Abstract
We determined the nucleotide sequence of the bla gene for the Acinetobacter calcoaceticus β-lactamase previously described as CARB-5. Alignment of the deduced amino acid sequence with those of known β-lactamases revealed that CARB-5 possesses an RTG triad in box VII, as described for the Proteus mirabilis GN79 enzyme, instead of the RSG consensus characteristic of the other carbenicillinases. Phylogenetic studies showed that these RTG enzymes constitute a new, separate group, possibly ancestors of the carbenicillinase family.
The assumption that carbenicillin-inactivating β-lactamases were a homogeneous cluster of enzymes confined to Pseudomonas aeruginosa strains (9) did not last long. First, some Pseudomonas-specific enzymes may have spread, on rare occasions, to various enterobacteria (19). Second, evidence is accumulating that, in addition to Proteus mirabilis in Japan (27) and other enterobacteria, strains of Vibrio cholerae, Alcaligenes xylosoxidans, Aeromonas hydrophila, and Acinetobacter calcoaceticus also produce such β-lactamases (SAR-1 and CARB-6 for V. cholerae and PSE-1, AER-1, and CARB-5 for A. xylosoxidans, A. hydrophila, and A. calcoaceticus, respectively). Major progress in the comprehension and classification of carbenicillinases is expected from structural data. Four main structures account for the diversity of carbenicillinases. The major structure is that of the CARB group, which includes CARB-1 (PSE-4), CARB-2 (PSE-1), CARB-3 (17, 18), CARB-4 (22, 25), CARB-6 (6), and P. mirabilis N29 β-lactamase (12). The three other enzyme structures do not belong, at this time, to any particular group: PSE-3 β-lactamase (5), P. mirabilis GN79 (24), and AER-1 β-lactamases (25). Sequence information is currently unavailable for two β-lactamases, CARB-5 and SAR-1, which have been defined as carbenicillinases due to their hydrolytic properties. Our aim was to determine the nucleotide sequence of the bla gene (CARB-5) from A. calcoaceticus strain A85-145, the resistance properties and enzyme characteristics of which have been previously published (21).
Clinical strains producing CARB-5.
Several clinical isolates of Acinetobacter calcoaceticus subsp. anitratus, resistant to ticarcillin alone but susceptible to ticarcillin in combination with clavulanic acid, have been found to produce a new β-lactamase, with a pI of 6.35 (14). Its molecular weight of 28,000 and substrate profile are typical of a carbenicillin-hydrolyzing enzyme. The enzyme was inhibited by anti-CARB-3 serum, p-chloromercuribenzoate, cloxacillin, clavulanic acid, and sulbactam (21).
Nucleotide sequence of blaCARB-5.
Several sets of oligonucleotide primer pairs specific for most CARB sequences were unsuccessfully used for amplification. Unexpected positive results were obtained with primers based on the sequence of the P. mirabilis GN79 bla gene (24). The nucleotide sequence of the structural gene was then determined by direct sequencing of PCR fragments amplified from chromosomal DNA extracted by the X-Trax procedure (6) with two pairs of synthetic primers (Table 1) comprising the whole reference sequence of the P. mirabilis GN79 gene. Amplification by PCR was performed at 44°C as described elsewhere (6).
TABLE 1.
Oligonucleotide primers used in PCR amplification
| Primer | Location (5′–3′)a | Sequence (5′–3′) |
|---|---|---|
| A1 | 301–320 | GTTAACTCATTATGAACGTA |
| A1′ | 866–852 | TGTTGTCGTGTCTCG |
| A2 | 667–680 | GGATGTCGCTCGCA |
| A2′ | 1212–1193 | TAAATCAGTTACGGCTATTC |
Oligonucleotide positions are given according their location on the GN79 bla gene sequence (24).
