Abstract
GABAA receptors mediate rapid responses to the neurotransmitter gamma-aminobutyric acid and are robust regulators of the brain and spinal cord neural networks that control locomotor behaviors, such as walking and swimming. In developing zebrafish, gross pharmacological blockade of these receptors causes hyperactive swimming, which is also a feature of many zebrafish epilepsy models. Although GABAA receptors are important to control locomotor behavior, the large number of subunits and homeostatic compensatory mechanisms have challenged efforts to determine subunit-selective roles. To address this issue, we mutated each of the 8 zebrafish GABAA α subunit genes individually and in pairs using a CRISPR-Cas9 somatic inactivation approach and, then, we examined the swimming behavior of the mutants at 2 developmental stages, 48 and 96 h postfertilization. We found that disrupting the expression of specific pairs of subunits resulted in different abnormalities in swimming behavior at 48 h postfertilization. Mutation of α4 and α5 selectively resulted in longer duration swimming episodes, mutations in α3 and α4 selectively caused excess, large-amplitude body flexions (C-bends), and mutation of α3 and α5 resulted in increases in both of these measures of hyperactivity. At 96 h postfertilization, hyperactive phenotypes were nearly absent, suggesting that homeostatic compensation was able to overcome the disruption of even multiple subunits. Taken together, our results identify subunit-selective roles for GABAA α3, α4, and α5 in regulating locomotion. Given that these subunits exhibit spatially restricted expression patterns, these results provide a foundation to identify neurons and GABAergic networks that control discrete aspects of locomotor behavior.
Keywords: locomotion, zebrafish, GABA receptors, CRISPR
Introduction
Neural networks in the vertebrate hindbrain and spinal cord rely upon a balance of excitatory and inhibitory neurotransmitter systems to orchestrate locomotion. Classically inhibitory, the neurotransmitter gamma-aminobutyric acid (GABA) is recognized as a key regulator of these circuits. GABA exerts its effects through two different classes of receptors, GABAA and GABAB. GABAB receptors are G-protein-coupled receptors, while GABAA receptors are ligand-gated ion channels that generate rapid responses to GABA. In mammalian systems, GABAA receptors exhibit remarkable diversity, with each receptor thought to form a heteropentamer containing various combinations of 19 different subunits: α1–6, β1–3, γ1–3, δ, ε, π, θ, and ρ1–3 (Sieghart and Sperk 2002; Simon et al. 2004; Chua and Chebib 2017). Each subunit is encoded by a discrete gene that is spatially and developmentally regulated to generate distinct, but sometimes overlapping, expression patterns (Laurie et al. 1992a, 1992b; Wisden et al. 1992). Several receptor subunits confer distinct biophysical and pharmacological properties, localize to synaptic or extrasynaptic sites, interact with specific cytoplasmic proteins, and contribute to different neuronal networks (Farrant and Nusser 2005; Jacob et al. 2008; Fritschy and Panzanelli 2014). While this incredible receptor heterogeneity is not fully understood, it could provide the opportunity to better understand, or even manipulate, distinct neuronal networks.
Several studies have used pharmacological blockade of GABAA receptors to reveal the central roles that these receptors play in regulating the initiation, rhythmicity, frequency, and duration of locomotor network output from the spinal cord. These studies have been performed in a variety of vertebrate systems. For example, in neonatal mice, application of GABAA receptor antagonists to the spinal cord has been shown to cause inappropriate, bilateral discharges and regulate the onset and duration of rhythmic activity (Hinckley et al. 2005). In lamprey, application of GABAA receptor antagonists to spinal cord networks increased the frequency of locomotor bursts and disrupted lengthwise coordination (Schmitt et al. 2004). In Xenopus tadpoles, GABAA receptor-mediated inhibition has been found to play both a tonic role in regulating the responsiveness to touch stimuli and a phasic role that stops swimming (Perrins et al. 2002; Lambert et al. 2004a, 2004b). Although these and several other studies illustrate the importance of GABAA receptors in controlling locomotor networks, antagonists that block the majority of receptor isoforms were used, therefore these studies are less informative about the roles of specific subunits.
Genetic inactivation of GABAA receptor subunits in mice has had only limited success in identifying subunits required to mediate locomotor behavior. Although several of the 19 subunits exhibit robust expression in portions of the brain and spinal cord that mediate locomotion, few gene deletions have been found to cause abnormal locomotor behavior (Rudolph and Möhler 2004; Vicini and Ortinski 2004; Smith and Rudolph 2012). Ablation of some GABAA receptor genes has been shown to cause changes in the expression of other subunits, which suggests that homeostatic adaptations may explain at least some of the deletions that show no or only subtle locomotor phenotypes (Peng et al. 2002; Kralic et al. 2006; Zeller et al. 2008; Panzanelli et al. 2011; Zhou et al. 2013; Fritschy and Panzanelli 2014).
Early larval-stage zebrafish, ∼2–10 days postfertilization (dpf), are a leading model for locomotor neural network analysis, and GABAA receptors regulate locomotion in this system. Bath application of GABAA receptor antagonists, such as pentylenetetrazole (PTZ), induce hyperactive swimming in the form of exaggerated movements and extended swimming episodes. This hyperactive swimming, and the presence of epileptiform discharges, is also established as an epileptic seizure model (Baraban et al. 2005; Baxendale et al. 2012; Cho et al. 2020). Zebrafish harbor an array of GABAA subunits similar to mammals, with 23 identified subunits (Cocco et al. 2017; Monesson-Olson et al. 2018; Sadamitsu et al. 2021); however, few mutants have been identified as important for locomotion. At 5 dpf, loss-of-function mutations in the broadly expressed γ2 subunit were reported to elicit hyperactive swimming that is behaviorally similar to PTZ exposure (Liao et al. 2019). Also, at 5 dpf, morpholino knockdown of α1 was reported to cause a hypomotile phenotype at 5 dpf but no larval behavioral phenotype was detected in α1 loss-of-function mutants (Samarut et al. 2018; Reyes-Nava et al. 2020). Instead, these mutants performed hyperactive swimming at 5 weeks postfertilization. The apparent discrepancy in these results is likely due to the different methods used to disrupt α1 expression. At 7 dpf, loss-of-function mutations in the widely expressed β3 subunit were observed to cause subtle increases in spontaneous swimming (Yang et al. 2019). However, in possible disagreement with these results, Griffin et al. did not detect a behavioral phenotype in β3 mutants at 8 dpf (Griffin et al. 2021). Since GABAA α subunits help form the GABA binding site, they are thought to be obligatory receptor components (Phulera et al. 2018; Zhu et al. 2018; Laverty et al. 2019; Masiulis et al. 2019). Thus, it is surprising that few α subunits have been identified that control embryonic or larval locomotor behavior. It is possible that, as in mammals, homeostatic compensation is able to conceal subtype-selective roles. Disrupting multiple subunits simultaneously could evade these mechanisms and reveal the α subunits required to regulate swimming behavior.
Here, we used an F0 CRISPR-Cas9, somatic mutation approach to screen the locomotor phenotypes of mutants in each of the 8 α subunits both individually and in combination at 2 different developmental stages: 48 and 96 h postfertilization (hpf). We found that disrupting select pairs of α subunits causes different types of hyperactive behavior at 48 hpf, which then dramatically decreased or was absent by 96 hpf. The absence of hyperactive behavior by 96 hpf was confirmed in selected F2 germline mutants and, correspondingly, electrophysiological recordings revealed brain activity indistinguishable from wild-type controls. These findings illustrate subunit selective roles of GABAA receptor α subunits in regulating locomotor behavior which, given their restricted expression patterns in larvae, serve as entry points to reveal cellular and circuit mechanisms that enable GABA to control locomotion.
Materials and methods
Zebrafish maintenance and breeding
Adult zebrafish were maintained according to standard procedures, with the zebrafish facility on a 14-h light/10-h dark cycle. Embryos and larvae were kept at 28.5°C in E3 media and staged according to morphological criteria (Kimmel et al. 1995; Parichy et al. 2009). All genetic manipulations and behavioral experiments were performed using a Tübingen (Tü) or Tüpfel longfin (TLF) genetic background. All animal procedures for this study were approved by the University of Massachusetts Amherst or the Amherst College Institutional Animal Care and Use Committees (IACUC) under assurance numbers 3551-1 and 3925-1, respectively, with the Office of Laboratory Animal Welfare.
Guide and Cas9 RNA preparation and microinjection
Single guide RNAs were designed using the online tool, ChopChop v3 (Labun et al. 2019) (Supplementary Table 1). ChopChop selects target sites for CRISPR-Cas9 using the NGG motif and ranks them based on efficiency (Montague et al. 2014). For each gene, two neighboring targets with high efficiency were selected within exons that would disrupt all known splice variants as assessed in ENSEMBL. CRISPR-Somatic Tissue Activity Tests (CRISPR-STAT) were used to assess mutational efficiency for both targets of a selected gene, and the target that yielded better results was used for phenotypic analysis. In some cases, neither of the initial targets was effectively mutated so two additional targets were selected.
