using the multitaper frequency transformation (“mtmfft”) method on a Hanning window with the default settings of a 1000 ms window length with 75% overlap for the frequency range of 1–50 Hz in steps of 1 Hz. The calculation of the power spectrum of the entire condition is a median operation, which allows the user to suppress the influence of outliers in the data across long periods that are common in naturalistic paradigms. These settings can manually be changed in the functions DEEP_main_8.m and in the sub-function DEEP_pWelch.m.
2.9. Step-9: Averaging PLVs or cross-frequency PLVs, TFRs and PSD values over dyads or participants
In this final step, the user can select data of dyads or participants to estimate the mean of the PLVs or cross-frequency PLVs, TFRs and PSDs for different conditions.
3. Results
We used a dual EEG dataset recorded from 70 8-month-old infants (36 girls, Mage= 8.29 months, SDage= 8.03 days) and their mothers (Mage= 33.40 years, SDage= 4.62 years). The sample included in the analyses consisted of 35 infants (22 girls, Mage= 8.27 months, SDage= 8.17 days) and their mothers (Mage= 34.28 years, SDage= 4.03 years). Our original study included several experimental conditions. In this paper, we analyzed two conditions: 1) resting state (45 s, marker S13) and 2) free play (150 s, marker S11). In the resting state condition, the infants and their mothers together watched an experimenter blowing bubbles as their EEG activity was recorded concurrently. In the free play condition, the mothers were instructed to play with their infants as they would do at home, without using toys. The mothers did not receive further instructions.
Previous studies (e.g., Muthukumaraswamy and Singh, 2011; Burgess, 2013) suggested that increases in power can lower error in phase estimation and give the appearance of increased phase locking. Though this has been primarily examined as an event-locked phenomenon, we were curious if this might also be a feature of continuous EEG recordings, which are common within developmental social neuroscience. Accordingly, we first examined whether systematic relationships could be observed between average theta and alpha power across the whole testing session (considered in the infant and adult separately) and averaged infant-adult theta and alpha cross-frequency PLVs. For comparison, we ran the same analyses with PLVs.
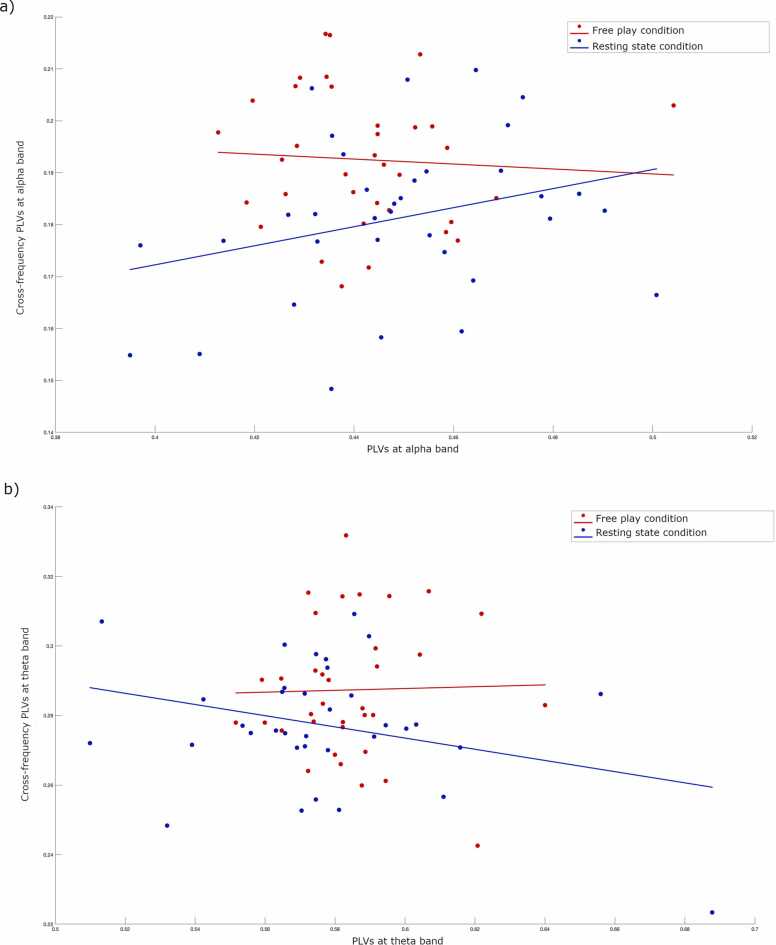
We found a significant relationship between infant-adult theta cross-frequency PLVs and infant theta power in the free play condition (rho=0.42, p = .01), but not in the resting state condition (rho=0.04, p = .82), indicating a positive association between infant theta power and infant-adult theta cross-frequency PLVs during free play. The relationships between infant-adult alpha cross-frequency PLVs and infant alpha power were not significant in any of the conditions. No consistent relationships between infant-adult cross-frequency PLVs and adult power were observed in either theta or alpha frequency bands. Similar to cross-frequency PLV findings, we observed a marginally significant relationship between infant theta power and infant-adult PLVs in the free play condition (rho= 0.31, p = .07) but not in the resting state condition (rho=0.24, p = .17). This increases confidence that, when considering averaged values across the whole testing session, obtained phase alignment values are largely independent of power (see Supplementary Materials S4-S5, Fig. 1a to d). However, one explanation for stronger power and phase locking associations for the free play condition compared with resting state condition may be that free play condition showed more rapid transient fluctuations in power, which could affect phase entrainment via the mechanisms suggested by Burgess (2013).
