Abstract
Accurate segmentation and tracking of cells in microscopy image sequences is extremely beneficial in clinical diagnostic applications and biomedical research. A continuing challenge is the segmentation of dense touching cells and deforming cells with indistinct boundaries, in low signal-to-noise-ratio images. In this paper, we present a dual-stream marker-guided network (DMNet) for segmentation of touching cells in microscopy videos of many cell types. DMNet uses an explicit cell marker-detection stream, with a separate mask-prediction stream using a distance map penalty function, which enables supervised training to focus attention on touching and nearby cells. For multi-object cell tracking we use M2Track tracking-by-detection approach with multi-step data association. Our M2Track with mask overlap includes short term track-to-cell association followed by track-to-track association to re-link tracklets with missing segmentation masks over a short sequence of frames. Our combined detection, segmentation and tracking algorithm has proven its potential on the IEEE ISBI 2021 6th Cell Tracking Challenge (CTC-6) where we achieved multiple top three rankings for diverse cell types. Our team name is MU-Ba-US, and the implementation of DMNet is available at, http://celltrackingchallenge.net/participants/MU-Ba-US/.
1. Introduction
The capacity of cells to exert forces on their environment and alter their shape as they move [3] is essential to many biological processes including the cellular immune response to infections [25], embryonic development [48], wound healing [8] and tumor growth [16]. Detecting cell shape and their changes over time as cells navigate the microenvironment are essential for understanding the multiple mechanisms guiding and regulating cell motility [74]. We propose an end-to-end pipeline for accurate cell detection, segmentation and tracking as shown in Figure 1.
Figure 1:
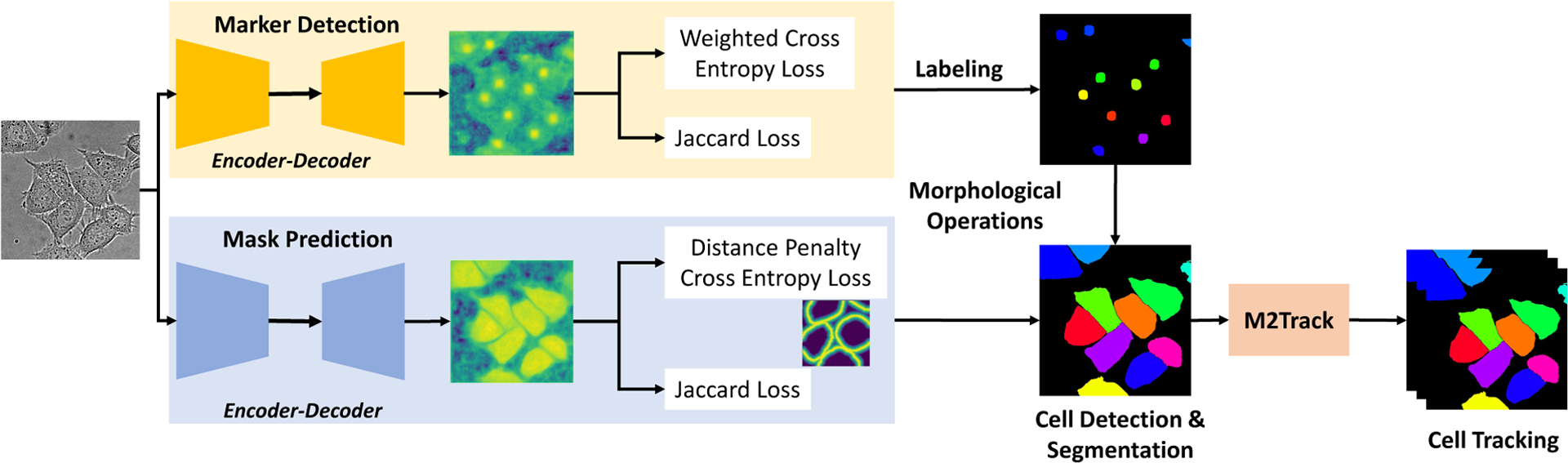
Overall pipeline with two stream DMNet for cell segmentation and M2Track for tracking.
Manually segmenting and tracking cells is an expensive, labor intensive and subjective (difficult to reproduce) task due to the need for deep expert domain knowledge and large amounts of image data acquired during live-cell studies. Automated methods and pipelines are needed to perform microscopy video analysis, particularly to segment, track, and characterize cells to accelarate scientific discovery and clinical adoption.
Over several decades, many classical computer vision methods and pipelines have been developed for automated cell detection and segmentation [8, 54, 23, 47]. More recently, various models have been developed for cell boundary prediction to handle segmentation of touching cells [63, 57, 63, 57, 40, 39, 27, 64]. However, accurate cell analysis under different protocols, imaging modalities and cell types remains challenging due to experimental variability, low signal-to-noise ratios, touching or overlapping cells, indistinct deforming boundaries particularly in high cell density cases, agile, unpredictable motion of individual cells, and dynamic interactions between cells.
Recently, deep-learning methods have shown tremendous success in many applications of computer vision including natural object image classification [18], aerial scene classification [13], feature tracking in wide area motion imagery [26], 3D point cloud classification [5], and particularly in various biomedical image analysis e.g. vessel segmentation [33], and malaria diagnosis [34] etc. If adapted, these methods offer promising solutions for cell detection and segmentation.
Cell tracking and lineage is the process of locating cells of interest in images and maintaining their identity over time across cell divisinos to analyze their spatio-temporal behavior (i.e. proliferation, mitosis, and apoptosis). Cell tracking plays an important role in biomedical research for tasks such as cell lineage-tracing [60, 6, 19] and high-throughput motion or behavior analysis [24, 32, 4, 67]. Analyzing cell behavior on live-cell videos requires robust cell tracking approaches to overcome the challenges that the videos have, such as, frequent cell deformations, non-distinct appearance, low image quality in term of contrast, resolution, and imaging acquisition artifacts.
A two-stage segmentation and tracking pipeline is proposed in this work to localize and track different cell types in time-lapse video sequences, as shown in Figure 1. The pipeline consists of two main modules: cell segmentation and cell tracking modules. The cell segmentation module is designed to precisely localize and segment different cells, and the tracking module uses a multi-step data association approach to efficiently track cells across frames. Our pipeline participated in CTC-6 with results on eight 2D datasets with different characteristics in term of cell shape, density, motion patterns, and microscopy modalities. Our results either outperformed the other methods that participated to the challenge or produced comparable results as described in the experimental results section.
