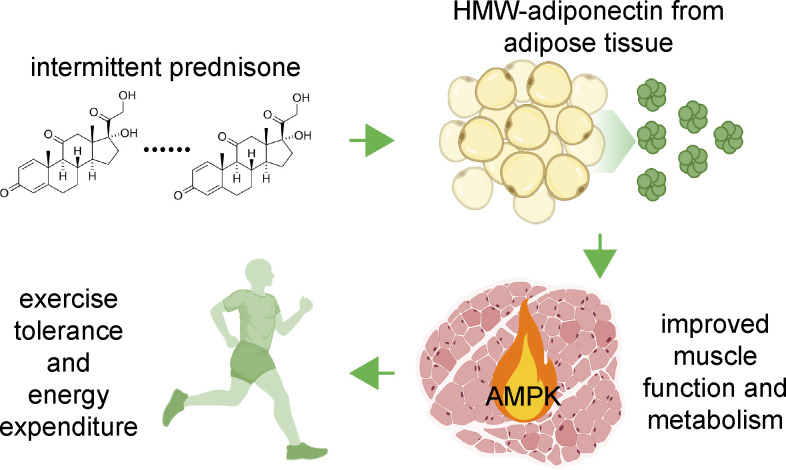
Chronic daily intake of exogenous glucocorticoids produces or exacerbates metabolic stress. In contrast, this study reports that intermittent, once-weekly dosing of the glucocorticoid prednisone promotes exercise tolerance and muscle metabolism in mice with diet-induced obesity through adiponectin elevation.
Abstract
The fat–muscle communication regulates metabolism and involves circulating signals like adiponectin. Modulation of this cross-talk could benefit muscle bioenergetics and exercise tolerance in conditions like obesity. Chronic daily intake of exogenous glucocorticoids produces or exacerbates metabolic stress, often leading to obesity. In stark contrast to the daily intake, we discovered that intermittent pulses of glucocorticoids improve dystrophic muscle metabolism. However, the underlying mechanisms, particularly in the context of obesity, are still largely unknown. Here we report that in mice with diet-induced obesity, intermittent once-weekly prednisone increased total and high-molecular weight adiponectin levels and improved exercise tolerance and energy expenditure. These effects were dependent upon adiponectin, as shown by genetic ablation of the adipokine. Upregulation of Adipoq occurred through the glucocorticoid receptor (GR), as this effect was blocked by inducible GR ablation in adipocytes. The treatment increased the muscle metabolic response of adiponectin through the CAMKK2–AMPK cascade. Our study demonstrates that intermittent glucocorticoids produce healthful metabolic remodeling in diet-induced obesity.
Graphical Abstract
Introduction
Weakness and exercise intolerance correlate with metabolic stress, blunted nutrient uptake, and mitochondrial impairment in obese muscle (Abdul-Ghani and DeFronzo, 2010). The fat–muscle communication is impaired in dietary obesity (Yang et al., 2014), and mechanisms to restore muscle sensitivity to this cross-talk are still poorly elucidated.
Glucocorticoid steroids such as prednisone are widely used immune suppressants and their chronic daily intake promotes metabolic stress and obesity (Nadal et al., 2017). Surprisingly, we found that, opposite to once-daily dosing, intermittent once-weekly prednisone dosing improved muscle mitochondrial function, lean mass, and insulin sensitivity in mice and humans with muscular dystrophy (Quattrocelli et al., 2019). However, whether this same benefit could be elicited in the setting of obesity, in the absence of dystrophic muscle, was unclear.
AMP-activated kinase (AMPK) is a key driver of carbohydrate and lipid metabolism in muscle (Hayashi et al., 1998). Muscle AMPK can be phosphorylated and activated by calcium/calmodulin dependent protein kinase 2 (CAMKK2), which is in turn activated by the adiponectin–adiponectin receptor 1 (ADIPOR1) signaling axis (Iwabu et al., 2010). Among many cytokines mediating the fat–muscle cross-talk, adiponectin stands out as the most abundant adipokine in serum and a crucial regulator of muscle health (Krause et al., 2019). Obesity is associated with lower adiponectin levels in adipose tissue and circulation, as well as adiponectin resistance in tissues driving energy expenditure, like muscles (Achari and Jain, 2017). However, there is still no consensus on the role of glucocorticoids in adiponectin regulation in adipose tissue (Sukumaran et al., 2012), as little is known about the direct effects of glucocorticoid regimens on adiponectin production and sensitivity.
Here, we report that intermittent glucocorticoids alleviate metabolic stress and exercise intolerance in mice with dietary obesity. The treatment upregulated total and high-molecular weight adiponectin levels in fat tissue and circulation, increasing adiponectin sensitivity and nutrient oxidation in the muscle through the CAMKK–AMPK axis. Adiponectin was required for the pro-metabolic effects of intermittent glucocorticoids, and the adipocyte-specific glucocorticoid receptor was required for treatment-driven adiponectin upregulation. In aggregate, our findings provide novel mechanisms through which intermittent dosing of glucocorticoid steroids promotes exercise tolerance in dietary obesity.
Results and discussion
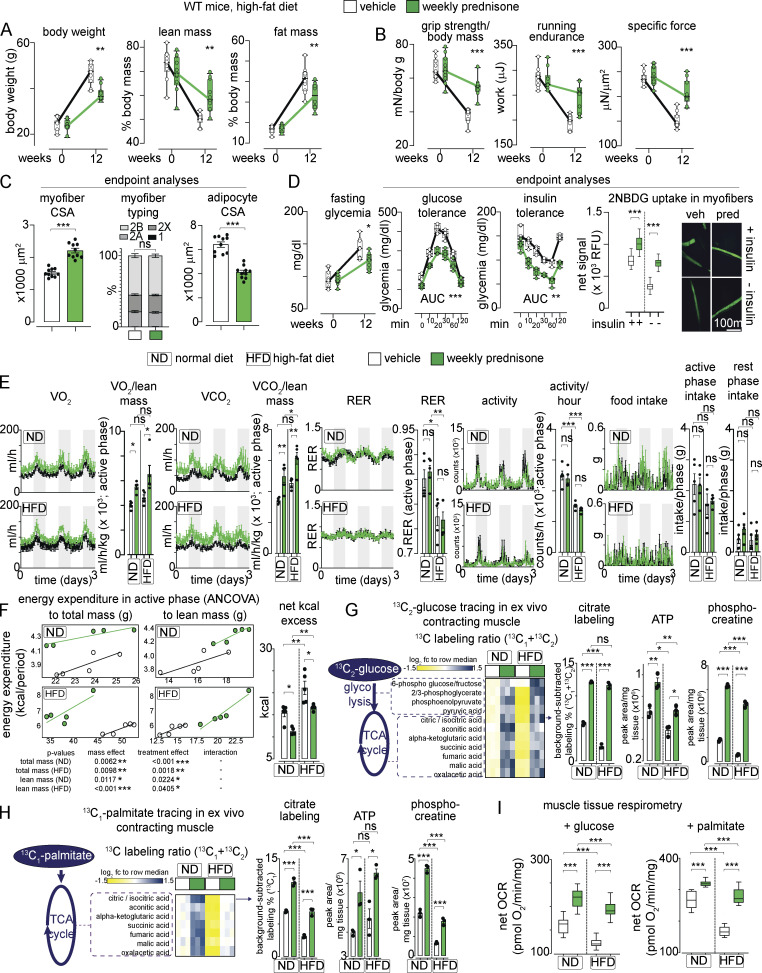
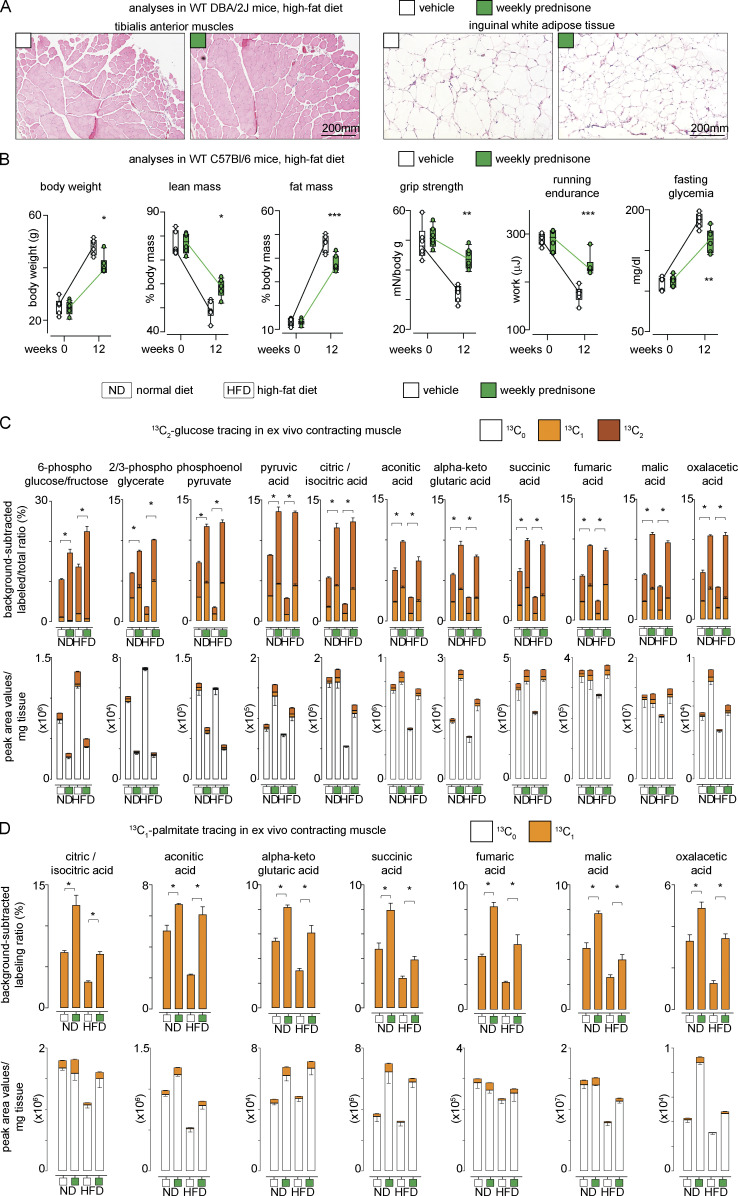
Intermittent prednisone improved muscle metabolic remodeling in dystrophic mice (Quattrocelli et al., 2019). We analyzed whether these effects extended to dietary obesity, where normal non-injured muscle undergoes nutrient stress. WT-mice were fed ad libitum with a high-fat diet (HFD; 60% kcal in fat) for 12 wk from the age of 8 wk. Initially, we used male mice from the DBA/2J background to match sex and genetic background from our previous studies with DBA/2J-mdx dystrophic mice (Quattrocelli et al., 2017; Quattrocelli et al., 2019). Mice were randomized to receive either once-weekly i.p. 1 mg/kg prednisone or the vehicle (n = 10 mice/group). Compared with the vehicle, the treatment reduced the overall increase in body weight at the endpoint, promoting a significant reduction in adiposity and, conversely, proportional retention of lean mass (Fig. 1 A). The treatment increased muscle performance in obese mice, as shown by improved values of grip strength/body mass, treadmill running endurance, and specific force of tibialis anterior muscle compared with the vehicle at endpoint (Fig. 1 B). Compared with the vehicle at the endpoint, the treatment increased myofiber cross-sectional area (CSA) without significant shifts in myofiber typing in muscle, while decreasing adipocyte CSA in white adipose tissue (Fig. 1 C and Fig. S1 A). Moreover, the treatment improved glycemia, glucose tolerance, and insulin tolerance, as well as glucose uptake in muscle, quantitated through ex vivo uptake assay of the fluorescent 2-NBDG glucose analog (Fig. 1 D). The effects were not dependent on the DBA/2J genetic background, as we replicated the treatment effects in age-matched C57BL/6J WT mice on HFD (n = 5 mice/group; Fig. S1 B). We then profiled whole-body energetics of the treated and control mice using indirect calorimetry at the end of diet/drug treatment. We included mice on a normal diet (ND) to discriminate regimen-specific effects independent from obesity. In both ND and HFD, the treatment increased overall metabolic rate, indicated by increased VO2 and VCO2, without significant effects on overall RER, activity, and food intake (Fig. 1 E). We used CalR (Mina et al., 2018) to analyze energy expenditure. The treatment increased energy expenditure in the active phase as shown by ANCOVA analyses with total mass or lean mass as co-variates (Fig. 1 F). Also, the treatment reduced the net energy excess calculated from kcal intake and expenditure (Fig. 1 F). Thus, intermittent prednisone mitigated obesity and exercise intolerance in the setting of HFD, and this effect correlated with increased energy expenditure.
