Abstract
DNA deaminase enzymes play key roles in immunity and have recently been harnessed for their biotechnological applications. In base editors (BEs), the combination of DNA deaminase mutator activity with CRISPR-Cas localization confers the powerful ability to directly convert one target DNA base into another. While efforts have been made to improve targeting efficiency and precision, all BEs to date utilize a constitutively active DNA deaminase. The absence of regulatory control over promiscuous deaminase activity remains a major limitation to accessing the widespread potential of BEs. Here, we reveal sites that permit splitting of DNA cytosine deaminases into two inactive fragments, whose reapproximation reconstitutes activity. These findings allow for the development of split-engineered base editors (seBEs), which newly enable small-molecule control over targeted mutator activity. We show that the seBE strategy facilitates robust regulated editing with BE scaffolds containing diverse deaminases, offering a generalizable solution for temporally controlling precision genome editing.
INTRODUCTION
Base editors involve the partnership of a catalytically-impaired Cas protein with a DNA deaminase1,2. Guided by a single-guide RNA (sgRNA), the Cas protein first unwinds the target DNA without introducing double-stranded DNA breaks3. The tethered DNA deaminase can then act on the exposed single-stranded DNA to induce C:G to T:A mutations in the case of AID/APOBEC cytosine base editors (CBEs) or A:T to G:C mutations with evolved TadA adenosine base editors (ABEs)4,5. In the case of CBEs, the fusion of one or more protein inhibitors of uracil repair (UGIs) further promotes C:G to T:A transitions over other outcomes6. Alternatively, more processive DNA deaminases can facilitate targeted diversification in place of precise transition mutations7,8.
In their physiological roles in immune defense, AID/APOBEC enzymes are highly regulated at multiple levels, including transcriptional control, alternative splicing, post-translational modifications, and through interaction partners9,10. Efficient regulation is imperative, as DNA deaminases also pose risks to the genome11. Mistargeting of AID and its APOBEC3 (A3) relatives promotes mutations and translocations present in a variety of cancers12–14. These known pathological activities help explain why BEs, which contain unregulated deaminases, have recently been shown to have significant sgRNA-independent off-target activities. Indeed, genome-wide transition mutations occur more frequently after CBE or ABE exposure, and transcriptome-wide mutations increase due to off-target deaminase activity on RNA15–20.
While next-generation BE variants have improved on-target profiles15,17,21,22, the risk of untargeted mutagenesis posed to the cell by a constitutively expressed and unregulated DNA deaminase has not yet been solved. Cas9 engineering has offered routes to gain regulatory control over nuclease activity23–25; however, most of these strategies have yet to be translated to BEs. Cas engineering, including Cas splitting strategies23–26, might help regulate sgRNA-dependent activities in BEs. However, most off-target activities seen in base editors are sgRNA-independent where aberrant deaminase activity can target genomic ssDNA intermediates or promote transcriptome-wide mutations. Recognizing that a solution to this challenge could significantly improve genome editing, we considered the possibility of employing split-protein methods to regulate the mutator activity of the deaminase itself24. Towards this goal, we set out to first determine sites in the DNA deaminase scaffold that allow splitting into two inactive fragments that can spontaneously reassemble into a functional enzyme. Subsequently, we exploit these sites to successfully engineer BEs to permit small molecule regulatory control over base editing activity.
RESULTS
AID tolerates domain insertion and enzyme splitting.
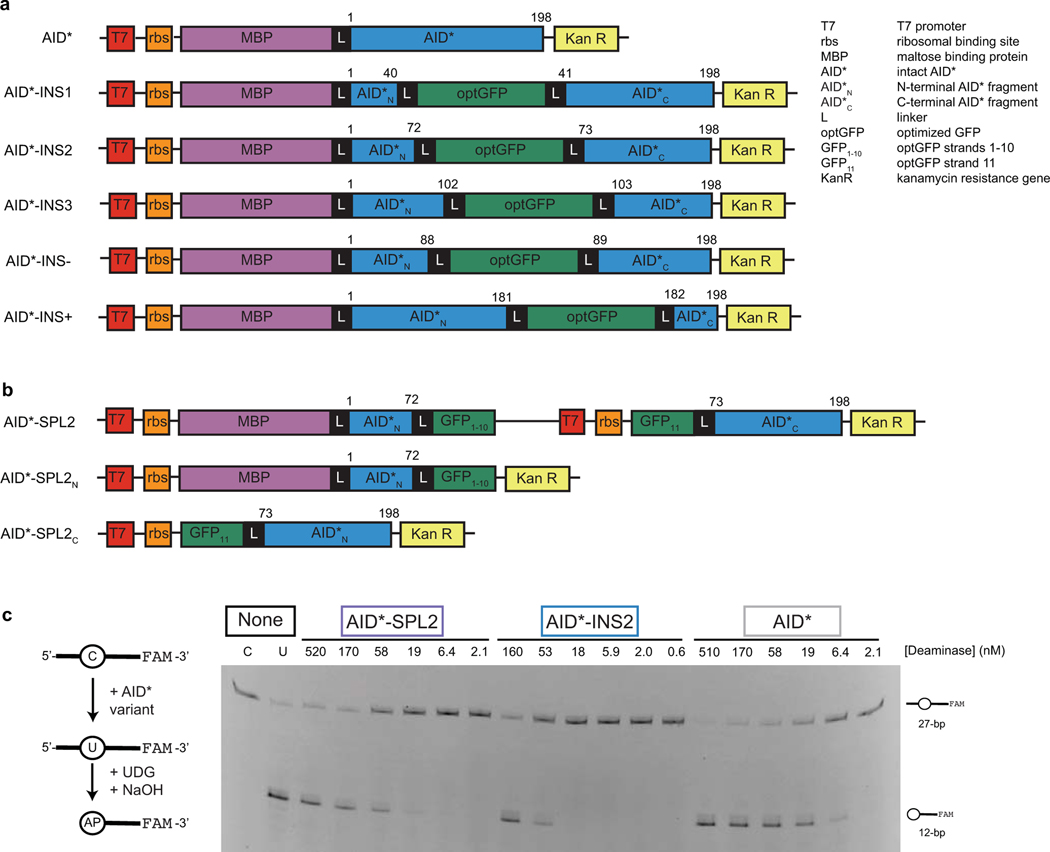
To advance towards a split DNA deaminase, we looked to precedents from the larger deaminase family that share a characteristic α/β deaminase fold27. The family includes pyrimidine salvage enzymes and double-stranded DNA deaminases (DddA) that have previously been split via rational manipulation of loop regions28,29, suggesting that splitting of AID/APOBEC enzymes might also be feasible. Our strategy involved two steps: first identifying sites that tolerate insertion of GFP, and second splitting GFP to test if the DNA deaminase can be spontaneously reconstituted from separate fragments (Fig. 1a). Building on the known structure of human AID30, we focused first on a variant containing a total of twelve hyperactivating mutations (AID*, see list of mutations in methods) that could help potentiate efficient genome editing31,32. We targeted five loops in AID* as distinct GFP insertion sites (Fig. 1b; Extended Data Fig. 1a). Three constructs (AID*-INS1–3) target loops in the core deaminase fold, each with an insertion of an evolved GFP variant (optGFP33). Additionally, we inserted optGFP into the dispensable34 C-terminal loop as a positive control (AID*-INS+) and into the active site loop (β3−α3) as an inactive negative control (AID*-INS-).
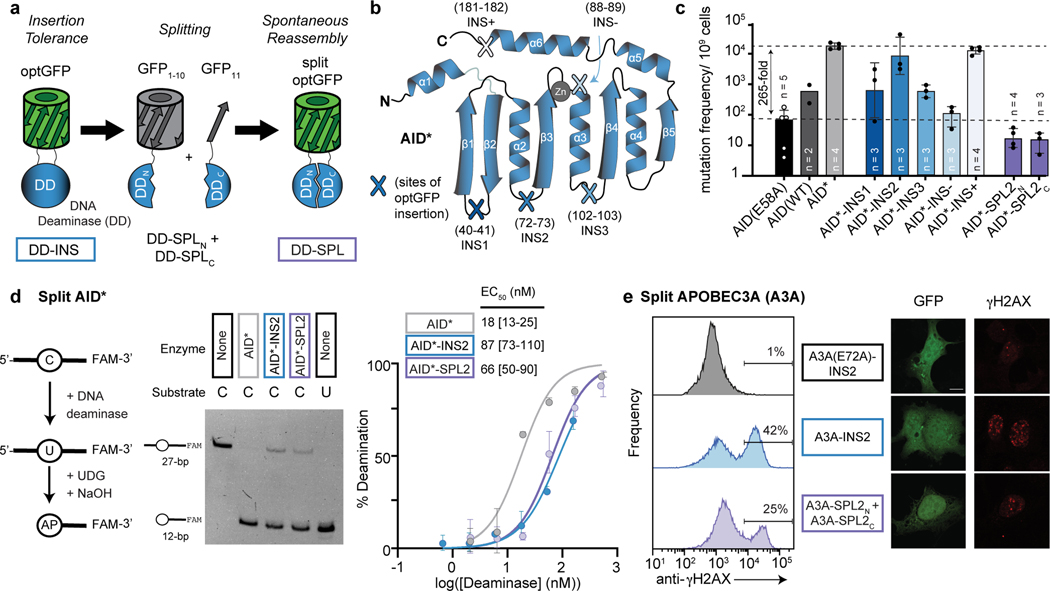
Figure 1. Split deaminases show activity in vitro and in cells.
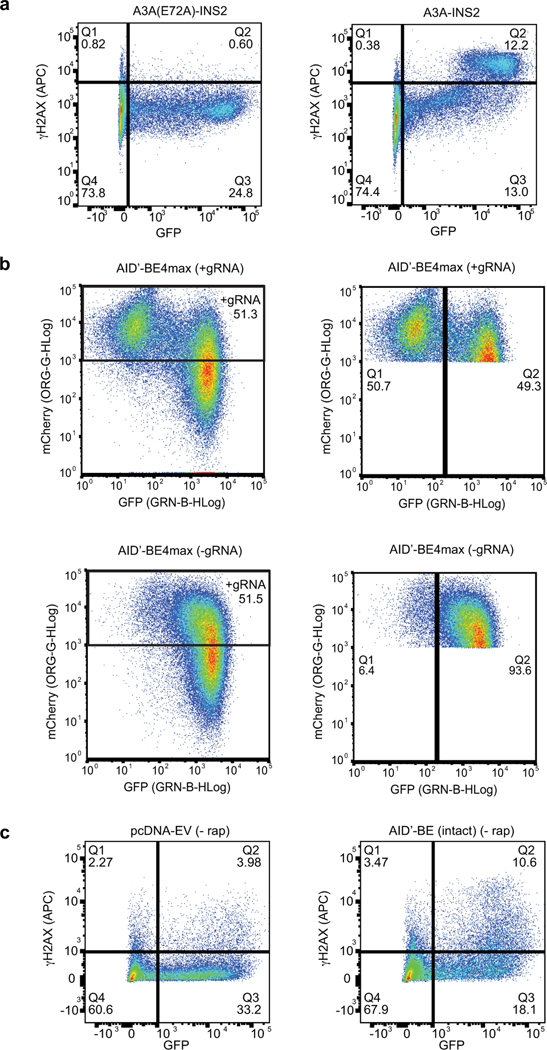
(a) Strategy for testing insertion tolerance followed by split tolerance of DNA deaminases (DD). Constructs with inserted optGFP (DD-INS) are split within optGFP to yield fragments DD-SPLN and DD-SPLC. Spontaneous GFP assembly (DD-SPL) upon co-expression can restore DNA deaminase activity by approximating split fragments. (b) Schematic showing the topology of the DNA deaminase fold, with the active site defined by Zn-interacting residues. Selected sites targeted for insertional mutagenesis in AID* are highlighted. (c) Mutation frequency, as measured by the frequency of acquired rifampin resistance upon expression of AID variants in E. coli. AID(E58A), catalytically inactive control. Each individual data point is indicated on the log-scale plot, with mean and standard deviation. (d) Left—an in vitro assay to measure deaminase activity on a 3’-fluorescein (FAM) labeled oligonucleotide substrate. UDG, uracil DNA glycosylase. AP, abasic site. Middle—a representative denaturing gel (100 nM DNA, 200 nM enzyme) showing substrate (C) and product (U) controls, highlights that the spontaneously-assembled AID*-SPL2 is active. Right—product formation was quantified as a function of enzyme concentration (n = 3) and fit to a sigmoidal dose-response curve to determine the amount of enzyme needed to convert half of the substrate (EC50) under these fixed reaction conditions with mean and 95% confidence interval shown. (e) HEK293T cells were transfected with catalytically inactive mutant A3A(E72A)-INS2, A3A-INS2, or co-transfected with A3A-SPL2N and A3A-SPL2C. After transfection, cells were stained for γH2AX and analyzed by flow cytometry for both GFP and γH2AX expression. Left—representative histogram of the subset of GFP positive cells. Right—representative immunofluorescent images of transfected U2OS cells. GFP staining indicates expression or split reconstitution, and γH2AX serves as a marker of active A3A-mediated DNA damage. A 10 µm scale bar is provided. Representative experiments were repeated independently three times and the results were reproducible.
To test for insertional tolerance, we expressed constructs in E. coli and measured deaminase activity with a rifampin-based mutagenesis assay. In this assay, DNA deaminase expression promotes untargeted mutagenesis of the bacterial genome, and the associated frequency of acquired rifampin resistance (RifR) is a well-established means to assess overall deaminase activity31,35. Using this approach, wild-type AID expression increases RifR 12-fold relative to a catalytically inactive mutant AID(E58A), while hyperactive AID* shows a 265-fold RifR increase (Fig. 1c). As predicted, AID*-INS- shows compromised mutator activity, while AID*-INS+ produces comparable activity to AID*. Turning to the core insertion variants, either β1−β2 (AID*-INS1) or α3−β4 (AID*-INS3) insertion was tolerated, but with significantly reduced activity. Promisingly, however, AID*-INS2 (α2−β3) showed activity comparable to intact AID* alone, suggesting that the enzyme scaffold is tolerant to the introduction of a protein domain at this location.
