Abstract
Copy number variation (CNV) is an important genetic mechanism that drives evolution and generates new phenotypic variations. To explore the impact of CNV on chicken domestication and breed shaping, the whole-genome CNVs were detected via multiple methods. Using the whole-genome sequencing data from 51 individuals, corresponding to six domestic breeds and wild red jungle fowl (RJF), we determined 19,329 duplications and 98,736 deletions, which covered 11,123 copy number variation regions (CNVRs) and 2,636 protein-coding genes. The principal component analysis (PCA) showed that these individuals could be divided into four populations according to their domestication and selection purpose. Seventy-two highly duplicated CNVRs were detected across all individuals, revealing pivotal roles of nervous system (NRG3, NCAM2), sensory (OR), and follicle development (VTG2) in chicken genome. When contrasting the CNVs of domestic breeds to those of RJFs, 235 CNVRs harboring 255 protein-coding genes, which were predominantly involved in pathways of nervous, immunity, and reproductive system development, were discovered. In breed-specific CNVRs, some valuable genes were identified, including HOXB7 for beard trait in Beijing You chicken; EDN3, SLMO2, TUBB1, and GFPT1 for melanin deposition in Silkie chicken; and SORCS2 for aggressiveness in Luxi Game fowl. Moreover, CSMD1 and NTRK3 with high duplications found exclusively in White Leghorn chicken, and POLR3H, MCM9, DOCK3, and AKR1B1L found in Recessive White Rock chicken may contribute to high egg production and fast-growing traits, respectively. The candidate genes of breed characteristics are valuable resources for further studies on phenotypic variation and the artificial breeding of chickens.
Keywords: chicken, copy number variation, evolution, domestication, breed-specific
Introduction
Since the days of Darwin, it has been recognized that a succession of livestock species leads to significant differences in behavior, morphology, and physiology in response to domestication compared with their wild ancestors (Charles Darwin, 1859). Deciphering the genetic basis, molecular mechanisms, and evolutionary driving forces of the complex traits that have been innovated or reshaped by human manipulation during livestock domestication and breed formation has long captured the interest of animal biologists in resource utilization and animal breeding. The development of genomic sequencing technologies provides more powerful tools for a comprehensive understanding of the genetic architectures and evolutionary trajectories of complicated traits in humans and animals (International Chicken Genome Sequencing, 2004; Scally et al., 2012). In addition to the single nucleotide polymorphisms (SNPs), structural variation has been recognized as another crucial factor for driving phenotypic variations, complex diseases, and developmental abnormalities, such as obesity, diabetes, psychiatric diseases, and cancers (Sebat et al., 2004; Feuk et al., 2006; Freeman et al., 2006; Redon et al., 2006). Copy number variation (CNV) is a major type of structural variation in the genome and generally defined by the insertion, duplication, or deletion of a relatively large size of DNA with length >50 bp; this process contributes to much more variability than SNPs (Iafrate et al., 2004; Freeman et al., 2006). Various molecular mechanisms have been proposed for the formation of CNVs, with non-allelic homologous recombination being regarded as a major source of structural variation in regions of extended homology (Hastings et al., 2009; Arlt et al., 2012). CNVs typically affect gene expression and phenotypic specialization directly through dosage compensation (Zhou et al., 2011), or indirectly through altering gene expression by reshaping the three-dimensional genome architecture and local DNA accessibility; thus, they influence the regulatory relationships or intensities between the regulatory elements and targeted genes (Beroukhim et al., 2016; Franke et al., 2016; Rice and Mclysaght, 2017).
CNVs play an important role in the evolutionary adaptation of an organism under both natural and artificial selection, affecting fitness and reproductive ability, which indicates a significant source of adaptive potential. The copy number of AMY1 is strongly correlated with the evolution of diets; specifically, individuals with high-starch diets have more AMY1 copies than those with low-starch diets (Perry et al., 2007). Minias et al. (2019) reported that the evolution of the MHC copy number in birds was driven by different selective pressures, such as by intra- and extracellular pathogens and parasites. Besides, the CNVs have been implicated in phenotypic variability of traits important for domestication and breed formation in many livestock species. The duplication of KIT is significantly associated with white coat color in pigs (Giuffra et al., 2002) and cattle (Durkin et al., 2012). The chicken pea-comb is caused by the duplication of SOX5 in intron 1 (Wright et al., 2009). The CNV of ZNF280AY was negatively correlated with male reproduction trait in Holstein and Simmental bulls (Pei et al., 2019).
Domestic chickens (Gallus gallus domesticus), which were initially domesticated from the red jungle fowl (RJF) subspecies Gallus gallus spadiceus in East Asia (Peters et al., 2016; Wang M.-S. et al., 2020), have undergone climate and environmental changes, artificial domestication, and commercial breeding for a long period. After thousands of years of domestication and selection, domestic chickens have been separated into several hundreds of distinct breeds spreading globally, and their appearance, behavior, growth, and reproduction traits vary from breed to breed. Hence, chickens are an excellent model to explore the evolution and domestication of animals and to identify the evolutionary spectrum of CNVs controlling domestication, breed formation, and important economic traits. However, most researches mainly focused on yielding CNV maps for chickens (Crooijmans et al., 2013; Tian et al., 2013; Yi et al., 2014; Sohrabi et al., 2018) or identifying CNVs based on some certain traits (Zhang et al., 2014; Xu et al., 2017). The evolutionary spectrum of CNVs (e.g., magnitudes, trajectories, and mechanisms) affecting important economic traits of livestock species during their domestication and breed formation remains largely elusive. Therefore, the main aim of the study is not merely yielding an exhaustive CNV map for chicken but also exploring the role of CNVs in evolution and domestication. What’s more, the exclusive CNVs of each breed were systematically analyzed to identify the genes contributing to breed-specific characteristics. This study offers a new perspective on the evolutionary spectrum of CNVs under artificial selection during chicken domestication and breed shaping, and the results may help reveal potential genetic mechanisms for some meaningful traits in chicken, accelerate breeding programs, and improve the quality and efficiency of production.
Materials and Methods
Sample Preparation and Sequencing
We collected 47 samples from 6 typical breeds, including 4 Chinese indigenous breeds [eight Xinghua (XH) chickens from Guangdong province, eight Luxi Game (LXG) fowl from Shandong province, eight Beijing You (YOU) chickens from Beijing, nine Silkie (SILK) chickens from Jiangxi province], and 2 introduced commercial breeds [eight Recessive White Rock (RW) chickens, six White Leghorn (WL) chickens]. SILK chicken is known for its fluffy plumage, dark blue bones and skin, and five toes on each foot. Besides, the SILK chicken is thought to have medicinal properties. LXG chicken is a famous gamecock breed. YOU chicken is a dual-purpose breed with a special appearance (crest on the head, beard under the lower jaw, feathers on both shanks, and five or more toes on each foot). XH chicken has slow growth, low production, and favorable meat quality. As for the two commercial breeds, WL chicken is famous for high egg production and RW chicken for high meat yield. Samples were randomly collected to avoid genetic affinity. Blood samples from 47 birds were used to construct libraries and were sequenced using Hiseq2000 platform. About 95–117 million clean paired-end reads with a length of 100 bp were produced for each sample. The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in the BIG Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, under the accession number PRJCA000093, and they are publicly accessible at http://bigd.big.ac.cn/gsa. Data of four wild chickens, Red Jungle Fowl (RJF), were downloaded from the Sequence Read Archive (SRA) database with accession number PRJNA241474 (Wang et al., 2015). In total, 51 individuals from 7 breeds were included for whole-genome analysis (Supplementary Table S1).
Profiling of Whole-Genome CNVs
We analyzed the whole-genome CNVs using four programs, namely mrFAST, CNVnator, BreakDancer, and Pindel, which have disparate algorithms. mrFAST algorithm is mainly used to identify duplicated segments and simultaneously predict absolute CNVs for the duplicated segments (Alkan et al., 2009). CNVnator is based on the read depth signals, representing greater challenges when calling retrotransposons, duplications, and balanced CNVs (Abyzov et al., 2011). CNVnator is able to discover CNVs in a vast range of sizes, from a few hundred bases to mega bases in length, in the whole genome. BreakDancer is a bioinformatics tool that relates paired-end read alignments from a test genome to the reference genome for the purpose of comprehensively and accurately detecting various types of structural variation including deletion, insertion, inversion, intra-chromosomal translocation, and inter-chromosomal translocation. While BreakDancer is not suitable for detecting small variants. Pindel is a split read-based pattern growth approach to detect CNVs. Pindel performs better in detecting small insertions and deletions and can only detect a limited number of large structural variations (> 1 kb) (Abyzov et al., 2011; Liu et al., 2020).