Next, we examined the relationships between cross-frequency PLVs and PLVs in the resting state and free play conditions at alpha and theta frequencies separately. Whereas we observed a marginally significant relationship between cross-frequency PLVs and PLVs in the resting state condition at alpha frequency (rho=0.32, p = .06), no other correlation reached significance (all ps >0.14). These findings are illustrated in Fig. 2a and b.
Fig. 2.
Scatterplots illustrating Spearman’s correlation results showing the associations between cross-frequency PLVs and PLVs. a) associations between cross-frequency PLVs and PLVs at alpha frequency in resting state and free play conditions. b) associations between cross-frequency PLVs and PLVs at theta frequency in resting state and free play conditions. Blue and red colors represent resting state and free play conditions, respectively. Lines indicate the direction of the relationship. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
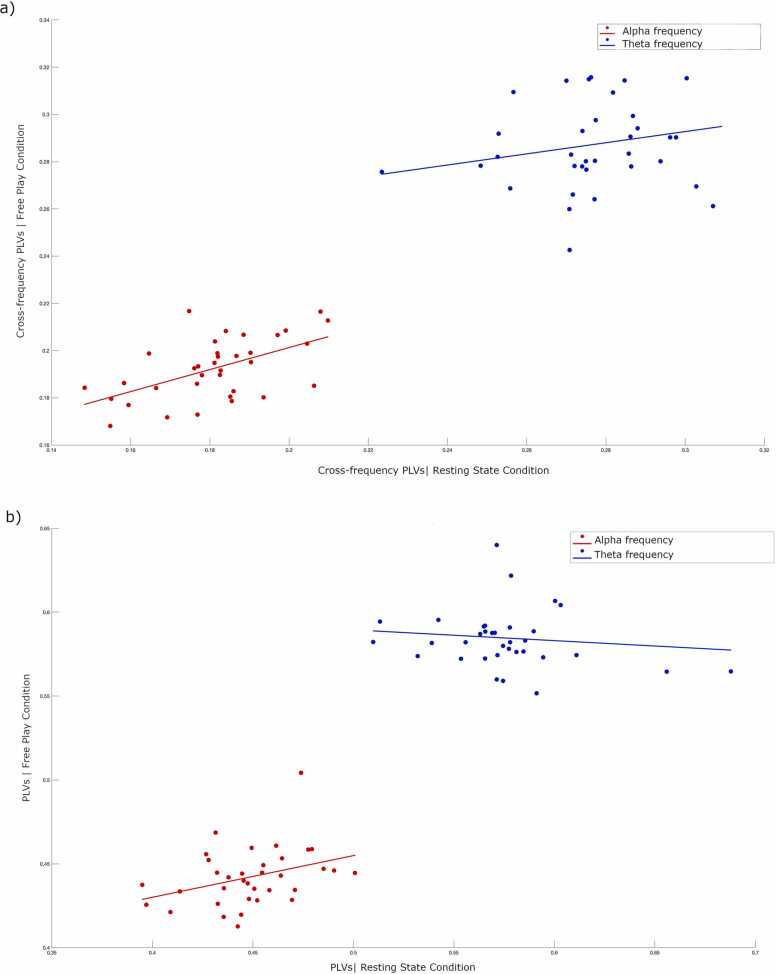
As an exploratory analysis, we examined the consistency in alpha and theta cross-frequency PLVs between the resting state and free play conditions. As shown in Fig. 3a and b, we observed a strong correlation between cross-frequency PLVs in free play and resting state conditions in the alpha band (rho=0.50, p < .01), but not in the theta band (rho=0.25, p = .14). Similarly, there was a significant relationship between PLVs in free play and resting state conditions in the alpha band (rho=0.37, p = .03) but not in theta band (rho= −0.11, p = .53).
