Abstract
The hexanucleotide repeat expansion in C9orf72 is a common cause of ALS/FTD and also rarely found in other psychiatric and neurodegenerative conditions. Alleles with >30 repeats are often considered an expansion, but the pathogenic repeat length threshold is still unclear. It is also unclear, whether intermediate repeat length alleles (often defined either as 7-30 or 20-30 repeats) have clinically significant effects. We determined the C9orf72 repeat length distribution in 3142 older Finns (aged 60-104). The longest non-expanded allele was 45 repeats. We found 7-45 repeats in 1036/3142 (33 %) individuals, 20-45 repeats in 56/3142 (1.8 %), 30-45 repeats in 12/3142 (0.38%) and expansion (>45 repeats) in 6/3142 (0.19%). There was no apparent clustering of neurodegenerative or psychiatric diseases in individuals with 30-45 repeats indicating that 30-45 repeats aren’t pathogenic. None of the six expansion carriers had a diagnosis of ALS/FTD but four had a diagnosis of a neurodegenerative or psychiatric disease. Intermediate length alleles (categorized as 7-45 and 20-45 repeats) did not associate with Alzheimer’s disease or cognitive impairment.
Keywords: Genetics, Alzheimer’s disease, Dementia, C9orf72, Cohort studies
1. Introduction
C9orf72 hexanucleotide repeat expansion is a major cause of sporadic and familial amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) (DeJesus-Hernandez et al., 2011, Renton et al., 2011) and it’s particularly common in Finland (Majounie et al., 2012). Expansions can span up to several thousand repeats, but the minimum length of pathogenic expansion is not known. Knowledge on the distribution of repeat lengths in the general population could help gain a better understanding of the threshold for pathogenicity. Perhaps the most widely used threshold for expansion is 30, defined in one of the original studies describing the C9orf72 repeat (Renton et al., 2011). Determining a threshold for expansion is complicated by the difficulty of reliably determining repeat lengths, and thresholds reflect more the limitation of used methods than actual alteration in biologic function. Southern blot is considered to be the golden standard of repeat length estimation but due to its high demand of good quality DNA, cost and work load, often other methods such as repeat-primed PCR (RP-PCR) are used.
Besides genuine expansions, “intermediate length” alleles (defined often as 7-30 or 20-30 repeats) have also been hypothesized to predispose to neurodegenerative diseases. In luciferase reporter assay, intermediate alleles showed decreased C9orf72 promoter activity as compared to short repeat alleles (Gijselinck et al., 2016). Moreover, adverse effects have been observed in a fly model with 30 repeats (Zhang et al., 2015) though the relevance of fly physiology to human pathophysiology in this context is unsure. As with expansion, the threshold for intermediate length repeats also varies in studies but allele length ≥7 has been shown to associate with the C9orf72 founder haplotype in the European population (van der Zee et al., 2013a). A recent review (Ng and Tan, 2017) summarized that most studies do not find association between intermediate length alleles and a variety of neurodegenerative diseases but intermediate length alleles might be associated with psychiatric symptoms. Most studies on intermediate length alleles focus on ALS/FTD spectrum, but only few on Alzheimer’s disease (AD) (Cacace et al., 2013a, Harms et al., 2013, Jiao et al., 2013, Kohli et al., 2013a, Xi et al., 2012).
In this study, we present C9orf72 hexanucleotide repeat length distribution in 3142 Finns and study the association of intermediate repeat alleles with AD and cognitive impairment.
2. Methods
2.1. Cohorts
We studied the C9orf72 hexanucleotide repeat length in 3161 individuals from four population-derived cohorts from the Helsinki-region, in Southern Finland. These were the Vantaa85+ study (Tanskanen et al., 2017) (n=469), Helsinki Birth cohort study (Barker et al., 2005, Eriksson et al., 2006, Kajantie et al., 2012, Lahti et al., 2014, Yliharsila et al., 2007) (n=1651), Helsinki Businessmen study (Strandberg et al., 2016) (n=666) and DEBATE study (Uusvaara et al., 2013) (n=375). We have previously used the same cohorts to study the association of TYROPB deletion and cognitive impairment (Kaivola et al., 2018). Information on AD, other dementia diagnoses, Mini Mental State Examination (MMSE) scores or lack of dementia was assessed from clinical records, death certificates, registry information or questionnaires. The source of information varied between cohorts. More detailed cohort descriptions and information on assessment of dementia are provided in the Supplementary Methods.
We studied the effect of intermediate repeat alleles in three settings: 1) individuals with AD vs. no AD, 2) individuals with cognitive impairment (MMSE score ≤24) vs. individuals with no cognitive impairment (MMSE score >24) and 3) individuals with cognitive impairment or any diagnosis of dementia vs. non-demented controls. All controls were ≥75 years. Repeat lengths 7-45 and 20-45 were used as the definition of intermediate length alleles.
2.2. Genetic analyses
We determined repeat lengths with RP-PCR as previously described (Renton et al., 2011) with minor changes to the protocol (Supplementary methods). Since it may be difficult to distinguish between an expansion and a long repeat in RP-PCR, we confirmed alleles with repeat length ≥20 and all putative expansions with over-the-repeat PCR. Samples with more than 30 repeats and the typical sawtooth pattern in RP-PCR, which only produced the smaller amplicon in over-the-repeat PCR were categorized as expansions (Supplementary Figure 1).
