ABSTRACT
Secondary bacterial infection is a common complication in severe influenza virus infections. During the H1N1 pandemic of 2009, increased mortality was observed among healthy young adults due to secondary bacterial pneumonia, one of the most frequent bacterial species being Streptococcus pneumoniae (Spn). Previous studies in mice and ferrets have suggested a synergistic relationship between Spn and influenza viruses. In this study, the ferret model was used to examine whether secondary Spn infection (strains BHN97 and D39) influence replication and airborne transmission of the 2009 pandemic H1N1 virus (H1N1pdm09). Secondary infection with Spn after H1N1pdm09 infection consistently resulted in a significant decrease in viral titers in the ferret nasal washes. While secondary Spn infection appeared to negatively impact influenza virus replication, animals precolonized with Spn were equally susceptible to H1N1pdm09 airborne transmission. In line with previous work, ferrets with preceding H1N1pdm09 and secondary Spn infection had increased bacterial loads and more severe clinical symptoms as compared to animals infected with H1N1pdm09 or Spn alone. Interestingly, the donor animals that displayed the most severe clinical symptoms had reduced airborne transmission of H1N1pdm09. Based on these data, we propose an asymmetrical relationship between these two pathogens, rather than a synergistic one, since secondary bacterial infection enhances Spn colonization and pathogenesis but decreases viral titers.
Keywords: Streptococcus pneumoniae, influenza virus, H1N1, secondary bacterial infection, viral titers, airborne transmission
Significance: replication and airborne transmission of respiratory viruses is influenced by many host and environmental parameters. The complex interplay between bacterial and viral coinfections on transmission of respiratory viruses has been understudied. In our study, we examined how Spn influences the replication and transmission of influenza virus. Our data support previous work demonstrating that infection with influenza virus benefits bacterial growth and transmission. Unexpectedly, we observed that secondary bacterial infection negatively impacts viral replication in the nasal cavity. These findings set the stage for further studies to understand how bacteria may negatively influence influenza viruses, and the impact of this asymmetrical relationship on the epidemiology of influenza viruses.
Introduction
Acute pneumonia occurs in 30%-40% of influenza A virus (IAV) hospitalizations and can be caused by the virus alone or in conjunction with secondary bacterial infection, most commonly Staphylococcus aureus and Streptococcus pneumoniae (Spn; Kalil and Thomas 2019). During the 2009 and 1918 H1N1 pandemics, secondary infection with Spn contributed to severity of disease, especially in young adults (Morens et al. 2008, Louie et al. 2009, Khardori 2010, Klein et al. 2016). Animal models to study the relationship between IAV and Spn have largely focused on immune responses, disease outcome, and the influence of IAV on Spn colonization (Diavatopoulos et al. 2010, Nakamura et al. 2011, Metzger and Sun 2013, Mina and Klugman 2014, Mina et al. 2015, Duvigneau et al. 2016). Given the epidemiological importance of viral–bacterial coinfections, there is a critical need for studies that examine how bacterial pathogens influence viral fitness within the host and its transmission to new hosts.
The prevailing model describes a synergism between IAV and Spn with regards to morbidity and mortality. Exacerbated disease during coinfection, compared to a single infection, is supported by epidemiological studies as well as studies in murine and ferret models of respiratory infection (Peltola et al. 2006, Morens et al. 2008, McCullers et al. 2010, Siegel et al. 2014). Studies in the murine model suggest that heightened disease results from a combination of dysregulation in immune responses, extensive epithelial damage, release of planktonic bacteria from biofilms, as well as increased bacterial colonization and dissemination (Marks et al.2013, Mina and Klugman 2014, Morens et al. 2008, Rudd et al. 2016, Siemens et al. 2017). Mechanistic studies indicate that increased bacterial colonization is a consequence of upregulation of bacterial receptors and IAV-induced increases in sialic acid, which serves as a nutrient source for Spn (Nita-Lazar et al. 2015, Siegel et al. 2014, Wren et al. 2017). Further, enhanced systemic dissemination of bacteria is facilitated by virus-induced cytokine release, oxidative stress, and pneumolysin release (Gonzalez-Juarbe et al. 2020, Nakamura et al. 2011). Many of these published studies focused primarily on the colonization, transmission, and pathogenesis of Spn with limited studies on the impact of viral replication and airborne dissemination.
In addition to colonization and disease, a few studies have also investigated the influence of coinfection on contact transmission. These studies, done primarily in infant mice, have shown that coinfection with IAV dramatically enhances Spn transmission with the efficiency of IAV being highly variable (Diavatopoulos et al. 2010, Short et al. 2012). One study further demonstrated that Spn infection preceding IAV inoculation can decrease IAV contact transmission (Ortigoza et al. 2018). However, mice in general are suboptimal models for airborne viral transmission (Bouvier 2015).
Ferrets are the gold standard animal model for IAV pathogenesis and transmission due to their susceptibility to human IAV strains, release of infectious virus into the air in aerosols of variable sizes, as well as similarity in lung physiology, infection kinetics, and clinical symptoms when compared to humans (Belser et al. 2013, 2020, Lakdawala et al. 2011, 2013, 2015). In addition, ferrets first infected with IAV have been shown to be susceptible to secondary colonization by Spn and to promote Spn airborne transmission (McCullers et al. 2010, Peltola et al. 2006). Recently, Rowe et al. (2020) have demonstrated that the nasal microbiota promotes airborne transmission of the 2009 H1N1 pandemic virus (H1N1pdm09) in the ferret model. Together, these studies support a model of synergism between Spn and IAV regarding host virulence, Spn colonization, and Spn transmission. However, the impact of Spn coinfection on IAV fitness in the host and airborne transmission has not been fully addressed, which may have important implications for the spread of IAV.
In this study, we examined the impact of secondary infections with Spn serotype 19F (strain BHN97) and serotype 2 (strain D39) on viral titers and airborne transmission of H1N1pdm09 in ferrets. We observed that secondary infection with Spn reduced viral titers in nasal washes of coinfected animals compared to H1N1pdm09 only infected ferrets. Moreover, the disease severity was negatively associated with airborne transmission in animals with secondary Spn infections. Finally, the negative impact of Spn on viral replication in the nasal cavity led us to test whether precolonization with Spn was protective against acquisition of airborne virus. While precolonization with bacteria was not protective against viral infections it surprisingly did not result in severe disease previously observed with Spn–IAV infections. In conclusion, we propose an asymmetrical model where IAV enhances colonization and pathogenesis of bacteria, while Spn negatively impacts viral titers and may also decrease airborne transmission.
Materials and methods
Cells, viruses, and bacteria
MDCK (Madin-Darby canine kidney, obtained from ATCC) were grown at 37°C in 5% CO2 in MEM medium (Sigma) containing 5% Fetal Bovine Serum (FBS, HyClone), penicillin/streptomycin, and L-glutamine. Reverse genetics plasmids of A/California/07/2009 were a generous gift from Dr Jesse Bloom (Fred Hutch Cancer Research Center, Seattle) and were rescued as previously described (Lakdawala et al. 2011). The viral titers were determined by tissue culture infectious dose 50 (TCID50) using the endpoint titration method on MDCK cells for H1N1pdm09. The bacterial strains used in this study were graciously provided by Dr Hasan Yesilkaya (wild type Spn serotype 2 strain D39; Andrew et al. 2018) and Dr Jason Rosch (wild type Spn serotype 19F strain BHN97; Rowe et al. 2019). Bacteria were grown from frozen stocks by streaking on TSA-II agar plates supplemented with 5% sheep blood (BD). Cultures were generated in fresh Columbia broth (Thermo Fisher) and incubated at 37°C and 5% CO2 without shaking. Pneumococci in nasal washes and tissues were determined by plating serial dilutions onto blood agar plates incubated at 37°C overnight.