First, we used mrFAST and mrCaNaVaR to exploit the whole-genome CNVs. The reference genome was first masked for repeat elements using RepeatMasker (Smit and Green, 2013) and Tandem Repeats Finder (Benson, 1999). mrFAST was implemented to align paired-end reads to the reference genome in a single mode, accommodating five edit distances, which reported all possible aligned locations for each read. mrCaNaVaR operated the whole-genome shotgun sequence algorithm, which utilized three categories of sliding windows to calculate the normalized read depth, identify large segmental duplications and deletions, and calibrate their absolute copy numbers. The raw read depth in each window was normalized based on its GC content via a LOESS-based smoothing technique. Segmental duplication and deletion regions were declared where at least six out of seven consecutive 5-kb non-masked windows with 1 kb sliding steps presented significantly increased and decreased read depths (mean ± 4*standard deviation); their boundaries were subsequently refined by 1 kb non-masked windows having increased and attenuated read depths (mean ± 2*standard deviation). The absolute copy numbers within 1 kb of non-overlapping non-masked windows were predicted based on the normalized read depths.
For CNVnator, BreakDancer, and Pindel, the clean paired-end reads were aligned to the indexed chicken reference genome (galGal4) by performing the maximal exact match algorithm in BWA (Li and Durbin, 2009), where reads were either uniquely mapped or randomly located at a place if they had multiple alignments. After alignment, we exploited BreakDancerMax to first classify the aligned read pairs into six types: normal, deletion, insertion, inversion, intra-chromosomal translocation, and inter-chromosomal translocation based on the separation distances and alignment orientation between the paired reads. Phred-style quality scores were required greater than 30, with sufficient mapping quality, and a confidence score of 90 was required. Only the deletion results were retained for further analysis. Pindel with split-reads approach was used with default parameters except that the cut off value for the number of supporting reads was set to 5. The deletion with length ≥50 bp was selected.
For CNVnator, the read depth signals were calculated as the count of mapped reads within consecutive non-overlapping bins and further corrected according to the corresponding GC content. A mean-shift technology was applied to partition these read depth signals into segments with presumably different underlying copy numbers. Based on these, the putative CNVs were further predicted by performing statistical significance tests, whose exact copy numbers were estimated as normalized read depths. Since the optimal bin size is crucial for CNV calling, it was once recommended as the one at which the ratio of the average read depth signal to its standard deviation is approximately 4–5 (Abyzov et al., 2011). The bin size in our study was determined as 500 bp. In order to avoid false discovery issues, we subsequently filtered out the CNVs with q0 >50% (zero mapping quality). And only the deletion results were retained for further analysis. We finally prepared the CNV dataset using mrFAST duplication result as duplication regions and integrating the deletion results supported by at least two of four softwares as deletion regions.
Population Genetics Analysis
We integrated the CNV into copy number variation region (CNVR) based on 1 bp overlap. If a CNVR was present in the sample, we labeled 1 for the region, and 0 for absent event. For example: sample 1 has a deletion of chr1:50500-51499, sample 2 has a deletion of chr1:50600- 52199, sample 3 has a deletion of chr1:53000-53999. Then the CNVR is chr1:50500-52199 and chr1:53000-53999. For the first CNVR, sample 1 and 2 present, but sample 3 does not. Then we performed principal component analysis (PCA) using princomp () under R environment (Team, 2021).
To explore the CNVR that shows stratification among populations, we calculated Vst among pairwise populations (Redon et al., 2006). We used a copy number of 1 kb repeat-free non-overlapping windows provided by mrFAST in CNVR. Vst was calculated by the following formula:
where is the total variance among all unrelated individuals, and are the variance within populations 1 and 2, respectively, and and are the population sizes.
The Vst value fluctuated between 0 and 1. The region where Vst = 0 exhibits no differentiation, whereas Vst = 1 means complete differentiation. When comparing one population with the other, if all the Vst values were above 0.4, we selected the region as a candidate breed-specific region.
Gene Annotation and Enrichment Analysis
We downloaded gene annotations from the Ensembl database with version 78. If there was overlap of at least 1 bp with CNV, the gene was annotated as CNV associated gene. We performed GO and KEGG enrichment analyses using DAVID (https://david.ncifcrf.gov/). The ingenuity pathway analysis (IPA, http://www.ingenuity.com) software was used to analyze the enrichment based on the IPA knowledgebase. Categories of disease and disorder, molecular and cellular functions, and physiological system development and functions are developed in IPA. The p-value was calculated by right-tailed Fisher’s exact tests.
Different Expression Analysis
Nine RNA sequencing data of cerebrum samples from LXG, SILK, RW, and WL male chickens (two for LXG, RW, and WL respectively and three for SILK) (Hou et al., 2020), as well as two transcriptomes of cerebrum samples from RJFs (one male and one female) with accession numbers SRR306710-306711 (Brawand et al., 2011) were downloaded and analyzed together. The clean sequencing data were mapped to the reference genome (galGal4) by TopHat v2.0.13 (Trapnell et al., 2009). The different expression level for the comparison of RJF versus all domesticated chickens was detected by HTSeq and DESeq with filters of Padj <0.01 and fold change (FC) >1.5 (Anders and Huber, 2010). We obtained the average expression level of differentially expressed genes (DEGs) for each breed based on the normalized count value provided by DESeq, and then we calculated the Pearson correlation coefficient with the average copy number of each breed.
Results
Identification of Whole-Genome Copy Number Variation
In this study, 47 birds from four Chinese indigenous breeds and two introduced commercial breeds were randomly selected and whole-genome sequenced (Supplementary Table S1). SILK, YOU, LXG, and XH chickens were sampled from Jiangxi, Beijing, Shandong, and Guangdong provinces, respectively. The two commercial breeds were RW and WL, representative of broiler and layer, respectively. Additionally, the genome sequence of four RJFs, which represented the wild chicken, was downloaded from the SRA database (Wang et al., 2015). Herein, whole-genome sequencing data from 51 individuals for seven breeds were included in our analysis. The sequencing data were endowed with an approximate coverage depth of 10× paired-end reads per individual and only autosome data were used (Supplementary Table S2).
To complementarily capture disparate aspects of CNVs, we applied multiple approaches that simultaneously characterize distinct properties of CNVs based on different principles. For duplication, the mrFAST method was performed, and an average of 379 duplications for each individual was detected. For deletion, we integrated four methods of mrFAST, CNVnator, BreakDancer, and Pindel, and short variations (< 50 bp) were removed. The average deletion numbers for each bird detected by four different methods were 54, 794, 2,282, and 2,278, respectively (Supplementary Table S2). Approximately 35.61% of these deletion events were discovered via at least two methods, and only 0.26% were shared by all four methods. For next analysis, only deletions supported by at least two methods were retained, with an average of 1,936 deletions per individual.
Then, we merged the above results of 19,329 duplications and 98,736 deletions to obtain CNVRs. A total of 11,123 CNVRs (length range: 68–2,802,702 bp) were elaborately identified, which represented 7% of the autosome genome, with the mean and median length of 6,748 bp and 986 bp, respectively. Most CNVRs (8,834) were denominated as “deletion,” and 1,911 were “duplication,” while the other 378 were “complex,” which included both “deletion” and “duplication” events (Supplementary Table S3). From the viewpoint of frequency spectrum that shared among CNVRs of the 51 individuals, we found 4,515 singletons, 5,121 lowly shared CNVRs with frequencies less than 20% excluding singletons, 1,200 medially shared with a frequency of 20–80%, 135 highly shared with a frequency of 80–99%, and 152 common CNVRs, resulting in a typical skewed distribution (Supplementary Figure S1). Meanwhile, the CNVR lengths presented a typical skewed distribution identically with a higher proportion of smaller CNVRs (Supplementary Figure S1), generally indicating a selective genetic load on CNVs.
Furthermore, we compared the CNVRs with the Ensembl BioMart v78 database (galGal4) to identify genes contained in those CNVRs and filtered the singleton CNV in the population to ensure a high quality of functional CNVs. A total of 2,636 protein-coding genes overlapped with duplications and deletions in CNVRs. Then, Kyoto Encyclopedia of Genes and Genomics (KEGG) pathway and Gene Ontology (GO) term analysis were performed using the DAVID database. Genes with copy number variations were mainly associated with axon guidance, ATP binding, GTPase activator activity, and phosphatidylinositol binding based on the GO term, and they were involved in vascular smooth muscle contraction, oocyte meiosis, and gap junction according to KEGG pathway analysis (Supplementary Table S4). This suggests that the functional shape of CNVs mainly modifies behavior, energy, and reproduction, which are typical target traits in chicken domestication.
Population Characteristics of Whole-Genome Copy Number Variation
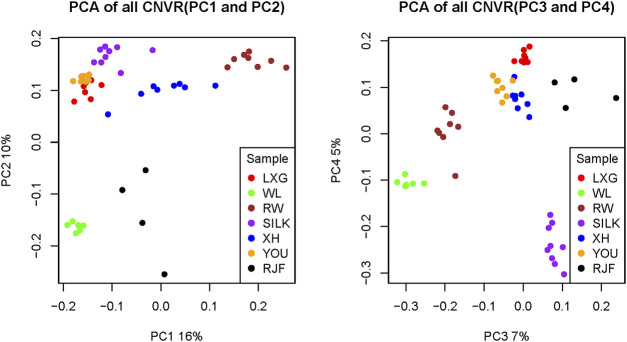
PCA was performed on all CNVRs detected in the seven populations to explore the population stratification based on the variations (Figure 1). By integrating PC1 and PC2, which interpreted 16% and 10% of the phenotypic variance respectively, the samples were primarily divided into four clusters. The first and second clusters represented the introduced meat dynamo RW and the introduced layer dynamo WL, respectively. The third cluster was the wild RJF from Yunnan and Hainan provinces of China. The fourth was a cluster of Chinese native breeds, which could also be separated from each other, except for YOU and LXG breeds. Based on PC3 and PC4, which described 7% and 5% of the total variance, respectively, these populations could also be distinguished apparently, and SILK chicken was further separated from other Chinese native breeds. These findings suggested that the CNV spectrum can substantially reflect breed differentiation in light of genetic divergence and artificial selection.