Template DNA for gRNA synthesis was generated using the PCR-based method described in (Shah et al. 2016). For in vitro transcription, we first generated an HPLC-purified scaffolding primer (5′-GATCCGCACCGACTCGGTCCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3′), which was common to all gRNA templateless PCRs. Next, for each target, we synthesized a unique primer sequence that contained a 5′ T7 binding site, the 20 nucleotide specific target (Supplementary Table 1), and a 3′ 20 nucleotide site of scaffolding homology (5′-AATTAATACGACTCACTATA-[20 nucleotide Target Sequence]-GTTTTAGAGCTAGAAATAGC-3′). The PCR contained: 0.4 units of Phusion High-Fidelity DNA Polymerase (New England Biolabs, M0530S), 13.4 μL ddH2O, 1 μL target specific primer (10 μM, IDT), 1 μL scaffolding primer (10 μM, IDT), 4 μL 5× Phusion HF, and 0.4 μL dNTPs (10 mM). Reactions were run in a thermocycler (BioRad) using the following conditions: 98°C for 30 s then 40 cycles of 98°C for 10 s, 60°C for 10 s, 72°C for 15 s, which was followed by 72°C for 10 min. The PCR was purified using the QIAGEN MinElute kit and used as a template for in vitro transcription reactions. Using 0.5–1 μg of purified PCR product, gRNAs were generated using the MEGAscript T7 Transcription kit (Thermofisher), purified via lithium chloride precipitation, and verified using a TAE denaturing gel. A nanodrop spectrometer was used to determine gRNA concentrations, which were then diluted to 200 ng/μl in RNase free water.
Cas9 mRNA was synthesized similar to Shah et al. (2016), but with the following changes. Purification of the linearized plasmid was performed using the E.Z.N.A. Cycle Pure Kit (Omega Bio-Tek), while purification of the Cas9 mRNA was by lithium chloride precipitation. Cas9 mRNA was diluted to 1200 ng/μL in RNase free water.
Microinjections were performed at the 1–4 cell stage into the yolk of embryonic zebrafish. The injection cocktail contained: 2 μL gRNA at 200 ng/μL, 2 μl Cas9 mRNA at 1200 ng/μL, 1 μL stop cassette primer (Gagnon et al. 2014) at 10 μM, 2 μL 0.05% phenol red, and 4 μL RNase-free water. Approximately 500 pL was injected per embryo. Mock injections, containing all cocktail ingredients except gRNAs, were not observed to have a significant effect compared to uninjected siblings, so uninjected sibling animals were used as the controls for most experiments.
Tyrosinase pigmentation analysis
Twenty-four hours after injection, embryos were dechorionated using forceps and screened for morphological abnormalities using a dissecting microscope (Zeiss). Morphologically abnormal fish were excluded from further analysis. At 48 and 96 hpf, larvae were anesthetized using 0.04% MS-222 (Tricaine) and pictures were taken using a Stemi 305 (Zeiss). All images were captured with the same resolution (1280 × 960), magnification, lighting conditions, and exposure time. Images were analyzed using a custom image processing pipeline implemented in Python (source code available upon request). Briefly, images were converted from color to grayscale and pixel intensities were scaled to fit between 0.0 (black) and 1.0 (white). A pixel intensity threshold was used to segment pigmentation from the background and other parts of the fish. The optimal threshold was empirically determined to be 0.7 multiplied by the mean pixel intensity computed across all images. The number of pixels with intensities falling below the threshold, representing pigmentation, were reported for each image. All images were processed using the same threshold value.
Behavioral analysis
Behavioral analysis was performed in a double-blind fashion. At 24 hpf, injected embryos had their chorions removed and were subject to morphological screening. At 48 hpf, escape responses to touch were examined, similar to as described in Friedrich et al. (2012) and McKeown et al. (2012). Briefly, light touch was applied to the head using a 3.22/0.16 g of force von Frey filament. Swimming responses were captured using a high-speed digital camera (XStream 1024, IDT Vision) mounted to a 35 mm lens (Nikon) at a frame rate of 250 Hz. n values refer to the number of animals, with each animal tested only once at each time point. The head-to-tail angle for each frame of the response was measured using custom software written in MATLAB (https://github.com/DownesLab/Zebrafish-Movement-Analyzer-ZeMovA- (last accessed: 18 February 2022)). C-bends were defined as any body flexion over 110°, while escape response duration was defined as beginning the frame before initial movement was observed and ending at the last frame of detected movement (McKeown et al. 2012). Larvae were kept in individual wells of 24-well plates and tested again at 96 hpf.
CRISPR-STAT
CRISPR-STAT analysis was performed similar to as described by Carrington et al. (2015). Primer pairs were designed to flank the targeting gRNA sites as determined in ChopChop span target sequences (Supplementary Table 1). Forward primers were tagged with a 6-FAM dye (Integrated DNA Technologies) which allowed for visualization in fragment analysis. Reverse primers were tagged with a PIG-tail adapter (5′-GTGTCTT-3′) to reduce stutter. DNA was extracted from 6 embryos using the Extract-N-Amp Tissue PCR Kit (Millipore-Sigma) and amplified using Amplitaq Gold Taq Polymerase (ThermoFisher). Reactions were run in a thermocycler (BioRad) using the following conditions: 94°C for 12 min then 40 cycles of 94°C for 30 s, 53°C for 30 s, 72°C for 30 s, which was followed by 72°C for 10 min. DNA was diluted 1:20 in ddH20 and run on an AB3730xl DNA Analyzer (Genewiz, South Plainfield, NJ, USA). Fragment analysis was performed using Geneious Software (Biomatters, Inc). Peaks were defined as signals exceeding 1,000 Relative Fluorescent Units (RFU). A gene target was considered successfully mutated if the peak ratio was 2 or above.
α3 and α4 mutant lines
To generate α3 and α4 mutant lines, CRISPR-Cas9 injected animals were raised to adulthood. CRISPR-STAT analysis was used to identify mosaic animals and these animals were crossed to a wild-type strain (TLF). Two α3 mutant alleles were identified in the F1 animals, a 7-base pair (bp) deletion, uma500, and an 18-bp insertion, uma501 (see Supplementary Fig. 2). A single α4 mutant allele was identified in the F1 animals, an 11 bp deletion, uma504.
Local field potential recording
Local field potential (LFP) recordings were obtained from zebrafish larvae at 96 hpf using a technique similar to that in Liu and Baraban (2019). Prior to each recording, larvae were paralyzed by immersion for 30–60 min in α-bungarotoxin (125 µM in dH20, Invitrogen, Waltham, MA, USA) and subsequently embedded dorsal side up in 2% low-melting point agarose in extracellular solution (130 mM NaCl, 10 mM HEPES, 2 mM KCl, 2 mM CaCl2, 1 mM MgCl2, pH 7.8). In some experiments, the convulsant agent PTZ (10 mM) was applied to induce ictal-like brain activity. For each recording, a glass microelectrode (6–12 MΩ) was filled with extracellular solution and inserted under visual guidance into the optic tectum. LFPs were recorded at 100X gain in current clamp mode using a Sutter Double IPA amplifier (Sutter Instruments, Novato, CA, USA). Voltage signals were low-pass filtered at 500 Hz–1 kHz and digitized at 5–10 kHz using SutterPatch Software (Sutter Instruments, Novato, CA, USA). Following acquisition, voltage traces were analyzed for ictal-like activity using NeuroMatic software (Rothman and Silver 2018). The number of ictal-like events in the first 30 min of recording was assessed by threshold detection (0.15 mV) from a moving 1-s baseline window using onset and peak features in the event detection program. For PTZ treated fish, 1 h was allocated for wash-on and only the first 30 min of recording after this wash-on period was analyzed.
Statistical analysis
To determine significant differences the following statistical tests and software were used. Welch’s t-test and Ordinary 1-way ANOVA were used as indicated. When t-tests were applied, F tests were used to compare variance. When ANOVAs were applied, multiple comparison tests were used where test groups were compared against wild-type controls. A Dunnett test was used to correct against familywise error. Statistical tests were performed and plots and figures were generated using Prism (GraphPad Software).