Animal ethics statement
Ferret transmission experiments were conducted at the University of Pittsburgh in compliance with the guidelines of the Institutional Animal Care and Use Committee (approved protocol #19075697). Humane endpoints for this study included body weight loss exceeding 20% (relative to weight at challenge) and a prolonged clinical activity score of 3 based on the system designed by Reuman (Reuman et al. 1989). Animals were sedated with approved methods for all procedures. Isoflurane was used for all nasal wash and survival blood draw, ketamine and xylazine were used for sedation for all terminal procedures followed by cardiac administration of euthanasia solution. Approved University of Pittsburgh DLAR staff administered euthanasia at time of sacrifice. After the first ferret rapidly succumbed to the H1N1pdm09-BHN97 coinfection and was found to have bacteria disseminated to all body sites, the remaining coinfected ferrets were subcutaneously administered 5 mg/kg Baytril, twice a day for 7 days.
Animals
Male ferrets, 5–6-months-old, were purchased from Triple F Farms (Sayre, PA). All ferrets were screened for antibodies against circulating influenza A and B viruses, as determined by hemagglutinin inhibition assay using the following antigens obtained through the International Reagent Resource, Influenza Division, WHO Collaborating Center for Surveillance, Epidemiology, and Control of Influenza, Centers for Disease Control and Prevention, Atlanta, GA: 2018–2019 WHO Antigen, Influenza A (H3) Control Antigen (A/Singapore/INFIMH-16- 0019/2016), BPL-Inactivated, FR-1606; 2014–2015 WHO Antigen, Influenza A(H1N1)pdm09 Control Antigen (A/California/07/2009 NYMC X-179A), BPL-Inactivated, FR-1184; 2018–2019 WHO Antigen, Influenza B Control Antigen, Victoria Lineage (B/Colorado/06/2017), BPL-Inactivated, FR-1607; 2015–2016 WHO Antigen, Influenza B Control Antigen, Yamagata Lineage (B/Phuket/3073/2013), BPL-Inactivated, and FR-1403.
Clinical symptoms such as weight loss, temperature, and activity score (Reuman et al. 1989) were recorded during each nasal wash procedure and other symptoms such as sneezing, coughing, lethargy, or nasal discharge were noted during any handling events. Animals were given A/D diet twice a day to entice eating once they reached 10% weight loss. A summary of clinical symptoms for each study are provided in Table S1 (Supporting Information).
Tissues and blood were collected to assess viral and bacterial loads. Tissues were weighed and homogenized in sterile PBS at 5% (nasal turbinate, spleen, and trachea) or 10% (lung) weight per volume. The soft palate was homogenized in 1 ml PBS. Bacterial loads were assessed by plating serial dilutions on blood agar plates.
Transmission studies
Our transmission caging setup is a modified Allentown ferret and rabbit bioisolator cage similar to those used in (Lakdawala et al. 2011, 2015). For each study, three or four donor ferrets were anesthetized by isofluorane and inoculated intranasally with 106 TCID50/ 500 ul of A/California/07/2009. For coinfection experiments, the IAV-infected donors were inoculated with 105, 106, or 107 CFUs/500 ul of Spn D39 or BHN97 at 48 h postinfection. At 6 h postbacterial infection, a recipient ferret was placed into the adjacent cage, which is separated by two staggered perforated metal plates welded together one inch apart. Nasal washes were collected from each donor and recipient every day for 13 days. To prevent accidental contact or fomite transmission by investigators, the recipient ferret was handled first and extensive cleaning of all chambers, biosafety cabinet, and temperature monitoring wands was performed between each recipient and donor and between each pair of animals. Sera was collected from donors and recipients upon completion of experiments to confirm seroconversion. Environmental conditions were monitored every hour using a HOBO UX100-011 data logger (Onset) and ranged between 20 and 22°C with 44%–50% relative humidity (Figure S3, Supporting Information). To ensure no accidental exposure during husbandry procedures, recipient animal sections of the cage were cleaned first then then infected side, three people participated in each husbandry event to ensure that a clean pair of hands handled bedding and food changes. One cage was done at a time and a 10-min wait time to remove contaminated air was observed before moving to the next cage. New scrappers, gloves, and sleeve covers were used on subsequent cage cleaning.
Serology assay
Analysis of neutralizing antibodies from ferret sera was performed as previously described in (Lakdawala et al. 2011). Briefly, the microneutralization assay was performed using 103.3 TCID50 of A/California/07/2009 virus incubated with 2-fold serial dilutions of heat-inactivated ferret sera. The neutralizing titer was defined as the reciprocal of the highest dilution of serum required to completely neutralize infectivity of 103.3 TCID50 of virus on MDCK cells. The concentration of antibody required to neutralize 100 TCID50 of virus was calculated based on the neutralizing titer dilution divided by the initial dilution factor, multiplied by the antibody concentration.
Spn ELISA
96-well ELISA plates (Immulon) were coated with 50 µl of 5 × 107 CFUs/ml of Spn in coating buffer (KPL) and incubated overnight at 4ºC. Plates were blocked for 1 h at room temperature with 150 µl PBS with 0.01% Tween-20, 3% normal goat serum, and 0.5% milk powder. Ferret sera was heat inactivated at 56°C for 30 min. A series of 2-fold serial dilutions were performed in blocking buffer and incubated on the ELISA plate for 2 h at room temperature. After washing with PBS-0.01% Tween-20, plates were incubated with peroxidase-conjugated goat antiferret IgG (Jackson). SureBlue TMB peroxidase substrate (KPL) was added to each well and the reaction stopped with 250 mM HCl. Absorbance was read at 450 nm.
Aerosol particle sampling
Aerosol sampling of three H1N1pdm09 alone and coinfected donors from both Spn D39 transmission studies was performed, as previously described (Lakdawala et al. 2011). Briefly, air samples were collected using cyclone-based air samplers (BC251) developed by the National Institute for Occupational Safety and Health (Blachere et al. 2009), which were placed on the outside of the donor side of the transmission cage. Samples were collected between 3 and 7 dpi as well as a day prior to infection to act as a baseline with a designated air sampler assigned to each ferret to reduce cross-contamination. Air was sampled for 1 h at a flow rate of 3.5 l/min, which was calibrated before each use using a flow meter (TSI 4100 series). The NIOSH BC251 samplers separate particles based upon size. Each sampler contained an empty 15 ml conical that collected particles greater than 4 μm, a 1.5-ml tube that collected particles between 1 and 4 μm and a 3 μm pore Fluoropore membrane filter (Millipore) to collect submicron particles.
RNA was collected with 500 μl of MagMAX Lysis/Binding Solution Concentrate (Applied Biosystems) in each collection tube with vigorous vortexing. For transmission study one, RNA was extracted using the MagMAX Total Nucleic Acid Isolation Kit, per manufacturer's recommendations. MagMAX Total RNA isolation Kit was used to collect RNA from transmission study two, per manufacturer's recommendations. The total amount of influenza RNA was quantified using Applied Biosystems Taqman one-step RT-PCR kit with primers (F-5′AGA TGA GTC TTC TAA CCG AGG TCG3′ and R – 5′GCA AAG ACA TCT TCA AGT CTC TG3′) and a probe (FAM-TCA GGC CCC CTC AAA GCC GA-[NFQ]) specific for the IAV M gene segment. RNA was extracted from known titers of H1N1pdm09 to act as a standard curve.
