Highlights
-
•
Increased sialoglycoprotein expression in high fibrosis regions and in TNBC.
-
•
Cancer cells are the primary biosynthetic source of sialoglycoproteins.
-
•
Immunosuppressive SIGLEC-positive myeloid cells are increased in TNBC.
-
•
Immunosuppressive Siglec ligands are increased in high fibrosis regions.
Keywords: Sialic acid, Sialoglycoproteins, Siglec receptors, Breast cancer, Fibrosis
Abstract
Tumors feature elevated sialoglycoprotein content. Sialoglycoproteins promote tumor progression and are linked to immune suppression via the sialic acid-Siglec axis. Understanding factors that increase sialoglycoprotein biosynthesis in tumors could identify approaches to improve patient response to immunotherapy. We quantified higher levels of sialoglycoproteins in the fibrotic regions within human breast tumor tissues. Human breast tumor subtypes, which are more fibrotic, similarly featured increased sialoglycoprotein content. Further analysis revealed the breast cancer cells as the primary cell type synthesizing and secreting the tumor tissue sialoglycoproteins and confirmed that the more aggressive, fibrotic breast cancer subtypes expressed the highest levels of sialoglycoprotein biosynthetic genes. The more aggressive breast cancer subtypes also featured greater infiltration of immunosuppressive SIGLEC7, SIGLEC9, and SIGLEC10-pos myeloid cells, indicating that triple-negative breast tumors had higher expression of both immunosuppressive Siglec receptors and their cognate ligands. The findings link sialoglycoprotein biosynthesis and secretion to tumor fibrosis and aggression in human breast tumors. The data suggest targeting of the sialic acid-Siglec axis may comprise an attractive therapeutic target particularly for the more aggressive HER2+ and triple-negative breast cancer subtypes.
Introduction
Many tumor types exhibit elevated levels of sialoglycoproteins that correlate with increased proliferation and metastasis, and poor prognosis [1], [2]. The glycocalyx, the layer of glycoproteins and proteoglycans that covers all cells, is abnormal on cancer cells and features increased sialic acid content [3]. We previously showed that a collective effect of glycocalyx dysregulation is increased bulkiness, defined as a denser, thicker glycocalyx and includes increases in number of glycoconjugates and glycoconjugate sites, and elaboration of glycan structures. A bulkier glycocalyx enhances integrin signaling, promotes cell cycle progression, increases metastatic potential, and sterically hinders macrophage phagocytosis [4], [5], [6], [7]. These findings indicate that the increased expression of glycoproteins including sialoglycoproteins in tumor cells likely play a causal role in tumor progression and aggression.
One mechanism by which sialoglycoproteins regulate tumor progression is by inducing immune suppression by acting as ligands to the Siglec family of receptors that are expressed on most immune cell types and contain immunoreceptor tyrosine-based inhibitory motif (ITIM) domains [8]. Siglec signaling—in particular Siglec-7, Siglec-9, Siglec-10, and Siglec-15 expressed on macrophages, monocytes, NK cells, and activated T cells—inhibits cancer cell killing in vitro and promotes tumor growth in vivo [9], [10], [11], [12], [13], [14], [15]. Shed sialoglycoproteins originating from the cancer cell glycocalyx are also immunosuppressive and promote differentiation of Siglec-7- and Siglec-9-positive monocytes to pro-tumor macrophages in pancreatic ductal adenocarcinoma (PDAC) [16]. These findings emphasize the critical role played by sialoglycoproteins in immune modulation and suggest that inhibition of the sialic acid-Siglec axis could improve cancer treatments including checkpoint inhibitor responses [17]. Nevertheless, factors that modulate sialoglycoprotein expression and their relationship to tumor aggression and anti-tumor immunity remain poorly defined.
Solid tumors are characterized by a desmoplastic response that associates with fibrosis, involving increased deposition, remodeling and crosslinking of extracellular matrix proteins. In breast cancer and PDAC, fibrosis associates with more aggressive subtypes [18], [19]. Tumor fibrosis can promote malignancy by fostering cancer cell growth and survival, enhancing invasion and migration, and inducing immune suppression [20], [21], [22]. Highly fibrotic cancers are often defined as immune deserts defined by poor infiltration of tumoricidal GzmB+CD8+ T cells [23]. What is not clear, however, are the mechanisms by which fibrosis induces immune suppression. Fibrosis induces multiple changes in tumors, including metabolic changes that lead to dysregulated anabolism of many different complex molecules, including glycoproteins [24].
We hypothesized that an association between fibrosis and increased production of immunosuppressive sialoglycoproteins could provide a functional link for fibrosis-induced immune suppression. In this study, we performed a comprehensive histological analysis and cellular census of human breast tumors to assess the link between fibrosis, tumor aggression, and the sialic acid-Siglec axis. We found that sialoglycans, including Siglec-7, Siglec-9, and Siglec-10 ligands, are produced by cancer cells and are increased in regions of high fibrosis and more aggressive breast cancer subtypes. In addition, more aggressive breast cancer subtypes feature greater infiltration of SIGLEC7-, SIGLEC9-, and SIGLEC10-positive myeloid cells. Our findings demonstrate an association of tumor fibrosis and tumor aggression with sialoglycoprotein expression and infiltration of immunosuppressive Siglec-positive myeloid cells. Additionally, our data predict that the highly aggressive triple-negative (TN) subtype as the most likely to benefit from therapeutic targeting of the sialic acid-Siglec axis.
Results
Section 1. Acidic glycoprotein content is elevated in high fibrosis regions and associates with breast cancer aggression
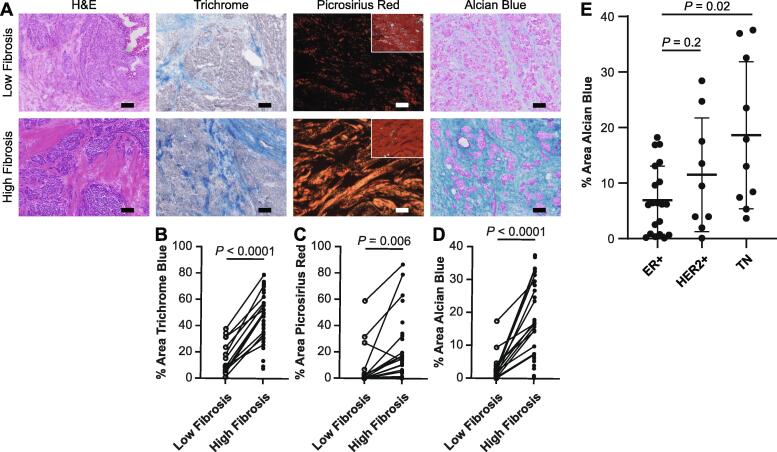
We first sought to investigate whether a link exists between acidic glycoprotein expression and fibrosis. We stained a cohort of human breast tumors (Table S1) for total collagen (Trichrome blue), fibrillar collagen (Picrosirius red), and polyacidic molecules including sialoglycoproteins, hyaluronic acid, and sulfated proteoglycans (Alcian blue). Serial tissue sections were stained and whole slide imaging (WSI) was used to evaluate acidic glycoprotein content in regions of varying levels of fibrosis. High and low fibrosis regions within each tumor were identified by a breast pathologist in parallel hematoxylin and eosin (H&E) stained tissue (Fig. 1A). These identified regions of high and low fibrosis defined by collagen abundance were confirmed by Trichrome blue and Picrosirius red staining (Fig. 1B-C). Analysis revealed that Alcian blue staining was significantly higher in the high fibrosis regions within the tumor tissue as compared to the low fibrosis regions. Sialidase pretreatment decreased Alcian blue staining (Fig. S1), indicating that the Alcian blue staining represents sialoglycoproteins. These findings suggest there may be a link between fibrosis and levels of sialoglycoproteins in human breast tumor tissue (Fig. 1D).
