Abstract
Obesity and poor diet often go hand-in-hand, altering metabolic signaling and thereby impacting breast cancer risk and outcomes. We have recently demonstrated that dietary patterns modulate mammary microbiota populations. An important and largely open question is whether the microbiome of the gut and mammary gland mediates the dietary effects on breast cancer. To address this, we performed fecal transplants between mice on control or high-fat diets (HFD) and recorded mammary tumor outcomes in a chemical carcinogenesis model. HFD induced pro-tumorigenic effects, which could be mimicked in animals fed a control diet by transplanting HFD-derived microbiota. Fecal transplants altered both the gut and mammary tumor microbiota populations, suggesting a link between the gut and breast microbiomes. HFD increased serum levels of bacterial lipopolysaccharide (LPS), and control diet-derived fecal transplant reduced LPS bioavailability in HFD-fed animals. In vitro models of the normal breast epithelium showed that LPS disrupts tight junctions (TJ) and compromises epithelial permeability. In mice, HFD or fecal transplant from animals on HFD reduced expression of TJ-associated genes in the gut and mammary gland. Furthermore, infecting breast cancer cells with an HFD-derived microbiome increased proliferation, implicating tumor-associated bacteria in cancer signaling. In a double-blind placebo-controlled clinical trial of breast cancer patients administered fish oil supplements before primary tumor resection, dietary intervention modulated the microbiota in tumors and normal breast tissue. This study demonstrates a link between the gut and breast that mediates the effect of diet on cancer.
Introduction
Breast cancer is the most prevalent cancer in women, with over 281,000 new cases diagnosed annually and over 42,000 women dying each year in the United States (1). Several studies have demonstrated a strong link between obesity and a greater risk of developing breast cancer by as much as 50% in postmenopausal women (2). Moreover, some studies have reported a three-fold higher breast cancer mortality rate in obese women. It is estimated that up to 3 out of 10 breast cancers might be prevented if women were not overweight, indicating an essential role of body weight in the etiology of breast cancer (3).
Several studies have highlighted the critical role of the gut microbiome in obesity. Mice predisposed to obesity have a shift in the ratio of two major phyla, Bacteroidetes: Firmicutes, which translates into an increased ability to harvest energy from indigestible dietary products, higher lipopolysaccharide (LPS) bioavailability, and promotion of chronic low-grade inflammation (4–7). Furthermore, the types of fat consumed by mice in obesity-inducing diets (OID) impact their gut microbiome population (4). In human obesity, decreased bacterial diversity was correlated with adiposity (5–7). The gut microbiome is a critical variable affecting disease development. Gut microbiome dysbiosis is associated with many other adverse health outcomes, including cancer, diabetes (types 1 and 2), asthma, allergies, and inflammatory bowel disease (5,8–12).
Increasing evidence suggests that the gut microbiome is implicated in cancer development and recurrence in various cancers, including colon, liver, lung, stomach, and skin (13). While most studies to date have explored the role of the microbiome in intestinal disease, preliminary research implicating the gut microbiome in breast cancer is emerging. In preclinical studies, pathogenic Helicobacter hepaticus infection promoted mammary adenocarcinoma in female C57BL/6 APCmin/+ mice reliant upon intact immunity pro-inflammatory tumor necrosis factor (TNF)-α (14–16). In clinical studies, postmenopausal breast cancer patients had decreased fecal microbial diversity compared with controls, supporting the role of the microbiome in breast cancer (17). Additional evidence of gut microbiome affecting breast health was seen in a clinical trial of orally administered Lactobacillus salivarius. Treatment with Lactobacillus reduced the incidence and severity of lactational mastitis (18). Moreover, recent clinical studies have noted fecal microbiota shifts in breast cancer patients that exhibit quality of life side effects; severe breast cancer-related fatigue correlated with increased fecal Bacteroides and Ruminococcus abundance when compared with breast cancer patients without fatigue (19).
There is also evidence of a breast mammary gland and tumor-specific microbiome (20–27). Human breast tissue samples displayed a low Bacteroidetes: Firmicutes ratio, similar to the gut ratio from obese individuals (24). We demonstrated in a non-human primate model that dietary patterns influence the normal mammary gland microbiome; Consumption of a Mediterranean diet led to a 10-fold increase in breast Lactobacillus abundance compared with Western diet-fed monkeys (23). Bile salt (originating from the intestine) exists in the aspirate of breast cyst fluid, which is further evidence for a gut-breast signaling axis that may affect breast health (28). In human breast cancer clinical studies, nipple aspirates from women with a history of breast cancer had elevated amounts of genus Alistipes microbes (20). Moreover, mammary gland tissue obtained from women with either benign or malignant breast tumors indicated reduced Lactobacillus abundance in mammary gland tissue from patients with malignant breast disease (22). More recently, elevated enterotoxigenic Bacteroides fragilis was observed in cancerous breast tissue, with B. fragilis toxin inducing breast epithelial hyperplasia. Both gut and breast enterotoxigenic B. fragilis (but not non-toxigenic B. fragilis) enhanced breast tumor growth and metastatic potential in a murine triple negative breast cancer, giving further evidence in support of a entero-mammary signaling axis (29). These data suggest an impact of mammary gland microbiota on breast cancer. We recently demonstrated that systemic neoadjuvant chemotherapy was associated with increased breast tumor Pseudomonas abundance (21). Administration of P. aeruginosa conditioned media to breast cancer cells potentiated chemotherapy responsiveness. These data suggest that bioactive compounds secreted by tumor-associated bacteria can directly influence breast cancer initiation and therapy efficacy. Overall, there is evidence that the microbiome plays an important role in mediating breast cancer risk and breast cancer outcomes. However, the mechanisms linking the microbiome to dietary-influences on breast cancer risk are largely unknown.
In the present study, we found that supplementing control diet-fed mice with high-fat diet (HFD)-derived microbiota in a fecal transplant model increased breast cancer risk. Moreover, fecal transplant shifted both the gut and breast tumor microbiota populations, suggesting a pivotal role of the mammary gland and tumor microbiota in promoting tumorigenesis. Administration of antibiotics reduced mammary carcinogenesis. Co-culturing 4T1 breast cancer cells with HFD-derived microbiota before injection into the mammary tissue or treating tumor-bearing mice with LPS enhanced tumor growth, which was not observed in untreated or 4T1 co-cultured with control diet-derived microbiota tumors. Furthermore, in a double-blind placebo-controlled clinical trial investigating oral fish oil supplements in breast cancer patients, fish oil consumption for approximately four weeks before tumor resection surgery significantly modified tumor-adjacent mammary gland and breast tumor microbiota populations, demonstrating that oral interventions can alter breast tumor microbiota.
Materials and Methods
Materials.
Lipopolysaccharide (LPS) was purchased from Sigma Aldrich (cat# L2630). Antibodies were obtained for lipoteichoic acid (LTA) (Gram-positive bacteria; Santa Cruz; cat# sc-57752), ZO-1 (Invitrogen; cat# 339100), claudin-1 (AbCam; cat# ab15098), Ki67 (Cell Signaling; cat#12202), F4/80 (Cell Signaling; cat# 70076), anti-human CD45 Antibody (Pacific Blue™-conjugated; BioLegend; cat# 368539), Cytokeratin (AlexaFluor®−488-conjugated; BioLegend; cat# 628608), and PE-conjugated mouse IgG1 (BioLegend; cat# 400111). AlexaFluor-488 and AlexaFluor-568 secondary antibodies were from Life Technologies (cat# A-11034 and A-11031). RT-PCR primers were purchased from IDT (see methods below for primer sequences). LPS ELISA and IHC antibody was purchased from LSBio (cat# LS-F17912–1 and LS-C375096). A secondary LPS antibody was purchased from MyBioSource (cat# MBS2012794) to confirm results. FITC-dextran (4-kDa) was purchased from Sigma Aldrich (cat# 46944–500MG-F). EPA was purchased from Cayman Chemicals.
Study approval and tissue procurement.
This study was approved by the institutional review board of the Wake Forest School of Medicine (IRB00023419) in accordance with HHS regulations for the protection of human research subjects. Subjects (n=22) were identified as participants in a prospective randomized, placebo-controlled phase II clinical trial of omega-3 PUFA dietary supplementation in patients with stage I-III breast carcinoma. Informed written consent was obtained from all the participants. Patients received either 2 grams of omega-3 PUFA (650 mg eicosapentaenoic acid (EPA) /450 mg docosahexaenoic acid (DHA)) or placebo (soybean oil) post orally daily from enrollment to surgery (range of 4–52 days; mean 14.5 days intervention). Breast tumor specimens were snap-frozen within 45–60 minutes from surgery. The pathologist’s visual inspection made a tumor or normal designation before submitting tumor and tumor-adjacent mammary gland samples to the Tumor Bank (21).
Animals.
Female 3-week-old BALB/c and female 4-week-old C57BL/6 mice were purchased from Jackson. All Teklad custom diets were purchased from Envigo. Mice were placed on a 10% fat/10% sucrose control diet (Control; TD.08806) or a 60% kcal from fat/ 10% sucrose lard-based diet (HFD; TD.06414). Mice were placed on a 45% kcal from the fat Western diet (TD.180300; omega-6:omega-3 PUFA ratio of 34.4) or a Western diet where 8% kcal from palm oil were replaced with fish oil (Western + fish oil diet; TD.180301; omega-6:omega-3 PUFA ratio of 6.2). The protocol was approved by the Animal Care and Use Committee of the Wake Forest School of Medicine, and all procedures were carried out in accordance with relevant guidelines and regulations.
Fecal transplant DMBA-mammary carcinogenesis model.
Female 3-week-old BALB/c mice were placed on control (n=40) or lard (n=45) diets. Subsets of mice from each diet were given antibiotics (streptomycin 5 mg/mL, colistin 1 mg/mL, and ampicillin 1mg/mL) in their drinking water (n=15 per diet) or a control diet-derived or lard diet-derived fecal gavage once weekly for the entire duration of the study (n=10 per fecal transplant condition). Fecal-derived slurries were formulated 25% per volume from fresh feces obtained from mice fed either the control diet or lard diet in saline. At 6 weeks of age, mice were treated with a 15 mg subcutaneous single injection of medroxyprogesterone acetate followed by weekly doses of 1 mg DMBA in peanut oil for 3 weeks to induce mammary tumorigenesis (30). Mice were palpated weekly for tumor formation for 16 weeks post-DMBA administration. See Figure 1A for the model schematic. Tumor-free survival, tumor multiplicity, tumor latency, and tumor weight were recorded. Plasma, tumors, and tissues from mammary glands, intestine, and liver were collected at the end of the study.
Figure 1.
