Abstract
The Zebrafish Information Network (zfin.org) is the central repository for Danio rerio genetic and genomic data. The Zebrafish Information Network has served the zebrafish research community since 1994, expertly curating, integrating, and displaying zebrafish data. Key data types available at the Zebrafish Information Network include, but are not limited to, genes, alleles, human disease models, gene expression, phenotype, and gene function. The Zebrafish Information Network makes zebrafish research data Findable, Accessible, Interoperable, and Reusable through nomenclature, curatorial and annotation activities, web interfaces, and data downloads. Recently, the Zebrafish Information Network and 6 other model organism knowledgebases have collaborated to form the Alliance of Genome Resources, aiming to develop sustainable genome information resources that enable the use of model organisms to understand the genetic and genomic basis of human biology and disease. Here, we provide an overview of the data available at the Zebrafish Information Network including recent updates to the gene page to provide access to single-cell RNA sequencing data, links to Alliance web pages, ribbon diagrams to summarize the biological systems and Gene Ontology terms that have annotations, and data integration with the Alliance of Genome Resources.
Keywords: Danio rerio, zebrafish, knowledgebase, database
Introduction
The Zebrafish Information Network (ZFIN, zfin.org) is the knowledgebase for the model organism Danio rerio (zebrafish). Since 1994, ZFIN has served the zebrafish research community by collecting, integrating, and making available zebrafish data. ZFIN’s biocurators, who have expertise in genetics, developmental, cellular, molecular, and evolutionary biology, have annotated over 16,000 zebrafish research publications for data that include genes, gene function, sequences, alleles, mutant and transgenic lines, human disease models, gene expression, phenotype, orthology, sequence targeting reagents (STR), and antibodies. Additionally, ZFIN supports the zebrafish research community by providing wiki resources to view antibody and protocol information, as well as ZFIN pages for researchers, laboratories, and companies.
ZFIN is the nomenclature authority for zebrafish genes and alleles, and provides official nomenclature support for the zebrafish community. This facilitates the discovery and knowledge integration of gene data, making these data accessible and reusable. The ZFIN Nomenclature Coordinator provides a core service of coordinating nomenclature with the HUGO Gene Nomenclature Committee (HGNC)1 and the Mouse Gene Nomenclature Committee (MGNC) to ensure that zebrafish nomenclature reflects a gene’s orthologous relationship to the mammalian (primarily human) genes. In addition, the ZFIN Nomenclature Coordinator works with the Zebrafish Nomenclature Committee (composed of active zebrafish researchers) to approve unique names and symbols for zebrafish loci and transgenic components used in research according to guidelines outlined at https://wiki.zfin.org/display/general/ZFIN+Zebrafish+Nomenclature+Guidelines.
ZFIN aims to make zebrafish research data Findable, Accessible, Interoperable, and Reusable (FAIR)2 by contributing to data annotation standards, the use of biomedical ontologies for data annotations, and persistent identifiers for annotations and metadata, as well as making data freely available at ZFIN.3–5 ZFIN has provided guidance to the zebrafish scientific community on minimum requirements for data submissions to ensure accurate integration with other data as well as encouraging FAIR standards.6,7 In an effort to support FAIR data principles further, ZFIN and the Mouse Genome Database,8 the Rat Genome Database,9 WormBase,10 FlyBase,11 and the Gene Ontology Consortium12 have collaborated to create the Alliance of Genome Resources (Alliance).13,14 The Alliance aims to provide a centralized multispecies platform that provides access to integrated, harmonized model organism data that facilitate the use of model organisms to understand the genetic and genomic basis of human biology and disease.15 Working in conjunction with the Alliance, ZFIN has continued work on data standardization, incorporating new data visualizations on the gene page, as well as automated textual gene descriptions, and providing zebrafish researchers access to multispecies gene expression, phenotype, human disease, and orthology data.