Fig. 1.
High fibrosis regions have greater acidic glycoprotein content. A. Representative images of patient-matched low and high fibrosis regions of human breast tumors stained with H&E, Trichrome, Picrosirius red (polarized image; inset brightfield), and Alcian Blue. Scale bar is 100 μm. B-D. Quantification of positively stained area of B. Trichrome, C. Picrosirius red, and D. Alcian blue in low and high fibrosis regions. E. Quantification of Alcian blue-positive area across whole tumor tissues by subtype. The mean is plotted and the error bars represent 1 standard deviation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Human breast cancers can be divided into histological subtypes that predict tumor aggression with ER+ reflecting lower grade, HER2+ reflecting higher grade, and TN reflecting highest grade. We reported previously that the level of tissue fibrosis associated with the more aggressive HER2+ and TN breast cancer subtypes [18]. To determine whether the correlation between acidic glycoproteins and tissue fibrosis reflected tumor aggression, we next compared the level of Alcian blue staining across whole tissues according to breast cancer subtype. Alcian blue staining was highest in TN tumors and lowest in ER+ tumors with the highest intensity of Alcian blue staining found in the regions within these tumors with the greatest intensity of fibrosis (Fig. 1E and S1). These data reveal a link between tumor aggression, fibrosis, and sialoglycoprotein content.
Section 2. Cancer cells express a sialoglycoprotein biosynthetic gene program that is the most abundant in the more aggressive breast cancer subtypes
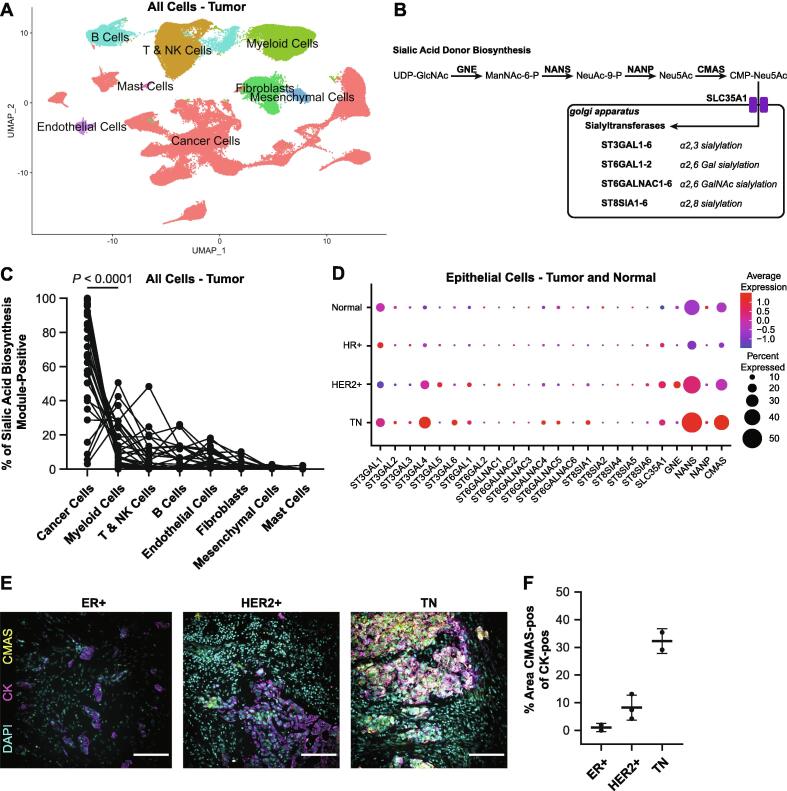
To understand the cellular source of the increased acidic glycoprotein in fibrotic and aggressive breast tumors, we quantified expression of sialoglycoprotein biosynthetic genes from a publicly available scRNAseq dataset [25]. We first clustered 34 breast tumors (20 HR+ including 19 ER+ and 1 PR+, 6 HER2+, and 8 TN) (Fig. 2A and S2) and extracted the epithelial cell clusters based on KRT19 expression (Fig. S2). We also performed the clustering and extraction of epithelial cell clusters based on KRT14 and KRT18 expression from 12 normal breast tissues (Fig. S3). We then merged the tumor and normal epithelial cell datasets. Our analysis revealed that cells clustered by malignancy and subtype in UMAP space (Fig. S4).
Fig. 2.
Sialoglycoprotein biosynthetic gene programs are found in cancer cells and elevated in aggressive breast cancers. A. UMAP projection of all cells from scRNAseq data of 34 breast tumors [20]. B. Diagram of sialylglycoprotein biosynthetic pathway. C. Plot of percent sialic acid biosynthesis module-positive (CMAS and NANS) by cell type for each patient. D. Dot plot showing expression of all sialoglycoprotein biosynthetic genes in epithelial cells extracted from normal breast, and HR+, HER2+, and TN tumors. E. Representative images of multicolor IF of CMAS and CK. Scale bar is 100 μm. F. Quantification of CMAS staining in CK-positive cells. For each subtype n = 2–3 patient samples were quantified. The mean is plotted and the error bars represent 1 standard deviation.
We then identified the cell clusters responsible for sialoglycoprotein biosynthesis based on gene expression. We evaluated expression of 20 sialyltransferase genes (ST3GAL1-6, ST6GAL1-2, ST6GALNAC1-6, ST8SIA1-6) and 5 sialic acid biosynthetic genes (SLC35A1, GNE, NANS, NANP, CMAS) per cluster for the tumor and normal tissues (Fig. 2B and S2-S3). We found elevated expression of CMAS, a N-acylneuraminate cytidylyltransferase and the last step in CMP-sialic acid biosynthesis, in cancer cells in tumors (Fig. S2) and in secretory L1-type luminal epithelial cells, marked by SPLI [26], in normal breast tissue (Fig. S3). In addition, multiple clusters had high expression of NANS, a sialic acid synthase, including cancer cells, myeloid cells, and B cells in tumors and basal epithelial cells, mesenchymal cells, and immune cells in normal breast. Importantly, limitations in sequencing depth associated with scRNAseq protocols are likely responsible for the low detected expression we quantified in these data sets for many of the sialoglycoprotein biosynthetic genes [27]. Nevertheless, within tumor tissues, cancer cells featured higher expression of the NANS and CMAS gene module, relative to all other cell clusters (Fig. 2C). These data suggest that cancer cells are likely the major source of the increased level of sialylation that characterizes human breast tumors.