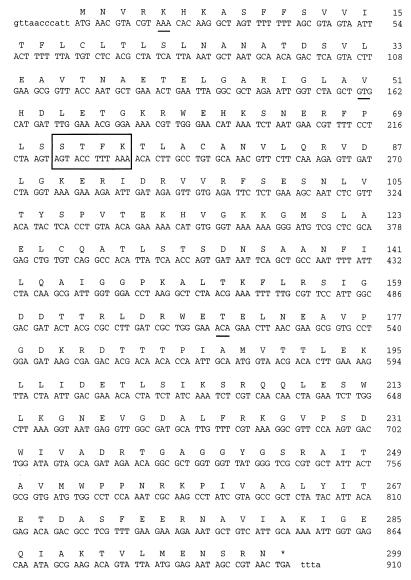
The nucleotide sequence of the PCR product (912 bp) contains a 905-bp open reading frame. The coding region is 897 bp (positions 12 to 908) and encodes a protein of 298 amino acids (Fig. 1).
FIG. 1.
Nucleotide and deduced amino acid sequence of A. calcoaceticus β-lactamase (CARB-5). The active site is boxed, and the differences from the P. mirabilis GN79 β-lactamase sequence are underlined.
CARB-5 is a class A β-lactamase.
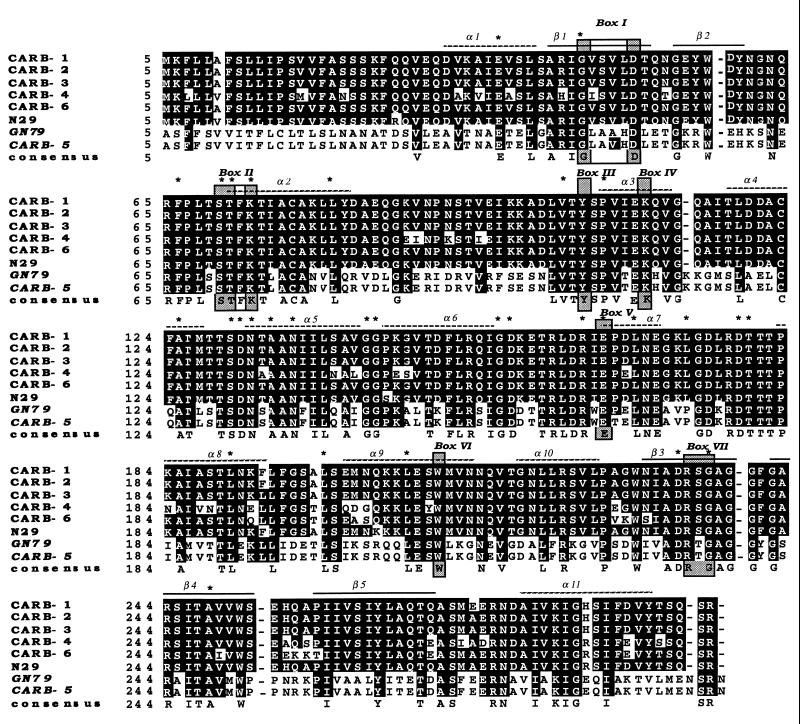
Analysis of the deduced amino acid sequence showed that specific motifs were very similar to those of class A β-lactamases: the bla active-site (STFK) tetrad at positions 72 to 75 (70 to 73 according to the standard numbering scheme of Ambler [2]) and the consensual boxes I to VI described by Joris et al. (15) were conserved (Fig. 2). In contrast, box VII consisted of an RTG triad instead of the conserved RSG motif specific to the carbenicillinases (3). All the residues specific to class A β-lactamases were conserved except that the Leu at position 177 (164 according to Ambler numbering) was replaced with a Pro, as reported for P. mirabilis GN79 (24).
FIG. 2.
Multiple sequence alignment of the amino acid sequences of the CARB-1, CARB-2, CARB-3, CARB-4, CARB-6, P. mirabilis N29 and GN79, and CARB-5 β-lactamases. The shaded boxes (I to VII) correspond to amino acids conserved in all penicillin-recognizing enzymes, as identified by Joris et al. (15). Alpha-helix and beta-barrel motifs are from the PC1 crystal structure (3, 10). Asterisks indicate the conserved residues specific for class A β-lactamases. Amino acid changes are indicated as black letters on a white background. Sequences are numbered according to the method of Ambler (2).