Gene page
Summary section update
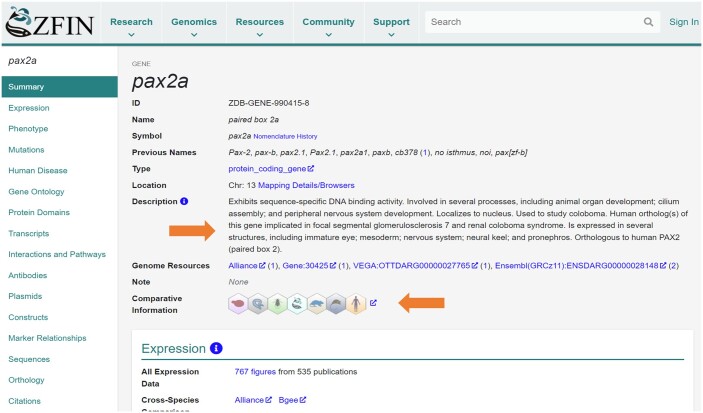
The gene page integrates and displays data pertinent to the gene, providing a comprehensive and current understanding of gene expression, mutant phenotypes, associated diseases, gene functions, alleles, and orthology. The layout and functionality of the gene page was recently updated to provide better navigation and usability.16 The ZFIN gene page has a layout similar to the Alliance gene page, providing users a comparable view and functionality that is helpful when navigating between sites. At the top of the gene page, the gene summary section provides basic information about the gene including symbol, links to nomenclature history, name, previously used aliases, textual gene description, and links to gene pages at other databases (Fig. 1). The textual gene description is new to the summary section of the gene page and is computationally generated and provided by the Alliance.17 In addition, comparative information was recently added to the summary section, displaying a hexagonal icon that represents the model organism databases of the Alliance. Clicking the icon directs users to the orthology section of the corresponding Alliance gene page, providing quick access to homologous gene information. The summary section is followed by sections that present data pertaining to the gene. A navigation pane on the left side of the page allows quick access to the data sections of interest. Currently, ZFIN contains data on 37,466 genes that are categorized as protein coding genes, ncRNA genes, or pseudogenes.
Fig. 1.
The ZFIN gene page. Web page providing information as it relates to a gene. The top portion provides general information about the gene, and the left navigation panel allows users to navigate directly to areas of interest. Arrows indicate new links or information provided by the Alliance.
Gene expression section update
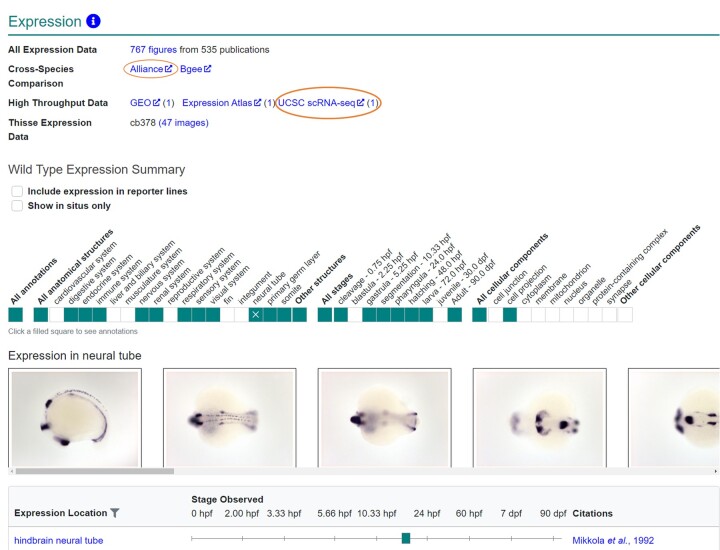
The “Expression” section on the gene page focuses primarily on the wild-type (WT) expression of the gene. This section begins with links to: (1) “All Expression Data” which links to a summary of gene expression in both WT and mutant fish; (2) external sites with cross-species comparison data; (3) links to high throughput data sets; and (4) Thisse large-scale WT screen18 (Fig. 2). Recently links to UCSC scRNAseq data were added to the high throughput data section, providing users access to the new single-cell RNA-seq atlas of zebrafish gene expression during organogenesis.19 Single-cell RNA-seq is a technique that has been increasingly used in zebrafish to understand transcriptional changes at the cellular level during development.20–22 These data, along with traditional in situ data that ZFIN annotates, help to provide an understanding of changes in gene expression during development. The WT expression summary provides a high-level visual summary ribbon that denotes the anatomical systems, stages, and GO cellular components (CCs) that have annotations and is discussed in more detail in the following section.
Fig. 2.
Expression section of the gene page. Section of the gene page providing information about where and when the gene is expressed. Links to all expression data, Alliance and Bgee for cross-species expression data, high throughput data, and Thisse18 expression data are provided. Circles indicate new links. The wild-type expression summary provides an overview of expression data, with color blocks indicating where annotations exist. Clicking on shaded boxes opens a table with more detailed gene expression information as shown.
ZFIN curates gene expression using gene symbols, genotypes and strains, the Zebrafish Experimental Conditions Ontology (ZECO),23 the Zebrafish Anatomical Ontology (ZFA),24 the Zebrafish Stage ontology (ZFS),24 GO cellular compartment ontology (GO-CC),25,26 and Spatial ontology (BSPO).27 Through the use of metadata identifiers and annotation standardization, ZFIN gene expression annotations comply with FAIR standards. ZFIN currently has 14,350 genes with expression data and has curated 216,696 zebrafish gene expression assays; of those 134,233 are in WT backgrounds with standard or control conditions. In addition, ZFIN has curated 31,964 transgenic reporter assays.