Fig. 3.
Scatterplots illustrating Spearman’s correlation results showing the associations between different conditions. a) cross-frequency PLVs in resting state and free play conditions in the alpha and theta bands. b) PLVs in resting state and free play conditions in the alpha and theta bands. Red and blue colors represent alpha and theta frequencies, respectively. Lines indicate the direction of the relationship. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
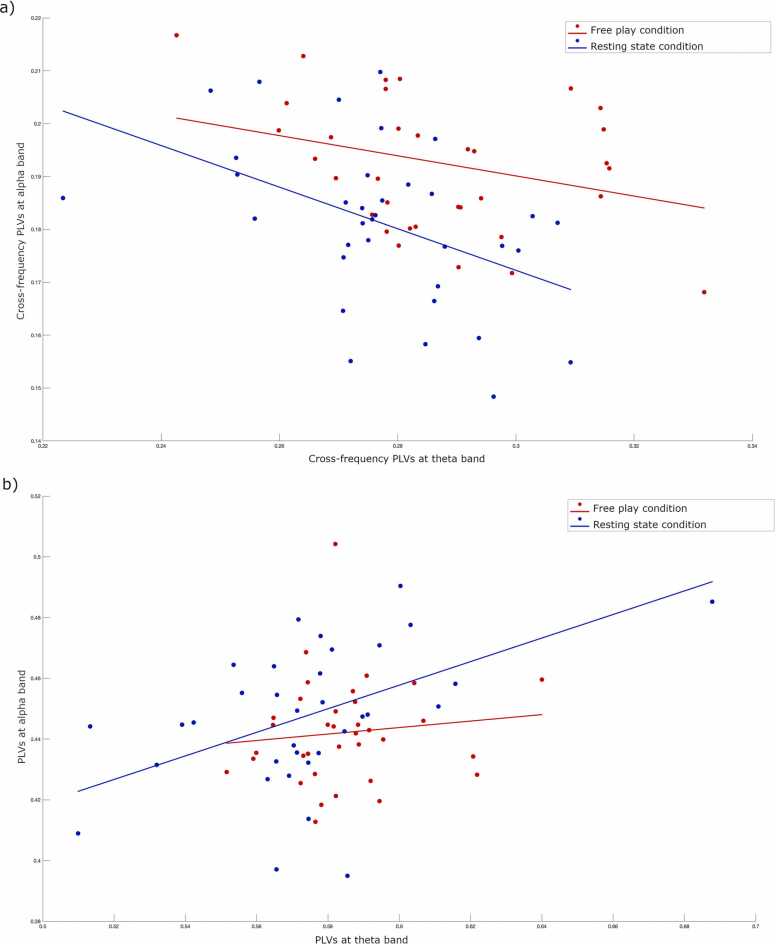
Also as an exploratory analysis, we examined the relationship between alpha cross-frequency PLVs and theta cross-frequency PLVs across conditions (see Fig. 4a and b). Here, we observed negative relationships for both the free play (rho= −0.27, p = .12) and resting state conditions, although only the latter relationship was significant (rho= −0.46, p < .01), such that stronger alpha cross-frequency PLVs were associated with lower theta cross-frequency PLVs. Regarding PLVs, we observed a relationship in the opposite direction (stronger alpha PLVs associated with increased theta PLVs), which was again only significant for the resting state condition (rho=0.49, p < . 01).
Fig. 4.
Scatterplots illustrating Spearman’s correlation results showing the associations between different frequency bands. a) alpha cross-frequency PLVs and theta cross-frequency PLVs in resting state and free play conditions. b) alpha PLVs and theta PLVs in resting state and free play conditions. Blue and red colors represent resting state and free play conditions, respectively. Lines indicate the direction of the relationship.