Nucleic and amino acid sequence analyses with the BLAST and FASTA (1) programs showed substantial homology (more than 99%) to the P. mirabilis GN79 sequence but only 43, 44, and 45% identity to the SHV (8), CARB (17, 18), and TEM (11) β-lactamases, respectively. Pairwise alignment between the CARB-5 and GN79 nucleic acid sequences revealed four mutations in the coding region (C to A at position 24, C to T at 163, G to T at 450, and C to A at 520) giving rise to three amino acid substitutions (Fig. 1 and 2): Gln to Lys at position 5 (2 according to Ambler), Ala to Val at position 51 (48 according to Ambler numbering), and Pro to Thr at position 170 (167 according to Ambler numbering). Multiple sequence alignment (28) of the CARB-5 amino acid sequence with the previously described sequences of the CARB-1, CARB-2 and CARB-3 (17, 18), CARB-4 (22, 25), CARB-6 (6), P. mirabilis N29 (12), and P. mirabilis GN79 (24) β-lactamases showed that the first amino acid change (Gln to Lys) was located outside the sequence of the other carbenicillinases, the second (Ala to Val) was common to all the carbenicillinase sequences, and the third (Pro to Thr) was unique among all aligned sequences (Fig. 2).
Although the CARB-5 enzyme contains the specific Arg 234 residue characteristic of carbenicillinases, CARB-5 seems to be only distantly related to these enzymes. Moreover, it also contains RTG box VII, reported for the first and only time for P. mirabilis GN79 (24), instead of the RSG usually encountered in carbenicillinases (3).
Phylogenetic analysis.
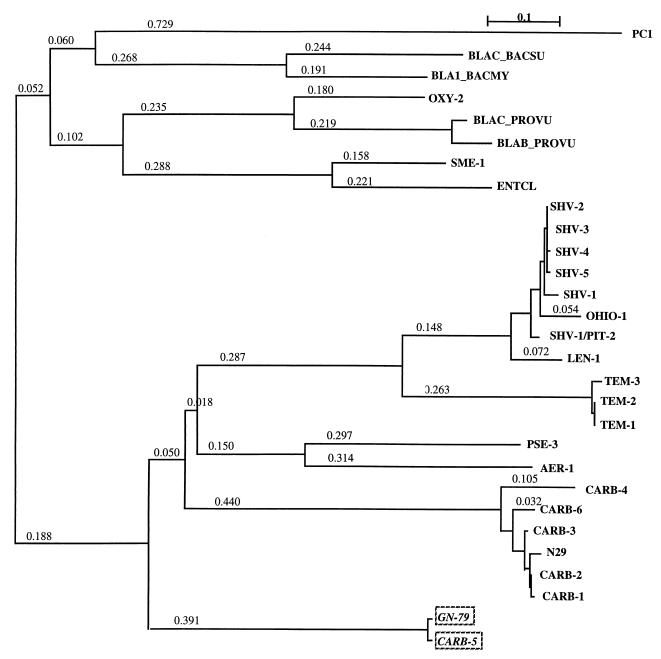
A phylogenetic tree was constructed from ClustalW multiple sequence alignment of 29 class A β-lactamases (Table 2), using the neighbor-joining method (23). This tree (Fig. 3) exhibited two major branches. One, including cephalosporinases, carbapenemases, and Staphylococcus aureus PC1 enzyme, constitutes the outgroup. The other ramifies into three main subbranches corresponding to three distinct groups. One of these contains the SHV and TEM enzymes and the second consists of the carbenicillinases according to the previously proposed scheme (4). Surprisingly, CARB-5 clustered with P. mirabilis GN79 to make up a third group distantly related to the other two groups. These results were confirmed by other methods (data not shown): PROTPARS (protein sequence parsimony method), PRODIST (protein distances) from the PHYLIP package of Joseph Felsenstein (Department of Genetics at the University of Washington), and Web Gene Bee services (Belozersky Institute, Moscow State University) (13). The reliability of the phylogenetic trees was estimated by bootstrapping (26). The node and branching leading to the CARB-5–GN79 cluster were confirmed in 997 and 1,000 of 1,000 bootstrap tests (data not shown). The general configuration of this tree differs slightly from that in Fig. 3 (neighbor-joining method), with PC1, BLAC_BACSU, and BLA1_BACMY making up a separate outgroup.