To summarize, our contributions are three-fold: (i) we developed DMNet a dual-stream marker guided network for accurate cell segmentation and detection, (ii) we designed M2Track a two level cell tracking module for associating detections and linking tracklets, and (iii) our proposed pipeline demonstrates state-of-the-art performance, scalability and robustness across cell types on the CTC-6 microscopy videos. The subsequent parts of this paper are organized as the following. Section 2 reviews the related work in cell segmentation and tracking. Section 3 describes our proposed approach and details in the design of cell segmentation and tracking. Section 4 presents quantitative results on CTC-6 followed by conclusions.
2. Related Work
2.1. Cell Segmentation
Early methods for cell segmentation include simple thresholding methods [37, 72], hysteresis thresholding [30], edge detection [70, 65], or shape matching [14, 68]. Some methods use sophisticated approaches based on region growing [44, 51, 15], machine learning [61, 10, 56] or energy minimization [52, 62, 73, 21, 45, 22, 20, 7]. For a more comprehensive review of earlier cell segmentation methods, please refer to [47, 53].
More recently, with the development of deep learning networks, many methods benefit from training neural networks with annotated data. Existing methods usually design models for cell boundary or border prediction to handle touching cells. [63] proposes to predict adapted thicker borders and smaller cells in the model to reduce the amount of merged cells. [64] designs a novel representation of cell borders, the neighbor distances, to segment touching cells of various types. [40] utilizes distance transforms with discrete boundaries for single cell nuclei, and [27] uses horizontal and vertical gradient maps. [39] tackles the label inconsistencies problem through encoding a center vector.
Different from these other methods working on various border prediction for handling clustered cells, we propose a dual-stream network to generate guided markers to help splitting cells for accurate cell detection and segmentation.
2.2. Cell Tracking
Cell tracking and behaviour understanding algorithms study individual cell movement, velocity, formation, mitosis, cell groups behavior and etc. Tracking methods can be categorized into two groups: tracking-by-detection [42, 45, 59] and tracking-by-model evolution. Tracking-by-detection, requires locating cells in advance on the entire sequences using segmentation [42, 49, 38], or detection algorithms [69, 12] followed by an association process to link detections in time to generate cell trajectories. Tracking-by-model evolution involves an initialization step to locate the cells of interest on the first frame followed by a per cell model evolution in time by using deformable models such as active contours to keep track of individual cell states (position, motion, shape, and orientation) in the following frames [28, 36, 29]. The most popular cell tracking methodology is tracking-by-detection [42, 45, 59]. Some methods use online mode [41], which implement track linking by associating detections between consecutive frames. In such cases, the information is gathered only from current and past frames. These methods tend to be sensitive to detection errors, and produce fragmented tracklets. While the offline models [43], exploit information from the whole time-lapse sequence (i.e. past, and future frames) and have longer and more reliable trajectories.
Our tracking-by-detection cell tracking module is adapted from our earlier works on multi-object tracking for video surveillance [1, 2]. Multi-cell M2Track module is used to track the cells detected by our DMNet segmentation module. The goal is to link the detected cells, recover from missed-detections by better data association using a fast intersection-over-union (IOU) mask matching, predict cell motion using Kalman filtering [31], and link tracklets by taking into account tracklet history such as velocity, motion, and spatial information. Our tracking module can explicitly handle cells entering and exiting the field of view, birth and death of cells, and mitosis.
3. Our Approach Using DMNet and M2Track
The overall pipeline is illustrated in Figure 1. There are two modules in our pipeline: cell segmentation module DMNet, and multi-cell tracking module, as described in the following parts.
3.1. DMNet: Detection and Segmentation
The cell detection and segmentation task is defined to find segmentation mask of each cell. There are two streams in the proposed DMNet, one stream is designed for cell marker detection, and the other is designed for cell mask prediction, as show in Figure 1.
Marker Detection Stream
The marker-based loss function Lmarker(·) is computed pixelwise with respect to the labeled marker annotations using a soft Jaccard and weighted cross-entropy loss functions,
| (1) |
where α and β are used to balance the Jaccard loss LJaccard and weighted cross-entropy loss Lwce. The Jaccard loss is,
| (2) |
Since the distribution of marker and non-marker pixels is highly biased, we use a class balanced cross-entropy loss, which is defined as,
| (3) |
where each prediction map in the mini-batch of marker detection stream is , of size R × C, denotes a predicted marker map (see Figure 2 (e)), yk is the groundtruth mask (see Figure 2 (b), yk is the binarized version of it). balance the marker/non-marker pixels to control the weight of positive over negative samples.