FIGURE 1.
The principal component analysis (PCA) plot of all CNVRs. (A) Samples were divided into four groups: commercial layer, commercial broiler, RJFs, and Chinese native breeds. (B) Samples were divided more detailly that SILK was further separated from the other Chinese native breeds. XH: Xinghua; LXG: Luxi Game fowl; YOU: Beijing You; SILK: Silkie; RW: Recessive White Rock; WL: White Leghorn; RJF: Red jungle fowl.
Ubiquitous Copy Number Variations in Chickens
The common copy number variations of chickens included in all the wild and domesticated individuals indicate that the genomic characteristics might be relevant to speciation of chicken or its ancestors. As mentioned, 152 CNVRs were commonly detected among all 51 individuals. Among these regions, 72 were highly represented duplication CNVRs and 172 corresponding genes were involved, with minimum and maximum copy numbers of 3 and 373, respectively. They were predominantly enrichment in feather keratin, sensory perception, metabolic, reproductive processes, muscle contraction, and immunity (Supplementary Table S5).
As expected, highly duplicated segments of keratin genes in terms of feather keratin and feather-keratin-like protein were observed in chromosomes 25 and 27, with copy numbers ranging from 3 to 90. MUC5B, a member of the mucin family, was discovered to be intensively duplicated with varied copy numbers ranging from 17 to 42. Additionally, four genes, MRPS35, SMARCD3, KHDRBS2, and ANO3 exhibited a minimum copy number of more than 30 and a maximum of over 60. NRG3 and NCAM2, which are associated with development and differentiation of nervous system, were also included, and their copy numbers ranged from 3 to 42 and 15 to 40, respectively. Besides, 3–5 copy numbers of VTG2 and 3–6 of GBE1 were identified.
Copy Number Variations Correlated With Chicken Domestication
The loci with high copy number differentiation between wild and domestic populations are considered as candidates driving adaptive selection. Therefore, the copy number variations between RJF and all the six domestic breeds were compared to identify the potential genes associated with chicken domestication. The method of Vst was used to estimate the population differentiation of copy numbers (Redon et al., 2006). A total of 235 CNVRs containing 255 protein-coding genes were discovered in the windows with 1% toppest Vst (> 0.2042) (Supplementary Table S6). The gene enrichment analysis by IPA showed that these copy number differentiated genes are predominantly involved in nervous system development, reproductive system development and function, cellular growth and proliferation, and immunity (p < 0.01) (Supplementary Table S7).
First, quite a few of genes which were involved in the development and function of nervous system were found to be highly differentiated in gene copy, including NRG3, BICD1, ANKRD11, SEMA3F, UNK, EPS15L1, KIF3C, FBXO41, MYT1, NR2E1, PSMC5, SEMA3F, SOX2, PTPRM, and RPS6KB1. A CNVR in exon 1 of NRG3 was observed to have an average copy number higher than 30 in RJFs, but the median copy number in domestic individuals was 16 (Supplementary Figure S2). Second, some genes associated with immunity or stress response, such as CBFA2T3, IL22RA1, HERPUD1, ARIH2, GATA3, TNFAIP8L1, CD86, CD8A, SMARCD2, and TNPO3, were also detected possessing differential copy numbers in genome. Besides, some other valuable genes correlated with energy metabolism, cell growth and proliferation or reproduction functions were also found, for instance, GBE1, ACAT, ACO2, HEP21, INHA, GPC6, MEI1, PTPRM, RPS6KB1, etc. GBE1 presented one copy number more in intron 7 in RJF than in all the other six domesticated chicken breeds (Supplementary Figure S2).
Breed-Specific Copy Number Variations
Considering that CNV may be correlated with the origin or breeding of different chicken breeds, we further compared each domestic breed with each of the other six breeds to determine the extremely specific regions. If the six Vst values of a region were over 0.4, it was considered as a breed-specific region. In total, 600 CNVRs harboring 90 annotated genes were found to be breed-specific (Supplementary Table S8).
RW chickens exhibited 64 breed-specific CNVRs and 39 protein-coding genes, showing the highest number of specific genes across all the six domestic breeds. Genes specific to RW chickens may be candidate genes for growth and muscle traits. For example, POLR3H, MCM9, ASF1A, MYH1B, and DOCK3 may contribute to rapid growth and plump muscle traits. Furthermore, WL chickens possess 63 regions and 19 genes. Many genes, i.e., GSK3B, CSMD1, NTRK3, and STRBP, may be associated with reproduction traits.
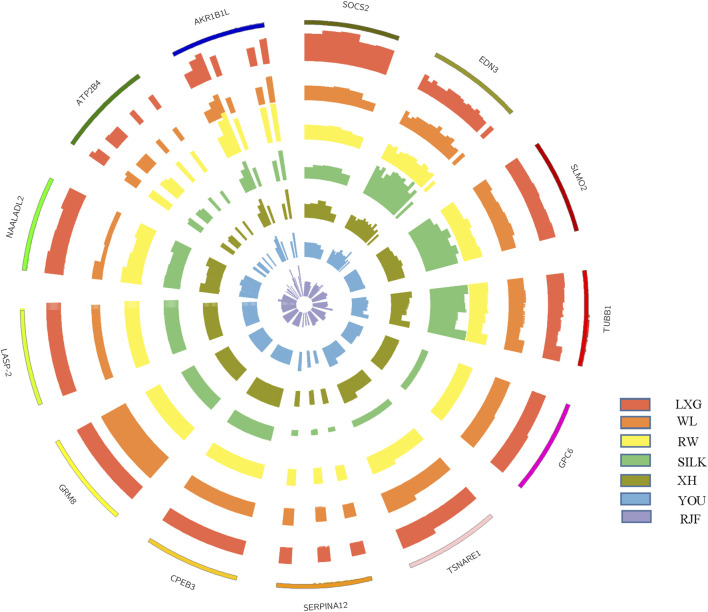
For the four Chinese native chicken breeds, LXG, SILK, YOU, and XH, we detected 32, 23, 17, and 2 regions, corresponding to 8, 15, 7, and 1 protein-coding gene, respectively. From the schematization of copy numbers for a part of genes and the CNV-involved gene segments that distinguished chicken breeds (Figure 2), we could notice the difference clearly. In LXG chickens, SOCS2 presented a copy number of about 4, which was significantly higher than that of the other breeds. EDN3 had around 4 copies exclusively in SILK chickens, and its neighboring genes SLMO2 and TUBB1 also had higher copy numbers than other breeds, which have been frequently targeted as tetra- or penta-ploid (Han et al., 2014; Yi et al., 2014). Moreover, SILK chickens have specially experienced one-copy deletion on GPC6, TSNARE1, and SERPINA12. For XH chickens, CPEB3 was the only detected breed-specific gene, showing one-copy higher particularly. It was reported that CPEB3 was related to pathway of oocyte meiosis (Wang Y. et al., 2020).
FIGURE 2.
The schematization for a part of copy number variation genes distinguished in different chicken breeds. The outermost circle indicates the CNV-involved gene regions, with the thickness implying the normal diploid. The other circles from outside to inside indicate the breeds of Luxi Game fowl (LXG), White Leghorn (WL), Recessive White Rock (RW), Silkie (SILK), Xinghua (XH), Beijing You (YOU), and red jungle fowl (RJF) in turn, whose thickness proportionally means the estimated copy number.
Correlation Between Copy Number Variation and Gene Expression in Cerebrum
Considering that variation in gene copy number is expected to change the RNA expression level, the DEGs between wild RJF and each of the other four domesticated breeds (LXG, SILK, RW, and WL) in cerebrum published by our team (Hou et al., 2020) were further explored. A total of 28 genes were shared among the domestication-related copy number differentiated genes. Additionally, the correlations between the gene expression and copy numbers were intersected (Supplementary Table S9). More than half (16/28) of the 28 overlaps presented a significant correlation between copy number and gene expression, with absolute Pearson’s correlation coefficients larger than 0.88 (p < 0.05). Among the significantly correlated genes, most (9/16) showed negative correlations.
As mentioned, 15 genes participating in the development and function of nervous system showed copy number variation specifically in RJF, among which seven (BICD1, ANKRD11, SEMA3F, UNK, EPS15L1, KIF3C, and FBXO41) were significantly differentially expressed in the cerebrum, with one (ANKRD11) upregulated and the other six downregulated in domesticated breeds compared with RJF. Meanwhile, except for BICD1, six showed negative correlation with copy number.