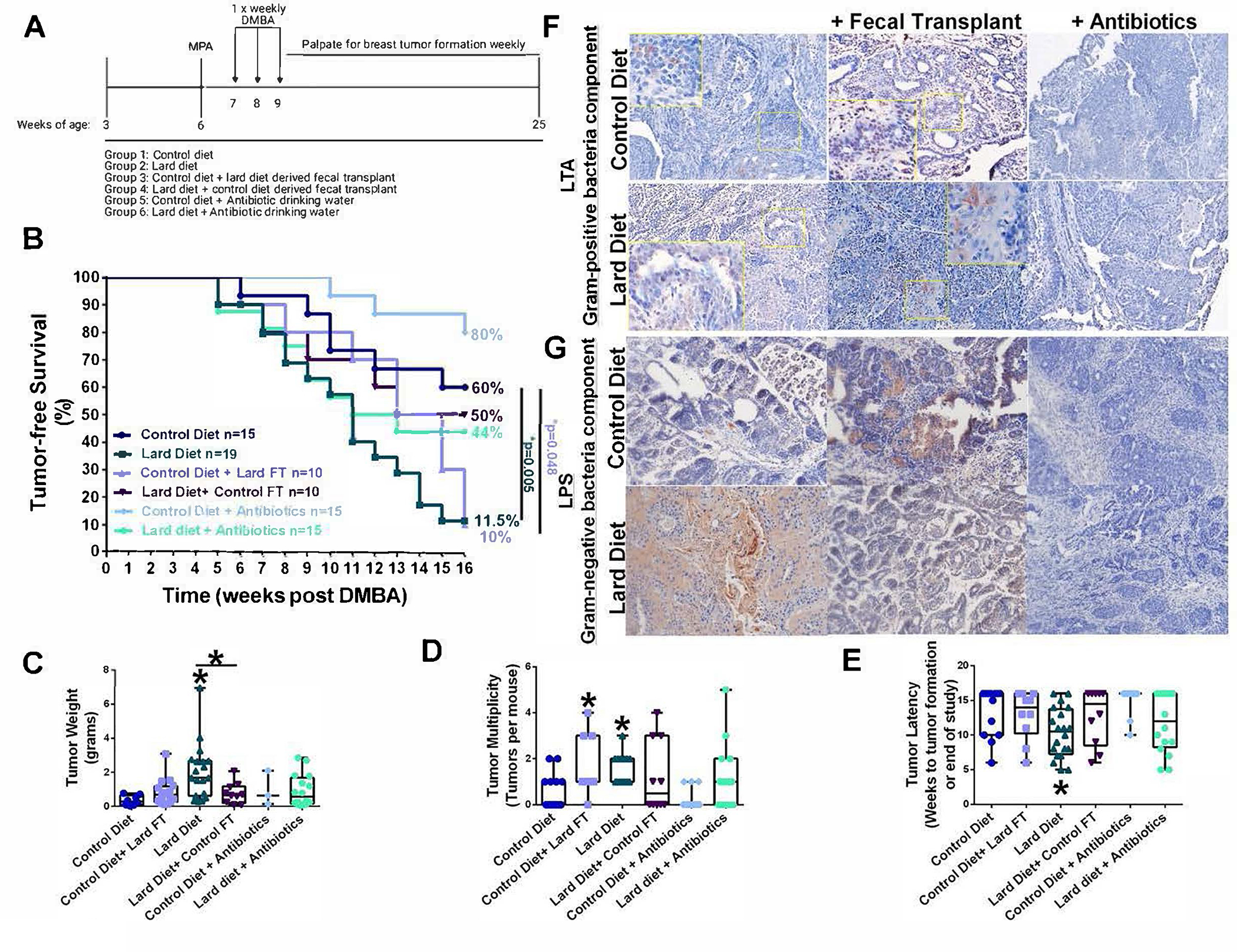
Modulating microbiota regulates dietary-influenced breast cancer risk. A. MPA-DMBA mammary carcinogenesis model schematic in mice. B. Tumor-free survival in mice fed a control or lard diet. A subset of mice on each diet was given broad-spectrum antibiotics administered in their drinking water. Another subset of mice on each diet was given weekly fecal transplant for the study duration. Groups are as follows: Control diet-fed mice, lard diet-fed mice, control diet-fed mice given a lard diet-derived fecal transplant, lard diet-fed mice given a control diet-derived fecal transplant, control diet-fed mice given antibiotics, and lard diet-fed mice given antibiotics. Breast tumors were induced by MPA-DMBA administration, and mice were palpated weekly for breast tumor formation. Comparison of survival curves was performed by Log-rank (Mantel-Cox) test. C. Tumor weight at the end of the study. n=3–20. *p<0.05. D. Tumor multiplicity at the end of the study n=10–19. *p<0.05. E. Tumor latency. n=10–19. *p<0.05. F. Breast tumor sections were stained with an antibody against LTA to identify gram-positive bacteria. G. Breast tumor sections were stained with an antibody against LPS to identify gram-negative bacteria.
Cell culture.
Cells were grown at 37°C in a humidified, 5% CO2:95% air atmosphere. 4T1 (CRL-2539) mouse mammary epithelial cells (mimicking stage IV breast cancer) and RAW 264.7 mouse macrophage cell lines were cultured in phenol red-containing RPMI medium supplemented with 10% fetal bovine serum (FBS) and defined as basal growth conditions. These cell lines were authenticated by IDEXX BioAnalytics using short tandem repeat (STR) analysis in February 2020 (Columbia, MO). HMT-3522 S1 (S1) non-neoplastic mammary epithelial cells were obtained from Dr. Mina Bissell (Lawrence Berkeley Laboratory) and propagated between passages 54 and 60 in H14 medium (31). For 3D cell culture experiments, acinar differentiation was achieved by culturing S1 cells on top of Matrigel (Corning, Corning NY, USA) in 8-well chambered slides (MilliporeSigma, Burlington MA, USA) for 10 days, as described (32).
Epithelial polarity and permeability assays.
To assess the effect of diet on epithelial polarity, differentiated S1 acini were treated with 1 μg/mL LPS or with 1% (v/v) H14 fecal-conditioned sterile media (FCM) derived from control or lard diet-fed mice. FCM was formulated by mixing 1 gram of feces from mice fed a control or lard diet in 10 mL of H14 medium. Feces-media mixtures were vortexed to obtain uniform suspensions and incubated for 1 hour in a 37°C water bath. Media were sterile filtered to remove bacteria contaminants but leave fecal-derived bacterial compounds and metabolites. Acini were treated twice, 11 and 13 days after cell seeding, and immunostained on day 14 to quantify the distribution of apical polarity markers ZO-1 and claudin-1, as described previously (33). Briefly, acini were permeabilized, fixed, and incubated overnight at 4°C with the primary antibodies in the presence of 10% goat serum. Secondary antibodies were added for 40 min at room temperature. Nuclei were counterstained with 4’, 6-diamidino-2-phenylindole (DAPI). Samples were mounted using ProLong antifade solution (ThermoFisher). Confocal imaging was performed with a Zeiss CLSM710 confocal microscope, using a 63x oil immersion objective (NA = 1.4), and 100 acini per replicate were assessed for polarity.
To assess the effect of LPS on trans-epithelial permeability, S1 cells were seeded at a density of 90,000 cells/cm2 on laminin-coated (Corning; 2.8 μg/cm2) inserts with 0.4 μm pore sizes to induce polarization of the cell monolayers. Cells were treated with LPS as above, and Evans blue-labeled albumin (EBA) was added on day-14 to the abluminal chamber for 1 hour. EBA concentration in the abluminal chamber was assessed by spectrophotometry at OD 650 nm. Measurements were done in triplicates.
Breast cancer cell microbiota co-culture model.
4T1 or RAW 264.7 cells were treated with 1:10 dilution of 10% fecal-derived conditioned media for 24 hours. Fecal-derived conditioned media was formulated by mixing 1 gram of feces from mice on different diets in 10 mL of basal growth RPMI media without antibiotics. Feces-media mixture was vortexed until uniformly suspended in media and incubated for 1 hour in a 37°C water bath. Unlike the above-described fecal-derived conditioned media utilized in the breast epithelial polarity and permeability assays, media was not sterile filtered to remove bacteria that enabled bacterial infection of breast cancer cells before in vivo implantation. Cells were rinsed three times with sterile PBS, cells were harvested, and 1 × 106 4T1 cells or 1 × 106 4T1 + 1 × 105 RAW 264.7 cells were injected into the right inguinal mammary fat pad of 8-week BALB/c mice. Mice bearing tumors generated from inoculation with untreated 4T1 cells were treated weekly with 5 mg/kg LPS by IP injection. Tumors were measured with calipers every 3 days for 4 weeks. At the end of the study, tumors were excised, weighed, and snap-frozen or embedded in paraffin for further analysis.
Intestinal permeability assay.
We fed 4-week old female C57BL/6 mice a Western or Western + Fish oil diet for 12-weeks. At 16 weeks of age, mice were overnight fasted and FITC-dextran intestinal permeability assay was performed (34). In brief, mice were administered 600 mg/kg 4-kDa FITC dextran. After 1 hour, mice were sacrificed and plasma collected. Plasma was diluted 1:5 in PBS, and plasma FITC-dextran concentrations will be calculated from a standard curve on a fluorescent plate reader (Cytation1; Agilent).
Colon thickness ultrasound measurements.
At the end of the study, the colon was excised from Western or Western + Fish oil diet-fed mice, rinsed clean, and filled with ultrasound gel. The colon was covered with additional gel immediately before scanning with the Vevo 2100 ultrasound system (Fujifilm’s VisualSonics) using a 40 MHz probe on transverse position. Using Vevo Lab software, images were obtained sequentially, starting from the distal region moving toward the proximal region. The images were analyzed using three images from each of the three regions (distal, middle, and proximal) for each animal were selected. The thickness of the colon was determined at 6 different points around the circumference of each image and averaged, and the average of these three images is reported as a single data point for each region of the colon.
RT-PCR.
RNA was extracted from the intestine and mammary glands tissues using Trizol following the manufacturer’s protocol. cDNA was synthesized from 5 μg of total RNA using Superscript first-strand RT-PCR reagents described by the manufacturer. qRT-PCR was then performed using the Sybr green kit. Primers were used for the following genes sequences: ZO-1 (CCTGTGAAGCGTCACTGTGT; CGCGGAGAGAGACAAGATGT), HPRT (CATAACCTGGTTCATCATCGC; TCCTCCTCAGACCGCTTTT), or Tuba1a (AGATGCCAAGCGACAAAACCA; AAAAAGCTGCCGGTAGGTTCC).
LPS endotoxin ELISA.
Snap-frozen plasma samples from mice were used at a 1:3 dilution to determine microbiota effects on circulating LPS bioavailability following the manufacture’s instructions. Snap-frozen mammary gland samples from Western or Western + Fish oil diet-fed C57BL/6 mice were homogenized in PBS, and 35 μg of protein was used to determine localized MG LPS bioavailability.
Immunohistochemistry.
Sections of paraffin-embedded 4T1 or DMBA-induced breast tumors were stained using antibodies recognizing LTA or LPS to identify the presence of a tumor-specific microbial population. Sections from 4T1 or DMBA-induced breast tumors were also stained using antibodies against Ki67 (proliferation) or F4/80 (macrophage marker). Staining was visualized by the Mantra Quantitative Pathology Image System with a 20x objective. Human breast tumor microarray was purchased from US Biomax, Inc. (BR1008a) that contained primary breast tumor tissue (n=50), breast tumor lymph node metastases (n=40), and mammary gland tissue (n=10). The TMA was stained against LTA or LPS using the Dako DAB chromagen IHC kit, and percent positive breast cancer cases were quantified. The TMA was also co-stained with an antibody against LTA (PE-red), infiltrating immune cells (CD45; PacificBlue-blue), and cell structural marker cytokeratin (Alexa Fluor 488-green) to visualize potential bacterial co-localization within immune cells. Human placebo or fish oil-treated patient tumor tissue from CCCWFU 98113 clinical trial were stained for LTA or LPS using the Dako DAB chromagen IHC kit, and percent positive breast cancer cases were quantified.
16S sequencing and statistics.
