Abstract
Aluminum (Al)-tolerant tobacco cell line ALT301 derived from SL (wild-type) hardly exhibits Al-triggered reactive oxygen species (ROS) compared with SL. Molecular mechanism leading to this phenotype was investigated comparatively with SL. Under normal growth condition, metabolome data suggested the activation of glycolysis and lactate fermentation but the repression of the tricarboxylic acid (TCA) cycle in ALT301, namely aerobic fermentation, which seemed to be transcriptionally controlled partly by higher expression of genes encoding lactate dehydrogenase and pyruvate dehydrogenase kinase. Microarray and gene ontology analyses revealed the upregulation of the gene encoding related to APETALA2.3 (RAP2.3)-like protein, one of the group VII ethylene response factors (ERFVIIs), in ALT301. ERFVII transcription factors are known to be key regulators for hypoxia response that promotes substrate-level ATP production by glycolysis and fermentation. ERFVIIs are degraded under normoxia by the N-end rule pathway of proteolysis depending on both oxygen and nitric oxide (NO), and NO is produced mainly by nitrate reductase (NR) in plants. In ALT301, levels of the NR gene expression (NIA2), NR activity and NO production were all lower compared with SL. Consistently, the known effects of NO on respiratory pathways were also repressed in ALT301. Under Al-treatment condition, NO level increased in both lines but was lower in ALT301. These results suggest that the upregulation of the RAP2.3-like gene and the downregulation of the NIA2 gene and resultant NO depletion in ALT301 coordinately enhance aerobic fermentation, which seems to be related to a higher capacity to prevent ROS production in mitochondria under Al stress.
Keywords: Aerobic fermentation, Aluminum-tolerant tobacco cell line, Group VII ethylene response factors, Nitrate reductase, Nitric oxide, Reactive oxygen species
Introduction
Aluminum (Al) is the third most abundant element in the earth’s crust and is dissolved in soil water as free ions under acidic conditions. Acidic soils are distributed in arable lands worldwide, and Al ions cause root growth inhibition especially in young seedlings, which are thought to be a major constraint in crop productivity in acidic soils (Kochian 1995).
Al-tolerant mechanisms in plants have been studied extensively, which includes the exclusion of Al ions from root apices based on Al-activated exudation of organic acids (Ma 2007, Ryan et al. 2011, Kochian et al. 2015). Molecular details of the exclusion mechanism have been well studied especially in wheat (Triticum aestivum) and Arabidopsis (Arabidopsis thaliana). In these species, Al-activated malate transporter (ALMT) plays a major role to tolerate Al toxicity by excluding Al ions with malate (Sasaki et al. 2004, Hoekenga et al. 2006). Although wheat ALMT1 is constitutively expressed (Sasaki et al. 2004, 2006), Arabidopsis ALMT1 is induced by Al or low pH together with a series of genes related to Al and proton (H+) tolerances, which are regulated by a transcription factor STOP1 (sensitive to proton Rhizotoxicity1) (Iuchi et al. 2007, Sawaki et al. 2009). Compared to wheat and Arabidopsis, rice (Oryza sativa) is more tolerant to Al and seems to have multiple genes involved in internal and external detoxification of Al, which are upregulated under Al stress by a transcription factor ART1 (Al resistance transcription Factor1) (Yamaji et al. 2009). ART1 is a zinc finger protein and a functional homolog of STOP1.
Non-chlorophyllic cultured tobacco cell lines serve as a model system of actively growing cells at root apex and have been investigated the responses to Al. Our study using cell lines of SL (derived from cv. Samsun) and BY-2 (derived from cv. bright yellow) revealed three types of toxic mechanisms of Al leading to cell death, involving the plasma membrane, mitochondria and the vacuole, respectively (Yamamoto 2019). The plasma membrane pathway was observed in SL treated with Al in nutrient medium, where enhancement of the iron (Fe)-mediated lipid peroxidation by Al leads to cell death. The mitochondrial pathway was observed in SL treated with Al in a simple solution containing calcium (Ca) and sucrose (Ca-sucrose solution), where Al causes mitochondrial dysfunction accompanying respiration inhibition, ATP depletion and the production of reactive oxygen species (ROS). The same symptoms were also observed at root apices in pea (Pisum sativum) (Yamamoto et al. 2002, Kobayashi et al. 2004). The vacuolar pathway was observed in BY-2 (but not in SL) treated with Al in a Ca-sucrose solution, where Al causes the upregulation of the VPEs encoding vacuolar processing enzymes leading to the collapse of vacuole. The same responses were observed at root apices in tobacco (cv. bright yellow) (Kariya et al. 2013, 2018).
In order to elucidate the Al-tolerant mechanism using the tobacco cell system, SL cells were mutagenized with ethyl methane sulfonate, and then, Al-tolerant cells were screened from the survivors after Al treatment in a Ca-sucrose solution (Devi et al. 2001). One of the Al-tolerant cell lines, ALT301, was investigated for Al responses comparatively with the parental line SL (wild type). The degrees of Al accumulation, Al-triggered citrate efflux (an exclusion mechanism of Al in tobacco) and callose production were similarly observed in both lines. However, ALT301 had constitutively higher amounts of ascorbate and glutathione (GSH) and exhibited cross-tolerance to hydrogen peroxide (H2O2), ferrous iron (Fe2+) and copper (Cu+). Taken together, it was concluded that an intracellular mechanism leading to higher antioxidant status seems to be related to tolerance to Al as well as H2O2, Fe2+ and Cu+ in ALT301 (Devi et al. 2001, 2003). Furthermore, it was found that Al-triggered ROS production was hardly observed in ALT301 (Yamamoto et al. 2002, Abdel-Basset et al. 2010), and that the ATP content was higher in ALT301 under treatment conditions without (control) or with Al, compared with SL (Yamamoto et al. 2002). Thus, ALT301 seems to have acquired some intracellular mechanisms to prevent the ROS production and ATP depletion associated with mitochondrial dysfunction.
In this study, in order to elucidate molecular mechanisms related to Al tolerance in ALT301, we first performed metabolome and transcriptome analyses in SL and ALT301 under normal growth conditions, and then, the findings were confirmed under Al-treatment conditions. We report several lines of evidence suggesting the constitutive enhancement of aerobic fermentation in ALT301. In addition, we found both the upregulation of the RAP2.3-like gene encoding ERFVII transcription factor and the downregulation of the NIA2 gene encoding nitrate reductase (NR) in ALT301 under normal growth and Al-treatment conditions, which could be primary events leading to the metabolic, transcriptional and physiological differences observed between the lines. Finally, we will discuss the physiological role of aerobic fermentation in preventing ROS production under Al stress.
Results
Growth patterns of SL and ALT301 under normal growth conditions
Supplementary Fig. S1 indicates growth curves of SL and ALT301 during 14-day culture under normal growth condition (see Materials and Methods). These cell lines exhibited different growth patterns. SL grew slowly for 3 d (lag phase), then exponentially for 6 d (log phase) and then reached a stationary phase. Compared with SL, ALT301 exhibited a longer lag phase (4 d) and then a longer log phase (10 d) at lower growth rates. ALT301 did not exhibit a stationary phase but kept logarithmic growth until day 14 even under high cell density.
Metabolite differences between SL and ALT301 under normal growth conditions
Metabolome analysis.
Metabolome analysis by use of our capillary electrophoresis-mass spectrometry (CE-MS) system identified 35 cationic metabolites and 27 anionic metabolites in SL and ALT301 under normal growth condition (Supplementary Table S1). The content values of the metabolites were fluctuated among three biological replicates in both cell lines; therefore, the differences of the mean values between the lines, if any, were not statistically significant in most of the metabolites (the results of statistical analysis, see Supplementary Table S1). However, under this limitation, we tried to compare the metabolites between the cell lines by the relative content values over SL. The metabolites were aligned in the order of their relative values from the highest to the lowest (Supplementary Fig. S2), and the metabolites indicating at least 2-fold changes were focused, as an arbitrary guide suggesting the difference between the lines. In cationic metabolites (Supplementary Fig. S2A), the metabolite of the highest ratio value was GSH, which consists with our previous finding indicating a higher GSH content in ALT301, compared with SL (Devi et al. 2003). The metabolite of the third highest value was allantoin, while the metabolite of the lowest value was allantoic acid, resulting a higher allantoin/allantoic acid ratio in ALT301. Allantoin often accumulates in stressed plants and has been reported to enhance tolerance to abiotic stresses by various mechanisms, including the reduction of ROS production via the enhancement of the activation of antioxidant enzymes (Nourimand and Todd 2016). In anionic metabolites (Supplementary Fig. S2B), some metabolites indicating higher ratio values were associated with glycolysis (fructose 1, 6-bisphosphate [FBP], lactate [Lac], pyruvate [PA]), while some metabolites indicating lower ratio values were associated with the tricarboxylic acid (TCA) cycle (fumarate [FA], malate [MA], citrate [CA], cis-aconitate [ACA]).
These findings prompted us to compare the intermediates of glycolysis and the TCA cycle in SL and ALT301 (the intermediates in central carbon metabolic pathways, see Supplementary Fig. S3). Fig. 1 depicts the contents of metabolites associated with glycolysis and the TCA cycle, together with the relative content value over SL of each metabolite. Concerning glycolysis, seven glycolysis intermediates and Lac were detected. Among them, three (FBP, PA, Lac) were higher (>2-fold), and all the rest were more or less higher (glucose 1-phosphate [G1P], glucose 6-phosphate [G6P], fructose 6-phosphate [F6P], 3-phosphoglycerate [3PGA]) or the same (glyceraldehyde 3-phophate [GA3P]), compared with SL (Fig. 1A). On the contrary, among the six TCA cycle intermediates detected, four intermediates (CA, ACA, FA, MA) were remarkably lower (≥5-fold) in ALT301, although two intermediates (2-oxoglutarate [2OG], succinate [SuA]) were higher (Fig. 1B). These results suggest that glycolysis flux is activated toward Lac fermentation, but that glycolysis flux toward the TCA cycle is repressed, in ALT301 under normal growth conditions.
Fig. 1.
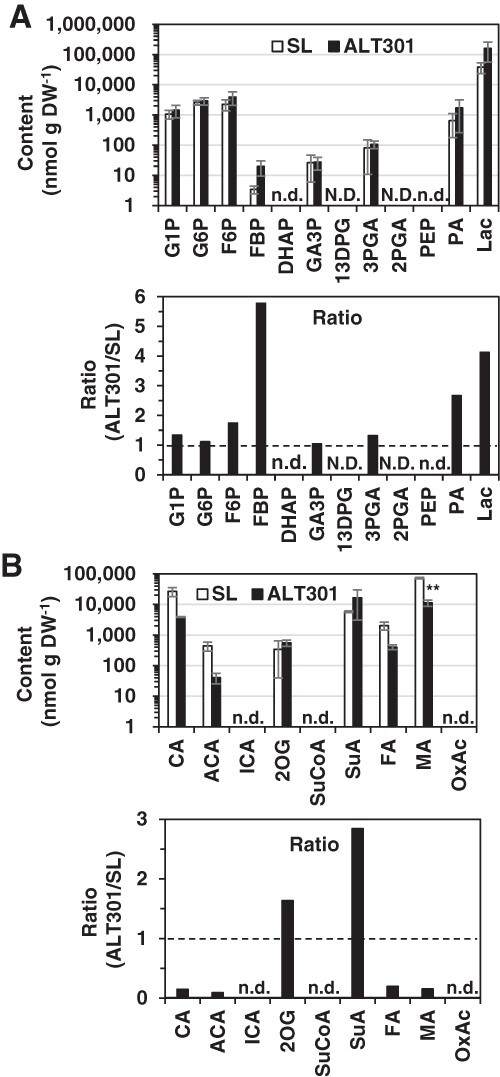
Comparison of contents of the metabolites associated with glycolysis (A) and the TCA cycle (B) between SL and ALT301 under normal growth condition. On the abscissa, the metabolites are aligned according to metabolic pathways (see Supplementary Fig. S3). Values are the content (nmol g dray weight [DW]−1; mean ± SE, n = 3 from three independent experiments) (top) and the ratio relative to SL (bottom) in which broken line indicates the ratio value of 1 (values and the results of statistical analysis, see Supplementary Table S1). Statistically significant difference between SL and ALT301 is indicated with asterisk (**P < 0.01, Welch’s t-test). N.D. means ‘not determined’ and n.d. means ‘not detected’. Abbreviations: DHAP, Dihydroxyacetone phosphate; 13DPG, 1,3-Diphospho-D-glycerate; 2PGA, 2-Phosphoglycerate; other metabolites, see Supplementary Table S1.