The limit of detection of the NIOSH BC251 samplers is unknown. After each use, the BC251 samplers were decontaminated with distilled water followed by two isopropanol washes through the air inlet and the other holes.
Results
Secondary infection with Spn BHN97 impacts H1N1pdm09 viral titers and overall pathogenesis
In the ferret model, H1N1pdm09 virus has consistently been reported to efficiently replicate in the nasal cavity and transmit through the air (Koster et al. 2012, Lakdawala et al. 2011, 2015, Maines et al. 2009, Paules et al. 2017, Pulit-Penaloza et al. 2018). Only a limited number of studies have investigated IAV replication and airborne transmission in the ferret model with secondary Spn infection (McCullers et al. 2010, Rowe et al. 2020). Thus, in this study, we utilized the ferret model to study how Spn impacts IAV replication and airborne transmission. We first selected Spn clinical strain BHN97 (19F serotype) for our analysis, as this serotype has increased its distribution in the human population after implementation of the pneumococcal vaccine and is closely related to the vaccine escape serotype 19A (Hsieh et al. 2013).
To directly compare the impact of secondary bacterial infection on IAV replication and transmission, we setup two parallel groups. In the first group, we intranasally infected ferrets with H1N1pdm09, then 2 days later, near the peak of viral shedding, intranasally administered 107 colony forming units (CFUs) of Spn BHN97 (Fig. 1A). A total of 6 h following Spn infection, naïve recipients were placed into the adjacent cage, which is separated from the donors by perforated metal plates. This cage setup prevents physical contact but allows for air flow from the donor to the recipient (Lakdawala et al. 2011, 2015, Le Sage et al. 2021) and serves to measure airborne, but not contact-dependent, transmission. In the second group, we followed the same design and timeline, but instead of secondary Spn infection we mock infected ferrets with PBS (Fig. 1B).
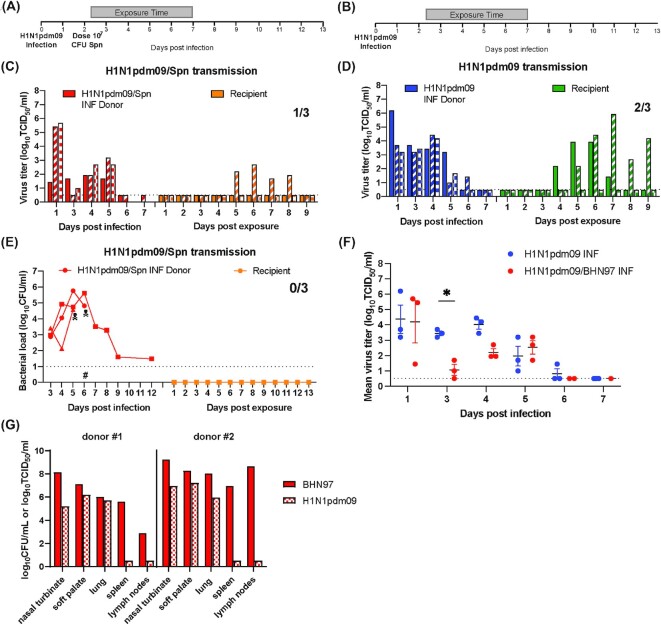
Figure 1.
Spn BHN97 is highly pathogenic during secondary bacterial infection and impacts H1N1pdm09 viral titers. (A) and (B) Experimental setup: donors (N = 6) were infected with 106 TCID50 H1N1pdm09 (A/CA/07/2009). A total of 2 days later, donors were either inoculated with 107 CFUs of Spn (BHN97; N = 3) (A) or mock infected with PBS (N = 3) (B). Recipients were placed in the adjacent cage 6 h after bacterial infection and exposed for 5 days (gray bar). (C) and (D) H1N1pdm09 titers and transmission in daily nasal washes. Titers in ferrets with secondary bacterial infection (C) and IAV alone (D), where each bar represents an individual animal. Limits of detection for bacteria and IAV are denoted by a dashed line. (E) Bacterial loads and transmission in daily nasal washes: bacterial loads from donors and recipients in panel (A) are indicated by each dot, which represents an individual animal. Skull symbol represents sudden death of a ferret; #, the beginning of antibiotic treatment. Red: donor animals and orange: recipient animals. (F) Daily viral titers for animals with secondary bacterial infection and single viral infections. Comparison of mean virus titer of donors during the first week of infection between concurrent H1N1pdm09 alone vs. secondary infection with Spn BHN97. Solid lines represent the mean ± SEM for each group per day. Two-way ANOVA analysis was used to determine statistically significant differences (*P< .05). (G) Tissue-specific bacterial loads and viral titers postmortality: density of Spn BHN97 (log10 CFU/ml) and titers of H1N1pdm09 (log10 TCID50/ml) from infected donors that succumbed to infection on day 5 (#1) and 6 (#2) postinfection.
To establish transmission efficiency and morbidity, we measured viral and bacterial titers from nasal secretions and assessed clinical symptoms on all surviving animals every day for 14 days post-H1N1pdm09 infection (12 days post-Spn infection). Transmission efficiency was based on the presence of bacteria or virus in the recipient nasal secretions or seroconversion for either pathogen on day 14 postinfection. We defined Spn colonization as detectable bacterial shedding in nasal secretions for more than 3 days.
Ferrets infected with both H1N1pdm09 and Spn BHN97 became colonized with Spn and shed bacteria starting 24 h after Spn inoculation, or 3 days post-H1N1pdm09 infection (dpi; Fig. 1E, red lines). H1N1pdm09 and Spn BHN97 coinfected donor animals experienced severe clinical symptoms including dehydration and labored breathing (Table S1, Supporting Information) and two ferrets succumbed to the infection and were found deceased on days 5 and 6 postinfection (Fig. 1E, skull symbol). Analysis of tissues and blood at time of necropsy revealed high titers of both H1N1pdm09 and Spn BHN97 in all respiratory organs assessed as well as bacteria in the spleen, lymph nodes, and blood (not shown) indicating that these animals were septic (Fig. 1G). Based on the observed clinical symptoms we developed a comprehensive rubric to assess clinical symptomology on ferrets including weight loss, dehydration, breathing, activity, and other symptoms (Table S2, Supporting Information). The intended recipient exposure time was 11 consecutive days, but due to the severe disease and death of these donors, the recipients for all pairs in both groups were separated after 5 days. In this study, we did not observe any detectable bacterial shedding (Fig. 1E, orange lines) or seroconversion (Table S1, Supporting Information) in the recipient animals, suggesting that Spn BHN97 was not transmitted through the air (Fig. 1E, orange lines).
Donors with secondary bacterial infections shed H1N1pdm09 in nasal secretions until 5 dpi (Fig. 1C, red bars), and H1N1pdm09 transmitted to one of three recipient animals (Fig. 1C, orange bars). The concurrent H1N1pdm09 only transmission experiment was performed with the same 5-day exposure time (Fig. 1B), which resulted in donors shedding virus until 6 dpi (Fig. 1D, blue bars) and two of three recipients shedding virus from 4 to 9 days postexposure (dpe; Fig. 1D, green bars). Seroconversion for H1N1pdm09 was observed only in animals that also shed virus (Table S1, Supporting Information).
To determine if secondary Spn infection affected IAV replication, the nasal secretions from donors infected only with H1N1pdm09 were titered at the same time as those from donors with secondary Spn infection (Fig. 1F). Comparison of the viral titer in the nasal secretions revealed similar titers on day 1 post-H1N1pdm09 infection, which was prior to Spn infection, as expected. Interestingly, in the animals subsequently infected with Spn, the H1N1pdm09 viral titers were significantly reduced on day 3 postinfection (Fig. 1F, *P < .05) and recovered to similar levels as the IAV only controls on day 5 postinfection. These data suggest that secondary Spn infection may diminish H1N1pdm09 replication in the upper respiratory tract (URT) of ferrets.