Mouse and human bacterial microbiome 16S sequencing were performed by Microbiome Insights Inc. (Vancouver, British Columbia), as previously described in (21). In brief, DNA was isolated from the feces using the MoBio Powersoil extraction kit. 16S rRNA genes were PCR- amplified with dual-barcoded primers targeting the V4 region, as previously described (35,36). Amplicons were sequenced with an Illumina MiSeq using the 250-bp paired-end kit (v.2). Bacterial sequences were denoised, taxonomically classified using Greengenes (v. 13_8), and clustered into similar operational taxonomic units (OTUs) with the mothur software package (v. 1.38) following the recommended protocol (https://www.mothur.org/wiki/MiSeq_SOP). MiSeq-generated Fastq files were quality-filtered and clustered into operational taxonomic units (OTUs) using the mothur software package [http://www.mothur.org]. High-quality reads were classified using Greengenes v. 13_8 as the reference database. The significance of diversity differences was tested using analysis of variance (ANOVA). We corrected for the paired nature of the experimental design in the fecal transplant study by restricting the permutations to mouse ID. Finally, we used DESeq2 to examine OTUs across treatment groups.
Statistics.
Data are presented as the mean ± standard deviation (SD). Statistical differences were evaluated by one-way analysis of variance (ANOVA) followed by Bonferroni post hoc tests to compared all groups to each other, or tumor-free survival data by log-rank Mantel-Cox test. Apical polarity measurements between control and lard FCM or control and LPS treated structures was evaluated by a two-tailed paired t-test. Correlations between tumor microbe content and tumor infiltrating leukocytes were summarized by Pearson correlation and expressed as r. Statistical significance was set at p < 0.05.
Results
Modulating the enteric microbiota shifts dietary-mediated risk of mammary tumors in a murine model of breast cancer.
Female mice were placed on a control diet or lard diet at weaning. A subset of mice from each group (n=10–20) was given antibiotics in their drinking water, and tumors were induced using a MPA/DMBA mammary carcinogenesis protocol (Figure 1A). One mouse consuming a lard diet on the MPA/DMBA protocol was removed before tumor formation (week 6) due to rapid weight loss and is indicated by a censored point in Figure 1B. As expected (30), the lard diet significantly reduced tumor-free survival and increased tumor weight and tumor multiplicity (Figure 1B–D). Broad-spectrum antibiotics did not result in significant changes in tumor characteristics. However, trends towards improved outcomes were apparent for animals treated with antibiotics: 80% of the mice on a control diet supplemented with antibiotics were tumor-free compared to 60% of mice fed the same diet without antibiotics. Similarly, antibiotics increased tumor-free survival from 11.5% to 44% for mice on a lard diet.
Giving broad-spectrum antibiotics results in a systemic ablation of the host microbiota, both commensal and pathogenic. As a more nuanced approach to understanding the impact of the microbiota on dietary-mediated breast cancer risk, we used weekly fecal transplants to effectively swap the enteric microbiome between dietary groups. To an equal degree, mice fed a lard diet and mice on a control diet receiving fecal transplants from lard-fed animals had significantly reduced tumor-free survival (10–12%) compared to mice fed control diet (60% tumor-free survival rate) (Figure 1B). Reciprocally, tumor-free survival of lard diet-fed mice receiving fecal transplants from animals on the control diet increased from 12% to 50% compared to the mice fed the same diet without a fecal transplant. Tumor weight was greater in the lard diet group versus both controls and the lard diet group given fecal transplants from the control diet group (Figure 1C). Similarly, tumor multiplicity (or the numbers of breast tumors per mouse) was significantly elevated in lard diet-fed mice and in control diet-fed mice that received fecal transplants from mice on the lard diet (Figure 1D). Tumor latency (or the weeks to initial tumor detection) was significantly shorter in the lard diet-fed animals (Figure 1E).
To determine whether fecal transplants had any effects on tumor bacterial content, mammary tumors were stained for bacterial outer membrane or cell wall components, such as lipoteichoic acid (LTA) as a marker of gram-positive bacteria (Figure 1F) or LPS as a marker or gram-negative bacteria (Figure 1G). Bacteria from the Firmicutes phyla are predominately gram-positive and detected by LTA positivity. Since consumption of a high-fat diet increases Firmicutes’ proportional abundance (37), elevated intratumoral gram-positive bacteria may indicate a dietary effect. Interestingly, differences in gram-positive and gram-negative bacteria localization were noted depending upon the treatment. Mammary tumors from mice fed a control diet or mice on a lard diet given a fecal transplant from the control group showed only a few cytoplasmic gram-positive bacteria staining in infiltrating immune-like cells (Figure 1F) and reduced LPS positivity (Figure 1G).
In contrast, mammary tumors from lard diet-fed mice and mice on a control diet given fecal transplants from the lard diet group displayed increased nuclear and cytoplasmic gram-positive bacteria staining in tumor epithelial cells and increased intracellular LPS immuno-reactivity. Antibiotic treatment in either diet ablated LPS and LTA staining. Tumor sections were also stained for Ki67 and F4/80 to determine microbiota effects on proliferation and infiltrating tumor-associated macrophages (Supplemental Figure S1A–B). Mammary tumors from mice consuming a lard diet or mice on a control diet given fecal transplants from the lard diet group displayed elevated Ki67 immuno-reactivity and elevated tumor-associated macrophages, suggesting diet-regulated microbiota mediate tumor inflammation and proliferation.
Fecal transplantation shifts the proportions of bacteria in the fecal and breast tumor microbiomes.
Both consumption of the lard diet and fecal swap of microbiota from lard-fed mice to control diet-fed mice reduced the proportional abundance of Bacteroidetes and increased Firmicutes abundance (Figure 2A). However, transplant of control diet-derived microbiota to lard diet-fed mice did not affect phylum level proportions of enteric bacteria, suggesting weekly control diet fecal transplant is insufficient to overcome high fat dietary influences at the phylum level. However, there were significant changes in beta diversity. Volcano plots generated from the 16S sequencing of feces indicated significant regulation of Akkermansia by fecal transplants in both diet groups (Figure 2B–C). In addition, bacteria from the butyrate-producing genus (Butyricimonas) and the Lachnospiraceae family were in higher abundance in the lard diet and lard diet fecal transplant groups (Figure 2C).
Figure 2.
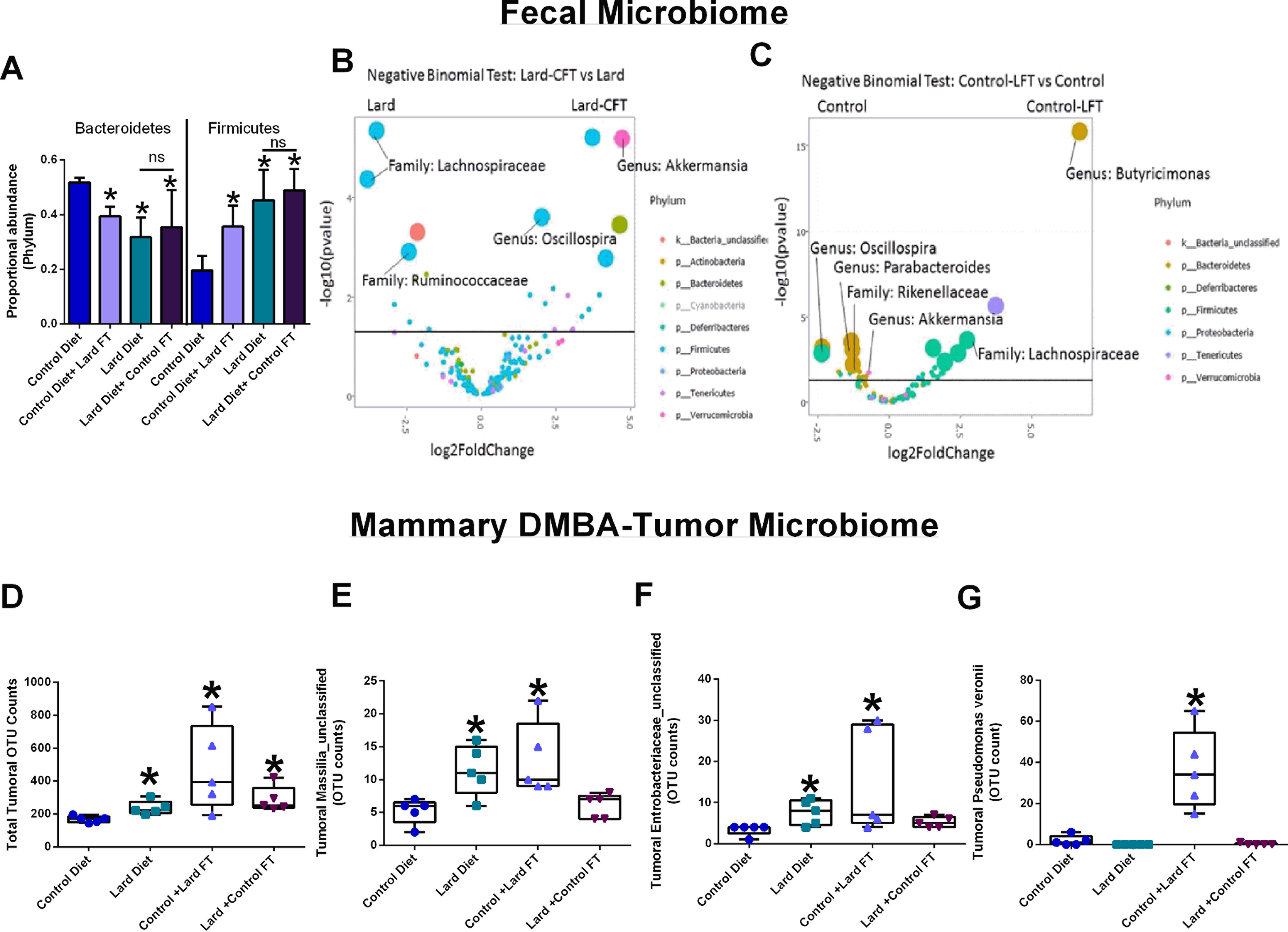
Fecal transplant shifts both gut and mammary microbiome. 16S sequencing of DNA isolated from fecal and breast tumor samples. A. Proportional abundance of Bacteroidetes and Firmicutes phyla in fecal samples collected from the different treatment groups. B. Volcano plots comparing microbiota population shifts in fecal samples collected from lard diet-fed and lard diet-fed mice administered weekly fecal transplants with feces from control diet-fed mice. C. Volcano plots comparing microbiota population shifts in fecal samples collected from control diet-fed and control diet-fed mice administered fecal transplants from the lard diet group. D. Total operational taxonomic units (OTU) counts in mammary tumors. Tumoral OTU counts of Massilia_unclassified (E), Enterobacteriaceae_unclassified (F), and Pseudomonas veronii (G) were identified in tumors from the different treatment groups. n=5; *p<0.05 relative to control diet.
16S sequencing of mammary tumors from this DMBA fecal transplant study indicates that all intervention groups displayed elevated total operational taxonomic units (OTU) counts in tumors compared to tumors from control diet-fed animals (Figure 2D), meaning that interventions modulated tumor total bacteria abundance. Lard diet-consuming mice and control diet-fed mice given a lard fecal transplant had increased tumoral Massilia_unclassified (Figure 2E) and Entrobacteriaceae_unclassified bacteria (Figure 2F), while control diet-fed mice given a lard fecal transplant showed elevated Pseudomonas veronii (Figure 2G). All three of these Proteobacteria taxa express LPS and maybe the populations identified by tumoral LPS IHC staining (Figure 1G).
Diet-regulated microbiota modulates epithelial cell polarity in the intestine and mammary gland.