Expression, phenotype, gene ontology ribbons
The expression, phenotype, and gene ontology (GO) sections of the gene page utilize a ribbon diagram to visually summarize which anatomical systems, developmental stages, or GO biological process (BP), molecular function (MF), or cellular compartment terms have annotations (Figs. 2–4). The ribbon diagrams were recently added to ZFIN gene pages and are adapted from the ribbon diagrams displayed on Alliance gene pages.16 The terms used in the expression and phenotype summary ribbon were chosen to support grouping of annotations and are either hierarchically high-level ontology terms that subsume many biological concepts or terms that have been used in many annotations in ZFIN. For example, the anatomy portion of the ribbon summary uses high-level anatomical system terms from the ZFA ontology that have child terms that describe the anatomical structures that are part of the indicated anatomical system. The ribbon summary also lists terms that may not be child terms of an anatomical system but are of particular interest to zebrafish researchers and have a high number of expression or phenotype annotations. These include “fin,” “integument,” “neural tube,” “primary germ layer,” and “somite.” The “other structures” term groups all other annotations that use terms that are not child terms of an anatomical system.
The terms used for the GO ribbon are high-level terms that were chosen through a collaboration with the GO Consortium and the Alliance based on annotation coverage for model species represented at the Alliance. These high-level terms cover broad aspects of the BP, MF, and CC branches of GO and have a subset tag, “goslim_agr,” to specify that the term is a member of the GO Alliance subset. A subset of an ontology, or slim, is a trimmed down version of the ontology that contains a subset of terms and is useful for providing a high-level view of the content of an ontology. The Alliance subset is named “GO slim AGR subset” and can be downloaded at http://geneontology.org/docs/download-ontology/#subsets. The expression, phenotype, and GO ribbon diagrams have similar functionality, with shaded boxes denoting the presence of annotations for the indicated system, stage, BP, MF, or cellular compartment. Clicking on the shaded box opens a table view that provides more detailed annotations.
Phenotype section update
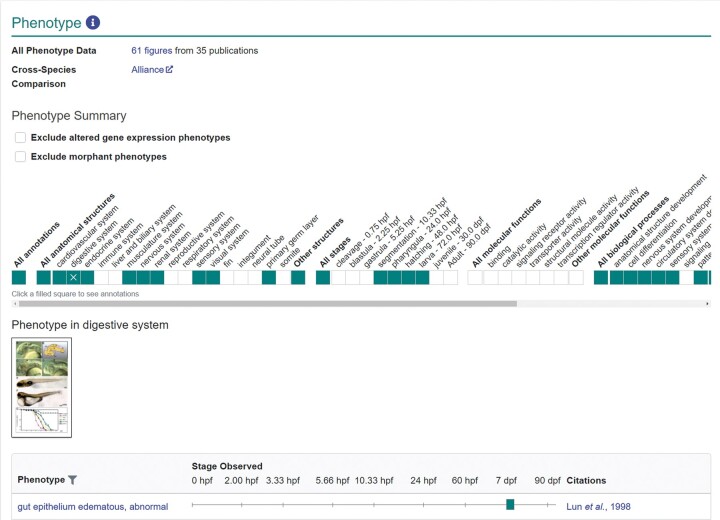
The “Phenotype” section of the gene page reports the phenotype of mutant and gene knockdown fish (Fig. 3). This section begins with a link to all phenotype data, which directs users to a phenotype figure summary page that lists the phenotype annotations with associated figures and publications for a gene. A new cross-species comparison section provides a link to the phenotype section of the Alliance gene page. The Phenotype Summary provides a high-level ribbon overview of the anatomical systems, stages, MFs, and BPs that have phenotype annotations.
Fig. 3.
Phenotype section of the gene page. Section of the gene page providing mutant and gene knockdown phenotype information. Links to Alliance gene page provided in Cross-Species Comparison section. The phenotype summary provides a high-level overview of systems, stages, MFs, and BPs that have annotations. Clicking shaded boxes opens a table where more detailed annotation information is provided, as shown. Image reproduced/adapted with permission. Lun K, Brand M. A series of no isthmus (noi) alleles of the zebrafish pax2.1 gene reveals multiple signaling events in development of the midbrain-hindbrain boundary. Development 1998 Aug;125(16):3049-62. doi: 10.1242/dev.125.16.3049. PMID: 9671579.