Pyridine nucleotides.
Nicotinamide adenine dinucleotide (NAD[H]) and nicotinamide adenine dinucleotidephosphate (NADP[H]) are necessary for energy metabolism including fermentations; therefore, the contents of pyridine nucleotides in cells are examined. Compared with SL, the total content of NADH and that of NADPH were higher in ALT301 (1.4-fold and 1.1-fold over SL, respectively; Supplementary Fig. S4), although the NAD+/NADH ratio was lower (0.8-fold; Supplementary Fig. S4A), the NADP+/NADPH ratio was higher (1.6-fold; Supplementary Fig. S4B) in ALT301.
Transcriptional differences between SL and ALT301 under normal growth conditions
Genes related to glycolysis and fermentation.
With the limited information on cDNA sequence available for tobacco genes on the National Center for Biotechnology Information (NCBI) website and little information on paralogous genes encoding isozymes, we performed expression analysis of the genes related to glycolysis and fermentation. First, we focused on the genes associated with the mobilization of sugars into glycolysis, which included the gene encoding sucrose transporter (NtSUT1) and the genes related to the invertase pathway (HXK [hexokinase]) and the sucrose synthase pathway (SUSY [sucrose synthase], FRK [fructokinase]) (see Supplementary Fig. S3). Fig. 2A depicts real-time reverse transcription polymerase chain reaction (RT-PCR) data indicating the upregulated gene (SUSY) and the downregulated genes (NtSUT1-L, HXK6, FRK2) in ALT301, compared with SL. The sucrose synthase pathway has been proposed to consume less energy than the invertase pathway (Huber and Akazawa 1986, Ricard et al. 1998, Stein and Granot 2019). Thus, higher expression of the SUSY seems to be beneficial to energy metabolism in ALT301. Enzyme activity of SUSY was also significantly higher in ALT301 (6.19 ± 0.49 nmol mg protein−1 min−1) than that in SL (3.96 ± 0.38 nmol mg protein−1 min−1) (mean ± SE, n = 6, P < 0.01). Concerning the genes related to glycolysis, we focused on the gene encoding pyruvate kinase (PK) that catalyzes one of the two steps for substrate-level ATP production in glycolysis (see Supplementary Fig. S3). ALT301 exhibited higher expression of PK than SL (Fig. 2A).
Fig. 2.
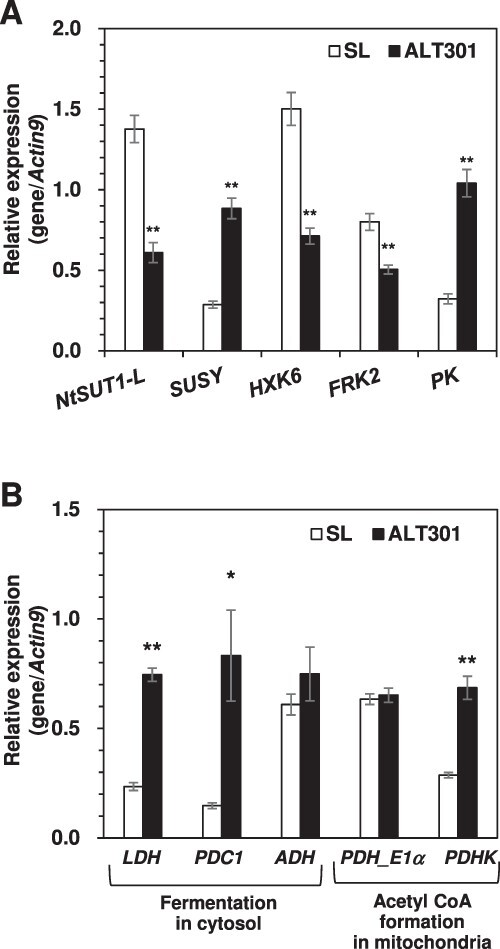
Expression of the genes related to glycolysis (A) and fermentation (B) in SL and ALT301 under normal growth condition. Real-time RT-PCR was performed in (A) with the genes encoding sucrose transporter (NtSUT1-L), the enzymes associated with the mobilization of sucrose into glycolysis (SUSY, HXK6 and FRK2), and an enzyme associated with ATP production in glycolysis (PK), and in (B) with the genes encoding the enzymes associated with three steps from pyruvate to Lac (LDH), ethanol (PDC1, ADH) and AcCoA (PDH_E1α, PDHK), respectively. The internal control was the Actin9 gene. Values are the mean ± SE (n = 5 from five independent experiments). Significant differences between SL and ALT301 are indicated with asterisks (*P = 0.05, **P = 0.01, Welch’s t-test).
Fig. 2B indicates the expression levels of the genes functioning at the branching point from pyruvate to either fermentations (LDH encoding lactate dehydrogenase; PDC1 encoding pyruvate decarboxylase; ADH encoding alcohol dehydrogenase) or the TCA cycle (PDH_E1α encoding pyruvate dehydrogenase E1α subunit that plays a key role in the function of the PDH complex; PDHK encoding PDH kinase that acts to inactivate PDH by phosphorylation) in SL and ALT301 under normal growth condition. The expression of LDH was higher in ALT301. The expression of PDC1 was also higher in ALT301, but that of ADH was similar in both lines. The expression level of PDH_E1α was similar in both lines, while the expression of PDHK was higher in ALT301, suggesting that the higher PDHK expression and resultant higher PDHK activity might lead to lower PDH activity in ALT301. In fact, ALT301 exhibited constitutively lower PDH activity under Al-treatment condition (see below ‘Enzyme activities related to pyruvate metabolism’). These results suggest that the metabolic shift from the TCA cycle to Lac fermentation in ALT301 is transcriptionally controlled by higher expressions of LDH and PDHK.
Transcriptome analyses.
Microarray analysis and gene ontology (GO) enrichment analysis were performed in order to reveal transcriptional differences between SL and ALT301 under normal growth condition and to find out regulation mechanism(s) of the metabolic and transcriptional changes in ALT301.
Using our microarray data (Supplemental Data Set 1), we identified 1,303 probes (680 upregulated and 623 downregulated with known functions) with changes of at least 2-fold in their differential transcript levels with significance (P < 0.05) in ALT301, compared with SL (note that some probes within the 1,303 probes carry the same TIGR ID and/or UniGene ID).
Within the limits of the probes on the Agilent microarray system, we surveyed the differentially expressed genes related to glycolysis, the TCA cycle and antioxidant systems (Supplementary Table S2). Concerning glycolysis, six genes encoding SUSY, INV, FRK, phosphoglyceromutase (catalyzing the conversion of 3-phosphoglycerate [3PGA] to 2-phosphoglycerate [2PGA]), PDC and ADH were upregulated in ALT301, while two genes encoding hexose transporter and HXK were downregulated. The expression patterns of some genes (SUSY, PDC, HXK) were consistent with our real-time RT-PCR data (Fig. 2). Concerning the TCA cycle, two genes encoding PDH and aconitase (catalyzing the conversion of CA to isocitrate [ICA] via aconitate) were upregulated (2.0-fold). On the other hand, one gene encoding 2-OG dehydrogenase E2 subunit (a part of 2-OG dehydrogenase complex catalyzing the conversion of 2-OG to succinyl-CoA [SuCoA]) was strongly downregulated (8.5-fold) in ALT301, which might be related to a slightly higher content of 2-OG in ALT301 (Fig. 1B).
Finally, concerning antioxidant systems (Halliwell and Gutteridge 2007), we found the upregulation of many genes encoding a wide range of antioxidant enzymes/proteins in ALT301 (peroxidases, catalase, superoxide dismutases, probable glutathione-S-transferase [GST], dehydroascorbate reductase, thioredoxin, feredoxin, glutaredoxin and peroxiredoxin), but also found the downregulation of several genes (peroxidases, GSH reductase, GSH synthetase, probable GST and thioredoxin).
Gene ontology enrichment analysis on the upregulated and the downregulated genes in ALT301.
Using our microarray data (Supplemental Data Set 1), we also identified 395 probes (256 upregulated and 139 downregulated with known functions) with changes of at least 3-fold in their differential transcript levels with significance (P < 0.01) in ALT301 over SL. The physiological and functional aspects of these differentially expressed genes were assessed by GO enrichment analysis using agriGO v2. For the upregulated genes, 45 GO terms were significantly enriched (Supplementary Table S3A). Among, 40 terms of the ‘biological process (P)’ category were further classified into four groups (I–IV), based on their hierarchical networks shown in Supplementary Fig. S5A. In group I, 12 significant GO terms were finally integrated into two GO terms at the bottom that were ‘defense response, incompatible interaction’ and ‘defense response to fungus’. Likewise, 22 terms in group II, 4 terms in group III and 3 terms in group IV were integrated into GO terms, ‘ethylene mediated signaling pathway’, ‘cellular amino acid derivative biosynthetic process’ and ‘cell wall modification’, respectively, at the bottoms (Supplementary Fig. S5A).
The upregulated genes annotated to these five GO terms located at the bottoms of the hierarchical network are listed in Supplementary Table S4A. In this list, we found several interesting genes related to ethylene signaling pathway such as the genes encoding chitinases (group I and II), PR47 (group II) and 1-amino cyclopropane-1-carboxylate oxidase (ACO) (group III) (more details, see below ‘Genes related to ethylene signaling pathway’).
Supplementary Table S4B lists the upregulated genes annotated to three significant GO terms of ‘molecular function (F)’ category and two significant GO terms of the ‘cellular component (C)’ category (see Supplementary Table S3A), which included the genes encoding chitinases, β-1,3-glucanase and cytochrome P450 (F category) and peroxidase and pectin methylesterase (C category).
On the contrary, for the downregulated genes in ALT301, only two GO terms (‘photosynthesis’ of the P category and ‘protein dimerization activity’ of the F category) were significantly enriched (Supplementary Table S3B). Supplementary Table S4C lists the downregulated genes annotated to these GO terms, which included the genes encoding chlorophyll a/b binding protein (Cab40) and photosystem II 23 kDa polypeptide (P category) and cryptochrome1 and plastid division regulator MinD (F category).
Genes related to ethylene signaling pathway.
As shown in the hierarchical image (Supplementary Fig. S5A), group II consisted of 22 GO terms that represented a half of the significant GO terms enriched for the upregulated genes in ALT301. These 22 GO terms were integrated into one GO term ‘ethylene mediated signaling pathway’, and this GO term was associated with four genes encoding three chitinases (basic chitinase, known as CHN48 [gene accession: S44869]; endo chitinase, known as CHN50 [M15173]; acidic chitinase, known as pathogenesis-related [PR]-P [X51426]) and PR47 (gene accession: AF154656) (Supplementary Table S4A, Group II). PR47 is also known as related to Apetala2.3 (RAP2.3)-like protein (gene accession: XM_016591614). The upregulation of these genes in ALT301 was confirmed by real-time RT-PCR (Fig. 3). In Arabidopsis, RAP2.3 is one of the group VII ethylene response factor (ERFVII) transcription factors. Arabidopsis encodes five ERFVIIs, consisting of three constitutively-expressed genes (RAP2.2, RAP2.3, RAP2.12) and two hypoxia-inducible genes (HYPOXIA RESPONSIVE ERF1 [HRE1], HRE2). ERFVIIs are involved in the adaptation to hypoxia redundantly by regulating a similar set of hypoxia response genes (e.g. SUS encoding sucrose synthase, PDC, ADH) that promote substrate-level ATP production by glycolysis through fermentative metabolism of pyruvate in Arabidopsis (Gibbs et al. 2011, 2015, Giuntoli and Perata 2018, Fukao et al. 2019).
Fig. 3.