Results
High-efficiency F0 somatic gene targeting using CRISPR-Cas9
To identify GABAA α subunits that control larval escape behavior, we sought an approach to rapidly screen through different loss-of-function mutant combinations. Injecting zebrafish embryos with a cocktail containing Cas9-encoding and guide RNAs (gRNAs) has been shown to mutate target genes with enough efficiency to cause biallelic disruption (Shah et al. 2015; Varshney et al. 2015; Wu et al. 2018). Phenotypes can often be observed in these F0 somatic mutants, which can save a great deal of time compared to analysis of F2 germline homozygous mutants. A disadvantage of this approach is that F0 somatic mutants are genetic mosaics, with different cells harboring different indels in the target gene, which may yield weaker phenotypes compared to those found in germline mutants. To confirm the efficiency of disrupting gene function using this approach, we targeted the tyrosinase (tyr) gene, similar to previous studies (Wu et al. 2018). Tyrosinase is essential for producing melanin and its disruption provides an easily observable loss of pigmentation. Phenylthiourea (PTU) is routinely used in zebrafish research to suppress melanin synthesis and it provides an effective method to evaluate CRISPR knockdown of tyr. We observed that injection of Cas9 and gRNAs targeting the tyr gene led to a substantial and persistent reduction in pigmentation in 48 and 96 hpf larvae, although not all melanin synthesis was eliminated compared to PTU-treated controls (Supplementary Fig. 1, a and b). Correspondingly, PCR analysis of the somatic mutants through CRISPR-STAT indicated that a variety of mutations were induced in the tyr gene (Supplementary Fig. 1c). Taken together, these results confirm that F0 somatic mutants provide a rapid and effective means to screen gene function.
GABAA receptors control early larval swimming behavior
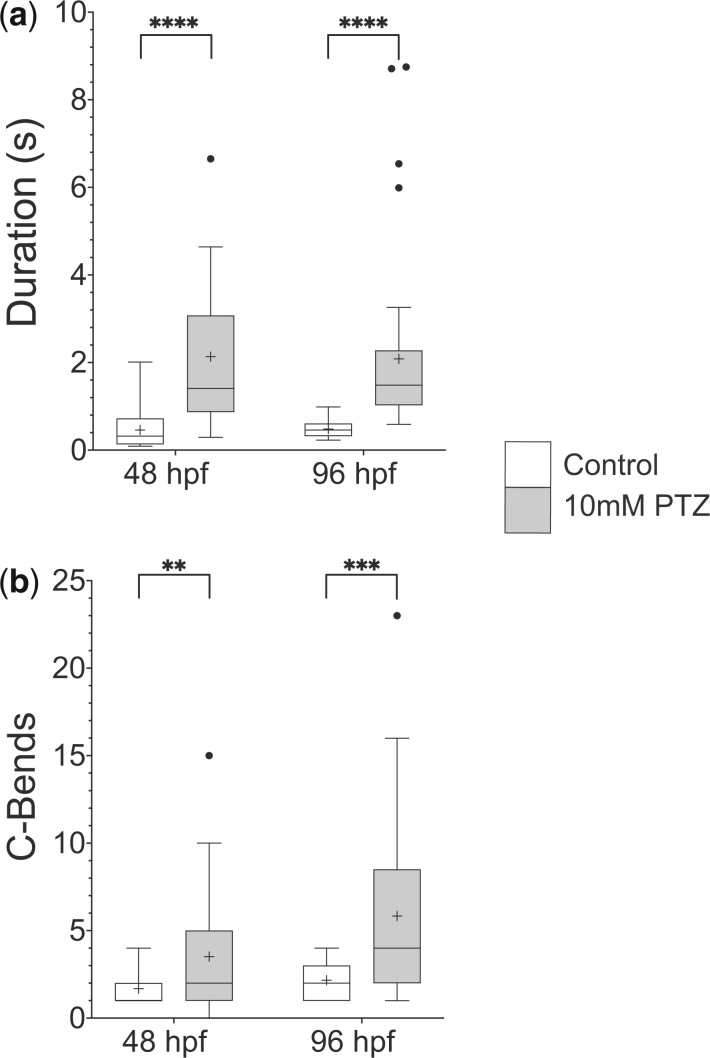
Although PTZ is known to elicit hyperactive swimming responses in fish older than 5 dpf, its effect on earlier larval stages is less clear. We focused behavioral analysis on 48 and 96 hpf larvae since these time points present the opportunity to analyze a more nascent zebrafish nervous system and the early stages of GABAA receptor control of locomotion. At 48 hpf, larvae are newly hatched and demonstrate burst swimming behavior, while 96 hpf larvae exhibit more mature swimming patterns (Brustein et al. 2003; McKeown et al. 2009; Roussel et al. 2020). Because acoustic and light responses are either absent or less robust at these early developmental stages, gentle touch was used to induce escape behavior. At both 48 and 96 hpf, larvae respond to touch to the head with a well-characterized C-start, which consists of an initial C-shaped body bend to reorient the animal away from the touch stimulus, followed by lower-amplitude body undulations that propel it several body lengths away (Eaton et al. 1977; Granato et al. 1996; O’Malley et al. 1996; Eaton et al. 2001; Kohashi et al. 2012). After PTZ exposure, hyperactive behavior is observed, and two prominent aspects of swimming performance are altered similar to older fish: larvae performed longer duration escape responses and these responses are interspersed with multiple C-shaped body bends (Fig. 1 and Supplementary Movies 1 and 2). These results indicate that GABAA receptors are essential to regulate swimming behavior during escape responses in early larval zebrafish, as has been previously shown for later stages of development.
Fig. 1.
PTZ exposure induces hyperactive swimming in early stage larval zebrafish. PTZ is a potent GABAA receptor antagonist. Bath application of 10 mM PTZ to both 48 and 96 hpf zebrafish caused an increase in a) swim duration shown in seconds and b) C-bends, defined as large-amplitude body bends over 110°. Box plots represent the 25% and 75% quartiles, with the median represented by a horizontal black line and the mean represented by a black plus sign within the box. Tukey’s whiskers were used. n = 20 and 39 for wild-type and PTZ treated larvae, respectively, **P < 0.01, ***P < 0.001, ****P < 0.0001 using unpaired Welch’s t-tests.
Mutation of pairs of α subunits induces hyperactive behavior at 48 hpf
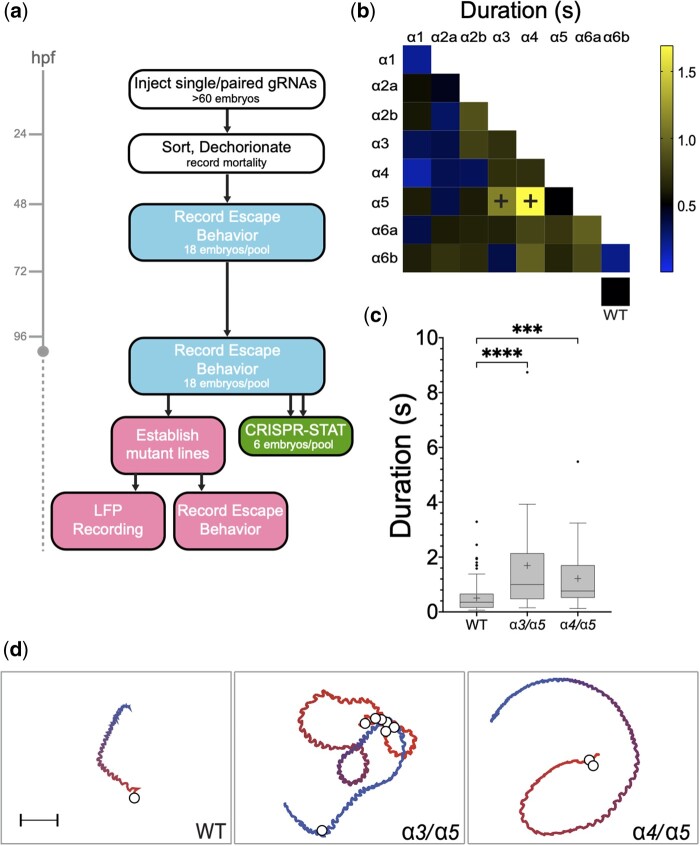
To identify GABAA α subunits that control locomotion, we generated F0 somatic mutations in each of the 8 subunits individually and in all pairwise combinations, then we analyzed touch-evoked behavior, focusing on escape response duration and body-bend amplitude (Fig. 2a). Of the 36 conditions examined, no individually mutated α subunit gave rise to abnormal response durations at 48 hpf; however, mutating pairs α3/α5 or α4/α5 resulted in increased swimming times (Fig. 2b and Supplementary Fig. 3a). Sibling controls exhibited an average swimming duration of 0.53 ± 0.03 s (n = 256, all n values here and subsequently refer to the number of larvae, each larva tested once). In contrast, larvae with mutations in α3/α5 and α4/α5 responded with an average of 1.27 ± 0.33 s (n = 18) and 1.69 ± 0.49 s (n = 18), respectively (Fig. 2, c and d, Supplementary Movies 3 and 4, and Supplementary Fig. 3a).