Next, we asked if sialoglycoprotein biosynthetic gene expression within the breast epithelial cells varied by clinical cancer subtype. We found elevated expression of NANS, CMAS, ST3GAL4, a beta-galactoside α2-3 sialyltransferase, and ST6GAL1, a beta-galactoside α2-6 sialyltransferase, in more aggressive tumor subtypes (i.e., HER2+ and TN tumors), relative to HR+ tumors and normal breast tissues (Fig. 2D). Interestingly, epithelial cells from HR+ tumors did not have elevated expression of any sialoglycoprotein biosynthetic genes, relative to HER2+ and TN tumors and normal breast epithelium. There was substantial interpatient heterogeneity in the expression of sialoglycoprotein biosynthetic genes (Fig. S5). We confirmed elevated expression of CMAS on the protein level using immunofluorescence staining of breast tumor sections. We observed the fraction of cancer cells, marked by pan-cytokeratin (CK-positive), that are CMAS-positive was highest for TN tumors and lowest for ER+ tumors, consistent with the scRNAseq results (Fig. 2E-F).
We next confirmed that an individual cell would have potential for sialoglycoprotein biosynthesis through coexpression of NANS and CMAS. We identified coexpression of NANS in 55% of CMAS-positive epithelial cells, and coexpression of CMAS in 31% of NANS-positive epithelial cells as CMAS-positive (Fig. S5). These data suggest that sialoglycoprotein biosynthesis may be elevated in the more aggressive breast cancer clinical subtype. The findings are also consistent with the increased Alcian blue staining we quantified in the more aggressive breast cancer subtypes (Fig. 1E).
Section 3. Tumor sialylation is increased in high fibrosis regions
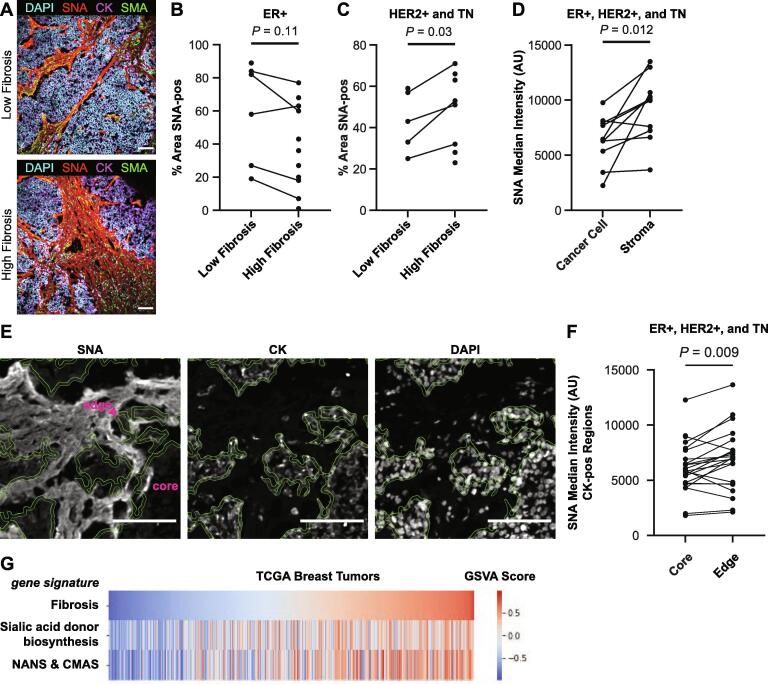
To understand if changes in sialic acid biosynthetic gene expression contributes to the changes in sialic acid content we and others observed in these human breast cancers, we next used quantitative lectin immunofluorescence. We first stained the human breast cancer tissue with Sambucus nigra lectin (SNA), which binds preferentially to α2,6-sialylated glycoproteins (Fig. 3A). For the less aggressive ER+ tumors, SNA staining did not change between low and high fibrosis regions (Fig. 3B). Interestingly however, in the more aggressive HER2+ and TN breast cancer subtypes, we quantified an increase in the SNA-positive area in high fibrosis regions as compared to the low fibrosis regions (Fig. 3C). Increased SNA staining in HER2+ and TN breast cancer subtypes is consistent with elevated expression of ST6GAL1 in these subtypes (Fig. 2D).
Fig. 3.
Sialic acid levels are elevated in the cancer cell glycocalyx and stroma within high fibrosis regions of aggressive breast tumors. A. Representative IF images of a TN tumor in high and low fibrosis regions stained with DAPI, SNA lectin, pan-CK, and SMA. Scale bar is 100 μm. B-C. Quantification of SNA-positive area in low and high fibrosis regions in B. ER+ and C. HER2+ and TN tumors. D. Quantification of SNA intensity in cancer cell and stromal regions. E. Representative IF images of SNA staining of tumor edge and core regions. The 5 μm thick region bounded by the green lines represents the tumor edge. DAPI and CK inserts are from the same image. Scale bar is 100 μm. F. Quantification of SNA intensity in tumor core and edge regions. G. Heatmap showing the expression of gene signatures in RNAseq data from breast tumors from TCGA. Patients were ordered by the fibrosis signature. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
SNA-positive regions were found throughout the tumor (Fig. 3A) and could represent sialoglycoproteins found in the glycocalyx, sialylated extracellular matrix proteins, sialylated secreted proteins, and sialylated shed proteins [28]. Based on lectin staining localization, we identified two regions containing sialoglycoproteins—cancer cell and the stromal regions. We used smooth muscle actin (SMA), which marks stromal fibroblasts to identify stromal regions [20]. SNA intensity was greatest in SMA-positive regions, relative to CK-positive regions (Fig. 3D). The elevated SNA staining in stromal regions was consistent with the elevated Alcian blue staining we quantified earlier in the stromal regions of the breast tumors (Fig. 1A).
Sialyation of the cancer cell compartment, which includes the glycocalyx, was determined by quantifying SNA intensity within CK-positive regions. To begin with we failed to observe any differences in sialylation of the cancer cell compartment between high and low fibrosis regions within the breast cancer tissue (Fig. S6). Accordingly, given that the tumor invasive front displays a strikingly reorganized collagenous stroma that we previously showed was stiffer [18], we next compared the SNA lectin staining of the cancer cell compartment between the tumor invasive front and tumor core. We classified CK-positive edge regions using a 5 μm circle erosion of CK-positive regions (Fig. 3E). The tumor front featured elevated SNA lectin staining, compared to the CK-positive core (Fig. 3F). These data reveal that the cancer cell compartment is less sialylated than proximal stromal regions, and that cancer cells on the tumor front feature a greater level of sialylation than cancer cells in the tumor core.
To confirm that the lectin staining results were not specific to SNA ligands, we also stained tumor tissues with MAL-II lectin, which binds preferentially to α2,3-sialylated glycoproteins. We observed a similar staining pattern between SNA and MAL-II lectins, suggesting that the sialoglycoprotein distribution does not vary by sialic acid linkage (Fig. S6).
The presence of sialoglycoprotein in fibrotic stroma and at the tumor edge suggests a causal link between fibrosis and sialoglycoprotein content in human breast tumors. To test this correlation, we queried breast tumor RNAseq data deposited in The Cancer Genome Atlas (TCGA) for evidence of any correlation of sialic acid biosynthesis gene programs with level of tumor fibrosis using a previously described fibrosis gene signature [29] (Fig. 3G). We found that the sialic acid donor biosynthesis gene program (GNE, NANS, NANP, CMAS, and SLC35A1) was positively correlated with the fibrosis gene signature (r = 0.465, P = 3.51e-60). The sialic acid biosynthesis module used in Fig. 2C, which consisted of NANS and CMAS, was even more highly correlated with the fibrosis gene signature (r = 0.566, P = 3.01e-94). These data support a causal link between fibrosis and sialic acid donor biosynthesis in breast cancer.