The smallest allele is here denoted as 2-3 repeats since 2 and 3 repeat alleles cannot be distinguished by RP-PCR. RP-PCR results may be inaccurate in distinguishing hetero- and homozygosity in certain allele combinations (especially 2-3/5 from 5/5 and 5/8 from 8/8). This can slightly increase the proportion of heterozygotes at the expense of homozygotes. Considering this uncertainty, we did not analyze cognitive measures separately in individuals homozygous for the intermediate alleles. There were no samples homozygous for the 20-45 repeat alleles in our cohorts. APOE was genotyped as previously described (Myllykangas et al., 1999).
2.3. Statistics
We used logistic regression to test for association between intermediate repeats in all three settings and used APOE ε4 carriership, age and sex as covariates. We applied Bonferroni correction to take account of multiple test settings and set the statistical significance to p=0.05/3=0.017. We used Kruskal-Wallis H test to determine if repeat length distributions differed between our four cohorts. All analyses were conducted with IBM SPSS statistics v.24 (IBM Corp. Released 2016. IBM SPSS Statistics for Windows, Version 24.0. Armonk, NY: IBM Corp.).
Assuming AD prevalence of 10 % in Finnish individuals aged over 75 years (Rahkonen et al., 2003), intermediate allele (≥7 repeats) frequency of 17 % based on our initial results, case-control ratio of 1:4, type 1 error rate 0.05 and relative risk of 1.5 with dominant effect, 201 cases were required for 80 % power (Purcell et al., 2003) (Genetic power calculator, http://zzz.bwh.harvard.edu/gpc/ accessed 4.6.2018). From the four cohorts, we identified 226 individuals with AD, 432 with MMSE ≤24, and 613 with any diagnosis of dementia or cognitive impairment.
2.4. Ethics
The study was approved by the Coordinating Ethics Committee of the Helsinki University Central Hospital. The Vantaa 85+ study was also approved by the Ethics Committee of the Health Centre of the City of Vantaa.
3. Results
3.1. Repeat length distribution
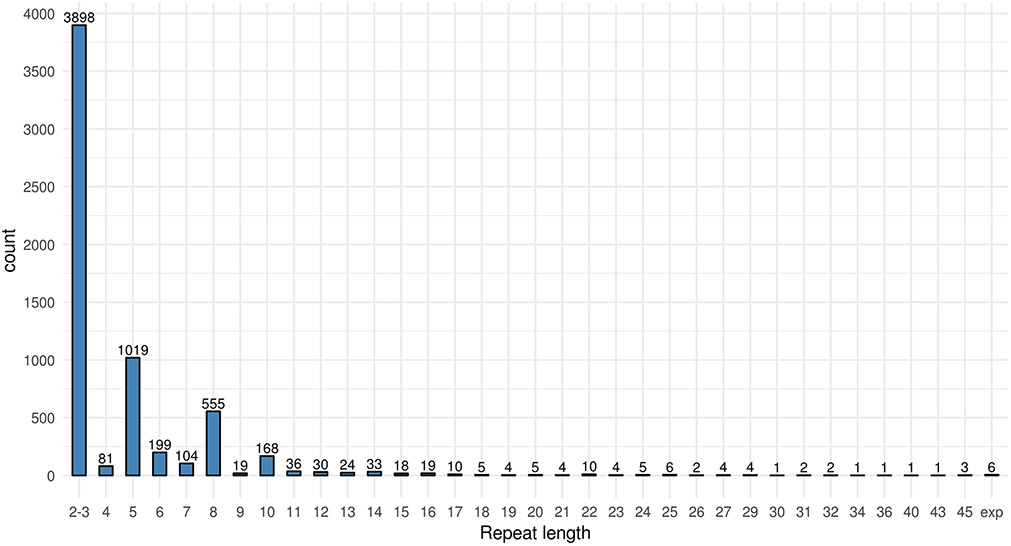
We successfully determined repeat lengths in 3142/3161 (99.3 %) individuals. There was no statistically significant difference in the repeat length distributions between cohorts (H(3)=2.129, p=0.55). Repeat length distribution of all alleles is shown in Figure 1. The repeat length distribution showed peaks in repeat numbers 2-3, 5, 8 and 10. The longest non-expanded allele we could characterize with repeat-primed PCR and amplify with over-the-repeat PCR was 45 repeats. The longer allele was in the range of 7-45 repeats in 1036/3142 (33 %) individuals, 20-45 in 56/3142 (1.8 %), 30-45 in 12/3142 (0.38%) and expansion (>45 repeats) in 6/3142 (0.19%). All expansion carriers were from the largest (HBCS) cohort. Summary of C9orf72 repeat lengths and case-control distributions per cohort is shown in Supplementary Table 1.
Figure 1.
Repeat length distribution of all alleles in 3142 Finns. “Exp” denotes expansion.
Interestingly, repeat lengths ≥20 were more common in Finland than reported elsewhere in previous large studies. To compare the frequency of ≥20 repeat lengths, we searched PUBMED for large population based (>1000 individuals) and case-control studies (>1000 controls) with C9orf72 repeat length distribution available. Besides the original articles describing the repeat, we identified nine articles (Beck et al., 2013, Cacace et al., 2013b, Fahey et al., 2014, He et al., 2015, Kohli et al., 2013b, Nuytemans et al., 2013, Rutherford et al., 2012, Theuns et al., 2014, van der Zee et al., 2013b) but only four had enough detailed information on ≥20 repeats for comparison. In these four studies, the prevalence of ≥20 repeats was significantly lower than in Finland (all allelic frequencies <0.56 % and all p<0.019, Fisher’s exact test) (Table 1). Of the nine studies, only two found expansion carriers in their control groups with carrier frequency of 1/5748 (p=0.0095 vs. our Finnish sample, Fisher’s exact test) (Theuns et al., 2014) and 11/7579 (p=0.60 vs. FIN) (Beck et al., 2013).