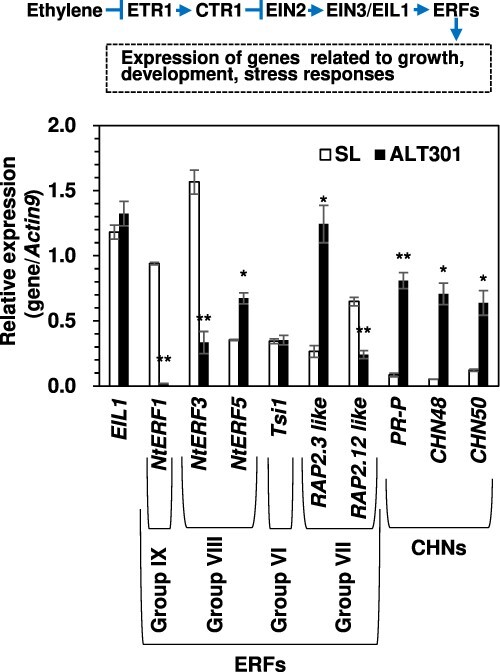
Expression of the genes encoding transcription factors (EIL, ERFs) and chitinases (CHNs) associated with ethylene signaling pathway in SL and ALT301 under normal growth condition. Gene expression analysis was performed by real-time RT-PCR. Values are the mean ± SE (n = 3–5 from three to five independent experiments, respectively). Significant differences between SL and ALT301 are indicated with asterisks (*P < 0.05, **P < 0.01, Welch’s t-test). Ethylene signaling pathway in Arabidopsis (Kazan 2015) is shown (top). Group names of the ERF genes are indicated, which is based on Rushton et al. (2008) and Nakano et al. (2006). Note that some of the ERF genes have several synonyms (see the National Center for Biotechnology Information [NCBI] database using the gene accessions listed in Supplementary Table S6). Abbreviations: ETR1, Ethylene Response1; CTR1, Constitutive Triple Response1; EIN2, Ethylene Insensitive2; EIN3, Ethylene Insensitive3; EIL1, Ethylene Insensitive-Like Protein1; ERF, Ethylene Response Factor. The gene Tsi1 encodes Tobacco stress-induced gene1.
Based on these lines of evidence, we compared the expression levels of the genes related to ethylene biosynthesis and ethylene signaling pathway in SL and ALT301. Concerning ethylene biosynthesis pathway, the expressions of ACSs encoding 1-aminocyclopropane-1-carboxylate synthase were either upregulated (ACS1) or downregulated (ACS2), while the expressions of ACOs were either similar (NtERF26) or upregulated (NtACO3) in ALT301, under normal growth condition (Supplementary Fig. S6A). However, the rate of ethylene production was found to be similar in SL and ALT301 (Supplementary Fig. S6B).
Concerning ethylene signaling pathway (see Fig. 3, top), expression of the EIL1 (Ethylene Insensitive-Like Protein1) was similar in both lines (Fig. 3). Among the ERF genes examined, two genes (NtERF5, RAP2.3-like) were upregulated and three genes (NtERF1, NtERF3, RAP2.12-like) were downregulated in ALT301, significantly, compared with SL.
These results suggest that the upregulation of the RAP2.3-like gene in ALT301 is neither due to an increase in ethylene evolution nor due to the enhancement of the ethylene signaling pathway.
Comparison of endogenous level of nitric oxide and nitric oxide-mediated processes between SL and ALT301 under normal growth conditions
Expression of the genes encoding NR, NR enzyme activity and the level of NO.
ERFVII proteins are the substrates of the N-end rule pathway of targeted proteolysis that is active in the presence of both molecular oxygen (O2) and nitric oxide (NO) (Gibbs et al. 2014). Therefore, under normoxia, if the upregulation of the RAP2.3-like and resultant increase in RAP2.3-like protein is involved in the metabolic shift to aerobic fermentation in ALT301, it seems to be also necessary to avoid the proteolysis of RAP2.3-like protein via the N-end rule pathway. In this context, one of the stabilization mechanisms could be a decline of NO production, and the NO production mainly depends on NR in most plants (Tejada-Jimenez et al. 2019). Real-time RT-PCR of two genes encoding NR in tobacco, NIA1 and NIA2, revealed the downregulation of NIA2 in ALT301 (37% of SL), but not NIA1, under normal growth condition (Fig. 4A). Consistently, NR activity in ALT301 was lower than that in SL (47% of SL) (Fig. 4B), and NO level was also lower in ALT301 (27% of SL) (Fig. 4C). These results suggest that the NO depletion in ALT301 is due to the downregulation of the NIA2 and resultant decrease in NR activity and that NO depletion in ALT301 could stabilize ERFVIIs (RAP2.3-like protein and others) under normoxia, compared with SL.
Fig. 4.
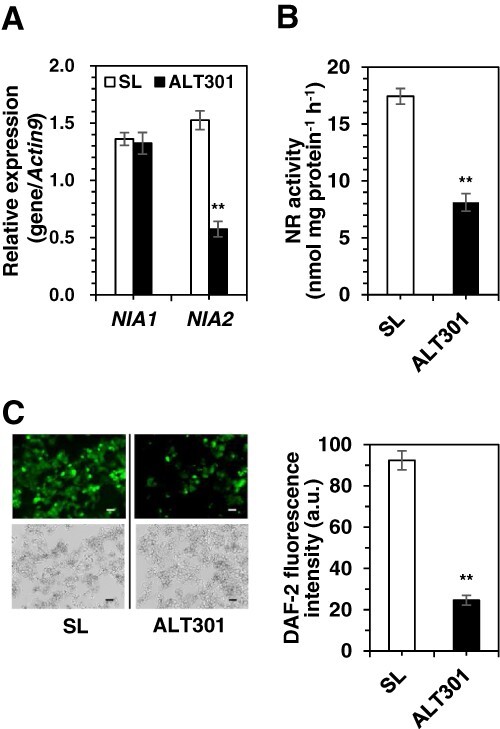
Expression of the genes (NIA1, NIA2) encoding nitrate reductase (NR) (A), NR activity (B), and the level of NO (C) in SL and ALT301 under normal growth condition. In (A), gene expression analysis was performed by real-time RT-PCR. In (A) and (B), significant differences between SL and ALT301 are indicated with asterisks (**P < 0.01, Welch’s t-test). Values are the mean ± SE (n = 6 from six independent experiments in A; n = 12 from four independent experiments in B). In (C), fluorescence image of NO in cells (top) obtained by staining with 4,5-diaminofluorescein diacetate (DAF-2 DA) and bright image of cells (bottom) are indicated. All the fluorescence images were obtained under the same exposure time (3 sec). The experiments were performed three times independently, and reproducible results were obtained. Here shows representative images. Bar indicates 100 μm. NO level was quantitatively determined as DAF-2 fluorescence intensity (mean gray value per cell area [150 μm x 150 μm]) (arbitrary unit). Significant differences between SL and ALT301 are indicated with asterisks (**P < 0.01 by Welch’s t-test). Values are the mean ± SE (n = 45 cell areas from three independent experiments).
On the other hand, another possible mechanism to stabilize ERFVII by escaping from the N-end rule pathway could be a decrease in exogenous oxygen supply to ALT301, compared with SL, under normal growth condition. However, it is less likely. First, both lines form small clumps but of similar sizes (Figs. 4C, 6A and 10) under suspension culture condition. Second, in order to supply oxygen, cells were cultured in an Erlenmeyer flask on a rotary shaker (see Materials and Methods). Under normal growth condition, the concentration of O2 in medium decreased with an increase in the cell density of SL and ALT301 to the same extent (Supplementary Fig. S7). Third, during a log phase, ALT301 exhibited a higher respiration rate (O2 uptake) than SL (see below ‘Respiration rate and expression of the genes encoding COXI and AOX’). Taken together, these findings suggest that both cell lines were grown under the condition of similar O2 supply.
Fig. 6.
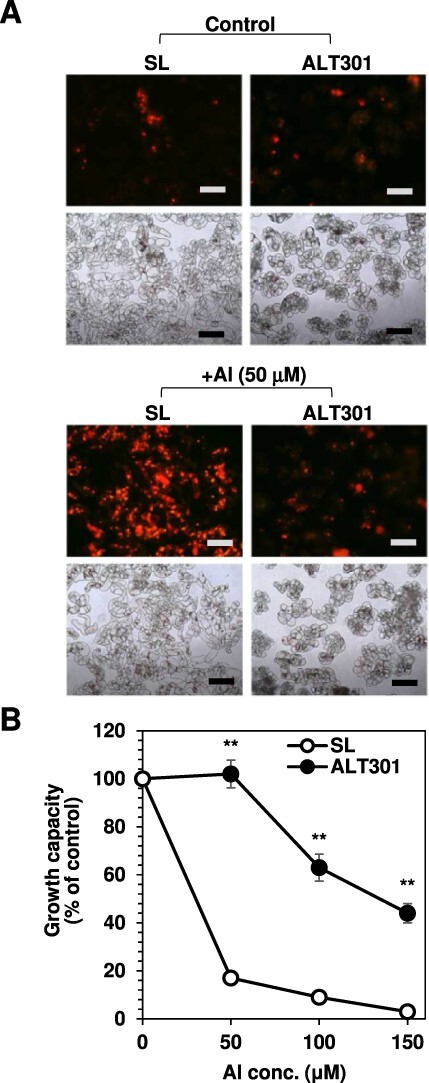
Comparison of ROS production (A) and growth capacity (B) between SL and ALT301 under Al-treatment condition. Cells were treated without (control) or with Al (+Al) for 18 h, and then ROS production and growth capacity were determined. In (A), fluorescence image of ROS production in cells (top) obtained by staining with dihydroethidium and bright image of cells (bottom) are indicated. All the fluorescence images were obtained under the same exposure time (1 sec). The experiments were repeated three times independently, and reproducible results were obtained. Here shows representative images. Bar indicates 200 μm. In (B), significant differences between SL and ALT301 are indicated with asterisks (**P < 0.01, Welch’s t-test). Values are the mean ± SE (n = 7 from seven independent experiments).
Fig. 10.
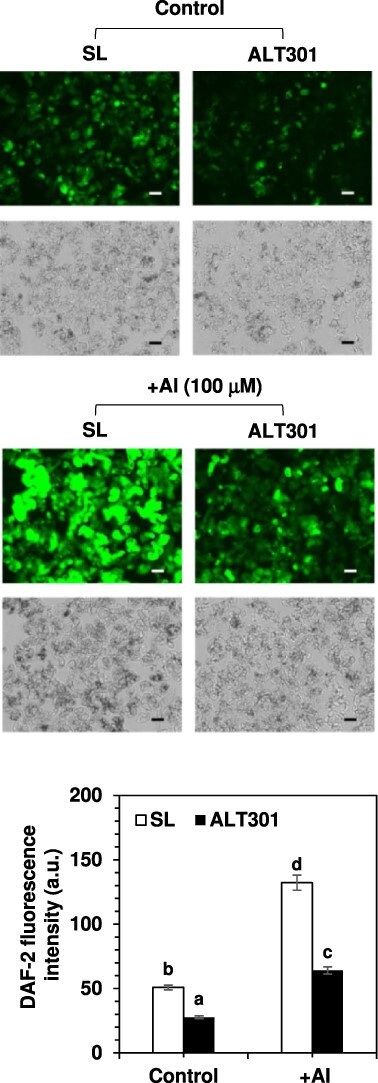
Comparison of NO production between SL and ALT301 under Al-treatment condition. Cells were treated without (control) or with 100 μM Al (+Al) for 18 h, and then NO production was determined. The fluorescence image of NO production in cells (top), which was obtained by staining with DAF-2 DA, is indicated with the bright image of cells (bottom). All the fluorescence images were obtained under the same exposure time (1/1.5 sec). The experiments were repeated three times independently, and reproducible results were obtained. Here shows representative images. Bar indicates 100 μm. NO level was quantitatively determined as DAF-2 fluorescence intensity (mean gray value per cell area [150 μm × 150 μm]) (arbitrary unit). Different letters indicate statistically significant differences among 4 samples (P < 0.05, Tukey–Kramer test). Values are the mean ± SE (n = 45 cell areas from three independent experiments).
Respiration rate and expression of the genes encoding cytochrome oxidase and alternative oxidase.
It has been reported that NO competitively inhibits the respiratory oxygen consumption by cytochrome pathway (COX) but not by alternative pathway (AOX) in plant mitochondria (Millar and Day 1996, Yamasaki et al. 2001) and that NO transcriptionally activates the AOX expression (Huang et al. 2002). Since ALT301 exhibited lower NO level than SL (Fig. 4C), the respiration rate was compared between SL and ALT301, together with the expression levels of the COXI encoding COXI subunit 1 and the AOX encoding alternative oxidase, during the culture for up to 14 d under normal growth condition (Fig. 5). During the log phase from day 4 to day 8 after a start of subculture (Fig. 5A), the respiration rate was higher in ALT301 (Fig. 5B). The COXI expression was similar between the lines (Fig. 5C), but the AOX expression was 2 to 3 times lower in ALT301 than that in SL (Fig. 5D). These results suggest that the lower level of NO production in ALT301 leads to both the higher rate of respiration and the lower level of the AOX expression, compared with SL.
Fig. 5.