Section 4. Aggressive breast tumor subtypes feature elevated infiltration of SIGLEC7, SIGLEC9, and SIGLEC10-positive myeloid cells
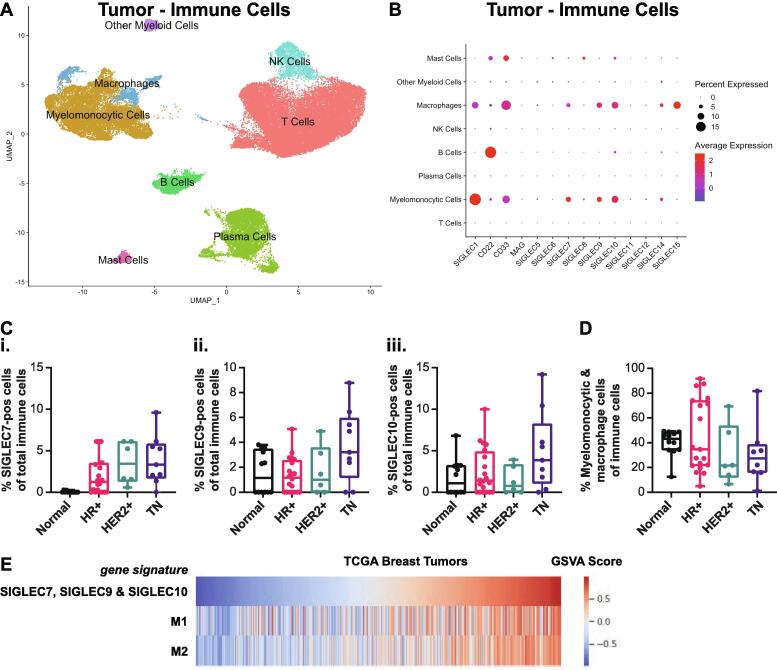
We next explored potential links between the increased sialoglycoprotein content we observed in the high fibrosis regions of the tumor stroma as compared to the tumor invasive edge to markers of immune modulation. We focused on tumor-infiltrating Siglec-positive cells which have been implicated in immune inhibition [17]. Human Siglecs are a family of 14 receptors that are primarily expressed on immune cells, bind to sialoglycoprotein ligands, and have activating and/or inhibiting signal transduction functions [30]. In breast cancer, inflammation and macrophage infiltration correlate positively with tumor aggression and with the more aggressive breast cancer histophenotypes [18]. To further assign the cell types that express SIGLEC genes, we applied the human breast cancer scRNAseq dataset [25] analyzed in Fig. 2 to quantify SIGLEC expression. We focus our analysis here on SIGLEC7, SIGLEC9, and SIGLEC10, which have the greatest evidence of pro-tumor functions in breast cancer [9], [11], [14], [16], [30], [31], [32]. After first confirming that SIGLEC expression is found primarily in immune cell types we thereafter extracted the data on the immune cells for further analysis (Fig. S7). We also performed the same analysis on the normal breast samples, and similarly found SIGLEC expression primarily in immune cells (Fig. S8). We assigned cell types to each cluster based on marker genes (Figs. S7-S8). Analysis revealed that myeloid cells were the predominant cell type with expression of SIGLEC7, SIGLEC9, and SIGLEC10 in the human breast tumors (Fig. S7). In normal breast tissues, we did not observe any SIGLEC7 expression, and identified myelomonocytic cells had the highest expression of SIGLEC9 and SIGLEC10 (Fig. S8), although a lower percentage of cells from this cluster were positive compared to the myeloid cell cluster in the tumor samples. We continued our analysis of SIGLEC expression within the extracted immune cells to further characterize SIGLEC-positive immune cells. We extracted and reclustered the immune cells from all tumor samples (Fig. 4A and S7) and found that SIGLEC7, SIGLEC9, and SIGLEC10 expression was highest in the myelomonocytic and macrophage cells in tumors (Fig. 4B). In normal breast tissue, we failed to observe SIGLEC7 expression in any of the immune cell types, and discerned lower expression of SIGLEC9 and SIGLEC10 within the myeloid population relative to breast tumors (Fig. S8). Recently, Siglec-15 has also been shown to have pro-tumor functions [15]. We observed high SIGLEC15 expression in the macrophage cluster, but not the myelomonocytic cluster.
Fig. 4.
SIGLEC7, SIGLEC9 and SIGLEC10-positive myeloid infiltration is increased in breast tumors and correlates with aggression. A. UMAP projection of immune cells from scRNAseq data of 34 breast tumors [20]. B. Dot plot of SIGLEC expression in immune cell clusters in breast tumors. C. Quantification of i. SIGLEC7, ii. SIGLEC9, and iii. SIGLEC10-positive inflammation as a percentage of total inflammation in normal breast and breast cancer subtypes. D. Quantification of myeloid infiltration as a percentage of total inflammation in normal breast and breast cancer subtypes. E. Heatmap showing the expression of gene signatures in RNAseq data from breast tumors from TCGA. Patients were ordered by the immunosuppressive Siglec signature.
The percentage of SIGLEC7-, SIGLEC9-, and SIGLEC10-positive cells of all immune cells increased with subtype aggression (Fig. 4C). Importantly, SIGLEC7 expression was not observed in normal human breast tissue, indicating that SIGLEC7-positive cells may be a feature of human breast cancer. The increased infiltration of SIGLEC7-, SIGLEC9-, and SIGLEC10-positive myeloid cells was not simply due to increased myeloid inflammation. The immune infiltration and myeloid infiltration as a percentage of total cells increased with breast cancer aggression subtype (Fig. S9), which we and others have previously reported [18]. However, the amount of infiltrating myeloid cells as a percentage of infiltrating immune cells did not vary by subtype in this dataset (Fig. 4D). We interpreted this to mean that the infiltrating myeloid cells in the more aggressive breast cancer subtype likely have increased SIGLEC7, SIGLEC9, and SIGLEC10 expression, relative to the myeloid cells in the less aggressive breast cancer subtype and to normal human breast tissue.
Prior work showed Siglec-7, Siglec-9, and Siglec-10 have immunosuppressive and pro-tumor functions using a functional-based approach [9], [11], [14], [16], [30], [31], [32]. Here, we sought to explore links between these Siglecs and an immunosuppressive phenotype using transcriptomic data. As illustrated in Fig. 3G, we queried breast tumor RNAseq data deposited in TCGA for evidence of a correlation between an immunosuppressive Siglec gene signature, consisting of SIGLEC7, SIGLEC9, and SIGLEC10, with previously described M1 (anti-tumor) and M2 (pro-tumor) macrophage gene signatures [29] (Fig. 4E). Macrophages are phenotypically plastic, and anti-tumor and pro-tumor macrophage classifications represent phenotypes of a spectrum of states [33], [34]. Although the immunosuppressive Siglec gene signature correlated positively with both the anti-tumor and pro-tumor gene signatures, we noted a stronger correlation with the pro-tumor gene signature (r = 0.758, P = 2.27e-206) as compared to the anti-tumor gene signature (r = 0.563, P = 3.68e-93). These data further support the classification of SIGLEC7, SIGLEC9, and SIGLEC10-positive myeloid cells as immunosuppressive.