Table 1.
Comparison of frequency of ≥20 repeats and expansions in population based datasets or large (n>1000) control samples from case-control studies. *Written personal communication from Professor Mead.
| Population | Number of individuals |
Number of ≥20 repeats (expansions excluded) |
Allelic frequency of ≥20 repeats (%) |
P-value vs. our sample |
Expansions in controls (n) |
Reference |
|---|---|---|---|---|---|---|
| Finnish | 3142 | 56 | 0.89 | 6 | This study | |
| British 1958 birth cohort | 7577 | 64* | 0.42 | 0.000051 | 11 | (Beck et al., 2013) |
| Irish | 1234 | 10 | 0.41 | 0.019 | 0 | (Fahey et al., 2014) |
| European/Asian/North American/Australian | 5886 | 61 (≥17 repeats) | 0.52 | 0.0045 | 1 | (Theuns et al., 2014) |
| North American | 1444 | 11 | 0.38 | 0.0078 | 0 | (Rutherford et al., 2012) |
3.2. Intermediate repeat length alleles, Alzheimer’s disease and cognitive impairment
3.2.1. Alzheimer’s disease
We identified 226 individuals with AD (68-97 years, mean age 86, 58% female, 33 % carried 7-45 repeats, 1.3% carried 20-45 repeats) and 893 controls (aged 75-101, mean age 83, 58% female, 33% carried 7-45 repeats, 1.3% carried 20-45 repeats). Intermediate repeat length alleles did not associate with AD when intermediate repeat threshold was ≥7 (p=0.90) or ≥20 (p=0.96). Age (p=3.8x10−9, odds ratio (OR) 1.07, 95 % confidence interval (CI) 1.05 - 1.10) and APOE ε4 carriership (p=5.5 x 10−10, OR 2.69, CI 1.97 - 3.68) were statistically significantly associated with AD but sex was not (p=0.45).
3.2.2. MMSE scores
When individuals were divided based only on MMSE score into cases (MMSE score ≤24, n=432, aged 61-99, mean age 85, 75% female, 27% carried 7-45 repeats, 1.6% carried 20-45 repeats) and controls (MMSE score ≥25, n=719, aged 75-98, mean age 80, 38% female, 33% carried 7-45 repeats, 1.3% carried 20-45 repeats), there was a nominally significant association between 7-45 repeats and better cognition (p=0.025, OR 0.71 for cognitive impairment, CI 0.53 - 0.96) but the association did not remain statistically significant after Bonferroni correction. This association disappeared when repeat length ≥20 was used as threshold for intermediate repeat allele (20-45 repeats, p=0.72). Age (p=5.0x10−20, OR 1.1, CI 1.1 - 1.2), female sex (9.8x10−14, OR 3.1, CI 2.3 - 4.1) and APOE ε4 carriership (p=0.00078, OR 1.7, CI 1.2 - 2.2) associated significantly with cognitive impairment.
3.2.3. Any diagnosis of dementia or cognitive impairment
We identified 613 cases (aged 62-101, mean age 86, 65% female, 29% carried 7-45 repeats and 1.3% carried 20-45 repeats) and 1115 controls (aged 75-101, mean age 83, 37% female, 34% carried 7-45 repeats, 1.5% carried 20-45 repeats). There was a nominally significant association between intermediate repeat lengths and protection from dementia or cognitive impairment (p=0.032, OR 0.78, CI 0.62 - 0.98) but this result was not statistically significant after Bonferroni correction and again, the association was lost when intermediate repeat threshold ≥20 was used (20-45 repeats, p=0.62). Age (p=5.6x10−18, OR 1.08, CI 1.06 - 1.1), female sex (p=1.1x10−24, OR 3.0, CI 2.5 - 3.8) and APOE ε4 carriership (p=4.0x10−7, OR 1.8, CI 1.4 - 2.2) associated significantly with dementia or cognitive impairment.
3.2.4. Neurodegenerative and psychiatric diagnoses
We did not find any evident clustering of neurodegenerative or psychiatric disease in the 12 (0.38%) individuals with 30-45 repeats (Table 2). There were six (0.19%) expansion carriers and none had diagnosis of ALS or FTD; two had died at ages of 60 and 68 years due to coronary artery disease. Of the four living individuals (aged 71-79 years), one had been diagnosed with Parkinson disease (G20, ICD-10) and another had a diagnosis of local brain atrophy (G31.0, ICD-10). A third had been diagnosed with “other alcoholic hallucinosis” (291.20, ICD-8) and a fourth with depressive neurosis and other neurosis (300.40 and 300.88, ICD-8). A summary of carriers of 30-45 repeats and expansions is shown in Table 2.
Table 2.