In the intestine, tight junctions (TJ) establish a seal within the epithelial cell layer, enabling selective transport of nutrients into the interstitial space. Microbiota can regulate tight junction proteins to influence tissue permeability (38,39). Accordingly, we measured the reduced expression of the TJ marker ZO-1 in intestinal tissue samples from the lard diet group and animals on a control diet that received fecal transplants from the lard diet group (Figure 3A). As an independent measure of intestinal permeability, we measured microbial lipopolysaccharide (LPS) bioavailability in snap-frozen plasma samples from the different treatment groups. Consumption of the lard diet significantly elevated circulating LPS concentrations compared to the plasma concentrations in control diet-fed mice. Reciprocally, administration of fecal transplants from the control diet group to animals consuming the lard diet reduced plasma LPS concentrations (Figure 3B). These results reinforce the well-established concept that high-fat diets have a negative impact on intestinal barrier function (40).
Figure 3.
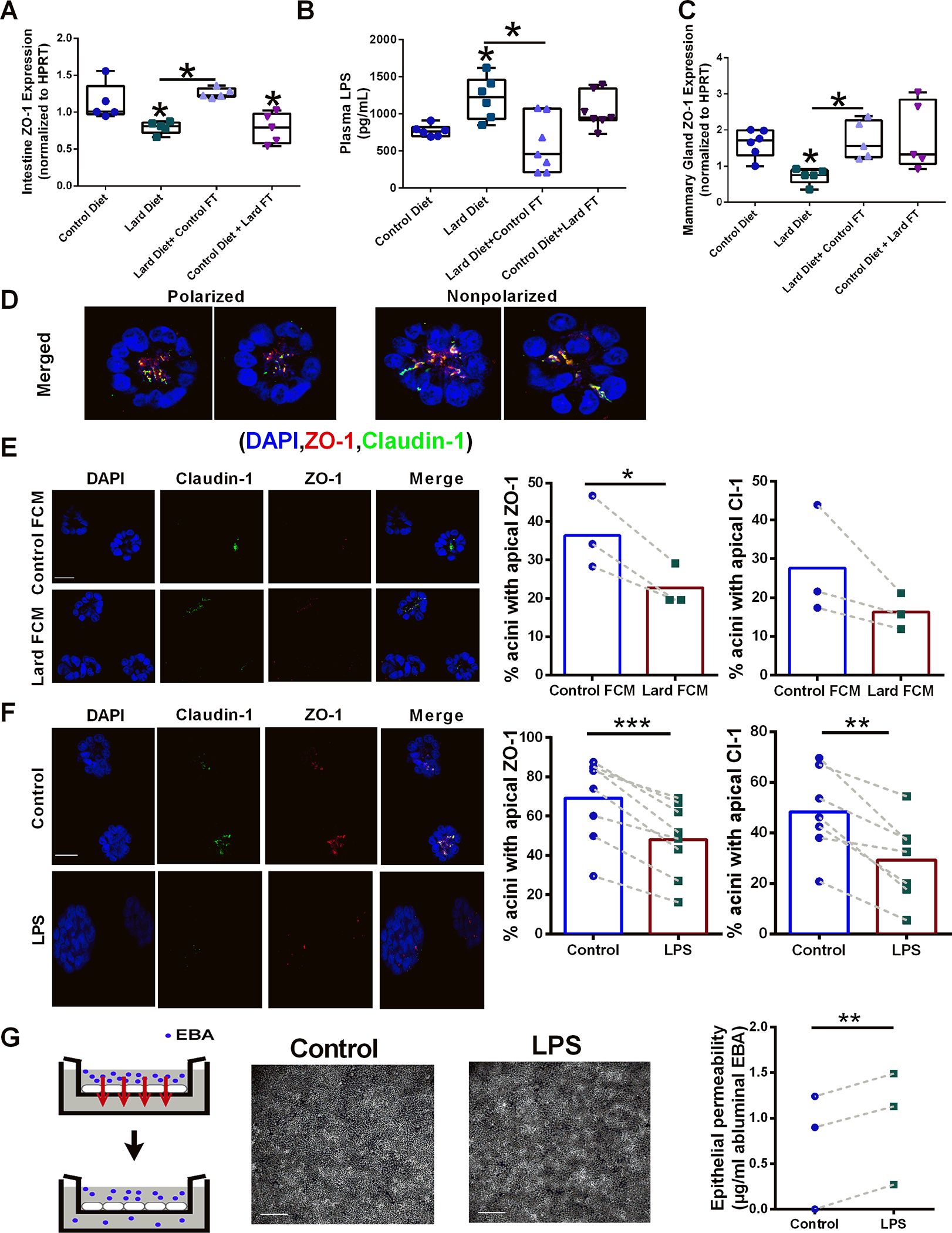
Diet-derived microbiota affects epithelial permeability and apical polarity markers. A. Relative ZO-1 gene expression normalized to HPRT in intestinal tissues from control diet-fed mice, lard diet-fed mice; lard diet-fed mice administered a control diet-derived fecal transplant, and control diet-fed mice administered a lard diet-derived fecal transplant n=5; *p<0.05. B. Circulating plasma LPS from control diet-fed mice, lard diet-fed mice, lard diet-fed mice administered a control diet-derived fecal transplant, and control diet-fed mice administered a lard diet-derived fecal transplant n=6;*p<0.05. C. Relative ZO-1 gene expression normalized to HPRT in mammary gland tissues from control diet-fed mice, lard diet-fed mice; lard diet-fed mice administered a control diet-derived fecal transplant, and control diet-fed mice administered a lard diet-derived fecal transplant n=6;*p<0.05. D. Examples of polarized and non-polarized mammary acini structures stained for the tight junction markers ZO-1 (red) and claudin-1 (green). Cell nuclei were counter-stained with DAPI (blue). Scale bars, 10 μm. E. Mammary acini structures were treated with control diet-derived or lard diet-derived sterile conditioned media, and the effects on apical polarity were measured by ZO1 and Claudin-1 localization n=3; *p<0.05. Scale bars, 100 μm. F. Mammary acini structures were treated with PBS (control) or 1 μM LPS, and the effects on apical polarity were measured by ZO1 and Claudin-1 localization n=6;*p<0.05. Scale bars, 100 μm. G. The effect of LPS on breast epithelial permeability was measured using a transwell assay. Monolayers of breast epithelial cells were treated with PBS or LPS, and diffusion of Evans blue albumin (EBA) to the lower compartment was quantified as a measure of permeability. n=3;*p<0.05. Microscopy images of DAPI-stained nuclei show that LPS treatment did not affect cell layer confluence. Scale bars, 200 μm.
Interestingly, in addition to changes in the intestine, mice on the lard diet had significantly reduced ZO-1 gene expression in mammary gland tissues compared with animals in the control diet group. Fecal transplantation reversed this effect (Figure 3C). Control fecal transplant from the lard group did not alter ZO-1 expression in the control diet group, suggesting that commensal microbiota is sufficient to maintain ZO-1 expression in the mammary gland in this context. These results go along with our previous observation that a high-fat diet disrupts epithelial cell polarity in the mammary gland (33) and uncover the microbiome as a regulator of this effect.
Elevated levels of pro-inflammatory LPS in the serum from the lard diet group may directly affect mammary epithelial cells.
To address whether microbial metabolites regulate breast epithelial polarity, we used 3D cultures of human breast epithelial cells recapitulating mammary gland units (acini). As shown in Figure 3D, this culture system recapitulates epithelial polarization, which can be quantified by immunostaining for TJ markers. We treated acini with fecal conditioned media or LPS to simulate breast epithelial cell exposure to bacterial compounds with pro-inflammatory activity. Acini treated with LPS or with lard diet-derived fecal conditioned medium displayed a reduction in apical staining of the TJ proteins ZO-1 and claudin-1 (Figure 3E–F), suggesting that loss of apical polarity may be driven by bioactive compounds produced from dysbiotic gut microbiota. Functionally, treatment of differentiated breast epithelial monolayers with LPS increased trans-epithelial permeability (Figure 3G), indicating that microbiota bioactive compounds may disrupt tissue architecture in the mammary gland. This observation is significant because the loss of epithelial polarity in normal tissues is considered necessary for tumor initiation (41). Increased mammary gland permeability caused by TJ dysfunction may also affect bacterial localization in the breast epithelium.
Human breast tumors contain bacteria outside of the immune milieu.
To determine whether bacteria are present in human breast tissues, we stained a human tissue microarray comprised of primary breast tumor samples, breast cancer lymph node metastatic lesions, and normal mammary gland tissue (adjacent to the tumors) using antibodies directed against LTA (Figure 4A) or LPS (Figure 4B). The majority of human breast tissue stained positive for LTA but not LPS-positive bacteria (Figure 4C). Interestingly, primary breast tumors and normal-adjacent mammary gland tissue had high (70–78% positive) LTA tissue staining, while only 35% of breast tumor lymph node metastases stained positive (Figure 4C). On the other hand, primary breast tumors and breast lymph node metastases displayed high (72–93% positive) LPS reactivity, while only 10% of normal-adjacent mammary gland tissue had high LPS positivity. To investigate whether intratumoral bacterial content influences immune cell infiltration, we co-stained human breast tumor tissue for CD45 (common leukocyte antigen; pan hematopoietic cell marker), LTA (gram-positive bacteria), and cytokeratin (breast tumor cytoskeleton marker). We show using immunofluorescence (Figure 4D) that the bacterial abundance in primary breast tumors varies widely. While gram-positive bacteria-laden cells and CD45+ tumor-infiltrating leukocytes positively correlated within tumor sections (Figure 4E; r=0.8945; n=20, p<0.001), gram-positive bacteria were not identified within CD45+ cells, suggesting that tumor immune cell recruitment was stimulated by bacteria presence within epithelial cells. The presence of bacteria within breast tumor tissue may influence breast tumorigenesis through immune recruitment activities or directly affecting cancer cell signaling.
Figure 4.