We conclude that in ferrets with preceding viral infection, Spn BHN97 can disseminate beyond the URT and cause high morbidity and mortality. Further, even with the high mortality rate, we observed reduced viral titers in ferret nasal washes with a secondary Spn infection relative to virus alone. However, given the lethality observed with coinfection of Spn BHN97 and H1N1pdm09, analysis of airborne transmission may have been compromised. Therefore, to investigate whether Spn influences H1N1pdm09 airborne transmission we repeated the experiment with a different Spn strain not expected to cause this high degree of mortality.
Secondary infection with Spn D39 reduces H1N1pdm09 viral titers in the URT
Selection of experimental design
Pathogenesis of Spn can be strain and dose dependent in animal models (D'Mello et al. 2020, Forbes et al. 2008, Mlacha et al. 2013), thus we performed a dose titration experiment with 105, 106, or 107 CFUs of Spn D39 to determine its pathogenesis (Figure S1, Supporting Information). Isolated in 1916, the model strain Spn D39 is a serotype 2 strain that has been used widely to study pneumococcal pathogenesis in murine models (Lanie et al. 2007). For these experiments, ferrets were intranasally infected with H1N1pdm09, then 2 days later, Spn D39 was administered by the same route with the three doses. While transmission of Spn did not occur with inoculation doses of 105 and 106 CFUs, an inoculation dose of 107 CFUs resulted in airborne transmission of the bacteria and seroconversion to H1N1pdm09 in the recipient animal. Airborne transmission of H1N1pm09 was also observed in the 106 CFUs pair. Importantly, Spn D39 was not lethal in the ferrets at all doses(Figure S1 and Table S1, Supporting Information). Therefore, our subsequent studies were completed in Spn D39 with a dose of 107 CFU/ml.
Secondary bacterial infection with Spn D39 decreases H1N1pdm09 titers in the ferret URT
The experiment with Spn BHN97 revealed that viral titers at day 3 were decreased in animals with a secondary bacterial infection (Fig. 1F). To investigate whether secondary infection with Spn D39 influenced viral titers, we performed two independent replicate studies. Combined, these two independent studies tested H1N1pdm09 titers in seven pairs of ferrets in the absence of secondary infections (virus only) and seven pairs of ferrets with both H1N1pdm09 and Spn D39. In both studies, donor ferrets were infected with H1N1pdm09, and 2 days later half of these donors were infected with 107 CFUs of Spn D39 (Fig. 2A and B). At 54 h postinfection (6 h postbacterial inoculation), recipient ferrets were placed into the transmission cage for an 11-day continuous exposure. Nasal secretions were collected from donor and recipient animals and assessed for Spn D39 and H1N1pdm09 titers. The viral titers and bacterial loads for each pair are shown in Figure S2 (Supporting Information).
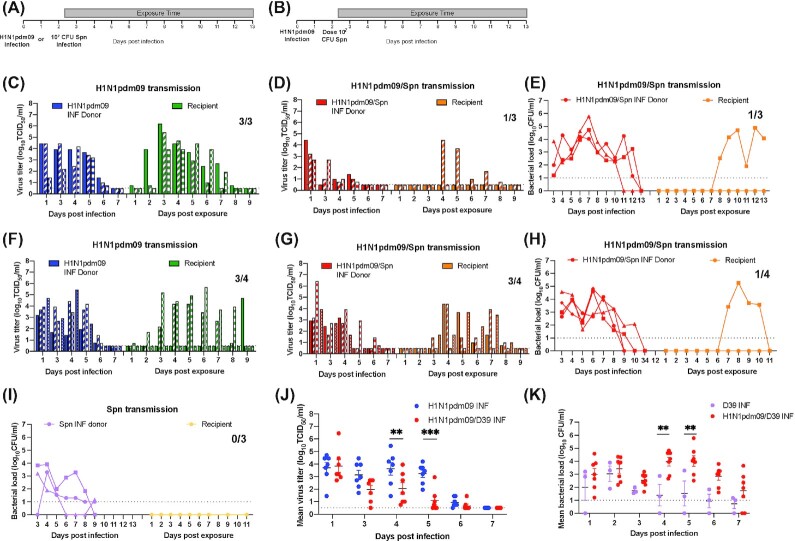
Figure 2.
Secondary infection with Spn D39 reduces viral titers and Spn D39 pathogenesis is exacerbated with preceding H1N1pdm09 infection. Experimental setup for airborne transmission of H1N1pdm or Spn D39 alone (A) or secondary infection with Spn D39 (B) in ferrets. A total of three or four donors were infected with 106 TCID50 H1N1pdm09 (A/CA/07/2009) or not infected for the Spn only group. A total of 2 days later, half of the donors were inoculated with 107 CFUs of Spn (D39; (D), (E), (G), and (H), red bars or lines and (I) purple lines). The other half did not receive Spn (C, blue bars) or (F, blue bars, which where mock infected with PBS). Recipients were placed in the adjacent cage 6 h later and exposed for 11 days (A and B, gray rectangle). Each bar represents viral titer for an individual animal and each dot represents the bacterial titer for an individual animal. Limit of detection is denoted by a dashed line. (J) Comparison of mean virus titer of donors during the first week of infection between concurrent H1N1pdm09 alone vs. coinfection with or D39. Limit of detection is denoted by a dashed line. Solid lines represent the mean ± SEM for each group per day. Two-way ANOVA analysis was used to determine statistically significant differences (**P < .005, ***P< .0005). (K) Comparison of mean bacterial load of donors during Spn D39 infection (N = 3) and coinfection with preceding H1N1pdm09 infection (N = 7). Limit of detection is denoted by a dashed line. Data represent the mean ± SEM for each group per day. Two-way ANOVA analysis was used to determine statistically significant differences (**P < .005).
Comparison of the viral titer in the nasal secretions of donor animals revealed similar titers on day 1 post-H1N1pdm09 infection, which was prior to Spn infection, as expected (Fig. 2J). Interestingly, the H1N1pdm09 viral titers were reduced on subsequent days after Spn infection. Viral titers in animals with a secondary Spn D39 infection were consistently lower than the H1N1pdm09 only infected donors on days 4 and 5 and did not recover to similar levels (Fig. 2J; **P< .005 and ***P< .0005, respectively). Together, these data demonstrate that secondary Spn D39 infection after a primary H1N1pdm09 infection can reduce H1N1pdm09 replication in the ferret nasal cavity.
A decrease in viral titers within the nasal secretions could also lead to less virus expelled into aerosols and thus influence transmission dynamics during secondary Spn infections. Environmental conditions between studies with regards to humidity and temperature were similar (Figure S3A and B, Supporting Information). We assessed the virus-laden aerosols every day after Spn coinfection using cyclone-based NIOSH samplers, as previously described (Lakdawala et al. 2011). Air sampling of the ferrets infected with H1N1pdm09 alone vs. those infected with both pathogens indicates that all animals expelled virus-laden aerosols larger than 4 μm in size, as detected by the presence of viral RNA and not infectious virus. Even those animals that failed to transmit the virus to recipient animals expelled virus-laden aerosols (Figure S3C and D, Supporting Information). Therefore, the decrease in viral load in the nasal wash during Spn secondary infection does not result in an appreciable decrease of detectable expelled aerosols containing viral RNA.