Fig. 2.
An F0 somatic mutation screen of GABAA receptor α subunits identifies mutations in α3/α5 and α4/α5 that increase swimming durations at 48 hpf. a) Overview of the GABAAR α subunit screen. Different colored boxes represent different aspects of the screen. b) Heat matrix of the 36 single and double F0 somatic mutant conditions showing mean startle response durations. The heatbar (right) indicates the average swim length. The box at lower right shows mock or uninjected controls. Ordinary one-way ANOVA revealed significant differences in swimming durations according to knock-down target, (N = 995 larvae total, 612 mutants with 13–26 larvae per condition, 383 wild-type siblings; [F(36,958) = 5.316, P < 0.0001]. A Dunnet’s post hoc test revealed significant pairwise differences between α3/α5 compared to wild-type and α4/α5 compared to wild-type (black plus signs), with those conditions exhibiting average swim durations of 1.27 ± 0.33 s (n = 18) and 1.69 ± 0.49 s, respectively. c) Boxplots of α3/α5 and α4/α5 somatic mutant swimming durations show the increased swimming durations compared to controls. ***P < 0.001, ****P < 0.0001 using Dunnett’s multiple comparison test. d) Traces of representative escape responses for wild-type, α3/α5 and α4/α5 somatic mutants. The color spectrum of each trace indicates the beginning (red) and end (blue) of the response, and white circles represent the location of C bends. The videos used to generate these traces are provided in the Supplementary material.
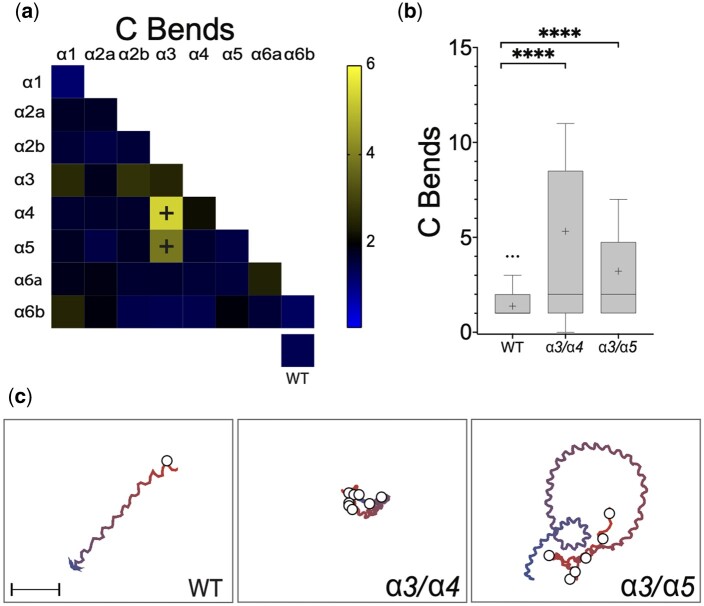
Mutations in α3, α4, and α5 also caused hyperactive increases in the number of C-shaped body bends at 48 hpf. Wild-type larvae exhibited an average of 1.37 ± 0.04 (n = 383) C-bends per escape response, while fish with mutations in α3/α4 and α3/α5 performed an average of 5.32 ± 1.89 (n = 18) and 3.88 ± 1.01 (n = 18) C-bends per escape response, respectively (Fig. 3, a and b and Supplementary Fig. 3b). Notably, mutations in pair α3/α4 increased the number of C-bends per escape response without causing significantly longer swimming durations, while mutations in pair α4/α5 caused an increase in swimming durations without a significant increase in C-bends. No mutant pairs without α3 were found to increase the number of high-amplitude body bends. Similarly, no mutant pairs without α5 were found to increase swimming durations. These observations suggest that α3 might play a dominant role in controlling the number of C-bends during an escape response, while α5 predominantly regulates swimming durations.
Fig. 3.
Somatic mutation of pairs α3/α4 or α3/α5 causes an increased number of large-amplitude body bends at 48 hpf. a) Heat matrix of the α subunit single and double somatic mutant conditions showing the mean numbers of C-bends per response. The heatbar (right) indicates the average swim duration. The box at lower right shows mock or uninjected controls. Ordinary one-way ANOVA revealed significant differences in large-amplitude body bends according to knock-down target, [F(36, 834) = 4.221, P < 0.0001]. A Dunnett’s post hoc test revealed significant pairwise differences between α3/α4 compared to wild-type and α3/α5 compared to wild-type (black plus signs), with those conditions exhibiting average C-bend per response of 5.32 and 3.88, respectively, compared to 1.37. b) Boxplots of α3/α4 and α3/α5 somatic mutant C-bends show the increased number of large amplitude body bends per swimming episode compared to sibling controls. ***P < 0.001, ****P < 0.0001 using Dunnett’s multiple comparison test. (C) Traces of representative escape responses for wild-type, α3/α4 and α3/α5. The color spectrum of each trace indicates the beginning (red) and end (blue) of the response, and white circles represent the location of C bends. The videos used to generate these traces are provided in the Supplementary material.
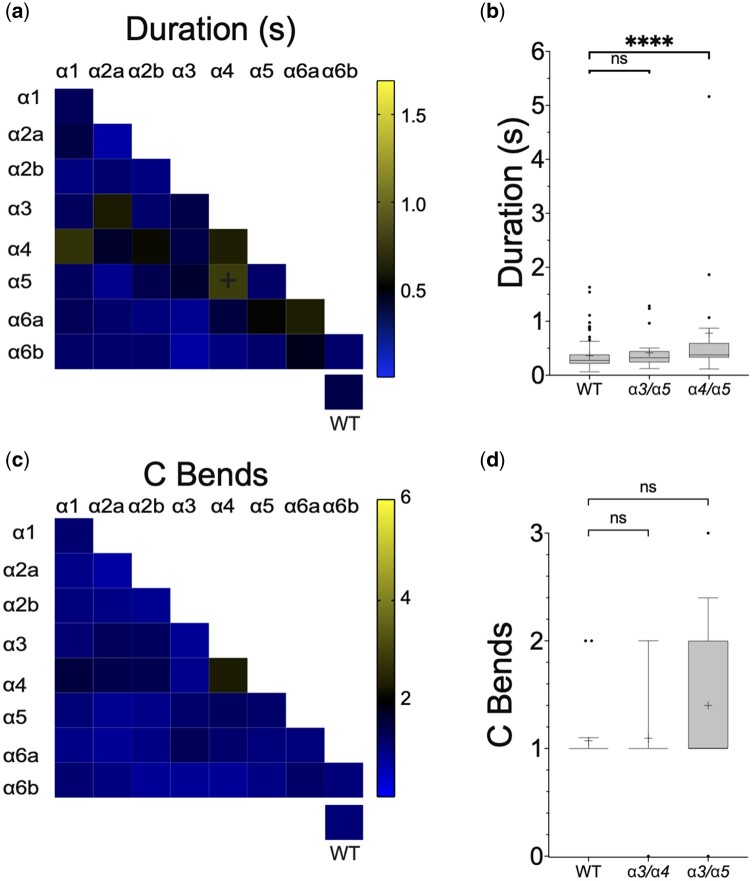
Hyperactive phenotypes at 48 hpf are absent or attenuated by 96 hpf
The hyperactive phenotypes observed at 48 hpf were absent or greatly reduced by 96 hpf. Mutations in pair α3/α5 resulted in significantly longer duration swimming episodes at 48 hpf, however, at 96 hpf the swimming behavior of these same animals was indistinguishable from controls (Fig. 4, a and b and Supplementary Fig. 3c). Mutations in pair α4/α5 did result in longer duration swimming responses at both 48 and 96 hpf; however, at the later time point, the difference compared to controls was far less [a difference of 1.18 ± 0.15 (n = 18) s at 48 hpf and 0.41 ± 0.07 (n = 18) s at 96 hpf compared to controls]. No other mutations caused abnormal swimming durations. Similarly, although mutations in pairs α3/α4 or α3/α5 induced excess C-bends at 48 hpf, neither these nor any other mutations were found to cause significantly more large-amplitude body flexions at 96 hpf (Fig. 4, c and d and Supplementary Fig. 3d).
Fig. 4.