Figure 2:
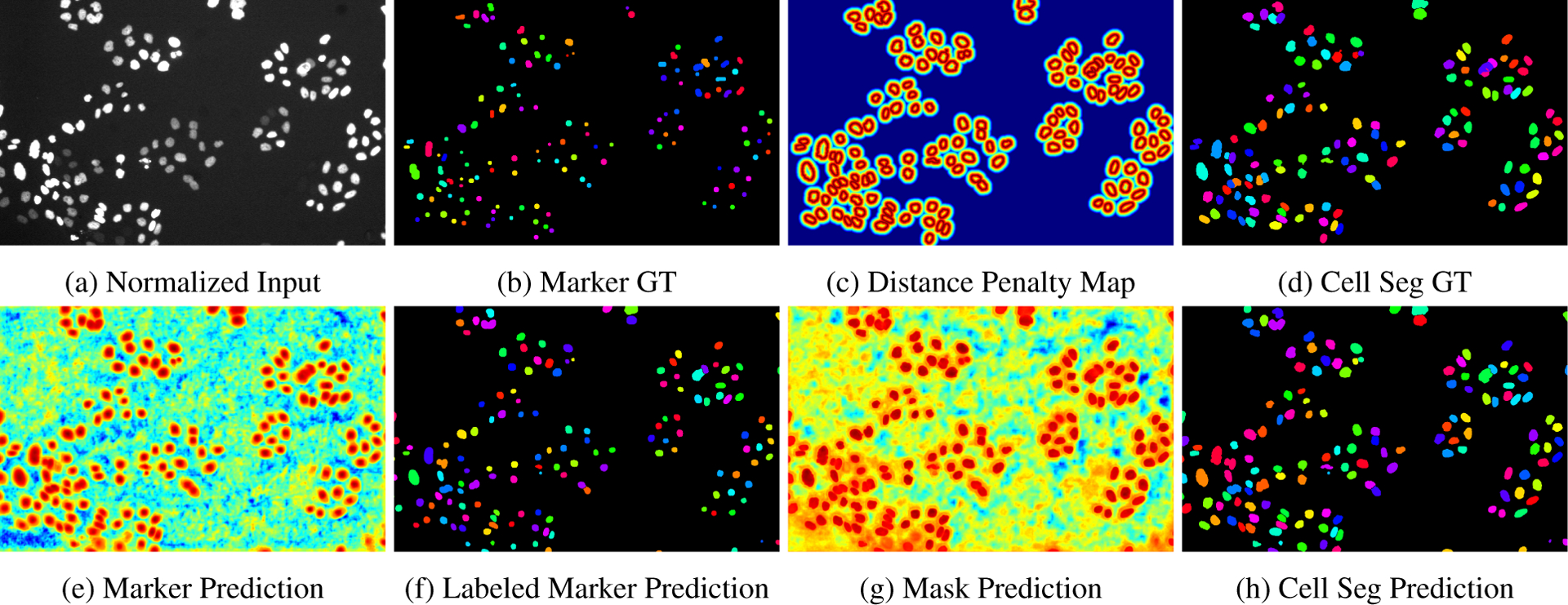
Illustration of intermediate results in the DMNet workflow for cell segmentation: (a) Normalized raw input image, (b) Marker Ground-Truth for the supervision of the marker detection stream, (c) Distance penalty map in Ldist, (d) Cell segmentation ground-truth showing cells with tracking ids (binarized version provides supervision for mask prediction stream), (e) Marker prediction output from marker detection stream, (f) Labeled predicted markers after thresholding and connected component labeling, (g) Mask prediction output from mask prediction stream, and (h) Cell segmentation prediction using mask and marker, after splitting cells using marker guided morphological watershed algorithm.
Mask Prediction Stream
For the mask prediction stream, the loss function Lmask is computed pixelwise with respect to the labeled mask segmentation annotations using a a soft Jaccard and distance penalized cross-entropy loss functions as,
| (4) |
The Ldist is defined as,
| (5) |
where Lce is the cross-entropy loss, and . Here each prediction map in the mini-batch of mask prediction stream is (see Figure 2 (g)), of size R × C. The cross-entropy loss is modified by a distance penalty map ϕ, which inverses and normalizes the distance transform map D. The Euclidean distance transform map is computed as,
| (6) |
where b(i, j) is the location of a background pixel (value 0) that is closest to corresponding input points x(i, j), where edge pixels of cells are 0, and remaining pixels are 1. Figure 2 (c) shows an example distance penalty map ϕ.
Cell Detection & Segmentation
During the inference, both markers and masks are generated, and then the morphological operation watershed [55] is applied to split cell mask guided by our generated markers.
Encoder-Decoder Backbone
For each stream, we use the same Convolutional Neural Network (CNN) structure HRNet [71, 66] as the CNN model to learn the marker localization and mask prediction map since the HRNet encodes rich representations of low-resolution and high-resolution information.
3.2. M2Track: Multi-Cell Tracking Module
Our multi-cell tracking module in Figure 3, tracks the detected cells estimated by the DMNet segmentation module. Tracking module is a multi-step cascade data association process. The cascade data association has two steps: first, short-term tracking which is frame-to-frame data association and matching using mask intersection over union (IOU) score. IOU computation speeds up for overcoming dense scenes with very large cell number. IOU mask score is used for matching current frame detections with previous frame trajectories using linear assignment optimization algorithm [17]. Followed by the second step, long-term tracking, which is called global data association step that connects cells at the track level using spatial and temporal clues to re-link fragmented tracklets. Several modules are used to improve the performance including gating strategy for reducing assignment complexity of ids by pruning improbable assignments; Kalman filter for recovering from miss-detections, and removing unreliable tracklets. For more details of the tracking algorithm, please refer to [1, 2].
Figure 3:
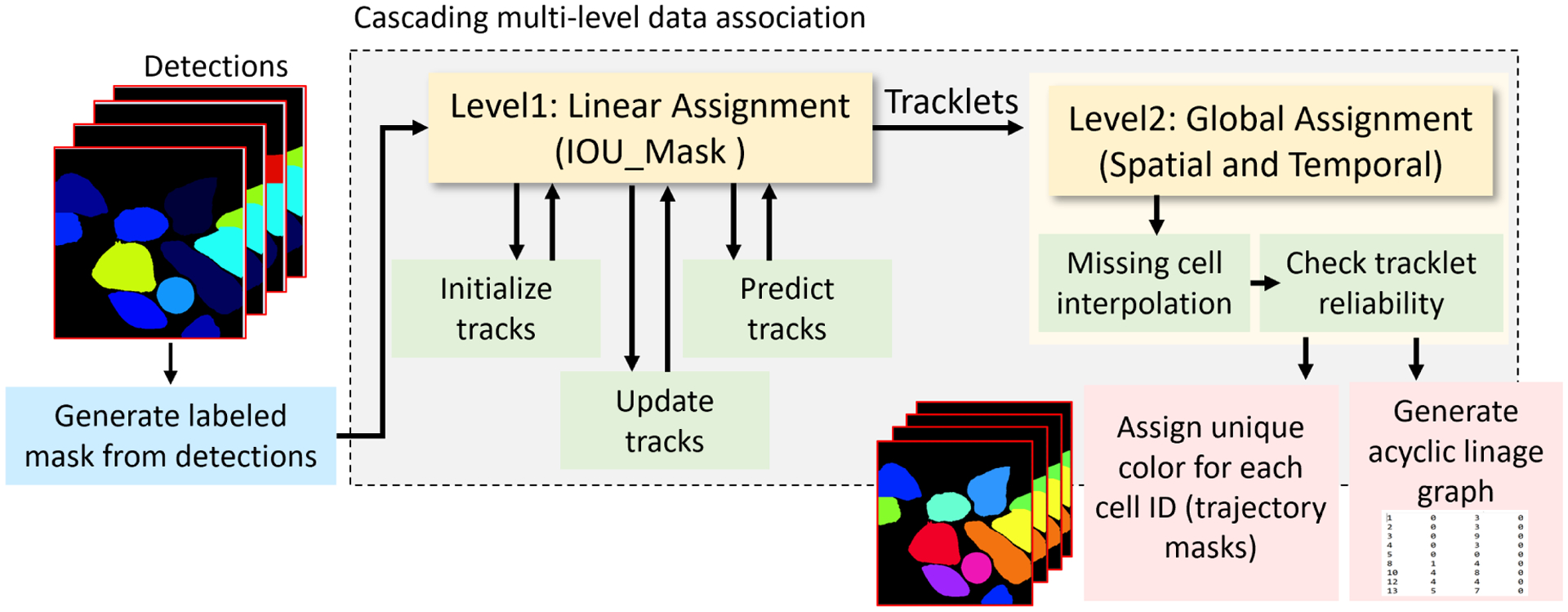
M2Track with intersection-over-union mask overlap matching for multi-cell tracking-by-detection. The two major modules are: Level 1 for managing frame-to-frame linear assignments between detected cells and handles entering and exiting cells, Level 2 for tracklet linking and missing or occluded cells.