TABLE 2.
Sequences used in phylogenetic studies
| Enzyme designation | Description (SwissProt or GenBank no.) |
|---|---|
| PC1 | Staphylococcus aureus PC1 (BLAC_STAAU) |
| BLAC_BACSU | Bacillus subtilis cephalosporinase (BLAC_BACSU) |
| BLA1_BACMY | Bacillus mycoides cephalosporinase (BLA1_BACMY) |
| OXY-2 | Klebsiella oxytoca cephalosporinase (BLA4_KLEOX) |
| BLAC_PROVU | Proteus vulgaris cephalosporinase (BLAC_PROVU) |
| BLAB_PROVU | Proteus vulgaris cefuroximase (BLAB_PROVU) |
| SME-1 | Carbapenemase (BLAN_SERMA) |
| ENTCL | Enterobacter cloacae carbapenemase (BLAN_ENTCL) |
| GN79 | Proteus mirabilis carbenicillinase (BLAC_PROMI) |
| CARB-1 | Pseudomonas aeruginosa carbenicillinase (PSE-4) (BLP4_PSEAE) |
| CARB-2 | Pseudomonas aeruginosa carbenicillinase (PSE-1) (BLP1_PSEAE) |
| CARB-3 | Pseudomonas aeruginosa carbenicillinase (BLC3_PSEAE) |
| CARB-4 | Pseudomonas aeruginosa carbenicillinase (PAU14749) |
| CARB-6 | Vibrio cholerae carbenicillinase (AF030945) |
| N29 | Proteus mirabilis carbenicillinase (D86225) |
| AER-1 | Aeromonas hydrophilia carbenicillinase (AHU14748) |
| PSE-3 | Rhodopseudomonas capsulata sp108 (unpublished data) |
| TEM-1 | Escherichia coli pBR322 (BLAT_ECOLI) |
| TEM-2 | BLAT_ECOLI |
| TEM-3 | Klebsiella pneumoniae penicillinase (KPBLATEM3) |
| LEN-1 | Klebsiella pneumoniae bla (BLAC_KLEPN) |
| OHIO-1 | Enterobacter cloacae SHV (BLA1_ENTCL) |
| SHV-1/PIT-2 | Escherichia coli SHV-1/PIT-2 (BLA1_ECOLI) |
| SHV-1 | Klebsiella pneumoniae SHV-1 (BLA1_KLEPN) |
| SHV-2 | Escherichia coli and Klebsiella pneumoniae SHV-2 (BLA2_ECOLI) |
| SHV-3 | Klebsiella pneumoniae SHV-3 (BLA3_KLEPN) |
| SHV-4 | Klebsiella pneumoniae SHV-4/CAZ-5 (BLA4_KLEPN) |
| SHV-5 | Klebsiella pneumoniae SHV-5 (BLA5_KLEPN) |
FIG. 3.
Dendrograms obtained from multiple alignment of 29 class A β-lactamases according to the neighbor-joining method (23). Branch length values represent relative phylogenetic distances.
CARB-5 (RTG-2) and P. mirabilis GN79 (RTG-1) β-lactamases are members of a new carbenicillinase group called RTG.