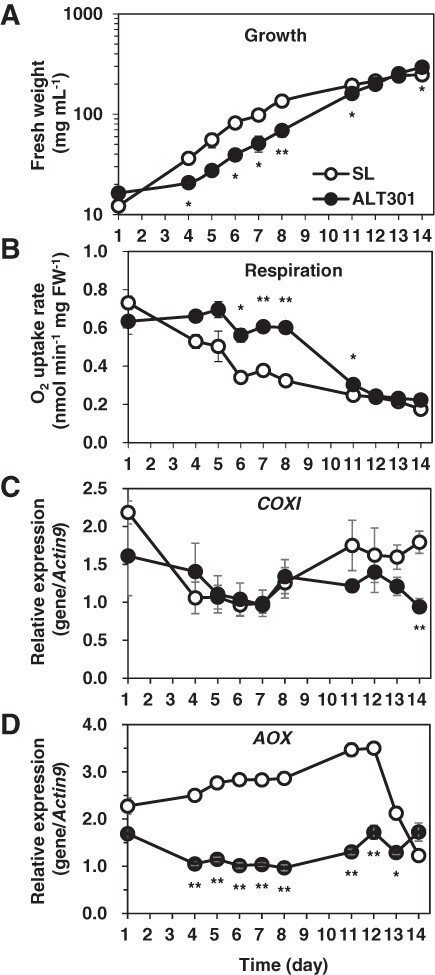
Time courses of growth (A), respiration (B), and expression of the genes related to respiratory oxygen consumption (COXI [C] and AOX [D]) in SL and ALT301 during culture under normal growth condition. At times indicated after a start of subculture, cells were withdrawn for determinations of FW (to monitor growth) and respiration (O2 uptake rate), simultaneously. Cells were also harvested for expression analyses of the genes by real-time RT-PCR. Significant differences between SL and ALT301 are indicated with asterisks (*P < 0.05, **P < 0.01, Welch’s t-test). Values are the mean ± SE (n = 3 from three independent experiments).
Metabolic and transcriptional differences in SL and ALT301 under Al-treatment conditions
ROS production and growth capacity in SL and ALT301 under Al-treatment condition.
Compared with SL, ALT301 hardly produced ROS under Al exposure (Fig. 6A) and maintained higher growth capacity after Al treatment (Fig. 6B). These characteristics of ALT301 have been maintained nearly for 20 years since our first reports (Devi et al. 2001, Yamamoto et al. 2002).
Metabolome analysis of the intermediates related to glycolysis and the TCA cycle.
Under the control treatment without Al, six glycolysis intermediates and Lac were detected and all of them were higher in ALT301 (Fig. 7A), whereas five out of the six TCA-cycle intermediates detected were lower in ALT301 (Fig. 7B), compared with SL. Thus, these results suggest the activation of glycolysis toward Lac fermentation but the repression of the TCA cycle in ALT301 under control treatment, as observed under normal growth condition (Fig. 1).
Fig. 7.
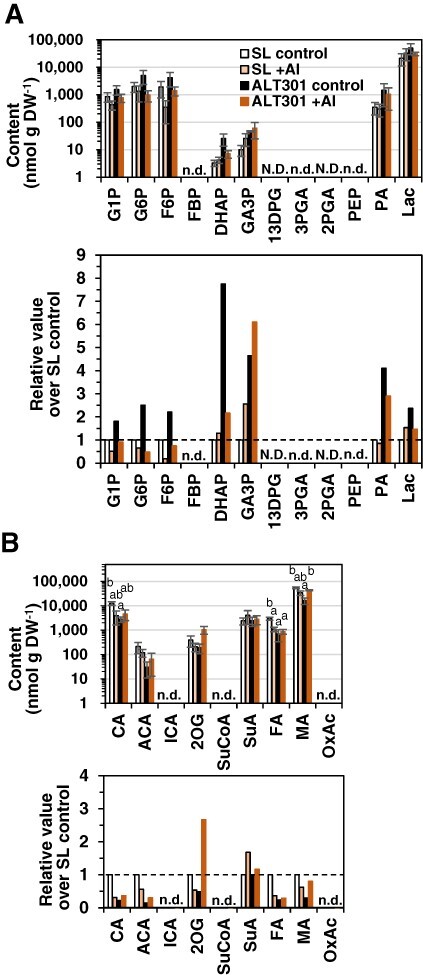
Comparison of contents of the metabolites associated with glycolysis (A) and the TCA cycle (B) between SL and ALT301 under Al-treatment condition. Cells were treated without (control) or with Al (+ Al, 50 μM) for 18 h. Values are the content (mean ± SE, n = 3 from three independent experiments) (top) and the ratio relative to SL control (bottom) in which broken line indicates the ratio value of one (values and the results of statistical analyses, see Supplementary Table S5). Statistically significant differences among four samples of each metabolite are indicated with different letters (P < 0.05, Tukey–Kramer test). N.D. means ‘not determined’ and n.d. means ‘not detected’.
On the other hand, Al exposure caused metabolic changes in both lines, but differently. Concerning glycolysis and fermentation, the metabolites on the first half of the glycolytic flow (G1P, G6P, F6P) were decreased but the second half (DHAP, GA3P, Lac) were increased in SL, whereas all the metabolites (except for GA3P) were decreased in ALT301 (Fig. 7A). The metabolites associated with the TCA cycle were generally decreased in SL (except for SuA) but increased in ALT301 (Fig. 7B).
Ethanol production.
Ethanol is also known to be fermentation endpoint in plants; therefore, ethanol content was determined in SL and ALT301. Both lines exhibited similar levels of ethanol under normal growth condition and control treatment without Al (Supplementary Fig. S8). Under Al exposure, ethanol content increased in both lines to similar extents with increases in Al dose (Supplementary Fig. S8A) and exposure time (Supplementary Fig. S8B), indicating that Al exposure enhances ethanol fermentation similarly in both lines.
Expression of the genes related to glycolysis, fermentation and ethylene signaling pathway, and the genes encoding NR (NIAs).
Concerning the genes related to glycolysis, SUSY and PK were not significantly upregulated in ALT301 under control treatment without Al exposure (Fig. 8A), although these genes were upregulated in ALT301 under normal growth conditions (Fig. 2), compared with SL. On the other hand, under Al exposure, the SUSY was upregulated in both lines, and the PK was upregulated in ALT301 (Fig. 8A).
Fig. 8.
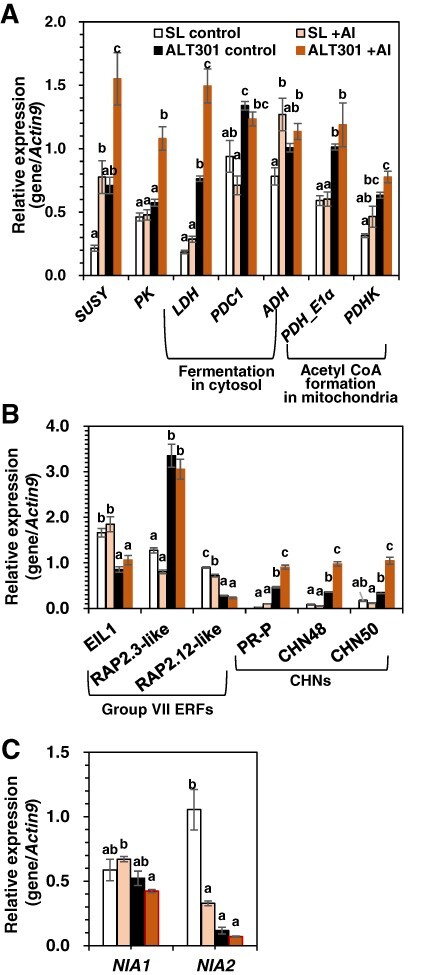
Expression of the genes related to glycolysis and fermentations (A), ethylene signaling (B) and the genes (NIA1, NIA2) encoding NR (C) in SL and ALT301 under Al-treatment condition. Cells were treated without (control) or with Al (+Al, 100 μM) for 18 h. Then, cells were harvested for expression analyses of the genes by real-time RT-PCR. Different letters in each gene analysis indicate statistically significant differences among four samples (P < 0.05, Tukey–Kramer test). Values are the mean ± SE (n = 4 from four independent experiments in A; n = 3 from three independent experiments in B; n = 3 from three independent experiments in C).
Concerning the genes related to fermentation, LDH, PDC1 and PDHK were upregulated in ALT301, compared with SL, under both control and Al treatments (Fig. 8A), as observed under normal growth conditions (Fig. 2B). On the other hand, PDH_E1α was upregulated in ALT301 under both control and Al treatments (Fig. 8A), which was not observed under normal growth conditions (Fig. 2B). Al exposure upregulated LDH in ALT301 and ADH in SL, respectively (Fig. 8A).
Concerning the genes related to the ethylene signaling pathway, EIL1 was downregulated in ALT301 under both control and Al treatments (Fig. 8B), which was not observed under normal growth conditions (Fig. 3). The RAP2.3-like, PR-P, CHN48 and CHN50 were all upregulated and RAP2.12-like was downregulated in ALT301 under both control and Al treatments (Fig. 8B), as observed under normal growth conditions (Fig. 3). Al exposure did not affect the expression levels of EIL1 and RAP2.3-like in both lines but upregulated PR-P, CHN48 and CHN50 in ALT301 and downregulated RAP2.12-like in SL (Fig. 8B).
Under control treatment without Al exposure, the expression level of NIA1 was similar in SL and ALT301 (Fig. 8C), as observed under normal growth conditions (Fig. 4A), and Al exposure did not affect the expression level in each line. By contrast, NIA2 was downregulated in ALT301 under control treatment (11% of SL) (Fig. 8C), as observed under normal growth conditions (Fig. 4A). Al exposure downregulated NIA2 in both lines, but the expression level was still lower in ALT301 (22% of SL), although the differences were not statistically significant.
Enzyme activities related to pyruvate metabolism.
In the glycolysis pathway, pyruvate is metabolized to three directions, Lac via LDH, ethanol via PDC followed by ADH and Acetyl-CoA (AcCoA) via PDH in mitochondria where AcCoA is necessary to start the TCA cycle (Fig. 9, right). Compared with SL, ALT301 exhibited constitutively higher LDH and lower PDH activities, regardless of the presence or absence of Al (Fig. 9A, B). Thus, it seems that the enhancement of Lac fermentation and the repression of the TCA cycle in ALT301 under control treatment (Fig. 7) as well as under normal growth condition (Fig. 1) are based on the activities of these key enzymes. By contrast, PDC and ADH activities were similar in both lines under control treatment (Fig. 9C, D) and similarly increased under Al exposure, although these activities were significantly higher in ALT301 at 100 μM Al. Thus, it seems that the enhancement of ethanol fermentation in both lines under Al exposure (Supplementary Fig. S8) is based on the increment of PDC and ADH activities.
Fig. 9.
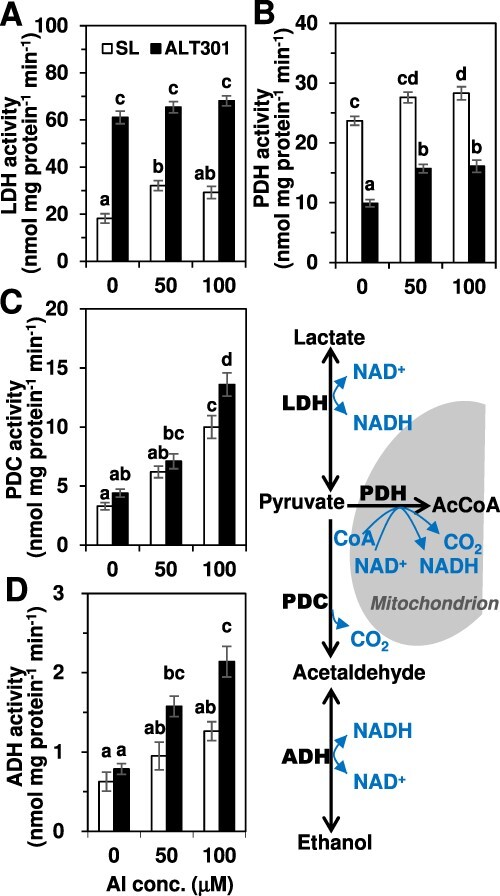
Comparison of enzyme activities of lactate dehydrogenase (LDH) (A), pyruvate dehydrogenase (PDH) (B), pyruvate decarboxylase (PDC) (C) and alcohol dehydrogenase (ADH) (D) between SL and ALT301 under Al-treatment condition. The four enzymes shown in the metabolic pathway at right were determined specific activities using cell-free extracts prepared from the cells treated with various concentrations of Al (0, 50 and 100 μM) for 24 h. Different letters in each figure indicate statistically significant differences among six samples (P < 0.05, Tukey–Kramer test). Values are the mean ± SE (n = 3 from three independent experiments).
Nitric oxide production.