Summary of individuals with ≥30 repeats. *Age of death, cause of death coronary artery disease. **Psychiatric diagnoses were not available. RP-PCR chromatograms are provided in Supplementary Figure 2
| Individual number |
Sex | Age at last information collection |
Repeat length (shorter/longer allele) |
Information on cognition |
|---|---|---|---|---|
| 1 | Female | 91 | 2/30 | MMSE score 26** |
| 2 | Male | 70 | 2/31 | MMSE score 29** |
| 3 | Female | 72 | 2/31 | No dementia diagnoses** |
| 4 | Male | 85 | 5/31 | MMSE score 28** |
| 5 | Male | 70 | 2/32 | MMSE score 30** |
| 6 | Male | 81 | 2/34 | MMSE score 29** |
| 7 | Female | 89 | 5/36 | MMSE score 21** |
| 8 | Male | 71 | 2/40 | MMSE score 30 / No dementia or psychiatric diagnoses |
| 9 | Male | 72 | 2/43 | No dementia or psychiatric diagnoses |
| 10 | Female | 77 | 2/45 | MMSE score 29/No dementia or psychiatric diagnoses |
| 11 | Male | 74 | 2/45 | MMSE score 29/No dementia or psychiatric diagnoses |
| 12 | Female | 73 | 8/45 | MMSE score 29/No dementia or psychiatric diagnoses |
| 13 | Male | 79 | 2/expansion | G20 (Parkinson disease, ICD-10), No psychiatric diagnoses. |
| 14 | Female | 60* | 2/expansion | 300.40(depressive neurosis, ICD-8), 300.88 (other neurosis, ICD-8). No dementia diagnoses. |
| 15 | Female | 71 | 2/expansion | G31.0 (local brain atrophy, ICD-10). No psychiatric diagnoses. |
| 16 | Male | 68* | 5/expansion | No dementia or psychiatric diagnoses |
| 17 | Female | 71 | 6/expansion | No dementia or psychiatric diagnoses |
| 18 | Female | 74 | 8/expansion | 291.20 (Alcoholic psychosis, ICD-8). No dementia diagnoses. |
4. Discussion:
The C9orf72 hexanucleotide repeat expansion is a major cause of ALS/FTD, however, what repeat lengths are pathogenic is still unclear. Our data on 3142 individuals shows that 30-45 repeats are found in 0.38% (1 per 262) of individuals and these individuals did not have any apparent clustering of neurodegenerative or psychiatric disease. Our data suggest that 30-45 repeat alleles should not be considered unequivocally pathogenic and in those individuals other risk factors than the C9orf72 expansion should be considered. The smallest expansions that segregate with combinations of ALS, FTD-ALS and “FTLD or dementia” are reportedly ca. 55-100 repeats as measured by Southern blot (Gijselinck et al., 2016). However, an individual with 70 repeats without neurodegeneration at the age of 90 has been reported (McGoldrick et al., 2018). More data is still needed for more precise estimation of the pathological threshold, which may actually turn out as a “grey zone” where disease penetrance is dependent on modulating cofactors.
Our data is in line with previous studies indicating a lack of major association between intermediate repeats and AD. Somewhat surprisingly, there was a nominally significant association between 7-45 repeat length alleles and better cognition. This association was not statistically significant in any of the individual cohorts and was only seen in the pooled dataset. This can be a spurious association, but nevertheless, it supports the hypothesis that ≥7 repeat alleles do not predispose to cognitive impairment or dementia. Across all our cohorts, there was heterogeneity in sex and age distributions, variation in socioeconomic status and educational history (one cohort consisted solely of men with high socioeconomic status) and there was no uniform way of defining cognition and dementia status and this information was collected from several sources. Despite these limitations, we found a clear association between APOE ε4 genotype and AD (p=5.5 x 10−10, OR 2.69), cognitive impairment (p=0.00078, OR 1.7) and overall dementia status (p=4.0x10−7, OR 1.8) which argues against a major bias in our definition of dementia. Furthermore, the Finnish national registers have been found to identify dementia cases accurately (Solomon et al., 2014).
None of the six expansion carriers we identified had a diagnosis of ALS or FTD but four had a diagnosis of a neurodegenerative or psychiatric disease. Two had died of coronary heart disease before the age of 70 and others were under 80 years. Therefore, it’s possible that some expansion carriers could have developed or may develop ALS/FTD in their later years.
Previous reports have shown that C9orf72 expansion is especially common cause of ALS/FTD in Finland (Majounie et al., 2012). In the present study, the expansion frequency in older Finns was only slightly higher than in the UK 1958 birth cohort. However, this UK population was considerably younger than our Finnish sample. The penetrance of the C9orf72 expansion is known to be strongly age-related. Nearly complete penetrance is observed by as late as 83 years. The mean age of onset varies somewhat with phenotype but is estimated to be around 58 years. Therefore, it can be expected that the expansion frequency in the UK cohort will markedly decrease by time (REF https://www.nature.com/articles/s41598-017-02364-1). In line with age-related penetrance, expansions were unequally distributed in our cohorts; all Finnish expansion carriers were from the HBCS cohort, whose participants were the youngest of the four cohorts (born 1934-1944, and registry information available up till December 31 of 2013, Supplementary methods). If we restrict the analysis to this cohort only, expansion prevalence would be 6/1644 (0.36% or 1 in 274) in Finland.