Phenotype annotations provide core insights into gene function and are utilized to facilitate the understanding of disease processes and outcomes. ZFIN annotates phenotype data using the Phenotype and Trait Ontology (PATO),28 ZFA, GO, and Chemical Entities of Biological Interest Ontology (ChEBI).29 A phenotype annotation is composed of the mutant or knockdown fish, the experimental conditions, the stage at which the phenotype is observed, the phenotype statement, and the associated publication. Phenotype statements are composed of entities and qualities assembled in the E + Q syntax30 using the aforementioned ontologies. To facilitate cross-species phenotype integration, ZFIN participates as a core member of the UPheno initiative, which aims to reconcile logical definitions across several model organism phenotype ontologies.31 This reconciliation work will benefit the Alliance as it moves toward providing ribbon diagrams for cross-species phenotype comparisons. ZFIN has curated 52,305 phenotype statements for single gene mutant or knockdown fish under standard or control conditions. There are 4,895 genes represented by 7,055 alleles with mutant phenotypes. Additionally, there are 2,241 genes that have no mutant alleles but have MO induced phenotypes and 245 genes that have phenotypes reported in animals injected with CRISPRs (CRISPants).
Mutations section update
The “Mutations” section of the gene page is intended to provide a high-level summary of the alleles and knockdown reagents that have been used to investigate gene function. As part of the gene page update, the tables in this section were updated from lists to data tables that provide more data and allow users to gain a better understanding of the allele or knockdown reagent without having to go to another web page for more information.32 The Mutations section has 2 tables, the “mutants” table, which summarizes information for the alleles of a gene, and “STR” table, which lists curated gene knockdown reagents such as morpholinos, CRISPRs, and TALENs. ZFIN facilitates FAIR standards for STR reagents by obtaining STR sequence information from publications, providing distinct nomenclature and identifiers, and reporting this information via user interfaces and download files. In addition, STR sequence targets are verified, and if errors are found authors are contacted to make corrections. Currently, ZFIN has records for 10,763 Morpholinos, 6,872 CRISPRs, and 813 TALENs. As the nomenclature authority for allele designations, ZFIN has made allele associations for 19,319 genes and has records for 53,522 alleles.
Human disease section update
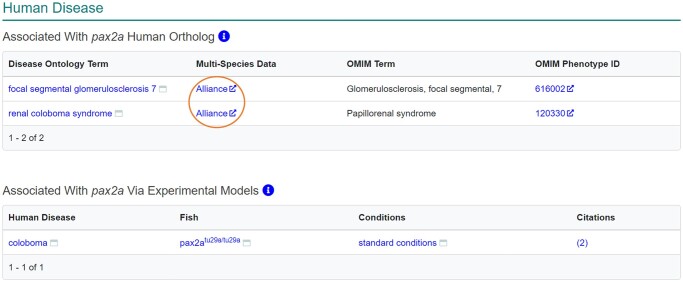
The “Human disease” section on the gene page provides 2 types of disease associations, diseases associated with zebrafish genes via orthology to human disease-causing genes and experimentally verified disease models (Fig. 4). The “Associated with the Human Ortholog” table lists disease ontology (DO) terms,33 which links to the ZFIN DO term page where more information can be found about the disease, as well as the OMIM disease name and links to the corresponding OMIM page.34 Recently, links to the Alliance Disease page were added to this table, directing users to the Alliance disease page that provides cross-species information about genes and alleles associated with disease as well as experimental models of disease. Information in the “Associated with Human Ortholog” table is produced by computational mappings of ZFIN curated orthologs to human genes and their disease associations from the genemap and mim2gene files from OMIM (https://omim.org/downloads/). This zebrafish gene to human gene/disease mapping is used to make associations between the ZFIN gene and the DO terms via DO term to OMIM disease mappings in the DO file.35 The “Associated Experimental Models” table lists curated experimental models of human disease. These models have been experimentally verified to model some or all aspects of a human disease. ZFIN currently has 1,900 curated disease models and 3,606 diseases associated with 5,154 zebrafish genes via orthology with 4,078 human genes. Individual DO term pages can be found via the search at the top of any ZFIN page.
Fig. 4.
Human disease section of the gene page. Provides information about human diseases associated with the gene via orthology to human genes and experimentally validated models of human disease. Links to Alliance disease pages (circled) provide access to multispecies disease information including associated genes, and experimental models.