Nucleotide and amino acid sequence features and biochemical properties of the CARB-5 and P. mirabilis GN79 enzymes (21, 24) exhibit a great degree of similarity. Although the homology of these sequences with known CARB structures is low, they can be related to the carbenicillinase group in regard to their biochemical properties. However, despite their sequence identities, the β-lactamase neutralization assay for CARB-5 with anti-CARB serum gave results in conflict with those obtained for GN79 in the original study. The P. mirabilis GN79 β-lactamase was not neutralized by serum raised against the P. mirabilis N29 β-lactamase, which fully neutralized PSE-1 and PSE-4 (27). In contrast, CARB-5 was inhibited by anti-CARB-3 serum that also reacts with other CARB enzymes, including PSE-1 and PSE-4. Moreover, this result accounted for the designation of CARB-5. Such discrepancy in the results may be due to different technical approaches: the neutralization of GN79 was studied in a dilute-liquid-phase assay, whereas CARB-5 was studied undiluted in a specific gel assay. The liquid-phase assay is known to be more specific than the gel assay, which is more sensitive. Indeed, neutralization in gel makes it possible for the three-dimensional structure of the β-lactamase to combine with low-affinity heterologous antibodies. Such unexpected cross-neutralization in the gel assay has also been reported for the TEM and SHV β-lactamases with anti-TEM-1 serum (20).
In the CARB-5 amino acid sequence, all residues specific to class A β-lactamases are conserved, as are six of seven boxes (15). The most striking feature is the presence of an RTG triad (box VII), as for GN79, instead of the RSG characteristic of the CARB family and unique among class A β-lactamases. The CARB-5 and GN79 enzymes seem to constitute a new carbenicillinase group.
RTG-1 and RTG-2 are possible ancestors of the carbenicillinase family.
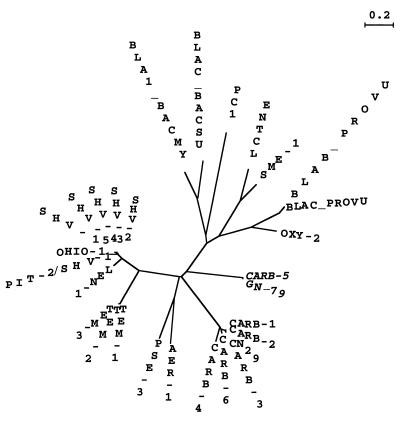
Phylogenetic trees (Fig. 3 and 4) clearly show the evolution of 29 β-lactamases. S. aureus PC1 and the two Bacillus cephalosporinases (BLAC_BACSU and BLA1_BACMY), all issued from gram-positive bacteria, appeared early in evolution. Analysis of 18 class A β-lactamases has shown the origin of the enzyme to be the actinomycetes, from which it migrated first into nonactinomycete gram-positive lines, such as Bacillus, and later into the gram-negative bacteria (16). This is consistent with the results of Huletsky et al. (11) and supports their suggestion that the β-lactamases from gram-negative and gram-positive bacteria constitute two distinct groups. This would imply that the bla genes of gram-positive and Bacillus species appeared early in evolution, followed by the PSE and CARB enzymes and later by the SHV and TEM enzymes found in enteric bacteria. If we consider the evolutionary representation of the phylogenetic tree (Fig. 4), CARB-5 and GN79 are tightly linked and appeared earlier in evolution than other CARB enzymes, which make a separate cluster.
FIG. 4.
Evolutionary representation of unrooted phylogenetic tree. The phylogenetic tree was obtained as described in the legend to Fig. 3.
Interestingly, CARB-5 and GN79 are chromosomally acquired genes. It has been suggested that β-lactamases evolved and spread from the ancestral source; Streptomyces chromosomal penicillin-recognizing enzymes would be the oldest known enzymes of this type (7). The LEN-1 chromosomal enzyme was previously thought to be the ancestor of the SHV family (11). A similar hypothesis could be put forward for the GN79 and CARB-5 group, which would then be the ancestors of the carbenicillinase family.
Thus, the carbenicillinase encoded in the A. calcoaceticus chromosome is a novel carbenicillinase, structurally related to the P. mirabilis GN79 enzyme. These two chromosomal enzymes may be considered to be ancestors of the carbenicillinase family.