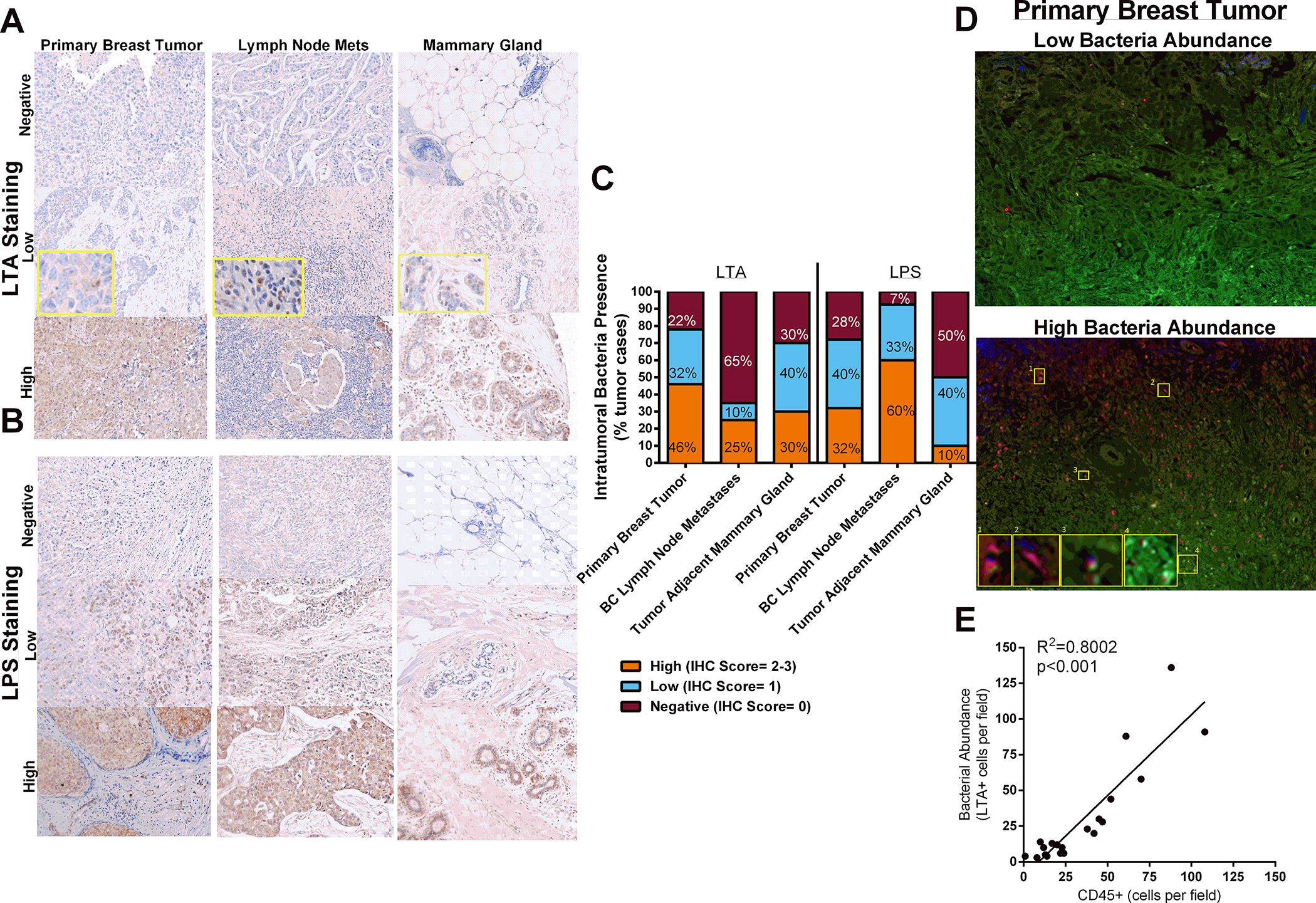
Bacteria localization in human tissue. A. Primary breast tumors, breast cancer lymph node metastases, and tumor-adjacent normal mammary gland tissue were stained with an antibody against gram-positive bacteria to indicate the presence of bacteria in human patient samples. B. Primary breast tumors, breast cancer lymph node metastases, and tumor-adjacent normal mammary gland tissue were stained with an antibody against LPS. C. LTA and LPS stained tissue were quantified. n=10–50. D. Breast tumor tissue microarray was stained against CD45 (blue), LTA (red), and pan-cytokeratin (green). E. Bacteria laden cells correlates with tumor-infiltrating leukocyte content (n=20; r=0.8945; p<0.001).
Diet-influenced microbiota modulates 4T1 mammary tumor growth in vivo.
Having detected bacteria in cancer cells in most of the breast tumors analyzed, we next assessed whether diet-regulated microbe presences within tumor epithelial cells affected cancer outcomes. Using non-sterile fecal-derived conditioned media, we purposefully infected 4T1 cancer cells before injection into mammary glands of female BALB/c mice to determine the effects of intracellular bacteria on tumor growth in vivo. 4T1 cells inoculated with lard diet fecal-derived microbiota potentiated primary tumor growth compared with tumor growth from mice bearing 4T1 cells inoculated with control diet fecal-derived microbiota or uninfected 4T1 cells (Figure 5A–C). A subset of mice bearing uninfected 4T1 tumors were treated weekly with LPS injections. We also measured increased primary tumor growth in these animals compared with tumors from mice bearing 4T1 cells inoculated with control diet fecal-derived microbiota or uninfected/untreated 4T1 cells (Figure 5D). While lard diet-derived oral fecal transplant in tumor-bearing mice had a modest effect on primary 4T1 tumor growth compared to control mice, tumors from lard diet-derived fecal transplant mice were smaller than tumors from mice bearing 4T1 cells inoculated with lard diet-derived microbiota; perhaps due to differential amounts of bacteria within primary tumor cells (Supplemental Figure S2A–B, G–H). Since microbiota mechanism of actions are often linked with immune cell programing and modulation of inflammation, we also included tumor groups bearing a mixture of 4T1 breast cancer cell and mouse macrophage cells (RAW 264.7 cell line) pre-inoculated with control or lard diet fecal-derived microbiota with 4T1 breast cancer cells prior to implantation (Supplemental Figure S2C–D, I). It is important to note that concurrent antibiotic treatment of mice bearing 4T1 cells inoculated with lard diet fecal-derived microbiota did not prevent tumor growth induced by lard diet fecal-derived microbiota most likely due to the inability of antibiotics to permeate the tumor, as shown by high levels of LTA and LPS immunoreactivity in these tumors (Supplemental Figure S2E, G–H). Pre-treatment of macrophage cells with lard-derived fecal conditioned media, but not control diet-derived microbiota, increased primary tumor growth with an associated change in tumor Ki67 immunoreactivity (Supplemental Figure S2F). Both pre-treatment with lard diet fecal-derived conditioned media to the cancer epithelial cells (4T1) or systemic treatment with LPS increased tumor proliferation as determined by Ki67 immunohistochemistry (Figure 5E). 4T1 cancer cells pre-inoculated with control diet-derived microbiota media did not affect tumor growth or proliferation, suggesting that commensal bacterial populations do not stimulate these processes. Tumor sections were also stained for LTA, LPS, and F4/80 to visualize bacterial localization and macrophage infiltration within the tissue. 4T1 tumors from mice injected with pre-inoculated control or lard diet fecal-derived microbiota cells displayed elevated gram-positive bacteria burden, LPS-positive bacteria, and macrophage infiltration compared to controls (Figure 5F–H).
Figure 5.
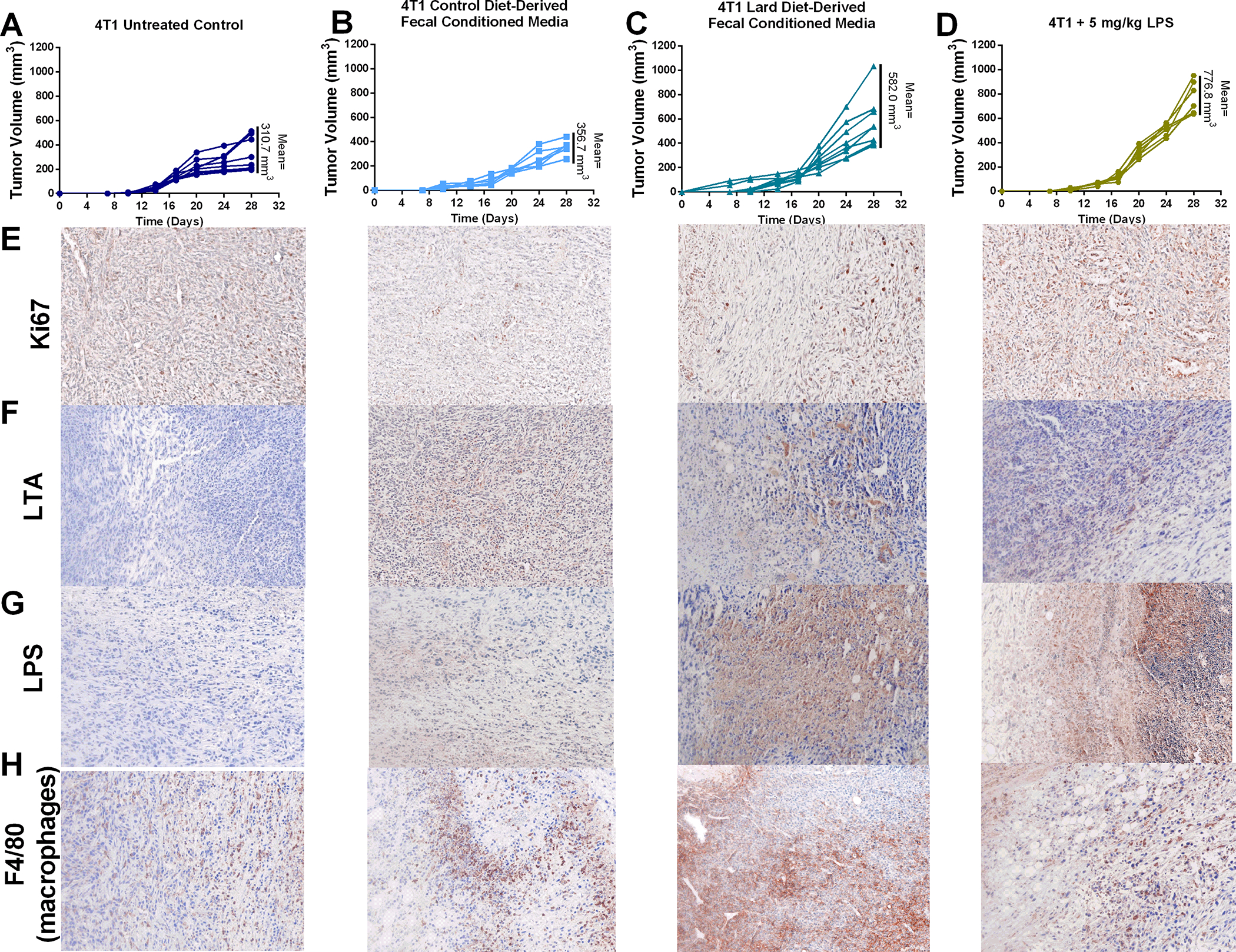
Intratumoral microbiota drives primary mammary tumor growth through modulation of proliferation and immune infiltrate. A. Uninfected 4T1 tumor volume B. Control diet-derived fecal conditioned media infected 4T1 breast tumor volume over time. C. Lard diet-derived fecal conditioned media infected 4T1 breast volume over time. D. Tumor volume over time of uninfected 4T1 tumor-bearing mice treated with 5 mg/kg LPS weekly. E. Tumor proliferation as determined by Ki67 immunoreactivity. F. Gram-positive bacteria content in 4T1 breast tumors. G. LPS positive bacteria content in 4T1 tumors. H. Infiltrating tumor-associated macrophage content in 4T1 tumors was determined by F4/80 immuno-reactivity. n=6–9.*p<0.05
Fish oil supplementation in a Western diet reduces intestinal permeability and MG LPS bioavailability.
The Western diet (45% kcal from fat; fat source corn oil, palm oil, milk-fat; 25% sucrose) is a more translationally relevant dietary pattern than the high-fat lard diet (60% kcal from fat, lard as fat source). Many deleterious health aspects linked with Western diet are thought to be mediated by the high pro-inflammatory omega-6:omega-3 PUFA ratio; therefore, fish oil supplements to lower the omega-6:omega-3 PUFA ratio is a commonly utilized intervention (42,43). Using C57BL/6 mice fed a Western diet with or without fish oil supplementation for 12 weeks, we show that the fish oil intervention lowers intestinal permeability as determined by a 24% reduction of plasma FITC-dextran (Figure 6A). Moreover, at the end of the study, excised colon thickness was measured by ultrasound. We found that fish oil supplementation to the Western diet was associated with a 14% increase in colon thickness compared with Western diet consuming animals (Figure 6B). In addition to this change in epithelial morphology, we measured elevated expression of the tight junction gene ZO-1 in intestinal tissue from the Western diet with fish oil supplementation compared to Western diet without supplement (Figure 6C). Taken together, these data suggest increased intestinal health by fish oil administration, which may decrease pro-inflammatory LPS bioavailability. To test this hypothesis, we performed an LPS ELISA on the plasma of animals on the Western diet with or without fish oil. We show that fish oil supplementation decreased systemic (plasma) LPS concentrations (Figure 6D).
Figure 6.