Analysis of influenza transmission dynamics in single viral infections versus secondary Spn D39 infections
Next, we used these two independently performed replicate studies to compare the transmission efficiency of H1N1pdm09 in the presence or absence of secondary Spn D39 infection. The timing of viral shedding, days 3–8 postexposure, was similar in all recipient animals infected with H1N1pdm09, demonstrating that secondary bacterial infection did not delay H1N1pdm09 transmission (Fig. 2C, D, F, and G, green and orange bars). Transmission of H1N1pdm09 in single infections was 3/3 in the first study and 3/4 in the second study (Fig. 2C and F, respectively), for a combined transmission efficiency of 6/7 (85%). This is consistent with the robust transmission of H1N1pdm09 to naïve ferrets reported by our group and many others (Itoh et al. 2009, Lakdawala et al. 2015, Le Sage et al. 2021, Munster et al. 2009). In contrast, in the first study, we observed a lower transmission efficiency in ferrets with a secondary Spn infection, the efficiency was 1/3 (Fig. 2D). However, in the second study the virus transmitted to 3/4 of the ferrets (Fig. 2G). In total, transmission of H1N1pdm09 was observed in 4/7 (57%) pairs with secondary Spn D39 infection compared to 6/7 (85%) in H1N1pdm09 only infected pairs. This difference in H1N1pdm09 transmission between ferret with single viral infections and Spn secondary infections was not statistically significant, nonetheless it highlights a possible trend towards decreased viral transmission in hosts with secondary Spn infection that merits further study.
Analysis of Spn transmission dynamics and pathogenesis with preceding H1N1pdm09 infection
To investigate whether preinfection with H1N1pdm09 influenced Spn D39 transmission and bacterial load, ferrets were inoculated with 107 CFUs Spn D39 in the absence of IAV in a separate experiment (Fig. 2I). Bacterial colonization of donors occurred in the absence of H1N1pdm09 infection with the donors shedding over multiple days (Fig. 2I, purple lines). No transmission events were detected in the Spn-only group recipients (Fig. 2I, yellow lines). In ferrets with secondary bacterial infections, all the donors became colonized with Spn D39 and shed bacteria on multiple dpi (Fig. 2E and H, red lines). Airborne transmission of D39 was observed in 2/7 (28%) of coinfected donors to naïve recipients (Fig. 2E and H, orange lines). Comparison of the Spn D39 load in donors from bacteria alone or coinfected group indicates that bacteria loads in nasal washes are greater in the presence of H1N1pdm09 at later times postinfection with significant differences at 4 and 5 dpi (Fig. 2K). Intriguingly, the temporal dynamics of transmission appear distinct for each pathogen. This is particularly clear in the first study where shedding of H1N1pdm09 from a recipient animal waned at day 8 postexposure when bacterial shedding was beginning (Figure S2B, Supporting Information). These data suggest that Spn and H1N1pdm09 may not be co-transmitted, or that Spn is only detected a few days after the transmission event. Sacrifice of recipients with a secondary infection (N = 2) at endpoint (days 15 or 12 postexposure) revealed no H1N1pdm09 virus in any organ tested, whereas high bacterial loads were found in most tissues tested, including respiratory tissues and the spleen (Figure S4, Supporting Information). Thus, bacterial systemic dissemination and severe morbidity was observed in recipients infected with both Spn and H1N1pdm09. While the low number of transmission events in general precludes a statistical analysis of these data, the trend fits well with existing data that suggests that preinfection with influenza viruses promotes Spn transmission and density (McCullers et al. 2010, Nakamura et al. 2011, Wren et al. 2014, 2017).
Coinfected donors with severe pathogenesis do not transmit H1N1pdm09
In addition to pathogen loads and transmission dynamics, we collected extensive data on symptoms. Cumulative disease symptoms were scored for all animals using a rubric that includes ferret activity, breathing, nasal discharge, weight loss, dehydration, and diarrhea (Table S2, Supporting Information). Consistent with published findings (McCullers et al. 2010, Peltola et al. 2006), we found clinical symptoms were more severe in animals with a secondary bacterial infection as compared to those infected with H1N1pdm09 or Spn alone. Specifically, the score for cumulative symptoms went averages of 1.3 and 1.9 in bacterial and viral single infections, respectively to 7.8 in animals with a secondary infection (Fig. 3A). H1N1pdm09 infected donors had significantly fewer symptoms as compared to coinfected H1N1pdm09/D39 infected donors (P-value .0076).
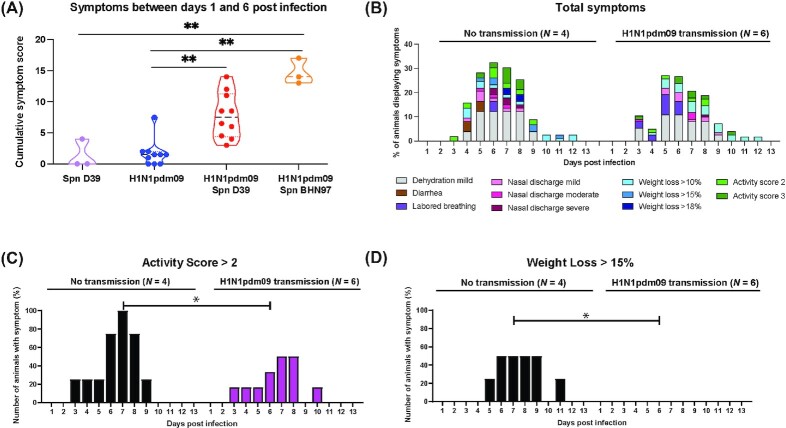
Figure 3.
Coinfection of H1N1pdm09 and Spn causes exacerbated disease. (A) Total number of symptoms between days 1 and 6 postinfection for donor ferrets infected with Spn D39 alone (N = 3), H1N1pdm09 alone (N = 10), coinfected with H1N1pdm09/Spn D39 (N = 10) or H1N1pdm09/Spn BHN97 (N = 3). Two-way ANOVA analysis was used to determine statistically significant differences (**P< .05). (B) Percent number of H1N1pdm09/Spn D39 coinfected donors displaying each symptom that either did not or did transmit the virus. (C) Percent of coinfected donor animals that exhibited an activity score greater than 2. Two-way ANOVA analysis was used to determine statistically significant differences (*P< .05). (D) Percent of coinfected donor animals that exhibited a greater than 15% weight loss. Two-way ANOVA analysis was used to determine statistically significant differences (*P < .05).
To assess whether clinical severity in donor animals impacted H1N1pdm09 airborne transmission, we sorted the data for all donor ferrets infected with both H1N1pdm09/Spn D39 (including those from the dose titration experiment) into two groups (Fig. 3B). Group 1 included the animals that did transmit H1N1pdm09 to their paired recipient (N = 6), while group 2 included the animals that did not (N = 4). We focused on Spn D39, because the lethality of Spn BHN97 in 2/3 donor ferrets limits clinical scores to the first 5 days. Interestingly, donors with secondary infections that did not transmit H1N1pdm09 displayed more severe symptoms than donors that transmitted the virus (Fig. 3B, a breakdown of each clinical category can be found in Figure S5 (Supporting Information). When considering each symptom individually there was a statistically significant association between activity score (above 2) and transmission (Fig. 3C, ANOVA P < .05) as well as extent of weight loss (above 15%) and transmission (Fig. 3D, ANOVA P< .05). Taken together, these data suggest that severe clinical symptom onset in donor animals may be associated with reduced airborne transmission of H1N1pdm09.
Spn D39 colonized recipient ferrets are susceptible to H1N1pdm09 infection but not severe disease.