Interestingly, we found that non-expanded repeat lengths ≥20 are roughly twice as common in Finland than in other reported populations (Table 1). This finding may have a connection with the high frequency of C9orf72 expansion in Finland and raises the question, whether the ≥20 repeat alleles could be prone to germline instability and expansion in the offspring. In this scenario, the larger intermediate alleles would form a pool from which new expansions can be derived, a mechanism shown in families with sporadic Huntington’s disease and in Huntington’s disease CAG repeat knock-in mice (Goldberg et al., 1993, Myers et al., 1993, Wheeler et al., 1999). As more than half of the Finnish ALS patients with the C9orf72 expansion are sporadic (Majounie et al., 2012) (Laaksovirta et al unpublished data) this hypothesis warrants further research by genotyping sporadic C9orf72 ALS cases’ parents when possible. Another approach would be to genotype paired DNA samples from blood and gametes (Martorell et al., 2004)
Supplementary Material
Acknowledgements
KK was funded by the Finnish Cultural Foundation and the UH Faculty of Medicine MD/PhD program. PJT has been supported by grants from the Sigrid Juselius Foundation and Helsinki University Hospital. This work was funded in part by the Intramural Research Program of the National Institute on Aging, National Institutes of Health (Z01-AG000949).
The Helsinki Birth Cohort Study has been funded by The Academy of Finland, the Finnish Diabetes Research society, Folkhälsan Research Foundation, Novo Nordisk Foundation, Finska Läkaresällskapet, Signe and Ane Gyllenberg Foundation, University of Helsinki, Ahokas Foundation, Juho Vainio Foundation, and Helsinki University Hospital.
The Helsinki Businessmen Study has been funded by VTR-funding of the HUS/Helsinki University Hospital (TYH 2014245; 2015211, Sisu) and the Academy of Finland (grant number 311492). The authors are grateful to Professor Simon Mead (National Prion Clinic, MRC Prion Unit at the University College London) for sharing detailed C9orf72 allele distribution in the UK 1958 birth cohort.
Footnotes
Disclosure statement
Bryan J. Traynor and Pentti J. Tienari hold patent on C9orf72 in diagnostics and treatment of ALS/FTD. The other authors have no actual or potential conflict of interest.
References
- Barker DJ, Osmond C, Forsen TJ, Kajantie E, Eriksson JG, 2005. Trajectories of growth among children who have coronary events as adults. N.Engl.J.Med 353, 1802–1809. [DOI] [PubMed] [Google Scholar]
- Beck J, Poulter M, Hensman D, Rohrer JD, Mahoney CJ, Adamson G, Campbell T, Uphill J, Borg A, Fratta P, Orrell RW, Malaspina A, Rowe J, Brown J, Hodges J, Sidle K, Polke JM, Houlden H, Schott JM, Fox NC, Rossor MN, Tabrizi SJ, Isaacs AM, Hardy J, Warren JD, Collinge J, Mead S, 2013. Large C9orf72 hexanucleotide repeat expansions are seen in multiple neurodegenerative syndromes and are more frequent than expected in the UK population. Am.J.Hum.Genet 92, 345–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cacace R, Van Cauwenberghe C, Bettens K, Gijselinck I, van der Zee J, Engelborghs S, Vandenbulcke M, Van Dongen J, Baumer V, Dillen L, Mattheijssens M, Peeters K, Cruts M, Vandenberghe R, De Deyn PP, Van Broeckhoven C, Sleegers K, 2013a. C9orf72 G4C2 repeat expansions in Alzheimer's disease and mild cognitive impairment. Neurobiol.Aging 34, 1712.e1–1712.e7. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R, 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 72, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson JG, Osmond C, Kajantie E, Forsen TJ, Barker DJ, 2006. Patterns of growth among children who later develop type 2 diabetes or its risk factors. Diabetologia. 49, 2853–2858. [DOI] [PubMed] [Google Scholar]
- Fahey C, Byrne S, McLaughlin R, Kenna K, Shatunov A, Donohoe G, Gill M, Al-Chalabi A, Bradley DG, Hardiman O, Corvin AP, Morris DW, 2014. Analysis of the hexanucleotide repeat expansion and founder haplotype at C9ORF72 in an Irish psychosis case-control sample. Neurobiol.Aging 35, 1510.e1–1510.e5. [DOI] [PubMed] [Google Scholar]
- Gijselinck I, Van Mossevelde S, van der Zee J, Sieben A, Engelborghs S, De Bleecker J, Ivanoiu A, Deryck O, Edbauer D, Zhang M, Heeman B, Baumer V, Van den Broeck M, Mattheijssens M, Peeters K, Rogaeva E, De Jonghe P, Cras P, Martin JJ, de Deyn PP, Cruts M, Van Broeckhoven C, 2016. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol.Psychiatry 21, 1112–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg YP, Kremer B, Andrew SE, Theilmann J, Graham RK, Squitieri F, Telenius H, Adam S, Sajoo A, Starr E, 1993. Molecular analysis of new mutations for Huntington's disease: intermediate alleles and sex of origin effects. Nat.Genet 5, 174–179. [DOI] [PubMed] [Google Scholar]