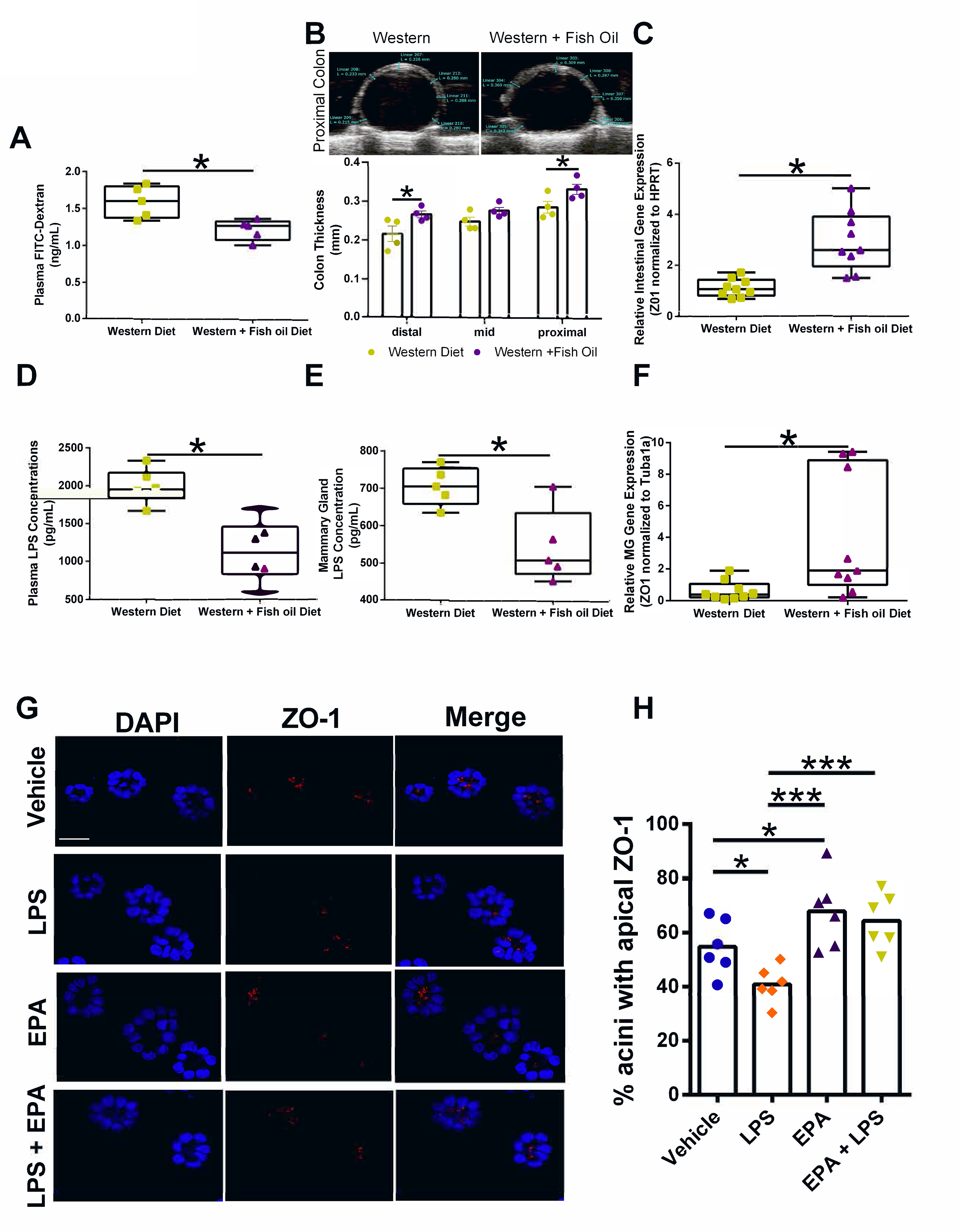
Fish oil supplementation modifies intestinal permeability, LPS bioavailability, and breast acini polarity. A. Plasma FITC-dextran concentrations in 16-week old female C57BL/6 mice fed a Western or Western + fish oil diet. n=5; *p<0.05. B. Colon thickness (proximal, mid, and distal colon measurements) determined by Vevo ultrasound in concentrations in 16-week old female C57BL/6 mice fed a Western or Western + fish oil diet. n=4; *p<0.05. C. Relative ZO-1 gene expression normalized to HPRT in intestinal tissues from Western or Western + fish oil supplemented n=9; *p<0.05. D. Plasma LPS concentration n=6, *p<0.05. E. Mammary gland LPS concentration. n=5, *p<0.05. F. Relative ZO-1 gene expression normalized to Tubu1a in mammary gland tissues from Western or Western + fish oil supplemented n=9; *p<0.05. G. Representative images of mammary acini structures treated with LPS, EPA, or a combination of LPS+EPA. Scale bars, 100 μm. H. Effects of n-3 PUFA on LPS apical polarity disruption measured by ZO1 localization. n=6; *p<0.05.
Fish oil supplementation in a Western diet reduces mammary gland LPS levels and improves epithelial polarity.
In addition to lowering systemic LPS, we found that the fish oil intervention also significantly decreased localized LPS levels in the MG (Figure 6E). This decrease in MG LPS was associated with elevated ZO-1 gene expression in the MG tissue (Figure 6F). While the addition of fish oil to the Western diet in our model clearly has a positive effect at the gut level, reducing systemic levels of pro-inflammatory LPS, omega-3 PUFA supplementation may also directly affect breast epithelial cells. To test this possibility, we exposed breast acini in 3D cell culture to combinations of the omega-3 PUFA EPA and LPS. The EPA levels used for these experiments correspond to levels achieved in the breast compartment in women taking fish oil supplements (42,44). Acini treated with EPA displayed a slightly increased proportion of structures with apical localization of ZO-1 compared to vehicle-treated cultures (Figure 6G–H), suggesting that fish oil administration may promote a healthy breast architecture.
In contrast, acini exposure to omega-6 PUFA arachidonic acid levels typical of a Western diet significantly reduced the proportion of acini with apical ZO-1 compared to vehicle (Supplementary Figure S3A–B). In accordance with Figure 3, LPS administration reduced apical staining of ZO-1 (Figure 6G–H). Co-administration of EPA and LPS prevented LPS-mediated disruption of apical polarity (Figure 6G–H), suggesting that fish oil supplementation may counteract negative signaling mediated by pro-inflammatory bioactive compounds produced from dysbiotic gut microbiota.
Oral fish oil supplements shift microbiota populations in human breast tumors and normal breast tissue.
Next, we leveraged an intervention trial with fish oil supplementation in breast cancer patients to assess if omega-3 PUFA can modulate tumor and breast microbial populations in humans. Analysis of 16S microbiome sequencing results obtained from matched tumors and tumor-adjacent normal breast tissue indicates that, while the types of microbes present at each site are similar, abundances differ between tumors and normal-adjacent tissue (Figure 7A). Breast tumor tissue contained elevated Lachnospiraceae (Figure 7B), and Ruminococcus (Figure 7C) compared to tumor-adjacent normal mammary tissue regardless of intervention, potentially implicating these microbes in breast tumorigenesis. Fish oil administration had no significant effect on Lachnospiraceae in tumor tissue. However, fish oil supplementation strongly reduced the proportional abundance of Ruminococcus (Figure 7D) in the tumors and normal-adjacent tissue samples. In patients who received placebo, Clostridiales_unclassified was elevated in normal tissue compared to the tumors, whereas the opposite was measured in patients who received the fish oil supplement (Figure 7E). Fish oil supplementation also reduced Bacteroidales_unclassified (Figure 7F) in the tumors and normal-adjacent tissue samples. We next stained breast tumor tissue from women treated with placebo or fish oil intervention during the window-of-opportunity trial with antibodies against LTA or LPS (Figure 7G–I). Breast tumors from women consuming fish oil display a modest reduction in LTA-positive bacteria and a marked ablation of intratumoral LPS-positive bacteria compared with tumor tissue from the placebo arm.
Figure 7.
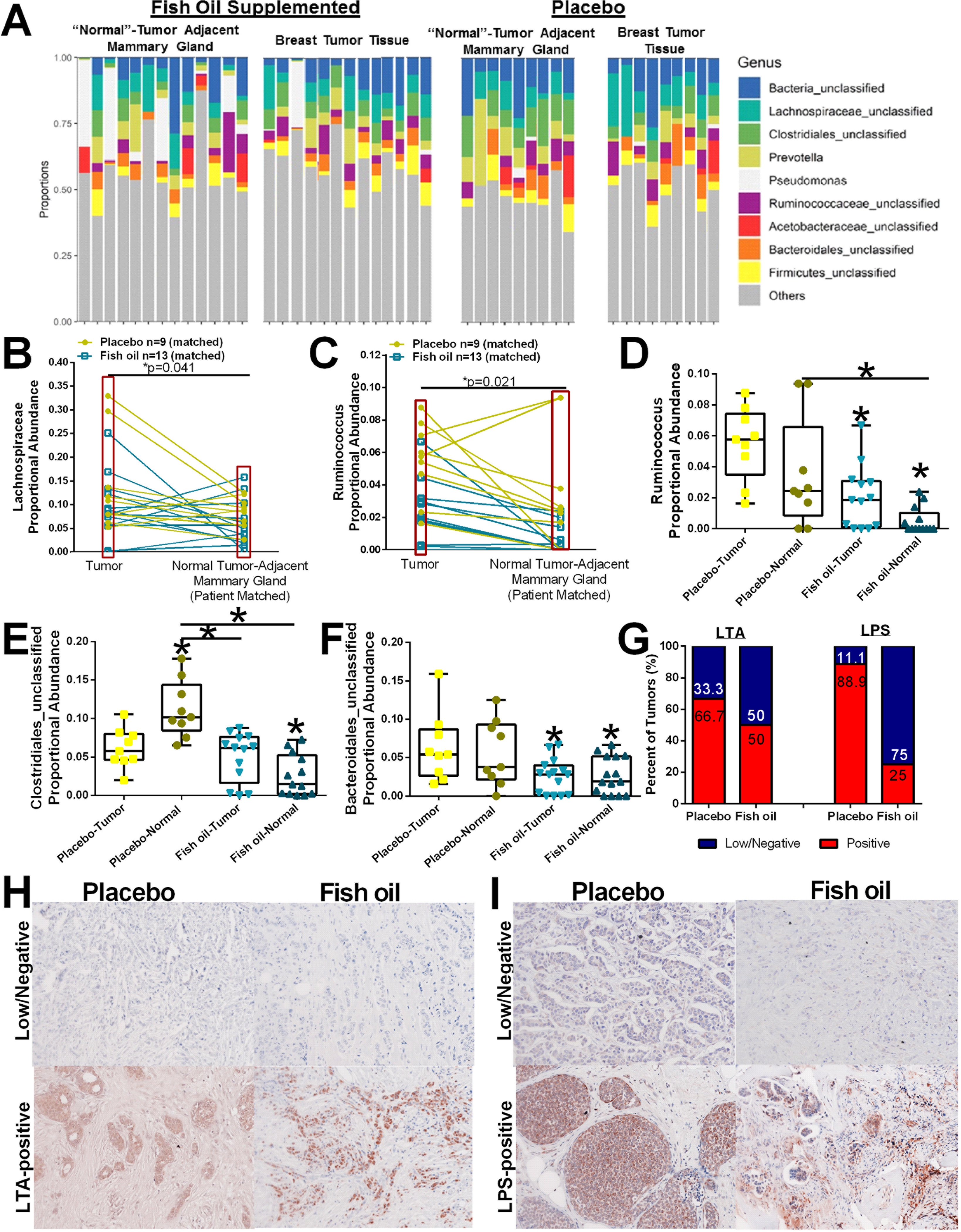
Oral administration of fish oil supplements shifts breast tumor and tumor-adjacent mammary gland tissue microbiota abundance. A. Relative abundance of bacterial genera in different breast tumor and tumor-adjacent mammary gland tissue samples obtained from breast cancer patients taking a placebo or fish oil is visualized by bar plots. Each bar represents a subject and each colored box a bacterial taxon. The height of a color box represents the relative abundance of that organism within the sample. “Other” represents lower abundance taxa. B. Proportional abundance of Lachnospiraceae (family) indicating breast tumors display elevated Lachnospiraceae compared with normal tumor-adjacent mammary tissue regardless of intervention. n=22;*p=0.04. C. Proportional abundance of Ruminococcus (genus) indicating breast tumors display elevated Ruminococcus compared with normal tumor-adjacent mammary tissue regardless of intervention. n=22;*p=0.02. D. Proportional abundance of Ruminococcus (genus) by tissue type and intervention. placebo-treated n=9; fish oil treated n=13; *p<0.05. Proportional abundance of Clostridiales_unclassified (E) and Bacteroidales_unclassified (F) by tissue type and intervention. placebo-treated n=9; fish oil treated n=13; *p<0.05. G. Percent of breast tumors that displayed high positivity (IHC score of 2 or 3) or low/negative (IHC score of 0 or 1) immune-reactivity against LTA or LPS. H. Representative IHC images of breast tumor sections stained against LTA to identify gram-positive bacteria content from placebo or fish oil supplemented women. I. Representative IHC images of breast tumor sections stained against LPS to identify gram-negative bacteria content from placebo or fish oil supplemented women.