Short-Term Tracking:
Short-term data association step, optimizes the associations of current detected cells Dt at frame t to the predicted track Tt−1 at frame t − 1, where the set of detecitons, Dt = {d1, d2, …., dN} is assigned to the previously tracked objects Tt−1 = {T1, T2, …., TM}, and Tt−1 is the set of predicted cell trajectories from previous cell motion history computed using Kalman filter with constant velocity model, N is the number of the detected cells at frame t, M is the number of tracked cells at frame t − 1. Mask IOU score is used to assign detection-to-track between following frames by minimizing a cost matrix using Munkres Hungarian algorithm [50] as:
| (7) |
where is an i row to j column entry on cost matrix representing the cost of assigning detection j to tracklet i at time t and its value represents the IOU between the area of i and j detect masks as:
| (8) |
with constraints,
Circular gating regions around the predicted track positions are used to eliminate highly unlikely associations to reduce computational cost, and to reduce false matches. Pairs of detection and tracks represent the results of minimum optimization. For each individual cell, a (one out of four) status (new track, linked track, lost track, and dead track) is assigned according to the assignment process. Since this step considers only information from consecutive frames, having false detections, occlusions, and matching ambiguities causes track fragmentation. Further step is important to improve the performance.
Long-Term Tracking:
Problems during object detection or data association process result in implicit fragmentation of cells. Long-term tracking is used to re-link fragmented trajectories to produce longer tracks. Using information across long video segments can make this process expensive. Optimizing hypotheses at the track level rather than the object level reduces the computational cost of data assignment by gating uncertain hypotheses. Spatial distances and temporal information are used for filtering.
4. Experimental Results
4.1. CTC-6 Dataset
Cell segmentation and tracking benchmark [11] consists of 2D and 3D time-lapse video sequences of fluorescent counterstained nuclei or cells moving on top or immersed in a substrate. The benchmark consists 20 different datasets (10 for (2D) and 10 for (3D)). They can be either contrast enhancing, or fluorescence microscopy recordings of live cells and organisms. Each dataset consists of two training and two testing videos. The training videos were provided with annotations, gold annotation (containing human-made reference annotations but not for all cells), and silver annotation (containing computer-generated reference annotations). The benchmark has different challenges: 1) Different appearances between datasets; 2) Low contrast between foreground and background; 3) The benchmark were taken in different light condition and different image acquisition environment; And 4) The ground-truth annotations for training set are not fully provided for gold annotations and not accurate for silver annotations.
We participated in ISBI 2021 CTC-6, with over thirty teams reporting results on the CTC website which is updated monthly. Not every method reported results for all datasets in the benchmark. We evaluated our pipeline on eight 2D datasets for cell segmentation and tracking. OPCSB is used for evaluating cell segmentation which is composed of the segmentation metric SEG, and the detection metric DET, as in:
| (9) |
OPCTB is used for cell tracking which is composed of segmentation metric SEG, and tracking metric TRA, as in:
For details of evaluation metrics, please refer to the CTC challenge website [11] and [46].
4.2. Implementation Details
Input images are pre-processed to enhance contrast using a z-score mapping. During training, the marker detection stream is trained with supervision using ground-truth of tracking markers, and segmentation mask is supervised by silver-truth of annotations. Both the marker localization and mask prediction streams were trained on eight 2D datasets (see Tables 1, 2, 3) and five 3D datasets that are not shown (Fluo-C2DL-MSC, Fluo-C3DH-H157, Fluo-C3DL-MDA231, Fluo-N3DH-CE, and Fluo-N3DH-CHO). When using 3D datasets, we used one frame or slice per volume with the most annotated labels for training. Input images are resized then cropped for training. Resize scale factor for each dataset are: Fluo-C2DL-MSC: 0.35, Fluo-C3DH-H157: 0.35, Fluo-C3DL-MDA231: 2, Fluo-N3DH-CE: 0.5, Fluo-N3DH-CHO: 0.6, PhC-C2DL-PSC: 3, BF-C2DL-MuSC: 0.75, BF-C2DL-HSC: 0.75. We crop patches with size of 256 × 256 from images in each dataset to train the networks, except BF-C2DL-HSC, BF-C2DL-MuSC which we crop patches of 512 × 512. Regular data augmentation strategies were used including rotation, flip, and scale from 0.8 to 1.5 for each sample. Hyperparameters are learning rate of 0.001 with Adam Optimizer for training both streams for 300 epochs with, α = 2.5 and β = 10.