Only α4/α5 somatic mutants continue to exhibit a hyperactive phenotype at 96 hpf. a) Heat matrix of the single and double F0 somatic mutant conditions showing evoked swimming response durations at 96 hpf. The heatbar (right) indicates the average swim duration. The box at lower right shows mock or uninjected controls. Ordinary one-way ANOVA revealed a significant difference in swimming duration dependent upon knock-down target [F(36,754) = 2.914, P < 0.0001]. A Dunnet’s post hoc test revealed only a significant pairwise difference between α4/α5 (black plus signs) and wild-type. b) A boxplot of α3/α5, α4/α5 mutant pairs and controls. ****P < 0.0001 using Dunnett’s multiple comparison test. Both mutant conditions showed increased swimming durations at 48 hpf, however only the α4/α5 pair was statistically significant at 96 hpf. Although significant, the α4/α5 swimming duration is reduced compared to 48 hpf. c) Heat matrix of C bends at 96 hpf. No significant differences were detected. d) Box plots show that conditions that demonstrated increased C bends at 48 hpf were not significantly elevated at 96 hpf.
α3 and α4 F2 germline mutants confirm that hyperactive phenotypes at 48 hpf are reduced or absent by 96 hpf
Previous studies have suggested that phenotypes observed in F0 somatic mutants can weaken during development, possibly due to the effects of mosaicism (Wu et al. 2018). To address whether this mechanism could explain the reduction in hyperactive phenotypes at 96 hpf, we generated F2 α3 and α4 germline mutants (Fig. 5a). We selected these subunits because, in the somatic screen at 48 hpf, both of these subunits were implicated in controlling swimming behavior. While no individually mutated subunit gave rise to significantly abnormal behavior, mutations in α3 came the closest to reaching significance [Ordinary one-way ANOVA, F(36, 978) = 5.23, P = 0.17 compared to P = 0.77–0.99 for other individually mutated subunits], and mutations in α3 when paired with mutations in α4 significantly increased the number of C-bends (see Fig. 3a and Supplementary Fig. 3b). In line with α3 selectively controlling high-amplitude body bends but not swimming duration, α3 germline mutants demonstrated more C-bends (P < 0.001) without a significant increase in swimming times at 48 hpf (P = 0.27) (Fig. 5b). The α3 germline mutant phenotype was more robust than the α3 somatic phenotype, likely due to the mosaicism of somatic mutants. Similar to the α4 somatic mutant result, α4 germline mutants did not exhibit significant increases in duration or the number of C-bends at 48 hpf. Also, confirming our observations using somatic mutants, at 96 hpf neither α3 nor α4 germline mutants were significantly different from sibling controls (P = 0.3879). These data indicate that the absent or weaker phenotypes observed in the somatic mutants at 96 hpf are not due to mosaicism.
Fig. 5.
α3 F2 germline mutants confirm the α3 F0 somatic mutant phenotype. a) A schematic of the α3 protein is shown based upon (Macdonald et al. 2010). The 4 transmembrane domains are indicated, along with the location of the zebrafish (Danio rerio) and human (Homo sapiens) α3 mutations. b) Box plots for α3 trans-heterozygous mutants and siblings swimming duration in seconds (left) and C-bends per response (right) at both 48 and 96 hpf. Unpaired Student’s t-test indicated that α3 mutants exhibit significantly more C-bends at 48 hpf but not at 96 hpf ****P < 0.0001. α3 mutant swimming durations were not significantly greater than sibling controls at either time point. c) Schematic representation of LFP recording setup from a 96 hpf larva. d) LFP traces (left) from wild-type (n = 3), siblings (n = 7), PTZ treated wild-type (n = 3), α3 heterozygotes (n = 6), and α3 trans-heterozygous mutants (n = 3). Ictal-like activity was only detected in PTZ treated fish compared to wild-type (right). Ordinary one-way ANOVA with Dunnet’s post hoc test, ****P < 0.0001.
Given that the behavior of α3 mutants was indistinguishable from wild-type controls at 96 hpf, we next asked whether LFP recordings from the larval brain (Fig. 5c) would provide a more sensitive measure of phenotype abnormalities than our behavioral assay. Consistent with previous findings (Liu and Baraban 2019), LFP recordings over a period of 30 min revealed frequent large and abnormal discharges of activity at 96 hpf when PTZ was applied. However, α3 germline mutants and controls had neuronal activities that were indistinguishable from one another, consistent with their functionally normal swimming behaviors (Fig. 5d).
Discussion
In this study, we performed an F0 somatic mutant screen to identify GABAA α subunits that regulate hyperactive swimming during early larval stages of zebrafish development. We presented evidence that combinations of α3, α4, and α5 play selective roles in mediating different aspects of hyperactive behavior at 48 hpf. F0 somatic mutations in pairs α3/α4 significantly increased the number of high amplitude body bends, while mutations in pairs α4/α5 significantly increased swimming duration, and mutation of pairs α3/α5 caused significant increases in both parameters. We found that hyperactivity caused by somatic disruption of GABAA α subunits is ostensibly reduced by 96 hpf, a result confirmed using germline α3 and α4 mutants using behavioral and/or electrophysiological assays. Taken together, these data lay a foundation to investigate how GABAA receptors establish and maintain control of escape behavior at neuronal and circuit levels.
GABAA receptor subunits likely control escape behavior through different cellular mechanisms
The GABAA α subunits α3, α4, and α5 regulate escape behavior at 48 hpf; however, the cellular mechanisms through which they exert their effects are not yet clear. The zebrafish hindbrain, especially the Mauthner cell and its homologs, play a central and well-studied role in C-starts, and these cells are regulated by GABA (Triller et al. 1997; Korn and Faber 2005; Burgess and Granato 2007; Kohashi et al. 2012; Roy and Ali 2014; Liu and Baraban 2019). α3, α4, and α5 are all expressed in discrete populations of hindbrain cells by 48 hpf, so it is possible their reduced expression dysregulates hindbrain circuits to generate hyperactive behavior (Monesson-Olson et al. 2018). Alternatively, each of these subunits is also expressed in distinct cell types in the spinal cord, so it is also possible that spinal cord GABAA α subunit disruption elicits hyperactive behavior. In future studies, it will be interesting to determine the relative contribution of the hindbrain vs. the spinal cord in generating the abnormal behaviors caused by reduced GABAA receptor function.
Whether through the hindbrain or spinal cord, α3, α4, and α5 likely control escape behavior through different neurons or subcellular mechanisms. In both the hindbrain and spinal cord at 48 hpf, α3 and α5 are expressed in overlapping domains, raising the possibility they are expressed in at least some of the same cells (Monesson-Olson et al. 2018; Sadamitsu et al. 2021). In both structures, α4 seems to be expressed in cells distinct from α3 and α5; therefore, its effects could be mediated by different neurons.
Even when expressed in the same cells, α3, α4, and α5 are probably expressed in different subcellular domains. In mammalian neurons, GABAA receptors have been shown to cluster in either synaptic or extrasynaptic domains to mediate phasic or tonic inhibition (Farrant and Nusser 2005; Jacob et al. 2008; Fritschy and Panzanelli 2014). α3-containing receptors are enriched at synapses, to mediate phasic inhibition, while α4- and α5-containing receptors are predominantly extrasynaptic, to provide tonic inhibition. Although the subcellular localizations of these subunits have not been directly investigated in zebrafish, the high degree of amino acid sequence similarity compared to those in mammals (87.9% 74.3%, and 79.1% identity with mice for α3, α4, and α5, respectively) suggests that their subcellular distributions are likely conserved, which would localize α3 toward synapses and α4 and α5 toward extrasynaptic domains. High-resolution expression analysis will be required to determine if this is, indeed, the case.
We did not find significant effects in response to somatic mutation of α1, α2a, α2b, α6a, or α6b; however, these subunits cannot be entirely ruled out from playing regulatory roles in controlling zebrafish escape behavior. F0 somatic mutations reduce but do not eliminate expression, so it is possible that further reducing the expression of these subunits could reveal locomotor phenotypes. In addition, our screen focused on two behavioral parameters at 2 developmental stages. Examining additional developmental stages, responses to other sensory stimuli, or other parameters, such as body bend frequency or frequency variability, could reveal roles for these subunits in controlling escape behavior.