4. Discussion
Recent shifts towards studying social perception and cognition using naturalistic paradigms pave the way for cutting-edge data recording techniques such as hyperscanning. These methods allow social neuroscientists to study dynamic interactions between two or more individuals in real-life settings. However, EEG hyperscanning results have not always replicated well (Konvalinka and Roepstorff, 2012). The root cause of this is unknown, however what is clear is the field's need for transparent and standardized data analysis methods to aid the replicability of findings. To our knowledge, there is only one pipeline, namely HyPyP, which aims to fulfill this urgent need in the field of dual EEG research (Ayrolles et al., 2021). However, there has been no attempt to develop and test data analysis pipelines that particularly address developmental EEG hyperscanning studies. Here, we present DEEP, which allows the analysis of dual EEG data obtained from developmental populations. By giving flexibility to users in selecting different parameters and settings for the adult and the infant data, DEEP optimizes the analysis of hyperscanning data recorded from participants at different age groups. Users can preprocess the data of two individuals followed by the calculations of inter-brain phase alignment indices such as PLVs or cross-frequency PLVs. DEEP also runs control analyses on randomly generated data to validate the results. To our knowledge, this is the first dual EEG pipeline that incorporates surrogate analysis steps. In addition to analyzing data concurrently obtained from infants/children and adults, DEEP can also be used to analyze EEG data simultaneously recorded from two adults or participants at similar ages.
We tested the pipeline with an infant-adult EEG hyperscanning dataset that included a resting state and a free play condition. One interesting observation was that inter-brain phase alignment values, as quantified by the cross-frequency PLVs in theta and alpha frequencies, were negatively associated, particularly in the resting state condition in which the mother and the infant jointly watched the experimenter blowing bubbles. Although more research is needed to understand what theta synchronization and alpha desynchronization represent in terms of inter-brain synchrony, this disjunction between theta and alpha frequencies is commonly observed in intra-individual measurements of attention and learning, where alpha power suppression (i.e., desynchronization) often goes along with theta power increases (i.e., synchronization) (e.g., Köster et al., 2019). Because this paper aims to introduce DEEP, we will examine these findings more in detail in separate papers.
As this is the first version of DEEP, there are still open issues that we aim to address in future versions. To start with, at the moment, DEEP only allows users to analyze EEG datasets acquired by the Brain Products EEG recording system. We will adjust the pipeline such that several EEG data structures can be read into the pipeline. In the meantime, we provide users with a data conversion method for BioSemi data, which can be found in Supplementary Materials S1. Moreover, the current version of the pipeline only processes dual EEG datasets such as the ones from infant/child and adult dyads or two adults. We strive to extend the pipeline such that data from three or more individuals could be examined, which would be particularly helpful in studies that are conducted in group settings (e.g., Dikker et al., 2017; Reinero et al., 2021).
Although the data of the infant and the adult are processed one by one with different parameters and settings defined by the user, roughly the same basic preprocessing steps are applied to both of the datasets. In future versions, we aim to modify the pipeline to enable separate preprocessing steps for the infant and adult data. For example, given that the channels in the outer ring of electrodes are particularly noisy for the infant data (Debnath et al., 2020), one can choose to remove the electrodes in the periphery for the infant data but not for the adult data, which could be particularly useful when running intra-brain analyses (e.g., see step-8). Similarly, given the differences in the efficacy of ICA methods to correct for eye movement artifacts in the adult and infant data, it would be useful to apply separate ICA processes. Moreover, it would be highly interesting to extend the pipeline by including interpersonal synchrony measures that address the directionality between the two signals (i.e., one predicts the other) such as Partial Directed Coherence (PDC; Baccalá and Sameshima, 2001), which has been previously used in developmental studies (e.g., Leong et al., 2017). Finally, in the future versions of the pipeline, we hope to implement further surrogate analyses methods such as the phase randomization approach to examine whether using different surrogate methods affects the results.