Table 1:
DMNet cell segmentation performance (OPCSB) on CTC-6 of March 2021. All reported results are from the CTC Challenge website. The first row is OPCSB and the second row is ranking compared to other submitted algorithms. Not all methods reported results for all datasets which are shown as NA. Since CALT-US did not report results for Fluo-C2DL-MSC we provide two sets of Rankings – 8 datasets and 7 datasets for equivalent comparison. Rank Sum is the sum of all the ranks across cell types. DMNet consistently outperforms other methods on 2D cell segmentation.
| Dataset | BF-C2DL-HSC | BF-C2DL-MuSC | DIC-C2DH-HeLa | Fluo-C2DL-MSC | Fluo-N2DH-GOWT1 | Fluo-N2DL-HeLa | PhC-C2DH-U373 | PhC-C2DL-PSC | Avg | Rank Sum(8,7) |
|---|---|---|---|---|---|---|---|---|---|---|
| KIT-Sch-GE [35] | 0.905 | 0.878 | 0.850 | 0.686 | 0.895 | 0.938 | 0.927 | 0.859 | 0.893 | |
| PURD-US [58] | 0.745 | 0.678 | 0.703 | 0.478 | 0.915 | 0.943 | 0.940 | 0.790 | 0.816 | |
| CALT-US [9] | 0.901 | 0.852 | 0.925 | - | 0.948 | 0.915 | 0.959 | 0.703 | 0.886 | |
| DMNet (Ours) | 0.835 | 0.860 | 0.864 | 0.602 | 0.939 | 0.954 | 0.949 | 0.826 | 0.890 |
Table 2:
DMNet cell tracking performance (OPCTB) on CTC-6 of March 2021. The first row is OPCTB and the second row is ranking compared to other submitted algorithms. All reported results are from the CTC Challenge website. Unreported results are shown as NA. DMNet consistently outperforms other methods on 2D cell tracking.
| Dataset | BF-C2DL-HSC | BF-C2DL-MuSC | DIC-C2DH-HeLa | Fluo-C2DL-MSC | Fluo-N2DH-GOWT1 | Fluo-N2DL-HeLa | PhC-C2DH-U373 | PhC-C2DL-PSC | Avg | Rank Sum |
|---|---|---|---|---|---|---|---|---|---|---|
| KIT-Sch-GE | 0.901 | 0.872 | 0.848 | 0.683 | 0.894 | 0.938 | 0.925 | 0.855 | 0.865 | |
| PURD-US | 0.716 | 0.670 | 0.684 | 0.479 | 0.914 | 0.941 | 0.939 | 0.783 | 0.766 | |
| CALT-US | NA | NA | NA | NA | NA | NA | NA | NA | NA | |
| DMNet (Ours) | 0.828 | 0.849 | 0.854 | 0.591 | 0.939 | 0.953 | 0.947 | 0.821 | 0.848 |
Table 3:
Performance of DMNet segmentation and tracking pipeline on CTC-6 of March 2021. For each performance metric, the first row is accuracy and the second row is ranking compared to all other submitted algorithms (as of March 2021). Top three performance of DMNet by cell type are bolded.
| Dataset | BF-C2DL-HSC | BF-C2DL-MuSC | DIC-C2DH-HeLa | Fluo-C2DL-MSC | Fluo-N2DH-GOWT1 | Fluo-N2DL-HeLa | PhC-C2DH-U373 | PhC-C2DL-PSC |
|---|---|---|---|---|---|---|---|---|
| OPCTB | 0.828 | 0.849 | 0.854 | 0.591 | 0.939 | 0.953 | 0.947 | 0.821 |
| OPCSB | 0.835 | 0.860 | 0.864 | 0.602 | 0.939 | 0.954 | 0.949 | 0.826 |
| DET | 0.971 | 0.979 | 0.926 | 0.681 | 0.946 | 0.985 | 0.975 | 0.945 |
| SEG | 0.699 | 0.742 | 0.802 | 0.522 | 0.931 | 0.923 | 0.923 | 0.708 |
| TRA | 0.957 | 0.957 | 0.907 | 0.661 | 0.946 | 0.983 | 0.972 | 0.933 |
4.3. Comparison on CTC-6 Benchmark
DMNet+M2Track performance is compared with stateof-the-art methods on Cell Segmentation and Cell Tracking Tasks. Since not every method reported results for all 2D datasets, we show the three most competitive methods KIT-Sch-GE [35], PURD-US [58] and CALT-US [9], which have results for almost all eight 2D cell microscopy videos.
DMNet is robust and achieves state-of-the-art cell detection and segmentation performance on all eight 2D CTC-6 datasets. In Table 1, we compare our DMNet with the state-of-the-art methods on CTC-6 challenge. We compute the rank sum of each method of OPCSB on all the datasets. Because CALT-US didn’t report results on Fluo-C2DL-MSC, we put NA in that column, and compute the rank sum on eight datasets and seven datasets respectively as shown in the last column of Table 1. Our DMNet achieves best results on all the 2D datasets with rank sum 60 (eight datsets) and 43 (seven datasets), which demonstrates the robustness and effectiveness on 2D cell segmentation. DMNet+M2Track is robust and achieves the state-of-the-art cell tracking performance on all eight 2D CTC datasets. In Table 2, we compute the rank sum of each method of OPCTB on all the datasets. Because CALT-US didn’t perform cell tracking, therefore it is empty in this table. Our DMNet + M2Track achieves best rank sum with 36, which demonstrates the robustness and effectiveness on 2D cell tracking task. Table 3 shows the ranks of our pipeline compared to the other participants on CTC-6. Our pipeline ranked in the top three on four out of the eight 2D cell type microscopy videos. Not every method provided results for all cell types, whereas DMNet+M2Track results are given for all videos.
Figure 4 shows the results of DMNet+M2Track segmentation and tracking pipeline for three cell types. The first column shows the Raw Input image for three cell types, which are typically low contrast, with dense, clustered cells and small object size. A z-score normalization is applied to the raw input image to remove outliers. The raw input is stretched to increase image contrast, as shown in the second column (Normalized Input). We show the groundtruth segmentation with tracking ids as Tracking GT in the third column. The fourth and fifth columns are the final marker detections and cell tracking predictions for all the cells in each frame of the video. We can clearly see in Figure 4 column (d) that DMNet accurately predicts and separates the cell markers. Hence, using labeled markers as guidance for the watershed algorithm to split the predicted cell masks results in consistently satisfactory cell segmentation results.