Section 5. Association of Siglec ligands with regions of fibrosis
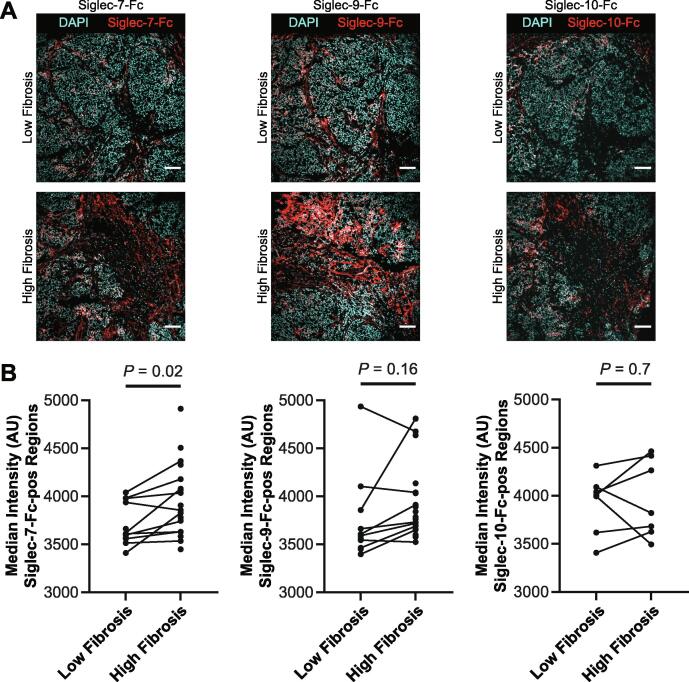
While Siglec receptors bind to sialylated ligands, they do possess unique substrate specificities [14], [31], [35], [36]. To better understand the location and quantify immunoinhibitory Siglec ligands within human breast tumors, we stained our cohort of breast cancer tissue with Siglec receptor-Fc fusion proteins (Fig. 5A). Siglec ligands stained positively in all breast cancer tissues. However, there was significant heterogeneity in Siglec ligand-positive area and intensity. As with the plant lectins, we observed higher staining intensity in stromal regions within the tumor tissue as compared to the cancer cell region. However, unlike the relatively uniform staining of SNA and MAL-II lectin in stromal regions, Siglec ligand staining was heterogeneous in these regions. These data demonstrate the different specificities between Siglecs and plant lectins.
Fig. 5.
Association of Siglec ligands with fibrosis. A. Representative images from a TN tumor of Siglec-7, Siglec-9, and Siglec-10 staining in low and high fibrosis regions. Scale bar is 100 μm. B. Quantification of median intensity within Siglec ligand-positive regions for Siglec-7-Fc, Siglec-9-Fc, and Siglec-10-Fc.
To clarify the relationship between Siglec ligand content and local fibrosis, we calculated the fraction of area with Siglec ligand-positive staining. There did not appear to be a difference in Siglec ligand-positive area between high and low fibrosis regions for the three Siglec-Fc reagents tested (Fig. S10). However, we did quantify higher intensity of Siglec ligand-positivity for Siglec-7 ligands in regions within the breast cancer tissue that had high levels of fibrosis (Fig. 5B). We similarly observed increased Siglec-9 ligand levels in regions within the breast tumors that had high amounts of fibrosis. However, we failed to observe any association between levels of Siglec-10 ligand and tumor tissue fibrosis. These data suggest there may be a relationship between the level of the ligands Siglec-7 and Siglec-9 and tissue fibrosis in human breast cancer.
Discussion
Our data establish an association between breast cancer fibrosis, sialoglycoprotein content and human breast cancer aggression as indicated by the HER2+ and TN subtypes. Increased sialylation is found in multiple tumor types, including breast, and increases metastatic potential in multiple mouse tumor models [17]. While recent studies have focused on the functional consequences of hypersialylation in tumors, less is known about the causes of increased sialylation. Our study provides evidence there may be a functional link between cancer fibrosis and the increased sialylation found in tumors. We found greater acidic glycoprotein content in high fibrosis regions and in more aggressive breast tumor subtypes. We also found increased sialylation in high fibrosis regions for HER2+ and TN tumors. Finally, we observed greater Siglec-7 ligand levels in high fibrosis regions.
We found that cancer cells are the main producers of sialoglycoprotein biosynthesis within tumors. In addition, cancer cells in TN tumors have elevated expression of the sialoglycoprotein biosynthesis genes ST3GAL4, NANS, and CMAS. Although we found increased expression of sialoglycoprotein biosynthesis genes in HER2+ and TN tumors, relative to ER+ tumors, we still observed high levels of tumor sialylation in ER+ tumors. One explanation for this observation is that the greater biosynthetic flux of sialoglycoproteins in TN tumors could be matched by greater consumption fluxes, such as greater recycling of surface sialoglycoproteins and greater cellular uptake of shed sialoglycoproteins.
While recent studies have elucidated the immune suppressive role of glycocalyx sialoglycans at immunological synapses, our data showed greater sialoglycans abundance in the stroma. Given the greater sialylation of the stroma, we propose that cancer cells secrete and shed sialoglycoproteins that become physiochemically adsorbed to the extracellular matrix in the tumor microenvironment. Soluble shed sialoglycoproteins can serve as Siglec ligands that drive monocyte differentiation to immune suppressive macrophages [16], which suggests that stromal sialoglycoproteins could serve as Siglec ligands and contribute to an immunosuppressive phenotype in the tumor microenvironment. Greater fibrosis involves increased collagen accumulation [22] and could provide for more binding sites for sialoglycoprotiens. More work is needed to attribute cancer cells as the source of stromal sialoglyoproteins and to elucidate the functional consequences of stromal sialylation in cancer.
Sialoglycoproteins suppress cytotoxic immune responses in tumors [32]. High expression of sialyl-Tn antigen is correlated with lower tumor infiltration of CD8+ T cells in human endometrial cancer [37], and therapeutic desialylation of EMT6 xenografts increases tumor infiltration of GZMB+ CD8+ cytotoxic T cells [38]. Mechanistically, immune inhibition by Siglec receptors requires the presence of both ligands and receptor-positive cells. We found that TN tumors have increased infiltration of SIGLEC7-, SIGLEC9-, and SIGLEC10-positive myeloid cells, which have been shown previously to have immunosuppressive functions [12], [13], [14], [16], [38]. Interesting, these TN tumors also feature greater synthesis of sialoglycoproteins, which could serve as Siglec ligands. Thus, among breast tumor subtypes, TN tumors may be best targeted with inhibitors of the sialic acid-Siglec axis.