A common practice in infant EEG data analysis for reducing artifacts is to reject contaminated EEG epochs manually or automatically (Hoehl et al., 2012). Since automatic methods are optimized for adult EEG data (Bigdely-Shamlo et al., 2015, da Cruz et al., 2018, Pedroni et al., 2019), they often fail to detect artifacts exceeding rejection thresholds in the infant EEG data. Therefore, manual procedures such as artifact rejection based on visual inspection were widely used by developmental researchers. However, this approach decreases the replicability of the methodology and the results (Debnath et al., 2020). Another important step in advancing the pipeline would be to integrate automatic ICA classification and detection algorithms that are specifically established and tested for developmental populations. For example, recently Marriott Haresign et al. (2021) adapted the widely used MARA system (for ICA classification of adult data) to address the characteristics of developmental EEG data recorded using naturalistic paradigms. The retrained “iMARA” classifier was shown to discriminate artifactual ICA components from neural ICA components with a 75% accuracy rate in infant EEG data. Integrating such algorithms will further expand the pipeline by enabling the automatic identification and removal of artifactual ICA components in the infant data.
5. Conclusions
Developmental labs vary in their approach to processing EEG data. Although it remains a challenge to use automatized processing tools for developmental data due to high interindividual differences, there has been some recent progress in establishing standardized tools to analyze developmental EEG data. In this paper, we present DEEP, a pipeline which allows researchers to analyze dual EEG data obtained from infant/child-adult dyads or two adults. DEEP promotes standardized and transparent data analyses approaches while accommodating a variety of data handling approaches remediating replication issues in the field. The pipeline includes several preprocessing steps followed by the calculation of PLVs and cross-frequency PLVs as indices of inter-brain phase alignment. DEEP also allows researchers to validate their findings by running control analysis for spurious hyper-correlations. We plan to expand DEEP further in the future. The most up-to-date version of the pipeline can be accessed at https://github.com/dmatthes1982/MPI_CBS-DEEP together with the code and descriptions. We also publish an example hyperscanning dataset from infants and adults. With DEEP, we aim to provide developmental social neuroscientists with an accessible tool to analyze EEG hyperscanning datasets. By making the codes publicly available together with an example dataset, we are committed to move the field one step further in the process of making science open and reproducible.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the families who participated in the study. We are thankful to Umut Can Vural for his assistance with data processing and the preparation of the DEEP user manual. We also thank Mohammed Alahmadi and Jessica Gärtner for their contribution to surrogate data scripts. This work is supported by the Max Planck Society. E.K. is funded by the Deutsche Forschungsgemeinschaft (DFG) (project number: 402789467).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dcn.2022.101104.
Contributor Information
Ezgi Kayhan, Email: kayhan@uni-potsdam.de.
Daniel Matthes, Email: daniel.matthes@htwk-leipzig.de.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Supplementary material
.
References
- Ayrolles A., Brun F., Chen P., Djalovski A., Beauxis Y., Delorme R., Bourgeron T., Dikker S., Dumas G. HyPyP: a hyperscanning python pipeline for inter-brain connectivity analysis. Soc. Cogn. Affect. Neurosci. 2021;16(1–2):72–83. doi: 10.1093/scan/nsaa141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhari A., Leck W.Q., Gabrieli G., Bizzego A., Rigo P., Setoh P., Bornstein M.H., Esposito G. Parenting stress undermines mother-child brain-to-brain synchrony: a hyperscanning study. Sci. Rep. 2019;9(1):1–9. doi: 10.1038/s41598-019-47810-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccalá L.A., Sameshima K. Partial directed coherence: a new concept in neural structure determination. Biol. Cybern. 2001;84(6):463–474. doi: 10.1007/PL00007990. [DOI] [PubMed] [Google Scholar]