Figure 4:
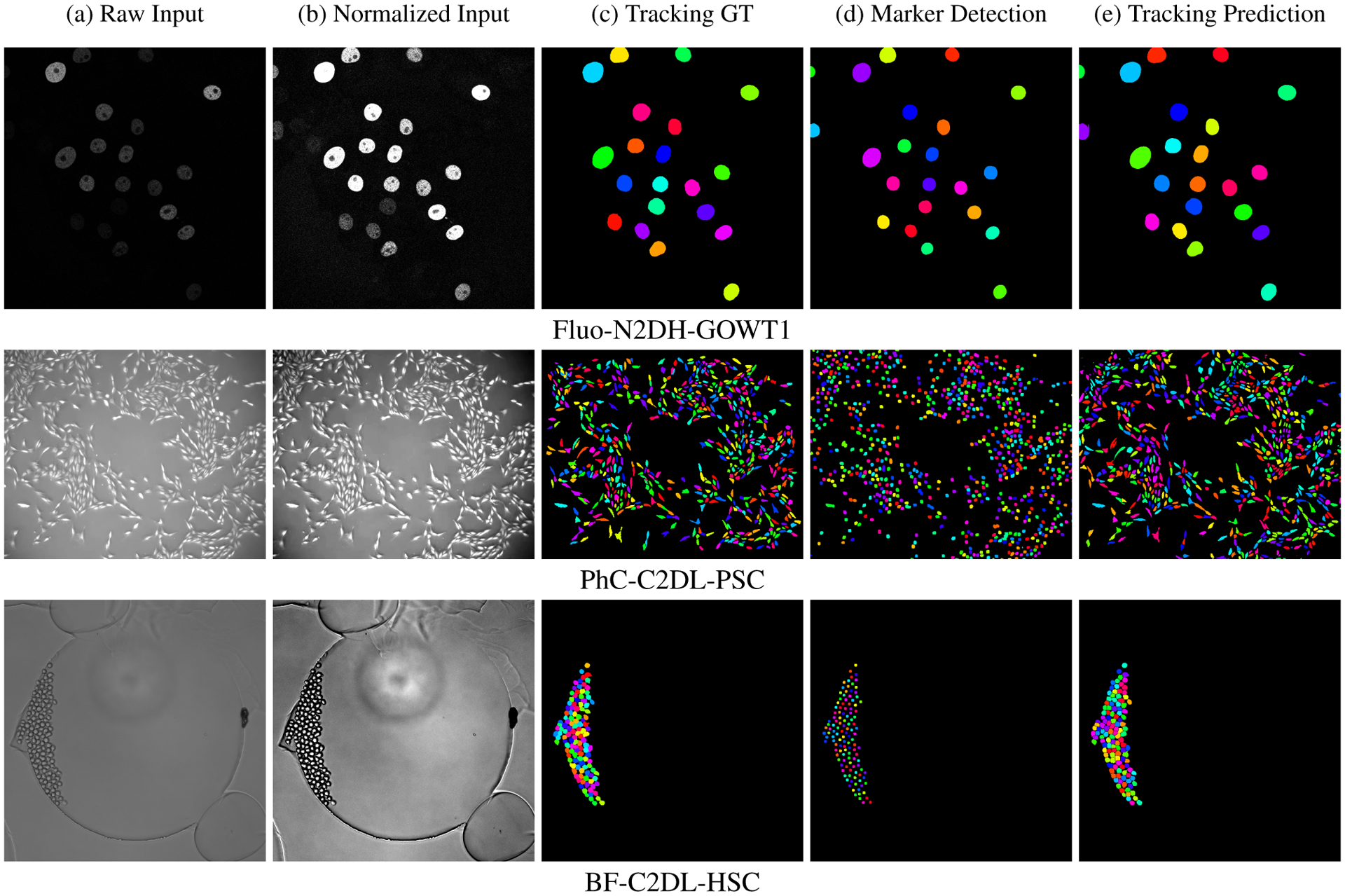
Visualization of DMNet+M2Track segmentation and tracking results for three cell types including Fluo-N2DH-GOWT1, PhC-C2DL-PSC and BF-C2DL-HSC exhibiting a range of cell sizes and densities.
5. Conclusions
The proposed DMNet and M2Track cell segmentation and tracking pipelines provide a common framework across a variety of cell types for high accuracy lineage estimation under challenging sample conditions of high cell density, touching or overlapping cells, deforming cell shape, variable size and indistinct boundaries. For cell segmentation, DMNet uses a dual-stream marker guided deep networks for detection and separation of touching cells. For cell tracking, our M2Track multi-object tracking pipeline generates accurate cell trajectories under challenging conditions (e.g high density, irregular shapes, and cell mitosis activity). DMNet+M2Track is among the best performing methods on the CTC-6 cell microscopy videos across a range of cell types with segmentation and tracking accuracies of over 82 percent (excluding Fluo-C2DL-MSC which has thin elongated mesenchymal stem cells). For 2D cell types our proposed approach has the best rank of all submitted methods in both the cell segmentation and cell tracking subtasks.
Acknowledgments
This work was partially supported by awards from U.S. NIH National Institute of Neurological Disorders and Stroke R01NS110915 and the U.S. Army Research Laboratory project W911NF-18-20285. Any opinions, findings, and conclusions or recommendations expressed in this publication are those of the authors and do not necessarily reflect the views of the U.S. Government or agency thereof.
References
- [1].Al-Shakarji Noor M, Bunyak Filiz, Seetharaman Guna, and Palaniappan Kannappan. Multi-object tracking cascade with multi-step data association and occlusion handling. In IEEE Int. Conf. Advanced Video and Signal Based Surveillance, 2018. [Google Scholar]
- [2].Al-Shakarji Noor M, Seetharaman Guna, Bunyak Filiz, and Palaniappan Kannappan. Robust multi-object tracking with semantic color correlation. In IEEE Int. Conf. Advanced Video and Signal Based Surveillance, 2017. [Google Scholar]
- [3].Ananthakrishnan Revathi and Ehrlicher Allen. The forces behind cell movement. Int. J. Biological Sciences, 3(5):303, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Asante Emilia, Hummel Devynn, Gurung Suman, Kassim Yasmin M, Al-Shakarji Noor, Palaniappan Kannappan, Sittaramane Vinoth, and Chandrasekhar Anand. Defective neuronal positioning correlates with aberrant motor circuit function in zebrafish. Frontiers in Neural Circuits, page 59, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bao R, Palaniappan K, Zhao Y, Seetharaman G, and Zeng W. GLSNet: Global and local streams network for 3D point cloud classification. In IEEE Applied Imagery Pattern Recognition Workshop (AIPR), 2019. [Google Scholar]
- [6].Bao Zhirong, Murray John I, Boyle Thomas, Ooi Siew Loon, Sandel Matthew J, and Waterston Robert H. Automated cell lineage tracing in Caenorhabditis elegans. Proceedings of the National Academy of Sciences, 103(8):2707–2712, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bensch Robert and Ronneberger Olaf. Cell segmentation and tracking in phase contrast images using graph cut with asymmetric boundary costs. In IEEE Int. Symp. on Biomedical Imaging (ISBI), pages 1220–1223, 2015. [Google Scholar]
- [8].Bunyak Filiz, Palaniappan Kannappan, Nath Sumit Kumar, Baskin TL, and Dong Gang. Quantitative cell motility for in vitro wound healing using level set-based active contour tracking. In IEEE Int. Symp. on Biomedical Imaging (ISBI), pages 1040–1043, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].CALT-US. http://celltrackingchallenge.net/participants/CALT-US.