Having demonstrated insertional tolerance, we next evaluated if the insertion-tolerant site could be used to split the DNA deaminase. We initially inserted optGFP because it can be used to split between the last two β-strands (β10-β11); while the split fragments are non-fluorescent, GFP can be spontaneously reconstituted upon co-expression of both fragments33. We therefore split AID*-INS2 between β10 and β11 of optGFP, resulting in a construct pair of AID*N-optGFP1–10 (AID*-SPL2N) and GFP11-AID*C (AID*-SPL2C) (Fig. 1a, Extended Data Fig. 1b). As predicted, neither AID* fragment alone showed an increase in RifR (Fig. 1c). As the kinetics of split optGFP reassembly are too slow for the RifR E. coli assay, we next co-expressed the AID*-SPL2N and AID*-SPL2C to address if the fragments could spontaneously reconstitute into an enzyme that would be active in vitro. Using a protein tag on one fragment, we first purified the reconstituted protein complex (AID*-SPL2) from E. coli and observed visible fluorescence, suggesting spontaneous GFP assembly. To then test for in vitro activity, we used an assay that can report on a single C→U change, based on fragmentation of a single-stranded DNA oligonucleotide (Fig. 1d, Extended Data Fig. 1c). We found that the reconstituted protein complex showed deaminase activity comparable to that of the AID*-INS2 and only ∼4-fold reduced from that of intact AID*. These results support the AID* α2−β3 loop as a split site for generating inactive deaminase fragments that can be reconstituted.
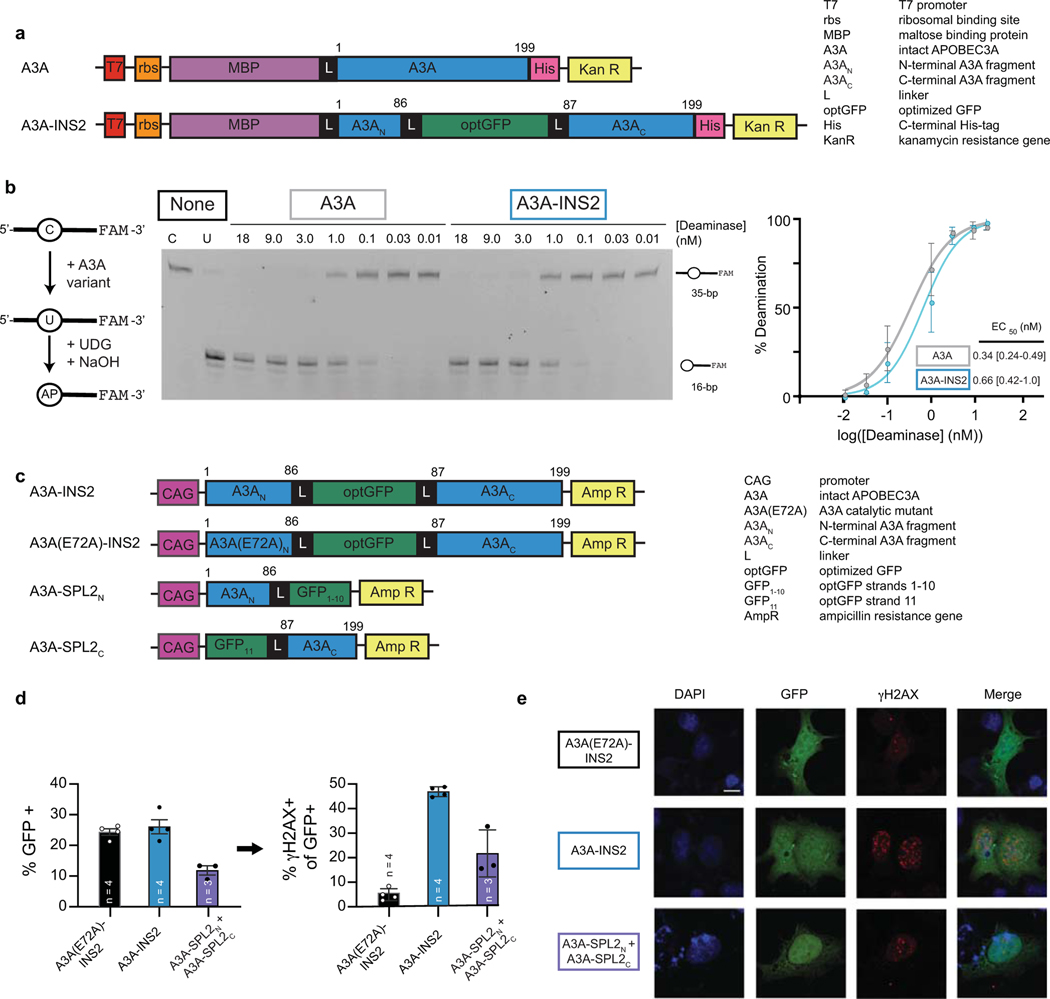
Enzyme splitting is generalizable to other DNA deaminases.
Given the shared structural architecture of AID/APOBEC family enzymes, we hypothesized that the α2−β3 loop might prove to be a generalizable split site. To this end, we examined if human APOBEC3A (A3A)21,36,37 could also be split into two inactive fragments that can be reconstituted. We first validated that A3A tolerated optGFP insertion at its α2−β3 loop in vitro (Extended Data Fig. 2a-b) and then examined its activity in mammalian cells. A3A expression can induce the DNA damage response (DDR), as detected by phosphorylation of histone variant H2AX (γH2AX)38. Accordingly, we analyzed the DDR in cells transfected with mammalian expression vectors containing an optGFP insertion in A3A, a catalytically inactive mutant (A3A-E72A), and the two split fragments (Extended Data Fig. 2c). Post-transfection, GFP+ cells expressing A3A-INS2 showed increased γH2AX relative to the catalytically inactive control. For cells co-expressing A3A split fragments, we readily observed both GFP reassembly and γH2AX by both flow cytometry and immunofluorescence microscopy (Fig. 1e, Extended Data Fig. 2d-e). These results support α2−β3 as a viable split site across the DNA deaminase family and highlight the feasibility of manipulating this site to achieve regulatory control over deaminase activity.
Small molecule control over base editing.
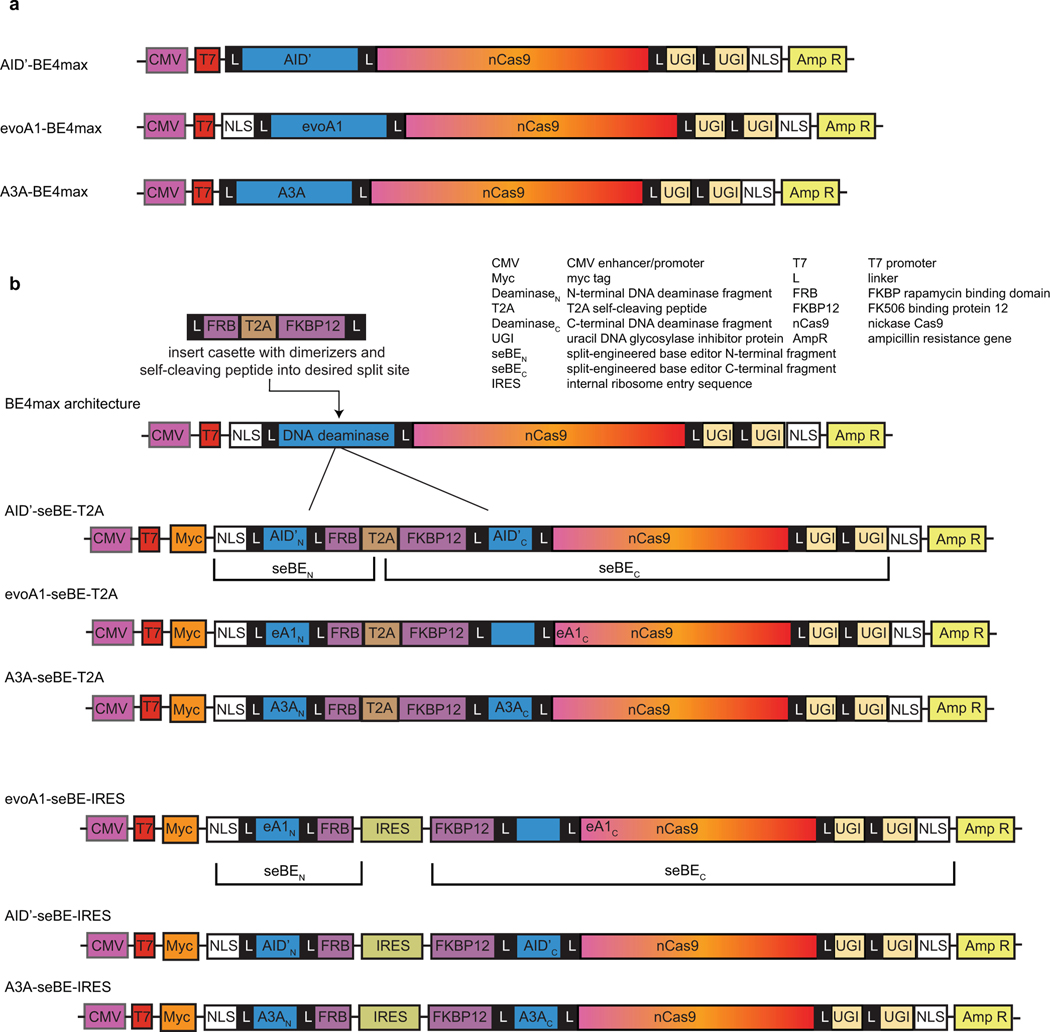
While split optGFP permits spontaneous reconstitution of DNA deaminase activity, our goal was to generate a controllable base editing system. We therefore next aimed to leverage our split sites in concert with chemically induced protein dimerization (CID) strategies to create seBEs. To achieve CID, we employed the common rapamycin-regulated heterodimerization of FK506 binding protein 12 (FKBP12) and FKBP rapamycin binding domain (FRB)39 (Fig. 2a). To explore the generalizability of the seBE strategy, we generated three distinct seBE variants in the scaffold of BE4max40, linking S. pyogenes Cas9 nickase (nCas9) and tandem UGIs to either an alternative hyperactive variant of human AID (AID’), evolved rat APOBEC1 (evoA1), or human A3A. The distinctive features of these deaminase variants permit exploration of different applications: AID is processive and primed for diversity generation7, evoA1 has been shown to be highly precise41, and A3A demonstrates high C to T conversion efficiency21,36,37. Starting from intact BE4max scaffolds, we created seBE constructs by inserting an artificial gene encoding FRB and FKBP12 at the loop between α2 and β3, with fragments separated by a T2A self-cleaving polypeptide (Fig. 2a, Extended Data Fig. 3a-b). The resulting constructs thus co-express two fragments: one containing the DNA deaminase N-terminus and FRB; the second containing FKBP12, the DNA deaminase C-terminus, nCas9, and two UGIs in series.
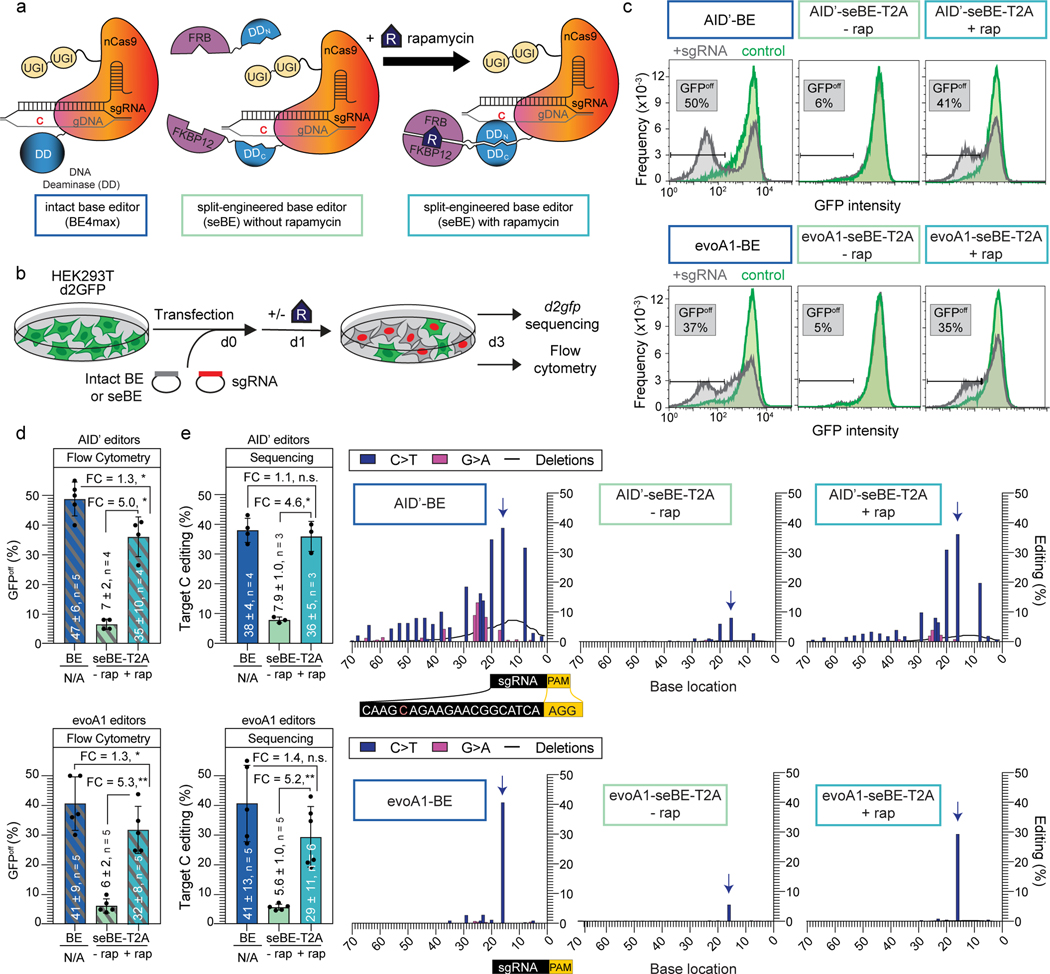
Figure 2. Split-engineered base editors represent a generalizable strategy to enable small-molecule-controlled editing.