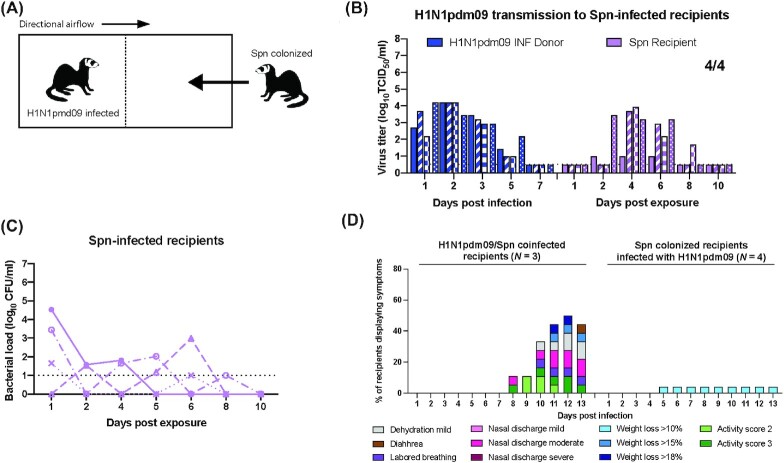
Our data demonstrate that secondary infection with Spn significantly decreases viral replication in the nasal cavity. Thus, we postulated that precolonization of Spn might prevent viral transmission by providing an inhospitable environment in the recipient for infection initiation. To determine whether Spn colonization was sufficient to prevent natural infection by H1N1pdm09, four ferrets were precolonized with Spn D39 to serve as recipients for H1N1pdm09 transmission (Fig. 4A). Nasal secretions were collected every other day and all recipients were shown to shed bacteria (Fig. 4C). Donors were infected with H1N1pdm09 and precolonized Spn recipients were placed in the adjacent cage 24 h later. Precolonized recipients were exposed to virally infected donors for 2 days, which we have previously shown results in effective transmission of H1N1pdm09 to all recipients (Le Sage et al. 2021, Paules et al. 2017). All recipients became infected with H1N1pdm09 and shed virus over multiple days between 2 and 8 dpe (Fig. 4B, purple bars), indicating that Spn D39 precolonization does not protect recipients from H1N1pdm09 natural infection. Strikingly, the precolonized Spn D39 recipients who subsequently became coinfected with H1N1pdm09 only displayed 10% weight loss (Fig. 4D). This is in contrast to the more severe symptoms (thick nasal discharge, weight loss of > 15%, and dissemination to beyond the respiratory track) associated with recipients infected from donors coinfected with H1N1pdm09 and then Spn (N = 3; Figures S2B, S2E, and S1D, Supporting Information). Moreover, bacterial loads in recipient animals were much lower than in animals with preceding H1N1pdm09 infections (Fig. 4C vs. orange lines in Fig. 2E, H and Figure S2D, Supporting Information). Taken together, these findings suggest that factors such as order of infection as well as anatomical sites of bacterial colonization and viral infection could influence the outcome of dual H1N1pdm09/Spn infections.
Figure 4.
Spn-colonized recipient ferrets are susceptible to H1N1pdm09 infection but not severe disease. (A) Schematic representation of cage setup, where recipients were inoculated with 107 CFUs of Spn (D39) 48 h prior (N = 4) to exposure. Donors (N = 4) were infected with 106 TCID50 H1N1pdm09 (A/CA/07/2009) and 24 h later recipients were placed in the adjacent cage and exposed for 2 days. (B) H1N1pdm09 titers from donor and recipient animals. Limit of detection is denoted by a dashed line. (C) Bacterial loads from recipients in panel (B) are indicated by each symbol, which represents an individual animal. (D) Percent number of H1N1pdm09/Spn D39 coinfected recipients from Figure S1D (Supporting Information), and Fig. 2D/2E and 2G/2H (N = 3) and Spn-colonized recipients infected with H1N1pdm09 from panels (B) and (C) (N = 4) displaying each symptom over the course of the experiment.
Discussion
Efficient replication and airborne transmission are critical for the spread and public health burden of respiratory pathogens. However, our understanding of how microbial communities contribute to the replication of virus within the patient and the spread of pathogenic bacteria and viruses through the air is limited. We demonstrate that infection with H1N1pdm09 and secondary infection of Spn decreases viral replication in the URT, increases disease severity, and possibly reduces airborne transmission of H1N1pdm09. In contrast, bacterial load in the URT was increased, and systemic dissemination and airborne transmission of Spn was observed. These data suggest an asymmetrical relationship between Spn and influenza virus.
During the 2009 H1N1 pandemic, severe infections were attributed to secondary bacterial pneumonia, commonly caused by Spn (Macintyre et al. 2018, Palacios et al. 2009). The increased disease severity upon secondary bacterial infection is strongly supported by animal models. Studies in mice and ferrets report increased morbidity and mortality in coinfected animals relative to single infection (Kash et al. 2011, Marks et al. 2013, McCullers et al. 2010, Mina et al. 2015, Peltola et al. 2006, Sharma-Chawla et al. 2016). Consistent with these previous data, we also observed enhanced disease as measured by increased weight loss, appearance of a thick layer of nasal discharge, diarrhea, shallow breathing, and severe dehydration that required administration of subcutaneous fluids in coinfected animals. Changes in bacterial colonization and host microbiota could impact host immune responses within a donor leading to severe pathology. Further studies are needed to determine whether a direct interplay between the pathogens or host responses are driving the severe disease associated with coinfection. Surprisingly, recipient animals colonized with Spn and then naturally infected with H1N1pdm09 through the air did not present with severe disease symptoms, which may indicate that the anatomical site where each pathogen initiates an infection may influence disease severity. Interestingly, we identified an association between disease symptoms and airborne transmission of H1N1pdm09. We found that coinfected donors that displayed more severe pathogenesis did not transmit H1N1pdm09 to their recipients as efficiently as donors with milder symptoms. These data put forward an important aspect of transmission heterogeneity that has been largely understudied, whereby transmissibility is related to disease severity.
Our data also revealed that secondary infection with Spn decreased H1N1pdm09 replication in the nasal cavity. This effect may be related to changes in the viral lifecycle induced by Spn. For example, presence of Spn at sites of viral replication could influence attachment and entry of influenza viruses into host cells, efficiency of viral RNA synthesis, or viral assembly and budding (Jones et al. 2020). It is already clear that changes in glycosylation of the host incurred during influenza virus infection influence subsequent Spn colonization, and similarly changes in glycosylation via Spn affect subsequent viral infection (Nita-Lazar et al. 2015, Ortigoza et al. 2018, Siegel et al. 2014, Wren et al. 2017). The detected reduction in viral titers is similar to recent data obtained from neonatal mice colonized with Spn prior to PR8 H1N1 infection (Ortigoza et al. 2018). Thus, direct or indirect interactions between Spn and influenza lead to decreased viral replication. However, the relation between bacteria and H1N1pdm09 may not be limited to streptococcus, a dysregulation in nasal microbiota could reduce susceptibility to influenza infections, as suggested by a recent study using topical antibiotics (Rowe et al. 2020).