- Harms M, Benitez BA, Cairns N, Cooper B, Cooper P, Mayo K, Carrell D, Faber K, Williamson J, Bird T, Diaz-Arrastia R, Foroud TM, Boeve BF, Graff-Radford NR, Mayeux R, Chakraverty S, Goate AM, Cruchaga C, NIA-LOAD/NCRAD Family Study Consortium, 2013. C9orf72 hexanucleotide repeat expansions in clinical Alzheimer disease. JAMA Neurol. 70, 736–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J, Tang L, Benyamin B, Shah S, Hemani G, Liu R, Ye S, Liu X, Ma Y, Zhang H, Cremin K, Leo P, Wray NR, Visscher PM, Xu H, Brown MA, Bartlett PF, Mangelsdorf M, Fan D, 2015. C9orf72 hexanucleotide repeat expansions in Chinese sporadic amyotrophic lateral sclerosis. Neurobiol.Aging 36, 2660.e1–2660.e8. [DOI] [PubMed] [Google Scholar]
- Jiao B, Guo JF, Wang YQ, Yan XX, Zhou L, Liu XY, Zhang FF, Zhou YF, Xia K, Tang BS, Shen L, 2013. C9orf72 mutation is rare in Alzheimer's disease, Parkinson's disease, and essential tremor in China. Front.Cell.Neurosci 7, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaivola K, Jansson L, Saarentaus E, Kiviharju A, Rantalainen V, Eriksson JG, Strandberg TE, Polvikoski T, Myllykangas L, Tienari PJ, 2018. Heterozygous TYROBP deletion (PLOSLFIN) is not a strong risk factor for cognitive impairment. Neurobiol.Aging 64, 159.e1–159.e4. [DOI] [PubMed] [Google Scholar]
- Kajantie E, Pietilainen KH, Wehkalampi K, Kananen L, Raikkonen K, Rissanen A, Hovi P, Kaprio J, Andersson S, Eriksson JG, Hovatta I, 2012. No association between body size at birth and leucocyte telomere length in adult life--evidence from three cohort studies. Int.J.Epidemiol 41, 1400–1408. [DOI] [PubMed] [Google Scholar]
- Kohli MA, John-Williams K, Rajbhandary R, Naj A, Whitehead P, Hamilton K, Carney RM, Wright C, Crocco E, Gwirtzman HE, Lang R, Beecham G, Martin ER, Gilbert J, Benatar M, Small GW, Mash D, Byrd G, Haines JL, Pericak-Vance MA, Zuchner S, 2013a. Repeat expansions in the C9ORF72 gene contribute to Alzheimer's disease in Caucasians. Neurobiol.Aging 34, 1519.e5–1519.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohli MA, John-Williams K, Rajbhandary R, Naj A, Whitehead P, Hamilton K, Carney RM, Wright C, Crocco E, Gwirtzman HE, Lang R, Beecham G, Martin ER, Gilbert J, Benatar M, Small GW, Mash D, Byrd G, Haines JL, Pericak-Vance MA, Zuchner S, 2013b. Repeat expansions in the C9ORF72 gene contribute to Alzheimer's disease in Caucasians. Neurobiol.Aging 34, 1519.e5–1519.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahti M, Eriksson JG, Heinonen K, Kajantie E, Lahti J, Wahlbeck K, Tuovinen S, Pesonen AK, Mikkonen M, Osmond C, Raikkonen K, 2014. Maternal Grand Multiparity and the Risk of Severe Mental Disorders in Adult Offspring. PLoS One. 9, e114679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majounie E, Renton AE, Mok K, Dopper EG, Waite A, Rollinson S, Chio A, Restagno G, Nicolaou N, Simon-Sanchez J, van Swieten JC, Abramzon Y, Johnson JO, Sendtner M, Pamphlett R, Orrell RW, Mead S, Sidle KC, Houlden H, Rohrer JD, Morrison KE, Pall H, Talbot K, Ansorge O, Chromosome 9-ALS/FTD Consortium, French research network on FTLD/FTLD/ALS, ITALSGEN Consortium, Hernandez DG, Arepalli S, Sabatelli M, Mora G, Corbo M, Giannini F, Calvo A, Englund E, Borghero G, Floris GL, Remes AM, Laaksovirta H, McCluskey L, Trojanowski JQ, Van Deerlin VM, Schellenberg GD, Nalls MA, Drory VE, Lu CS, Yeh TH, Ishiura H, Takahashi Y, Tsuji S, Le Ber I, Brice A, Drepper C, Williams N, Kirby J, Shaw P, Hardy J, Tienari PJ, Heutink P, Morris HR, Pickering-Brown S, Traynor BJ, 2012. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 11, 323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martorell L, Gamez J, Cayuela ML, Gould FK, McAbney JP, Ashizawa T, Monckton DG, Baiget M, 2004. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology. 62, 269–274. [DOI] [PubMed] [Google Scholar]
- McGoldrick P, Zhang M, van Blitterswijk M, Sato C, Moreno D, Xiao S, Zhang AB, McKeever PM, Weichert A, Schneider R, Keith J, Petrucelli L, Rademakers R, Zinman L, Robertson J, Rogaeva E, 2018. Unaffected mosaic C9orf72 case: RNA foci, dipeptide proteins, but upregulated C9orf72 expression. Neurology. 90, e323–e331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers RH, MacDonald ME, Koroshetz WJ, Duyao MP, Ambrose CM, Taylor SA, Barnes G, Srinidhi J, Lin CS, Whaley WL, 1993. De novo expansion of a (CAG)n repeat in sporadic Huntington's disease. Nat.Genet 5, 168–173. [DOI] [PubMed] [Google Scholar]