Figure 1.
Intermittent prednisone improves exercise tolerance in obesity. WT mice were fed a HFD for 12 wk and treated with weekly i.p. injections of either vehicle or 1 mg/kg prednisone. (A) During diet-induced obesity onset, treatment reduced accrual of body weight and fat mass, while improving lean mass retention. (B) Treatment increased strength, running endurance, and specific muscle force (tibialis anterior) after HFD regimen. (C) Treatment increased the average CSA of myofibers without significant changes in relative myofiber abundance in tibialis anterior muscle, while decreasing adipocyte CSA in white adipose tissue. (D) Treatment attenuated hyperglycemia and improved profiles from glucose tolerance and insulin sensitivity tests. Right: Isolated myofibers from treated mice showed increased capacity of 2NBDG (fluorescent glucose analog) uptake ex vivo. 2NBDG signal was normalized to signal from negative control myofibers (cytochalasin-treated). (E) Metabolic cage traces from mice on normal or HFD show that treatment increased overall metabolic rate, indicated by increased VO2 and VCO2, without significant effects on overall RER, activity, and food intake. (F) Treatment increased energy expenditure in the active phase as shown by ANCOVA analyses with total mass or lean mass as co-variates. Treatment reduced net calorie excess in both diets. (G) Isotope tracing in ex vivo contracting muscle showed that treatment increased glucose catabolism toward mitochondrial energy production in muscle, as shown by gain of 13C labeling of citrate and other intermediates of glycolysis and TCA cycle, as well as increased unlabeled levels of ATP and phosphocreatine in the same samples. (H) Analogous trends were obtained with 13C-palmitate. (I) Tissue respirometry on muscle biopsies showed that treatment increased basal respiration in the presence of either glucose or palmitate. Line plots, single values with min–max box-plots, and connected medians; histograms, single values and mean ± SEM box-plots, Tukey distribution. All panels report data verified in ≥2 independent experiments. n = 10 mice/group (A and B), 5 mice/group (C–F); 3 mice/group (D and E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; A–C (myofiber typing), D (glycemia): two-way ANOVA; C–I: one-way Welch’s ANOVA with Tukey multiple comparison.
Figure S1.
Additional supporting data related to Fig. 1. (A) Representative histological images for CSA analyses presented in Fig. 1 C. (B) Weekly prednisone effects on adiposity, exercise tolerance and fasting glycemia were recapitulated in WT C57BL6/J mice. (C and D) Histograms of 13C labeling and total metabolite levels of analyses shown in Fig. 1, G–H heatmaps. Line plots, single values with min-max boxplots and connected medians; histograms, single values and mean ± SEM.; boxplots, Tukey distribution. All panels report data verified in at least two independent experiments. n = 5 mice/group (A), 3 mice/group (B and C). *, P < 0.05; **, P < 0.01; ***, P < 0.001; (B and C) two-way ANOVA; (D) one-way Welch’s ANOVA with Tukey multiple comparison.
We then investigated nutrient utilization in the muscle. The absence of RER changes suggested increased metabolism of both glucose and lipids; hence, we probed muscle oxidative catabolism for both glucose and palmitate in nutrient-specific conditions. To exclude extra-muscular systemic effects, we performed 13C profiling and mass-spec in ex vivo isolated contracting muscle. We adapted reported conditions (Kerner et al., 2014) to isolate quadriceps muscles with repeated isometric contractions in the presence of 13C-labeled nutrients, i.e., 1,2-13C2-glucose and 1-13C1-palmitate. Each nutrient-specific catabolic cascade was analyzed quantitating the relative enrichment in appropriate 13C-labeled intermediates and TCA cycle metabolites in a non–steady state, as previously shown in the mdx muscle (Quattrocelli et al., 2019). Compared with the vehicle in ND and HFD, the treatment increased relative 13C labeling downstream of 1,2-13C2-glucose in glycolytic and TCA cycle intermediates, exemplified by citrate labeling (Fig. 1 G). The same samples showed increased levels of unlabeled ATP and phosphocreatine in treated versus control muscles (Fig. 1 G). Analogous trends were found in the presence of 1-13C-palmitate (Fig. 1 H). Enrichment and absolute values for labeled/unlabeled metabolites depicted in heatmaps are reported in Fig. S1, C and D. Moreover, we confirmed that the treatment increased mitochondrial respiration in muscle as compared to the vehicle after ND and HFD, quantitating basal muscle tissue respirometry with nutrient-specific inhibitors (Fig. 1 I). Thus, intermittent prednisone increased nutrient oxidative catabolism in the normal muscle.
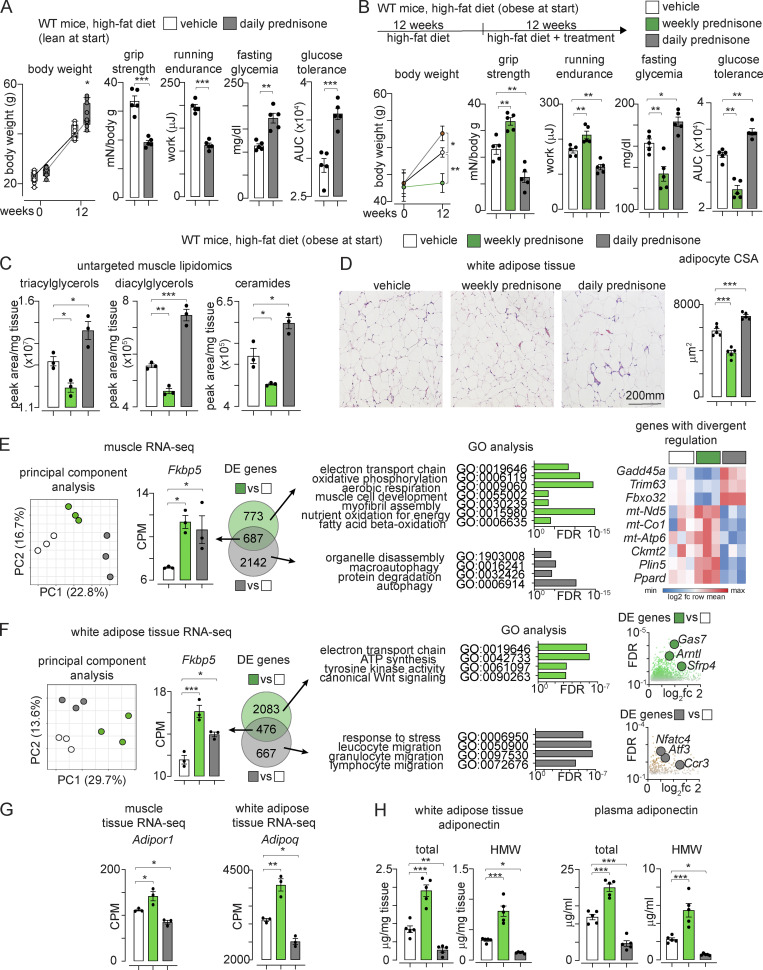
To evaluate whether the prednisone effects were specific to the intermittent dosing, we assayed the metabolic effects of a daily prednisone regimen, since we observed dimorphic metabolic effects of glucocorticoids in dystrophic muscle based on dosing frequency (Quattrocelli et al., 2019). Indeed, daily dosing of the same prednisone dose used for the intermittent regimen (1 mg/kg i.p.) exacerbated weight accrual, weakness, exercise intolerance, and hyperglycemia induced by HFD in mice that were lean at the start (Fig. 2 A).
Figure 2.
The positive effects of prednisone regimens on exercise tolerance and glucose homeostasis are dependent on dosing intermittence. (A) Opposite to once-weekly regimen, once-daily prednisone (12-wk-long treatment) worsened weakness and hyperglycemia during diet-induced obesity development. (B) Once-weekly prednisone (12-wk-long treatment) decreased weight accrual and improved strength, aerobic performance, fasting glucose, and glucose tolerance in mice that were already obese at the start of treatment and were maintained on HFD during treatment. Conversely, once-daily prednisone induced opposite effects. (C) Untargeted mass-spec profiling of muscle lipids revealed that weekly prednisone decreased triacylglycerols, diacylglycerols, and ceramides, while daily prednisone promoted their accumulation in muscle. (D) Weekly prednisone reduced adipocyte CSA, while daily prednisone increased it. (E) RNA-seq in muscle from obese mice showed regimen-specific global clustering of sample transcriptomes. The canonical glucocorticoid reporter Fkbp5 was among the DE genes shared by both regimens. However, among regimen-specific DE genes, we found that GO pathways of mitochondrial metabolism and muscle hypertrophy were specifically enriched with weekly prednisone. Conversely, GO pathways related to muscle wasting were specifically enriched with daily prednisone. Indeed, the expression of atrophy markers Gadd45a, Trim63, and Fbxo32 was decreased with weekly prednisone and increased with daily regimen. Conversely, expression of oxidative metabolism agonists mt-Nd5, mtCo1, mt-Atp6, Ckmt2, Plin5, and Ppard was increased by intermittent and decreased by daily dosing. (F) Regimen-divergent transcriptional effects were evident also in adipose tissue of obese mice. Weekly prednisone upregulated several pathways and genes linked to positive metabolic remodeling of adipose tissue, including Gas7, Sfrp4, and Arntl. The pro-obesity remodeling induced by daily prednisone correlated with the upregulation of pro-inflammatory genes like Ccr3 and adipose stress markers like Atf3 and Nfatc4, notable suppressors of adiponectin expression. (G) RNA-seq datasets revealed that weekly prednisone upregulated adiponectin (Adipoq) in adipose tissue and its receptor (Adipor1) in muscle, suggesting possible activation of this fat–muscle axis. (H) ELISAs confirmed the opposite effects of weekly versus daily prednisone on total and HMW adiponectin in adipose tissue and plasma. Line plots, single values with min–max box-plots, and connected medians; histograms, single values, and mean ± SEM. All panels report data verified in at least two independent experiments. n = 5 mice/group (A, B, D, and H); 3 mice/group (C and E–G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; A and B (body weight): two-way ANOVA; (A–H) one-way Welch’s ANOVA with Tukey multiple comparison.
We then analyzed whether the regimen-specific effects of intermittent versus daily prednisone were maintained in mice that were already obese before drug exposure. We fed WT mice HFD for 12 wk and then started a 12-wk-long treatment with the vehicle, intermittent (once-weekly) or daily prednisone, keeping the mice on HFD. Opposite to daily dosing, intermittent prednisone blunted weight accrual and improved strength, treadmill endurance, and glucose homeostasis in mice with pre-established obesity (Fig. 2 B). Untargeted mass-spec profiling of lipids revealed that intermittent dosing decreased intramuscular levels of triacylglycerols, diacylglycerols and ceramides, and muscle lipids that typically accumulate in metabolic diseases (Goodpaster and Wolf, 2004), while daily regimen worsened their accumulation (Fig. 2 C). Also, compared to the vehicle, intermittent dosing decreased adipocyte CSA, while daily dosing increased it (Fig. 2 D). Therefore, the favorable metabolic effects of prednisone were specific to the intermittent dosing even in mice already obese before treatment.