Discussion
In this study, whole-genome CNVs were explored in RJF and six domestic chicken breeds. The particularity for each breed predominantly contributed to their inner divergences, for example, high egg performance for WL, high meat yield for RW, fluffy plumage, black skin, and medical purpose for SILK, game fighting for LXG, low production and favorable meat quality for XH, and special appearances for YOU chickens. By combining multiple methods, four for deletions and one for duplications, we systematically and exquisitely analyzed the copy number information in 51 individuals and obtained a comprehensive map of genomic variations for chickens. A total of 19,329 duplications and 98,736 deletions were detected, covering 11,123 CNVRs and 7% of the autosome genome.
PCA results with both duplication and deletion CNVRs revealed that the seven chicken populations were well divided into four parts according to the degree of artificial selection pressure and the purpose of domestication: commercial broiler, commercial layer, Chinese native breeds, and their ancestor RJF. These results were consistent with the population stratification pattern based on SNPs (Hou et al., 2020), revealing that CNV is a powerful and effective tool for distinguishing samples and that CNVs may also participate in the driving of chicken domestication.
Genes Relating to Chicken Evolution
As an important genetic source, copy number variation can provide a good perspective for exploring the track of animal evolution. Therefore, 72 high duplication CNVRs (corresponding to 172 genes) detected in all individuals were analyzed to explore chicken evolution, which might be characterized as the genomic features specialized in chicken species.
As expected, feather keratin (FK or F-KER), olfactory receptor (OR), and neuregulin 3 (NRG3) were detected CNVs as previous studies reported. Keratin is a main protein that makes up avian feathers (Ramakrishnan et al., 2018). FK or F-KER was observed with intensive copy number duplication in current study, which is highly consistent with the results of Hillier et al. (2004). OR genes, which plays a pivotal role in the sense of smell among vertebrates, exhibited a copy number around 20 in this study. Zhou et al. (2019) reported more than 90 OR genes in chicken, which was still much fewer than in mammals like humans (396), dogs (857), cats (865), cattle (881), and pigs (1113) (Bu et al., 2019). The contraction of OR genes in chicken genome may be responsible for the relatively poor sense of smell for bird species. NRG3 was found to be associated with the development and differentiation of nervous system. Abe et al. (2017) also detected copy number variations and frameshift deletion of NRG3 in chicken.
Besides, some genes, such as MUC5B, NCAM2, and VTG2, were discovered with copy number differences in all individuals. MUC5B is a major contributor to the lubricating and viscoelastic properties of whole saliva, normal lung mucus, and cervical mucus (Roy et al., 2014; Käsdorf et al., 2017). The gene NCAM2 has been proposed to contribute to neurodevelopmental disorders in humans (Winther et al., 2012). Furthermore, we identified that VTG2, which is the yolk precursor protein expressed in females of nearly all oviparous organisms, possessed 3–5 copies across all individuals. The presence of multiple copies of VTG2 in the genome was due to the gene family expansion from common ancestor (Biscotti et al., 2018) and occurred prior to the most recent common ancestor of birds (Brawand et al., 2008; Finn et al., 2009).
Genes Involved in Chicken Domestication
About 150 years ago, Darwin first introduced the concept of domestication syndrome, that domestic animals tended to share some common characteristics, such as more docility and frequent estrus cycles (Darwin, 2010; Wilkins et al., 2014). The domestication genes that cause genetic differentiation between wild and domestic populations appear in the early stages of domestication (Larson et al., 2014). We compared the diversity of CNVs between RJF and six domestic chicken populations to identify genes that may further illuminate the genetic basis of chicken domestication. A total of 255 protein-coding genes, predominantly involved in the development of nervous, tissue and reproductive system, were detected.
Previous reports demonstrated that neuronal development and behavior modification are under strong selection during long-term domestication progresses (Albert et al., 2008; Sanchez-Villagra et al., 2016; Zhang et al., 2018). More than 15 genes associated with the development and function of the neural system exhibited copy number variations between domestic breeds and RJF. Seven out of these genes were differentially expressed in cerebrum between domestic breeds and RJF. Except for ANKRD11, NRG3, and MYT1, the other genes were detected copy number variation for the first time. NRG3 showed high copy number deletion in domestic chickens, which is consistent with previous study (Abe et al., 2017). As mentioned, NRG3 has been implicated in severe neurological disorders with developmental origins (Meier et al., 2013; Li et al., 2020). ANKRD11 showed copy number deletion in people with autism spectrum disorder compared to healthy persons (Marshall et al., 2008). The knockdown of ANKRD11 exhibited markedly reduced neurons dendrite outgrowth (Ka and Kim, 2018). The overexpression of MYT1 was suggested to help reduce anxiety in rats and has been associated with intellectual disability previously (Bahi and Dreyer, 2017). BICD1, UNK, and KIF3C are essential for neuron differentiation during development. BICD1, a conserved gene in Drosophila, C. elegans (Aguirre-Chen et al., 2011), and human (Baens and Marynen, 1997), is highly expressed in the developing central and peripheral nervous systems and plays an important role in neuronal homeostasis as a regulator of neurotrophin signaling (Terenzio et al., 2014). Loss of UNK alters the number of neural stem cells and neural progenitors resulting in increased neurogenesis (Maierbrugger et al., 2020). KIF3C, one of the kinesin-related motor subunits of the KIF3 family, is selectively expressed in the nervous system during embryonic development (Navone et al., 2001). FBXO41 and EPS15L1 are associated with the defects in central nervous system (Mukherjee et al., 2015; King et al., 2019; Milesi et al., 2019). SEMA3F, the class 3 subfamily of semaphorins, was found to contribute to the development of neuronal circuitry related to anxiety and fear responses in SEMA3F-knockout mice (Milesi et al., 2019). Besides, the transcription factors of nuclear receptor TLX (NR2E1) might be regarded as a key regulator of neural development and differentiation (Matsushita et al., 2014; Sun et al., 2017).
The domestication of wild animals is a process of human civilization and food production. Therefore, improving production performances is an important part for chicken domestication. More than nine genes correlated with energy metabolism, reproduction function or cell growth and proliferation were screened. Three representative genes, GBE1, HEP21, and INHA, were first found to have copy number variation between domesticated and wild chickens. GBE1 is involved in the metabolism of carbohydrate, presenting one copy deletion in all six domesticated chicken breeds. This gene was reported to be related with abdominal fat traits and presented higher expression levels in fast-growing chickens than in slow-growing chickens (Claire D'andre et al., 2013) or in fat chicken line than in lean chickens (Jin et al., 2017). HEP21, a gene unique to poultry, has doubled copy number variation in domesticated breeds. The expression of HEP21 was associated with the development and function of chicken oviduct (Lim and Song, 2014), and SNPs in the HEP21 gene have been reported to be related with sexual maturity (Chen et al., 2020). INHA protein is secreted by the avian granulosa cells of preovulatory follicles, and its gene expression level changed with the follicle development (Cui et al., 2019).
Improving disease resistance and immunocompetence of livestock is a precondition for security and abundance of food in the domestication of animals. At least ten genes were found to participate in the pathways of inflammatory and immune response. CBFA2T3, a factor in human acute myeloid leukemia, promotes myeloid differentiation (Steinauer et al., 2020). In cattle, this gene is found under selection pressure, which might be related to physiological adaptations to the environment and disease resistance (Ben-Jemaa et al., 2020). IL22RA1 is one of the receptors for IL-22, an important cytokine involved in host defense and inflammatory responses (Gaudino et al., 2020). The expression of ARIH2 is significantly altered in chicken immune organs after vaccine immunization (Wu et al., 2019). The biological function of TNFAIP8 has been extensively investigated and was reported to play vital roles in modulating inflammation and immunity (Lou and Liu, 2011). GATA3 was shown to regulate the differentiation and proliferation of T-cell (Ho et al., 2009). And HERPUD1 is found differentially expressed in chickens under stress (Guo Y. et al., 2020).
Breed-Specific Genes
Obviously, CNV plays a crucial role in chicken domestication and provides us a new perspective for studying the genetic mechanism underlying complex phenotypes. Therefore, copy number information exclusive to each domestic breed was screened to explore the genes contributing to breed characteristics. Some genes were discovered by previous study, such as EDN3, SOCS2, HOXB7, and HOXB8, but most genes relating to breed characteristics were identified for the first time. Among the discovered genes, some are already verified contributing to appearance, growth or reproduction traits in chicken, and the others, whose biological function are discussed based on studies in human or other animals, are worthy of further researches.
For RW chickens, the significantly high copy number duplications of POLR3H, MCM9, DOCK3, AKR1B1L, ASF1A, ATP2B4, and MYH1B may contribute to the fast-growing traits. Knocking down any of the three genes, that is, POLR3H, MCM9, and DOCK3, would cause delayed development or short growth (Lutzmann et al., 2012; Qin et al., 2015; Iwata-Otsubo et al., 2018; Franca et al., 2019; Guo T. et al., 2020). AKR1B1L, a member of the aldo/keto reductase superfamily, was related to body length and fat deposition in cattle (An et al., 2020) and chickens (Claire D'andre et al., 2013). ASF1A was reported to be a candidate gene for muscle weight and meat quality (Liu et al., 2013; Zhang et al., 2020). ATP2B4 is one of ATPase family that regulates intracellular calcium homeostasis (De Koning et al., 2001) and plays an important role in bone development (Wu et al., 2004; Kim et al., 2012). Furthermore, MYH1B was also demonstrated to participate in the early development of breast muscle or regeneration of muscle fibers in broilers (Pampouille et al., 2018).