- Myllykangas L, Polvikoski T, Sulkava R, Verkkoniemi A, Crook R, Tienari PJ, Pusa AK, Niinisto L, O'Brien P, Kontula K, Hardy J, Haltia M, Perez-Tur J, 1999. Genetic association of alpha2-macroglobulin with Alzheimer's disease in a Finnish elderly population. Ann.Neurol 46, 382–390. [PubMed] [Google Scholar]
- Ng ASL, Tan EK, 2017. Intermediate C9orf72 alleles in neurological disorders: does size really matter?. J.Med.Genet 54, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuytemans K, Bademci G, Kohli MM, Beecham GW, Wang L, Young JI, Nahab F, Martin ER, Gilbert JR, Benatar M, Haines JL, Scott WK, Zuchner S, Pericak-Vance MA, Vance JM, 2013. C9ORF72 intermediate repeat copies are a significant risk factor for Parkinson disease. Ann.Hum.Genet 77, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Cherny SS, Sham PC, 2003. Genetic Power Calculator: design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 19, 149–150. [DOI] [PubMed] [Google Scholar]
- Rahkonen T, Eloniemi-Sulkava U, Rissanen S, Vatanen A, Viramo P, Sulkava R, 2003. Dementia with Lewy bodies according to the consensus criteria in a general population aged 75 years or older. J.Neurol.Neurosurg.Psychiatry 74, 720–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Holtta-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chio A, Restagno G, Borghero G, Sabatelli M, ITALSGEN Consortium, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ, 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 72, 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford NJ, Heckman MG, Dejesus-Hernandez M, Baker MC, Soto-Ortolaza AI, Rayaprolu S, Stewart H, Finger E, Volkening K, Seeley WW, Hatanpaa KJ, Lomen-Hoerth C, Kertesz A, Bigio EH, Lippa C, Knopman DS, Kretzschmar HA, Neumann M, Caselli RJ, White CL 3rd, Mackenzie IR, Petersen RC, Strong MJ, Miller BL, Boeve BF, Uitti RJ, Boylan KB, Wszolek ZK, Graff-Radford NR, Dickson DW, Ross OA, Rademakers R, 2012. Length of normal alleles of C9ORF72 GGGGCC repeat do not influence disease phenotype. Neurobiol.Aging 33, 2950.e5–2950.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon A, Ngandu T, Soininen H, Hallikainen MM, Kivipelto M, Laatikainen T, 2014. Validity of dementia and Alzheimer's disease diagnoses in Finnish national registers. Alzheimers Dement. 10, 303–309. [DOI] [PubMed] [Google Scholar]
- Strandberg TE, Salomaa V, Strandberg AY, Vanhanen H, Sarna S, Pitkala K, Rantanen K, Savela S, Pienimaki T, Huohvanainen E, Stenholm S, Raikkonen K, Tilvis RS, Tienari PJ, Huttunen J, 2016. Cohort Profile: The Helsinki Businessmen Study (HBS). Int.J.Epidemiol 45, 1074–1074h. [DOI] [PubMed] [Google Scholar]
- Tanskanen M, Makela M, Notkola IL, Myllykangas L, Rastas S, Oinas M, Lindsberg PJ, Polvikoski T, Tienari PJ, Paetau A, 2017. Population-based analysis of pathological correlates of dementia in the oldest old. Ann.Clin.Transl.Neurol 4, 154–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theuns J, Verstraeten A, Sleegers K, Wauters E, Gijselinck I, Smolders S, Crosiers D, Corsmit E, Elinck E, Sharma M, Kruger R, Lesage S, Brice A, Chung SJ, Kim MJ, Kim YJ, Ross OA, Wszolek ZK, Rogaeva E, Xi Z, Lang AE, Klein C, Weissbach A, Mellick GD, Silburn PA, Hadjigeorgiou GM, Dardiotis E, Hattori N, Ogaki K, Tan EK, Zhao Y, Aasly J, Valente EM, Petrucci S, Annesi G, Quattrone A, Ferrarese C, Brighina L, Deutschlander A, Puschmann A, Nilsson C, Garraux G, LeDoux MS, Pfeiffer RF, Boczarska-Jedynak M, Opala G, Maraganore DM, Engelborghs S, De Deyn PP, Cras P, Cruts M, Van Broeckhoven C, GEO-PD Consortium, 2014. Global investigation and meta-analysis of the C9orf72 (G4C2)n repeat in Parkinson disease. Neurology. 83, 1906–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uusvaara J, Pitkala KH, Kautiainen H, Tilvis RS, Strandberg TE, 2013. Detailed cognitive function and use of drugs with anticholinergic properties in older people: a community-based cross-sectional study. Drugs Aging. 30, 177–182. [DOI] [PubMed] [Google Scholar]
- van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, Philtjens S, Vandenbulcke M, Sleegers K, Sieben A, Baumer V, Maes G, Corsmit E, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, de Mendonca A, Miltenberger-Miltenyi G, Pereira S, Pimentel J, Nacmias B, Bagnoli S, Sorbi S, Graff C, Chiang HH, Westerlund M, Sanchez-Valle R, Llado A, Gelpi E, Santana I, Almeida MR, Santiago B, Frisoni G, Zanetti O, Bonvicini C, Synofzik M, Maetzler W, Vom Hagen JM, Schols L, Heneka MT, Jessen F, Matej R, Parobkova E, Kovacs GG, Strobel T, Sarafov S, Tournev I, Jordanova A, Danek A, Arzberger T, Fabrizi GM, Testi S, Salmon E, Santens P, Martin JJ, Cras P, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Philtjens S, Sleegers K, Baumer V, Maes G, Corsmit E, Cruts M, Van Broeckhoven