All these features lead us to suggest the name of RTG carbenicillinases for this new enzyme group based on the unique RTG triad. The first reference enzymes would be RTG-1 (P. mirabilis GN79 β-lactamase) and RTG-2 (CARB-5).
Nucleotide sequence accession number.
The RTG-2/CARB-5 β-lactamase sequence has been submitted to GenBank. Its accession number is AF135373.
Acknowledgments
We thank L. Gilly for her efficient technical help, F. Letourneur (ICGM) for nucleotide sequencing, and C. Valencien (INFOBIOGEN) and M. Assous for fruitful help and discussions about phylogeny.
REFERENCES
- 1.Altschul S F, Thomas L M, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ambler R P. The structure of β-lactamases. Philos Trans R Soc Lond Biol. 1980;289:321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- 3.Boissinot M, Levesque R C. Nucleotide sequence of the PSE-4 carbenicillinase gene and correlations with the Staphylococcus aureus PC1 β-lactamase crystal structure. J Biol Chem. 1990;265:1225–1230. [PubMed] [Google Scholar]
- 4.Bush K, Jacoby G A, Medeiros A A. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob Agents Chemother. 1995;39:1211–1233. doi: 10.1128/aac.39.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell J I A, Scahill S, Gibson T, Ambler R P. The phototrophic bacterium Rhodopseudomonas capsulata sp108 encodes an indigenous class A beta-lactamase. Biochem J. 1989;260:803–812. doi: 10.1042/bj2600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choury D, Aubert G, Szajnert M-F, Azibi K, Delpech M, Paul G. Characterization and nucleotide sequence of CARB-6, a new carbenicillin-hydrolyzing β-lactamase from Vibrio cholerae. Antimicrob Agents Chemother. 1999;43:297–301. doi: 10.1128/aac.43.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Couture F, Lachapelle J, Levesque R C. Phylogeny of LCR-1 and OXA-5 with class A and class D β-lactamases. Mol Microbiol. 1992;6:1693–1705. doi: 10.1111/j.1365-2958.1992.tb00894.x. [DOI] [PubMed] [Google Scholar]
- 8.Garbarg-Chenon A, Godard V, Labia R, Nicolas J-C. Nucleotide sequence of SHV-2 β-lactamase gene. Antimicrob Agents Chemother. 1990;34:1444–1446. doi: 10.1128/aac.34.7.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hedges R W, Matthew M. Acquisition by Escherichia coli of plasmid-borne β-lactamases normally confined to Pseudomonas spp. Plasmid. 1979;2:269–278. doi: 10.1016/0147-619x(79)90045-3. [DOI] [PubMed] [Google Scholar]
- 10.Herzberg O. Refined crystal structure of β-lactamase from Staphylococcus aureus PC1 at 2Å resolution. J Mol Biol. 1991;217:701–719. doi: 10.1016/0022-2836(91)90527-d. [DOI] [PubMed] [Google Scholar]
- 11.Huletsky A, Couture F, Levesque R C. Nucleotide sequence and phylogeny of SHV-2 β-lactamase. Antimicrob Agents Chemother. 1990;34:1725–1732. doi: 10.1128/aac.34.9.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito Y, Hirano T. Carbenicillin-hydrolysing penicillinase mediated by a plasmid of Proteus mirabilis and its relationship to the PSE-type enzymes of Pseudomonas aeruginosa. J Appl Microbiol. 1997;83:175–180. doi: 10.1046/j.1365-2672.1997.00203.x. [DOI] [PubMed] [Google Scholar]
- 13.Iushmanov S V, Chumakov K M. Algorithms for constructing phylogenetic trees of maximum topological similarity. Mol Genet Microbiol Vir. 1988;3:9–15. . (In Russian.) [PubMed] [Google Scholar]