For WL chickens, the copy number deletion of CSMD1 and NTRK3 may be involved in the outstanding reproduction traits, because NTRK3 and CSMD1 have been reported to play crucial roles in follicle development, ovarian quality, and infertility in human and chicken (Lachman et al., 2007; Nilsson et al., 2009; Lee et al., 2019).
In SILK chickens, which are famous for their black skin and bone, four genes (EDN3, SLMO2, TUBB1, and GFPT1) exhibited a significant copy number increase, which may be responsible for melanin formation and deposition. EDN3, a well-known candidate for melanocytic proliferation and maintenance, has been reported to be strongly related to dermal pigmentation in SILK chickens (Shinomiya et al., 2012). SLMO2 and TUBB1 have also been discovered with around 4 and 5 copy number duplications, respectively, and they showed significantly high expression in both skin and muscle tissues in SILK chickens (Dorshorst et al., 2011). GFPT1 mutation has been reported to specifically affect the production of melanin in zebrafish (Yang et al., 2007).
LXG chickens, a typical cockfighting breed characterized by aggressiveness, strong bone strength, and powerful muscle, exclusively exhibited duplications on SOCS2, PRDM5, CRADD, and SORCS2. SOCS2 (Lorentzon et al., 2005) and PRDM5 (Galli et al., 2012) may contribute to the high bone mineral density. And the increased copy number and gene expression variation of SOCS2 in gamecocks were validated by Bi et al. (2017). The one more copy number increase of CRADD in LXG may be responsible for fast muscle growth trait (Horvat and Medrano, 1998; Smith et al., 2000). Meanwhile, a CNV region (GGA: 80,834,764–80,851,024 bp) overlapping the gene SORCS2 was particularly detected in LXG. SORCS2 was suggested to be a candidate gene for the aggressive behavior of LXG, because knockdown of SORCS2 would significantly decrease the expression of dopamine receptor genes and nerve growth factor (Li et al., 2016).
YOU chicken is a Chinese native breed with special appearance characteristics, including crests, beards, feathered legs, and polydactyly. Recently, Yang et al. (2020) deciphered that the copy number variation in HOXB7 and HOXB8 is responsible for the beard trait, which coincides exactly with the CNV detected in our study, especially exhibiting two copy number increase in YOU chicken.
Conclusion
This study provides genome-wide CNV information in one wild chicken breed (RJF), two commercial breeds, and four Chinese local breeds to synthetically understand the process of chicken domestication and to exclusively identify the genes contributing to breed characteristics. A total of 11,123 CNVRs with 19,329 duplications and 98,736 deletions were detected, covering 7% of the autosome genome and overlapping 2,636 protein-coding genes. The PCA results revealed that CNV is a powerful method for exploring evolution and domestication. In chicken evolution, genes related to nervous system, sensory, and follicle development were supposed to have pivotal roles. Meanwhile, cognitive and behavior changes, reproduction function, metabolism, and immunity are under strong selection during long-term animal domestication process. What’s more, some valuable genes contributing to fast growth, high reproduction, and distinct breed characteristics were also identified in this paper, which deserve further investigations. Taken together, the present study provides numerous copy number information for chickens and it is a valuable resource to facilitate genetic and functional investigation of domestication and economic traits in chickens.
Acknowledgments
The authors are thankful to Dr. Ying Yu in China Agricultural University, Dr. Guiping Zhao in Institute of Animal Sciences, Chinese Academy of Agricultural Sciences, Dr. Xiquan Zhang in Huanan Agricultural University, and Dr. Jianbin Li in Shandong Academy of Agricultural Sciences, for their kindly help and donation of chicken samples for the purpose of scientific research.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Ethics Statement
The animal study was reviewed and approved by all the experimental procedures involving animals were conducted in accordance with the animal welfare guidelines of ARRIVE guidelines (Percie Du Sert et al., 2020), and were approved by the Animal Experimental Ethics Board in Beijing Institute of Genomics, Chinese Academy of Science (permit number: 2017-YJ-01). In addition, all methods were carried out in accordance with relevant guidelines and regulations in the declaration.
Author Contributions
XB and YH initiated the study and designed the project. XC, XB, ZY, QC, and YH collected the samples. QC, YH, and HL supervised the study. XB, BZ, and YH performed the bioinformatics. XC, QC, and YH summarized the results. XC, XB, QC, and YH drafted the manuscript. All authors read and approved the final manuscript.
Funding
This work is financially supported by the Beijing Municipal Science and Technology Project (D171100007817003), Innovation Program of Beijing Academy of Agriculture and Forestry Sciences (KJCX20200101), and the China Agriculture Research System of MOF and MARA (CARS-41).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2022.830393/full#supplementary-material
Abbreviations
CNV, Copy number variation; CNVR, Copy number variation region; DEGs, Differentially expressed genes; GO, Gene ontology; IPA, Ingenuity pathways analysis; KEGG, Kyoto encyclopedia of genes and genomes; LXG, Luxi game; PCA, Principal component analysis; RJF, Red jungle fowl; RW, Recessive white rock; SILK, Silkie; SNP, Single nucleotide polymorphism; SRA, Sequence read archive; WL, White leghorn; XH, Xinghua; YOU, Beijing YOU.
References
- Abe H., Aoya D., Takeuchi H.-A., Inoue-Murayama M. (2017). Gene Expression Patterns of Chicken Neuregulin 3 in Association with Copy Number Variation and Frameshift Deletion. BMC Genet. 18, 69. 10.1186/s12863-017-0537-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abyzov A., Urban A. E., Snyder M., Gerstein M. (2011). CNVnator: an Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 21, 974–984. 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Chen C., Bülow H. E., Kaprielian Z. (2011). C. elegans Bicd-1, Homolog of the Drosophila Dynein Accessory Factor Bicaudal D, Regulates the Branching of PVD Sensory Neuron Dendrites. Development (Cambridge, England). 138, 507–518. 10.1242/dev.060939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert F. W., Shchepina O., Winter C., Römpler H., Teupser D., Palme R., et al. (2008). Phenotypic Differences in Behavior, Physiology and Neurochemistry between Rats Selected for Tameness and for Defensive Aggression towards Humans. Horm. Behav. 53, 413–421. 10.1016/j.yhbeh.2007.11.010 [DOI] [PubMed] [Google Scholar]
- Alkan C., Kidd J. M., Marques-Bonet T., Aksay G., Antonacci F., Hormozdiari F., et al. (2009). Personalized Copy Number and Segmental Duplication Maps Using Next-Generation Sequencing. Nat. Genet. 41, 1061–1067. 10.1038/ng.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B. X., Xu L., Xia J. W., Wang X. Q., Miao J., Chang T. P., et al. (2020). Multiple Association Analysis of Loci and Candidate Genes that Regulate Body Size at Three Growth Stages in Simmental Beef Cattle. Bmc Genet. 21, 32. 10.1186/s12863-020-0837-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Huber W. (2010). Differential Expression Analysis for Sequence Count Data. Genome Biol. 11, R106. 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt M. F., Wilson T. E., Glover T. W. (2012). Replication Stress and Mechanisms of CNV Formation. Curr. Opin. Genet. Development. 22, 204–210. 10.1016/j.gde.2012.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baens M., Marynen P. (1997). A Human Homologue (BICD1) of theDrosophila Bicaudal-DGene. Genomics. 45, 601–606. 10.1006/geno.1997.4971 [DOI] [PubMed] [Google Scholar]
- Bahi A., Dreyer J.-L. (2017). Viral-mediated Overexpression of the Myelin Transcription Factor 1 (MyT1) in the Dentate Gyrus Attenuates Anxiety- and Ethanol-Related Behaviors in Rats. Psychopharmacology. 234, 1829–1840. 10.1007/s00213-017-4588-7 [DOI] [PubMed] [Google Scholar]
- Ben-Jemaa S., Mastrangelo S., Lee S.-H., Lee J. H., Boussaha M. (2020). Genome-wide Scan for Selection Signatures Reveals Novel Insights into the Adaptive Capacity in Local North African Cattle. Sci. Rep. 10, 19466. 10.1038/s41598-020-76576-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. (1999). Tandem Repeats Finder: a Program to Analyze DNA Sequences. Nucleic Acids Res. 27, 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R., Zhang X., Meyerson M. (2016). Copy Number Alterations Unmasked as Enhancer Hijackers. Nat. Genet. 49, 5–6. 10.1038/ng.3754 [DOI] [PubMed] [Google Scholar]
- Bi H., Yi G., Yang N. (2017). Increased Copy Number of SOCS2 Gene in Chinese Gamecocks. Poult. Sci. 96, 1041–1044. 10.3382/ps/pew391 [DOI] [PubMed] [Google Scholar]