Gene ontology section
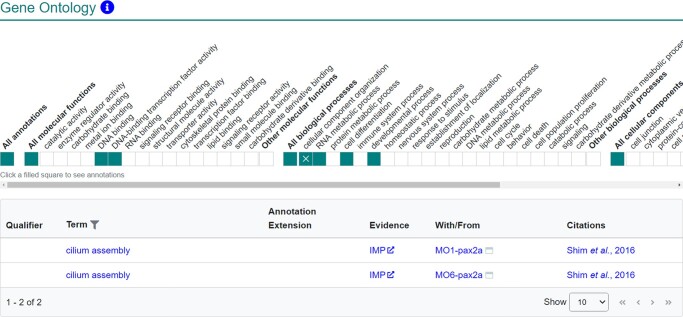
The “Gene ontology” section of the gene page reports the roles of a gene or gene product as described by the gene ontology (GO) term annotations for the gene. ZFIN manually produces GO annotations while curating publications.36 These annotations are created by associating a gene or gene product with GO terms that describe MFs that the gene product enables, the BP the gene product is involved in, and the CC where the gene product performs its function. ZFIN also downloads and displays GO annotations that are produced based on electronic annotation from InterPro2GO and UniProt.37,38 The GO section was recently updated to include a high-level summary ribbon view of the MF, BP, and CC annotations for a gene. Clicking on shaded boxes opens a table with more detailed GO annotations that include the annotated GO term, evidence code, with/from field, and citation (Fig. 5). ZFIN has 23,047 genes with GO annotations.
Fig. 5.
Gene ontology section of the gene page. This section provides an overview of the GO MF, BP, and CC annotations for a gene. Shaded boxes indicate presence of data; clicking shaded boxes, as shown, opens a table where more detailed annotation information is provided.
Orthology and other data sections
The “Orthology” section of the gene page provides an overview of the orthology relationships between zebrafish, human, and mouse genes. An expert curator using conserved synteny, gene family tree analysis, and amino acid alignments has vetted orthology displayed in this section. The primary resources used in the process of determining orthology and gene nomenclature are Ensembl,16 NCBI,39 and Panther.40 This level of orthology verification is necessary due to the whole genome duplication in the teleost lineage41 that causes many computational algorithms used for orthology to misidentify co-orthologs of mammalian genes. This section has new links to the orthology section of the zebrafish gene pages at the Alliance where the displays highlight computationally derived orthology and paralogy.
The other sections on the gene page provide additional information about the gene and links to related data both at ZFIN and other sites. The protein domains section contains 2 tables which provide data from InterPro.42 The domain, family, and site summary table lists protein binding sites, domains, and families with links to InterPro and the Domain Details Per Protein table denotes which domain/site/family is associated with each specific Uniprot entry.43 The transcript section links to several genome browsers and displays transcript diagrams, links to ZFIN transcript pages, and withdrawn transcripts. ZFIN links to SignaFish for interactions when known.44 Antibodies targeting the gene, plasmids with the coding region at Addgene, and constructs containing the gene are linked in their respective sections. There is also a section that links BACs, ESTs, and cDNAs associated with the gene as well as a section that links to sequences at GenBank and UniProt.
ZFIN data at the Alliance
ZFIN is a founding member of the Alliance of Genome Resources (alliancegenome.org),14 which has the primary mission of developing and maintaining comparative genome information resources that facilitate the use of multiple model organisms to understand the genetic and genomic basis of human biology and disease. As of the Alliance 4.1 release (August 2021, see Agapite et al. this issue), ZFIN contributes the following data to the Alliance: genome data, WT expression, phenotype data, mutant and transgenic alleles, variants, disease models, and orthology. Through the integration of model organism gene data, the Alliance provides comprehensive gene orthology data. The Alliance utilizes ZFIN data, and other model organism database data, to produce species-specific genome browsers that display the genome, genes, and variants. In addition, the Alliance uses ZFIN data to produce zebrafish-specific gene and allele pages, and provides links to data pages at ZFIN including the gene, allele, fish, and disease pages. ZFIN’s InterMine-based data mining database, ZebrafishMine, has been replaced by the InterMine instance developed at the Alliance, AllianceMine (https://www.alliancegenome.org/alliancemine).4,45,46 AllianceMine has gene, orthology, allele, gene ontology, and disease ontology data and search templates for zebrafish and other model organisms. AllianceMine will continue to be updated with additional ZFIN data and data types with each Alliance release. As discussed previously, reciprocal links are provided from ZFIN pages to appropriate Alliance data pages. The Alliance makes ZFIN data available along with data from all the participating model organism databases via either swagger APIs from the Alliance (https://www.alliancegenome.org/api/swagger-ui/) or as download files (https://www.alliancegenome.org/downloads).