- [10].Castilla Carlos, Maška Martin, Sorokin Dmitry V, Meijering Erik, and Ortiz-de Solórzano Carlos. 3-d quantification of filopodia in motile cancer cells. IEEE Transactions on Medical Imaging, 38(3):862–872, 2018. [DOI] [PubMed] [Google Scholar]
- [11].Cell Tracking Challenge. http://celltrackingchallenge.net.
- [12].Chamanzar Alireza and Nie Yao. Weakly supervised multi-task learning for cell detection and segmentation. In IEEE Int. Symp. Biomedical Imaging, pages 513–516, 2020. [Google Scholar]
- [13].Cheng Gong, Han Junwei, and Lu Xiaoqiang. Remote sensing image scene classification: Benchmark and state of the art. Proceedings of the IEEE, 105(10):1865–1883, 2017. [Google Scholar]
- [14].Cicconet Marcelo, Geiger Davi, and Gunsalus Kristin C. Wavelet-based circular hough transform and its application in embryo development analysis. In VISAPP (1), pages 669–674, 2013. [Google Scholar]
- [15].Cliffe Adam, Doupé David P, Sung HsinHo, Lim Isaac Kok Hwee, Ong Kok Haur, Cheng Li, and Yu Weimiao. Quantitative 3d analysis of complex single border cell behaviors in coordinated collective cell migration. Nature Communications, 8(1):1–13, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Condeelis John and Pollard Jeffrey W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell, 124(2):263–266, 2006. [DOI] [PubMed] [Google Scholar]
- [17].Crouse David F. On implementing 2D rectangular assignment algorithms. IEEE Transactions on Aerospace and Electronic Systems, 52(4):1679–1696, 2016. [Google Scholar]
- [18].Deng Jia, Dong Wei, Socher Richard, Li Li-Jia, Li Kai, and Fei-Fei Li. ImageNet: A large-scale hierarchical image database. In IEEE Conf. on Computer Vision and Pattern Recognition, pages 248–255, 2009. [Google Scholar]
- [19].Ding Yunfeng, Liu Yonghong, Lee Dong-Kee, Tong Zhangwei, Yu Xiaobin, Li Yi, Xu Yong, Lanz Rainer B, O’Malley Bert W, and Xu Jianming. Cell lineage tracing links ERα loss in Erbb2-positive breast cancers to the arising of a highly aggressive breast cancer subtype. Proceedings of the National Academy of Sciences, 118(21), 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dufour Alexandre, Shinin Vasily, Tajbakhsh Shahragim, Guillén-Aghion Nancy, Olivo-Marin J-C, and Zimmer Christophe. Segmenting and tracking fluorescent cells in dynamic 3-D microscopy with coupled active surfaces. IEEE Transactions on Image Processing, 14(9):1396–1410, 2005. [DOI] [PubMed] [Google Scholar]
- [21].Dufour Alexandre, Thibeaux Roman, Labruyere Elisabeth, Guillen Nancy, and Olivo-Marin Jean-Christophe. 3-D active meshes: Fast discrete deformable models for cell tracking in 3-D time-lapse microscopy. IEEE Transactions on Image Processing, 20(7):1925–1937, 2010. [DOI] [PubMed] [Google Scholar]
- [22].Dzyubachyk Oleh, Van Cappellen Wiggert A, Essers Jeroen, Niessen Wiro J, and Meijering Erik. Advanced level-set-based cell tracking in time-lapse fluorescence microscopy. IEEE Trans. Medical Imaging, 29(3):852–867, 2010. [DOI] [PubMed] [Google Scholar]
- [23].Ersoy Ilker, Bunyak Filiz, Higgins John M, and Palaniappan Kannappan. Coupled edge profile active contours for red blood cell flow analysis. In IEEE Int. Symp. on Biomedical Imaging (ISBI), pages 748–751, 2012. [Google Scholar]
- [24].Ersoy Ilker, Bunyak Filiz, Palaniappan Kannappan, Sun Mingzhai, and Forgacs Gabor. Cell spreading analysis with directed edge profile-guided level set active contours. In International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), pages 376–383, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Evans Rachel, Patzak Irene, Svensson Lena, De Filippo Katia, Jones Kristian, McDowall Alison, and Hogg Nancy. Integrins in immunity. J. Cell Science, 122(2):215–225, 2009. [DOI] [PubMed] [Google Scholar]
- [26].Gao K, AliAkbarpour H, Seetharaman G, and Palaniappan K. DCT-based local descriptor for robust matching and feature tracking in wide area motion imagery. IEEE Geoscience and Remote Sensing Letters, pages 1–5, 2020. [Google Scholar]
- [27].Graham Simon, Vu Quoc Dang, Raza Shan E Ahmed, Azam Ayesha, Tsang Yee Wah, Kwak Jin Tae, and Rajpoot Nasir. Hover-net: Simultaneous segmentation and classification of nuclei in multi-tissue histology images. Medical Image Analysis, 58:101563, 2019. [DOI] [PubMed] [Google Scholar]
- [28].Hafiane Adel, Bunyak Filiz, and Palaniappan Kannappan. Level set-based histology image segmentation with regionbased comparison. Proceedings Microscopic Image Analysis with Applications in Biology, 2008. [Google Scholar]
- [29].Hafiane Adel, Bunyak Filiz, and Palaniappan Kannappan. Evaluation of level set-based histology image segmentation using geometric region criteria. In IEEE Int. Symp. on Biomedical Imaging (ISBI), pages 1–4, 2009. [Google Scholar]
- [30].Henry Katherine M, Pase Luke, Ramos-Lopez Carlos Fernando, Lieschke Graham J, Renshaw Stephen A, and Reyes-Aldasoro Constantino Carlos. Phagosight: An open-source matlab® package for the analysis of fluorescent neutrophil and macrophage migration in a zebrafish model. PLOS One, 8(8):e72636, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Kalman RE. A new approach to linear filtering and prediction problems. J. Basic Engineering, 82(1):35–45, 1960. [Google Scholar]
- [32].Kassim Yasmin M, Al-Shakarji NoorM, Asante Emilia, Chandrasekhar Anand, and Palaniappan Kannappan. Dissecting branchiomotor neuron circuits in zebrafish—toward high-throughput automated analysis of jaw movements. In IEEE Int. Symp. on Biomedical Imaging (ISBI), pages 943–947, 2018. [Google Scholar]
- [33].Kassim YM, Glinskii OV, Glinsky VV, Huxley VH, Guidoboni G, and Palaniappan K. Deep U-Net regression and hand-crafted feature fusion for accurate blood vessel segmentation. In IEEE International Conference on Image Processing, pages 1445–1449, Aug 2019. [Google Scholar]
- [34].Kassim Yasmin M, Palaniappan Kannappan, Yang Feng, Poostchi Mahdieh, Palaniappan Nila, Maude Richard J, Antani Sameer, and Jaeger Stefan. Clustering-based dual deep learning architecture for detecting red blood cells in malaria diagnostic smears. IEEE Journal of Biomedical and Health Informatics, 25(5):1735–1746, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].KIT-Sch-GE. http://celltrackingchallenge.net/participants/KIT-Sch-GE.