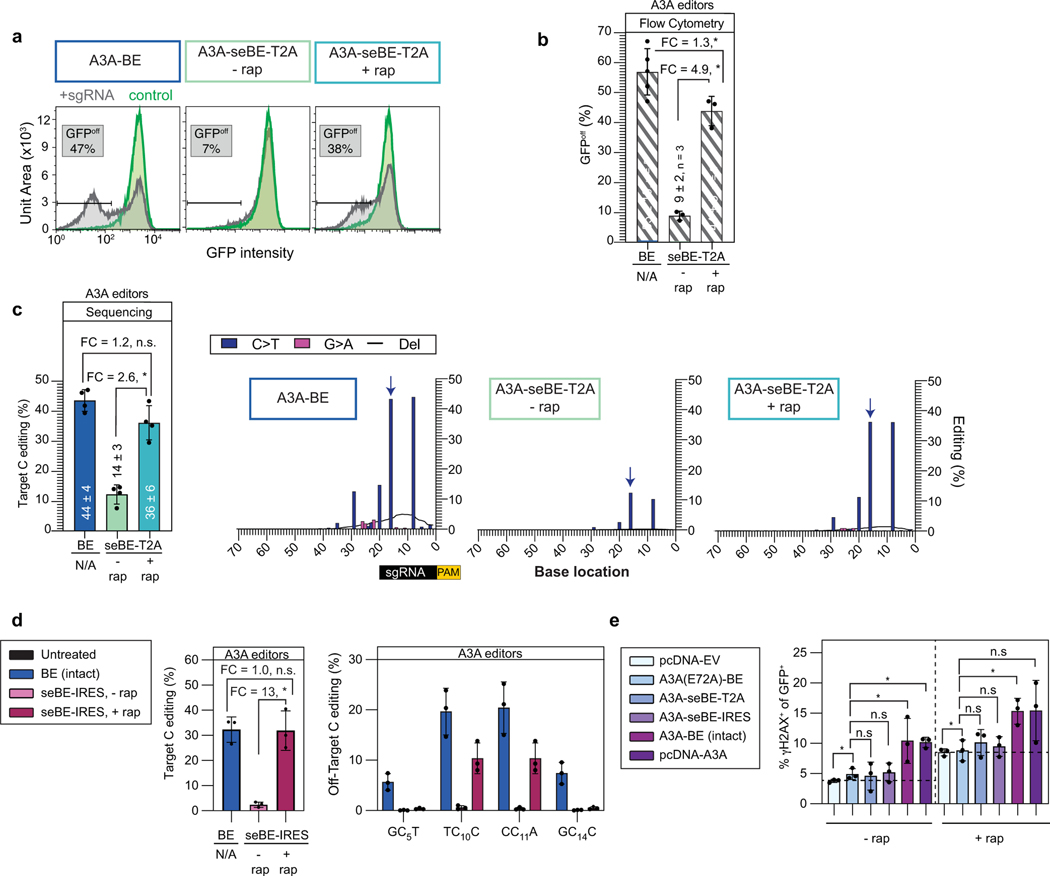
(a) Schematics of a traditional intact base editor in the BE4max scaffold and the split-engineered base editor (seBE) strategy, including chemically induced dimerization of FRB and FKBP12 by rapamycin. (b) Editing efficiency can be evaluated in a HEK293T cell line containing a single copy of integrated, constitutively expressed d2gfp. The presence of d2gfp-targeting sgRNA can introduce a stop codon (Q158*) and abrogate fluorescence to generate GFPoff cells, which can be tracked by either flow cytometry or deep-sequencing of the locus. (c) Representative flow cytometry histograms associated with transfection of intact or seBE constructs in the presence or absence of rapamycin. (d) Mean and standard deviation for quantification of GFPoff cells by flow cytometry, with individual data points shown. Two-sided Mann-Whitney test was performed to compare intact and split base editor GFPoff % (*p ≤ 0.05; **p ≤ 0.01) Exact p-values provided as statistical source data files. (e) Left—deep sequencing results demonstrating C to T conversion efficiency of the Q158 target cytosine under conditions identical to (d). The mean and standard deviation are noted, with individual data points shown. Fold-change (FC) is the ratio of mean values for the higher versus the lower condition in each comparison. Two-sided Mann-Whitney test was performed (n.s., not significant; *p ≤ 0.05; **p ≤ 0.01) Right—editing footprints across the d2gfp locus for each condition. The full targeting sequencing is provided with the sgRNA protospacer (black) starting 20 bp from the PAM (yellow) and the target C highlighted in red. In the editing footprint, the target cytosine base within the Q158 codon is noted with a blue arrow. Data represent position-wise averages of three or more biological replicates, with individual replicate data provided in Supplementary Table 1. Exact p-values provided as statistical source data files.
To measure editing efficiency, we derived a HEK293T reporter cell line with a single copy of destabilized GFP (d2gfp) stably integrated (Fig. 2b). When d2gfp is targeted, successful base editing generates a nonsense mutation at Q158 measurable by flow cytometry (GFPoff) (Fig. 2c-d). For the intact AID’-BE4max, minimal GFPoff cells were observed in the absence of a targeting sgRNA, but editing was highly efficient in its presence (49 ± 6%). With AID’-seBE-T2A, targeting sgRNA, and no rapamycin, we observed near background levels of GFPoff (7 ± 2%). Upon rapamycin addition, we observed robust GFP inactivation (36 ± 7%) indicative of successful CID. These observed patterns were mirrored with evoA1 and A3A seBE constructs, which generated rapamycin-dependent detection of GFPoff cells to levels approaching those of intact BEs (Fig. 2c-d; Extended Data Fig. 4a-b).
To more rigorously assess activity, we deep-sequenced the d2gfp locus for each condition to profile editing footprints (Fig. 2e). For intact AID’-BE4max, the target cytosine within the Q158 codon showed the highest editing percentage within the locus (38 ± 4%). However, clones also harbored multiple bystander mutations, including deletions (7.6 ± 1.4%) and G→A mutations, suggesting editor activity on the sgRNA target strand and showcasing the known processive behavior of AID7,42. For AID’-seBE-T2A, we observed low levels of editing at the target base in the absence of rapamycin (7.9 ± 1.0%) and marked elevation in its presence (36 ± 5%). The mutational footprint of the seBE appeared similar to the intact editor, albeit with fewer cumulative deletions (2.2 ± 0.3%). We also observed controllable editing in the evoA1 series, with the distinction that these editors are more precise rather than processive (Supplementary Table 1). With evoA1-seBE-T2A, rapamycin addition induced editing 5.2-fold, reaching a level (29 ± 11%) approaching that of the intact evoA1-BE4max (41 ± 13%). Rapamycin-dependent editing also extended to the A3A-based editors (Extended Data Fig. 4c), demonstrating that small-molecule-regulated base editing is generalizable across multiple seBE constructs.
Alternative expression strategies tune regulatory control.
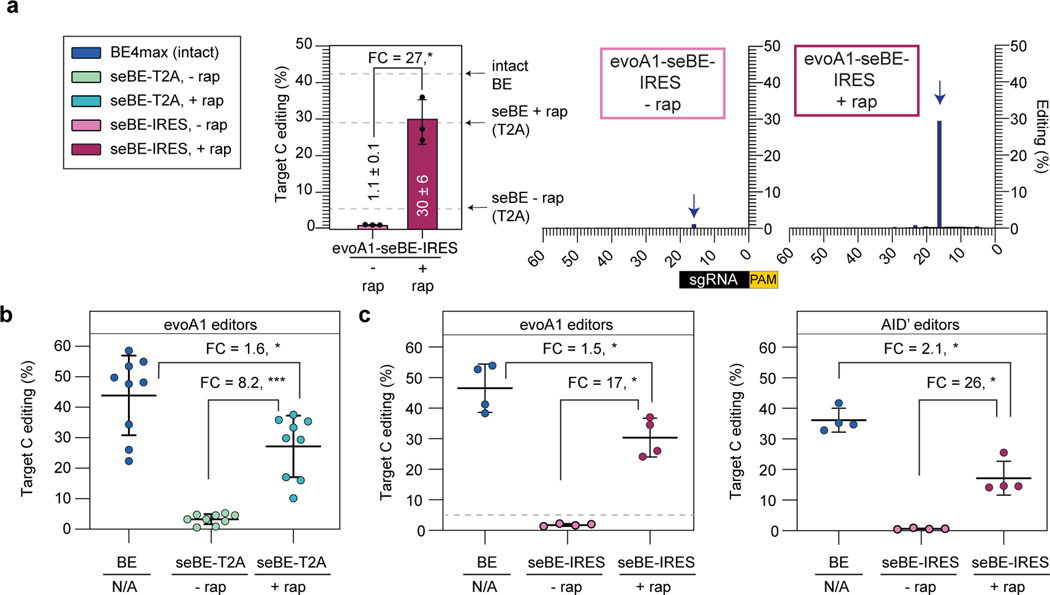
A strength of the seBE strategy is that the system is well poised for modifications to alter either the nature or the degree of regulatory control. For example, we noted that while editing was readily induced by rapamycin with seBEs, low-level activity was still observable in the absence of rapamycin. We hypothesized that this editing could have resulted from incomplete ribosome skipping with the T2A self-cleaving peptide, which would yield an intact editor. To further increase the dynamic range of small-molecule inducible editing, we generated an enhanced bicistronic vector for the evoA1-seBE construct. In evoA1-seBE-IRES, the seBEN and seBEC polypeptides were expressed separately from two independent translation start sites: one associates with the CMV promoter and the other from an internal ribosome entry sequence (IRES) (Extended Data Fig. 3b). Indeed, sequencing analysis revealed that the seBE-IRES construct greatly reduced editing in the absence of rapamycin (1.1 ± 0.1%, Fig. 3a) compared to the T2A construct (5.6 ± 1.0%, Fig. 2e). Meanwhile, rapamycin-dependent editing remained robust (30 ± 6%) and precise (Fig. 3a). Thus, increasing the stringency with which split fragments are separately expressed readily permits 27-fold inducible control over base editing of d2gfp.
Figure 3. Split-engineered base editors permit efficient editing across genomic sites.
(a) d2gfp-HEK293T cells were edited using evoA1-seBE-IRES, where split protein fragments are independently translated. Left—deep sequencing results demonstrating C to T conversion efficiency of the Q158 target cytosine with and without rapamycin. Bars indicate means and error bars indicate standard deviations of three biological replicates. Two-sided Mann-Whitney test was performed (*p ≤ 0.05), exact p-values provided as statistical source data files. The dotted line represents the mean values for the intact evoA1-BE4max and evoA1-seBE-T2A with and without rapamycin from Fig. 2e for comparison. Right—editing footprints across the d2gfp locus for each condition. Data represent position-wise averages of three biological replicates, with individual replicate data provided in Supplementary Table 1. (b) Cells were edited with evoA1-BE4max or evoA1-seBE-T2A in the absence or presence of rapamycin. The editing efficiency across nine genomic loci involving epigenetic regulators and two well-established target sites are shown. Each data point represents three biological replicates at each target site. The mean and standard deviation across all sites is noted, with individual data points shown. The fold-change (FC) is the ratio of mean values for the higher versus the lower condition in each comparison. Two-sided Mann-Whitney test was performed (*p ≤ 0.05; ***p ≤ 0.001). Exact p-values provided in statistical source data files. The individual loci editing data are provided in Extended Data Fig. 5a. (c) Cells were edited with intact BE4max or seBE-IRES constructs in the absence or presence of rapamycin. Left—cells treated with evoA1-based constructs; Right—cells treated with AID’-based constructs. The editing efficiency across three genomic loci involving epigenetic regulators and one well-established target site (EMX1) are shown. Each data point represents three biological replicates. The fold-change (FC) is the ratio of mean values for the higher versus the lower condition. Two-sided Mann-Whitney test was performed (*p ≤ 0.05). Exact p-values provided in statistical source data files. For evoA1-seBE-IRES, the dotted line represents the mean values for the evoA1-seBE4-T2A for the same four loci without rapamycin (from Fig. 3b) for comparison. The individual loci editing data are provided in Extended Data Fig. 5b-c.
seBEs permit inducible editing across broad genomic targets.