We profiled quadriceps muscle and inguinal white adipose tissue of treated mice through RNA-seq. In muscle, the principal component analysis showed regimen-specific global clustering of sample transcriptomes. The canonical glucocorticoid reporter Fkbp5 was among the differentially expressed (DE) genes shared by both regimens. However, among regimen-specific DE genes, we found that gene ontology (GO) pathways of mitochondrial metabolism and muscle hypertrophy were specifically enriched with weekly prednisone. Conversely, GO pathways related to muscle wasting were specifically enriched with daily prednisone. Indeed, the expression of Gadd45a, Trim63, and Fbxo32, canonical markers of muscle atrophy (Ebert et al., 2012; Latres et al., 2005), was decreased with weekly prednisone and increased with daily regimen. Conversely, expression of oxidative metabolism markers mt-Nd5, mtCo1, mt-Atp6, Ckmt2, Plin5, and Ppard was increased by intermittent prednisone and decreased by daily prednisone (Fig. 2 E). Regimen-divergent transcriptional effects were evident also in adipose tissue. GO pathways related to oxidative metabolism, tyrosine kinase activity, and Wnt signaling were enriched after intermittent prednisone. Within these pathways, the treatment upregulated several genes linked to metabolic reactivation of adipose tissue, including Gas7, Sfrp4, and Arntl (Guo et al., 2012; Yang et al., 2009; Zhang et al., 2020). The pro-obesity remodeling induced by daily prednisone correlated with the upregulation of pro-inflammatory genes like Ccr3 and adipose stress markers like Atf3 and Nfatc4, notable suppressors of adiponectin expression (Kim et al., 2006; Fig. 2 F). Regarding the possible activation of the fat–muscle adiponectin axis, RNA-seq datasets revealed that intermittent prednisone upregulated adiponectin (Adipoq) in the adipose tissue and its receptor (Adipor1) in the muscle, while daily prednisone induced opposite effects (Fig. 2 G). ELISAs confirmed the opposite effects of intermittent versus daily prednisone on total and high-molecular-weight (HMW) adiponectin in adipose tissue and plasma (Fig. 2 H). HMW adiponectin is the most metabolically active adiponectin isoform, and its levels inversely correlate with the incidence of type-2 diabetes and its associated cardiovascular risks (Horakova et al., 2015; Zhu et al., 2010). Thus, the pro-metabolic effects of prednisone on muscle and adipose tissues were specific to intermittent dosing and correlated with adiponectin expression and levels.
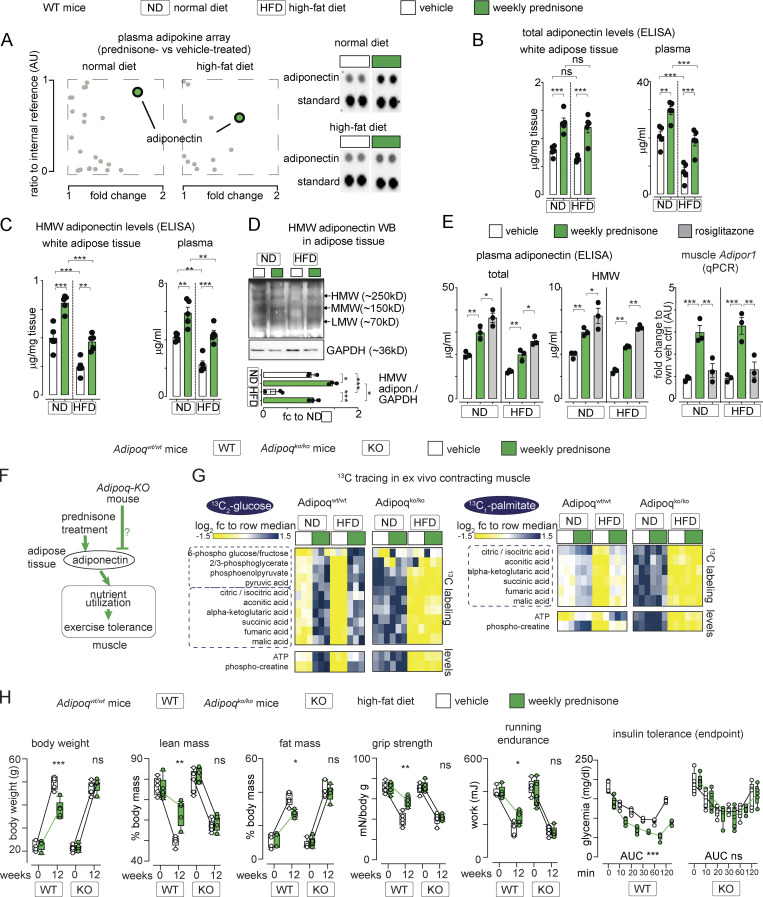
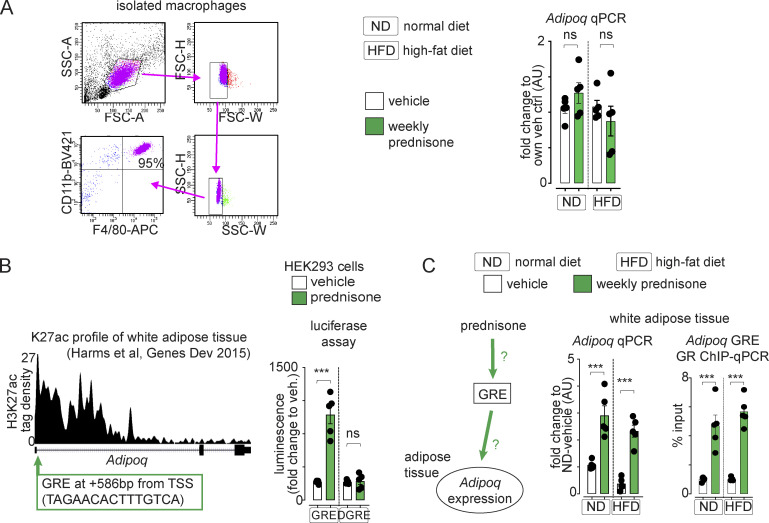
We sought to confirm the effect of intermittent prednisone on adiponectin through an unbiased approach. We assayed the serum of obese animals (12 wk HFD) through a 38-cytokine profiler array, controlling the experiment with ND mice. Compared with the vehicle, adiponectin was the top enriched adipokine in mice treated with intermittent prednisone after ND and HFD (Fig. 3 A). ELISAs in adipose tissue and plasma confirmed the treatment effect on total and HMW adiponectin in ND and HFD mice that were lean at the start (Fig. 3, B and C). Treatment effects on HMW adiponectin levels were also confirmed through WB (Fig. 3 D). Adiponectin overexpression by macrophages was reported to be beneficial in obesity (Luo et al., 2010). In that study, peritoneal macrophages were analyzed as resident macrophages, and we also isolated peritoneal macrophages to probe treatment effects on adiponectin expression. Cytometry confirmed enrichment (95%) for CD11b+/F4/80+ cells in our macrophage pool. Compared with the vehicle, we did not detect significant changes by intermittent prednisone on macrophage adiponectin expression in ND and HFD mice (Fig. S2 A), suggesting that the treatment effect on adiponectin production was mainly dependent on adipose tissue. Moreover, we compared intermittent prednisone (once-weekly 1 mg/kg) to rosiglitazone (daily 10 mg/kg) for the stimulation of the adiponectin axis, as the rosiglitazone regimen stimulates adiponectin production in obese mice (Wong et al., 2011). It must be noted that to match the 4-wk-long rosiglitazone treatment, the intermittent prednisone treatment in this experiment lasted only 4 wk and not 12 wk. In both ND and HFD, the adiponectin increase with intermittent prednisone was slightly lower than with rosiglitazone. However, weekly prednisone outperformed rosiglitazone in upregulating the adiponectin receptor Adipor1 in muscle, underscoring the coordinated fat–muscle axis activation with intermittent glucocorticoids (Fig. 3 E). Thus, intermittent prednisone increased total and HMW adiponectin levels in the adipose tissue and circulation in normal and obese mice.
Figure 3.
Adiponectin production is required for intermittent prednisone effects on muscle performance. (A) Serum adipokine arrays identified adiponectin as a top upregulated hit after weekly prednisone regimens in both normal and HFD conditions. (B and C) After 12 wk of treatment, total and HMW adiponectin levels were elevated in fat tissue and circulation of mice on normal and HFDs. (D) WB confirmation of HMW adiponectin trends in adipose tissue. (E) Weekly prednisone upregulated adiponectin to a modestly lower level than rosiglitazone. However, compared to rosiglitazone, treatment significantly upregulated muscle Adipor1, underscoring the treatment effects on the fat–muscle axis. (F–H) Body-wide ablation of adiponectin (Adipoq-KO) blocked the effects of weekly prednisone on nutrient oxidation (G) and adiposity and exercise tolerance (H) seen in the Adipoq-WT littermates. Line plots, single values with min-max boxplots and connected medians; histograms, single values and mean ± SEM. All panels report data verified in at least two independent experiments. n = 5 mice/group (A–C and H), 3 mice/group (D, E, and G). *, P < 0.05; **, P < 0.01; ***, P < 0.001; (B–E) one-way Welch’s ANOVA with Tukey multiple comparison; (H) two-way ANOVA. Source data are available for this figure: SourceData F3.
Figure S2.
Additional supporting data related to Fig. 3 and Fig. 4. (A) Peritoneal macrophages were highly enriched (95%) in CD11b+/F4/80+ cells. We could not detect significant changes of treatment on Adipoq expression by peritoneal macrophages in mice on either normal or high-fat diets. (B) H3K27ac ChiP-seq datasets from white adipose tissue showed a canonical GRE juxtaposed to the TSS of the Adipoq gene locus. The regulatory region was responsive to prednisone stimulation in transfected HEK293T cells but not in the absence of the GRE sequence (ΔGRE). (C) Weekly prednisone treatment upregulated Adipoq expression and GR occupancy of probed Adipoq GRE after both normal and high-fat diets. Histograms, single values and mean ± SEM. All panels report data verified in at least two independent experiments. n = 5 mice/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way Welch’s ANOVA with Tukey multiple comparison.
We then tested the extent to which adiponectin mediated the metabolic benefits of intermittent prednisone. We used transgenic mice, the Adipoq-KO mice, for adiponectin genetic ablation (Ma et al., 2002; Fig. 3 F). Adipoq-KO male mice (n = 5/group) and their Adipoq-WT male littermates (n = 3/group) were randomized to receive ND or HFD, and the vehicle versus once-weekly 1 mg/kg i.p. prednisone for 12 wk from the age of 8 wk. At the endpoint, treatment failed to increase nutrient oxidation in the muscle in Adipoq-KO mice, while the treatment effects were recapitulated in Adipoq-WT mice, as assessed by 13C tracing (Fig. 3 G). Accordingly, treatment failed to improve adiposity, exercise tolerance, and insulin tolerance with HFD in Adipoq-KO mice, while the treatment effects were recapitulated in Adipoq-WT mice (Fig. 3 H). Therefore, intermittent prednisone promoted muscle metabolism and exercise tolerance through adiponectin.
We analyzed whether intermittent prednisone increased adiponectin (encoded by Adipoq) production through transcriptional activation by the cognate drug receptor, the glucocorticoid receptor (GR; Presman et al., 2010). We identified a putative canonical GR element (GRE) adjacent to the transcription start site of Adipoq from H3K27ac ChIP-seq data in adipose tissue (GEO accession no. GSE63964; Harms et al., 2015). Luciferase assay in HEK293 cells confirmed that the GRE-containing region was activated by prednisone, and this activation was blunted when the GRE was specifically removed (Fig. S2 B). Adipoq upregulation in adipose tissue by intermittent prednisone correlated with increased GRE occupancy by GR (Fig. S2 C).