- Bevilacqua D., Davidesco I., Wan L., Chaloner K., Rowland J., Ding M., Poeppel D., Dikker S. Brain-to-brain synchrony and learning outcomes vary by student–teacher dynamics: evidence from a real-world classroom electroencephalography study. J. Cogn. Neurosci. 2019;31(3):401–411. doi: 10.1162/jocn_a_01274. [DOI] [PubMed] [Google Scholar]
- Bigdely-Shamlo N., Mullen T., Kothe C., Su K.-M., Robbins K.A. The PREP pipeline: standardized preprocessing for large-scale EEG analysis. Front. Neuroinformatics. 2015;9:16. doi: 10.3389/fninf.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A.P. On the interpretation of synchronization in EEG hyperscanning studies: a cautionary note. Front. Hum. Neurosci. 2013;7:881. doi: 10.3389/fnhum.2013.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cruz J.R., Chicherov V., Herzog M.H., Figueiredo P. An automatic pre-processing pipeline for EEG analysis (APP) based on robust statistics. Clin. Neurophysiol. 2018;129(7):1427–1437. doi: 10.1016/j.clinph.2018.04.600. [DOI] [PubMed] [Google Scholar]
- Debnath R., Buzzell G.A., Morales S., Bowers M.E., Leach S.C., Fox N.A. The Maryland analysis of developmental EEG (MADE) pipeline. Psychophysiology. 2020;57(6) doi: 10.1111/psyp.13580. [DOI] [PubMed] [Google Scholar]
- Dikker S., Michalareas G., Oostrik M., Serafimaki A., Kahraman H.M., Struiksma M.E., Poeppel D. Crowdsourcing neuroscience: Inter-brain coupling during face-to-face interactions outside the laboratory. NeuroImage. 2021;227 doi: 10.1016/j.neuroimage.2020.117436. [DOI] [PubMed] [Google Scholar]
- Dikker S., Wan L., Davidesco I., Kaggen L., Oostrik M., McClintock J., Rowland J., Michalareas G., Bavel J.J.V., Ding M., Poeppel D. Brain-to-brain synchrony tracks real-world dynamic group interactions in the classroom. Curr. Biol. 2017;27(9):1375–1380. doi: 10.1016/j.cub.2017.04.002. [DOI] [PubMed] [Google Scholar]
- Dumas G., Nadel J., Soussignan R., Martinerie J., Garnero L. Inter-brain synchronization during social interaction. PLOS ONE. 2010;5(8) doi: 10.1371/journal.pone.0012166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabard-Durnam L.J., Mendez Leal A.S., Wilkinson C.L., Levin A.R. The Harvard automated processing pipeline for electroencephalography (Happe): standardized processing software for developmental and high-artifact data. Front. Neurosci. 2018;12:97. doi: 10.3389/fnins.2018.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallotti M., Frith C.D. Social cognition in the we-mode. Trends Cogn. Sci. 2013;17(4):160–165. doi: 10.1016/j.tics.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Georgieva S., Lester S., Noreika V., Yilmaz M.N., Wass S., Leong V. Toward the understanding of topographical and spectral signatures of infant movement artifacts in naturalistic EEG. Front. Neurosci. 2020;14:352. doi: 10.3389/fnins.2020.00352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gramfort A., Luessi M., Larson E., Engemann D.A., Strohmeier D., Brodbeck C., Goj R., Jas M., Brooks T., Parkkonen L., Hämäläinen M. MEG and EEG data analysis with MNE-Python. Front. Neurosci. 2013;7:267. doi: 10.3389/fnins.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa C., Ikeda T., Yoshimura Y., Hiraishi H., Takahashi T., Furutani N., Hayashi N., Minabe Y., Hirata M., Asada M., Kikuchi M. Mu rhythm suppression reflects mother-child face-to-face interactions: a pilot study with simultaneous MEG recording. Sci. Rep. 2016;6(1):1–8. doi: 10.1038/srep34977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata M., Ikeda T., Kikuchi M., Kimura T., Hiraishi H., Yoshimura Y., Asada M. Hyperscanning MEG for understanding mother–child cerebral interactions. Front. Hum. Neurosci. 2014;8:118. doi: 10.3389/fnhum.2014.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehl S., Markova G. Moving developmental social neuroscience toward a second-person approach. PLOS Biol. 2018;16(12) doi: 10.1371/journal.pbio.3000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehl S., Wahl S. Recording infant ERP data for cognitive research. Dev. Neuropsychol. 2012;37(3):187–209. doi: 10.1080/87565641.2011.627958. [DOI] [PubMed] [Google Scholar]