ZFIN technical implementation
The ZFIN technical stack is substantially unchanged since our last publication.16 In brief the ZFIN web architecture is primarily written in Java using the Spring Framework and served by JSP in Apache/Tomcat. The ZFIN user interface is implemented using React, GWT, Angular, jQuery, and plain JavaScript. Groovy, SQL, and Perl are used to process and load bulk data. Hibernate serves as the object-relational mapping library from Java to the relational PostgreSQL database. ZFIN uses both Solr and Java/Spring to support our search interfaces. Data from papers are entered via a web-based curation interface primarily written in GWT with a few implementations of AngularJS interfaces. The community wiki is powered by Atlassian Confluence software (http://www.atlassian.com/software/confluence/). A detailed and browsable view of the current ZFIN data model can be found at http://zfin.org/schemaSpy/index.html. Our current plan is to move from our servers to cloud servers to provide dynamic response to load and smooth our server costs.
Data availability
As the knowledgebase for the zebrafish model organism research community, one of the main goals for ZFIN is to make the data that ZFIN annotates and integrates as accessible as possible. As discussed earlier, ZFIN makes integrated data available on the Gene Page. ZFIN also produces data-specific web pages for alleles, fish, antibodies, constructs, STR, clones, publications, and ontology terms where information that is more detailed can be found for each data type. In addition, data at ZFIN can be searched using the single box search as well as dedicated search forms for specific data types. For computational exploration of data, ZFIN maintains a downloads page that provides access to data-specific download files (https://zfin.org/downloads) and archives https://zfin.org/downloads/archive. ZFIN also can provide custom download files upon request. Data provided in download files are FAIR compliant denoting the date the file was generated, as well as all identifiers, symbols, names, and ontology terms. As previously noted, ZFIN data are also available via AllianceMine, API, and download files at the Alliance.
Funding
This study was supported by National Human Genome Research Institute at the US National Institutes of Health [U41 HG002659 (ZFIN) and U24 HG010859 (Alliance of Genome Resources)].
Conflicts of interest
None declared.
Literature Cited
- 1. Povey S, Lovering R, Bruford E, Wright M, Lush MWain, H.. The HUGO Gene Nomenclature Committee (HGNC). Hum Genet. 2001;109:678–680. [DOI] [PubMed] [Google Scholar]
- 2. Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. Comment: the FAIR guiding principles for scientific data management and stewardship. Sci Data. 2016;3(1). doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Köhler S, Doelken SC, Ruef BJ, Bauer S, Washington N, Westerfield M, Gkoutos G, Schofield P, Smedley D, Lewis SE, et al. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research. 2013;2:30. doi: 10.12688/f1000research.2-30.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Slyke CE, Bradford YM, Howe DG, Fashena DS, Ramachandran S, Ruzicka L, ZFIN Staff. Using ZFIN: Data Types, Organization, and Retrieval. In: Methods in Molecular Biology, NIH Public Access; 2018. p. 307–347. [DOI] [PMC free article] [PubMed]
- 5. Vasilevsky NA, Brush MH, Paddock H, Ponting L, Tripathy SJ, LaRocca GM, Haendel MA.. On the reproducibility of science: unique identification of research resources in the biomedical literature. PeerJ. 2013;1:e148. doi: 10.7717/peerj.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Howe DG, Frazer K, Fashena D, Ruzicka L, Bradford Y, Ramachandran S Ruef BJ, Van Slyke C, Singer A, Westerfield W. Data extraction, transformation, and dissemination through ZFIN. Methods Cell Biol. 2011;104:311–325. doi: 10.1016/B978-0-12-374814-0.00017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howe DG, Bradford YM, Eagle A, Fashena D, Frazer K, Kalit P, Mani P, Martin R, Moxon ST, Paddock H, et al. A scientist’s guide for submitting data to ZFIN. Methods Cell Biol. 2016;135:451–481. doi: 10.1016/bs.mcb.