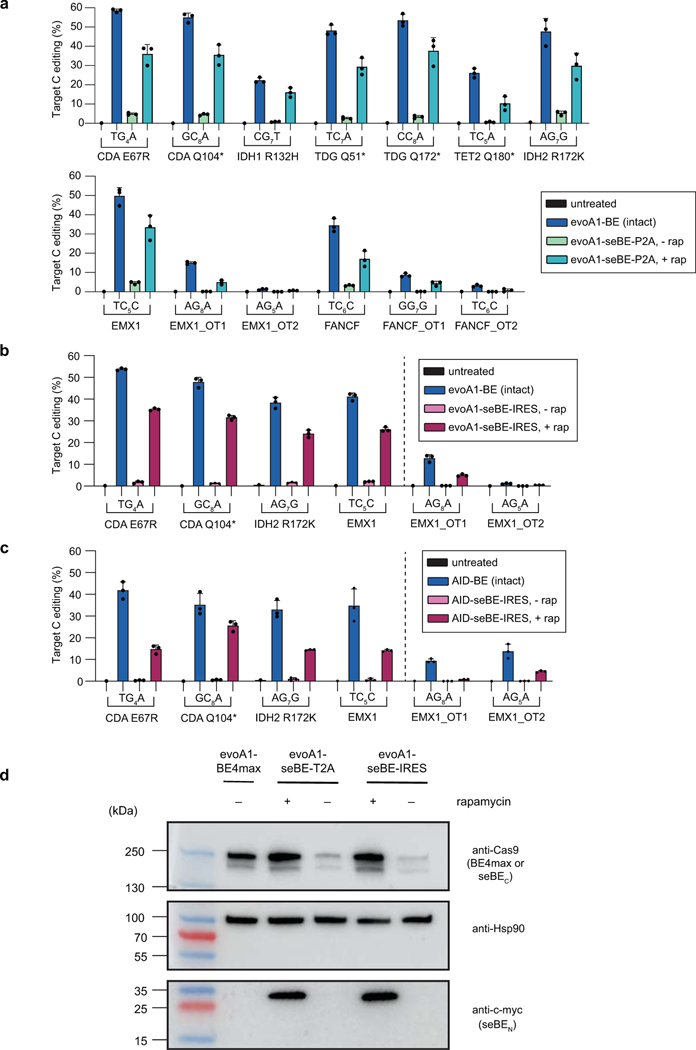
Building on the observation of inducible editing using the d2gfp assay, we next explored whether seBEs can similarly permit controllable editing for a broader array of genomic sites with different characteristics. We first focused our analysis on the evoA1-based constructs, given their observed precision and frequent application in the field. We targeted seven loci involving epigenetic regulators and two well-established target sites, which span different sequence context and mutations that can variably generate stop codons or activity-altering point mutations. Across these sites, the intact evoA1-BE4max average on-target editing efficiency was 44% (Fig. 3b, Extended Data Fig. 5a). For evoA1-seBE-T2A in the absence of rapamycin, on-target editing across sites was detectable but low (mean ∼3.3%). Upon CID with rapamycin, base editing activity was significantly induced across sites (mean ∼27%). On average, base editing was induced 8.2-fold by rapamycin and reached 64% of the editing efficiency of unregulated intact editors.
Given the improved dynamic range of the IRES constructs in the d2gfp assay, we next explored the robustness and inducibility of their editing at a subset of alternative genomic loci with both evoA1 and AID’-based editors. Across sites, in this experimental comparison the average on-target editing efficiency of the intact evoA1-BE4max was 47% (Fig. 3c, Extended Data Fig. 5b). For evoA1-seBE-IRES, background on-target editing (1.8%) in the absence of rapamycin was reduced relative to that observed with the T2A (4.8%) against the same targets. Upon the addition of rapamycin, base editing was induced 17-fold (mean ∼30%), reaching 64% of the editing efficiency achieved with the intact editor across sites. Using the same approach, we next assayed AID’-based editors, given their distinct sequence preference and the wider editing window compared to previously reported BEs. Across sites, AID’-BE4max average on-target editing efficiency was 36% (Fig. 3c, Extended Data Fig. 5c). Strikingly, for AID’-seBE-IRES in the absence of rapamycin, on-target editing was reduced to 0.7%. Upon the addition of rapamycin, base editing was induced 26-fold (mean ∼17%), reaching 47% of the editing efficiency achieved with the intact editor.
To explore the mechanism underlying improved control with seBE constructs, we examined protein expression in cells transfected with an EMX1-targeting sgRNA and evoA1-BE4max, evoA1-seBE-T2A, or evoA1-seBE-IRES constructs in the absence or presence of rapamycin (Extended Data Fig. 5d). As expected, the intact evoA1-BE4max was stably expressed. By contrast, in the absence of rapamycin, the split fragments from both evoA1-seBE-T2A and evoA1-seBE-IRES were barely detectable, suggesting the fragments are unstable in the absence of dimerization, a feature which could aid in reversibility. Accordingly, in the presence of rapamycin, both split fragments can be readily detected at levels comparable to the intact editor. Thus, both the low background and high-inducibility of seBE constructs can be explained by the formation of a stable and active base editor complex dependent upon chemically induced dimerization.
Assessing seBEs off-target effects.
Noting the high degree of inducible control for on-target editing, we next aimed to evaluate the impact of seBEs on off-target editing. BEs are associated with different classes of off-target editing. Analogous to traditional Cas9 genome editing systems, BEs can bind to off-target genomic sites with similarity to the target sgRNA protospacer. A subset of these binding events can lead to sgRNA-dependent base editing upon DNA deaminase action (Fig. 4a). Unlike traditional Cas9 genome-editing systems, BEs are also associated with sgRNA-independent off-target editing, whereby DNA deaminases can act on transiently exposed genomic ssDNA or on cellular RNA. We hypothesized that the control of base editing achieved by seBEs would minimize off-target activities in the absence of rapamycin and significantly reduce off-target activities upon CID, relative to intact BEs.
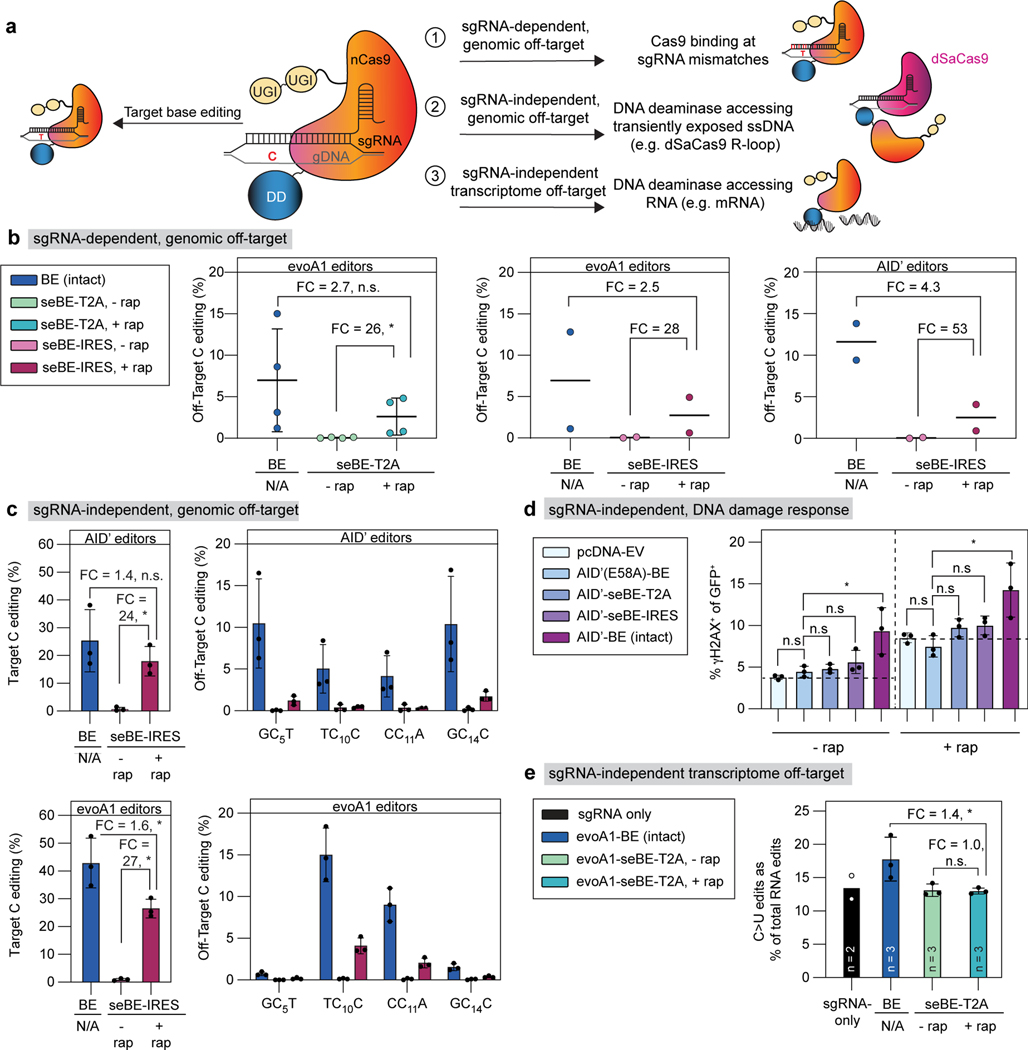
Figure 4. Split-engineered base editors decrease off-target effects.
(a) Schematic showing on-target base editing and potential sources of off-target mutagenesis. (b) Left—sgRNA-dependent off-target genomic editing was quantified in cells transfected with evoA1-BE4max or evoA1-seBE-T2A and sgRNAs targeting either EMX1 or FANCF. The mean and standard deviation from four biological replicates are shown, with individual loci editing data in Extended Data Fig. 5a. Two-sided Mann-Whitney test was performed (n.s., not significant; *p≤ 0.05, exact p-values provided in statistical source data files. Center and right—sgRNA-dependent off-target genomic editing was quantified in cells transfected with evoA1 (center) or AID’ (right) intact or seBE-IRES constructs and EMX1-targeting sgRNA. Two biological replicates and the average are shown with individual loci editing data in Extended Data Fig. 5b-c. No statistical analysis was performed due to sample size. (c) sgRNA-independent off-target genomic editing was quantified in cells transfected with AID’ (top) or evoA1 (bottom) intact or seBE-IRES constructs and EMX1-targeting sgRNA, along with dSaCas9-sgRNA targeting a different locus. Left—EMX1 on-target editing. Right—off-target editing at the locus opened by dSaCas9. The mean and standard deviation from three biological replicates are shown. Two-sided Mann-Whitney test was performed (n.s., not significant; *p ≤ 0.05, exact p-values provided in statistical source data files). Editing footprints and replicates are provided in Supplementary Fig. 4. (d) Quantification of γH2AX upon base editor expression in the absence and presence of rapamycin. HEK293T cells transfected with noted constructs and an EMX1-targeting sgRNA vector expressing GFP. Cells were either maintained with or without rapamycin and stained for γH2AX. The fraction of GFP and γH2AX positive cells are shown, with mean and standard deviation from three biological replicates. Two-sided Mann-Whitney test was performed (n.s., not significant; *p ≤ 0.05, exact p-values provided in statistical source data files). (e) The frequencies of C>U edits as a fraction of total RNA edits unique to transfected cells are shown. The mean of biological replicates and standard deviation (if n=3) are shown, with catalog of RNA edits in Extended Data Fig. 6 and Supplementary Table 2. Two-sided Mann-Whitney test was performed (n.s., not significant; *p ≤ 0.05, exact p-values provided in statistical source data files).
To probe for sgRNA-dependent off-target effects, we first analyzed well-established genomic off-target sites for both the EMX1 and FANCF-targeting sgRNAs15,43 (Fig. 4b, Extended Data Fig. 5a). While sgRNA-dependent off-target editing was readily detected at all four sites with the intact editor, off-target editing was absent without rapamycin for evoA1-seBE-T2A and reached only 37% of the level observed with intact evoA1-BE4max upon addition of rapamycin. Extending our analysis to IRES constructs, we evaluated both the evoA1 and AID’-seBEs at EMX1-associated off-target sites. As with the T2A constructs, editing at sgRNA-dependent off-target sites was absent without rapamycin for both IRES constructs, and reached only 40% (evoA1-seBE-IRES) and 23% (AID’-seBE-IRES) of the levels reached by their corresponding intact editors upon addition of rapamycin (Fig. 4b).
Unlike sgRNA-dependent genomic off-target effects, DNA deaminase-dependent off-target activity in the genome is stochastic, making it more difficult to readily detect. Accordingly, a method known as the R-loop assay has been developed to amplify the signal from sgRNA-independent genomic off-target deamination at a specific locus44–46. In this assay, HEK293T cells are co-transfected with three plasmids encoding (1) intact S. pyogenes Cas9 (SpCas9)-derived BE or the seBE-IRES construct, (2) an EMX1-targeting sgRNA, and (3) a catalytically inactive S. aureus Cas9 (dSaCas9) with an SaCas9 sgRNA targeting an unrelated genomic locus. The dSaCas9 artificially opens but does not cleave genomic DNA at the SaCas9 sgRNA targeting site, creating a long-lived R loop with ssDNA. The deaminase from the SpCas9-associated base editor may then act at this ssDNA, independent of EMX1-targeting. In this assay, on-target editing efficiency at EMX1 for evoA1-BE4max and AID’-BE4max were 43% and 25% respectively, mirroring previous experiments in the absence of the added dSaCas9 construct. On-target editing was similarly unaffected with seBE-IRES constructs. For evoA1-seBE-IRES and AID’-seBE-IRES, in the absence of rapamycin, on-target editing was low (mean 1% and 0.7%, respectively); in its presence, on-target editing was induced, reaching 27% for evoA1-seBE-IRES and 18% for AID’-seBE-IRES (Fig. 4c). Using this system, off-target sgRNA-independent deamination at the dSaCas9 R-loop site was readily detectable for intact evoA1-BE4max (mean ∼6.6%) and AID’-BE4max (mean 7.5%) constructs. For evoA1-seBE-IRES and AID’-seBE-IRES constructs in the absence of rapamycin, off-target editing was near background (mean 0.1% and 0.2% respectively). In the presence of rapamycin, off-target activity was detectable but substantially decreased when compared to intact BEs by 3.9-fold (mean ∼1.7%) and 8.1-fold (mean ∼0.9%). The observed trends also all held true for A3A-based editors (Extended Data Fig. 4d), indicating that, as with sgRNA-dependent off-target editing, sgRNA-independent off-target editing appears suppressed in the absence of rapamycin and substantially reduced upon chemically induced dimerization.