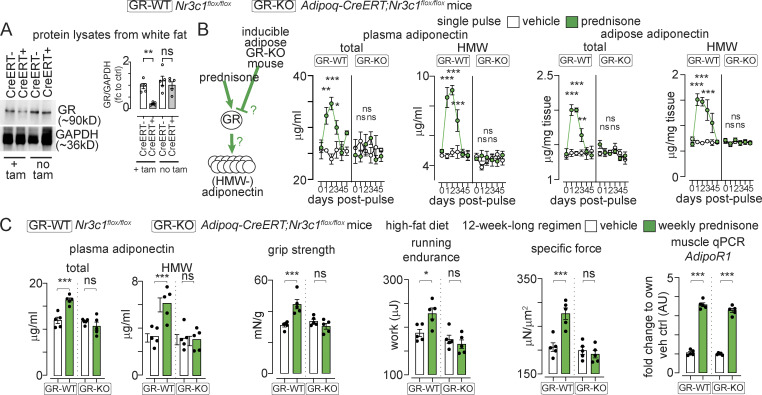
To further validate these findings, we generated a mouse model to induce GR ablation in postnatal adipose tissue. We derived Adipoq-CreERT+/−, Nr3c1flox/flox mice from Adipoq-CreERT+/− (Jeffery et al., 2015) and Nr3c1flox/flox mice (Mittelstadt et al., 2012), respectively. At 8 wk of age, we induced ablation of adipocyte GR (encoded by Nr3c1) through a sequence of i.p. tamoxifen injections (20 mg/kg/d for 5 d) followed by 14 d on tamoxifen-containing chow (40 mg/kg) and 2 d of regular chow (tamoxifen washout). This strategy ablated ∼85% of GR in white adipose tissue, while no changes were observed in the absence of tamoxifen (Fig. 4 A). We compared Adipoq-CreERT+/− and Nr3c1flox/flox (GR-KO) to Nr3c1flox/flox (GR-WT) littermates after both received tamoxifen. After a single pulse of i.p. prednisone, GR-WT mice showed a transient elevation in total adiponectin in the adipose tissue and circulation, while this elevation was blocked in GR-KO mice. Analogous trends were found with HMW adiponectin (Fig. 4 B). Moreover, we treated GR-KO and GR-WT mice with a 12-wk-long regimen of intermittent prednisone while on HFD. Although muscle Adipor1 was comparably upregulated by treatment in both genotypes, treatment failed to upregulate adiponectin and exercise tolerance in GR-KO mice, while treatment effects were recapitulated in GR-WT littermates (Fig. 4 C). Thus, intermittent prednisone elevated adiponectin through the adipose GR.
Figure 4.
Adipose GR is required for adiponectin elevation by intermittent prednisone. (A) WB confirming inducible GR ablation in adipose tissue of fat GR-KO mice but not fat GR-WT littermates. (B) Total and HMW-adiponectin levels are transiently increased after a single prednisone pulse in fat GR-WT mice, while this effect is lost in fat GR-KO mice. (C) Analogous trends were obtained after 12 wk of intermittent prednisone during HFD exposure. This correlated with treatment effects on muscle performance recapitulated in fat GR-WT mice and blocked in fat GR-KO mice, despite treatment-induced Adipor1 upregulation in muscle in both mice. All panels report data verified in at least two independent experiments. Line plots, mean values with SEM; histograms, single values and mean ± SEM. n = 5 mice/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; (A and C) one-way Welch’s ANOVA with Tukey multiple comparison; (B) two-way ANOVA with Tukey multiple comparison for time points. Source data are available for this figure: SourceData F4.
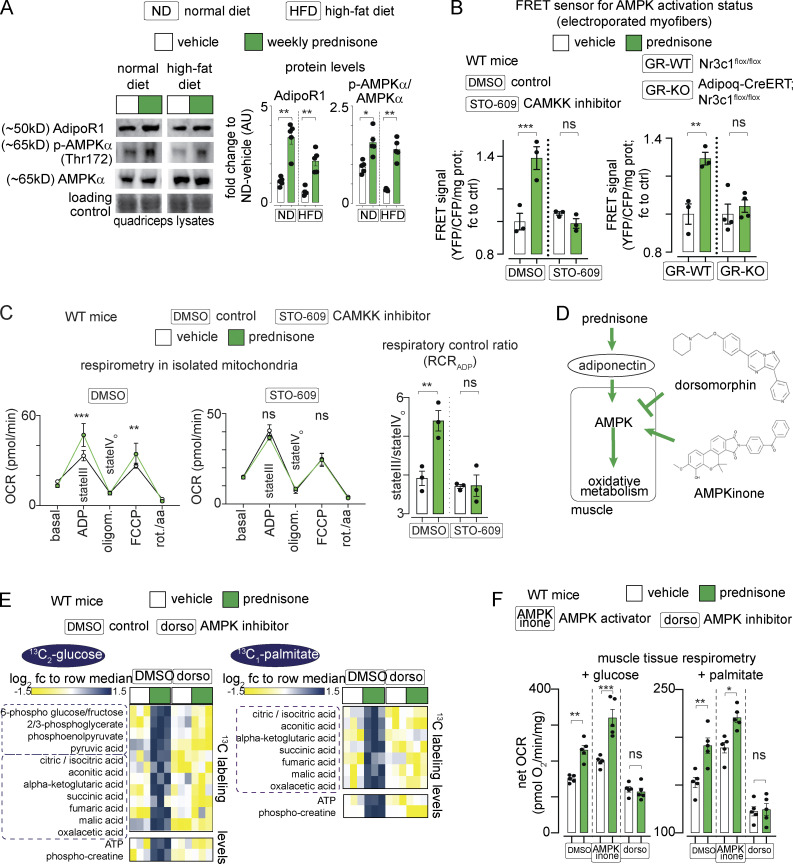
In the muscle, adiponectin activates its receptor AdipoR1, which activates CAMKK2, which in turn phosphorylates and activates AMPK (Iwabu et al., 2010). Compared with the vehicle in ND and HFD, intermittent prednisone increased the protein levels of AdipoR1 and phosphorylated AMPKα (Fig. 5 A). We tested the effects on AMPK activity through the FRET sensor AMPKAR-EV (Konagaya et al., 2017). FRET-based ratiometric fluorometry in electroporated flexor digitorum brevis (FDB) muscles showed that at 24 h postpulse, prednisone increased the AMPK activation signal in muscle with an ∼40% signal gain, which is consistent with previously reported values for AMPK activation in muscle with this construct (Konagaya et al., 2017). Moreover, co-injection with STO-609, selective inhibitor of CAMKK2 (Hawley et al., 2005), blunted the prednisone-induced gain of AMPK activity (Fig. 5 B, left). Analogously, the prednisone pulse increased AMPK activity in the muscle of GR-WT but not GR-KO mice, correlating with the loss of drug-driven adiponectin elevation in these mice (Fig. 5 B, right). Consistent with AMPK activation, at 24 h postpulse prednisone increased muscle mitochondrial respiration, measured as respiratory control ratio (RCR) in isolated mitochondria, but not after co-injection with STO-609 (Fig. 5 C). Thus, a prednisone pulse activates muscle AMPK through the adiponectin–CAMKK2 axis.
Figure 5.
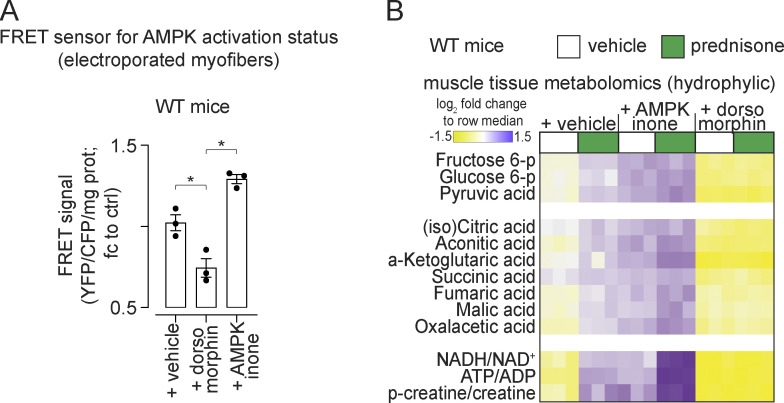
Pulsatile prednisone activates the AMPK response to adiponectin in muscle. (A) Representative blots showing that intermittent prednisone increased the protein levels of total ADIPOR1 and phosphorylated AMPK in both normal and obese muscle. (B) FRET sensor in electroporated myofibers showed increased AMPK activity in muscle after a prednisone pulse. In WT mice, this effect was blunted by co-injection with the CAMKK inhibitor STO-609 (left). In line with GR-driven adiponectin elevation, this effect in muscle was blunted by fat-restricted GR ablation (right). (C) A prednisone pulse increased mitochondrial RCR in isolated muscle mitochondria and this effect was blunted by the CAMKK inhibitor STO-609. (D) Treatment effects on muscle AMPK were challenged with small-molecule AMPK modulators dorsomorphin (non-selective AMPK inhibitor) and AMPKinone (specific AMPK activator). (E) Dorsomorphin blocked the gain in nutrient oxidation and aerobic energy production induced by a prednisone pulse, shown here with 13C tracing in isolated muscle. (F) Muscle tissue respirometry showed that AMPKinone had additive effects on glucose- and palmitate-fueled respiration, while dorsomorphin abrogated prednisone-induced gains. Histograms, single values, and mean ± SEM. All panels report data verified in at least two independent experiments. n = 5 mice/group (A and F), n = 3 mice/group (B–E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way Welch’s ANOVA with Tukey multiple comparison; C (OCR curves): two-way ANOVA with Tukey multiple comparison for time points. Source data are available for this figure: SourceData F5.
The involvement of AMPK was further challenged through co-injection of the non-specific inhibitor dorsomorphin (Yang et al., 2018) and the specific activator AMPKinone (Oh et al., 2010) in WT mice (Fig. 5 D). Dorsomorphin and AMPKinone effects on muscle AMPK were confirmed through FRET (Fig. S3 A). At 24 h after injection, 13C labeling assays showed that dorsomorphin blunted the effects of prednisone on nutrient oxidation in muscle (Fig. 5 E). Respirometry assays showed that AMPKinone and prednisone had additive effects on glucose- or palmitate-fueled basal respiration in the muscle tissue, while dorsomorphin blunted the prednisone effects (Fig. 5 F). Untargeted metabolomics of muscle tissue showed additive effects of AMPKinone and prednisone on levels of glucose-6-phosphate (glucose uptake), pyruvate (glycolysis), TCA cycle intermediates, ATP:ADP, and phosphorylated:total creatine, whereas dorsomorphin blocked these effects (Fig. S3 B). Thus, pulsatile prednisone stimulated the AMPK response to adiponectin in the muscle.
Figure S3.
Additional supporting data related to Fig. 5. (A) Validation of dorsomorphin and AMPKinone effects on AMPK activity in the muscle through the FRET sensor at 24-h after exposure. (B) Untargeted metabolomics showed that prednisone and AMPKinone (AMPK activation) had additive effects on hexose phosphorylation, pyruvate, TCA cycle intermediates and bioenergetics (NADH/NAD+; ATP/ADP; creatine phosphorylation). Dorsomorphin (AMPK antagonist) blunted the prednisone-driven gains. Data verified in at least two independent experiments. n = 3 mice/group. *, P < 0.05; **, P < 0.01; ***, P < 0.001; one-way Welch’s ANOVA with Tukey multiple comparison.