- Hoehl S., Wahl S., Michel C., Striano T. Effects of eye gaze cues provided by the caregiver compared to a stranger on infants’ object processing. Dev. Cogn. Neurosci. 2012;2(1):81–89. doi: 10.1016/j.dcn.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehl S., Wiese L., Striano T. Young infants’ neural processing of objects is affected by eye gaze direction and emotional expression. PLOS ONE. 2008;3(6) doi: 10.1371/journal.pone.0002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyniak C.P., Quiñones-Camacho L.E., Camacho M.C., Chin J.H., Williams E.M., Wakschlag L.S., Perlman S.B. Adversity is linked with decreased parent-child behavioral and neural synchrony. Dev. Cogn. Neurosci. 2021;48(6) doi: 10.1016/j.dcn.2021.100937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Casas B., Tomlin D., Anen C., Camerer C.F., Quartz S.R., Montague P.R. Getting to know you: reputation and trust in a two-person economic exchange. Science. 2005;308(5718):78–83. doi: 10.1126/science.1108062. [DOI] [PubMed] [Google Scholar]
- Konvalinka I., Roepstorff A. The two-brain approach: how can mutually interacting brains teach us something about social interaction? Front. Hum. Neurosci. 2012;6:215. doi: 10.3389/fnhum.2012.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster M., Langeloh M., Hoehl S. Visually entrained theta oscillations increase for unexpected events in the infant brain. Psychol. Sci. 2019;30(11):1656–1663. doi: 10.1177/0956797619876260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruppa J.A., Reindl V., Gerloff C., Oberwelland Weiss E., Prinz J., Herpertz-Dahlmann B., Konrad K., Schulte-Rüther M. Brain and motor synchrony in children and adolescents with ASD: an fNIRS hyperscanning study. Soc. Cogn. Affect. Neurosci. 2021;16(1):103–116. doi: 10.1093/scan/nsaa092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaux J.-P., Rodriguez E., Martinerie J., Varela F.J. Measuring phase synchrony in brain signals. Hum. Brain Mapp. 1999;8(4):194–208. doi: 10.1002/(SICI)1097-0193(1999)8:4<194::AID-HBM4>3.0.CO;2-C. https://doi.org/10.1002%2F(SICI)1097-0193(1999)8%3A4%3C194%3A%3AAID-HBM4%3E3.0.CO%3B2-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong V., Byrne E., Clackson K., Georgieva S., Lam S., Wass S. Speaker gaze increases information coupling between infant and adult brains. Proc. Natl. Acad. Sci. USA. 2017;114(50):13290–13295. doi: 10.1073/pnas.1702493114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, V., Noreika, V., Clackson, K., Georgieva, S., Brightman, L., Nutbrown, R.,. & Wass, S. (2019). Mother-infant interpersonal neural connectivity predicts infants’ social learning. PsyArXiv.〈https://psyarxiv.com/gueaq〉.
- Liu D., Liu S., Liu X., Zhang C., Li A., Jin C., Chen Y., Wang H., Zhang X. Interactive brain activity: review and progress on EEG-based hyperscanning in social interactions. Front. Psychol. 2018;9:1862. doi: 10.3389/fpsyg.2018.01862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott Haresign, I., Phillips, E., Whitehorn, M., Noreika, V., Jones, E., Leong, V., & Wass, S.V. (2021). Automatic classification of ICA components from infant EEG using MARA. bioRxiv. 10.1101/2021.01.22.427809. [DOI] [PMC free article] [PubMed]
- Marshall P.J., Bar-Haim Y., Fox N.A. Development of the EEG from 5 months to 4 years of age. Clin. Neurophysiol.: Off. J. Int. Fed. Clin. Neurophysiol. 2002;113(8):1199–1208. doi: 10.1016/s1388-2457(02)00163-3. [DOI] [PubMed] [Google Scholar]
- Michel C., Kaduk K., Ni Choisdealbha À., Reid V.M. Event-related potentials discriminate familiar and unusual goal outcomes in 5-month-olds and adults. Dev. Psychol. 2017;53(10):1833. doi: 10.1037/dev0000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.G., Vrtička P., Cui X., Shrestha S., Hosseini S.M.H., Baker J.M., Reiss A.L. Inter-brain synchrony in mother-child dyads during cooperation: an fNIRS hyperscanning study. Neuropsychologia. 2019;124:117–124. doi: 10.1016/j.neuropsychologia.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthukumaraswamy S.D., Singh K.D. A cautionary note on the interpretation of phase-locking estimates with concurrent changes in power. Clin. Neurophysiol. 2011;122(11):2324–2325. doi: 10.1016/j.clinph.2011.04.003. [DOI] [PubMed] [Google Scholar]