Overexpression of an isolated DNA deaminase can cause genomic toxicity by creating DSBs (Fig. 4a), which activates the DDR as detectable by accumulation of γH2AX. Intact BEs also contain an unregulated and constitutively active DNA deaminase, along with nCas9, which could further increase DNA damage via nicks. To assess DNA damage from these sources in the setting of seBEs, we next analyzed γH2AX expression in cells transfected with base editor constructs and an EMX1-targeting sgRNA in the absence and presence of rapamycin (Fig. 4d). Focusing first on transfected cells in the absence of rapamycin, expression of a catalytically inactive AID(E58A)-BE led to levels of γH2AX similar to those with transfection of an empty vector control (mean 4.5% and 3.8%, respectively). Notably, transfection of the intact editors is associated with a significant increase in γH2AX (mean 9.4%), while both AID’-seBE-T2A and AID’-seBE-IRES show no significant increase above the catalytically inactive control. In the presence of rapamycin, all samples showed higher levels of γH2AX due to the known rapamycin-related suppression of DSB repair47. However, all constructs followed the same trend, with only intact BEs significantly increasing genomic toxicity. The results with AID’-based editors were mirrored with A3A editors (Extended Data Fig. 4e) where the intact A3A-BE4max editor and the isolated A3A domain, but not A3A-seBEs, induce similar increased levels of γH2AX accumulation.
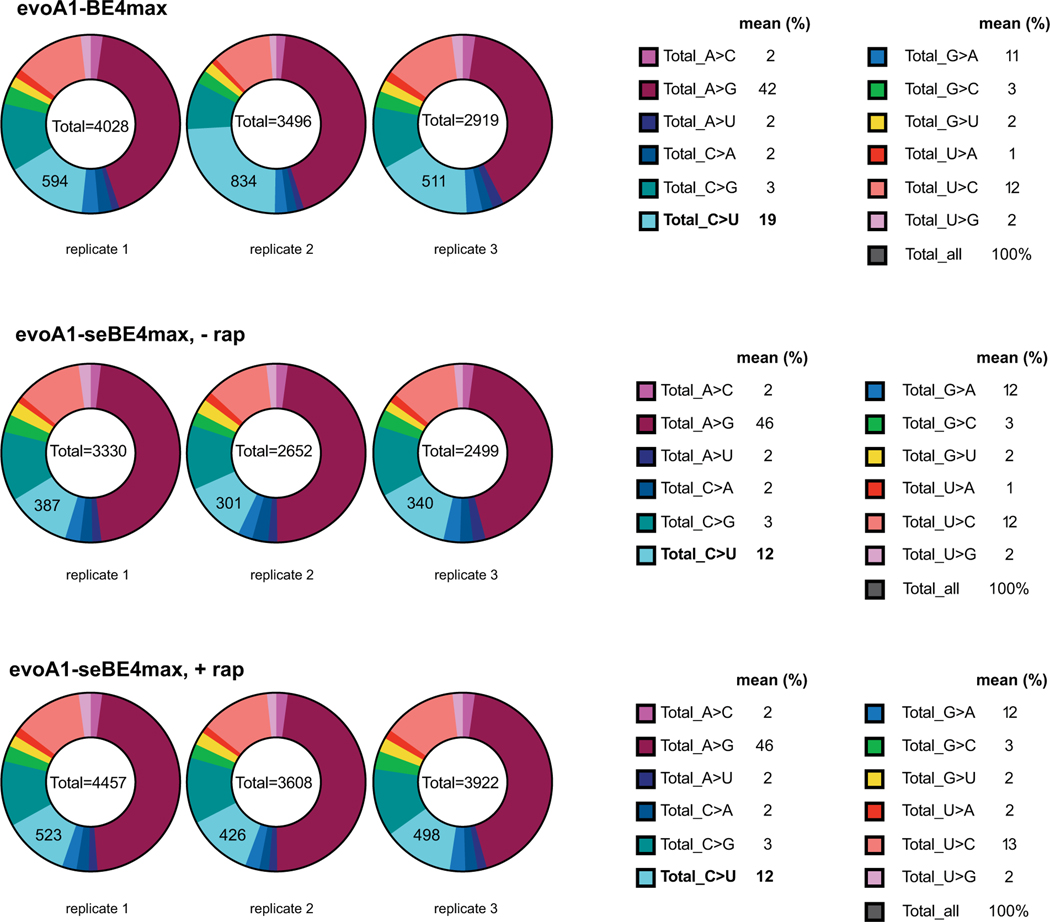
Intact BEs have also been shown to cause sgRNA-independent transcriptome-wide C-to-U deamination of RNA (Fig. 4a). To probe off-target activity in the transcriptome, we performed RNA-seq on samples undergoing d2gfp editing without enrichment or sorting. While transcriptome-wide mutations with intact evoA1-BE4max were lower than those previously reported with BE3-based editors19, the intact BE showed a profile distinct from the evoA1-seBE-T2A (Fig. 4e). Importantly, expression of the seBE-T2A construct did not increase C-to-U mutations when compared to a sgRNA-only transfected control either in the presence or absence of rapamycin. By contrast, with the intact editor we noted a 1.6-fold higher fraction of C-to-U mutations compared to the sgRNA-only or seBE transfected controls (Fig. 4e, Extended Data Fig. 6, Supplementary Table 2). Taken together, the sgRNA-dependent and the three orthogonal sgRNA-independent off-target assays all highlight a consistent pattern, whereby seBE off-target activities are substantially reduced relative to intact editors in the presence of rapamycin and not detectable in its absence.
seBEs permit temporal control over base editing.
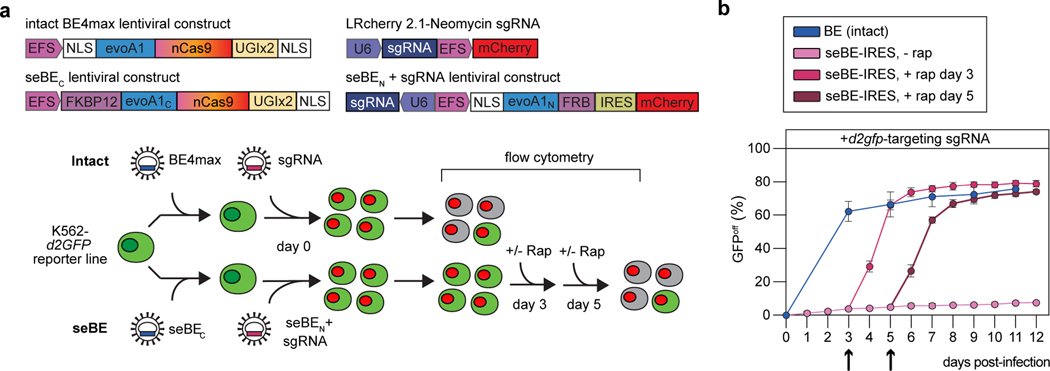
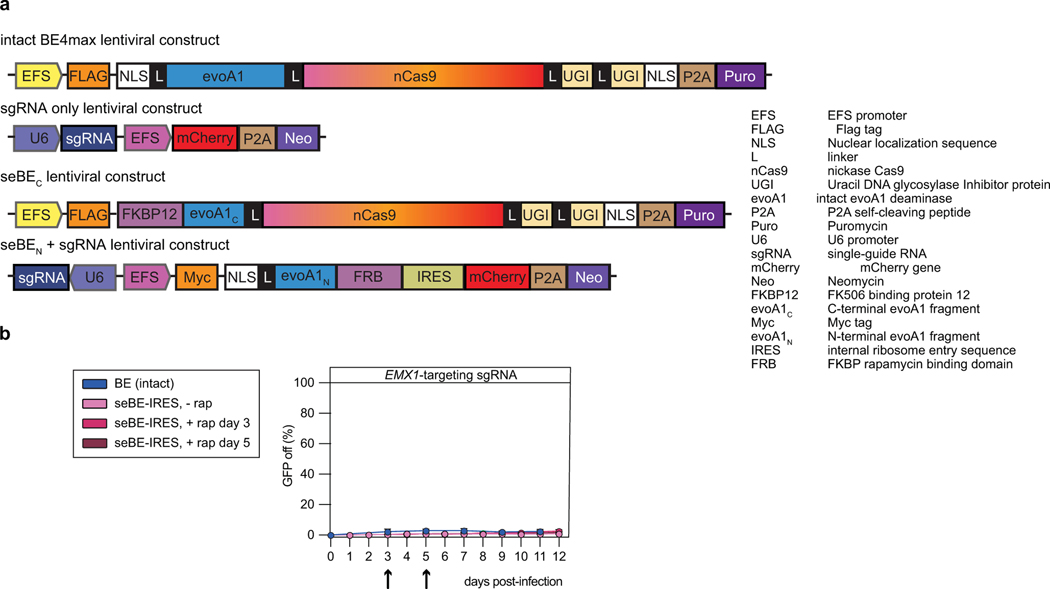
In addition to reducing off-target activity, small-molecule activation of seBEs offers the potential to manipulate the timing of targeted genome editing in living cells. Temporal control over base editing would allow for genome changes to be introduced when desired (e.g. at particular stages in development or at critical steps in pathogenesis). To evaluate if the seBE complex could be used to lie dormant in a cell line until base editing is induced by rapamycin, we employed a K562 leukemia reporter cell line with a single copy of stably integrated d2GFP. Cells were infected with intact evoA1-BE4max lentivirus, followed by a sgRNA targeting either d2gfp or EMX1 as a control (Fig. 5a, Extended Data Fig. 7a). As with our HEK293T reporter cell line, successful d2gfp editing generates a nonsense mutation and GFP inactivation (GFPoff) can be tracked over time by flow cytometry. In cells with evoA1-BE4max, unregulated and rapid editing occurs upon introduction of a d2GFP-targeting sgRNA but not an EMX1-targeting sgRNA, with 62% loss of d2GFP after 3 days, reaching a maximum editing of 76% (Fig. 5b, Extended Data Fig. 7b). When cells are instead infected with lentivirus encoding the seBE fragments and a d2gfp-targeting sgRNA, minimal editing is observed through 12 days in the absence of rapamycin. To examine the inducibility of editing, we added rapamycin at day 3 after infection and observed rapid accumulation of GFPoff cells, reaching 74% loss after 3 days of rapamycin. Similar kinetics of GFP inactivation can be observed by selecting a later time point, with addition of rapamycin at day 5 resulting in a 57% loss of d2GFP after 3 days to reach a maximum of 74%. Importantly, when cells were infected with an EMX1-targeting sgRNA, they did not demonstrate a decrease in d2GFP fluorescence, highlighting the specificity of targeted, controllable genome editing (Extended Data Fig. 7b). Our results show that seBEs offer strong temporal control, lying dormant in a cell in the absence of rapamycin and upon induction, performing base editing at a similar rate and efficiency as intact BEs.
Figure 5. seBEs permit temporal control over base editing.
(a) Top—shown are the lentiviral constructs for introducing the intact BE4max or the seBE fragments along with mCherry expressing sgRNA constructs. Bottom—editing efficiency was evaluated in a K562 cell line containing a single copy of integrated, constitutively expressed d2gfp. Editing of the d2gfp locus results in loss of GFP fluorescence which can be tracked by flow cytometry either in the presence or absence of added rapamycin. (b) Quantification of GFPoff cells by flow cytometry for cells with intact BE4max or seBE-IRES with no rapamycin or rapamycin (25 nM) added at either day 3 or day 5 (marked with arrow) and then maintained continuously. Mean and standard deviation are noted, with individual data points shown (n = 3).
DISCUSSION
In sum, we have demonstrated a generalizable strategy for small-molecule regulation of DNA deaminase activity. Although we focus on BE applications, these split sites could also be used to study conditional control over endogenous AID/APOBEC deaminases, as in antibody somatic hypermutation or cancer mutagenesis. Given that the α2−β3 loop tolerates insertion of either split GFP or FKBP/FRB, we anticipate extensions to other CID strategies, such as those using non-immunomodulatory rapalogs, readily reversible abscisic acid, or photo-inducible protein dimerization systems25. Each of these post-translational strategies offer some distinctive advantages over translational control, with the potential for more rapid onset and layered tight regulatory control over activity in subcellular location, space, or time24,48. seBEs are also anticipated to function with editor scaffolds beyond BE4max, including those using other Cas proteins49 or different deaminases with altered editing windows or DNA/RNA discrimination. Splitting the deaminase halves between two different RNA-guided targeting modules could also minimize sgRNA-dependent off-target activities, akin to recently developed split dsDNA deaminase editors (split DddA)29 or the dimeric Cas9-FokI heterodimerization systems50. Additionally, split Cas9, although not addressing the challenge of unregulated deaminase activity, leverages a strategy for differential nuclear-cytoplasmic localization of split fragments26 that could be incorporated with seBEs to further suppress activity in the absence of rapamycin. We conclude by noting that small-molecule inducible seBEs are poised to permit editing in more complex settings, including in vivo, in order to achieve needed spatial and temporal control over base editing.
METHODS
Design and cloning of intact and split DNA deaminase constructs
Complete DNA sequences for plasmids used are provided in the Supplementary Sequences document. Relevant primers used for cloning are listed in Supplementary Table 3.