The duration of dietary interventions modifies the proportional abundance of select microbes in the tumor and normal-adjacent mammary gland tissue.
The window-of-opportunity clinical trial utilizes the time between diagnosis and surgical resection of the primary tumor; therefore, women assigned to the fish oil interventional arm of the study were on supplements for different lengths of time. Within the fish oil intervention, we grouped women in two categories: short-term supplementation (≤14 days; mean = 10 days) or long-term fish oil administration (≥17 days; mean of 28 days) (Supplemental Figure S4A). Patient BMI did not differ between groups or treatment times. However, it is important to note that the average BMI for patients on this clinical trial indicates a predominately overweight and obese study population (Supplemental Figure S4B). Administration of fish oil for approximately 4 weeks potentiated shifts within the tumor and tumor-adjacent mammary tissue. Within tumor-adjacent mammary tissue, long-term administration of the fish oil supplement elevated the proportional abundance of Lactobacillus and decreased Pseudomonas microbes (Supplemental Figure S4C–D). It is important to note that the elevated Pseudomonas content observed in some samples may simply be a contaminant artifact. Alternatively, the Pseudomonas detected in several of the short-term interventional [but not in the long-term interventional] samples may actually represent introduction of a foreign microbe by biopsy. The longer period between biopsy and surgery may allow the body to combat the introduction of the microbe, which may be why elevated Pseudomonas is not observed in the long-term administration samples. Further research is needed to explore the potential of microbes being introduced by biopsy. Long-term supplementation with fish oil within the tumor tissue significantly reduced Ruminococcus abundance (Supplemental Figure S4C–D). These data suggest that simple dietary supplementation with omega-3 PUFA can rapidly change tissue microbiome populations.
Discussion
Breast tumor tissue and mammary gland tissue were identified to have a microbial component (21–27,45,46); however, the influence of diet and diet-mediated microbial populations within the mammary compartment on breast tumorigenesis is under-explored. As a proof-of-concept, we induced breast tumorigenesis in control or lard diet-fed mice given antibiotics in their drinking water or fecal transplants. Long-term antibiotic administration led to a decreased tumor incidence trend in both control diet and lard diet-fed animals, implicating microbiota and inflammation in breast tumor initiation. Antibiotic administration did not affect tumor weight or tumor multiplicity, suggesting broad-spectrum microbiota inhibition did not modulate tumor progression or inter-animal homogeneity of carcinogen-induced tumor response. Original epidemiological studies reported increased breast cancer risk in patients with increasing cumulative days of antibiotic use (47); however, a more recent study extending the timeline to a 9 year follow-up window with approximately 2.1 million women reported weak associations between antibiotic usage and breast cancer. These associations could be explained by uncontrolled confounding factors such as the underlying inflammatory diseases being treated by antibiotics (48). Our pre-clinical data, lacking possible confounding effects from baseline infections, indicate that antibiotics do not increase breast cancer risk but may reduce breast tumorigenesis, albeit long-term antibiotic usage is not advised nor translatable to the clinic. Therefore, our fecal transplantation model represents a more clinically relevant translational model that better mechanistically defines the impact of dietary-modulated gut microbiota on breast carcinogenesis. Furthermore, we show that administering lard diet-derived fecal transplants to control diet-fed mice is sufficient to increase breast cancer risk in a carcinogen-induced mammary tumorigenesis model, implicating diet-modulated microbiota in breast cancer risk.
Studies associating alterations in the gut microbiome with breast cancer are now emerging. Bobin-Dubigeon and colleagues performed 16S sequencing on stool samples collected prior to treatment or surgery in early stage breast cancer patients (49). Patients with stage II and III breast cancer showed elevated proportional abundance of Clostridium and Lachnospiraceae family members in feces compared with samples from patients with stage 0 and I breast cancer, suggesting an interplay between the gut microbiome and breast cancer progression. We observed the upregulation of Lachnospiraceae in lard diet-fed mice. Lachnospiraceae populations were also regulated in our fecal transplant model, suggesting that diet modulates gut microbiome populations that could promote breast cancer. We also show elevated Lachnospiraceae proportional abundance within human breast tumor tissue compared to normal tumor-adjacent breast tissue, further implicating these microbes in breast cancer’s etiology.
We also observed changes in the breast tumor microbiome induced by diet and fecal transplants. The concept of an entero-mammary transmission route as a potential active mechanism to transfer live bacteria and bacterial metabolites from the gastrointestinal tract to the mammary gland through the mesenteric lymph nodes has been proposed (50–52). Pathological conditions that disrupt the gut barrier increase bacterial translocation from the gut to other tissue types, supporting a “leaky gut” model (53–55). Our data indicate disruption of both the gut and mammary gland tight junction protein expression by in vivo fecal transplant, in vitro lard-diet fecal derived conditioned media, and bacterial-derived products (such as LPS). TJ protein localization disruption was confirmed by diet-derived fecal conditioned media and bacterial-derived products in a 3-D breast acini model. We found increased circulating LPS in plasma from animals fed lard or Western diets and in mice from a control diet that were administered a lard diet-derived fecal transplant. These results suggest changes in gut permeability caused by the microbiota, elevating bacterial metabolite bioavailability. Coupled with the observed elevated bacterial OTU counts in tumors from lard diet-fed and mice administered a fecal transplant, the data indicate that gut dysbiosis induced by Western and HFD-induced microbiota facilitates bacterial translocation to the mammary compartment. In further support, the literature suggests elevated plasma LPS in obese subjects compared with non-obese patient plasma samples (56), suggesting that the obesity-modulated leaky gut model is also observed in humans. We previously demonstrated that a habitual diet could modulate microbiota populations in normal non-cancerous breast tissue from a non-human primate model (23). The mammary gland microbiome sequencing results were compared with the gut microbiota populations in a subset of monkeys on each diet (57,58). Interestingly, the effect of diet on the fecal microbiome compared with the mammary gland microbiome displayed some similarities and several divergences. Increased Lactobacillus and decreased Ruminococcus populations were observed in both mammary gland and feces of Mediterranean diet-fed monkeys (23,57), which provided further evidence supporting some connection between the gut and breast microbiota populations.
We previously demonstrated that patient obesity shifts breast tumor microbiota populations; obese patients displayed increased tumoral Enterobacteriaceae compared with non-obese patient tumor microbial abundance (21). Enterobacteriaceae is a family of bacteria predominately LPS expressing and found in the human intestinal tract. Increased Enterobacteriaceae OTUs were observed in the guts of overweight women (59). We also observe increased Enterobacteriaceae OTUs in mouse DMBA mammary tumors from lard-fed mice and control diet-fed mice administered a lard fecal transplant. Taken together, these data suggest that obesity and HFD consumption shifts breast tumor microbiota similarly in mice and humans.
There has been increasing evidence demonstrating the differences in microbiota between breast tumor tissues and non-malignant breast tissues (22,60). Our human breast cancer patient data from women administered a placebo or a daily supplement of fish oil clinical trial indicates that while the normal tumor adjacent mammary gland and the tumor tissue display some similarities in the types of bacteria present, they differ in the proportional abundance of these microbes. Breast tumors displayed elevated Lachnospiraceae and Ruminococcus populations than in the normal tumor-adjacent mammary gland tissue, suggesting potential selection advantage of some microbes within the tumor microenvironment that may play a role in breast cancer cell signaling. Furthermore, we demonstrated that oral supplementation of fish oil led to microbial changes within the normal breast and tumor compartments.
More mechanistic studies need to be performed in order to discover the interconnectivity between microbiota between the gut and breast tissue. Our work here, and work performed by others (29), support the gut/breast signaling axis hypothesis by indicating differential gram-positive bacteria localization, LPS-expressing bacterial localization, and bacterial 16S OTUs mammary gland and breast tumors by diet. Our study has limitations. In particular, our mammary carcinogenesis model did not include heat-inactivated fecal transplant groups, nor animals that underwent fecal transplantations with concurrent antibiotic administration. Inclusion of these groups in follow-up experiments will clarify the relative role of live microbes versus other compounds present in fecal transplant that may influence cancer risk. Further experimentation into the gut/breast signaling axis is required to fully understand this dynamic process and the impact on breast carcinogenesis. However, our data supports the hypothesis that diet changes or supplementing patients with probiotics could lead to improved breast cancer outcomes, including more prolonged disease-free survival and a decrease in breast cancer incidence. Importantly, this is a modifiable clinical variable that is highly actionable.
In conclusion, we demonstrated that the gut-mammary signaling axis modulates dietary influences on breast cancer risk. We found that fecal transplants impact both the gut and breast tumor microbiomes and cancer outcomes in our mammary carcinogenesis model. Using fecal conditioned cell culture media, we showed that dietary-influenced microbiota modulates primary breast tumor growth. We also demonstrated in human breast tumor samples that bacteria exist outside of immune cells and are modulated by oral dietary intervention. This work provides essential evidence on the critical roles that gut and mammary microbiomes, regulated by diet, play in breast cancer risk and progression, highlighting the potential of interventions to correct dysbiosis in prevention and treatment.
Supplementary Material
Statement of Significance:
This study demonstrates that diet shifts the microbiome in the gut and the breast tumor microenvironment to affect tumorigenesis, and oral dietary interventions can modulate the tumor microbiota in breast cancer patients.
Acknowledgments:
This work was funded by the Chronic Disease Research Fund (to KLC), an American Cancer Society Research Scholar grant (RSG-16–204-01-NEC to KLC and 133727-RSG-19–150-01-LIB to DSP), a grant from the Susan G. Komen Foundation (CCR18547795 to KLC), the American Institute for Cancer Research (208537 to GLK), NIH NCI (R01CA253329 to KLC and R21CA249349 to DSP), ASTRO-BCRF Career Development Award (637969 to DSP), and Breakthrough Awards from the Department of Defense Breast Cancer Research Program (W81XWH-20–1-0014 to KLC and BC170905 to PAV). IRACDA PRIME K12 fellowship, 1K12-GM102773 (PI-Howlett) to MUR. This study was supported by the Wake Forest Baptist Comprehensive Cancer Center’s TTPSR supported by the National Cancer Center’s Comprehensive Cancer Support Grant award number P30CA012197. The authors thank Dr. Monica Jenks for technical assistance.