Under control treatment without Al, NO production was lower in ALT301 (54% of SL; Fig. 10) as observed under normal growth conditions (Fig. 4). Al exposure enhanced NO production in both lines (2.6-fold in SL and 2.3-fold in ALT301), but the NO level was still lower in ALT301 (48% of SL; Fig. 10).
Discussion
Fig. 11 summarizes the findings in this study together with previous findings, which are aligned according to our proposed pathway starting from both the upregulation of the RAP2.3-like gene and the downregulation of the NIA2 gene toward the Al-tolerant phenotype in ALT301.
Fig. 11.
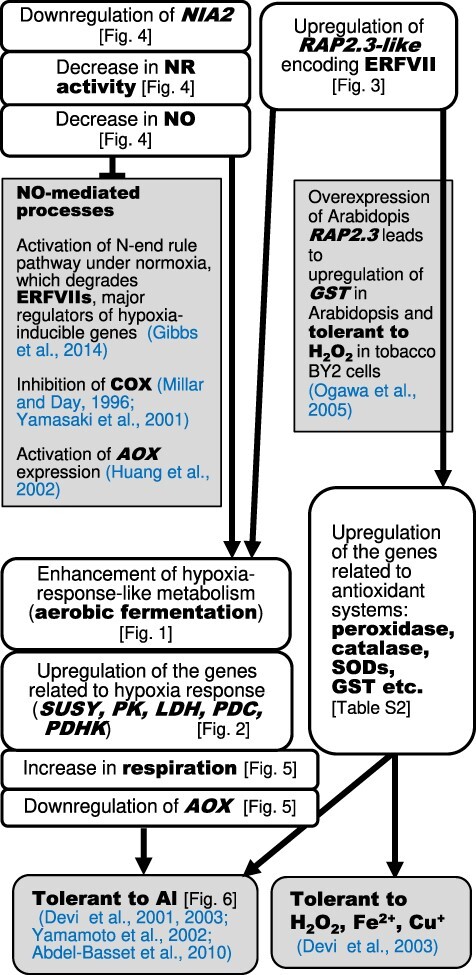
Summary of the events constitutively expressed in ALT301 (Al-tolerant) compared with SL (wild type), and the proposed metabolic pathways leading to tolerant phenotype to Al and oxidative stress. This study revealed several events activated or repressed in ALT301, compared with SL, which are described in white boxes (the corresponding data, see Figs and Table indicated in brackets). The plausible cause-and-effect relationship between the events and the tolerant phenotype to Al, H2O2, Fe2+ and Cu2+ in ALT301 are proposed, based on previous findings that are indicated in gray boxes with references.
Al-tolerant cell line ALT301 exhibits aerobic fermentation
Comparative metabolome analysis between SL (wild type) and ALT301 under normal growth conditions revealed an enhancement of glycolytic flux and a shift of pyruvate metabolism from the TCA cycle to Lac fermentation in ALT301 under aerobic conditions, indicating aerobic fermentation (Fig. 1). The aerobic fermentation in ALT301 seems to be based, at least in part, on constitutively higher expression of LDH and PDHK under normal growth as well as Al-treatment conditions (Figs. 2B, 8A), and the resultant higher LDH and lower PDH activities (Fig. 9).
Transcriptional regulation of aerobic fermentation involves the upregulation of the RAP2.3-like gene and the downregulation of the NIA2 gene in ALT301
Forty-five GO terms were significantly enriched for the upregulated genes in ALT301, compared with SL, under normal growth condition (Supplementary Table S3A). The hierarchical image of the 40 GO terms of the ‘biological process’ category revealed that more than a half of the GO terms were integrated into one GO term indicating ‘ethylene mediated signaling pathway’ (Supplementary Fig. S5A) that was associated with four genes encoding three chitinases and RAP2.3-like protein (Supplementary Table S4A, Group II). The upregulations of these genes in ALT301 were confirmed by real-time RT-PCR under both normal growth and Al-treatment conditions (Figs. 3, 8B). The RAP2.3 encodes ERFVII in Arabidopsis. ERFVIIs are plant-specific transcription factors and important regulators of a wide range of plant growth, development and responses to abiotic and biotic stresses. One universal function attributed to ERFVIIs in higher plants is to coordinate response to oxygen deficiency (Gibbs et al. 2015, Giuntoli and Perata 2018, Fukao et al. 2019). In Arabidopsis, ERFVIIs act redundantly as principal activators of hypoxia-responsive genes including SUS, ADH and PDC under low-oxygen conditions (Licausi et al. 2011, Gibbs et al. 2011, Gasch et al. 2016) and enhance ethanol fermentation. Compared with SL, ALT301 under normal growth conditions exhibits the enhancement of Lac fermentation (Fig. 1) and constitutively higher expression of SUSY, PK, LDH, PDC1 and PDHK genes (Fig. 2). In general, these genes have been reported as hypoxic responsive genes in plants (Mustroph et al. 2010), although all the hypoxia-responsive genes are not necessarily regulated by ERFVIIs. Therefore, the upregulation of the RAP2.3-like gene could be related, at least in part, to the upregulation of these hypoxia-responsive genes in ALT301.
However, since ERFVIIs are functional under hypoxia but are degraded under normoxia by the N-end rule pathway of targeted proteolysis, the upregulation of RAP2.3-like and resultant increase in RAP2.3-like protein might not be enough to escape from the N-end rule pathway under normoxia, so ALT301 might have a stabilization mechanism of ERFVII proteins under normoxia. This is the case. Because the N-end rule pathway depends on both O2 availability and endogenous NO level, the absence of either gas permits the accumulation of ERFVIIs. In Arabidopsis, the constitutively synthesized ERFVIIs (RAP2.2, RAP2.3, RAP2.12) are stabilized under hypoxia (O2 depletion) and activate hypoxia-responsive genes (Licausi et al. 2011, Gibbs et al. 2011, Gasch et al. 2016). On the other hand, the main source of NO in plants is NR (Tejada-Jimenez et al. 2019) and blocking NO production through the genetic knockout of the NR encoding gene is sufficient to stabilize ERFVIIs even under normoxia in Arabidopsis (Gibbs et al. 2014). In our study, compared with SL, ALT301 exhibited the downregulation of the NIA2 gene, a decrease in NR activity and a decrease in NO level (Fig. 4), suggesting that the downregulation of the NIA2 and resultant decrease in NR activity leads to NO depletion in ALT301, which could stabilize ERFVIIs including RAP2.3-like protein under normoxia.
It remains to be elucidated how the upregulation of RAP2.3-like and the downregulation of NIA2 are controlled in ALT301. The genome sequencing of the NIA2 and the RAP2.3-like in SL and ALT301 indicated that the sequence of the coding region as well as the sequences of 5ʹ-flanking (8 kb) and the 3ʹ-flanking (5 kb) regions of each gene were the same in these lines (our unpublished results).
Mechanism of tolerance to oxidative stress based on the upregulation of the RAP2.3-like gene in ALT301
In addition to hypoxia, ERFVIIs also enhance tolerance to oxidative stress (Gibbs et al. 2015). Papdi et al. (2015) reported that estradiol-induced overexpression of all RAP2 genes (RAP2.2, RAP2.3, RAP2.12) conferred tolerance to oxidative stress (H2O2) and osmotic stress under normoxia in Arabidopsis. Ogawa et al. (2005) reported that ectopic overexpression of the gene encoding Arabidopsis ethylene-responsive element binding protein (AtEBP [locus tag: AT3G16770], also known as RAP2.3) in tobacco BY-2 cells conferred tolerance to H2O2 and heat treatments and that transgenic Arabidopsis plants overexpressing the AtEBP (RAP2.3) exhibited upregulation of the GST6 gene. On the other hand, our previous study revealed that ALT301 was more tolerant to oxidative stress caused by H2O2 and transition metals (Fe2+, Cu+) (Devi et al. 2003) and that ALT301 exhibited the upregulation of the genes encoding antioxidant enzymes (peroxidases, catalase, SODs, probable GST, dehydroascorbate reductase) (Supplementary Table S2), compared with SL. Thus, it is likely that the upregulation of the RAP2.3-like gene in ALT301 (Fig. 3) also leads to higher expression of at least a few genes encoding antioxidant proteins in ALT301, compared with SL.
Prevention mechanism of ROS production in mitochondria based on both aerobic fermentation and a decrease in NO level in ALT301
Since the electron transport chain in the mitochondrial inner membrane is a major site of ROS production (Semenza 2007), the metabolic shift to aerobic fermentation in ALT301 could be one of the mechanisms to reduce ROS production in ALT301 under normoxia. Similar metabolic shift under aerobic conditions has been reported in human cancer cells as ‘the Warburg effect’ or aerobic glycolysis (Vander Heiden et al. 2009, Nagao et al. 2019). Compared with normal differentiated cells that rely primarily on mitochondrial oxidative phosphorylation to produce ATP, most cancer cells produce ATP predominantly through accelerated glycolysis followed by Lac fermentation under normoxic condition. The coordinated shift in metabolism from oxidative phosphorylation to glycolysis is transcriptionally regulated by the hypoxia-inducible factor 1 (HIF-1), a master transcription factor affecting the adaptive response to hypoxia by regulating a wide range of hypoxia-responsive genes including the upregulation of genes encoding lactate dehydrogenase A (LDHA) and pyruvate dehydrogenase kinase-1 (PDK1). The HIF-1α, the main regulatory subunit of HIF-1 activity, is degraded under normoxia by O2-dependent hydroxylation of purine residues followed by ubiquitination and degradation through the 26S proteasome but the HIF-1 functions in cancer cells under normoxia by several mechanisms to increase the accumulation of the HIF-1α subunit (Nagao et al. 2019). The upregulations of both the LDHA and the PDK1 have been identified as one of critical adaptations to prevent excess ROS production via mitochondrial respiratory chain (Semenza 2007). Furthermore, aerobic glycolysis in animals is thought to be an inefficient way to generate ATP but was proposed to be adapted to facilitate the uptake and incorporation of nutrients into biomass needed to produce a new cell in cancer cells as well as all proliferating cells (Vander Heiden et al. 2009).
By contrast, aerobic fermentation enhanced in ALT301 is associated with a higher respiration rate (Fig. 5B) and a higher ATP content (Yamamoto et al. 2002), compared with SL. The differences observed between human cancer cells and tobacco ALT301 cells could be due to the involvement of NO in tobacco cells. In plants, NO plays a key role for controlling aerobic fermentation negatively by the degradation of ERFVIIs via activation of the N-end rule pathway of proteolysis (Gibbs et al., 2014, 2015) and also affects mitochondrial electron flow differently to animals. Plant mitochondria possess two respiratory electron transport pathways, the cytochrome pathway and plant-specific alternative pathway, with COX and AOX, respectively, as a terminal electron acceptor. NO competitively inhibits oxygen consumption via COX but not via AOX (Millar and Day 1996, Yamasaki et al. 2001). The inhibitory effect of NO on COX decreases the extent of the membrane potential (∆ψ) generated by the H+ translocation during the electron transport process and suppresses ATP synthesis in mitochondria (Yamasaki et al. 2001). In addition, it has been proposed that the inhibitory effect of NO on COX generates ROS due to the over-reduction of ubiquinone pool. On the other hand, NO, as a signaling molecule, induces the AOX1a expression in Arabidopsis suspension cells (Huang et al. 2002), suggesting the participation of AOX to counteract the toxic effect of NO on COX, although, unlike the COX, AOX is not coupled to H+ translocation necessary for ATP production. Consistently, ALT301 exhibited lower level of NO (Fig. 4C), higher respiration rate (Fig. 5B) and lower expression of the AOX gene (Fig. 5D) during the log phase under normal growth condition, compared with SL. In addition, ATP content was higher in ALT301 than that in SL (Yamamoto et al. 2002).
Taken together, it is likely that the lower production of NO in ALT301 seems to repress the NO-mediated processes including the activation of the N-end rule pathway and the inhibition of the COX, which leads to aerobic fermentation (Fig. 1) and an increase in the respiration rate (Fig. 5B) as well as an increase in ATP content (Yamamoto et al. 2002). These conclusions are supported by phenotypic similarity between ALT301 and the transgenic tobacco plant, LNR-H, which lacked NR activity in roots due to the nia1nia2 double mutation of the NR loci (Hänsch et al. 2001). Compared with wild-type roots, LNR-H roots contained higher Lac under both aerated and anoxic growth conditions. Furthermore, under aerated conditions, LNR-H roots exhibited a higher respiration rate as well as a higher content of ATP than wild type (2003a Stoimenova et al. , 2003b).