Methods
Human breast tumor tissues
Tumor tissues were obtained from patients with breast cancer from University of California, San Francisco, (UCSF) or Duke University Medical Center between 2010 and 2020. Tissue specimens were flash frozen in OCT (Tissue-Tek) by slow immersion in liquid nitrogen or placement on dry ice and stored at − 80 °C until ready for sectioning. Samples were stored and analyzed with deidentified labels to protect patient data in accordance with the procedures outlined in the Institutional Review Board Protocol #10–03832, approved by the UCSF Committee of Human Resources and the Duke University IRB (Pro00054515). HER2+ subtype included all HER2+ patients regardless of ER status.
Picrosirius red staining and quantification
Frozen OCT tissue sections were fixed in 4% PFA for 10 min at RT, and then washed three times with PBS for 5 min each. Tissues were then stained with 0.1% picrosirius red (Direct Red 80, Sigma-Aldrich, 365,548 and picric acid solution, Sigma-Aldrich, P6744) for 1 hr and counterstained with Weigert’s hematoxylin (Thermo Scientific, 88,028 and 88029) for 10 min at RT. Polarized light images were acquired using an Olympus IX81 microscope fitted with an analyser (U-ANT) and a polarizer (U-POT, Olympus) oriented parallel and orthogonal to each other. Images were quantified using an ImageJ macro to determine percentage area coverage per field of view using one to five fields of view per tissue region. The ImageJ macro is available at https://github.com/northcottj/picrosirius-red.
Trichrome blue staining
Frozen OCT tissue sections were fixed in 4% PFA for 10 min at RT, and then washed three times with PBS for 5 min each. Tissues were then stained with Masson Trichrome Stain (Thermo Scientific) according to the manufacturer’s instructions.
Alcian blue staining
Frozen OCT tissue sections were fixed in 4% PFA for 10 min at RT, and then washed three times with PBS for 5 min each. Tissues were then placed in alcian blue (Sigma, B8428) for 1 hr and counterstained with nuclear fast red (Vector, H-3403) for 5 min at RT. Whole-slide images were obtained with a ZEISS Axio Scan.Z1 digital slide scanner equipped with CMOS and color cameras, with a 10x objective.
Quantification of Trichrome blue and Alcian blue staining
For whole-slide images of trichrome blue and alcian blue staining, one to three images at 1218x809 pixels were selected per tissue region. Each image was color deconvoluted using https://github.com/landinig/IJ-Colour_Deconvolution2 [39] and a minimal threshold was set for blue pixels and maintained for all images. The area of region that was covered by the minimal threshold was calculated and one to three images per tissue region were then pooled and averaged.
Sialidase treatment of tumor sections
Frozen OCT tissue sections were fixed in 4% PFA for 10 min at RT, and then washed three times with PBS for 5 min each. Sections were then incubated with 0.25 U/mL Vibrio cholerae sialidase (Roche #11080725001) in glycosidase buffer (50 mM sodium acetate, 5 mM CaCl2, pH 5.5), or glycosidase buffer alone, in a humid chamber at 37 °C for 1 h. Slides were then washed with PBS and stained with Alcian blue as above.
scRNAseq analysis
We took advantage of a recently published human breast cancer atlas [25] that was publicly available on the Gene Expression Omnibus (GSE161529). We predominantly focused our SeuratV4 analysis [40] on merged data for 34 distinct breast tumors representing different disease subtypes, that we compared to merged data for 12 normal mammary glands. Prior to merging, we first processed each file individually in Seurat and retained cells based on RNA features, removing outliers that were always below 200 and generally above 5,000, as well as any cells exhibiting >10% mitochondrial DNA. The UMAP plot resulting from the merged data was produced using 20 principal components during the dimensional reduction step and with a resolution of 0.1. Cluster annotation proceeded via attributing to clusters the expression of core classification genes (e.g., B cells = IGKC, IGLC2, MS4A1; Cancer/Epithelial cells = KRT19, KRT18, KRT8; Endothelial cells = VWF, CLDN5, PECAM1; Fibroblast cells = COL1A1, COL1A2, TAGLN; Mast cells = TPSAB1, TPSB2, Kit; Mesenchymal cells = IGFBP7, ACTA2, TAGLN, PDGFRB; Myeloid cells = APOE, LYZ, CD72; T & NK cells = CD3D, GMZA, NKG7). Of note, the mesenchymal cell cluster was left broadly annotated due to varied gene expression, although it does demonstrate some features attributable to pericytes. In instances where cells expressing SIGLEC genes were enumerated on a per sample basis, immune cells were first gated by virtue of them having PTPRC (CD45) expression > 0.5 and then the resulting cells were enumerated using SIGLEC expression > 0. In analyses where the epithelial cells were counted for their expression of SiaSynthesis genes, we first gated the cells via KRT19 expression > 0.5 and next quantified those expressing individual SiaSynthesis genes > 0. The analysis of the normal mammary gland samples proceeded in a manner akin to our tumor analysis.
Immunoflourescence staining
Frozen OCT tissue 10 μm sections were fixed in 4% PFA for 10 min at RT, and then washed three times with PBS for 5 min each. Tissue sections were permeabilized with 0.25% v/v Triton-X-100 in PBS for 5 min at room temperature. Tissues sections were then blocked with 1x Carbo-Free blocking solution (Vector Laboratories # SP-5040–125) for 1 h at room temperature. Primary antibodies were diluted in 1x Carbo-Free blocking solution and incubated overnight at 4 °C. Tissue sections were washed three times with PBS and stained with fluorescently labelled secondary antibodies diluted in 1x Carbo-Free blocking solution for 1 h at room temperature. Tissue sections were washed three times with PBS, counterstained with DAPI, mounted, and imaged. Primary antibodies used and dilution ratios are: anti-panCK (Biolegend #914204; 1:100), anti-CMAS (Sigma-Aldrich #HPA039905; 1:300), SNA-biotin (Vector Laboratories #B-1305–2; 1:200), anti-SMA (Cell Signaling Technology #19245 s; 1:200), MAL-II biotin (Vector Laboratories #B-1265–1; 1:100). For Siglec-Fc reagents, which were used to image Siglec ligands, a pre-complexing protocol [38] was used where 1 μg/mL Siglec-Fc reagent was incubated with 1 μg/mL donkey anti-human-DyLight488 (Thermo-Fisher #PISA510126) in 1x Carbo-Free blocking solution before being added to the tissue sections at the secondary labeling step. Siglec-Fc reagents were purchase from R&D Systems: Siglec-7-Fc (#1138SL050) Siglec-9-Fc (#1139SL050), and Siglec-10-Fc (#2130SL050).
Immunofluorescence imaging and analysis
IF-stained breast tumor sections were imaged on an inverted Nikon Ti2-E microscope equipped with a CREST X-Light V2 large field of view spinning disk confocal. For each slide, 3–6 representative images were collected for each high and low fibrosis regions. Images were collected using a 20x or 40x objective. IF micrographs were batch analyzed using custom pipelines built using the NIS-Elements General Analysis (GA3) tool.
Analyses of RNAseq data deposited in TCGA database
For RNAseq analysis, HTseq count files were acquired from the TCGA-BRCA repository from 1,222 patient samples. Lowly expressed genes were filtered out if average expression was less than 1 CPM, which resulted in inclusion of 16,974 genes. Normalization was performed using calcNormFactors in edgeR. Pathway enrichment was done using GSVA. Pearson correlation 'r' and 'p' values are reported.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We dedicate this paper to the memory of Zena Werb for her mentorship and support. We thank Natasha Korets for histology support. We would also like to thank Kelly Kersten, Jessica Stark, and Lam-Kiu Fong for helpful discussions.