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, Anagnostopoulos A, Asabor R, Baldarelli RM, Beal JS, Bello SM, et al. ; the Mouse Genome Database Group. Mouse Genome Database (MGD) 2019. Nucleic Acids Res. 2019b;47(D1):D801–D806. doi: 10.1093/nar/gky1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Laulederkind SJF, Hayman GT, Wang SJ, Smith JR, Petri V, Hoffman MJ, De Pons J, Tutaj MA, et al. A primer for the rat genome database (RGD). Methods Mol Biol. 2018;1757:163–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee RYN, Howe KL, Harris TW, Arnaboldi V, Cain S, Chan J, Chen WJ, Davis P, Gao S, Grove C, et al. WormBase 2017: molting into a new stage. Nucleic Acids Res. 2018;46(D1):D869–D874. doi: 10.1093/nar/gkx998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. ; the FlyBase Consortium. FlyBase 2.0: the next generation. Nucleic Acids Res. 2019;47(D1):D759–D765. doi: 10.1093/nar/gky1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carbon S, Douglass E, Dunn N, Good B, Harris NL, Lewis SE, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, et al. The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47:D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Agapite J, Albou L-P, Aleksander S, Argasinska J, Arnaboldi V, Attrill H, Bello SM, Blake JA, Blodgett O, Bradford YM, et al. ; The Alliance of Genome Resources Consortium. Alliance of Genome Resources Portal: unified model organism research platform. Nucleic Acids Res. 2020;48(D1):D650–D658. doi: 10.1093/nar/gkz813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bult CJ, Blake JA, Calvi BR, Cherry JM, DiFrancesco V, Fullem R, Howe KL, Kaufman T, Mungall C, Perrimon N, et al. The alliance of genome resources: building a modern data ecosystem for model organism databases. Genetics. 2019a;213:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Howe DG, Blake JA, Bradford YM, Bult CJ, Calvi BR, Engel SR, Kadin JA, Kaufman TC, Kishore R, Laulederkind SJF, et al. Model organism data evolving in support of translational medicine. Lab Anim. 2018;47(10):277–289. doi: 10.1038/s41684-018-0150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Howe DG, Ramachandran S, Bradford YM, Fashena D, Toro S, Eagle A, Frazer K, Kalita P, Mani P, Martin R, et al. The zebrafish information network: major gene page and home page updates. Nucleic Acids Res. 2021b;49(D1):D1058–D1064. doi: 10.1093/nar/gkaa1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kishore R, Arnaboldi V, Van Slyke CE, Chan J, Nash RS, Urbano JM, Dolan ME, Engel SR, Shimoyama M, Sternberg PW, et al. Automated generation of gene summaries at the Alliance of Genome Resources. Database. 2020;2020:baaa037. doi: 10.1093/database/baaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Thisse B, Pflumio S, Fürthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis (NIH R01 RR15402). ZFIN Direct Data Submission; 2001. https://zfin.org/ZDB-PUB-010810-1
- 19. Farnsworth DR, Saunders LM, Miller AC.. A single-cell transcriptome atlas for zebrafish development. Dev Biol. 2020;459(2):100–108. doi: 10.1016/j.ydbio.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF.. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. 2018;360(6392):eaar3131. doi: 10.1126/science.aar3131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang Q, Iyer S, Lobbardi R, Moore JC, Chen H, Lareau C, Hebert C, Shaw ML, Neftel C, Suva ML, et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J Exp Med. 2017;214(10):2875–2887. doi: 10.1084/jem.20170976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM.. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science. 2018;360(6392):981–987. doi: 10.1126/science.aar4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bradford YM, Van Slyke CE, Toro S, Ramachandran S. The zebrafish experimental conditions ontology: systemizing experimental descriptions in ZFIN. In: CEUR Workshop Proceedings; August 2016, Corvallis, OR.
- 24. Van Slyke CE, Bradford YM, Westerfield M, Haendel MA.. The zebrafish anatomy and stage ontologies: representing the anatomy and development of Danio rerio. J Biomed Semantics. 2014;5(1):12.doi: 10.1186/2041-1480-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Carbon S, Douglass E, Good BM, Unni DR, Harris NL, Mungall CJ, Basu S, Chisholm RL, Dodson RJ, Hartline E, et al. ; The Gene Ontology Consortium. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dahdul WM, Cui H, Mabee PM, Mungall CJ, Osumi-Sutherland D, Walls RL, Haendel MA.. Nose to tail, roots to shoots: spatial descriptors for phenotypic diversity in the biological spatial ontology. J Biomed Semantics. 2014;5(1):34–13. doi: 10.1186/2041-1480-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gkoutos GV, Green ECJ, Mallon AM, Hancock JM, Davidson D.. Using ontologies to describe mouse phenotypes. Genome Biol. 2004;6(1):R8.doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hastings J, Owen G, Dekker A, Ennis M, Kale N, Muthukrishnan V, Turner S, Swainston N, Mendes P, Steinbeck C, et al. ChEBI in 2016: improved services and an expanding collection of metabolites. Nucleic Acids Res. 2016;44(D1):D1214–D1219. doi: 10.1093/nar/gkv1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Washington NL, Haendel MA, Mungall CJ, Ashburner M, Westerfield M, Lewis SE.. Linking human diseases to animal models using ontology-based phenotype annotation. PLoS Biol. 2009;7(11):e1000247. doi:10.1371/journal.pbio.1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matentzoglu N, Balhoff JP, Bello SM, Boerkoel CF, Bradford YM, Carmondy LC, Cooper LD, Grove CA, Harris NL, Kohler S, et al. Phenotype Ontologies Traversing All the Organisms (POTATO) Workshop Aims to Reconcile Logical Definitions across Species; August, 2018, Corvallis, OR. doi: 10.5281/ZENODO.2382757. [DOI] [Google Scholar]