Zebrafish likely employ robust homeostatic mechanisms across development
The hyperactive phenotypes observed at 48 hpf were all absent or greatly reduced by 96 hpf. This result was surprising since PTZ readily elicits hyperactive behavior at all time points after 48 hpf, demonstrating that GABAA receptors play critical roles in regulating locomotion across a wide variety of developmental stages. A likely explanation is that zebrafish employ robust homeostatic compensation. The mechanisms that underlie this compensation are probably multifaceted. The teleost lineage has undergone genome duplication such that there are many duplicated genes in zebrafish, including several GABAA receptor subunits (Amores et al. 1998; Postlethwait et al. 1998; Monesson-Olson et al. 2018; Sadamitsu et al. 2021). The expression of homologous genes or simply genes with similar sequence motifs can be recruited through transcriptional adaptation, which is thought to be triggered by nonsense mediated decay (El-Brolosy et al. 2019). Mutations that cause frame-shifts and premature stop codons can cause nonsense mediated decay, therefore at least some of the mutations generated in this study probably induced transcriptional adaptation. Pairs of α subunits were mutated to uncover adaptations within the α subunit subfamily, however, transcriptional responses could involve other GABAA receptor subunits. At the network level, neurons could switch the neurotransmitter they release or entire circuits could be reconfigured to maintain excitation–inhibition balance as has been observed in developing frogs and some α subunit knock-out mice (Schneider Gasser et al. 2007; Panzanelli et al. 2011). Another possible factor is the developmental change in chloride gradients that convert the actions of glycine and GABA from excitatory to inhibitory (Ben-Ari 2002; Reynolds et al. 2008; Zhang et al. 2010). However, arguing against this possibility, locomotor neurons in both the spinal cord and hindbrain demonstrate synaptic inhibitory responses as early as 26 hpf (Ali et al. 2000; Pietri et al. 2009; Knogler and Drapeau 2014). Although different neurons can maturate and alter their chloride gradients at different developmental times, the fact that spinal cord and hindbrain locomotor neurons demonstrate inhibitory responses well before the time points examined in this study suggests that altered chloride gradients are unlikely to account for the reductions in hyperactive behavior that were observed. No matter the mechanism, our results identify a narrow window, between 48 and 96 hpf, across which putative adaptation occurs in developing zebrafish. The short time period, relatively small nervous system, ex utero development, genetic resources, and high-resolution brain and spinal cord atlases make larval zebrafish an outstanding system to further investigate homeostatic mechanisms activated by GABAA receptor mutation.
Zebrafish α3 mutants as a possible epilepsy model
In addition to their roles in modulating locomotion, GABAA receptors are widely viewed as central factors in the development, progression, and treatment of epilepsy syndromes (Olsen and Avoli 1997; Treiman 2001; Cherubini 2012; Walker and Kullmann 2012). Abnormalities in GABAA receptor inhibition are observed in genetic and acquired epilepsies, drugs that block these receptors cause seizures, and drugs that enhance GABAA receptor inhibition are potent anticonvulsants. In zebrafish, PTZ application is an established seizure model, and larval or juvenile loss-of-function mutations α1, γ2, and β3 have been proposed to model the epilepsies caused by mutations in their human orthologs (Baraban et al. 2005; Baxendale et al. 2012; Samarut et al. 2018; Liao et al. 2019; Yang et al. 2019; Cho et al. 2020; Reyes-Nava et al. 2020; Griffin et al. 2021). Here, we showed that mutation of α3 causes hyperactive behavior in early zebrafish larvae. Human loss-of-function variants in α3 are associated with a rare, severe epileptic encephalopathy, raising the possibility that zebrafish α3 mutants at least partially model this disorder (Niturad et al. 2017). Hyperactive swimming, in and of itself, is not necessarily a seizure, however, it is possible the α3 mutant has validity as an epilepsy model. Similar to the work done to assess human germ-line mutations in the Ras/MAPK pathway in both zebrafish and Drosophila, mutation of zebrafish α3 could disrupt molecular mechanisms similar to those in α3 associated epileptic encephalopathy, but in a different cellular context, one which gives rise to hyperactive swimming (Jindal et al. 2017). In this way, zebrafish α3 mutants could furnish a useful readout assay to uncover genetic or pharmacological interactions that are relevant to human α3 pathogenic mutations. Given the proven effectiveness of larval zebrafish for high-throughput small molecule screens, for example, α3 mutant hyperactive behavior could serve as a screening platform to identify novel anticonvulsant drugs (Griffin et al. 2017, 2020; Lam and Peterson 2019; Patton et al. 2021).
Data availability
Zebrafish lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are present within the article, figures, and tables. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.18430457.
Acknowledgments
The authors thank Abhay Mittal for developing kinematic analysis software; Saige Calkins, Caroline Martin, and Oshiomah Oyageshio for excellent fish care; and Marie Abate, Sean Doherty, Ana Dolan, and Chinemerem Nwokemodo-Ihejirika for technical assistance. They also thank Dr. Kelly Anne McKeown, other members of the Downes and Trapani labs, and the entire University of Massachusetts Amherst zebrafish community for thoughtful discussion.
Funding
This work was funded by the National Science Foundation (IOS 1456866) to Gerald B. Downes and Josef G. Trapani.
Conflicts of interest
All authors declare no conflicting interests.
Literature cited
- Ali DW, Drapeau P, Legendre P.. Development of spontaneous glycinergic currents in the mauthner neuron of the zebrafish embryo. J Neurophysiol. 2000;84(4):1726–1736. doi: 10.1152/jn.2000.84.4.1726. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282(5394):1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Baraban SC, Taylor MR, Castro PA, Baier H.. Pentylenetetrazole induced changes in zebrafish behavior, neural activity and c-fos expression. Neuroscience. 2005;131(3):759–768. doi: 10.1016/j.neuroscience.2004.11.031. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y. Excitatory actions of gaba during development: the nature of the nurture. Nat Rev Neurosci. 2002;3(9):728–739. 10.1038/nrn920 [DOI] [PubMed] [Google Scholar]
- Baxendale S, Holdsworth CJ, Meza Santoscoy PL, Harrison MRM, Fox J, Parkin CA, Ingham PW, Cunliffe VT.. Identification of compounds with anti-convulsant properties in a zebrafish model of epileptic seizures. Dis Model Mech. 2012;5(6):773–784. doi: 10.1242/dmm.010090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brustein E, Saint-Amant L, Buss RR, Chong M, McDearmid JR, Drapeau P.. Steps during the development of the zebrafish locomotor network. J Physiol Paris. 2003;97(1):77–86. doi: 10.1016/j.jphysparis.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Burgess HA, Granato M.. Sensorimotor gating in larval zebrafish. J Neurosci. 2007;27(18):4984–4994. doi: 10.1523/JNEUROSCI.0615-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington B, Varshney GK, Burgess SM, Sood R.. CRISPR-STAT: an easy and reliable PCR-based method to evaluate target-specific sgRNA activity. Nucleic Acids Res. 2015;43(22):e157. doi: 10.1093/nar/gkv802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini E. Phasic GABAA-mediated inhibition. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies. (4th ed, Chap. 8). Bethesda (MD): Oxford University Press; 2012. p. 97–110. [Google Scholar]
- Cho S-J, Park E, Baker A, Reid AY.. Age bias in zebrafish models of epilepsy: what can we learn from old fish? Front Cell Dev Biol. 2020;8:573303. doi: 10.3389/fcell.2020.573303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HC, Chebib M.. GABAA receptors and the diversity in their structure and pharmacology. Adv Pharmacol San Pharmacol. 2017;79:1–34. doi: 10.1016/bs.apha.2017.03.003. [DOI] [PubMed] [Google Scholar]
- Cocco A, Rönnberg AMC, Jin Z, André GI, Vossen LE, Bhandage AK, Thörnqvist P-O, Birnir B, Winberg S.. Characterization of the γ-aminobutyric acid signaling system in the zebrafish (Danio rerio Hamilton) central nervous system by reverse transcription-quantitative polymerase chain reaction. Neuroscience. 2017;343:300–321. doi: 10.1016/j.neuroscience.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Eaton RC, Bombardieri RA, Meyer DL.. The Mauthner-initiated startle response in teleost fish. J Exp Biol. 1977;66(1):65–81. doi: 10.1242/jeb.66.1.65. [DOI] [PubMed] [Google Scholar]