Chronic daily intake of glucocorticoids promotes obesity and exercise intolerance (Thomson et al., 2007). Here, we report data indicating that intermittent once-weekly intake of 1 mg/kg prednisone induces opposite metabolic effects in a murine model of dietary obesity, promoting exercise tolerance and muscle metabolic function. The intermittent regimen stimulated those healthful effects through adiponectin production by adipose tissue, increasing the muscle metabolic response to the adipokine.
The effects on global metabolic homeostasis and exercise tolerance were independent of the genetic background of test mice, as they were recapitulated in WT mice from DBA/2J and C57BL/6J strains, as well as from WT littermates of Adipoq-KO and adipose GR-KO mice (mixed B6-129 backgrounds). In our experiments, several parameters were, as expected, strongly impacted by diet-induced obesity compared to ND per se, including muscle bioenergetics and total-HMW adiponectin. However, our study was mainly focused on treatment effects, i.e., intermittent prednisone versus vehicle. Indeed, we found that the treatment effects on adiponectin elevation and muscle metabolism were replicated in both normal and obese mice and therefore independent of the global changes induced by HFD. We focused on diet-induced obesity, and similar effects may be expected in genetic obesity models like ob/ob and db/db models, although dedicated studies are needed.
Treatment-dependent increase in adiponectin production was dependent on adipocyte GR activation, and we identified a putative regulatory GRE juxtaposed to the adiponectin gene TSS. The transgenic model we used was a conditional GR-KO model in adult adipose tissue (Adipoq-CreERT2) and is therefore different from the constitutive GR-KO (Adipoq-Cre) previously reported (Shen et al., 2017). In that model, adipose tissue and overall homeostasis were likely adapted to GR ablation from fetal or postnatal development. Also, adiponectin production or sensitivity was not assessed. Nevertheless, that study showed that adiposity was induced by HFD independent of GR activity in adipocytes (Shen et al., 2017). This is convergent with our findings in obese mice, where we found opposing effects of intermittent versus daily prednisone on adiposity, muscle metabolism and adiponectin. Adiponectin elevation was specific to pulsatile or intermittent prednisone, whereas daily prednisone decreased adiponectin. These effects of daily prednisone correlated with the upregulation of known repressors of adiponectin expression, Atf3 and Nfatc4 (Kim et al., 2006), in adipose tissue. These repressors were not activated by intermittent prednisone, thereby suggesting a daily-prednisone-specific mechanism reversing the adiponectin elevation induced by pulsatile or intermittent prednisone.
In the muscle, intermittent prednisone induced the upregulation of the AMPK response to adiponectin. Our strategy of AMPK inhibition through dorsomorphin has the obvious limitation of additional non-AMPK-specific effects of this molecule, including BMP pathway regulation (Dasgupta and Seibel, 2018). Nonetheless, the dorsomorphin-related findings were consistent with the trends obtained with the highly specific CAMKK2 inhibitor STO-609 and opposite to the additive effects obtained with AMPKinone, specific activator of AMPK phosphorylation, in vitro and in vivo (Oh et al., 2010). Moreover, these trends were confirmed with the AMPK FRET sensor in electroporated myofibers. Genetic interventions on regulators of hormonal muscle responses will further specify the mechanisms elicited by exogenous glucocorticoids in the muscle.
The 13C labeling assays in ex vivo contracting muscles showed that the overall rates of both glucose and fatty acid oxidation were improved by intermittent prednisone in both ND and HFD. Due to the conditions intrinsic to this assay, e.g., isolated muscle and single nutrient excess, the observed effects on nutrient oxidation from diet and treatment might be more acute than in vivo, where the Randle cycle partially compensates for nutrient imbalance during the early obesity stage (Felber, 1990). However, our findings with respirometry, glucose uptake, and intramuscular lipid levels suggest the intriguing possibility that intermittent prednisone promotes metabolic flexibility in the muscle, which is affected in obesity (Goodpaster and Sparks, 2017).
In conclusion, our study reported that intermittent prednisone promoted a virtuous fat-muscle communication through adiponectin. These findings pave the way for adjuvant drug strategies to restore adiponectin sensitivity and exercise tolerance in conditions of metabolic stress.
Materials and methods
Animal handling and treatments
Mice were housed in a pathogen-free facility in accordance with the American Veterinary Medical Association and under protocols fully approved by the Institutional Animal Care and Use Committee at Northwestern University Feinberg School of Medicine (#ISO00011692) and Cincinnati Children’s Hospital Medical Center (#2020-0008). Consistent with the ethical approvals, all efforts were made to minimize suffering. Euthanasia was performed through carbon dioxide inhalation followed by cervical dislocation and heart removal. Mice were maintained on a 14 h/10 h light/dark cycle and diet/pharmacological treatments were initiated at ∼8 wk of age. Male mice were used for all strains. Mice were obtained and interbred from Jackson Laboratories. Experiments with WT mice were performed on mice from the DBA/2J background (JAX #000671) and the C57BL/6J background (JAX #000664). Experiments with Adipoq-KO and -WT mice were performed on littermate mice obtained from interbred heterozygous mutants after three mating rounds on a mixed B6-129 background (JAX #008195). Experiments with conditional GR ablation in fat tissue were performed on littermate mice from Adipoq-CreERT+/-;Nr3c1flox/flox × Nr3c1flox/flox matings, which were derived from transgenic founders (JAX #021021, #024671) after three mating rounds on a mixed B6-129 background. Gene ablation was induced with tamoxifen right before the start of drug treatments using a combination of i.p. (20 mg/kg/d for 5 d; #T5648; Sigma-Aldrich) and chow-mediated (40 mg/kg from 48 h prior to start; #130860; Envigo) administration (Khalil et al., 2017). ND consisted of ad libitum Mouse Breeder Sterilizable Diet (#7904; Harlan Teklad), while HFD consisted of ad libitum Research Diet #D12492 (60% kcal in fat content). Weekly prednisone treatment consisted of once-weekly i.p. injection of 1 mg/kg prednisone (#P6254; Sigma-Aldrich; Quattrocelli et al., 2019). Daily prednisone treatment consisted of once-daily i.p. injection of 1 mg/kg prednisone. The injectable solution was diluted from a 5 mg/ml stock in DMSO (#D2650; Sigma-Aldrich) to 50 μl volume. Vehicle injections consisted of the same ratio of DMSO:saline used for the injections prepared from the prednisone stock and depending on mouse weight. Importantly, the frequency of vehicle injection for the vehicle cohort matched the highest frequency of other treatments: in comparison with only intermittent prednisone, vehicle was also injected once-weekly; in comparison with both intermittent and daily prednisone regimens, vehicle was injected once-daily. For in vivo acute treatments, dorsomorphin (#21207; Cayman Chemical) was injected i.p. at 0.2 mg/kg (Yang et al., 2018); AMPKinone (#10631; Cayman Chemical) was injected i.p. at 10 mg/kg (Oh et al., 2010); and STO-609 was injected i.p. at a dose of 600 nmol/kg (York et al., 2017). Rosiglitazone (Cat# HY-17386; Fisher Scientific) treatment consisted of once-daily i.p. injections with a dose of 10 mg/kg for 4 wk (Wong et al., 2011). Injections were administered at the beginning of the light-phase (∼ZT0; ∼6 am) and tissues were harvested 24 h after single-pulse or last chronic treatment. All in vivo, ex vivo, and postmortem analyses were conducted blinded to the treatment group.
Lean/fat mass, histology, and functional analyses
Magnetic resonance imaging (MRI) scans to determine fat and lean mass ratios (percentage of total body weight) were conducted in non-anesthetized, non-fasted mice at ∼ZT8 (∼2 pm) using the EchoMRI-100H Whole Body Composition analyzer (EchoMRI). Mice were weighed immediately before MRI scan. Myofiber and adipocyte (inguinal white adipose tissue) cross-sectional areas were obtained from histology analyses at the endpoint. Excised tissues were fixed in 10% formaldehyde (cat. #245-684; Fisher Scientific) at room temperature for ∼24 h and then stored at +4°C before processing. Seven micrometer sections from the center of paraffin-embedded muscles or adipose tissues were stained with H&E (cat. #12013B, 1070C; Newcomer Supply). CSA quantitation was conducted on 400 myofibers/adipocytes per tissue per mouse. Imaging was performed using an Axio Observer A1 microscope (Zeiss), using 10× and 20× (short-range) objectives. Images were acquired through Gryphax software (version 1.0.6.598; Jenoptik) and quantitated through ImageJ (Schneider et al., 2012).
For myofiber typing, sections were incubated with primary antibodies BA-F8 (1:10), SC-71 (1:30), and BF-F3 (1:10; all by Developmental Studies Hybridoma Bank) overnight at 4°C. Then, sections were incubated with secondary antibodies AlexaFluor350 anti-IgG2b, AlexaFluor488 anti-IgG1, and AlexaFluor594 anti-IgM (Cat #A21140, A21121, 1010111; Life Technologies). Type 1 fibers stained blue, type 2A stained green, type 2× showed no staining, and type 2B stained red. Myofiber types were then quantitated over at least five serial sections and quantitated as percentage of total counted myofibers.
Forelimb grip strength was monitored using a meter (#1027SM; Columbus Instruments) blinded to treatment groups. Animals performed 10 pulls, with a 5-s rest on a flat surface between pulls. Grip strength was expressed as force normalized to body weight. Running endurance was tested on a motorized treadmill with plastic non-electrified resting posts (#1050RM; Columbus Instruments). Speed was accelerated at 1 m/min2 starting at 1 m/min, and the individual test was interrupted when the subject spent >30 s resting. Immediately before sacrifice, in situ tetanic force from tibialis anterior muscle was measured using a Whole Mouse Test System (Cat #1300A; Aurora Scientific) with a 1N dual-action lever arm force transducer (300C-LR; Aurora Scientific) in anesthetized animals (0.8 liter/min of 1.5% isoflurane in 100% O2). Specifications of tetanic isometric contraction were as follows: initial delay, 0.1 s; frequency, 200 Hz; pulse width, 0.5 ms; duration, 0.5 s; and stimulation, 100 mA (Quattrocelli et al., 2019). Muscle length was adjusted to a fixed baseline of ∼50 mN resting tension for all muscles/conditions. Running endurance was analyzed as weight normalized work (μJ), i.e., body weight (g) × distance2 (m2)/time2 (s2). All analyses were conducted blinded to treatment.
Tolerance tests
Glucose was measured in the blood (first drop from tail venipuncture) in individual cages after 8 h fasting at ∼ZT8 (∼2 pm) with an AimStrip Plus glucometer system (Germaine Laboratories) and expressed as mg/dl values. Mice were injected with either 1 g/kg glucose (#D8375-1g; Millipore Sigma) or 0.5 U/kg insulin (#12585014; Thermo Fisher Scientific) in 200 μl intraperitoneal injections, and glucose was then monitored by tail venipuncture at 0 min, 10 min, 20 min, 30 min, 60 min, and 120 min after injection. All tests were conducted blinded to treatment groups.
FRET assays
The plasmid encoding the AMPK FRET sensor (Addgene #105241; Konagaya et al., 2017) was electroporated in FDB myofibers, and FRET analyses were conducted on live myofibers at 7 d after electroporation and 24 h after i.p. drug injections. Live intact myofibers from both FDBs per mouse were pooled in a single well and assayed in 100 μl Ringer’s solution at +37°C at plate reader (Synergy HTX multi-mode 96-well plate reader; BioTek). FRET was performed using 433 nm as the excitation wavelength, and the emission wavelengths were 475 for CFP and 530 for YFP. Data were analyzed as FRET-YFP fluorescence normalized to CFP emission. All FRET analyses were conducted blinded to treatment groups.