- Biscotti M. A., Barucca M., Carducci F., Canapa A. (2018). New Perspectives on the Evolutionary History of Vitellogenin Gene Family in Vertebrates. Genome Biol. Evol. 10, 2709–2715. 10.1093/gbe/evy206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D., Soumillon M., Necsulea A., Julien P., Csárdi G., Harrigan P., et al. (2011). The Evolution of Gene Expression Levels in Mammalian Organs. Nature. 478, 343–348. 10.1038/nature10532 [DOI] [PubMed] [Google Scholar]
- Brawand D., Wahli W., Kaessmann H. (2008). Loss of Egg Yolk Genes in Mammals and the Origin of Lactation and Placentation. Plos Biol. 6, e63. 10.1371/journal.pbio.0060063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu P., Jian Z., Koshy J., Shen Y., Yue B., Fan Z. (2019). The Olfactory Subgenome and Specific Odor Recognition in forest Musk Deer. Anim. Genet. 50, 358–366. 10.1111/age.12796 [DOI] [PubMed] [Google Scholar]
- Charles Darwin M. A. (1859). On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray. [PMC free article] [PubMed] [Google Scholar]
- Chen B., Liang G., Zhu X., Tan Y., Xu J., Wu H., et al. (2020). Gene Expression Profiling in Ovaries and Association Analyses Reveal as a Candidate Gene for Sexual Maturity in Chickens. Anim. : open access J. MDPI. 10, 181. 10.3390/ani10020181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claire D'andre H., Paul W., Shen X., Jia X., Zhang R., Sun L., et al. (2013). Identification and Characterization of Genes that Control Fat Deposition in Chickens. J. Anim. Sci. Biotechnol. 4 (1), 43. 10.1186/2049-1891-4-43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooijmans R. P., Fife M. S., Fitzgerald T. W., Strickland S., Cheng H. H., Kaiser P., et al. (2013). Large Scale Variation in DNA Copy Number in Chicken Breeds. BMC Genomics. 14, 398. 10.1186/1471-2164-14-398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Z., Liu L., Zhao X., Ran J., Wang Y., Yin H., et al. (2019). Analysis of Expression and Single Nucleotide Polymorphisms of INHA Gene Associated with Reproductive Traits in Chickens. Biomed. Res. Int. 2019, 8572837. 10.1155/2019/8572837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. (2010). The Variation of Animals and Plants under Domestication. Cambridge University Press. Vol. 2. [Google Scholar]
- De Koning D. J., Harlizius B., Rattink A. P., Groenen M. A., Brascamp E. W., Van Arendonk J. A. (2001). Detection and Characterization of Quantitative Trait Loci for Meat Quality Traits in Pigs. J. Anim. Sci. 79, 2812–2819. 10.2527/2001.79112812x [DOI] [PubMed] [Google Scholar]
- Dorshorst B., Molin A.-M., Rubin C.-J., Johansson A. M., Strömstedt L., Pham M.-H., et al. (2011). A Complex Genomic Rearrangement Involving the Endothelin 3 Locus Causes Dermal Hyperpigmentation in the Chicken. Plos Genet. 7, e1002412. 10.1371/journal.pgen.1002412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin K., Coppieters W., Drögemüller C., Ahariz N., Cambisano N., Druet T., et al. (2012). Serial Translocation by Means of Circular Intermediates Underlies Colour Sidedness in Cattle. Nature. 482, 81–84. 10.1038/nature10757 [DOI] [PubMed] [Google Scholar]
- Feuk L., Carson A. R., Scherer S. W. (2006). Structural Variation in the Human Genome. Nat. Rev. Genet. 7, 85–97. 10.1038/nrg1767 [DOI] [PubMed] [Google Scholar]
- Finn R., Kolarevic J., Kongshaug H., Nilsen F. (2009). Evolution and Differential Expression of a Vertebrate Vitellogenin Gene Cluster. BMC Evol. Biol. 9, 2. 10.1186/1471-2148-9-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franca M. M., Han X., Funari M. F. A., Lerario A. M., Nishi M. Y., Fontenele E. G. P., et al. (2019). Exome Sequencing Reveals the POLR3H Gene as a Novel Cause of Primary Ovarian Insufficiency. J. Clin. Endocrinol. Metab. 104, 2827–2841. 10.1210/jc.2018-02485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke M., Ibrahim D. M., Andrey G., Schwarzer W., Heinrich V., Schöpflin R., et al. (2016). Formation of New Chromatin Domains Determines Pathogenicity of Genomic Duplications. Nature. 538, 265–269. 10.1038/nature19800 [DOI] [PubMed] [Google Scholar]
- Freeman J. L., Perry G. H., Feuk L., Redon R., Mccarroll S. A., Altshuler D. M., et al. (2006). Copy Number Variation: New Insights in Genome Diversity. Genome Res. 16, 949–961. 10.1101/gr.3677206 [DOI] [PubMed] [Google Scholar]
- Galli G. G., Honnens De Lichtenberg K., Carrara M., Hans W., Wuelling M., Mentz B., et al. (2012). Prdm5 Regulates Collagen Gene Transcription by Association with RNA Polymerase II in Developing Bone. Plos Genet. 8, e1002711. 10.1371/journal.pgen.1002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudino S. J., Beaupre M., Lin X., Joshi P., Rathi S., Mclaughlin P. A., et al. (2020). IL-22 Receptor Signaling in Paneth Cells Is Critical for Their Maturation, Microbiota Colonization, Th17-Related Immune Responses, and Anti-Salmonella Immunity. Mucosal Immunol. 14 (2), 389–401. 10.1038/s41385-020-00348-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuffra E., Törnsten A., Marklund S., Bongcam-Rudloff E., Chardon P., Kijas J. M. H., et al. (2002). A Large Duplication Associated with Dominant white Color in Pigs Originated by Homologous Recombination between LINE Elements Flanking KIT. Mamm. Genome. 13, 569–577. 10.1007/s00335-002-2184-5 [DOI] [PubMed] [Google Scholar]
- Guo T., Zheng Y., Li G., Zhao S., Ma J., Qin Y. (2020). Novel Pathogenic Mutations in Minichromosome Maintenance Complex Component 9 (MCM9) Responsible for Premature Ovarian Insufficiency. Fertil. sterility. 113, 845–852. 10.1016/j.fertnstert.2019.11.015 [DOI] [PubMed] [Google Scholar]
- Guo Y., Jiang R., Su A., Tian H., Zhang Y., Li W., et al. (2020). Identification of Genes Related to Effects of Stress on Immune Function in the Spleen in a Chicken Stress Model Using Transcriptome Analysis. Mol. Immunol. 124, 180–189. 10.1016/j.molimm.2020.06.004 [DOI] [PubMed] [Google Scholar]
- Han R., Yang P., Tian Y., Wang D., Zhang Z., Wang L., et al. (2014). Identification and Functional Characterization of Copy Number Variations in Diverse Chicken Breeds. BMC Genomics. 15, 934. 10.1186/1471-2164-15-934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J., Lupski J. R., Rosenberg S. M., Ira G. (2009). Mechanisms of Change in Gene Copy Number. Nat. Rev. Genet. 10, 551–564. 10.1038/nrg2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier L. W., Miller W., Birney E., Warren W., Hardison R. C., Ponting C. P., et al. (2004). Sequence and Comparative Analysis of the Chicken Genome Provide Unique Perspectives on Vertebrate Evolution. Nature. 432, 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Ho I.-C., Tai T.-S., Pai S.-Y. (2009). GATA3 and the T-Cell Lineage: Essential Functions before and after T-Helper-2-Cell Differentiation. Nat. Rev. Immunol. 9, 125–135. 10.1038/nri2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvat S., Medrano J. F. (1998). A 500-kb YAC and BAC Contig Encompassing the High-Growth Deletion in Mouse Chromosome 10 and Identification of the MurineRaidd/CraddGene in the Candidate Region. Genomics. 54, 159–164. 10.1006/geno.1998.5540 [DOI] [PubMed] [Google Scholar]
- Hou Y., Qi F., Bai X., Ren T., Shen X., Chu Q., et al. (2020). Genome-wide Analysis Reveals Molecular Convergence Underlying Domestication in 7 Bird and Mammals. BMC Genomics. 21 (1), 204. 10.1186/s12864-020-6613-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate A. J., Feuk L., Rivera M. N., Listewnik M. L., Donahoe P. K., Qi Y., et al. (2004). Detection of Large-Scale Variation in the Human Genome. Nat. Genet. 36, 949–951. 10.1038/ng1416 [DOI] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing, C (2004). Sequence and Comparative Analysis of the Chicken Genome Provide Unique Perspectives on Vertebrate Evolution. Nature. 432, 695–716. 10.1038/nature03154 [DOI] [PubMed] [Google Scholar]
- Iwata-Otsubo A., Ritter A. L., Weckselbatt B., Ryan N. R., Burgess D., Conlin L. K., et al. (2018). DOCK3 -related Neurodevelopmental Syndrome: Biallelic Intragenic Deletion of DOCK3 in a Boy with Developmental Delay and Hypotonia. Am. J. Med. Genet. 176, 241–245. 10.1002/ajmg.a.38517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P., Wu X., Xu S., Zhang H., Li Y., Cao Z., et al. (2017). Differential Expression of Six Genes and Correlation with Fatness Traits in a Unique Broiler Population. Saudi J. Biol. Sci. 24, 945–949. 10.1016/j.sjbs.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ka M., Kim W.-Y. (2018). ANKRD11 Associated with Intellectual Disability and Autism Regulates Dendrite Differentiation via the BDNF/TrkB Signaling Pathway. Neurobiol. Dis. 111, 138–152. 10.1016/j.nbd.2017.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Käsdorf B. T., Weber F., Petrou G., Srivastava V., Crouzier T., Lieleg O. (2017). Mucin-Inspired Lubrication on Hydrophobic Surfaces. Biomacromolecules. 18, 2454–2462. 10.1021/acs.biomac.7b00605 [DOI] [PubMed] [Google Scholar]
- Kim H. J., Prasad V., Hyung S.-W., Lee Z. H., Lee S.-W., Bhargava A., et al. (2012). Plasma Membrane Calcium ATPase Regulates Bone Mass by fine-tuning Osteoclast Differentiation and Survival. J. cell Biol. 199, 1145–1158. 10.1083/jcb.201204067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C. R., A. A. Quadros A. R., Chazeau A., Saarloos I., van der Graaf A. J., Verhage M., et al. (2019). Fbxo41 Promotes Disassembly of Neuronal Primary Cilia. Sci. Rep. 9, 8179. 10.1038/s41598-019-44589-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachman H. M., Pedrosa E., Petruolo O. A., Cockerham M., Papolos A., Novak T., et al. (2007). Increase inGSK3β Gene Copy Number Variation in Bipolar Disorder. Am. J. Med. Genet. 144B, 259–265. 10.1002/ajmg.b.30498 [DOI] [PubMed] [Google Scholar]