- Eaton RC, Lee RKK, Foreman MB.. The Mauthner cell and other identified neurons of the brainstem escape network of fish. Prog Neurobiol. 2001;63(4):467–485. doi: 10.1016/S0301-0082(00)00047-2. [DOI] [PubMed] [Google Scholar]
- El-Brolosy MA, Kontarakis Z, Rossi A, Kuenne C, Günther S, Fukuda N, Kikhi K, Boezio GLM, Takacs CM, Lai S-L, et al. Genetic compensation triggered by mutant mRNA degradation. Nature. 2019;568(7751):193–197. doi: 10.1038/s41586-019-1064-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrant M, Nusser Z.. Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat Rev Neurosci. 2005;6(3):215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Lambert AM, Masino MA, Downes GB.. Mutation of zebrafish dihydrolipoamide branched-chain transacylase E2 results in motor dysfunction and models maple syrup urine disease. Dis Model Mech. 2012;5(2):248–258. doi: 10.1242/dmm.008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritschy J-M, Panzanelli P.. GABAA receptors and plasticity of inhibitory neurotransmission in the central nervous system. Eur J Neurosci. 2014;39(11):1845–1865. doi: 10.1111/ejn.12534. [DOI] [PubMed] [Google Scholar]
- Gagnon JA, Valen E, Thyme SB, Huang P, Akhmetova L, Ahkmetova L, Pauli A, Montague TG, Zimmerman S, Richter C, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9(5):e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, van Eeden FJ, Schach U, Trowe T, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg CP, Jiang YJ, et al. Genes controlling and mediating locomotion behavior of the zebrafish embryo and larva. Dev Camb Engl. 1996;123(1):399–413. [DOI] [PubMed] [Google Scholar]
- Griffin A, Anvar M, Hamling K, Baraban SC.. Phenotype-based screening of synthetic cannabinoids in a dravet syndrome zebrafish model. Front Pharmacol. 2020;11:464. doi: 10.3389/fphar.2020.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin A, Carpenter C, Liu J, Paterno R, Grone B, Hamling K, Moog M, Dinday MT, Figueroa F, Anvar M, et al. Phenotypic analysis of catastrophic childhood epilepsy genes. Commun Biol. 2021;4(1):1–13. doi: 10.1038/s42003-021-02221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin A, Hamling KR, Knupp K, Hong S, Lee LP, Baraban SC.. Clemizole and modulators of serotonin signalling suppress seizures in dravet syndrome. Brain J Brain. 2017;140(3):669–683. doi: 10.1093/brain/aww342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinckley C, Seebach B, Ziskind-Conhaim L.. Distinct roles of glycinergic and GABAergic inhibition in coordinating locomotor-like rhythms in the neonatal mouse spinal cord. Neuroscience. 2005;131(3):745–758. doi: 10.1016/j.neuroscience.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Jacob TC, Moss SJ, Jurd R.. GABA(A) receptor trafficking and its role in the dynamic modulation of neuronal inhibition. Nat Rev Neurosci. 2008;9(5):331–343. doi: 10.1038/nrn2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindal GA, Goyal Y, Yamaya K, Futran AS, Kountouridis I, Balgobin CA, Schüpbach T, Burdine RD, Shvartsman SY.. In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc Natl Acad Sci USA. 2017;114(3):510–515. doi: 10.1073/pnas.1615651114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF.. Stages of embryonic development of the zebrafish. Dev Dyn off Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Knogler LD, Drapeau P.. Sensory gating of an embryonic zebrafish interneuron during spontaneous motor behaviors. Front Neural Circuits. 2014;8:121. doi: 10.3389/fncir.2014.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohashi T, Nakata N, Oda Y.. Effective sensory modality activating an escape triggering neuron switches during early development in zebrafish. J Neurosci. 2012;32(17):5810–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn H, Faber DS.. The Mauthner cell half a century later: a neurobiological model for decision-making? Neuron. 2005;47(1):13–28. doi: 10.1016/j.neuron.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Kralic JE, Sidler C, Parpan F, Homanics GE, Morrow AL, Fritschy J-M.. Compensatory alteration of inhibitory synaptic circuits in cerebellum and thalamus of gamma-aminobutyric acid type A receptor alpha1 subunit knockout mice. J Comp Neurol. 2006;495(4):408–421. doi: 10.1002/cne.20866. [DOI] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E.. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47(W1):W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P-Y, Peterson RT.. Developing zebrafish disease models for in vivo small molecule screens. Curr Opin Chem Biol. 2019;50:37–44. doi: 10.1016/j.cbpa.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert TD, Howard J, Plant A, Soffe S, Roberts A.. Mechanisms and significance of reduced activity and responsiveness in resting frog tadpoles. J Exp Biol. 2004a;207(Pt 7):1113–1125. doi: 10.1242/jeb.00866. [DOI] [PubMed] [Google Scholar]
- Lambert TD, Li W-C, Soffe SR, Roberts A.. Brainstem control of activity and responsiveness in resting frog tadpoles: tonic inhibition. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2004b;190(4):331–342. doi: 10.1007/s00359-004-0505-8. [DOI] [PubMed] [Google Scholar]
- Laurie DJ, Seeburg PH, Wisden W.. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. II. Olfactory bulb and cerebellum. J Neurosci. 1992a;12(3):1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie DJ, Wisden W, Seeburg PH.. The distribution of thirteen GABAA receptor subunit mRNAs in the rat brain. III. Embryonic and postnatal development. J Neurosci. 1992b;12(11):4151–4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverty D, Desai R, Uchański T, Masiulis S, Stec WJ, Malinauskas T, Zivanov J, Pardon E, Steyaert J, Miller KW, et al. Cryo-EM structure of the human α1β3γ2 GABA A receptor in a lipid bilayer. Nature. 2019;565(7740):516–520. doi: 10.1038/s41586-018-0833-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M, Kundap U, Rosch RE, Burrows DRW, Meyer MP, Ouled Amar Bencheikh B, Cossette P, Samarut É.. Targeted knockout of GABA-A receptor gamma 2 subunit provokes transient light-induced reflex seizures in zebrafish larvae. Dis Model Mech. 2019;12(11):dmm040782. doi: 10.1242/dmm.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Baraban SC.. Network properties revealed during multi-scale calcium imaging of seizure activity in zebrafish. eNeuro. 2019;6(1):ENEURO.0041-19.2019. doi: 10.1523/ENEURO.0041-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald RL, Kang J-Q, Gallagher MJ.. Mutations in GABAA receptor subunits associated with genetic epilepsies. J Physiol. 2010;588(Pt 11):1861–1869. doi: 10.1113/jphysiol.2010.186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiulis S, Desai R, Uchański T, Serna Martin I, Laverty D, Karia D, Malinauskas T, Zivanov J, Pardon E, Kotecha A, et al. GABAA receptor signalling mechanisms revealed by structural pharmacology. Nature. 2019;565(7740):454–459. doi: 10.1038/s41586-018-0832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown KA, Downes GB, Hutson LD.. Modular laboratory exercises to analyze the development of zebrafish motor behavior. Zebrafish. 2009;6(2):179–185. doi: 10.1089/zeb.2008.0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeown KA, Moreno R, Hall VL, Ribera AB, Downes GB.. Disruption of Eaat2b, a glutamate transporter, results in abnormal motor behaviors in developing zebrafish. Dev Biol. 2012;362(2):162–171. doi: 10.1016/j.ydbio.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monesson-Olson B, McClain JJ, Case AE, Dorman HE, Turkewitz DR, Steiner AB, Downes GB.. Expression of the eight GABAA receptor α subunits in the developing zebrafish central nervous system. PLoS One. 2018;13(4):e0196083. doi: 10.1371/journal.pone.0196083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E.. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niturad CE, Lev D, Kalscheuer VM, Charzewska A, Schubert J, Lerman-Sagie T, Kroes HY, Oegema R, Traverso M, Specchio N, et al. ; EuroEPINOMICS Consortium. Rare GABRA3 variants are associated with epileptic seizures, encephalopathy and dysmorphic features. Brain J Brain. 2017;140(11):2879–2894. doi: 10.1093/brain/awx236. [DOI] [PubMed] [Google Scholar]
- Olsen RW, Avoli M.. GABA and epileptogenesis. Epilepsia. 1997;38(4):399–407. doi: 10.1111/j.1528-1157.1997.tb01728.x. [DOI] [PubMed] [Google Scholar]
- O'Malley DM, Kao YH, Fetcho JR.. Imaging the functional organization of zebrafish hindbrain segments during escape behaviors. Neuron. 1996;17(6):1145–1155. doi: 10.1016/S0896-6273(00)80246-9. [DOI] [PubMed] [Google Scholar]