Funding
This work was supported by an American Cancer Society–2017 Seattle Gala Paddle Raise Postdoctoral Fellowship (PF-18-118-01-CDD to KJM), the Program for Breakthrough Biomedical Research, which is partially funded by the Sandler Foundation to KJM, and NIH National Cancer Institute funding (R35CA242447-01A1 and R01CA227942 to VMW; R01CA222508-01 to VMW and ESH; K00CA212132 to EB).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2022.100105.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Cohen M., Elkabets M., Perlmutter M., Porgador A., Voronov E., Apte R.N., Lichtenstein R.G. Sialylation of 3-methylcholanthrene-induced fibrosarcoma determines antitumor immune responses during immunoediting. J. Immunol. Baltim. Md. 2010;1950(185):5869–5878. doi: 10.4049/jimmunol.1001635. [DOI] [PubMed] [Google Scholar]
- 2.Varki A., Kannagi R., Toole B., Stanley P. In: Essent. Glycobiol. 3rd ed. Varki A., Cummings R.D., Esko J.D., Stanley P., Hart G.W., Aebi M., Darvill A.G., Kinoshita T., Packer N.H., Prestegard J.H., Schnaar R.L., Seeberger P.H., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor (NY): 2015. Glycosylation changes in cancer. http://www.ncbi.nlm.nih.gov/books/NBK453023/ (accessed December 23, 2021) [Google Scholar]
- 3.Tarbell J.M., Cancel L.M. The glycocalyx and its significance in human medicine. J. Intern. Med. 2016;280:97–113. doi: 10.1111/joim.12465. [DOI] [PubMed] [Google Scholar]
- 4.Paszek M.J., DuFort C.C., Rossier O., Bainer R., Mouw J.K., Godula K., Hudak J.E., Lakins J.N., Wijekoon A.C., Cassereau L., Rubashkin M.G., Magbanua M.J., Thorn K.S., Davidson M.W., Rugo H.S., Park J.W., Hammer D.A., Giannone G., Bertozzi C.R., Weaver V.M. The cancer glycocalyx mechanically primes integrin-mediated growth and survival. Nature. 2014;511:319–325. doi: 10.1038/nature13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes J.M., Kaushik S., Bainer R.O., Sa J.K., Woods E.C., Kai F., Przybyla L., Lee M., Lee H.W., Tung J.C., Maller O., Barrett A.S., Lu K.V., Lakins J.N., Hansen K.C., Obernier K., Alvarez-Buylla A., Bergers G., Phillips J.J., Nam D.-H., Bertozzi C.R., Weaver V.M. A tension-mediated glycocalyx–integrin feedback loop promotes mesenchymal-like glioblastoma. Nat. Cell Biol. 2018;20:1203–1214. doi: 10.1038/s41556-018-0183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods E.C., Kai F., Barnes J.M., Pedram K., Pickup M.W., Hollander M.J., Weaver V.M., Bertozzi C.R. A bulky glycocalyx fosters metastasis formation by promoting G1 cell cycle progression. ELife. 2017;6 doi: 10.7554/eLife.25752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imbert P.R.C., Saric A., Pedram K., Bertozzi C.R., Grinstein S., Freeman S.A. An Acquired and Endogenous Glycocalyx Forms a Bidirectional “Don’t Eat” and “Don’t Eat Me” Barrier to Phagocytosis. Curr. Biol. 2021;31:77–89.e5. doi: 10.1016/j.cub.2020.09.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munkley J., Elliott D.J. Hallmarks of glycosylation in cancer. Oncotarget. 2016;7:35478–35489. doi: 10.18632/oncotarget.8155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jandus C., Boligan K.F., Chijioke O., Liu H., Dahlhaus M., Démoulins T., Schneider C., Wehrli M., Hunger R.E., Baerlocher G.M., Simon H.-U., Romero P., Münz C., von Gunten S. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J. Clin. Invest. 2014;124:1810–1820. doi: 10.1172/JCI65899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stanczak M.A., Siddiqui S.S., Trefny M.P., Thommen D.S., Boligan K.F., von Gunten S., Tzankov A., Tietze L., Lardinois D., Heinzelmann-Schwarz V., von Bergwelt-Baildon M., Zhang W., Lenz H.-J., Han Y., Amos C.I., Syedbasha M., Egli A., Stenner F., Speiser D.E., Varki A., Zippelius A., Läubli H. Self-associated molecular patterns mediate cancer immune evasion by engaging Siglecs on T cells. J. Clin. Invest. 2018;128:4912–4923. doi: 10.1172/JCI120612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hudak J.E., Canham S.M., Bertozzi C.R. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat. Chem. Biol. 2014;10:69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibarlucea-Benitez I., Weitzenfeld P., Smith P., Ravetch J.V. Siglecs-7/9 function as inhibitory immune checkpoints in vivo and can be targeted to enhance therapeutic antitumor immunity. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2107424118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Läubli H., Pearce O.M.T., Schwarz F., Siddiqui S.S., Deng L., Stanczak M.A., Deng L., Verhagen A., Secrest P., Lusk C., Schwartz A.G., Varki N.M., Bui J.D., Varki A. Engagement of myelomonocytic Siglecs by tumor-associated ligands modulates the innate immune response to cancer. Proc. Natl. Acad. Sci. USA. 2014;111:14211–14216. doi: 10.1073/pnas.1409580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barkal A.A., Brewer R.E., Markovic M., Kowarsky M., Barkal S.A., Zaro B.W., Krishnan V., Hatakeyama J., Dorigo O., Barkal L.J., Weissman I.L. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 2019;572:392–396. doi: 10.1038/s41586-019-1456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J., Sun J., Liu L.N., Flies D.B., Nie X., Toki M., Zhang J., Song C., Zarr M., Zhou X.u., Han X., Archer K.A., O’Neill T., Herbst R.S., Boto A.N., Sanmamed M.F., Langermann S., Rimm D.L., Chen L. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019;25(4):656–666. doi: 10.1038/s41591-019-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez E., Boelaars K., Brown K., Eveline Li R.J., Kruijssen L., Bruijns S.C.M., van Ee T., Schetters S.T.T., Crommentuijn M.H.W., van der Horst J.C., van Grieken N.C.T., van Vliet S.J., Kazemier G., Giovannetti E., Garcia-Vallejo J.J., van Kooyk Y. Sialic acids in pancreatic cancer cells drive tumour-associated macrophage differentiation via the Siglec receptors Siglec-7 and Siglec-9. Nat. Commun. 2021;12:1270. doi: 10.1038/s41467-021-21550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams O.J., Stanczak M.A., von Gunten S., Läubli H. Targeting sialic acid–Siglec interactions to reverse immune suppression in cancer. Glycobiology. 2018;28:640–647. doi: 10.1093/glycob/cwx108. [DOI] [PubMed] [Google Scholar]