Based on the lines of evidence described above, prevention mechanisms of ROS production in ALT301 are proposed in Fig. 12. Compared with SL, ALT301 seems to manage electron transport chain in mitochondria more safely (less ROS production) and more effectively (higher ATP production) by the two coordinated mechanisms: (i) the reduction of substrate supply to the TCA cycle by a shift of pyruvate metabolism from AcCoA production in mitochondria to Lac fermentation in cytosol, and (ii) the increase in the respiration rate via the activation of COX activity of COX and the repression of AOX due to the downregulation of the AOX gene. Both mechanisms are based on a decrease in NO level and resultant repression of the NO-mediated processes including the N-end rule pathway and the respiration pathways, respectively. Under Al exposure, ROS production was enhanced in SL but seldom in ALT301 (Fig. 6A), and the ROS production was associated with respiration inhibition in SL (Yamamoto et al. 2002). Al exposure also enhanced the NO production in both cell lines, but the NO level was less in ALT301 (Fig. 10) that could be partly due to a lower expression of the NIA2 in ALT301 (Fig. 8C). Thus, although it remains to be elucidated how Al triggers ROS production as well as NO production, the central carbon metabolic pathways constitutively expressed in ALT301 (Fig. 12) seem to be effective for the prevention of ROS production under Al exposure.
Fig. 12.
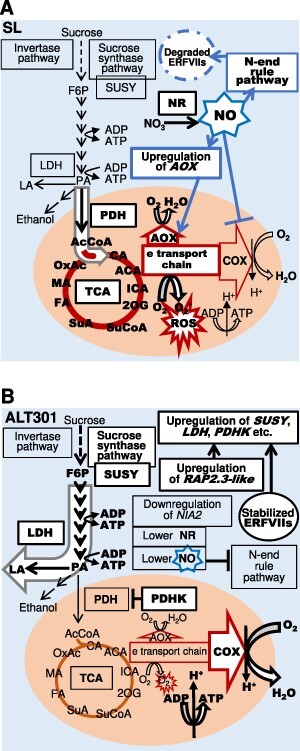
Working hypothesis of central carbon metabolic pathways in SL (A) and ALT301 (B) and prevention mechanisms of ROS production in mitochondria in ALT301. Under normal growth condition, compared with SL (A), ALT301 (B) exhibits the downregulation of the NIA2 gene and the upregulation of the RAP2.3-like gene. Here, we assume that the lower NO level due to the downregulation of the NIA2 gene and resultant lower activity of NR stabilizes ERFVIIs including the RAP2.3-like protein. Then, the stabilized ERFVIIs activate a wide range of genes including hypoxia-responsive genes such as SUSY, LDH and PDHK, which leads to the enhancement of the SUSY pathway, the enhancement of lactate fermentation and the inhibition of PDH activity leading to the limitation of pyruvate entry into the TCA cycle, even under normoxia. On the other hand, NO is known to inhibit the oxygen consumption by COX and to activate AOX expression. These NO effects should be observed more extensively in SL, and it is likely that the imbalance between the electron supply from the TCA cycle and the electron consumption by COX on the electron transport chain enhances ROS production. Under this condition, in ALT301, the enhancement of aerobic fermentation reduces the electron supply form the TCA cycle, while lower NO level activate COX activity and reduce AOX activity. Thus, compared with SL, ALT301 seems to have a higher respiration rate and higher capacity to produce ATP with less generation of ROS. The events enhanced in either cell line (metabolism, gene expression, enzyme activity) are highlighted with a white background. The processes of activation and inhibition are indicated with an arrow and a line with a bar (⊥), respectively. The NO-mediated processes are indicated by blue lines.
It remains to be clarified whether the Al-triggered NO burst is related to Al toxicity as suggested in this study or Al tolerance as suggested previously (Wang et al. 2010, Sun et al. 2014). In this context, the role of NR in the NO burst should be also clarified. In Arabidopsis seedlings, the nia1nia2 double mutant was reported to be more sensitive to Al (Wang et al. 2010) or more tolerant to Al (Li et al. 2020).
Other possible physiological roles of aerobic fermentation in plants
As a plant-specific function of fermentation, the Lac and ethanol fermentations in plant cells have been predicted to be the members of the pH-regulating metabolic pathway that consists of phosphoenolpyruvate carboxylase, malate dehydrogenase, malic enzyme, PDC, ADH and LDH (called ‘the alternative glycolytic and fermentation pathway’; Sakano 1998). Therefore, the constitutive enhancement of Lac fermentation in ALT301 might be involved in the prevention mechanism of acidosis in the cytoplasm. The pH-regulating pathway and another pH-regulating pathway, called the γ-aminobutyric acid shunt, are enhanced under the control of STOP1, which protect Arabidopsis from H+ and Al toxicity (Sawaki et al. 2009). In this study, the expression level of the NtSTOP1 gene determined by semiquantitative RT-PCR was similar between SL and ALT301 under normal growth condition (data not shown), suggesting that NtSTOP1 is not involved in the metabolic shift to Lac fermentation and Al tolerance in ALT301.
In plants, aerobic fermentation has been reported in a few cases. For example, tobacco pollen carries out efficient ethanolic fermentation concomitantly with a high rate of respiration during pollen maturation and tube growth (Mellema et al. 2002). The metabolism seems to fit to pollen tube growth that is a fast tip-growing cell system demanding for increasing biomass and high-energy status. As far as we know, the mechanism to enhance aerobic fermentation in pollen has not been fully elucidated yet, but might be controlled negatively by the level of NO produced by NR, as proposed in ALT301. Interestingly, Prado et al. (2004) reported that little NO was found in the cytosol of the pollen tube tip in lily (Lilium longiflorum) and suggested that the rate and orientation of pollen tube growth are negatively regulated by NO levels at the pollen tube tip.
Finally, it is worth noting that the responses to Al in SL (wild type) were quite similar to the responses to hypoxia in plants, such as an increase in fermentation product (ethanol; Supplementary Fig. S8) and the upregulation of SUSY and ADH (Fig. 8A) as well as decreases in the respiration rate and ATP content (Yamamoto et al. 2002). In addition, the productions of callose (Devi et al. 2001), NO (Fig. 10) and ROS (Fig. 6) observed in SL under Al stress have been also reported in plants under hypoxia (Piršelová and Matušíková 2013, Pucciariello and Perata 2017). Although the molecular mechanisms underlying this similarity remain to be resolved in the future, it is tempting to speculate that the pre-establishment of the hypoxia-response-like metabolism in ALT301 under normal growth condition might be beneficial to survive when cells encounter Al stress.
Conclusion
Comparative analyses of the metabolome and transcriptome between SL (wild type) and ALT301 (Al-tolerant) tobacco cell lines revealed the enhancement of aerobic fermentation, the upregulation of the RAP2.3-like gene, the downregulation of the NIA2 gene and a decrease in NO level in ALT301 under normal growth and Al-treatment conditions. These events could be involved coordinately in the prevention mechanism of ROS production in mitochondria under Al stress. Further study of Al responses focusing on the production mechanisms of NO and ROS and the possible interactions between NO, ROS and mitochondrial electron transport chain should give a clue to control Al toxicity and tolerance in plant cells.
Materials and Methods
Cell lines, culture conditions under normal growth and Al treatment and determinations of physiological factors
Two nonchlorophyllic cell lines, SL (derived from pith callus of N. tabacum L. cv Samsun; Nakamura et al. 1988) and ALT301 (Al-tolerant line derived from SL; Devi et al. 2001) were grown as described (Yamamoto et al. 1994) in a modified version of Murashige–Skoog medium (nutrient medium) in an Erlenmeyer flask (≤30 mL medium in 100 mL flask) on a rotary shaker operated at 100 revolutions per min to supply oxygen at 25oC in the dark, which was defined as normal growth condition in this study. Fresh weight (FW) of the cells in culture was determined by harvesting the cells (5 mL aliquot) on a filter paper by vacuum filtration. For determination of growth curve, a portion of the full culture at a cell density of ∼200 mg FW per mL was transferred into a new culture medium. The initial cell density (mg FW per mL) was adjusted to 10 in SL and 16 in ALT301. Then, the cell density was monitored every day for up to 14 d. During the log phase of growth, the mass doubling time was determined as described previously (Sameeullah et al. 2013) and was used for the comparison of growth rates between lines. Cell culture indicating a cell density of ∼60 mg FW per mL during the log phase under normal growth condition was used for all the experiments performed in this study, unless otherwise indicated.
For determination of oxygen uptake by the cells, cells (5 mL aliquot) were harvested and measured FW, and then, the cells (100 mg FW) were resuspended in 2 mL of nutrient medium and the consumption of oxygen was determined immediately by the use of a Clark-type oxygen electrode apparatus (Oxygraph, Hansatech Instruments, King’s Lynn, UK) under continuous mixing condition at 25oC.
Oxygen concentration in the medium of cell culture under normal growth condition was determined by the use of a Clark-type oxygen electrode apparatus as follows: when the shaker was stopped, most of the cells in culture were immediately precipitated on the bottom of the flask and the medium (5 mL aliquot) was gently withdrawn from the culture supernatant and was centrifuged (750 g for 3 min at 25oC) in order to remove remaining cells from the medium. The resultant supernatant was transferred to a tube and was incubated at 25oC for 10 min to adjust the medium temperature at 25oC. Then, 1-mL aliquot was transferred to the chamber of the oxygen electrode apparatus and, after 3 min for stabilization, oxygen concentration was measured at 25oC without stirring. For control, nutrient medium without cells was also maintained in a flask on a rotary shaker. The medium (5 mL aliquot) was withdrawn, treated and measured oxygen concentration under the same conditions as cell culture described above.
Al treatment was performed as described previously (Yamamoto et al. 2002, Kariya et al. 2013) with minor modifications. Briefly, cells at the log phase under normal growth condition were withdrawn, washed and then cultured without (control) or with AlCl3 at a cell density of 10 mg FW per mL in treatment medium (Ca-sucrose solution containing 3 mM CaCl2, 3% sucrose and 20 mM 2-morpholinoethanesulfonic acid [MES] at pH 5.0) for up to 24 h. The growth capacity (or growth capability) of Al-treated cells, compared with control cells, was determined as the extent of growth of Al-treated cells relative to that of control cells after subsequent culture in Al-free nutrient medium.
Metabolite analysis with CE-MS
Cells at the log phase under normal growth condition or after Al treatment were frozen in liquid nitrogen and lyophilized. Metabolites were extracted from the lyophilized samples, and the contents of metabolites were quantified by anion and cation analysis using CE-MS as described previously (Nakamura et al. 2012) with some modifications (more details, see Supplemental Protocol S1).
Quantitative determinations of small molecules (pyridine nucleotides, ethylene and ethanol)
Contents of the small molecules were determined in cells at the log phase under normal growth condition. The contents of NAD(H) and NADP(H) in cells were determined by enzymatic methods as described (Queval and Noctor 2007), using a multi-detection microplate reader (POWERSCAN HT, DS Pharma Biomedical, Osaka, Japan).
For quantification of ethylene produced by cells, the cells harvested on a filter paper (1 g FW) were transferred into a syringe or a chamber, and the samples were left standing under air tight condition at 20 ˚C for 1 h. Then, 1 mL of headspace gas from the syringe or the chamber was injected into a gas chromatograph (model GC-8A, Shimazu, Kyoto, Japan) equipped with an activated alumina column and flame ionization detector, and the ethylene evolution rate was determined as described previously (Hiwasa et al. 2003).
Ethanol content was determined in cells after treatment without (control) or with Al. Quantification of ethanol in cells was performed using the ethanol assay kit (ab65343; Abcam). Cell-free extracts were prepared by disruption of cells in a sonicator on ice, and then, the content of ethanol was determined fluorometrically following the manufacture’s protocol, using a fluorescence spectrophotometer (model F4500, Hitachi, Tokyo, Japan).
Real-time quantitative RT-PCR
Isolation of total RNA from cells and the preparation of the first strand cDNA were carried out as described previously (Kariya et al. 2013). Real-time RT-PCR was carried out using LightCycler (Roche, Penzberg, Upper Bavaria, Germany) with the gene-specific primer sets (Supplementary Table S6) and master mix (either Thunderbird SYBR qPCR mix [Toyobo, Osaka, Japan] or KOD SYBR qPCR mix [Toyobo]) according to the manufacturer’s protocol. For each gene, the quantification of the transcript levels was performed using the LightCycler software (Fit Points Method) with a standard curve generated by a serial dilution of the cDNA sample. The Actin9 was used as an internal control to normalize the amounts of total transcripts in each sample.