- Panzanelli P, Gunn BG, Schlatter MC, Benke D, Tyagarajan SK, Scheiffele P, Belelli D, Lambert JJ, Rudolph U, Fritschy J-M.. Distinct mechanisms regulate GABAA receptor and gephyrin clustering at perisomatic and axo-axonic synapses on CA1 pyramidal cells. J Physiol. 2011;589(Pt 20):4959–4980. doi: 10.1113/jphysiol.2011.216028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE.. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn off Dyn. 2009;238(12):2975–3015. doi: 10.1002/dvdy.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Zon LI, Langenau DM.. Zebrafish disease models in drug discovery: from preclinical modelling to clinical trials. Nat Rev Drug Discov. 2021;20(8):611–628. doi: 10.1038/s41573-021-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietri T, Manalo E, Ryan J, Saint-Amant L, Washbourne P.. Glutamate drives the touch response through a rostral loop in the spinal cord of zebrafish embryos. Dev Neurobiol. 2009;69(12):780–795. 10.1002/dneu.20741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Hauer B, Mihalek RM, Homanics GE, Sieghart W, Olsen RW, Houser CR.. GABA(A) receptor changes in delta subunit-deficient mice: altered expression of alpha4 and gamma2 subunits in the forebrain. J Comp Neurol. 2002;446(2):179–197. doi: 10.1002/cne.10210. [DOI] [PubMed] [Google Scholar]
- Perrins R, Walford A, Roberts A.. Sensory activation and role of inhibitory reticulospinal neurons that stop swimming in hatchling frog tadpoles. J Neurosci. 2002;22(10):4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phulera S, Zhu H, Yu J, Claxton DP, Yoder N, Yoshioka C, Gouaux E.. Cryo-EM structure of the benzodiazepine-sensitive α1β1γ2S tri-heteromeric GABAA receptor in complex with GABA. eLife. 2018;7:e39383. doi: 10.7554/eLife.39383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Yan YL, Gates MA, Horne S, Amores A, Brownlie A, Donovan A, Egan ES, Force A, Gong Z, et al. Vertebrate genome evolution and the zebrafish gene map. Nat Genet. 1998;18(4):345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- Reyes-Nava NG, Yu H-C, Coughlin CR II, Shaikh TH, Quintana AM.. Abnormal expression of GABAA receptor subunits and hypomotility upon loss of gabra1 in zebrafish. Biology Open. 2020;9(4): bio051367. 10.1242/bio.051367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Brustein E, Liao M, Mercado A, Babilonia E, Mount DB, Drapeau P.. Neurogenic role of the depolarizing chloride gradient revealed by global overexpression of KCC2 from the onset of development. J Neurosci. 2008;28(7):1588–1597. doi: 10.1523/JNEUROSCI.3791-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JS, Silver RA.. NeuroMatic: an integrated open-source software toolkit for acquisition, analysis and simulation of electrophysiological data. Front Neuroinform. 2018;12:14. doi: 10.3389/fninf.2018.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussel Y, Paradis M, Gaudreau SF, Lindsey BW, Bui TV.. Spatiotemporal transition in the role of synaptic inhibition to the tail beat rhythm of developing larval zebrafish. eNeuro. 2020;7(1):ENEURO.0508-18.2020. doi: 10.1523/ENEURO.0508-18.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy B, Ali DW.. Multiple types of GABAA responses identified from zebrafish Mauthner cells. Neuroreport. 2014;25(15):1232–1236. doi: 10.1097/WNR.0000000000000258. [DOI] [PubMed] [Google Scholar]
- Rudolph U, Möhler H.. Analysis of GABAA receptor function and dissection of the pharmacology of benzodiazepines and general anesthetics through mouse genetics. Annu Rev Pharmacol Toxicol. 2004;44:475–498. doi: 10.1146/annurev.pharmtox.44.101802.121429. [DOI] [PubMed] [Google Scholar]
- Sadamitsu K, Shigemitsu L, Suzuki M, Ito D, Kashima M, Hirata H.. Characterization of zebrafish GABAA receptor subunits. Sci Rep. 2021;11(1):6242. doi: 10.1038/s41598-021-84646-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarut É, Swaminathan A, Riché R, Liao M, Hassan-Abdi R, Renault S, Allard M, Dufour L, Cossette P, Soussi-Yanicostas N, et al. γ-Aminobutyric acid receptor alpha 1 subunit loss of function causes genetic generalized epilepsy by impairing inhibitory network neurodevelopment. Epilepsia. 2018;59(11):2061–2074. doi: 10.1111/epi.14576. [DOI] [PubMed] [Google Scholar]
- Schmitt DE, Hill RH, Grillner S.. The spinal GABAergic system is a strong modulator of burst frequency in the lamprey locomotor network. J Neurophysiol. 2004;92(4):2357–2367. doi: 10.1152/jn.00233.2004. [DOI] [PubMed] [Google Scholar]
- Schneider Gasser EM, Duveau V, Prenosil GA, Fritschy J-M.. Reorganization of GABAergic circuits maintains GABAA receptor-mediated transmission onto CA1 interneurons in alpha1-subunit-null mice. Eur J Neurosci. 2007;25(11):3287–3304. doi: 10.1111/j.1460-9568.2007.05558.x. [DOI] [PubMed] [Google Scholar]
- Shah AN, Davey CF, Whitebirch AC, Miller AC, Moens CB.. Rapid reverse genetic screening using CRISPR in zebrafish. Nat Methods. 2015;12(6):535–540. doi: 10.1038/nmeth.3360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AN, Moens CB, Miller AC.. Targeted candidate gene screens using CRISPR/Cas9 technology. Methods Cell Biol. 2016;135:89–106. doi: 10.1016/bs.mcb.2016.01.008. [DOI] [PubMed] [Google Scholar]
- Sieghart W, Sperk G.. Subunit composition, distribution and function of GABA(A) receptor subtypes. Curr Top Med Chem. 2002;2(8):795–816. doi: 10.2174/1568026023393507. [DOI] [PubMed] [Google Scholar]
- Simon J, Wakimoto H, Fujita N, Lalande M, Barnard EA.. Analysis of the set of GABA(A) receptor genes in the human genome. J Biol Chem. 2004;279(40):41422–41435. doi: 10.1074/jbc.M401354200. [DOI] [PubMed] [Google Scholar]
- Smith KS, Rudolph U.. Anxiety and depression: mouse genetics and pharmacological approaches to the role of GABA(A) receptor subtypes. Neuropharmacology. 2012;62(1):54–62. doi: 10.1016/j.neuropharm.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiman DM. GABAergic mechanisms in epilepsy. Epilepsia. 2001;42 (suppl 3):8–12. doi: 10.1046/j.1528-1157.2001.042suppl.3008.x. [DOI] [PubMed] [Google Scholar]
- Triller A, Rostaing P, Korn H, Legendre P.. Morphofunctional evidence for mature synaptic contacts on the Mauthner cell of 52-hour-old zebrafish larvae. Neuroscience. 1997;80(1):133–145. doi: 10.1016/s0306-4522(97)00092-4. [DOI] [PubMed] [Google Scholar]
- Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, et al. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25(7):1030–1042. doi: 10.1101/gr.186379.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicini S, Ortinski P.. Genetic manipulations of GABAA receptor in mice make inhibition exciting. Pharmacol Ther. 2004;103(2):109–120. doi: 10.1016/j.pharmthera.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Walker MC, Kullmann DM.. Tonic GABAA receptor-mediated signaling in epilepsy. In: Noebels JL, Avoli M, Rogawski MA, Olsen RW, Delgado-Escueta AV, editors. Jasper’s Basic Mechanisms of the Epilepsies. (4th ed., Chap. 9). Bethesda (MD): Oxford University Press; 2012. p. 111–121. [Google Scholar]
- Wisden W, Laurie DJ, Monyer H, Seeburg PH.. The distribution of 13 GABAA receptor subunit mRNAs in the rat brain. I. Telencephalon, diencephalon, mesencephalon. J Neurosci. 1992;12(3):1040–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RS, Lam II, Clay H, Duong DN, Deo RC, Coughlin SR.. A rapid method for directed gene knockout for screening in G0 zebrafish. Dev Cell. 2018;46(1):112–125.e4. doi: 10.1016/j.devcel.2018.06.003. [DOI] [PubMed] [Google Scholar]
- Yang X, Jounaidi Y, Mukherjee K, Fantasia RJ, Liao EC, Yu B, Forman SA.. Drug- selective anesthetic insensitivity of zebrafish lacking γ-Aminobutyric acid type A receptor β3 subunits. Anesthesiology. 2019;131(6):1276–1291. doi: 10.1097/ALN.0000000000002963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Wei H, Xia Y, Du J.. Development of light response and GABAergic excitation-to-inhibition switch in zebrafish retinal ganglion cells. J Physiol. 2010;588(Pt 14):2557–2569. doi: 10.1113/jphysiol.2010.187088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller A, Crestani F, Camenisch I, Iwasato T, Itohara S, Fritschy JM, Rudolph U.. Cortical glutamatergic neurons mediate the motor sedative action of diazepam. Mol Pharmacol. 2008;73(2):282–291. doi: 10.1124/mol.107.038828. [DOI] [PubMed] [Google Scholar]
- Zhou C, Huang Z, Ding L, Deel ME, Arain FM, Murray CR, Patel RS, Flanagan CD, Gallagher MJ.. Altered cortical GABAA receptor composition, physiology, and endocytosis in a mouse model of a human genetic absence epilepsy syndrome. J Biol Chem. 2013;288(29):21458–21472. doi: 10.1074/jbc.M112.444372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Noviello CM, Teng J, Walsh RM, Kim JJ, Hibbs RE.. Structure of a human synaptic GABA A receptor. Nature. 2018;559(7712):67–72. doi: 10.1038/s41586-018-0255-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Zebrafish lines are available upon request. The authors affirm that all data necessary for confirming the conclusions of this article are present within the article, figures, and tables. Supplementary material is available at figshare: https://doi.org/10.25386/genetics.18430457.