- 32. Schriml LM, Mitraka E, Munro J, Tauber B, Schor M, Nickle L, Felix V, Jeng L, Bearer C, Lichenstein R, et al. Human Disease Ontology 2018 update: classification, content and workflow expansion. Nucleic Acids Res. 2019;47(D1):D955–D962. doi: 10.1093/nar/gky1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A.. OMIM.org: online Mendelian Inheritance in Man (OMIM®), an Online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43(D1):D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bradford YM, Toro S, Ramachandran S, Ruzicka L, Howe DG, Eagle A, Kalita P, Martin R, Taylor Moxon SA, Schaper K, et al. Zebrafish models of human disease: gaining insight into human disease at ZFIN. ILAR J. 2017;58(1):4–16. doi: 10.1093/ilar/ilw040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2007;36(Database):D768–D72. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dimmer EC, Huntley RP, Alam-Faruque Y, Sawford T, O'Donovan C, Martin MJ, Bely B, Browne P, Mun Chan W, Eberhardt R, et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 2012;40(D1):D565–D570. doi: 10.1093/NAR/GKR1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang H-Y, Dosztányi Z, El-Gebali S, Fraser M, et al. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 2017;45(D1):D190–D199. doi: 10.1093/nar/gkw1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Howe KL, Achuthan P, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, Bhai J, et al. Ensembl 2021. Nucleic Acids Res. 2021a;49(D1):D884–D891. doi: 10.1093/nar/gkaa942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Agarwala R, Barrett T, Beck J, Benson DA, Bollin C, Bolton E, Bourexis D, Brister JR, Bryant SH, Canese K, et al. ; NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2018;46(D1):D8–D13. doi: 10.1093/nar/gkx1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A.. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Postlethwait J, Amores A, Cresko W, Singer A, Yan YL.. Subfunction partitioning, the teleost radiation and the annotation of the human genome. Trends Genet. 2004;20(10):481–490. doi: 10.1016/j.tig.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 42. Blum M, Chang H-Y, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, et al. The InterPro protein families and domains database: 20 years on. Nucleic Acids Res. 2021;49(D1):D344–D354. doi: 10.1093/nar/gkaa977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bateman A, Martin M-J, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, et al. The UniProt Consortium; UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Csályi K, Fazekas D, Kadlecsik T, Türei D, Gul L, Horváth B, Módos D, Demeter A, Pápai N, Lenti K, et al. SignaFish: a Zebrafish-specific signaling pathway resource. Zebrafish. 2016;13(6):541–544. doi: 10.1089/zeb.2016.1277. [DOI] [PubMed] [Google Scholar]
- 45. Kalderimis A, Lyne R, Butano D, Contrino S, Lyne M, Heimbach J, Hu F, Smith R, Stepan R, Sullivan J, Micklem G. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42. doi: 10.1093/nar/gku301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smith RN, Aleksic J, Butano D, Carr A, Contrino S, Hu F, Lyne M, Lyne R, Kalderimis A, Rutherford K, et al. Databases and ontologies InterMine: a flexible data warehouse system for the integration and analysis of heterogeneous biological data. 2012;28(23):3163–3165. doi: 10.1093/bioinformatics/bts577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
As the knowledgebase for the zebrafish model organism research community, one of the main goals for ZFIN is to make the data that ZFIN annotates and integrates as accessible as possible. As discussed earlier, ZFIN makes integrated data available on the Gene Page. ZFIN also produces data-specific web pages for alleles, fish, antibodies, constructs, STR, clones, publications, and ontology terms where information that is more detailed can be found for each data type. In addition, data at ZFIN can be searched using the single box search as well as dedicated search forms for specific data types. For computational exploration of data, ZFIN maintains a downloads page that provides access to data-specific download files (https://zfin.org/downloads) and archives https://zfin.org/downloads/archive. ZFIN also can provide custom download files upon request. Data provided in download files are FAIR compliant denoting the date the file was generated, as well as all identifiers, symbols, names, and ontology terms. As previously noted, ZFIN data are also available via AllianceMine, API, and download files at the Alliance.