Microarray analyses
Comparison of gene expression in SL and ALT301 at the log phase under normal growth condition was performed. Total RNA was extracted from three biological replicates of each cell line. RNA sample was labeled with Cy3 fluorescent dye using the Low Input Quick Amp Labeling Kit, One-color (Agilent Technologies, Santa Clara, CA, USA), following the manufacturer’s protocol. Labeled cRNA was purified using the RNeasy Mini Kit (Qiagen, Hilden, Germany) and then hybridized to Agilent Tobacco Oligo DNA Microarray (4 × 44k; Agilent Technologies), and scanned using an Agilent Microarray Scanner (G2565CA, Agilent Technologies). To analyze image and signal intensity, raw data of the scanning were processed with the software GeneSpring GX 10 (Agilent Technologies) and were normalized intensities. The P values for assessment of significance of differentially expressed genes between the lines were calculated using the values of raw signal intensity by t-test using MeV 4.9.0. The differentially expressed genes with the significance (P < 0.05) showing ≥ 2-fold changes were investigated (Supplemental Data Set 1).
For the differentially expressed genes with the significance (P < 0.01) showing ≥ 3-fold changes, the functional enrichment analyses of the GO terms were carried out, using agriGO v2 (Tian et al. 2017; http://systemsbiology.cau.edu.cn./agriGOv2/index.php, accessed on 14 March, 2019) using singular enrichment analysis with the Agilent tobacco genome array ID as the background/Reference in the default-setting mode.
Enzyme activities (SUSY, NR, LDH, PDC, ADH and PDH)
Sucrose synthase activity was measured in the direction of sucrose degradation as described previously (Zrenner et al. 1995). Cells were washed with 50 mM HEPES-KOH (pH 7.4) twice and were frozen in liquid nitrogen and ground in a mortar. Under the assay condition, the quantification of UDPG was performed with UDP-glucose dehydrogenase (Sigma-Aldrich, St. Louis, MO, USA) by measuring NAD+ reduction at 340 nm using a multi-detection microplate reader (POWERSCAN HT, DS Pharma Biomedical).
Nitrate reductase activity was measured as described previously (Scheible et al. 1997) with some modifications. We used extraction buffer (100 mM HEPES-KOH [pH 7.5], 2 mM EDTA, 2 mM DTT, 1% polyvinylpyrolidone) and assay buffer (100 mM HEPES-KOH [pH 7.5], 50 mM KNO3, 1 mM NADH and 2 mM EDTA). The reaction was stopped by adding zinc acetate.
For measurements of LDH, PDC and ADH activities, preparation of cell-free extracts and determination of enzyme activities were performed as described previously (Bouny and Saglio 1996) with some modifications. Briefly, cells were washed with extraction buffer (50 mM Tris-HCl [pH 7.5], 10 mM sodium borate, 5 mM DTT, 15% glycerol) and were immediately frozen in liquid nitrogen. The frozen cells were thawed in the extraction buffer, sonicated on ice and, then, centrifuged. The supernatant was desalted by centrifugation on a Sephadex G-25 column and was used for each enzyme assay. Under the assay conditions, NADH oxidation (LDH, PDC) and NAD+ reduction (ADH) were recorded by measuring the change in absorbance at 340 nm using a spectrophotometer (model UV-1600, Shimazu).
Preparation of the fraction containing mitochondria from tobacco cells and determination of PDH complex activity were performed as described previously (Camp and Randall 1985, Taylor et al. 1992, Jan et al. 2006) with some modifications. Cells were disrupted in a glass homogenizer with extraction buffer containing of 0.4 M mannitol, 25 mM 3-morpholinopropanesulfonic acid (MOPS) (pH7.8), 1 mM ethylene glycol tetraacetic acid (EGTA) and 4 mM cysteine. The homogenate was centrifuged at 2,000 g for 10 min. The supernatant was centrifuged at 10,000 g for 15 min to pellet mitochondrial fraction. The pellet was resuspended with the extraction buffer and centrifuged at 2,000 g for 15 min. The suspension of the pellet was used for the assay of PDH complex activity, without further purification step of mitochondria. PDH activity was determined according to Camp and Randall (1985) by measuring NAD+ reduction spectrophotometrically at 340 nm using a spectrophotometer (UV-1600).
Detections of NO and ROS
Detection of NO in cells was performed using 4,5-diaminofluorescein diacetate (DAF-2 DA) as described (Vicente et al. 2017) with some modifications. Cells were suspended in 10 mM HEPES-KOH (pH 7.4) containing 10 μM DAF-2DA (GORYO Chemical, Sapporo, Japan) and were shaken gently for 30 min in the dark. Cells were then harvested, washed and resuspended in the same buffer. Cells were visualized using a fluorescence microscope (model BZ-X700, KEYENCE, Osaka, Japan) with excitation at 470 nm and emission at 525 nm. The NO level in cells was determined as DAF-2 fluorescence intensity (mean gray value per cell area [150 μm × 150 μm]) with Fiji software. Cell areas were randomly selected first in a bright image of each micrograph (three areas per micrograph) and then the mean gray value of the corresponding areas in a fluorescence image was analyzed.
Detection of ROS in cells was performed using dihydroethidium as described previously (Yamamoto et al. 2002). The cells were visualized using a fluorescence microscope (model Akiotron, Carl Zeiss, Oberkochen, Germany) with a filter set No. 9 (excitation 450–490 nm, emission at 520 nm or more).
Statistical analyses
All data are shown as the mean ± SE, n ≥ 3, unless otherwise indicated. The means of two groups were compared by Welch’s t-test at 5 or 1% levels of significance using Microsoft Excel 2016. Means of three or more groups were compared by Tukey–Kramer test, which was performed with R version 3.6.0.
Supplementary Material
Acknowledgements
We would like to thank to Dr. S. Rama Devi for her pioneering studies on Al tolerance mechanism in ALT301 and to Ms Kazue Komatsu, Masako Matsumoto, Michiyo Ariyoshi (Okayama University) for their skilled assistance in experiments. This work was supported by the Japan Society for the Promotion of Science and the Ohara Foundation for Agricultural Science.
Contributor Information
Yoshiyuki Tsuchiya, Institute of Plant Science and Resources, Okayama University, Chuo 2-20-1, Kurashiki, Okayama 710-0046, Japan.
Takuji Nakamura, Lowland Crop Rotation System Group, Division of Lowland Farming Research, Hokkaido Agricultural Research Center (HARC), NARO, 1 Hitsujigaoka, Toyohira-ku, Sapporo 062-8555, Japan.
Yohei Izumi, Institute of Plant Science and Resources, Okayama University, Chuo 2-20-1, Kurashiki, Okayama 710-0046, Japan.
Keiki Okazaki, Central Region Agricultural Research Center, NARO (CARC/NARO), 2-1-18 Kannondai, Tsukuba, Ibaraki 305-8666, Japan.
Takuro Shinano, Laboratory of Plant Nutrition, Graduate School of Agriculture, Hokkaido University, N9, W9, Kitaku, Sapporo, Hokkaido 060-8589, Japan.
Yasutaka Kubo, Graduate School of Environmental and Life Science, Okayama University, Tsushima, Okayama 700-8530, Japan.
Maki Katsuhara, Institute of Plant Science and Resources, Okayama University, Chuo 2-20-1, Kurashiki, Okayama 710-0046, Japan.
Takayuki Sasaki, Institute of Plant Science and Resources, Okayama University, Chuo 2-20-1, Kurashiki, Okayama 710-0046, Japan.
Yoko Yamamoto, Institute of Plant Science and Resources, Okayama University, Chuo 2-20-1, Kurashiki, Okayama 710-0046, Japan.
Supplementary Data
Supplementary data are available at PCP online.
Data Availability
The microarray data reported in this article have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number (GSE158255).
Funding
This work was funded by a Grant-in-Aid for Scientific Research [grant no. 21580078 and 24380039 to Y.Y.] of the Japan Society for the Promotion of Science (JSPS).
Disclosures
The authors have no conflicts of interest to declare.
References
- Abdel-Basset R., Ozuka S., Demiral T., Furuichi T., Sawatani I., Baskin T.I., et al. (2010) Aluminium reduces sugar uptake in tobacco cell cultures: a potential cause of inhibited elongation but not of toxicity. J. Exp. Bot. 61: 1597–1610.doi: 10.1093/jxb/eraq027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouny J.M. and Saglio P.H. (1996) Glycolytic flux and hexokinase activities in anoxic maize root tips acclimated by hypoxic pretreatment. Plant Physiol. 111: 187–194.doi: 10.1104/pp.111.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp P.J. and Randall D.D. (1985) Purification and characterization of the pea chloroplast pyruvate dehydrogenase complex: a source of acetyl-CoA and NADH for fatty acid biosynthesis. Plant Physiol. 77: 571–577.doi: 10.1104/pp.77.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devi S.R., Yamamoto Y. and Matsumoto H. (2001) Isolation of aluminum-tolerant cell lines of tobacco in a simple calcium medium and their responses to aluminum. Physiol. Plant. 112: 397–402.doi: 10.1034/j.1399-3054.2001.1120313.x. [DOI] [PubMed] [Google Scholar]
- Devi S.R., Yamamoto Y. and Matsumoto H. (2003) An intracellular mechanism of aluminum tolerance associated with high antioxidant status in cultured tobacco cells. J. Inorg. Biochem. 97: 59–68.doi: 10.1016/S0162-0134(03)00182-X. [DOI] [PubMed] [Google Scholar]
- Fukao T., Barrera-Figueroa B.E., Juntawong P. and Peña-Castro J.M. (2019) Submergence and waterlogging stress in plants: a review highlighting research opportunities and understudied aspects. Front. Plant Sci. 10: 340.doi: 10.3389/fpls.2019.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch P., Fundinger M., Müller J.T., Lee T., Bailey-Serres J. and Mustroph A. (2016) Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in Arabidopsis. Plant Cell 28: 160–180.doi: 10.1105/tpc.15.00866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Conde J.V., Berckhan S., Prasad G., Mendiondo G.M. and Holdsworth M.J. (2015) Group VII ethylene response factors coordinate oxygen and nitric oxide signal transduction and stress responses in plants. Plant Physiol. 169: 23–31.doi: 10.1104/pp.15.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Lee S.C., Md Isa N., Gramuglia S., Fukao T., Bassel G.W., et al. (2011) Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479: 415–418.doi: 10.1038/nature10534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs D.J., Md Isa N., Movahedi M., Lozano-Juste J., Mendiondo G.M., Berckhan S., et al. (2014) Nitric oxide sensing in plants is mediated by proteolytic control of group VII ERF transcription factors. Mol. Cell 53: 369–379.doi: 10.1016/j.molcel.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuntoli B. and Perata P. (2018) Group VII ethylene response factors in Arabidopsis: regulation and physiological roles. Plant Physiol. 176: 1143–1155.doi: 10.1104/pp.17.01225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. and Gutteridge J.M.C. (2007) Free Radicals in Biology and Medicine. 4th edn. Oxford University Press, New York, NY. [Google Scholar]
- Hänsch R., Fessel D.G., Witt C., Hesberg C., Hoffmann G., Walch-Liu P., et al. (2001) Tobacco plants that lack expression of functional nitrate reductase in roots show changes in growth rates and metabolite accumulation. J. Exp. Bot. 52: 1251–1258.doi: 10.1093/jexbot/52.359.1251. [DOI] [PubMed] [Google Scholar]
- Hiwasa K., Kinugasa Y., Amano S., Hashimoto A., Nakano R., Inaba A., et al. (2003) Ethylene is required for both the initiation and progression of softening in pear (Pyrus communis L.) fruit. J. Exp. Bot. 54: 771–779.doi: 10.1093/jxb/erg073. [DOI] [PubMed] [Google Scholar]
- Hoekenga O.A., Maron L.G., Piñeros M.A., Cançado G.M., Shaff J., Kobayashi Y., et al. (2006) AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 9738–9743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., von Rad U. and Durner J. (2002) Nitric oxide induces transcriptional activation of the nitric oxide-tolerant alternative oxidase in Arabidopsis suspension cells. Planta 215: 914–923.doi: 10.1007/s00425-002-0828-z. [DOI] [PubMed] [Google Scholar]