The negative association between secondary Spn infection and H1N1pdm09 titers led us to investigate whether prior colonization of Spn could reduce the susceptibility to H1N1pdm09 infection through the air. We observed that precolonization with Spn did not protect recipients from acquiring H1N1pdm09 through the air and shed both pathogens in nasal washes. However, in contrast to secondary Spn infections, these animals did not present with exacerbated disease and bacterial loads did not increase upon IAV infection. Additionally, viral titers were not decreased in the nasal secretions compared to donor animals. These differences may be related to the diverse pathology associated with each pathogen. A primary influenza virus infection disrupts the protective epithelial cell lining and immunity in the respiratory tract, which opens the tissue for Spn colonization (Short et al. 2014, Siemens et al. 2017). In contrast, colonization with Spn may not have the same effect on immunity or integrity of airway epithelial cells that will influence viral replication. Further, given the prevalence of Spn colonization in children, it is tempting to consider how these findings relate to pediatric viral–bacterial infections. However, any parallels between our finding in the ferret model and children must be made cautiously, since our studies were conducted in adult ferret, who displayed low levels of Spn colonization. Differences between adult and pediatric immune systems as well as differences in the extent of colonization of Spn in human hosts vs. the ferret model could impact the severity of disease and pathogen transmission efficiencies.
In conclusion, our study highlights how bacteria may influence replication and airborne transmission of H1N1pdm09 virus. In addition, we have demonstrated that disease severity may be negatively correlated with influenza transmission and that colonizing Spn may need to change their niche or expression pattern before they contribute to increase morbidity associated with secondary bacterial infections. These results shift our thinking of bacterial–viral interactions and open the doors to a new understanding of microbial interactions, disease severity and transmission.
Funding
We would like to thank William G Lindley (National Institute for Occupational Safety and Health) for use of the cyclone-based air samplers. This work was supported by the National Institute of Allergy and Infectious Diseases (HHSN272201400007C (Rothman PI) S.S.L. (subcontractor)) ; and the NIAID (R01AI139077 to NLH). Additional funding for S.S.L. includes the American Lung Association Biomedical Research grant.
Authors' contributions
K.M.B., V.L., N.L.H., and S.S.L. designed the experiments, analyzed, and interpreted the data, and wrote the manuscript. K.M.B., V.L., A.J.F., J.E.J., G.H.P., and A.J.A. performed the experiments. S.S.C. and J.W.R. contributed to resources and analysis. All authors edited and approved the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
We are also grateful for additional support from the Eberly Family Trust (NLH).
Contributor Information
Karina Mueller Brown, Department of Biological Sciences, Carnegie Mellon University, 4400 Fifth Avenue, Pittsburgh, PA 15213, USA.
Valerie Le Sage, Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, 450 Technology Drive, Bridgeside Point II, Pittsburgh, PA 15219, USA.
Andrea J French, Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, 450 Technology Drive, Bridgeside Point II, Pittsburgh, PA 15219, USA.
Jennifer E Jones, Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, 450 Technology Drive, Bridgeside Point II, Pittsburgh, PA 15219, USA.
Gabriella H Padovani, Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, 450 Technology Drive, Bridgeside Point II, Pittsburgh, PA 15219, USA.
Annika J Avery, Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, 450 Technology Drive, Bridgeside Point II, Pittsburgh, PA 15219, USA.
Stacey Schultz-Cherry, Department of Infectious Diseases, St Jude Children's Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105, USA.
Jason W Rosch, Department of Infectious Diseases, St Jude Children's Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105, USA.
N Luisa Hiller, Department of Biological Sciences, Carnegie Mellon University, 4400 Fifth Avenue, Pittsburgh, PA 15213, USA.
Seema S Lakdawala, Department of Microbiology and Molecular Genetics, University of Pittsburgh School of Medicine, 450 Technology Drive, Bridgeside Point II, Pittsburgh, PA 15219, USA; Center for Vaccine Research, University of Pittsburgh School of Medicine, 3501 Fifth Avenue, Pittsburgh, PA 15213, USA.
Conflicts of interest statement
None declared.
References
- Andrew PW, Manzoor I, Abdullah ITet al. . Rgg-Shp regulators are important for pneumococcal colonization and invasion through their effect on mannose utilization and capsule synthesis. Sci Rep. 2018;8:1–15. DOI: 10.1038/s41598-018-24910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Maines TR, Katz JMet al. . Considerations regarding appropriate sample size for conducting ferret transmission experiments. Fut Microbiol. 2013;8:961–5. [DOI] [PubMed] [Google Scholar]
- Belser JA, Pulit-Penaloza JA, Maines TR. Ferreting out influenza virus pathogenicity and transmissibility: past and future risk assessments in the ferret model. Cold Spring Harb Perspect Med. 2020;10:1–15. DOI: 10.1101/cshperspect.a038323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachere FM, Lindsley WG, Pearce TAet al. . Measurement of airborne influenza virus in a hospital emergency department. Clin Infect Dis. 2009;48:438–40. DOI: 10.1086/596478. [DOI] [PubMed] [Google Scholar]
- Bouvier NM. Animal models for influenza virus transmission studies: a historical perspective. Curr Opin Virol. 2015;13:101–8. DOI: 10.1016/j.coviro.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Mello A, Riegler AN, Martínez Eet al. . An in vivo atlas of host–pathogen transcriptomes during Streptococcus pneumoniae colonization and disease. Proc Natl Acad Sci. 2020;117:33507–18. DOI: 10.1073/PNAS.2010428117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diavatopoulos DA, Short KR, Price JTet al. . Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 2010;24:1789–98. DOI: 10.1096/fj.09-146779. [DOI] [PubMed] [Google Scholar]
- Duvigneau S, Sharma-Chawla N, Boianelli Aet al. . Hierarchical effects of pro-inflammatory cytokines on the post-influenza susceptibility to pneumococcal coinfection. Sci Rep. 2016;6:1–11. DOI: 10.1038/srep37045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes ML, Horsey E, Hiller NLet al. . Strain-specific virulence phenotypes of Streptococcus pneumoniae assessed using the Chinchilla laniger model of otitis media. PLoS ONE. 2008;3:e1969. DOI: 10.1371/journal.pone.0001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Juarbe N, Riegler AN, Jureka ASet al. . Influenza-induced oxidative stress sensitizes lung cells to bacterial-toxin-mediated necroptosis. Cell Rep. 2020;32: 108062. DOI: 10.1016/j.celrep.2020.108062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Lin TL, Chang KYet al. . Expansion and evolution of Streptococcus pneumoniae serotype 19A ST320 clone as compared to its ancestral clone, Taiwan19F-14 (ST236). J Infect Dis. 2013;208:203–10. DOI: 10.1093/infdis/jit145. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Shinya K, Kiso Met al. . In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature. 2009;460:1021–5. DOI: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JE, Le Sage V, Lakdawala SS. Viral and host heterogeneity and their effects on the viral life cycle. Nat Rev Microbiol. 2021;19:272–82. DOI: 10.1038/s41579-020-00449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalil AC, Thomas PG. Influenza virus-related critical illness: pathophysiology and epidemiology. Crit Care. 2019;23:1–7.. DOI: 10.1186/s13054-019-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash JC, Walters KA, Davis ASet al. . Lethal synergism of 2009 pandemic h1n1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. MBio. 2011;2:1–9. DOI: 10.1128/mBio.00172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khardori NM. Clinical aspects of pandemic 2009 influenza A (H1N1) virus infection. Yearbook Med. 2010;2010:83–4. DOI: 10.1016/s0084-3873(10)79684-x. [Google Scholar]