- 18.Acerbi I., Cassereau L., Dean I., Shi Q., Au A., Park C., Chen Y.Y., Liphardt J., Hwang E.S., Weaver V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol. 2015;7:1120–1134. doi: 10.1039/c5ib00040h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laklai H., Miroshnikova Y.A., Pickup M.W., Collisson E.A., Kim G.E., Barrett A.S., Hill R.C., Lakins J.N., Schlaepfer D.D., Mouw J.K., LeBleu V.S., Roy N., Novitskiy S.V., Johansen J.S., Poli V., Kalluri R., Iacobuzio-Donahue C.A., Wood L.D., Hebrok M., Hansen K., Moses H.L., Weaver V.M. Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat. Med. 2016;22:497–505. doi: 10.1038/nm.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Winkler J., Abisoye-Ogunniyan A., Metcalf K.J., Werb Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020;11:5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maller O., Drain A.P., Barrett A.S., Borgquist S., Ruffell B., Zakharevich I., Pham T.T., Gruosso T., Kuasne H., Lakins J.N., Acerbi I., Barnes J.M., Nemkov T., Chauhan A., Gruenberg J., Nasir A., Bjarnadottir O., Werb Z., Kabos P., Chen Y.-Y., Hwang E.S., Park M., Coussens L.M., Nelson A.C., Hansen K.C., Weaver V.M. Tumour-associated macrophages drive stromal cell-dependent collagen crosslinking and stiffening to promote breast cancer aggression. Nat. Mater. 2021;20:548–559. doi: 10.1038/s41563-020-00849-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piersma B., Hayward M.K., Weaver V.M. Fibrosis and cancer: A strained relationship. Biochim. Biophys. Acta BBA - Rev. Cancer. 2020;1873 doi: 10.1016/j.bbcan.2020.188356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruosso T., Gigoux M., Manem V.S.K., Bertos N., Zuo D., Perlitch I., Saleh S.M.I., Zhao H., Souleimanova M., Johnson R.M., Monette A., Ramos V.M., Hallett M.T., Stagg J., Lapointe R., Omeroglu A., Meterissian S., Buisseret L., den Eynden G.V., Salgado R., Guiot M.-C., Haibe-Kains B., Park M. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Invest. 2019;129:1785–1800. doi: 10.1172/JCI96313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evers T.M.J., Holt L.J., Alberti S., Mashaghi A. Reciprocal regulation of cellular mechanics and metabolism. Nat. Metab. 2021;3:456–468. doi: 10.1038/s42255-021-00384-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pal B., Chen Y., Vaillant F., Capaldo B.D., Joyce R., Song X., Bryant V.L., Penington J.S., Di Stefano L., Tubau Ribera N., Wilcox S., Mann G.B., kConFab, Papenfuss A.T., Lindeman G.J., Smyth G.K., Visvader J.E. A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J. 2021;40 doi: 10.15252/embj.2020107333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nguyen Q.H., Pervolarakis N., Blake K., Ma D., Davis R.T., James N., Phung A.T., Willey E., Kumar R., Jabart E., Driver I., Rock J., Goga A., Khan S.A., Lawson D.A., Werb Z., Kessenbrock K. Profiling human breast epithelial cells using single cell RNA sequencing identifies cell diversity. Nat. Commun. 2018;9:2028. doi: 10.1038/s41467-018-04334-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haque A., Engel J., Teichmann S.A., Lönnberg T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017;9:75. doi: 10.1186/s13073-017-0467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinho S.S., Reis C.A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer. 2015;15:540–555. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 29.A.J. Combes, B. Samad, J. Tsui, N.W. Chew, P. Yan, G.C. Reeder, D. Kushnoor, A. Shen, B. Davidson, A.J. Barczac, M. Adkisson, A. Edwards, M. Naser, K.C. Barry, T. Courau, T. Hammoudi, R.J. Arguëllo, A.A. Rao, A.B. Olshen, T.I. Consortium, C. Cai, J. Zhan, K.C. Davis, R.K. Kelley, J.S. Chapman, C.E. Attreya, A. Patel, A.I. Daud, P. Ha, A.A. Diaz, J.R. Kratz, E.A. Collisson, G.K. Fragiadakis, D.J. Erle, A. Boissonnas, S. Asthana, V. Chan, M.F. Krummel, A Pan-Cancer Census of Dominant Tumor Immune Archetypes, 2021. https://doi.org/10.1101/2021.04.26.441344.
- 30.Macauley M.S., Crocker P.R., Paulson J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014;14:653–666. doi: 10.1038/nri3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beatson R., Tajadura-Ortega V., Achkova D., Picco G., Tsourouktsoglou T.-D., Klausing S., Hillier M., Maher J., Noll T., Crocker P.R., Taylor-Papadimitriou J., Burchell J.M. The mucin MUC1 modulates the tumor immunological microenvironment through engagement of the lectin Siglec-9. Nat. Immunol. 2016;17:1273–1281. doi: 10.1038/ni.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith B.A.H., Bertozzi C.R. The clinical impact of glycobiology: targeting selectins, Siglecs and mammalian glycans. Nat. Rev. Drug Discov. 2021;20(3):217–243. doi: 10.1038/s41573-020-00093-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cassetta L., Pollard J.W. Tumor-associated macrophages. Curr. Biol. CB. 2020;30:R246–R248. doi: 10.1016/j.cub.2020.01.031. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez H., Hagerling C., Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Dev. 2018;32:1267–1284. doi: 10.1101/gad.314617.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wisnovsky S., Möckl L., Malaker S.A., Pedram K., Hess G.T., Riley N.M., Gray M.A., Smith B.A.H., Bassik M.C., Moerner W.E., Bertozzi C.R. Genome-wide CRISPR screens reveal a specific ligand for the glycan-binding immune checkpoint receptor Siglec-7. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2015024118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alphey M.S., Attrill H., Crocker P.R., van Aalten D.M.F. High resolution crystal structures of Siglec-7. Insights into ligand specificity in the Siglec family. J. Biol. Chem. 2003;278(5):3372–3377. doi: 10.1074/jbc.M210602200. [DOI] [PubMed] [Google Scholar]
- 37.Ohno S., Ohno Y., Nakada H., Suzuki N., Soma G.-I., Inoue M. Expression of Tn and Sialyl-Tn Antigens in Endometrial Cancer: Its Relationship with Tumor-produced Cyclooxygenase-2, Tumor-infiltrated Lymphocytes and Patient Prognosis. Anticancer Res. 2006;26:4047–4053. [PubMed] [Google Scholar]
- 38.Gray M.A., Stanczak M.A., Mantuano N.R., Xiao H., Pijnenborg J.F.A., Malaker S.A., Miller C.L., Weidenbacher P.A., Tanzo J.T., Ahn G., Woods E.C., Läubli H., Bertozzi C.R. Targeted glycan degradation potentiates the anticancer immune response in vivo. Nat. Chem. Biol. 2020;16:1376–1384. doi: 10.1038/s41589-020-0622-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Landini G., Martinelli G., Piccinini F., Xu J. Colour deconvolution: stain unmixing in histological imaging. Bioinforma. Oxf. Engl. 2021;37(10):1485–1487. doi: 10.1093/bioinformatics/btaa847. [DOI] [PubMed] [Google Scholar]
- 40.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E.P., Jain J., Srivastava A., Stuart T., Fleming L.M., Yeung B., Rogers A.J., McElrath J.M., Blish C.A., Gottardo R., Smibert P., Satija R. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.