- Larson G., Piperno D. R., Allaby R. G., Purugganan M. D., Andersson L., Arroyo-Kalin M., et al. (2014). Current Perspectives and the Future of Domestication Studies. Proc. Natl. Acad. Sci. 111, 6139–6146. 10.1073/pnas.1323964111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. S., Rusch J., Lima A. C., Usmani A., Huang N., Lepamets M., et al. (2019). Rare Mutations in the Complement Regulatory Gene CSMD1 Are Associated with Male and Female Infertility. Nat. Commun. 10, 4626. 10.1038/s41467-019-12522-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and Accurate Short Read Alignment with Burrows-Wheeler Transform. Bioinformatics. 25, 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Liu L., Lin W., Zhou Y., Zhang G., Du X., et al. (2020). NRG3 Contributes to Cognitive Deficits in Chronic Patients with Schizophrenia. Schizophrenia Res. 215, 134–139. 10.1016/j.schres.2019.10.060 [DOI] [PubMed] [Google Scholar]
- Li Z., Zheng M., Abdalla B. A., Zhang Z., Xu Z., Ye Q., et al. (2016). Genome-wide Association Study of Aggressive Behaviour in Chicken. Sci. Rep. 6, 30981. 10.1038/srep30981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim W., Song G. (2014). Pivotal Roles for Hormonally Regulated Expression of the HEP21 Gene in the Reproductive Tract of Chickens for Oviduct Development and in Ovarian Carcinogenesis. Domest. Anim. Endocrinol. 48, 136–144. 10.1016/j.domaniend.2014.03.003 [DOI] [PubMed] [Google Scholar]
- Liu R., Sun Y., Zhao G., Wang F., Wu D., Zheng M., et al. (2013). Genome-wide Association Study Identifies Loci and Candidate Genes for Body Composition and Meat Quality Traits in Beijing-You Chickens. PLoS One. 8, e61172. 10.1371/journal.pone.0061172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Zhang M., Sun J., Chang W., Sun M., Zhang S., et al. (2020). Comparison of Multiple Algorithms to Reliably Detect Structural Variants in Pears. BMC genomics. 21, 61. 10.1186/s12864-020-6455-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorentzon M., Greenhalgh C. J., Mohan S., Alexander W. S., Ohlsson C. (2005). Reduced Bone mineral Density in SOCS-2-Deficient Mice. Pediatr. Res. 57, 223–226. 10.1203/01.pdr.0000148735.21084.d3 [DOI] [PubMed] [Google Scholar]
- Lou Y., Liu S. (2011). The TIPE (TNFAIP8) Family in Inflammation, Immunity, and Cancer. Mol. Immunol. 49, 4–7. 10.1016/j.molimm.2011.08.006 [DOI] [PubMed] [Google Scholar]
- Lutzmann M., Grey C., Traver S., Ganier O., Maya-Mendoza A., Ranisavljevic N., et al. (2012). MCM8- and MCM9-Deficient Mice Reveal Gametogenesis Defects and Genome Instability Due to Impaired Homologous Recombination. Mol. Cell. 47, 523–534. 10.1016/j.molcel.2012.05.048 [DOI] [PubMed] [Google Scholar]
- Maierbrugger K. T., Sousa-Nunes R., Bateman J. M. (2020). The mTOR Pathway Component Unkempt Regulates Neural Stem Cell and Neural Progenitor Cell Cycle in the Drosophila central Nervous System. Developmental Biol. 461, 55–65. 10.1016/j.ydbio.2020.01.006 [DOI] [PubMed] [Google Scholar]
- Marshall C. R., Noor A., Vincent J. B., Lionel A. C., Feuk L., Skaug J., et al. (2008). Structural Variation of Chromosomes in Autism Spectrum Disorder. Am. J. Hum. Genet. 82, 477–488. 10.1016/j.ajhg.2007.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita F., Kameyama T., Kadokawa Y., Marunouchi T. (2014). Spatiotemporal Expression Pattern of Myt/NZF Family Zinc finger Transcription Factors during Mouse Nervous System Development. Dev. Dyn. 243, 588–600. 10.1002/dvdy.24091 [DOI] [PubMed] [Google Scholar]
- Meier S., Strohmaier J., Breuer R., Mattheisen M., Degenhardt F., Mühleisen T. W., et al. (2013). Neuregulin 3 Is Associated with Attention Deficits in Schizophrenia and Bipolar Disorder. Int. J. Neuropsychopharmacol. 16, 549–556. 10.1017/s1461145712000697 [DOI] [PubMed] [Google Scholar]
- Milesi C., Alberici P., Pozzi B., Oldani A., Beznoussenko G. V., Raimondi A., et al. (2019). Redundant and Nonredundant Organismal Functions of EPS15 and EPS15L1. Life Sci. alliance. 2 (1), e201800273. 10.26508/lsa.201800273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minias P., Pikus E., Whittingham L. A., Dunn P. O. (2019). Evolution of Copy Number at the MHC Varies across the Avian Tree of Life. Genome Biol. Evol. 11, 17–28. 10.1093/gbe/evy253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee C., Holubowska A., Schwedhelm-Domeyer N., Mitkovski M., Lee S.-J., Kannan M., et al. (2015). Loss of the Neuron-specific F-Box Protein FBXO41 Models an Ataxia-like Phenotype in Mice with Neuronal Migration Defects and Degeneration in the Cerebellum. J. Neurosci. 35, 8701–8717. 10.1523/jneurosci.2133-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navone F., Consalez G. G., Sardella M., Caspani E., Pozzoli O., Frassoni C., et al. (2001). Expression of KIF3C Kinesin during Neural Development and In Vitro Neuronal Differentiation. J. Neurochem. 77, 741–753. 10.1046/j.1471-4159.2001.00277.x [DOI] [PubMed] [Google Scholar]
- Nilsson E., Dole G., Skinner M. K. (2009). Neurotrophin NT3 Promotes Ovarian Primordial to Primary Follicle Transition. Reproduction (Cambridge, England). 138, 697–707. 10.1530/REP-09-0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampouille E., Berri C., Boitard S., Hennequet-Antier C., Beauclercq S. A., Godet E., et al. (2018). Mapping QTL for white Striping in Relation to Breast Muscle Yield and Meat Quality Traits in Broiler Chickens. BMC genomics. 19, 202. 10.1186/s12864-018-4598-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei S. W., Qin F., Li W. H., Li F. D., Yue X. P. (2019). Copy Number Variation of ZNF280AY across 21 Cattle Breeds and its Association with the Reproductive Traits of Holstein and Simmental Bulls. J. Dairy Sci. 102, 7226–7236. 10.3168/jds.2018-16063 [DOI] [PubMed] [Google Scholar]
- Percie Du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M. T., Baker M., et al. (2020). The ARRIVE Guidelines 2.0: Updated Guidelines for Reporting Animal Research. Plos Biol. 18, e3000410. 10.1371/journal.pbio.3000410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., Dominy N. J., Claw K. G., Lee A. S., Fiegler H., Redon R., et al. (2007). Diet and the Evolution of Human Amylase Gene Copy Number Variation. Nat. Genet. 39, 1256–1260. 10.1038/ng2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J., Lebrasseur O., Deng H., Larson G. (2016). Holocene Cultural History of Red Jungle Fowl (Gallus gallus) and its Domestic Descendant in East Asia. Quat. Sci. Rev. 142, 102–119. 10.1016/j.quascirev.2016.04.004 [DOI] [Google Scholar]
- Qin Y., Jiao X., Simpson J. L., Chen Z.-J. (2015). Genetics of Primary Ovarian Insufficiency: New Developments and Opportunities. Hum. Reprod. Update. 21, 787–808. 10.1093/humupd/dmv036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan N., Sharma S., Gupta A., Alashwal B. Y. (2018). Keratin Based Bioplastic Film from Chicken Feathers and its Characterization. Int. J. Biol. macromolecules. 111, 352–358. 10.1016/j.ijbiomac.2018.01.037 [DOI] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K. R., Feuk L., Perry G. H., Andrews T. D., et al. (2006). Global Variation in Copy Number in the Human Genome. Nature. 444, 444–454. 10.1038/nature05329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice A. M., Mclysaght A. (2017). Dosage Sensitivity Is a Major Determinant of Human Copy Number Variant Pathogenicity. Nat. Commun. 8, 14366. 10.1038/ncomms14366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M. G., Livraghi-Butrico A., Fletcher A. A., Mcelwee M. M., Evans S. E., Boerner R. M., et al. (2014). Muc5b Is Required for Airway Defence. Nature. 505, 412–416. 10.1038/nature12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Villagra M. R., Geiger M., Schneider R. A. (2016). The Taming of the Neural Crest: a Developmental Perspective on the Origins of Morphological Covariation in Domesticated Mammals. R. Soc. Open Sci. 3, 160107. 10.1098/rsos.160107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A., Dutheil J. Y., Hillier L. W., Jordan G. E., Goodhead I., Herrero J., et al. (2012). Insights into Hominid Evolution from the gorilla Genome Sequence. Nature. 483, 169–175. 10.1038/nature10842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., et al. (2004). Large-scale Copy Number Polymorphism in the Human Genome. Science. 305, 525–528. 10.1126/science.1098918 [DOI] [PubMed] [Google Scholar]
- Shinomiya A., Kayashima Y., Kinoshita K., Mizutani M., Namikawa T., Matsuda Y., et al. (2012). Gene Duplication of Endothelin 3 Is Closely Correlated with the Hyperpigmentation of the Internal Organs (Fibromelanosis) in Silky Chickens. Genetics. 190, 627–638. 10.1534/genetics.111.136705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A., Green P. (2013). RepeatMasker Open-4.0. Available at: http://www.repeatmasker.org .