C, van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Philtjens S, Theuns J, Sleegers K, Baumer V, Maes G, Cruts M, Van Broeckhoven C, Engelborghs S, De Deyn PP, Cras P, Engelborghs S, De Deyn PP, Vandenbulcke M, Vandenbulcke M, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, Synofzik M, Maetzler W, Muller Vom Hagen J, Schols L, Synofzik M, Maetzler W, Muller Vom Hagen J, Schols L, Heneka MT, Jessen F, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, de Mendonca A, Miltenberger-Miltenyi G, Pereira S, Firmo C, Pimentel J, Sanchez-Valle R, Llado A, Antonell A, Molinuevo J, Gelpi E, Graff C, Chiang HH, Westerlund M, Graff C, Kinhult Stahlbom A, Thonberg H, Nennesmo I, Borjesson-Hanson A, Nacmias B, Bagnoli S, Sorbi S, Bessi V, Piaceri I, Santana I, Santiago B, Santana I, Helena Ribeiro M, Rosario Almeida M, Oliveira C, Massano J, Garret C, Pires P, Frisoni G, Zanetti O, Bonvicini C, Sarafov S, Tournev I, Jordanova A, Tournev I, Kovacs GG, Strobel T, Heneka MT, Jessen F, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, Jessen F, Matej R, Parobkova E, Danel A, Arzberger T, Maria Fabrizi G, Testi S, Ferrari S, Cavallaro T, Salmon E, Santens P, Cras P, European Early-Onset Dementia Consortium, 2013a. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum.Mutat 34, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Engelborghs S, Philtjens S, Vandenbulcke M, Sleegers K, Sieben A, Baumer V, Maes G, Corsmit E, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, de Mendonca A, Miltenberger-Miltenyi G, Pereira S, Pimentel J, Nacmias B, Bagnoli S, Sorbi S, Graff C, Chiang HH, Westerlund M, Sanchez-Valle R, Llado A, Gelpi E, Santana I, Almeida MR, Santiago B, Frisoni G, Zanetti O, Bonvicini C, Synofzik M, Maetzler W, Vom Hagen JM, Schols L, Heneka MT, Jessen F, Matej R, Parobkova E, Kovacs GG, Strobel T, Sarafov S, Tournev I, Jordanova A, Danek A, Arzberger T, Fabrizi GM, Testi S, Salmon E, Santens P, Martin JJ, Cras P, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Theuns J, Philtjens S, Sleegers K, Baumer V, Maes G, Corsmit E, Cruts M, Van Broeckhoven C, van der Zee J, Gijselinck I, Dillen L, Van Langenhove T, Philtjens S, Theuns J, Sleegers K, Baumer V, Maes G, Cruts M, Van Broeckhoven C, Engelborghs S, De Deyn PP, Cras P, Engelborghs S, De Deyn PP, Vandenbulcke M, Vandenbulcke M, Borroni B, Padovani A, Archetti S, Perneczky R, Diehl-Schmid J, Synofzik M, Maetzler W, Muller Vom Hagen J, Schols L, Synofzik M, Maetzler W, Muller Vom Hagen J, Schols L, Heneka MT, Jessen F, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, de Mendonca A, Miltenberger-Miltenyi G, Pereira S, Firmo C, Pimentel J, Sanchez-Valle R, Llado A, Antonell A, Molinuevo J, Gelpi E, Graff C, Chiang HH, Westerlund M, Graff C, Kinhult Stahlbom A, Thonberg H, Nennesmo I, Borjesson-Hanson A, Nacmias B, Bagnoli S, Sorbi S, Bessi V, Piaceri I, Santana I, Santiago B, Santana I, Helena Ribeiro M, Rosario Almeida M, Oliveira C, Massano J, Garret C, Pires P, Frisoni G, Zanetti O, Bonvicini C, Sarafov S, Tournev I, Jordanova A, Tournev I, Kovacs GG, Strobel T, Heneka MT, Jessen F, Ramirez A, Kurzwelly D, Sachtleben C, Mairer W, Jessen F, Matej R, Parobkova E, Danel A, Arzberger T, Maria Fabrizi G, Testi S, Ferrari S, Cavallaro T, Salmon E, Santens P, Cras P, European Early-Onset Dementia Consortium, 2013b. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum.Mutat 34, 363–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler VC, Auerbach W, White JK, Srinidhi J, Auerbach A, Ryan A, Duyao MP, Vrbanac V, Weaver M, Gusella JF, Joyner AL, MacDonald ME, 1999. Length-dependent gametic CAG repeat instability in the Huntington's disease knock-in mouse. Hum.Mol.Genet 8, 115–122. [DOI] [PubMed] [Google Scholar]
- Xi Z, Zinman L, Grinberg Y, Moreno D, Sato C, Bilbao JM, Ghani M, Hernandez I, Ruiz A, Boada M, Moron FJ, Lang AE, Marras C, Bruni A, Colao R, Maletta RG, Puccio G, Rainero I, Pinessi L, Galimberti D, Morrison KE, Moorby C, Stockton JD, Masellis M, Black SE, Hazrati LN, Liang Y, van Haersma de With J, Fornazzari L, Villagra R, Rojas-Garcia R, Clarimon J, Mayeux R, Robertson J, St George-Hyslop P, Rogaeva E, 2012. Investigation of c9orf72 in 4 neurodegenerative disorders. Arch.Neurol 69, 1583–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yliharsila H, Kajantie E, Osmond C, Forsen T, Barker DJ, Eriksson JG, 2007. Birth size, adult body composition and muscle strength in later life. Int.J.Obes.(Lond) 31, 1392–1399. [DOI] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu SL, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD, 2015. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 525, 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.