For bacterial studies with AID*, the parent pET41 plasmid with AID* combines three different sets of previously described30–32 mutations that increase activity or solubility (K10E, F42E, T82I, D118A, R119G, K120R, A121R, H130A, R131E, F141Y, F145E, and E156G) in a construct with an N-terminal maltose binding protein tag (MBP). The plasmids named AID*-INS contain an insertion of optGFP flanked by linkers at each position within a specified loop of AID*. The N-terminal fragment of AID (AID*N) and C-terminal fragment of AID (AID*C) were generated by PCR amplification from the AID* parent plasmid with primers listed in Supplementary Table 3a. A sequence containing linker-optGFP-linker was obtained as a gene fragment (Integrated DNA Technologies, IDT) and amplified with primers provided in Supplementary Table 3a, which add flanking regions that permit overlap extension PCR. Overlap extension PCR was performed to fuse the three fragments encoding AID*N, linker-optGFP-linker, and AID*C, using 10 cycles of amplification without primers to permit fusion of fragments, followed by amplification of the entire AID*N-optGFP-AID*C sequence with the outer primers. PCR products from the overlap extension PCR were TA cloned (Invitrogen). Sequence-confirmed inserts were then digested with SalI and AvrII and ligated into the digested parent plasmid with T4 DNA ligase (NEB). The control plasmids containing unmutated AID (AID-WT) or the catalytically inactive mutant AID(E58A), were previously reported31.
For bacterial studies with split AID*, the AID*-SPL2N and AID*-SPL2C constructs were created using AID*-INS2 as a scaffold in the pET41 backbone. To create AID*-SPL2N, the parent plasmid (AID*-INS2) was digested with KpnI and AvrII to remove the C-terminal region of AID*. Then, an oligonucleotide cassette containing a stop codon (TAG) was ligated into the digested vector. To create AID*-SPL2C, the parent plasmid (AID*-INS2) was digested with XbaI and KpnI to remove AID*-SPL2N. Then, a cassette containing a start codon (ATG) was ligated into the digested vector. The AID*-SPL2 plasmid, co-expressing the N-terminal and C-terminal fragments from separate promoters was created using AID*-INS2 as a scaffold. A gene fragment was synthesized containing the C-terminal region of AID*-SPL2N, the transcriptional terminator, the T7 RNA polymerase promoter, and the N-terminal region of AID*-SPL2C. This fragment was ligated into a KpnI/AvrII digested AID*-INS2 parent vector.
For bacterial expression of A3A constructs with insertion of optGFP, cloning was performed in the scaffold of MBP-A3A-His-pET41 backbone51,52 (Addgene #109231). The appropriate optGFP-containing insert was synthesized as a gene fragment (IDT), digested with EagI/AvrII (NEB), and ligated into the similarly digested parent plasmid.
For mammalian expression of A3A constructs, plasmids were cloned into a pLEXm backbone. A3A-INS2, A3A-SPL2N, and A3A-SPL2C were amplified from the pET41 construct, adding flanking regions of overlap with the pLEXm plasmid backbone. The final plasmids were then constructed using Gibson Assembly Master Mix (NEB), merging the amplified gene fragments with the EcoRI/XhoI (NEB) digested parent vector. The catalytically inactive variant A3A(E72A)-INS2 was created using Q5 Site-Directed Mutagenesis Kit (NEB).
Design and cloning of intact and split base editor constructs
For mammalian base editing constructs, the intact or split-engineered constructs were cloned into the scaffold of pCMV_BE4max (Addgene #112093), which contains rat APOBEC1. The parent plasmid contains a NotI restriction site. An additional XmaI restriction site was added into pCMV_BE4max using the Q5 Site-Directed Mutagenesis Kit (NEB) to facilitate cloning. This parent plasmid for construct construction was noted to have two point mutations that were propagated into further constructs, one in the flexible linker from nCas9 to the first UGI and a second correlating with a E11K change in the first UGI subunit. The E11K is located opposite of the UDG binding site in UGI and unlikely to impact activity53. The deaminase sequences were amplified from their respective pET41 plasmids, introducing a region of overlap. AID’ differs from AID* in that it contains a smaller subset of mutations, including K10E, T82I, D118A, R119G, K120R, A121R, and E156G. For AID, catalytically inactive constructs were made with Q5 Site-Directed Mutagenesis Kit (NEB) yielding the AID(E58A)-BE4max constructs. For the A3A, the catalytically inactive A3A(E72A) construct was first generated in the pET41 framework and then transferred into the base editor construct as above.
To facilitate cloning of seBE-T2A constructs, gene fragments were synthesized (IDT) containing DeaminaseN-FRB, the T2A self-cleaving peptide between the two fragments, and FKBP12-DeaminaseC. The associated strategy for linkers between domains was derived from that recently employed to split human TET254. Using the gene fragments, all BE4max and seBE-T2A plasmids were then constructed using Gibson Assembly Master Mix (NEB), merging the relevant gene fragments with the NotI/XmaI digested vector. Notably the intact AID’-BE4max and A3A-BE4max lack the N-terminal NLS present in BE4max vectors. A3A-seBE contains a missense mutation (M13I), which does not appear to impact activity.
The seBE-IRES constructs, where the two split protein fragments are independently translated, were cloned into the scaffolds of evoA1-, AID’- and A3A-seBE-T2A constructs. The IRES sequence fragment was amplified from control plasmid (Addgene #105594)55 with Phusion High-Fidelity DNA Polymerase (NEB). The vector backbones of seBE-T2A constructs were amplified, excluding the T2A sequence. The vector and IRES sequence fragment were then joined using the In-Fusion HD Cloning system (TBUSA).
To generate a constitutive all-in-one dead SaCas9 (dSaCas9) system where both dSaCas9 and its targeting-sgRNA are independently translated, the SaCas9 expression vector (Addgene #164563) was used as a template. Q5 Site-Directed Mutagenesis Kit (NEB) was first used to make a catalytically inactive Staphylococcus aureus SaCas9 (D10A, N580A). The P2A sequence was then removed and replaced with an IRES sequence fragment using the In-Fusion HD Cloning system (TBUSA) as described above.
To generate intact and split base editor constructs in lentiviral vectors, PCR fragments were amplified with Phusion High-Fidelity DNA Polymerase (Thermo Scientific) and joined using the In-Fusion HD Cloning system (TBUSA). For the intact BE lentiviral construct, the coding sequence (NLS-evoA1-nCas9–2×UGI-NLS) was amplified from pCMV_BE4max (Addgene #112093)40 and cloned into the scaffold of pLenti-FNLS-P2A-Puro (Addgene #110841)56. For the lentiviral seBEc construct, the coding sequence (NLS-FKBP12-evoA1C-nCas9–2×UGI-NLS) was amplified from the evoA1-seBE-IRES pCMV construct (above) and cloned into the scaffold of pLenti-FNLS-P2A-Puro (Addgene #110841)56. For the cloning of LRcherry2.1-Neomycin vector, the P2A-Neomycin sequence was incorporated into LRCherry2.1 (Addgene #108099)57 in the same reading frame with EFS-mCherry. For the lentiviral seBEN construct, the coding sequence (Myc-NLS-evoA1N-FRB-IRES) was amplified from the evoA1-seBE-IRES pCMV construct (above) and cloned into the scaffold of LRCherry2.1-Neomycin vector.
The sgRNA expression plasmids were constructed using oligonucleotide cassettes for cloning. Briefly, the primers listed in the Supplementary Table 3b were annealed and phosphorylated using T4 Polynucleotide Kinase (NEB) according to the manufacturer’s instructions and further purified using the Oligo Clean and Concentrator kit (Zymo Research). Next, LRcherry2.1 plasmid57, LRG plasmid (Addgene #65656)58, LRCherry2.1-Neomycin plasmid, LRcherry2.1-seBEN-P2A-Neomycin plasmid, or the dSaCas9-sgRNA plasmid were incubated with Esp3I (Thermo Fisher Scientific) at 37 °C for 2 hours to remove a short filler sequence, and further agarose gel purified. The sgRNA cassettes were then ligated in place of the filler using T4 DNA ligase (NEB).
Bacterial DNA deaminase rifampin mutagenesis assay
The previously reported rifampin mutagenesis assay34 was adapted to measure the mutation frequency of various DNA deaminases. Plasmids encoding a deaminase variant were transformed into BL21(DE3) E. coli that harbor a plasmid encoding uracil DNA glycosylase inhibitor (UGI) (chloramphenicol resistant). Overnight cultures were grown in LB with kanamycin (30 ng/mL) and chloramphenicol (25 ng/mL) from single colonies and diluted to an OD600 of 0.2. Cells were then grown for 1 hour at 37 °C before inducing deaminase expression with 1mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 4 hours of additional growth, serial dilutions were separately plated on Luria Bertani (LB) agar plates containing rifampin (100 μg/mL) and plasmid-selective antibiotics. The mutation frequencies were then calculated by the ratio of rifampin-resistant (RifR) colonies relative to the total colony forming units.
In vitro DNA deaminase oligonucleotide assay
For in vitro assays, purified intact, optGFP-inserted, or split DNA deaminases were expressed in BL21(DE3) E. coli that co-express the Trigger Factor (TF) chaperone, as previously described34. Briefly, 600 mL cultures were grown to an OD600 of 0.6 at 37 ºC. Cultures were shifted to 16 ºC for 16 hours after induction with 1 mM IPTG. For AID variants, the pelleted cells were resuspended in wash buffer containing 50 mM Tris-Cl (pH 7.5) 150 mM NaCl, and 10% glycerol, and lysed through sonication. The soluble fraction was filtered after high-speed centrifugation and incubated with 3 mL of Amylose Resin (NEB) for 1 hour at 4 ºC. The resin was washed extensively prior to elution with wash buffer plus 10 mM maltose. Total protein was quantified by comparison to a BSA standard curve. For A3A variants, the pelleted cells were resuspended in wash buffer containing 50 mM Tris-Cl (pH 7.5) 150 mM NaCl, 10% glycerol, and 25 mM imidazole, and lysed through sonication. The soluble fraction was filtered after high-speed centrifugation and incubated with 3 mL of HisPur cobalt resin (Thermo) for 1 hour at 4 ºC. The resin was washed extensively prior to elution with wash buffer with 150 mM imidazole.
For the in vitro assay, a 3’-fluorescein (FAM)-labeled oligonucleotide substrate was used containing a single cytosine, along with a product control oligonucleotide containing uracil at the same location. For AID variants, the oligonucleotide substrate was co-incubated with 3-fold dilutions of the purified AID variant (520 nM to 0.6 nM) and 25U of uracil DNA glycosylase (NEB). The reaction was performed in 20 mM Tris-HCl (pH 8.0), 1 mM DTT and 1 mM EDTA at 37°C for 1 hour. For A3A, the oligonucleotide substrate was co-incubated with 3-fold dilutions of the purified A3A variant (18 nM to 10 pM) and 25U of uracil DNA glycosylase. The reaction was performed in 35 mM succinic acid, sodium dihydrogen phosphate, and glycine (SPG) buffer (pH 5.5) and 0.1% Tween-20 at 37°C for 30 min. Deamination reactions were terminated by incubation at 95°C for 10 min. The samples were heat denatured by using 2X bromophenol blue loading dye containing 0.6 M NaOH to cleave abasic sites, 0.03 M EDTA, and 95% Formamide. Samples were run on a preheated 20% acrylamide/Tris-Borate-EDTA(TBE)/urea gel at 50°C, and imaged using FAM filters on a Typhoon imager (GE Healthcare). Product formation was quantified using ImageJ by taking the ratio of substrate to product under each condition. Product formation as a function of enzyme concentration was fit to a sigmoidal dose-response curve and used to determine the EC50, defined as the amount of enzyme that converts 50% of the substrate to product under the fixed reaction conditions.
γH2AX staining of mammalian cells
HEK293T cells used for flow cytometry were cultured in Dulbecco’s Modified Eagle Medium (DMEM) media (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C with 5% CO2 and cells were periodically tested to be mycoplasma negative. The cells were transfected with A3A-INS2 or A3A(E72A)-INS2, or co-transfected with A3A-SPL2N and A3A-SPL2C for 24 hours prior to harvest and staining with γH2AX antibody (BD Pharmigen, 647) and flow cytometry analysis. Cells were gated on FITC and APC using the Fortessa Flow Cytometer (BD Biosciences), and results were analyzed using FlowJo. The gating strategy is exemplified in Extended Fig. 8a.
U2OS cells used for immunofluorescent studies were cultured in Dulbecco’s Modified Eagle Medium (DMEM) media (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C with 5% CO2 and cells were periodically tested to be mycoplasma negative. U2OS cells plated on coverslips were transiently transfected with A3A-INS, A3A(E72A)-INS2 or co-transfected with A3A-SPL2N and A3A-SPL2C constructs for 24 hours prior to incubation with γH2AX antibody (Millipore Sigma) and immunofluorescent staining with Alexa Fluor 568 (Invitrogen) and DAPI. Stained cells were imaged with a Nikon A1R confocal microscope and analyzed using ImageJ.
Base editing assay using d2GFP inactivation by flow cytometry in HEK293T cells
HEK293T cells were lentivirally-transduced with a constitutively expressed destabilized GFP (d2GFP) reporter (derived from Addgene #14760) and selected for individual clones that contained a single copy of integrated d2gfp. The HEK293T d2GFP cells were maintained as above, seeded on 24-well plates, and transfected at approximately 60% confluency. 660 ng of intact BE4max or seBE4max constructs and 330 ng of LRcherry2.1 sgRNA expression plasmids were transfected using 1.5 µL of Lipofectamine 2000 CD (Invitrogen) per well according to manufacturer’s protocol. Negative control samples include LRcherry2.1 plasmid lacking a protospacer (labeled as no sgRNA samples). The d2gfp-targeting sgRNA exposes a window where base editing can result in the introduction of a Q158X nonsense mutation in d2gfp. For seBE experiments, 24 hours after transfection, rapamycin (Research Products International) was added to select wells at a final concentration of 200 nM. This concentration was continuously maintained until the end of the experiment. Transfected cells were harvested at day 3 after transfection, ensuring single-cell suspension. The percentage of d2GFP-negative and mCherry-positive (sgRNA+) cells was determined by flow cytometry with Guava Easycyte 10HT instrument (Millipore). Flow cytometry analysis was performed using FlowJo Software Version 10.7.1 (FloJo, LCC). The gating strategy is exemplified in Extended Fig. 8b.