- Smith J., Paton I. R., Horvat S., Medrano J. F., Burt D. W. (2000). Mapping the RAIDD Gene of Chicken (Gallus gallus): Identification of a Region Homologous to the Mouse High-Growth Region. Mamm. Genome. 11, 706–709. 10.1007/s003350010140 [DOI] [PubMed] [Google Scholar]
- Sohrabi S. S., Mohammadabadi M., Wu D.-D., Esmailizadeh A. (2018). Detection of Breed-specific Copy Number Variations in Domestic Chicken Genome. Genome. 61, 7–14. 10.1139/gen-2017-0016 [DOI] [PubMed] [Google Scholar]
- Steinauer N., Guo C., Zhang J. (2020). The Transcriptional Corepressor CBFA2T3 Inhibits All-Trans-Retinoic Acid-Induced Myeloid Gene Expression and Differentiation in Acute Myeloid Leukemia. J. Biol. Chem. 295, 8887–8900. 10.1074/jbc.RA120.013042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Cui Q., Shi Y. (2017). Nuclear Receptor TLX in Development and Diseases. Curr. Top. Dev. Biol. 125, 257–273. 10.1016/bs.ctdb.2016.12.003 [DOI] [PubMed] [Google Scholar]
- Team R. C. (2021). R: A Language and Environment for Statistical Computing. R. Found. Stat. Comput [Google Scholar]
- Terenzio M., Golding M., Russell M. R. G., Wicher K. B., Rosewell I., Spencer‐Dene B., et al. (2014). Bicaudal‐ D 1 Regulates the Intracellular Sorting and Signalling of Neurotrophin Receptors. Embo J. 33, 1582–1598. 10.15252/embj.201387579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Wang Y., Gu X., Feng C., Fang S., Hu X., et al. (2013). Copy Number Variants in Locally Raised Chinese Chicken Genomes Determined Using Array Comparative Genomic Hybridization. BMC genomics. 14, 262. 10.1186/1471-2164-14-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L., Salzberg S. L. (2009). TopHat: Discovering Splice Junctions with RNA-Seq. Bioinformatics (Oxford, England). 25, 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.-S., Li Y., Peng M.-S., Zhong L., Wang Z.-J., Li Q.-Y., et al. (2015). Genomic Analyses Reveal Potential Independent Adaptation to High Altitude in Tibetan Chickens. Mol. Biol. Evol. 32, 1880–1889. 10.1093/molbev/msv071 [DOI] [PubMed] [Google Scholar]
- Wang M.-S., Thakur M., Peng M.-S., Jiang Y., Frantz L. A. F., Li M., et al. (2020). 863 Genomes Reveal the Origin and Domestication of Chicken. Cell Res. 30, 693–701. 10.1038/s41422-020-0349-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Chen C. Z., Fu X. H., Liu J. B., Peng Y. X., Wang Y. J., et al. (2020). CPEB3 Regulates the Proliferation and Apoptosis of Bovine Cumulus Cells. Anim. Sci. J. 91, e13416. 10.1111/asj.13416 [DOI] [PubMed] [Google Scholar]
- Wilkins A. S., Wrangham R. W., Fitch W. T. (2014). The "domestication Syndrome" in Mammals: a Unified Explanation Based on Neural Crest Cell Behavior and Genetics. Genetics. 197, 795–808. 10.1534/genetics.114.165423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther M., Berezin V., Walmod P. S. (2012). NCAM2/OCAM/RNCAM: Cell Adhesion Molecule with a Role in Neuronal Compartmentalization. Int. J. Biochem. Cell Biol. 44, 441–446. 10.1016/j.biocel.2011.11.020 [DOI] [PubMed] [Google Scholar]
- Wright D., Boije H., Meadows J. R. S., Bed'hom B., Gourichon D., Vieaud A., et al. (2009). Copy Number Variation in Intron 1 of SOX5 Causes the Pea-Comb Phenotype in Chickens. Plos Genet. 5, e1000512. 10.1371/journal.pgen.1000512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Xu S., Zhang W., Liu Y., Wang Q., Man C. (2019). Arih2 Gene Influences Immune Response and Tissue Development in Chicken. Biosci. Rep. 39 (10), BSR20190933. 10.1042/BSR20190933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhu Z., Yerle M., Wang H. L., Wang H., Gu M., et al. (2004). Radiation Hybrid Mapping of Four Genes (MYBPC1,LUM,ZRF1andATP2B4) Expressed in Embryo Skeleton Muscle to Pig Chromosomes 5 and 9. Anim. Genet. 35, 472–473. 10.1111/j.1365-2052.2004.01195.x [DOI] [PubMed] [Google Scholar]
- Xu L., He Y., Ding Y., Sun G., Carrillo J. A., Li Y., et al. (2017). Characterization of Copy Number Variation's Potential Role in Marek's Disease. Int. J. Mol. Sci. 18, 1020. 10.3390/ijms18051020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.-T., Hindes A. E., Hultman K. A., Johnson S. L. (2007). Mutations in Gfpt1 and Skiv2l2 Cause Distinct Stage-specific Defects in Larval Melanocyte Regeneration in Zebrafish. Plos Genet. 3, e88. 10.1371/journal.pgen.0030088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. X., Zhou H., Ding J. M., He C., Niu Q., Gu C. J., et al. (2020). Copy Number Variation in HOXB7 and HOXB8 Involves in the Formation of Beard Trait in Chickens. Anim. Genet. 51, 958–963. 10.1111/age.13011 [DOI] [PubMed] [Google Scholar]
- Yi G., Qu L., Liu J., Yan Y., Xu G., Yang N. (2014). Genome-wide Patterns of Copy Number Variation in the Diversified Chicken Genomes Using Next-Generation Sequencing. BMC Genomics. 15, 962. 10.1186/1471-2164-15-962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Du Z.-Q., Dong J.-Q., Wang H.-X., Shi H.-Y., Wang N., et al. (2014). Detection of Genome-wide Copy Number Variations in Two Chicken Lines Divergently Selected for Abdominal Fat Content. BMC Genomics. 15, 517. 10.1186/1471-2164-15-517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Shen L.-Y., Xu Z.-C., Kramer L. M., Yu J.-Q., Zhang X.-Y., et al. (2020). Haplotype-based Genome-wide Association Studies for Carcass and Growth Traits in Chicken. Poult. Sci. 99, 2349–2361. 10.1016/j.psj.2020.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Jia Y., Almeida P., Mank J. E., Van Tuinen M., Wang Q., et al. (2018). Whole-genome Resequencing Reveals Signatures of Selection and Timing of Duck Domestication. Gigascience. 7 (4), giy027. 10.1093/gigascience/giy027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Tu H. M., Yu H. R., Zheng S., Dai B., Price M., et al. (2019). The Draft Genome of the Endangered Sichuan Partridge (Arborophila rufipectus) with Evolutionary Implications. Genes. 10. 10.3390/genes10090677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Lemos B., Dopman E. B., Hartl D. L. (2011). Copy-Number Variation: The Balance between Gene Dosage and Expression in Drosophila melanogaster . Genome Biol. Evol. 3, 1014. 10.1093/gbe/evr023 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.