- Huber S.C. and Akazawa T. (1986) A novel sucrose synthase pathway for sucrose degradation in cultured sycamore cells. Plant Physiol. 81: 1008–1013.doi: 10.1104/pp.81.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Koyama H., Iuchi A., Kobayashi Y., Kitabayashi S., Kobayashi Y., et al. (2007) Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA 104: 9900–9905.doi: 10.1073/pnas.0700117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan A., Nakamura H., Handa H., Ichikawa H., Matsumoto H. and Komatsu S. (2006) Gibberellin regulates mitochondrial pyruvate dehydrogenase activity in rice. Plant Cell Physiol. 47: 244–253.doi: 10.1093/pcp/pci241. [DOI] [PubMed] [Google Scholar]
- Kariya K., Demiral T., Sasaki T., Tsuchiya Y., Turkan I., Sano T., et al. (2013) A novel mechanism of aluminum-induced cell death involving vacuolar processing enzyme and vacuolar collapse in tobacco cell line BY-2. J. Inorg. Biochem. 128: 196–201.doi: 10.1016/j.jinorgbio.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Kariya K., Tsuchiya Y., Sasaki T. and Yamamoto Y. (2018) Aluminium-induced cell death requires upregulation of NtVPE1 gene coding vacuolar processing enzyme in tobacco (Nicotiana tabacum L.). J. Inorg. Biochem. 181: 152–161.doi: 10.1016/j.jinorgbio.2017.09.008. [DOI] [PubMed] [Google Scholar]
- Kazan K. (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20: 219‐229.doi: 10.1016/j.tplants.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y., Yamamoto Y. and Matsumoto H. (2004) Studies on the mechanism of aluminum tolerance in pea (Pisum sativaum L.) using aluminum-tolerant cultivar ‘Alaska’ and aluminum-sensitive cultivar ‘Hyogo’. Soil Sci. Plant Nutr. 50: 197–204.doi: 10.1080/00380768.2004.10408468. [DOI] [Google Scholar]
- Kochian L.V. (1995) Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46: 237–260.doi: 10.1146/annurev.pp.46.060195.001321. [DOI] [Google Scholar]
- Kochian L.V., Piñeros M.A., Liu J. and Magalhaes J.V. (2015) Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 66: 571–598.doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- Li D., Ma W., Wei J., Mao Y., Peng Z., Zhang J., et al. (2020) Magnesium promotes root growth and increases aluminum tolerance via modulation of nitric oxide production in Arabidopsis. Plant Soil 457: 83–95.doi: 10.1007/s11104-019-04274-9. [DOI] [Google Scholar]
- Licausi F., Kosmacz M., Weits D.A., Giuntoli B., Giorgi F.M., Voesenek L.A.C.J., et al. (2011) Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479: 419–422.doi: 10.1038/nature10536. [DOI] [PubMed] [Google Scholar]
- Ma J.F. (2007) Syndrome of aluminum toxicity and diversity of aluminum resistance in higher plants. Int. Rev. Cytol. 264: 225–252. [DOI] [PubMed] [Google Scholar]
- Mellema S., Eichenberger W., Rawyler A., Suter M., Tadege M. and Kuhlemeier C. (2002) The ethanolic fermentation pathway supports respiration and lipid biosynthesis in tobacco pollen. Plant J. 30: 329–336.doi: 10.1046/j.1365-313X.2002.01293.x. [DOI] [PubMed] [Google Scholar]
- Millar A.H. and Day D.A. (1996) Nitric oxide inhibits the cytochrome oxidase but not the alternative oxidase of plant mitochondria. FEBS Lett. 398: 155–158. [DOI] [PubMed] [Google Scholar]
- Mustroph A., Lee S.C., Oosumi T., Zanetti M.E., Yang H., Ma K., et al. (2010) Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 152: 1484–1500.doi: 10.1104/pp.109.151845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagao A., Kobayashi M., Koyasu S., Chow C.C.T. and Harada H. (2019) HIF-1-dependent reprogramming of glucose metabolic pathway of cancer cells and its therapeutic significance. Int. J. Mol. Sci. 20: 238.doi: 10.3390/ijms20020238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura C., Van Telgen H.J., Mennes A.M., Ono H. and Libbenga K.R. (1988) Correlation between auxin resistance and the lack of a membrane-bound auxin binding protein and a root-specific peroxidase in Nicotiana tabacum. Plant Physiol. 88: 845–849.doi: 10.1104/pp.88.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T. and Shinshi H. (2006) Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140: 411–432.doi: 10.1104/pp.88.3.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Yamamoto R., Hiraga S., Nakayama N., Okazaki K., Takahashi T., et al. (2012) Evaluation of metabolite alteration under flooding stress in soybeans. JARQ 46: 237–248. [Google Scholar]
- Nourimand M. and Todd C.D. (2016) Allantoin increases cadmium tolerance in Arabidopsis via activation of antioxidant mechanisms. Plant Cell Physiol. 57: 2485‐2496.doi: 10.1093/pcp/pcw162. [DOI] [PubMed] [Google Scholar]
- Ogawa T., Pan L., Kawai-Yamada M., Yu L.H., Yamamura S., Koyama T., et al. (2005) Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol. 138: 1436–1445.doi: 10.1104/pp.105.063586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papdi C., Pérez-Salamó I., Joseph M.P., Giuntoli B., Bögre L., Koncz C., et al. (2015) The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor-VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 82: 772–784.doi: 10.1111/tpj.12848. [DOI] [PubMed] [Google Scholar]
- Piršelová B. and Matušíková I. (2013) Callose: the plant cell wall polysaccharide with multiple biological functions. Acta Physiol. Plant 35: 635–644.doi: 10.1007/s11738-012-1103-y. [DOI] [Google Scholar]
- Prado A.M., Porterfield D.M. and Feijó J.A. (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714.doi: 10.1242/dev.01153. [DOI] [PubMed] [Google Scholar]
- Pucciariello C. and Perata P. (2017) New insights into reactive oxygen species and nitric oxide signalling under low oxygen in plants. Plant Cell Environ. 40: 73–482.doi: 10.1111/pce.12715. [DOI] [PubMed] [Google Scholar]
- Queval G. and Noctor G. (2007) A plate reader method for the measurement of NAD, NADP, glutathione and ascorbate in tissue extracts: application to redox profiling during Arabidopsis rosette development. Anal. Biochem. 363: 58–69.doi: 10.1016/j.ab.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Ricard B., Toai T.V., Chourey P. and Saglio P. (1998) Evidence for the critical role of sucrose synthase for anoxic tolerance of maize roots using a double mutant. Plant Physiol. 116: 1323–1331.doi: 10.1104/pp.116.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton P.J., Bokowiec M.T., Han S., Zhang H., Brannock J.F., Chen X., et al. (2008) Tobacco transcription factors: novel insights into transcriptional regulation in the Solanaceae. Plant Physiol. 147: 280–295.doi: 10.1093/jxb/eraq272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P.R., Tyerman S.D., Sasaki T., Furuichi T., Yamamoto Y., Zhang W.H., et al. (2011) The identification of aluminium-resistance genes provides opportunities for enhancing crop production on acid soils. J. Exp. Bot. 62: 9–20.doi: 10.1093/jxb/eraq272. [DOI] [PubMed] [Google Scholar]
- Sakano K. (1998) Revision of biochemical pH-stat: involvement of alternative pathway metabolisms. Plant Cell Physiol. 39: 467–473.doi: 10.1093/oxfordjournals.pcp.a029393. [DOI] [Google Scholar]
- Sameeullah M., Sasaki T. and Yamamoto Y. (2013) Sucrose transporter NtSUT1 confers aluminum tolerance on cultured cells of tobacco (Nicotiana tabacum L.). Soil Sci. Plant Nutr. 59: 756–770.doi: 10.1080/00380768.2013.830230. [DOI] [Google Scholar]
- Sasaki T., Ryan P.R., Delhaize E., Hebb D.M., Ogihara Y., Kawaura K., et al. (2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol. 47: 1343–1354.doi: 10.1093/pcp/pcl002. [DOI] [PubMed] [Google Scholar]
- Sasaki T., Yamamoto Y., Ezaki B., Katsuhara M., Ahn S.J., Ryan P.R., et al. (2004) A wheat gene encoding an aluminum-activated malate transporter. Plant J. 37: 645–653.doi: 10.1111/j.1365-313X.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- Sawaki Y., Iuchi S., Kobayashi Y., Kobayashi Y., Ikka T., Sakurai N., et al. (2009) STOP1 regulates multiple genes that protect Arabidopsis from proton and aluminum toxicities. Plant Physiol. 150: 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheible W.R., Lauerer M., Schulze E.D., Caboche M. and Stitt M. (1997) Accumulation of nitrate in the shoot acts as a signal to regulate shoot-root allocation in tobacco. Plant J. 11: 671–691.doi: 10.1046/j.1365-313X.1997.11040671.x. [DOI] [Google Scholar]
- Semenza G.L. (2007) Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem. J. 405: 1–9.doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- Stein O. and Granot D. (2019) An overview of sucrose synthases in plants. Front. Plant Sci. 10: 95.doi: 10.3389/fpls.2019.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoimenova M., Hänsch R., Mendel R., Gimmler H. and Kaiser W.M. (2003a) The role of nitrate reduction in the anoxic metabolism of roots I. Characterization of root morphology and normoxic metabolism of wild type tobacco and a transformant lacking root nitrate reductase. Plant Soil 253: 145–153.doi: 10.1023/A:1024563507275. [DOI] [Google Scholar]
- Stoimenova M., Libourel I., Ratcliffe R. and Kaiser W.M. (2003b) The role of nitrate reduction in the anoxic metabolism of roots II. Anoxic metabolism of tobacco roots with or without nitrate reductase activity. Plant Soil 253: 155–167.doi: 10.1023/A:1024591116697. [DOI] [Google Scholar]
- Sun C., Lu L., Liu L., Liu W., Yu Y., Liu X., et al. (2014) Nitrate reductase-mediated early nitric oxide burst alleviates oxidative damage induced by aluminum through enhancement of antioxidant defenses in roots of wheat (Triticum aestivum). New Phytol. 201: 1240–1250.doi: 10.1111/nph.12597. [DOI] [PubMed] [Google Scholar]
- Taylor A.E., Cogdell R.J. and Lindsay J.G. (1992) Immunological comparison of the pyruvate dehydrogenase complexes from pea mitochondria and chloroplasts. Planta 188: 225–231.doi: 10.1007/BF00216817. [DOI] [PubMed] [Google Scholar]
- Tejada-Jimenez M., Llamas A., Galván A. and Fernández E. (2019) Role of nitrate reductase in NO production in photosynthetic eukaryotes. Plants (Basel) 8: 56.doi: 10.3390/plants8030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., et al. (2017) agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45: W122–W129.doi: 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden M.G., Cantley L.C. and Thompson C.B. (2009) Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033.doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente J., Mendiondo G.M., Movahedi M., Peirats-Llobet M., Juan Y.-T., Shen Y.-Y., et al. (2017) The Cys-Arg/N-end rule pathway is a general sensor of abiotic stress in flowering plants. Curr. Biol. 27: 1–8.doi: 10.1016/j.cub.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-H., Huang J.-J. and Bi Y.-R. (2010) Nitrate reductase-dependent nitric oxide production is involved in aluminum tolerance in red kidney bean roots. Plant Sci. 179: 281–288.doi: 10.1016/j.plantsci.2010.05.014. [DOI] [Google Scholar]
- Yamaji N., Huang C.F., Nagao S., Yano M., Sato Y., Nagamura Y., et al. (2009) A zinc finger transcription factor ART1 regulates multiple genes implicated in aluminum tolerance in rice. Plant Cell 21: 3339–3349.doi: 10.1105/tpc.109.070771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y. (2019) Aluminum toxicity in plant cells: mechanisms of cell death and inhibition of cell elongation. Soil Sci. Plant Nutr. 65: 41–55.doi: 10.1080/00380768.2018.1553484. [DOI] [Google Scholar]
- Yamamoto Y., Kobayashi Y., Devi S.R., Rikiishi S. and Matsumoto H. (2002) Aluminum toxicity is associated with mitochondrial dysfunction and the production of reactive oxygen species in plant cells. Plant Physiol. 128: 63–72.doi: 10.1104/pp.010417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Rikiishi S., Chang Y.-C., Ono K., Kasai M. and Matsumoto H. (1994) Quantitative estimation of aluminum toxicity in cultured tobacco cells: correlation between aluminum uptake and growth inhibition. Plant Cell Physiol. 35: 575–583.doi: 10.1093/oxfordjournals.pcp.a078632. [DOI] [Google Scholar]
- Yamasaki H., Shimoji H., Ohshiro Y. and Sakihama Y. (2001) Inhibitory effects of nitric oxide on oxidative phosphorylation in plant mitochondria. Nitric Oxide 5: 261–270.doi: 10.1006/niox.2001.0353. [DOI] [PubMed] [Google Scholar]
- Zrenner R., Salanoubat M., Willmitzer L. and Sonnewald U. (1995) Evidence of the crucial role of sucrose synthase for sink strength using transgenic potato plants (Solanum tuberosum L.). Plant J. 7: 97–107.doi: 10.1046/j.1365-313X.1995.07010097.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray data reported in this article have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO series accession number (GSE158255).