Genomic DNA was also collected from cells using the DNeasy Blood & Tissue Kit (Qiagen) according to manufacturer’s instructions for amplification across the d2gfp locus and deep sequencing as described in the “DNA library preparation and sequencing” section below. Total RNA was isolated using Direct-zol™ RNA Miniprep Plus kit (Zymo Research #R2072) following the manufacturer’s protocol for sequencing as described below. For RNA-seq analysis, negative control transfections included d2gfp-targeting LRcherry2.1 plasmid without any base editor construct.
Base editing of various genomic loci
For editing of diverse genomic loci, HEK293T cells (lacking the single copy d2gfp) were used and maintained as above. The transfection protocol was performed as described above, with the exception that different sgRNAs were used for targeting of other loci. In each case, the sgRNAs expose a window where base editing can result in the introduction of point mutations in DNA modifying enzymes that lead to either missense or nonsense mutations. As with the d2GFP editing assay, 24 hours after transfection, rapamycin (Research Products International) was added to select wells at a final concentration of 200 nM. This concentration was continuously maintained until the end of the experiment. Transfected cells were harvested at day 3 after transfection, ensuring single-cell suspension. Genomic DNA was collected using the DNeasy Blood & Tissue Kit (Qiagen) according to manufacturer’s instructions for sequencing analysis as described in the “DNA library preparation and sequencing” section below.
Western blot
To analyze protein expression during a base editing experiments, the transfection and base editing protocol was performed as described above using intact BE4max or seBE constructs and the EMX1-targeting sgRNA plasmid. At the end of the experiment, cells were resuspended in CytoBuster Protein Extraction Reagent (Millipore Sigma) for lysis according to manufacturer’s instructions. Protein concentration was quantified by Qubit Protein Assay Kit (ThermoFisher), and 40 µg of total protein was loaded into a 4–15% Mini-Protean TGX Precast Protein Gel (BioRad). After electrophoresis, the iBlot Dry Blotting System (ThermoFisher) was used for transfer onto PVDF. The membrane was then blocked with 5% (w/v) low fat milk, 20 mM Tris-HCl, 10 mM NaCl and 0.1% Tween-20 (TBST) and incubated at 4°C with the appropriate primary antibody overnight: Myc-Tag (9B11) Mouse mAb (Cell Signaling) at 1:2000 dilution, anti-Cas (7A9–3A3) (Cell Signaling) at 1:1000 or Hsp90α/β (F-8) at 1:200 (Santa Cruz Biotechnology). The next day, the membranes were washed in 1X TBST and incubated in blocking buffer at 4°C for 1 hour with m-IgGκ BP-HRP secondary antibody (Santa Cruz Biotechnology). The membranes were imaged using Immobilon Western Chemiluminescent HRP Substrate (Millipore Sigma).
R-loop assay
HEK293T cells were seeded on 24-well plates, and transfected at ∼60% confluency. 400 ng of intact BE4max or seBE constructs, 200 ng of EMX1-targeting LRcherry2.1 sgRNA plasmid, and 400 ng of dSaCas9 expression plasmid were co-transfected using 1.5 µL of Lipofectamine 2000 CD (Invitrogen) per well according to manufacturer’s protocol. For seBE experiments, 24 hours after transfection, rapamycin was added to select wells at a final concentration of 200 nM and maintained until the end of the experiment when transfected cells were harvested 3 days after transfection. Genomic DNA was collected from cells using the DNeasy Blood & Tissue Kit (Qiagen) and both the EMX1 and SaCas9-targeted locus (Chr 9: 21036–21332) were amplified and then deep sequenced as described on the “DNA library preparation and sequencing” section below.
γH2AX staining of base edited cells
For γH2AX analysis of intact BE4max and seBE constructs in the presence or absence of rapamycin, the transfection protocol was performed on HEK293T cells seeded on 6-well plates and transfected at approximately 60% confluency. Parallel analysis of empty vector (pcDNA-EV) or the isolated DNA deaminase domains was carried out. Intact BE4max or seBE constructs and LRcherry2.1-EMX1 sgRNA expression plasmids were transfected in a 2:1 ratio using Lipofectamine 2000 CD (Invitrogen) per well according to manufacturer’s protocol. For seBE experiments, 24 hours after transfection, rapamycin was added to select wells at a final concentration of 200 nM and maintained until the end of the experiment. Cells were harvested 3 days after transfection and stained with γH2AX antibody (BD Pharmigen, 647) for flow cytometry analysis. Cells were gated on FITC and APC using the Fortessa Flow Cytometer (BD Biosciences), and results were analyzed using FlowJo. The gating strategy is exemplified in Extended Data Fig. 8c.
DNA library preparation and sequencing
Target loci of interest were PCR-amplified from 100 ng genomic DNA (primer pairs in Supplementary Table 3b) using KAPA HiFi HotStart Uracil+ Ready Mix (Kapa Biosystems) or Phusion High-Fidelity DNA Polymerase (New England Biolabs, NEB). PCR products were purified via QIAquick PCR Purification Kit (Qiagen).
Some samples were deep-sequenced by Amplicon-EZ Next Generation Sequencing (Genewiz). Alternatively, indexed DNA libraries were prepared using the NEBNext Ultra II DNA Library Prep Kit for Illumina with the following specifications. After adapter ligation and 4 cycles of PCR enrichment, indexed amplicon concentration was quantified by Qubit dsDNA HS Assay Kit (ThermoFisher), and size distribution was determined on a Bioanalyzer 2100 (Agilent) with the DNA 1000 Kit (Agilent). Indexed PCR amplicons were pooled together in an equimolar ratio for paired-end sequencing by MiSeq (Illumina) with the 300-cycle MiSeq Reagent Nano Kit v2 (Illumina). Raw reads were automatically demultiplexed by MiSeq Reporter. Demultiplexed read qualities were evaluated by FastQC v0.11.9 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Low-quality sequence (Phred quality score <28) and adapters were trimmed via Trim Galore v0.6.5 (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) prior to analysis with CRISPResso259. Sequencing yielded ∼13,000 median aligned reads per sample (5th percentile ∼4,000, 95th percentile ∼63,000). The reported data (Figs. 2–4) represent the frequency of editing at the target base alone, with complete analysis across the sgRNA region provided in Supplementary Figs. 1-4 for all sites other than for d2gfp, which is provided in Supplementary Table 1.
RNA sequencing
Total RNA, isolated as described above, was analyzed for quality using the RNA 6000 Nano Bioanalyzer kit (Agilent). Only RNA with an RNA integrity number (RIN) ≥ 8 was used for subsequent library construction. RNA-seq was performed on 0.5–1.0 μg of total RNA according to the Genewiz Illumina Hi-seq protocol for poly(A)-selected samples (2 × 150 bp pair-end sequencing, 350M raw reads per lane). The resulting reads were analyzed using the RADAR pipeline (RNA-editing Analysis-pipeline to Decode All twelve-types of RNA-editing events49). For each sgRNA-only sample respectively (n = 2), RNA edits that were present in other samples were removed and unique editing events present in the sgRNA-only sample were used for comparison against editing events present in other samples but not in the sgRNA-only sample (Supplementary Table 2). For the base editing samples, the average percentage of C to U edits from analyzing against each sgRNA-only sample are plotted. The analysis of distribution of editing events (Extended Data Fig. 6) was performed by removing any edits found in either of the sgRNA-only samples.
Lentiviral base editing assay using d2GFP inactivation by flow cytometry in K562 cells
K562 chronic myeloid leukemia cells were grown in RPMI-1640 (Gibco) with 10% bovine calf serum. Human embryonic kidney 293T cells were cultured in DMEM (Corning) with 10% bovine calf serum. Both cell culture media were supplemented with 1% penicillin/streptomycin, and cell lines were incubated at 37℃ with 5% CO2 and were periodically tested to be mycoplasma-negative.
For lentivirus production, HEK293T cells were seeded at ∼50% confluency in 10-cm plate and were transfected the next day (at ∼90% confluency). For each viral production, 10 μg of the plasmid of interest, 5 μg of VSV-G, and 7.5μg of psPAX2 (Addgene#12260) were transfected using 80 μL of polyethylenimine (Polysciences, PEI 25000) and 500 μL of Opti-MEM (Gibco). The media was changed with ∼6 mL fresh DMEM 6–8 hrs after transfection. Lentivirus was harvested several times within 48 hrs of transfection, filtered with a 0.45 μm PVDF filter (Millipore) and stored at −80℃ for long term use.
For lentivirus transduction, K562 cells were transduced with lentivirus using 8 μg/mL Polybrene (Sigma #H9268) and centrifuged at 650× g for 25 min at room temperature. The cells were incubated at 37 ℃ overnight and replaced with fresh media ∼15 hrs post transduction. Antibiotics were added with appropriate concentration 1 day post-infection with corresponding antibiotics (10 μg/mL blasticidin, 2 μg/mL puromycin, 1 mg/mL G418).
For the d2gfp disruption assay, the destabilized GFP (d2GFP) reporter (derived from Addgene #14760)60 was first transduced into K562 cells. The K562 d2gfp reporter cell line was then first transduced with either the intact BE4max or seBEC lentivirus, and then with the sgRNA-only or seBEC + sgRNA lentivirus. Cell lines were selected with their corresponding antibiotics. As with with HEK293T reporter cell line, the percentage of d2GFP-negative and mCherry-positive (sgRNA+) cells was determined by flow cytometry with Guava Easycyte 10HT instrument (Millipore). For the intact BE4max, flow cytometry analysis was performed on day 3 after transduction of the sgRNA vectors and every other day until day 11. For the seBE experiments, 1×105 cells were seeded in 24-well plate as the day 0 sample. GFP measurements were then taken every 24 hrs until day 12. Starting at either day 3 or day 5, rapamycin was added to select wells at a final concentration of 25 nM and maintained continuously until the end of the experiment. The same volume of DMSO was also added to another well as control (without rapamycin). Flow cytometry analysis was performed using FlowJo Software Version 10.7.1 (FloJo, LCC), and the fold change of GFP+ cells in mCherry+ population (normalized to day 0) was used for analysis.
Data availability
High-throughput RNA sequencing data will be deposited at the Gene Expression Omnibus (GEO) database (accession number pending). Individual amplicon sequencing data is available as Supplementary Information and raw reads will be available upon request. Novel plasmids used in this study will be made available from Addgene. Nucleic acid sequences of all constructs used in this study are provided in a note at the end of the Supplementary Information.
Extended Data
Extended Data Fig. 1. Intact, inserted, and split DNA deaminase constructs with AID*.
(a) Construct schematics for AID* and AID*-INS variants (analogous to those in Fig. 1b) that were used to determine the impact of optGFP insertion in E. coli. Numbers above the constructs represent the amino acid sequence of the deaminase itself. (b) Construct schematics for co-expression or individual expression of AID*-SPL2N and AID*-SPL2C fragments in E. coli. (c) Left—an in vitro assay to measure deaminase activity on a 3’-fluorescein (FAM) labeled oligonucleotide substrate. UDG, uracil DNA glycosylase. Right—a representative denaturing gel (100 nM DNA, variable enzyme concentration) is shown, along with substrate and product controls (C and U, respectively). Analysis of product formation with three independent replicates was used to define the dose-response curve provided in Fig. 1d.
Extended Data Fig. 2. Intact, inserted, and split DNA deaminase constructs with A3A.
(a) Construct schematics for A3A and A3A-INS2 variants used to determine the impact of optGFP insertion in E. coli. Numbers above the constructs represent the amino acids of the deaminase itself. (b) Left—an in vitro assay to measure deaminase activity on a 3’-fluorescein (FAM) labeled oligonucleotide substrate. UDG, uracil DNA glycosylase. AP, abasic site. Middle—a representative denaturing gel (100 nM DNA, variable enzyme concentration) is shown, along with unreacted substrate and product controls (C and U, respectively). Right—product formation was quantified as a function of enzyme concentration (n = 3) and fit to a sigmoidal dose-response curve to determine the amount of enzyme needed to convert half of the substrate (EC50) under these fixed reaction conditions with 95% confidence interval shown. Each data point represents the mean and standard deviation across three biological replicates. (c) Construct schematics for mammalian expression of A3A-INS2, A3A(E72A)-INS2, and A3A-SPL2 variants used to determine the impact of optGFP insertion on the DNA damage response in HEK293T cells (d) HEK293T cells were transfected with catalytically inactive mutant A3A(E72A)-INS2, A3A-INS2, or co-transfected with A3A-SPL2N and A3A-SPL2C. After transfection, cells were stained for γH2AX and sorted for both GFP and γH2AX expression. The bar plot depicts frequency of GFP+ or GFP+/γH2AX+ cells after transfection of HEK293T cells with the indicated constructs, corresponding to the representative histogram shown in Fig. 1e. The mean, standard deviation, and individual observations from independent biological replicated are shown. (e) Representative immunofluorescent images of transfected U2OS cells are shown, corresponding to Fig. 1e. DAPI stain highlights the nucleus, GFP staining shows insert expression or split reconstitution, and γH2AX serves as a marker of active A3A-mediated DNA damage. A 10 µm scale bar is provided. Representative experiments were repeated independently three times and the results were reproducible.
Extended Data Fig. 3. Intact and split-engineered base editor constructs.
(a) Parent construct schematics for intact BE4max scaffold editors with AID’, evoA1, and A3A. (b) Construct schematics for split-engineered seBE-T2A editors with AID’, evoA1, and A3A. Constructs were created by insertion of a cassette that splits the intact deaminase into two fragments, separated by a self-cleaving T2A peptide. At the bottom is the alternative seBE construct strategy, where the T2A fragment is replaced with an internal ribosome entry sequence (IRES) that leads to expression of two independently translated split protein fragments (denoted as seBEN and seBEC) with no need for protease processing.