Footnotes
Conflict of Interests Statement: The authors declare they have no competing financial interests and no conflict of interests to disclose.
References:
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7–33 [DOI] [PubMed] [Google Scholar]
- 2.Rock CL, Demark-Wahnefried W. Nutrition and survival after the diagnosis of breast cancer: a review of the evidence. J Clin Oncol 2002;20:3302–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petrelli JM, Calle EE, Rodriguez C, Thun MJ. Body mass index, height, and postmenopausal breast cancer mortality in a prospective cohort of US women. Cancer Causes Control 2002;13:325–32 [DOI] [PubMed] [Google Scholar]
- 4.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Backhed F. Crosstalk between Gut Microbiota and Dietary Lipids Aggravates WAT Inflammation through TLR Signaling. Cell Metab 2015;22:658–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley RE, Backhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci U S A 2005;102:11070–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature 2009;457:480–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006;444:1027–31 [DOI] [PubMed] [Google Scholar]
- 8.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med 2014;20:159–66 [DOI] [PubMed] [Google Scholar]
- 9.Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, et al. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell Host Microbe 2012;12:611–22 [DOI] [PubMed] [Google Scholar]
- 10.Petersen C, Round JL. Defining dysbiosis and its influence on host immunity and disease. Cell Microbiol 2014;16:1024–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013;155:1451–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garrett WS. Cancer and the microbiota. Science 2015;348:80–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, et al. The Human Microbiome and Cancer. Cancer Prev Res (Phila) 2017;10:226–34 [DOI] [PubMed] [Google Scholar]
- 14.Rao VP, Poutahidis T, Fox JG, Erdman SE. Breast cancer: should gastrointestinal bacteria be on our radar screen? Cancer Res 2007;67:847–50 [DOI] [PubMed] [Google Scholar]
- 15.Rao VP, Poutahidis T, Ge Z, Nambiar PR, Boussahmain C, Wang YY, et al. Innate immune inflammatory response against enteric bacteria Helicobacter hepaticus induces mammary adenocarcinoma in mice. Cancer Res 2006;66:7395–400 [DOI] [PubMed] [Google Scholar]
- 16.Rao VP, Poutahidis T, Ge Z, Nambiar PR, Horwitz BH, Fox JG, et al. Pro-inflammatory CD4+ CD45RB(hi) lymphocytes promote mammary and intestinal carcinogenesis in Apc(Min/+) mice. Cancer Res 2006;66:57–61 [DOI] [PubMed] [Google Scholar]
- 17.Goedert JJ, Jones G, Hua X, Xu X, Yu G, Flores R, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernandez L, Cardenas N, Arroyo R, Manzano S, Jimenez E, Martin V, et al. Prevention of Infectious Mastitis by Oral Administration of Lactobacillus salivarius PS2 During Late Pregnancy. Clin Infect Dis 2016;62:568–73 [DOI] [PubMed] [Google Scholar]
- 19.Chan A, Shwe M, Koh SAQ, Zhu C, Chan YS, Tan SH, et al. Gut microbiome alterations in breast cancer survivors with cancer-related fatigue. Journal of Clinical Oncology 2018;36:e22178–e [Google Scholar]
- 20.Chan AA, Bashir M, Rivas MN, Duvall K, Sieling PA, Pieber TR, et al. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci Rep 2016;6:28061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiba A, Bawaneh A, Velazquez C, Clear KYJ, Wilson AS, Howard-McNatt M, et al. Neoadjuvant Chemotherapy Shifts Breast Tumor Microbiota Populations to Regulate Drug Responsiveness and the Development of Metastasis. Mol Cancer Res 2020;18:130–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hieken TJ, Chen J, Hoskin TL, Walther-Antonio M, Johnson S, Ramaker S, et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci Rep 2016;6:30751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shively CA, Register TC, Appt SE, Clarkson TB, Uberseder B, Clear KYJ, et al. Consumption of Mediterranean versus Western diet leads to distinct mammary gland microbiome populations: Implications for breast cancer. Cell Reports 2018;25:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Urbaniak C, Cummins J, Brackstone M, Macklaim JM, Gloor GB, Baban CK, et al. Microbiota of human breast tissue. Appl Environ Microbiol 2014;80:3007–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith A, Pierre JF, Makowski L, Tolley E, Lyn-Cook B, Lu L, et al. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci Rep 2019;9:11940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H, Altemus J, Niazi F, Green H, Calhoun BC, Sturgis C, et al. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017;8:88122–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Urbaniak C, Gloor GB, Brackstone M, Scott L, Tangney M, Reid G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl Environ Microbiol 2016;82:5039–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Javitt NB, Budai K, Miller DG, Cahan AC, Raju U, Levitz M. Breast-gut connection: origin of chenodeoxycholic acid in breast cyst fluid. Lancet 1994;343:633–5 [DOI] [PubMed] [Google Scholar]
- 29.Parida S, Wu S, Siddharth S, Wang G, Muniraj N, Nagalingam A, et al. A Procarcinogenic Colon Microbe Promotes Breast Tumorigenesis and Metastatic Progression and Concomitantly Activates Notch and beta-Catenin Axes. Cancer Discov 2021 [DOI] [PubMed] [Google Scholar]
- 30.Sumis A, Cook KL, Andrade FO, Hu R, Kidney E, Zhang X, et al. Social isolation induces autophagy in the mouse mammary gland: link to increased mammary cancer risk. Endocr Relat Cancer 2016;23:839–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Briand P, Petersen OW, Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol 1987;23:181–8 [DOI] [PubMed] [Google Scholar]
- 32.Vidi PA, Bissell MJ, Lelievre SA. Three-dimensional culture of human breast epithelial cells: the how and the why. Methods Mol Biol 2013;945:193–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tenvooren I, Jenks MZ, Rashid H, Cook KL, Muhlemann JK, Sistrunk C, et al. Elevated leptin disrupts epithelial polarity and promotes premalignant alterations in the mammary gland. Oncogene 2019;38:3855–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woting A, Blaut M. Small Intestinal Permeability and Gut-Transit Time Determined with Low and High Molecular Weight Fluorescein Isothiocyanate-Dextrans in C3H Mice. Nutrients 2018;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 2013;79:5112–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 2009;75:7537–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen YY, et al. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009;137:1716–24 e1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Toth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 2014;6:263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009;58:1091–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohr MW, Narasimhulu CA, Rudeski-Rohr TA, Parthasarathy S. Negative Effects of a High-Fat Diet on Intestinal Permeability: A Review. Adv Nutr 2020;11:77–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin-Belmonte F, Perez-Moreno M. Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer 2011;12:23–38 [DOI] [PubMed] [Google Scholar]
- 42.Fabian CJ, Kimler BF, Hursting SD. Omega-3 fatty acids for breast cancer prevention and survivorship. Breast Cancer Res 2015;17:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabian CJ, Kimler BF, Phillips TA, Box JA, Kreutzjans AL, Carlson SE, et al. Modulation of Breast Cancer Risk Biomarkers by High-Dose Omega-3 Fatty Acids: Phase II Pilot Study in Premenopausal Women. Cancer Prev Res (Phila) 2015;8:912–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jackson PA, Husberg C, Hustvedt SO, Calder PC, Khan J, Avery H, et al. Diurnal rhythm of plasma EPA and DHA in healthy adults. Prostaglandins Leukot Essent Fatty Acids 2020;154:102054. [DOI] [PubMed] [Google Scholar]
- 45.Meng S, Chen B, Yang J, Wang J, Zhu D, Meng Q, et al. Study of Microbiomes in Aseptically Collected Samples of Human Breast Tissue Using Needle Biopsy and the Potential Role of in situ Tissue Microbiomes for Promoting Malignancy. Front Oncol 2018;8:318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020;368:973–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Velicer CM, Heckbert SR, Lampe JW, Potter JD, Robertson CA, Taplin SH. Antibiotic use in relation to the risk of breast cancer. JAMA 2004;291:827–35 [DOI] [PubMed] [Google Scholar]
- 48.Friedman GD, Oestreicher N, Chan J, Quesenberry CP Jr., Udaltsova N, Habel LA. Antibiotics and risk of breast cancer: up to 9 years of follow-up of 2.1 million women. Cancer Epidemiol Biomarkers Prev 2006;15:2102–6 [DOI] [PubMed] [Google Scholar]
- 49.Luu TH, Michel C, Bard JM, Dravet F, Nazih H, Bobin-Dubigeon C. Intestinal Proportion of Blautia sp. is Associated with Clinical Stage and Histoprognostic Grade in Patients with Early-Stage Breast Cancer. Nutr Cancer 2017;69:267–75 [DOI] [PubMed] [Google Scholar]
- 50.Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, et al. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 2007;119:e724–32 [DOI] [PubMed] [Google Scholar]
- 51.Jimenez E, Fernandez L, Maldonado A, Martin R, Olivares M, Xaus J, et al. Oral administration of Lactobacillus strains isolated from breast milk as an alternative for the treatment of infectious mastitis during lactation. Appl Environ Microbiol 2008;74:4650–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Andres J, Jimenez E, Chico-Calero I, Fresno M, Fernandez L, Rodriguez JM. Physiological Translocation of Lactic Acid Bacteria during Pregnancy Contributes to the Composition of the Milk Microbiota in Mice. Nutrients 2017;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng C, Wei H, Yu H, Xu C, Jiang S, Peng J. Metabolic Syndrome During Perinatal Period in Sows and the Link With Gut Microbiota and Metabolites. Front Microbiol 2018;9:1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ortiz S, Zapater P, Estrada JL, Enriquez P, Rey M, Abad A, et al. Bacterial DNA translocation holds increased insulin resistance and systemic inflammatory levels in morbid obese patients. J Clin Endocrinol Metab 2014;99:2575–83 [DOI] [PubMed] [Google Scholar]
- 55.Mokkala K, Roytio H, Munukka E, Pietila S, Ekblad U, Ronnemaa T, et al. Gut Microbiota Richness and Composition and Dietary Intake of Overweight Pregnant Women Are Related to Serum Zonulin Concentration, a Marker for Intestinal Permeability. J Nutr 2016;146:1694–700 [DOI] [PubMed] [Google Scholar]
- 56.Troseid M, Nestvold TK, Rudi K, Thoresen H, Nielsen EW, Lappegard KT. Plasma lipopolysaccharide is closely associated with glycemic control and abdominal obesity: evidence from bariatric surgery. Diabetes Care 2013;36:3627–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagpal R, Shively CA, Appt SA, Register TC, Michalson KT, Vitolins MZ, et al. Gut Microbiome Composition in Non-human Primates Consuming a Western or Mediterranean Diet. Front Nutr 2018;5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Newman TM SC, Register TC, Appt SE, Yadav H, Colwell RR, Fanelli B, Dadlani M, Graubics, Nguyen U, Ramamoorthy S, Uberseeder B, Clear KYJ, Wilson AS, Reeves KD, Chappell MC, Tooze JA, Cook KL. Diet, obesity, and the gut microbiome as determinants modulating metabolic outcomes in a non-human primate model. Microbiome 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clarke SF, Murphy EF, Nilaweera K, Ross PR, Shanahan F, O’Toole PW, et al. The gut microbiota and its relationship to diet and obesity: new insights. Gut Microbes 2012;3:186–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson KJ, Ingle JN, Tang X, Chia N, Jeraldo PR, Walther-Antonio MR, et al. A comprehensive analysis of breast cancer microbiota and host gene expression. PLoS One 2017;12:e0188873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