- Nastase S.A., Gazzola V., Hasson U., Keysers C. Measuring shared responses across subjects using intersubject correlation. Soc. Cogn. Affect. Neurosci. 2019;14(6):667–685. doi: 10.1093/scan/nsz037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Schleihauf H., Kayhan E., Matthes D., Vrtička P., Hoehl S. The effects of interaction quality on neural synchrony during mother-child problem solving. Cortex. 2020;124:235–249. doi: 10.1016/j.cortex.2019.11.020. [DOI] [PubMed] [Google Scholar]
- Nguyen T., Schleihauf H., Kungl M., Kayhan E., Hoehl S., Vrtička P. Interpersonal neural synchrony during father–child problem solving: an fNIRS hyperscanning study. Child Dev. 2021;92(4):565–580. doi: 10.1111/cdev.13510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noreika V., Georgieva S., Wass S., Leong V. 14 challenges and their solutions for conducting social neuroscience and longitudinal EEG research with infants. Infant Behav. Dev. 2020;58 doi: 10.1016/j.infbeh.2019.101393. [DOI] [PubMed] [Google Scholar]
- Pedroni A., Bahreini A., Langer N. Automagic: standardized preprocessing of big EEG data. NeuroImage. 2019;200:460–473. doi: 10.1016/j.neuroimage.2019.06.046. [DOI] [PubMed] [Google Scholar]
- Perone S., Gartstein M.A., Anderson A.J. Dynamics of frontal alpha asymmetry in mother-infant dyads: Insights from the still face paradigm. Infant Behav. Dev. 2020;61 doi: 10.1016/j.infbeh.2020.101500. [DOI] [PubMed] [Google Scholar]
- Piazza E.A., Cohen A., Trach J., Lew-Williams C. Neural synchrony predicts children’s learning of novel words. Cognition. 2021;214 doi: 10.1016/j.cognition.2021.104752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piazza E.A., Hasenfratz L., Hasson U., Lew-Williams C. Infant and adult brains are coupled to the dynamics of natural communication. Psychol. Sci. 2020;31(1):6–17. doi: 10.1177/0956797619878698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiñones‐Camacho L.E., Fishburn F.A., Camacho M.C., Hlutkowsky C.O., Huppert T.J., Wakschlag L.S., Perlman S.B. Parent–child neural synchrony: a novel approach to elucidating dyadic correlates of preschool irritability. J. Child Psychol. Psychiatry. 2020;61(11):1213–1223. doi: 10.1111/jcpp.13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl V., Gerloff C., Scharke W., Konrad K. Brain-to-brain synchrony in parent-child dyads and the relationship with emotion regulation revealed by fNIRS-based hyperscanning. NeuroImage. 2018;178:493–502. doi: 10.1016/j.neuroimage.2018.05.060. [DOI] [PubMed] [Google Scholar]
- Reindl, V., Wass, S., Leong, V., Scharke, W., Wistuba, S., Wirth, C.L., Konrad, K., & Gerloff, C. (2021). Synchrony of mind and body are distinct in mother-child dyads. bioRxiv. 10.1101/2021.02.21.432077. [DOI] [PubMed]
- Reinero D.A., Dikker S., Van Bavel J.J. Inter-brain synchrony in teams predicts collective performance. Soc. Cogn. Affect. Neurosci. 2021;16(1–2):43–57. doi: 10.1093/scan/nsaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saby J.N., Marshall P.J. The utility of EEG band power analysis in the study of infancy and early childhood. Dev. Neuropsychol. 2012;37(3):253–273. doi: 10.1080/87565641.2011.614663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria L., Noreika V., Georgieva S., Clackson K., Wass S., Leong V. Emotional valence modulates the topology of the parent-infant inter-brain network. NeuroImage. 2020;207 doi: 10.1016/j.neuroimage.2019.116341. [DOI] [PubMed] [Google Scholar]
- Schilbach L., Timmermans B., Reddy V., Costall A., Bente G., Schlicht T., Vogeley K. Toward a second-person neuroscience. Behav. Brain Sci. 2013;36(4):393–414. doi: 10.1017/S0140525×12000660. [DOI] [PubMed] [Google Scholar]
- Wahl S., Michel C., Pauen S., Hoehl S. Head and eye movements affect object processing in 4-month-old infants more than an artificial orientation cue. Br. J. Dev. Psychol. 2012;31:212–230. doi: 10.1111/bjdp.12001. [DOI] [PubMed] [Google Scholar]
- Wass S.V., Noreika V., Georgieva S., Clackson K., Brightman L., Nutbrown R., Covarrubias L.S., Leong V. Parental neural responsivity to infants’ visual attention: how mature brains influence immature brains during social interaction. PLOS Biol. 2018;16(12) doi: 10.1371/journal.pbio.2006328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material