Metabolic cages, 2-NBDG uptake, and tissue respirometry
VO2 (ml/h), VCO2 (ml/h), RER (VCO2/VO2), activity (XY + XZ counts), food intake (sum of food g consumed per time period), and energy expenditure (kcal/h) were assessed via indirect calorimetry using the TSE Automated Phenotyping System PhenoMaster (TSE system) at the NU Comprehensive Metabolic Core. Data collection started 24 h after prednisone or vehicle injection and lasted for 5 d. Results are expressed as raw values over 3 d and average values per 12-h phases (active phase, dark phase; rest phase, light phase). ANCOVA analysis of energy expenditure was performed using CalR (https://calrapp.org, accessed January 2021) using lean and total masses as co-variates (Mina et al., 2018). Calorie excess calculations consisted of total calorie intake, as calculated from calorie density of regular or high-fat chow, minus the calorie spent, as calculated from energy expenditure values.
2-NBDG uptake assay in live myofibers was conducted on freshly isolated FDB myofibers using 50 μM 2-NBDG (#11046; Cayman Chemical) with/without 85 μM insulin (#12585014; Thermo Fisher Scientific), using the previously reported procedures (Quattrocelli et al., 2019). Data were analyzed as net fluorescence values, i.e., relative fluorescent units calculated as intra-myofiber fluorescence after incubation and subtracted for average fluorescence in negative controls (10 μM cytochalasin B, GLUT channels inhibitor; #C6762; Millipore Sigma).
Basal tissue OCR values were obtained from basal rates of oxygen consumption of muscle biopsies at the Seahorse XF96 Extracellular Flux Analyzer platform (Agilent Technologies) using previously detailed conditions (Quattrocelli et al., 2019). Basal OCR was calculated as baseline value (average of three consecutive reads) minus value after rotenone/antimycin addition (average of three consecutive reads). Basal OCR values were normalized to total protein content and assayed in each well after the Seahorse run through homogenization and Bradford assay. Nutrients: 10 mM glucose and 1 mM palmitate-BSA (#G7021, #P0500; Millipore-Sigma); inhibitors: 0.5 μM rotenone + 0.5 μM antimycin A (Agilent Technologies).
RCR values were obtained from isolated mitochondria from the muscle tissue. Quadriceps were harvested from the mouse and cut up into very fine pieces. The minced tissue was placed in a 15-ml conical tube (#188261; USA Scientific) and 5 ml of MS-EGTA buffer with 1 mg Trypsin (#T1426-50MG; Sigma-Aldrich) was added to the tube. The tube was quickly vortexed and the tissue was left submerged in the solution. After 2 min, 5 ml of MS-EGTA buffer with 0.2% BSA (#A-421-250; Goldbio) was added to the tube to stop the trypsin reaction. MS-EGTA buffer: Mannitol: #M0214-45; ChemProducts, Sucrose: #100892; Millipore, Hepes: #15630-080; Gibco, EGTA: #E14100-50.0; RPI. The tube was inverted several times to mix and then set to rest. Once the tissue had mostly settled to the bottom of the tube, 3 ml of buffer was aspirated and the remaining solution and tissue were transferred to a 10-ml glass tissue homogenizer (#89026-382; Avantor). Once sufficiently homogenized, the solution was transferred back into the 15-ml conical tube and spun in the centrifuge at 1,000 g for 5 min at 4°C. After spinning, the supernatant was transferred to a new 15-ml conical tube. The supernatant in the new tube was then centrifuged at 12,000 g for 10 min at 4°C to pellet the mitochondria. The supernatant was discarded from the pellet and the pellet was then resuspended in 7 ml of MS-EGTA buffer and centrifuged again at 12,000 g for 10 min at 4°C. After spinning, the supernatant was discarded and the mitochondria were resuspended in 1 ml of Seahorse medium (#103335-100; Agilent Technologies) supplemented with 10 μl of 5 mM pyruvate (#P2256-100G; Sigma-Aldrich) and 10 μl of 5 mM malate (#20765; Cayman Chemical). After protein quantitation using a Bradford assay (#5000001; Bio-Rad Laboratories), 2.5 μg mitochondria were dispensed at 180 μl total volume per well in and let to equilibrate for 1 h at 37°C. Then, 20 μl of 5 mM ADP (#01905; Sigma-Aldrich), 50 μM Oligomycin (#495455-10 MG; Milipore), 100 μM Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (#C3463; TCI), and 5 μM Rotenone (#557368-1GM; Milipore)/Antimycin A (#A674-50MG; Sigma-Aldrich) were added to drug ports A, B, C, and D respectively to yield final concentrations of 0.5 mM, 50, 10, and 0.5 μM. At baseline and after each drug injection, samples were read three consecutive times. RCR was calculated as the ratio between state III (OCR after ADP addition) and uncoupled state IV (OCR after oligomycin addition). All metabolic cage, 2-NBDG, and Seahorse measurements were conducted blinded to treatment groups.
Unlabeled and labeled metabolite quantitation in muscle
Unlabeled muscle metabolomics was performed on hydrophylic metabolite fraction from whole quadriceps muscles and subsequent LC-MS analysis at the NU Metabolomics Core, following previously reported methods (Quattrocelli et al., 2019). Labeled metabolite analysis was performed on freshly explanted whole quadriceps muscle undergoing isometric contractions in the presence of labeled nutrient, insulin, and oxygen, as previously detailed (Quattrocelli et al., 2019). Briefly, 13C tracing from nutrients in the muscle was performed by adapting reported conditions (Kerner et al., 2014) to our muscle stimulus settings used to probe the muscle force (see below). Immediately after sacrifice, quadriceps muscles were dissected and immobilized on a Sylgard-coated well of a 12-multiwell plate using two 27-g needles at the muscle extremities. The well was pre-filled with 1× Ringers’ solution (146 mM NaCl, 5 mM KCl, 2 mM CaCl2, 1 mM MgCl2, and 10 mM Hepes, pH 7.4) containing 25 mU/ml insulin (only for glucose preps; Cat #RP-10908; Thermo Fisher Scientific) and the appropriate 13C nutrient, and kept at 37°C on a heated pad. The nutrient solution was constantly bubbled with 95% O2/5% CO2 line (∼2 psi). After 5 min equilibration in solution, electrodes were inserted at the muscle extremities, securing them to the holder needles. Using a Whole Mouse Test System (Cat #1300A; Aurora Scientific), 20 contractions (1×/min) were induced with the following specifications: initial delay, 0.1 s; frequency, 200 Hz; pulse width, 0.5 ms; duration, 0.5 s; 100 mA stimulation. Muscles were then removed from the 13C nutrient solution, quickly rinsed in nutrient-free Ringers’ solution, dried, and immediately flash-frozen. Muscle metabolites were then extracted and analyzed as per metabolomics procedures (LC-MS, see above), and mass resolution was carried out on pre-determined metabolites, while control energetics (ATP, phosphocreatine) were analyzed from simultaneous quantitation from the LC-MS system. Labeled nutrients: 10 mM 1,2-13C2-glucose, 1 mM 1-13C-palmitate (BSA conjugated; #453188, #292125; Sigma-Aldrich). Energy output, e.g., ATP, phospho-creatine, was monitored as unlabeled metabolite quantities in the same samples analyzed for labeled metabolites. Metabolite labeling ratio was calculated on peak area/mg tissue values subtracting the background 13C labeling ratio obtained from muscles exposed to unlabeled nutrients (same reagents used for respirometry) and expressed as percentage of total metabolite. Cumulative labeling percentage per metabolite was calculated by dividing the sum of all labeled derivatives by the sum of all unlabeled and labeled derivatives. Unlabeled metabolite levels were analyzed as peak area normalized to wet tissue weight (weight before cryo-pulverization). All analyses were conducted blinded to treatment.
Muscle lipidomics
Quadriceps muscle tissue was collected from mice, flash frozen in liquid nitrogen, cryopulverized at a Retsch CryoMill, and ∼50 mg was weighed out and placed in a 2-ml tube (1620-2700; USA Scientific) for lipidomics. To each tube, 1 ml of ice-cold chloroform/methanol (2:1; Cat #43685; Alfa Aesar; Cat#A454-4; Fisher Scientific) was added along with 10 µL of 0.1 mM BHT (B1095; Spectrum). Next, 50 μl of the internal lipid standard was added to each tube except for the blank calibrator. The samples were then homogenized by placing the tubes in the Qiagen TissuLyzer II at room temperature for two 2-min intervals with a 30-s rest in between. Each sample was then sonicated for 5 s at room temperature. Samples were centrifuged for 10 min at 4,000 g and the supernatant was transferred to a new 2-ml tube. Each tissue pellet was re-extracted by adding 500 μl of ice-cold chloroform/methanol (2:1) and sonicating again at room temperature for 5 s. The samples were spun again at 4,000 g for 10 min and the supernatant was collected and combined with the first extract. The combined extracts were then transferred to the Mass-Spec Core (CCHMC) for drying and analysis. The untargeted lipidomics analysis was conducted on a Q Exactive plus hybrid quadrupole-Orbitrap mass spectrometer interfaced with Vanquish ultra-high performance liquid chromatography (UHPLC) system (Thermo Fisher Scientific). A gradient mobile phase was used with a binary solvent system, which changed from 60% solvent A to 57% solvent A over 2 min, then to 50% solvent A at 2.1 min, then to 46% solvent A over 9.9 min, and then, after a change to 30% at 12.1 min, to 1% solvent A over 5.9 min, then to 60% solvent A at 18.1 min, and this was held for 2 min. The total run time was 20 min, and the flow rate was 0.4 ml/min. Solvent A consisted of acetonitrile/water(60/40) with 10 mM ammonium formate and 0.1% formic acid; solvent B consisted of isopropanol/acetonitrile (90/10) with 10 mM ammonium formate and 0.1% formic acid. The injection volume was 5 μl for both the negative and positive ion modes. An Acquity CSH C18 UPLC column (2.1 × 100 mm, 1.7 µm; Waters) was used for separation. The column temperature was set at 55°C. The ESI source was operated in the following parameters: spray voltage is 2.5 KV, capillary temperature, 350°C; sheath gas flow rate, 35; and auxiliary gas heater temperature, 325°C. Data were acquired using full MS scan (mass scan range 150–1,500 m/z, AGC target 3e6, maximum IT 100 ms, resolution 140,000) and collision-induced dissociation-based data dependent on MS/MS (resolution 17,500, AGC target 1e5, maximum IT 50 ms, loop count 15, top n = 15, isolation window 1.0 m/z, stepped NCE 20, 40, 60). Lipid annotation was conducted by searching against in-house lipidomics database using retention time, accurate mass, and fragmentation ion pattern in combination with major public spectral libraries including, LIPID MAPS structure database (LMSD; Sud et al., 2007) and HMDB (Wishart et al., 2018). Lipid profiling through mass-spec was performed blinded to treatment groups.
Adipokine array and adiponectin ELISA
Serum adipokine profiling was performed on 50 μl serum using reagents and procedures from the Proteome Profiler Mouse Adipokine Array kit (#ARY013; R&D). Data were analyzed as ratio to internal standard and treated-versus-control ratio. Mouse total adiponectin was quantitated through reagents and procedures from the Quantikine ELISA kits (#MRP300; R&D), while high-molecular-weight adiponectin was quantitated through the HMW Adiponectin ELISA kit (#638-13079; BioVendor) using internal standards and recommended interpolation to calculate μg/ml (serum) or pg/mg (tissues) values. All assays were conducted blinded to treatment groups.