Extended Data Fig. 4. Split-engineered base editors with A3A enable small-molecule-controlled editing.
Editing efficiency is evaluated in a d2gfp-HEK293T cell line. The presence of d2gfp-targeting sgRNA can introduce a stop codon (Q158*) and abrogate fluorescence to generate GFPoff cells, which can be tracked by either flow cytometry or deep-sequencing. Two-sided Mann-Whitney test was performed on all comparisons between means in this figure (n.s., not significant; *p ≤ 0.05, exact p-values provided as statistical source data files). (a) Representative flow cytometry histograms associated with transfection of intact or seBE-T2A constructs with or without rapamycin. (b) Mean, standard deviation and individual data points shown for quantification of GFPoff cells by flow cytometry (c) Left—deep sequencing results demonstrating C to T conversion efficiency at the target cytosine. The mean, standard deviation and three biological replicates are shown. Fold-change (FC) is the ratio of mean values for the higher versus lower condition. Right—editing footprints across the d2gfp locus for A3A-BE4max and A3A-seBE-T2A in the absence or presence of rapamycin. The numbers mark the distance from the PAM site. The target cytosine base is noted with a blue arrow. Data represent position-wise averages of three biological replicates, with individual replicate data provided in Supplementary Table 1. (d) sgRNA-independent off-target genomic editing in cells transfected with intact A3A-BE4 or A3A-seBE-IRES and sgRNA targeting EXM1, along with dSaCas9-sgRNA targeting a different locus. Left—EMX1 on-target editing. Right—off-target editing at the locus opened by dSaCas9. The mean, standard deviation, and three biological replicates are shown. Editing footprints for each locus and replicates are provided in Supplementary Fig. 4. (e) Quantification of γH2AX upon base editor expression with or without rapamycin. HEK293T cells were transfected with an empty vector (pcDNA-EV), catalytically inactive mutant A3A(E72A)-BE4max, A3A-seBE-T2A, A3A-seBE-IRES, intact A3A-BE4max, or isolated A3A domain (pcDNA-A3A) and an EMX1-targeting sgRNA expressing GFP. Cells were either maintained with or without rapamycin and then stained for γH2AX. Shown are the fraction of GFP and γH2AX positive cells. The mean, standard deviation, and three biological replicates are shown. individual observations are shown (n = 3). Two-sided Mann-Whitney test was performed (n.s., not significant; *p ≤ 0.05, exact p-values provided as statistical source data files).
Extended Data Fig. 5. Base editing efficiency in human cells at diverse loci and off-target sites.
HEK293T cells were untreated or transfected with intact BE4max or seBE constructs in the absence or presence of rapamycin. C or G describes whether the coding (C) or non-coding (G) strand cytosine is targeted, respectively, with the subscript denoting the position of the sgRNA targeted by the base editor. Shown are experiments with control constructs and (a) evoA1-seBE-T2A (corresponding to Fig. 3b and Fig. 4b), (b) evoA1-seBE-IRES (corresponding to Fig. 3c and Fig. 4b), and (c) AID’-seBE-IRES (corresponding to Fig. 3c and Fig. 4b). Bars indicate mean and error bars indicate standard deviations of n = 3 biological replicates. Complete editing footprints for each locus and replicate in (a-c) are provided in Supplementary Figs. 1-3. (d) Western blot of cells transfected with evoA1-based constructs in the absence and presence of rapamycin. The Cas9 antibody probes the intact editor and the seBEC fragment (which are of similar size); Hsp90 antibody serves as a loading control; c-myc antibody probe the N-terminal tag of the seBEN fragment. Representative experiment was repeated independently two times and the results were reproducible.
Extended Data Fig. 6. Split engineered base editors show low transcriptome-wide C to U mutations.
Total RNA was analyzed using the RADAR pipeline (RNA-editing Analysis-pipeline to Decode All twelve-types of RNA-editing events). The unique edits for each sample were cataloged by removing edits contained in either of the sgRNA-only samples. Left—shown the pie charts indicating the type of edit in each of the three independent replicates. Right—mean fractions of specific edits across the three replicates are provided.
Extended Data Fig. 7. Lentiviral constructs for intact and split-engineered base editor editing in K562 cells.
(a) Lentiviral construct schematics for intact BE4 max, sgRNA-only, seBEC and seBEN+sgRNA are shown. (b) Quantification of GFPoff cells by flow cytometry for cells with EMX1-targeting sgRNA along with intact BE4max or seBE-IRES. Cells were either treated with no rapamycin or with rapamycin (25 nM) added at either day 3 or day 5 (marked with arrow) and then maintained continuously. Mean and standard deviation are noted, with individual data points shown (n = 3).
Extended Data Fig. 8. Gating strategies for flow cytometry plots.
For all samples, viable cells were selected based on a forward scatter vs side scatter plot. Viable cells were then analyzed by the gating strategies shown. (a) Gating strategy for intact, inserted, and split A3A constructs shown in Figure 1e and Extended Data Figure 2d. (b) Gating strategy for base editing constructs generating GFPOFF cells shown in Figure 2c and Extended Data Figure 4a. Left - After sorting for live cells, a second gate for efficiently transfected cells was applied. Right – the GFPOFF population was then quantified. (c) Gating strategy for AID’- and A3A-based BE4max and seBE4max constructs shown in Figure 4d and Extended Data Figure 4e.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to M. Weitzman and K. Musunuru for helpful discussions. This work was in part supported by the Penn Center for Genomic Integrity (PCGI) and the US National Institutes of Health (NIH) through R01-GM138908 and R01-HG010646 (to R.M.K.). K.N.B is an NSF Graduate Research Fellow. N.H.E. was supported by NIH T32-GM007170 and F30-HG011578. A.M.G. was supported by NIH K08-CA212299.
Footnotes
COMPETING FINANCIAL INTERESTS
K.N.B, J.S., and R.M.K through the University of Pennsylvania have filed a patent application on aspects of this work. R.M.K. is on the Scientific Advisory Board for Life Edit, Inc.
REFERENCES
- 1.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Rees HA & Liu DR Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet. 19, 770–788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jinek M et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaudelli NM et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Komor AC et al. Improved base excision repair inhibition and bacteriophage Mu Gam protein yields C:G-to-T:A base editors with higher efficiency and product purity. Sci. Adv. 3, eaao4774 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu LD et al. Intrinsic Nucleotide Preference of Diversifying Base Editors Guides Antibody Ex Vivo Affinity Maturation. Cell. Rep. 25, 884–892.e3 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Ma Y et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat. Methods 13, 1029–1035 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Green AM & Weitzman MD The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities. DNA Repair (Amst) 83, 102700 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feng Y, Seija N, D I Noia JM & Martin A AID in Antibody Diversification: There and Back Again. Trends Immunol. 41, 586–600 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siriwardena SU, Chen K & Bhagwat AS Functions and Malfunctions of Mammalian DNA-Cytosine Deaminases. Chem. Rev. 116, 12688–12710 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns MB et al. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature 494, 366–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbiani DF & Nussenzweig MC Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu. Rev. Pathol. 8, 79–103 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Roberts SA et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 45, 970–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim D et al. Genome-wide target specificities of CRISPR RNA-guided programmable deaminases. Nat. Biotechnol. 35, 475–480 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Zuo E et al. Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou C et al. Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature 571, 275–278 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Kim D, Kim DE, Lee G, Cho SI & Kim JS Genome-wide target specificity of CRISPR RNA-guided adenine base editors. Nat. Biotechnol. 37, 430–435 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Grunewald J et al. Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base editors. Nature 569, 433–437 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S et al. Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Grunewald J et al. CRISPR DNA base editors with reduced RNA off-target and self-editing activities. Nat. Biotechnol. (2019). [DOI] [PMC free article] [PubMed]
- 22.Rees HA, Wilson C, Doman JL & Liu DR Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv. 5, eaax5717 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nuñez JK, Harrington LB & Doudna JA Chemical and Biophysical Modulation of Cas9 for Tunable Genome Engineering. ACS Chem. Biol. 11, 681–688 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Lim SA & Wells JA Split enzymes: Design principles and strategy. Methods Enzymol. 644, 275–296 (2020). [DOI] [PubMed] [Google Scholar]
- 25.Gangopadhyay SA et al. Precision Control of CRISPR-Cas9 Using Small Molecules and Light. Biochemistry 58, 234–244 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zetsche B, Volz SE & Zhang F A split-Cas9 architecture for inducible genome editing and transcription modulation. Nat. Biotechnol. 33, 139–142 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer LM, Zhang D, Rogozin IB & Aravind L Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 39, 9473–9497 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ear PH & Michnick SW A general life-death selection strategy for dissecting protein functions. Nat. Methods 6, 813–816 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Mok BY et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 583 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qiao Q et al. AID Recognizes Structured DNA for Class Switch Recombination. Mol. Cell 67, 361–373.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gajula KS et al. High-throughput mutagenesis reveals functional determinants for DNA targeting by activation-induced deaminase. Nucleic Acids Res. 42, 9964–9975 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M, Yang Z, Rada C & Neuberger MS AID upmutants isolated using a high-throughput screen highlight the immunity/cancer balance limiting DNA deaminase activity. Nat. Struct. Mol. Biol. 16, 769–776 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cabantous S, Terwilliger TC & Waldo GS Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23, 102–107 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Kohli RM et al. A portable hotspot recognition loop transfers sequence preferences from APOBEC family members to activation-induced cytidine deaminase. J. Biol. Chem. 284, 22898–22904 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang M, Rada C & Neuberger MS A high-throughput assay for DNA deaminases. Methods Mol. Biol. 718, 171–184 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Zong Y et al. Efficient C-to-T base editing in plants using a fusion of nCas9 and human APOBEC3A. Nat. Biotechnol. (2018). [DOI] [PubMed]
- 37.Gehrke JM et al. An APOBEC3A-Cas9 base editor with minimized bystander and off-target activities. Nat. Biotechnol. 36, 977–982 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landry S, Narvaiza I, Linfesty DC & Weitzman MD APOBEC3A can activate the DNA damage response and cause cell-cycle arrest. EMBO Rep. 12, 444–450 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voß S, Klewer L & Wu YW Chemically induced dimerization: reversible and spatiotemporal control of protein function in cells. Curr. Opin. Chem. Biol. 28, 194–201 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Koblan LW et al. Improving cytidine and adenine base editors by expression optimization and ancestral reconstruction. Nat. Biotechnol. 36, 843–846 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thuronyi BW et al. Continuous evolution of base editors with expanded target compatibility and improved activity. Nature biotechnology 37, 1070–1079 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mak CH, Pham P, Afif SA & Goodman MF A mathematical model for scanning and catalysis on single-stranded DNA, illustrated with activation-induced deoxycytidine deaminase. J. Biol. Chem. 288, 29786–29795 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai SQ et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–197 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doman JL, Raguram A, Newby GA & Liu DR Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol. (2020). [DOI] [PMC free article] [PubMed]
- 45.Jin S et al. Rationally Designed APOBEC3B Cytosine Base Editors with Improved Specificity. Mol. Cell 79, 728–740.e6 (2020). [DOI] [PubMed] [Google Scholar]
- 46.Wang L et al. Eliminating base-editor-induced genome-wide and transcriptome-wide off-target mutations. Nat. Cell Biol. 23, 552–563 (2021). [DOI] [PubMed] [Google Scholar]
- 47.Chen H et al. The mTOR inhibitor rapamycin suppresses DNA double-strand break repair. Radiat. Res. 175, 214–224 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearce S & Tucker CL Dual Systems for Enhancing Control of Protein Activity through Induced Dimerization Approaches. Adv. Biol. (Weinh) 5, e2000234 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X et al. Cas12a Base Editors Induce Efficient and Specific Editing with Low DNA Damage Response. Cell. Rep. 31, 107723 (2020). [DOI] [PubMed] [Google Scholar]
- 50.Tsai SQ et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 32, 569–576 (2014).stylefix [DOI] [PMC free article] [PubMed] [Google Scholar]
METHODS REFERENCES
- 51.Schutsky EK et al. Nondestructive, base-resolution sequencing of 5-hydroxymethylcytosine using a DNA deaminase. Nat. Biotech. 36, 1083–1090 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schutsky EK, Nabel CS, Davis AKF, DeNizio JE & Kohli RM APOBEC3A efficiently deaminates methylated, but not TET-oxidized, cytosine bases in DNA. Nucleic Acids Res. 45, 7655–7665 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mol CD et al. Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA. Cell 82, 701–708 (1995). [DOI] [PubMed] [Google Scholar]
- 54.Lee M et al. Engineered Split-TET2 Enzyme for Inducible Epigenetic Remodeling. J. Am. Chem. Soc. 139, 4659–4662 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y et al. A TFIID-SAGA Perturbation that Targets MYB and Suppresses Acute Myeloid Leukemia. Cancer. Cell. 33, 13–28.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zafra MP et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–893 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tarumoto Y et al. LKB1, Salt-Inducible Kinases, and MEF2C Are Linked Dependencies in Acute Myeloid Leukemia. Mol. Cell 69, 1017–1027.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi J et al. Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol. 33, 661–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clement K et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol. 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matsuda T & Cepko CL Controlled expression of transgenes introduced by in vivo electroporation. Proc. Natl. Acad. Sci. U. S. A. 104, 1027–1032 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput RNA sequencing data will be deposited at the Gene Expression Omnibus (GEO) database (accession number pending). Individual amplicon sequencing data is available as Supplementary Information and raw reads will be available upon request. Novel plasmids used in this study will be made available from Addgene. Nucleic acid sequences of all constructs used in this study are provided in a note at the end of the Supplementary Information.