- 14.Joly-Guillou M L, Bergogne-Bérézin E, Moreau N. Enzymatic resistance to β-lactam and aminoglycosides in Acinetobacter calcoaceticus. J Antimicrob Chemother. 1987;20:773–776. doi: 10.1093/jac/20.6.773. [DOI] [PubMed] [Google Scholar]
- 15.Joris B, Ghuysen J M, Dive G, Renard A, Dideberg O, Charlier P, Frère J M, Kelly J A, Boyington J C, Moews P C, Knox J R. The active-site-serine penicillin-recognizing enzymes as members of the Streptomyces R61 DD-peptidase family. Biochem J. 1988;250:313–324. doi: 10.1042/bj2500313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirby R. Evolutionary origin of class A and class C β-lactamases. J Mol Evol. 1992;34:345–350. doi: 10.1007/BF00160242. [DOI] [PubMed] [Google Scholar]
- 17.Labia R, Guionie M, Barthélémy M, Philippon A. Properties of three carbenicillin-hydrolyzing β-lactamases (CARB) from Pseudomonas aeruginosa: identification of a new enzyme. J Antimicrob Chemother. 1981;7:49–56. doi: 10.1093/jac/7.1.49. [DOI] [PubMed] [Google Scholar]
- 18.Lachapelle J, Dufresnes J, Levesque R C. Characterization of the blaCARB-3 gene encoding the carbenicillinase-3 β-lactamase of Pseudomonas aeruginosa. Gene. 1991;102:7–12. doi: 10.1016/0378-1119(91)90530-o. [DOI] [PubMed] [Google Scholar]
- 19.Medeiros A A, Hedges R W, Jacoby G A. Spread of a “Pseudomonas-specific” β-lactamase to plasmids of enterobacteria. J Bacteriol. 1982;149:700–707. doi: 10.1128/jb.149.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paul G, Barthélémy M, Philippon A, Peduzzi J, Gilly L, Labia R, Névot P. Immunological comparison of constitutive β-lactamase of Gram-negative bacteria by neutralization in zymogram gels: properties of anti-TEM-1 and anti-TEM-2 sera. Ann Inst Pasteur/Microbiol (Paris) 1988;139:435–451. [PubMed] [Google Scholar]
- 21.Paul G, Joly-Guillou M L, Bergogne-Bérézin E, Nevot P, Philippon A. Novel carbenicillin-hydrolyzing β-lactamase (CARB-5) from Acinetobacter calcoaceticus var. anitratus. FEMS Microbiol Lett. 1989;59:45–50. doi: 10.1111/j.1574-6968.1989.tb03080.x. [DOI] [PubMed] [Google Scholar]
- 22.Philippon A M, Paul G C, Thabaut A P, Jacoby G A. Properties of a novel carbenicillin-hydrolyzing β-lactamase (CARB-4) specified by an IncP-2 plasmid from Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1986;29:519–520. doi: 10.1128/aac.29.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 24.Sakurai Y, Tsukamoto K, Sawai T. Nucleotide sequence and characterization of a carbenicillin-hydrolyzing penicillinase gene from Proteus mirabilis. J Bacteriol. 1991;173:7038–7041. doi: 10.1128/jb.173.21.7038-7041.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanschagrin F, Bejaoui N, Levesque R C. Structure of CARB-4 and AER-1 carbenicillin-hydrolyzing β-lactamases. Antimicrob Agents Chemother. 1998;42:1966–1972. doi: 10.1128/aac.42.8.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford D L, Olsen G J. Phylogenetic interference. In: Hillis D M, Moritz C, Mable B K, editors. Molecular systematics. 2nd ed. Sunderland, Mass: Sinauer Associates, Inc.; 1996. pp. 407–514. [Google Scholar]
- 27.Takahashi I, Tsukamoto K, Harada M, Sawai T. Carbenicillin-hydrolyzing penicillinases of Proteus mirabilis and the PSE-type penicillinases of Pseudomonas aeruginosa. Microbiol Immunol. 1983;27:995–1004. doi: 10.1111/j.1348-0421.1983.tb02934.x. [DOI] [PubMed] [Google Scholar]
- 28.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]