Western blot (WB), qPCR, ChIP-qPCR, and luciferase assays
Protein analysis was performed on ∼50 μg total lysates from whole quadriceps muscles homogenized in PBS supplemented with 1 mM CaCl2, 1 mM MgCl2 (#C1016, #M8266; Sigma-Aldrich), and protease and phosphatase inhibitors (#04693232001, #04906837001; Roche). Blocking and stripping solutions: StartingBlock and RestorePLUS buffers (#37543, #46430; Thermo Fisher Scientific). Primary antibodies (all diluted 1:1,000 for O/N incubation at +4°C): rabbit anti-Adipor1 (#A1509; ABclonal); rabbit anti-AMPK (#A1229; ABclonal); rabbit anti-phospho-AMPK (#AP0116; ABclonal); rabbit anti-GR (#A2164; ABclonal); mouse anti-GAPDH (#AC002; ABclonal); and rabbit anti-adiponectin (#A2543; ABclonal). Secondary antibody (diluted 1:5,000 for 1-h incubation at room temperature): donkey anti-rabbit or anti-mouse (#sc-2313, #sc-2318; Santa Cruz Biotechnology). Counterstain for loading control was performed with ponceau (#P7170; Sigma-Aldrich). Blots were developed with SuperSignal Pico (cat. #34579; Thermo Fisher Scientific) using the iBrightCL1000 developer system (cat. #A32749; Thermo Fisher Scientific) with automatic exposure settings. WB gels and membranes were run/transferred in parallel and/or stripped for multiple antibody-based staining for densitometry analyses. Protein density was analyzed using the Gel Analysis tool in ImageJ software (Schneider et al., 2012) and expressed as fold changes to control samples.
For RT-qPCR assays, total RNA was extracted from cryo-pulverized quadriceps muscles with Trizol (#15596026; Thermo Fisher Scientific) and 1 μg RNA was reverse-transcribed using 1X qScript Supermix (#95048; QuantaBio). RT-qPCRs were conducted in threeplicates using 1X Sybr Green Fast qPCR mix (#RK21200; ABclonal) and 100 nM primers at a CFX96 qPCR machine (Bio-Rad Laboratories; thermal profile: 95°C, 15 s; 60°C, 30 s; 40×; melting curve). Primers were selected among validated primer sets from the MGH PrimerBank; IDs: 6753884a1, 33342282a1, 6996917a1, 6678359a1, 6857793a1, 25058536a1, 29788764a1, 6671519a1, and 31982423a1.
ChIP-qPCR was conducted on chromatin immunoprecipitated from freshly isolated inguinal white adipose tissue using the antibody rabbit anti-GR (#A2164; ABclonal) and previously described procedures (Quattrocelli et al., 2019). Signal was quantitated as percentage of input, and IgG-immunoprecipitated (#ab27472; Abcam) chromatin was assayed as negative control. Primers for Adipoq GRE: Fw, 5′-ACAGGAGAAGCAGGCAGAGA-3′; Rev, 5′-GCAGACAGATGTCCCCAACT-3′.
Luciferase plasmids containing the Adipoq GRE fragments were obtained by cloning genomic sequences in the pGL4.23 backbone (#E8411; Promega) using the NheI-XhoI sites upstream of the minimal promoter site. Fragments were cloned conserving the genomic orientation with regards to transcriptional orientation, adding KpnI and XhoI tails to the appropriate extremities via Phusion PCR (#M0530; New England Biolabs). Wild type and mutated fragments (Δ sites) were amplified from ad-hoc synthesized DNA oligonucleotides, using genomic sequences from the C57BL/6J genomic background (wt sequence, 5′-AAAAAAGCTAGCACAGGAGAAGCAGGCAGAGAGGGCAGCCCGAGGAAGTCCAGTTGAGGTCAGACTGTAGGATCTGTCTCTATCACTTAGTTCTACTGAGCAGATATGCACGGAGCATGCGCTGTAGAACACTTTGTCAGGGCTGTGAGTGAAAACATGGGTGTGCTCGGAGGAGTTGGGGACATCTGTCTGCCTCGAGAAAAAA-3′). HEK293 cells (#CRL-1573; ATCC) were transfected with Lipofectamine 3000 (#L3000015; Thermo Fisher Scientific) and luciferase activity was assayed 48 h after transfection using protocol and reagents from the Dual Luciferase Assay Kit (Cat #1910; Promega) instructions. Luminescence was recorded at the Synergy HTX multi-mode 96-well plate reader (BioTek). Raw values were normalized to Renilla luciferase and vehicle controls.
RNA-seq
RNA-seq datasets of muscle and adipose tissues from obese mice after vehicle, weekly, or daily prednisone treatments are available on the GEO database with accession number GSE189774.
RNA-seq was conducted on RNA extracted from quadriceps muscle and inguinal white adipose tissue. Total RNA was extracted as detailed above and re-purified using the RNeasy Mini Kit (cat. #74104; Qiagen). RNA-seq was performed at the DNA Core (CCHMC) blinded to treatment groups. Then, 150–300 ng of total RNA determined by Qubit (cat. #Q33238; Invitrogen) high-sensitivity spectrofluorometric measurement was poly-A selected and reverse transcribed using Illumina’s TruSeq stranded mRNA library preparation kit (cat. #20020595; Illumina). Each sample was fitted with one of the 96 adapters containing a different eight-base molecular barcode for high-level-multiplexing. After 15 cycles of PCR amplification, the completed libraries were sequenced on an Illumina NovaSeq 6000, generating 20 million or more high-quality 100 base long paired-end reads per sample. A quality control check on the fastq files was performed using FastQC. Upon passing basic quality metrics, the reads were trimmed to remove adapters and low-quality reads using default parameters in Trimmomatic (Bolger et al., 2014; Version 0.33). The trimmed reads were then mapped to a 10 mm reference genome using default parameters with strandness (R for single-end and RF for paired-end) option in Hisat2 (Kim et al., 2015; Version 2.0.5). In the next step, transcript/gene abundance was determined using kallisto (Bray et al., 2016; Version 0.43.1). We first created a transcriptome index in kallisto using Ensembl cDNA sequences for the reference genome. This index was then used to quantify transcript abundance in raw counts and counts per million. DE genes, FDR < 0.05 was quantitated through DESeq2 (Anders et al., 2013). PCA was conducted using ClustVis (Metsalu and Vilo, 2015). Heatmaps were imaged with TreeView3 (Saldanha, 2004). Gene ontology pathway enrichment was conducted using the Gene Onthology analysis tool (Ashburner et al., 2000).
Macrophage isolation
Resident, nonmanipulated macrophages were harvested from the peritoneal cavity immediately after sacrifice. After soaking the abdomen with 70% ethanol, a small incision was made along the midline with sterile scissors. The abdominal skin was manually retracted to expose the intact peritoneal wall. Then, 10 ml cold PBS was injected through the peritoneal wall along the mouse’s left side using 21-G needle. Abundant precaution was taken to not puncture the intestine and other internal organs. To maximize the number of cells harvested, the peritoneal cavity was gently massaged or shaken from the sides after injecting PBS. Using the same syringe and needle, the fluid from the peritoneum was aspirated. The collected peritoneal fluid was dispensed into a 15-ml conical tube. Then, the tube was centrifuged for 10 min at 400 g at 4°C. The supernatant was discarded and the cell pellet was resuspended in cold DMEM-F12-10 media. Cells were kept on ice throughout the process. Generally, around ∼3 × 106 total peritoneal cells were collected per mouse. For flow cytometry characterization, after collection, the peritoneal fluid samples were centrifuged at 400 g for 10 min at 4°C and resuspended in 1× HBSS supplemented with 2% BGS and 2 mM EDTA (cat. #88284; Thermo Fisher Scientific). Cells were incubated with fluorophore-conjugated primary antibodies for 20 min at 4°C with gentle rotation, washed twice with 1× HBSS and analyzed using a BD LSRFortessa running BD FACSDiVa V.8.0 software (BD Biosciences) and using the following laser configuration: violet (405 nm) and red (635 nm) to detect fluorophore-conjugated antibodies (APC anti-mouse F4/80 and BV421 anti-mouse CD11b; Cat# 123116, 101251; BioLegend). Analysis and quantitation were performed using FACSDiVa V.8.0 software (BD Biosciences). Macrophage isolations and analyses were conducted blinded to treatment.
Statistics
Statistical analyses were performed using Prism software v8.4.1 (Graphpad). The Pearson-D’Agostino normality test was used to assess data distribution normality. When comparing two groups, two-tailed Student’s t-test with Welch’s correction (unequal variances) was used. When comparing three groups of data for one variable, one-way ANOVA with Tukey multi-comparison was used. When comparing data groups for more than one related variable, two-way ANOVA was used. For ANOVA and t-test analyses, a P < 0.05 was considered significant. When the number of data points was <10, data were presented as single values (dot plots, histograms). Tukey distribution bars were used to emphasize data range distribution in analyses pooling larger data point sets per group (typically >10 data points). Analyses pooling data points over time were presented as line plots connecting medians of box plots showing the distribution of all data per time point.
Online supplemental material
Fig. S1 shows representative histology images for Fig. 1C, additional analyses in WT mice from the C57BL/6 background and extended labeling data for the mass-spec heatmaps from Fig. 1, G and H. Fig. S2 shows sorting strategy and qPCR data from macrophages, and extended data regarding Adipoq GRE regulation with a prednisone pulse. Fig. S3 shows additional FRET and untargeted metabolomics data for the dorsomorphin-AMPKinone experiment.
Supplementary Material
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
Acknowledgments
Mass-spec analyses were performed by the Metabolomics Core Facility at Robert H. Lurie Comprehensive Cancer Center of Northwestern University.
Funding was provided by National Institutes of Health grants DK121875 (M. Quattrocelli), HL158531 (M. Quattrocelli), Cincinnati Children’s Hospital Medical Center (CCHMC) Trustee Award (M. Quattrocelli), CCHMC Heart Institute Translational Grant (M. Quattrocelli), National Institutes of Health grant AG049665 (N.S. Chandel), National Institutes of Health grant AR052646 (E.M. McNally), and National Institutes of Health grant HL061322 (E.M. McNally).
Author contributions: Conceptualization: M. Quattrocelli, G.D. Barish, N.S. Chandel, and E.M. McNally. Methodology, Investigation, Formal Analysis: M. Quattrocelli, M. Wintzinger, K. Miz, M. Panta, and A.D. Prabakaran. Supervision and Funding Acquisition: M. Quattrocelli and E.M. McNally. Writing—original draft: M. Quattrocelli. Writing—review & editing: M. Quattrocelli, M. Wintzinger, K. Miz, M. Panta, A.D. Prabakaran, G.D. Barish, N.S. Chandel, and E.M. McNally.