- Kimaro Mlacha SZ, Romero-Steiner S, Dunning Hotopp JCet al. . Phenotypic, genomic, and transcriptional characterization of Streptococcus pneumoniae interacting with human pharyngeal cells. BMC Genomics. 2013;14: 383. DOI: 10.1186/1471-2164-14-383383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EY, Monteforte B, Gupta Aet al. . The frequency of influenza and bacterial coinfection: a systematic review and meta-analysis. Influenza Other Respir Vir. 2016;10:394–403.. DOI: 10.1111/irv.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster F, Gouveia K, Zhou Yet al. . Exhaled aerosol transmission of pandemic and seasonal H1N1 influenza viruses in the ferret. PLoS ONE. 2012;7:1–14. DOI: 10.1371/journal.pone.0033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Jayaraman A, Halpin Ret al. . The soft palate is an important site of adaptation for transmissible influenza viruses. Nature. 2015;526:122–5. DOI: 10.1038/nature15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Lamirande EW, Suguitan ALet al. . Eurasian-origin gene segments contribute to the transmissibility, aerosol release, and morphology of the 2009 pandemic H1N1 influenza virus. PLoS Pathog. 2011;7:e1002443. DOI: 10.1371/journal.ppat.1002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Shih AR, Jayaraman Aet al. . Receptor specificity does not affect replication or virulence of the 2009 pandemic H1N1 influenza virus in mice and ferrets. Virology. 2013;446:349–56. DOI: 10.1016/j.virol.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanie JA, Ng WL, Kazmierczak KMet al. . Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol. 2007;189:38–51. DOI: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sage V, Jones JE, Kormuth KAet al. . Pre-existing heterosubtypic immunity provides a barrier to airborne transmission of influenza viruses. PLoS Pathog. 2021;17:1–16. DOI: 10.1371/JOURNAL.PPAT.1009273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie JK, Acosta M, Winter Ket al. . Factors associated with death or hospitalization due to pandemic 2009 influenza A(H1N1) infection in California. JAMA. 2009;302:1896–902. DOI: 10.1001/jama.2009.1583. [DOI] [PubMed] [Google Scholar]
- Macintyre CR, Chughtai AA, Barnes Met al. . The role of pneumonia and secondary bacterial infection in fatal and serious outcomes of pandemic influenza a (H1N1) pdm09. BMC Infect Dis. 2018;18:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Jayaraman A, Belser JAet al. . Transmission and pathogenesis of swine-origin 2009 A(H1N1) influenza viruses in ferrets and mice. Science. 2009;325:484–7. DOI: 10.1126/science.1177238.Transmission. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks LR, Davidson BA, Knight PRet al. . Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. MBio. 2013;4:1–13. DOI: 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA, McAuley JL, Browall Set al. . Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J Infect Dis. 2010;202:1287–95. DOI: 10.1086/656333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger DW, Sun K. Immune dysfunction and bacterial coinfections following influenza. J Immunol. 2013;191:2047–52. DOI: 10.4049/jimmunol.1301152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina MJ, Klugman KP. The role of influenza in the severity and transmission of respiratory bacterial disease. Lancet Respir Med. 2014;2:750–63. DOI: 10.1016/S2213-2600(14)70131-6.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina MJ, Klugman KP, Rosch JWet al. . Live attenuated influenza virus increases pneumococcal translocation and persistence within the middle ear. J Infect Dis. 2015;212:195–201. DOI: 10.1093/infdis/jiu804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: Implications for pandemic influenza preparedness. J Infect Dis. 2008:198:962–70. DOI: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Wit EDe, Brand JM a Van Denet al. . Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;503:481–3. DOI: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Davis KM, Weiser JN. Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J Clin Invest. 2011;121:3657–65. DOI: 10.1172/JCI57762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nita-Lazar M, Banerjee A, Feng Cet al. . Desialylation of airway epithelial cells during influenza virus infection enhances pneumococcal adhesion via galectin binding. Mol Immunol. 2015;65:1–16. DOI: 10.1016/j.molimm.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortigoza MB, Blaser SB, Zafar MAet al. . An infant mouse model of influenza virus transmission demonstrates the role of virus-specific shedding, humoral immunity, and sialidase expression by colonizing Streptococcus pneumoniae. Am Soc Microbiol. 2018;9:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios G, Hornig M, Cisterna Det al. . Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS ONE. 2009;4:1–5. DOI: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paules CI, Lakdawala S, McAuliffe JMet al. . The hemagglutinin A stem antibody MEDI8852 prevents and controls disease and limits transmission of pandemic influenza viruses. J Infect Dis. 2017;216:356–65. DOI: 10.1093/infdis/jix292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola VT, Boyd KL, McAuley JLet al. . Bacterial sinusitis and otitis media following influenza virus infection in ferrets. Infect Immun. 2006;74:2562–7. DOI: 10.1128/IAI.74.5.2562-2567.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Jones J, Sun Xet al. . Antigenically diverse swine origin H1N1 variant influenza viruses exhibit differential ferret pathogenesis and transmission phenotypes. J Virol. 2018;92:1–16. DOI: 10.1128/jvi.00095-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Yochay G, Raz M, Dagan Ret al. . Nasopharyngeal carriage of Streptococcus pneumoniae by adults and children in community and family settings. Clin Infect Dis. 2004;38:632–9. DOI: 10.1086/381547. [DOI] [PubMed] [Google Scholar]
- Reuman PD, Keely S, Schiff GM. Assessment of signs of influenza illness in the ferret model. J Virol Methods. 1989;24:27–34. DOI: 10.1016/0166-0934(89)90004-9. [DOI] [PubMed] [Google Scholar]
- Rowe HM, Karlsson E, Echlin Het al. . Bacterial factors required for transmission of Streptococcus pneumoniae in mammalian hosts. Cell Host Microbe. 2019;25:884–91. DOI: 10.1016/j.chom.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Livingston B, Margolis Eet al. . Respiratory bacteria stabilize and promote airborne transmission of influenza A virus. MSystems. 2020;5:1–11. DOI: 10.1128/msystems.00762-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudd JM, Ashar HK, Chow VTet al. . Lethal synergism between influenza and Streptococcus pneumoniae. J Infect Pulmon Dis. 2016:2:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma-Chawla N, Sender V, Kershaw Oet al. . Influenza A virus infection predisposes hosts to secondary infection with different Streptococcus pneumoniae serotypes with similar outcome but serotype-specific manifestation. Infect Immun. 2016;84:3445–57. DOI: 10.1128/IAI.00422-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short KR, Kroeze EJBVFouchier RAMet al. . Pathogenesis of influenza-induced acute respiratory distress syndrome. Lancet Infect Dis. 2014;14:57–69. DOI: 10.1016/S1473-3099(13)70286-X. [DOI] [PubMed] [Google Scholar]
- Short KR, Reading PC, Wang Net al. . Increased nasopharyngeal bacterial titers and local inflammation facilitate transmission of Streptococcus pneumoniae. MBio. 2012;3:1–7. DOI: 10.1128/mBio.00255-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel SJ, Roche AM, Weiser JN. Influenza promotes pneumococcal growth during coinfection by providing host sialylated substrates as a nutrient source. Cell Host Microbe. 2014;16:55–67. DOI: 10.1016/j.chom.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens N, Oehmcke-Hecht S, Mettenleiter Tet al. . Port d'Entrée for respiratory infections - does the influenza a virus pave the way for bacteria?. Front Microbiol. 2017;8:1–17. DOI: 10.3389/fmicb.2017.02602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren JT, Blevins LK, Pang Bet al. . Influenza A virus alters pneumococcal nasal colonization and middle ear infection independently of phase variation. Infect Immun. 2014;82:4802–12. DOI: 10.1128/IAI.01856-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren JT, Blevins LK, Pang Bet al. . Pneumococcal neuraminidase A (NanA) promotes biofilm formation and synergizes with influenza A virus in nasal colonization and middle ear infection. Infect Immun. 2017;85:1–10. DOI: 10.1128/IAI.01044-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.