References
- Abdul-Ghani, M.A., and DeFronzo R.A.. 2010. Pathogenesis of insulin resistance in skeletal muscle. J. Biomed. Biotechnol. 2010:476279. 10.1155/2010/476279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achari, A.E., and Jain S.K.. 2017. Adiponectin, a therapeutic target for obesity, diabetes, and endothelial dysfunction. Int. J. Mol. Sci. 18:1321. 10.3390/ijms18061321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S., McCarthy D.J., Chen Y., Okoniewski M., Smyth G.K., Huber W., and Robinson M.D.. 2013. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 8:1765–1786. 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- Ashburner, M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. 2000. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25:25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M., Lohse M., and Usadel B.. 2014. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 30:2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, N.L., Pimentel H., Melsted P., and Pachter L.. 2016. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34:525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Dasgupta, B., and Seibel W.. 2018. Compound C/Dorsomorphin: Its use and misuse as an AMPK inhibitor. Methods Mol. Biol. 1732:195–202. 10.1007/978-14939-7598-3_12 [DOI] [PubMed] [Google Scholar]
- Ebert, S.M., Dyle M.C., Kunkel S.D., Bullard S.A., Bongers K.S., Fox D.K., Dierdorff J.M., Foster E.D., and Adams C.M.. 2012. Stress-induced skeletal muscle Gadd45a expression reprograms myonuclei and causes muscle atrophy. J. Biol. Chem. 287:27290–27301. 10.1074/jbc.M112.374777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felber, J.P.. 1990. Significance of the Randle-Mechanism in the etiology of diabetes type II. Horm. Metab. Res. Suppl. 22:11–17 [PubMed] [Google Scholar]
- Goodpaster, B.H., and Sparks L.M.. 2017. Metabolic flexibility in health and disease. Cell Metabol. 25:1027–1036. 10.1016/j.cmet.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodpaster, B.H., and Wolf D.. 2004. Skeletal muscle lipid accumulation in obesity, insulin resistance, and type 2 diabetes. Pediatr. Diabetes. 5:219–226. 10.1111/j.1399-543x.2004.00071.x [DOI] [PubMed] [Google Scholar]
- Guo, B., Chatterjee S., Li L., Kim J.M., Lee J., Yechoor V.K., Minze L.J., Hsueh W., and Ma K.. 2012. The clock gene, brain and muscle Arnt-like 1, regulates adipogenesis via Wnt signaling pathway. FASEB J. 26:3453–3463. 10.1096/fj.12-205781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, M.J., Lim H.W., Ho Y., Shapira S.N., Ishibashi J., Rajakumari S., Steger D.J., Lazar M.A., Won K.J., and Seale P.. 2015. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 29:298–307. 10.1101/gad.252734.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley, S.A., Pan D.A., Mustard K.J., Ross L., Bain J., Edelman A.M., Frenguelli B.G., and Hardie D.G.. 2005. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metabol. 2:9–19. 10.1016/j.cmet.2005.05.009 [DOI] [PubMed] [Google Scholar]
- Hayashi, T., Hirshman M.F., Kurth E.J., Winder W.W., and Goodyear L.J.. 1998. Evidence for 5′ AMP-activated protein kinase mediation of the effect of muscle contraction on glucose transport. Diabetes. 47:1369–1373. 10.2337/diab.47.8.1369 [DOI] [PubMed] [Google Scholar]
- Horakova, D., Azeem K., Benesova R., Pastucha D., Horak V., Dumbrovska L., Martinek A., Novotny D., Svagera Z., Hobzova M., et al. 2015. Total and high molecular weight adiponectin levels and prediction of cardiovascular risk in diabetic patients. Int. J. Endocrinol. 2015:545068. 10.1155/2015/545068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwabu, M., Yamauchi T., Okada-Iwabu M., Sato K., Nakagawa T., Funata M., Yamaguchi M., Namiki S., Nakayama R., Tabata M., et al. 2010. Adiponectin and AdipoR1 regulate PGC-1alpha and mitochondria by Ca(2+) and AMPK/SIRT1. Nature. 464:1313–1319. 10.1038/nature08991 [DOI] [PubMed] [Google Scholar]
- Jeffery, E., Church C.D., Holtrup B., Colman L., and Rodeheffer M.S.. 2015. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol. 17:376–385. 10.1038/ncb3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner, J., Minkler P.E., Lesnefsky E.J., and Hoppel C.L.. 2014. Fatty acid chain elongation in palmitate-perfused working rat heart: Mitochondrial acetyl-CoA is the source of two-carbon units for chain elongation. J. Biol. Chem. 289:10223–10234. 10.1074/jbc.M113.524314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, H., Kanisicak O., Prasad V., Correll R.N., Fu X., Schips T., Vagnozzi R.J., Liu R., Huynh T., Lee S.J., et al. 2017. Fibroblast-specific TGF-beta-Smad2/3 signaling underlies cardiac fibrosis. J. Clin. Invest. 127:3770–3783. 10.1172/jci94753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D., Langmead B., and Salzberg S.L.. 2015. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 12:357–360. 10.1038/nmeth.3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, H.B., Kong M., Kim T.M., Suh Y.H., Kim W.H., Lim J.H., Song J.H., and Jung M.H.. 2006. NFATc4 and ATF3 negatively regulate adiponectin gene expression in 3T3-L1 adipocytes. Diabetes. 55:1342–1352. 10.2337/db05-1507 [DOI] [PubMed] [Google Scholar]
- Konagaya, Y., Terai K., Hirao Y., Takakura K., Imajo M., Kamioka Y., Sasaoka N., Kakizuka A., Sumiyama K., Asano T., and Matsuda M.. 2017. A highly sensitive FRET biosensor for AMPK exhibits heterogeneous AMPK responses among cells and organs. Cell Rep. 21:2628–2638. 10.1016/j.celrep.2017.10.113 [DOI] [PubMed] [Google Scholar]
- Krause, M.P., Milne K.J., and Hawke T.J.. 2019. Adiponectin-consideration for its role in skeletal muscle health. Int. J. Mol. Sci. 20:1528. 10.3390/ijms20071528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latres, E., Amini A.R., Amini A.A., Griffiths J., Martin F.J., Wei Y., Lin H.C., Yancopoulos G.D., and Glass D.J.. 2005. Insulin-like growth factor-1 (IGF-1) inversely regulates atrophy-induced genes via the phosphatidylinositol 3-kinase/Akt/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway. J. Biol. Chem. 280:2737–2744. 10.1074/jbc.m407517200 [DOI] [PubMed] [Google Scholar]
- Luo, N., Liu J., Chung B.H., Yang Q., Klein R.L., Garvey W.T., and Fu Y.. 2010. Macrophage adiponectin expression improves insulin sensitivity and protects against inflammation and atherosclerosis. Diabetes. 59:791–799. 10.2337/db09-1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, K., Cabrero A., Saha P.K., Kojima H., Li L., Chang B.H., Paul A., and Chan L.. 2002. Increased beta -oxidation but no insulin resistance or glucose intolerance in mice lacking adiponectin. J. Biol. Chem. 277:34658–34661. 10.1074/jbc.c200362200 [DOI] [PubMed] [Google Scholar]
- Metsalu, T., and Vilo J.. 2015. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 43:W566–W570. 10.1093/nar/gkv468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina, A.I., LeClair R.A., LeClair K.B., Cohen D.E., Lantier L., and Banks A.S.. 2018. CalR: A web-based analysis tool for indirect calorimetry experiments. Cell Metabol. 28:656–666.e1. 10.1016/j.cmet.2018.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstadt, P.R., Monteiro J.P., and Ashwell J.D.. 2012. Thymocyte responsiveness to endogenous glucocorticoids is required for immunological fitness. J. Clin. Invest. 122:2384–2394. 10.1172/JCI63067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadal, A., Quesada I., Tuduri E., Nogueiras R., and Alonso-Magdalena P.. 2017. Endocrine-disrupting chemicals and the regulation of energy balance. Nat. Rev. Endocrinol. 13:536–546. 10.1038/nrendo.2017.51 [DOI] [PubMed] [Google Scholar]
- Oh, S., Kim S.J., Hwang J.H., Lee H.Y., Ryu M.J., Park J., Kim S.J., Jo Y.S., Kim Y.K., Lee C.H., et al. 2010. Antidiabetic and antiobesity effects of Ampkinone (6f), a novel small molecule activator of AMP-activated protein kinase. J. Med. Chem. 53:7405–7413. 10.1021/jm100565d [DOI] [PubMed] [Google Scholar]
- Presman, D.M., Alvarez L.D., Levi V., Eduardo S., Digman M.A., Marti M.A., Veleiro A.S., Burton G., and Pecci A.. 2010. Insights on glucocorticoid receptor activity modulation through the binding of rigid steroids. PLoS One. 5:e13279. 10.1371/journal.pone.0013279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocelli, M., Barefield D.Y., Warner J.L., Vo A.H., Hadhazy M., Earley J.U., Demonbreun A.R., and McNally E.M.. 2017. Intermittent glucocorticoid steroid dosing enhances muscle repair without eliciting muscle atrophy. J. Clin. Invest. 127:2418–2432. 10.1172/JCI91445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrocelli, M., Zelikovich A.S., Jiang Z., Peek C.B., Demonbreun A.R., Kuntz N.L., Barish G.D., Haldar S.M., Bass J., and McNally E.M.. 2019. Pulsed glucocorticoids enhance dystrophic muscle performance through epigenetic-metabolic reprogramming. JCI Insight. 4:e132402. 10.1172/jci.insight.132402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha, A.J.. 2004. Java Treeview--extensible visualization of microarray data. Bioinformatics. 20:3246–3248. 10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, Y., Roh H.C., Kumari M., and Rosen E.D.. 2017. Adipocyte glucocorticoid receptor is important in lipolysis and insulin resistance due to exogenous steroids, but not insulin resistance caused by high fat feeding. Mol. Metabol. 6:1150–1160. 10.1016/j.molmet.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sud, M., Fahy E., Cotter D., Brown A., Dennis E.A., Glass C.K., Merrill A.H. Jr., Murphy R.C., Raetz C. R. H., Russell D.W., and Subramaniam S.. 2007. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 35:D527–D532. 10.1093/nar/gkl838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran, S., Dubois D.C., Jusko W.J., and Almon R.R.. 2012. Glucocorticoid effects on adiponectin expression. Vitam Horm. 90:163–186. 10.1016/B978-012-398313-8.00007-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, S.P., Stump C.S., Romayne Kurukulasuriya L., and Sowers J.R.. 2007. Adrenal steroids and the metabolic syndrome. Curr. Hypertens. Rep. 9:512–519. 10.1007/s11906-007-0093-4 [DOI] [PubMed] [Google Scholar]
- Wishart, D.S., Feunang Y.D., Marcu A., Guo A.C., Liang K., Vazquez-Fresno R., Sajed T., Johnson D., Li C., Karu N., et al. 2018. HMDB 4.0: The human metabolome database for 2018. Nucleic Acids Res. 46:D608–D617. 10.1093/nar/gkx1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, W.T., Tian X.Y., Xu A., Yu J., Lau C.W., Hoo R.L.C., Wang Y., Lee V.W.Y., Lam K.S.L., Vanhoutte P.M., and Huang Y.. 2011. Adiponectin is required for PPARγ-mediated improvement of endothelial function in diabetic mice. Cell Metabol. 14:104–115. 10.1016/j.cmet.2011.05.009 [DOI] [PubMed] [Google Scholar]
- Yang, X., Bi P., and Kuang S.. 2014. Fighting obesity: When muscle meets fat. Adipocyte. 3:280–289. 10.4161/21623945.2014.964075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Deignan J.L., Qi H., Zhu J., Qian S., Zhong J., Torosyan G., Majid S., Falkard B., Kleinhanz R.R., et al. 2009. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat. Genet. 41:415–423. 10.1038/ng.325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Liu Y., Li M., Wu H., Wang Y., You Y., Li P., Ding X., Liu C., and Gong J.. 2018. Predictive and preventive significance of AMPK activation on hepatocarcinogenesis in patients with liver cirrhosis. Cell Death Dis. 9:264. 10.1038/s41419-018-0308-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, B., Li F., Lin F., Marcelo K.L., Mao J., Dean A., Gonzales N., Gooden D., Maity S., Coarfa C., et al. 2017. Pharmacological inhibition of CaMKK2 with the selective antagonist STO-609 regresses NAFLD. Sci. Rep. 7:11793. 10.1038/s41598-017-12139-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., Guan H., Fu Y., Wang X., Bai L., Zhao S., and Liu E.. 2020. Effects of SFRP4 overexpression on the production of adipokines in transgenic mice. Adipocyte. 9:374–383. 10.1080/21623945.2020.1792614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, N., Pankow J.S., Ballantyne C.M., Couper D., Hoogeveen R.C., Pereira M., Duncan B.B., and Schmidt M.I.. 2010. High-molecular-weight adiponectin and the risk of type 2 diabetes in the ARIC study. J. Clin. Endocrinol. Metabol. 95:5097–5104. 10.1210/jc.2010-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.