- [36].Kolla Likhitha, Kelly Michael C, Mann Zoe F, et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nature Communications, 11(1):1–16, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lerner Boaz, Clocksin William F, Dhanjal Seema, Hultén Maj A, and Bishop Christopher M. Automatic signal classification in fluorescence in situ hybridization images. Cytometry Part A, 43(2):87–93, 2001. [DOI] [PubMed] [Google Scholar]
- [38].Li Huiying, Zhao Xiaoqing, Su Anyang, Zhang Haitao, Liu Jingxin, and Gu Guiying. Color space transformation and multi-class weighted loss for adhesive white blood cell segmentation. IEEE Access, 8:24808–24818, 2020. [Google Scholar]
- [39].Li Jiahui, Hu Zhiqiang, and Yang Shuang. Accurate nuclear segmentation with center vector encoding. In International Conference on Information Processing in Medical Imaging, pages 394–404. Springer, 2019. [Google Scholar]
- [40].Li Xieli, Wang Yuanyuan, Tang Qisheng, Fan Zhen, and Yu Jinhua. Dual u-net for the segmentation of overlapping glioma nuclei. IEEE Access, 7:84040–84052, 2019. [Google Scholar]
- [41].Lou Xinghua and Hamprecht Fred A. Structured learning for cell tracking. In NIPS, volume 2, page 6, 2011. [DOI] [PubMed] [Google Scholar]
- [42].Lugagne Jean-Baptiste, Lin Haonan, and Dunlop Mary J. DeLTA: automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Computational Biology, 16(4):e1007673, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Magnusson Klas EG, Jaldén Joakim, Gilbert Penney M, and Blau Helen M. Global linking of cell tracks using the viterbi algorithm. IEEE Transactions on Medical Imaging, 34(4):911–929, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Malpica Norberto, De Solórzano Carlos Ortiz, Vaquero Juan José, Santos Andrés, Vallcorba Isabel, García-Sagredo José Miguel, and Del Pozo Francisco. Applying watershed algorithms to the segmentation of clustered nuclei. Cytometry Part A, 28(4):289–297, 1997. [DOI] [PubMed] [Google Scholar]
- [45].Maška Martin, Daněk Ondřej, Garasa Saray, Rouzaut Ana, Munoz-Barrutia Arrate, and Ortiz-de Solorzano Carlos. Segmentation and shape tracking of whole fluorescent cells based on the Chan-Vese model. IEEE Transactions on Medical Imaging, 32(6):995–1006, 2013. [DOI] [PubMed] [Google Scholar]
- [46].Matula Pavel, Maška Martin, Sorokin Dmitry V, Matula Petr, Ortiz-de Solórzano Carlos, and Kozubek Michal. Cell tracking accuracy measurement based on comparison of acyclic oriented graphs. PLOS One, 10(12):e0144959, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Meijering Erik. Cell segmentation: 50 years down the road [life sciences]. IEEE Signal Processing Magazine, 29(5):140–145, 2012. [Google Scholar]
- [48].Montell Denise J. Morphogenetic cell movements: Diversity from modular mechanical properties. Science, 322:1502–1505, 2008. [DOI] [PubMed] [Google Scholar]
- [49].Moshkov Nikita, Mathe Botond, Kertesz-Farkas Attila, Hollandi Reka, and Horvath Peter. Test-time augmentation for deep learning-based cell segmentation on microscopy images. Scientific Reports, 10(1):1–7, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Munkres J. Algorithms for the assignment and transportation problems. Journal of the Society for Industrial and Applied mathematics, 5(1):32–38, 1957. [Google Scholar]
- [51].de Solorzano C Ortiz, Rodriguez E Garcia, Jones Arthur, Pinkel Dan, Gray Joe W, Sudar Damir, and Lockett Stephen J. Segmentation of confocal microscope images of cell nuclei in thick tissue sections. Journal of Microscopy, 193(3):212–226, 1999. [DOI] [PubMed] [Google Scholar]
- [52].de Solorzano C Ortiz, Malladi R, Lelievre SA, and Lockett SJ. Segmentation of nuclei and cells using membrane related protein markers. J. Microscopy, 201(3):404–415, 2001. [DOI] [PubMed] [Google Scholar]
- [53].Ortiz-de Solórzano Carlos, Munoz-Barrutia Arrate, Meijering Erik, and Kozubek Michal. Toward a morphodynamic model of the cell: Signal processing for cell modeling. IEEE Signal Processing Magazine, 32(1):20–29, 2014. [Google Scholar]
- [54].Palaniappan K, Bunyak F, Nath S, and Goffeney J. Parallel processing strategies for cell motility and shape analysis. In High-throughput Image Reconstruction and Analysis, pages 39–87, 2009. [Google Scholar]
- [55].Parvati K, Rao Prakasa, and Das M Mariya. Image segmentation using gray-scale morphology and marker-controlled watershed transformation. Discrete Dynamics in Nature and Society, 2008, 2008. [Google Scholar]
- [56].Payer Christian, Štern Darko, Neff Thomas, Bischof Horst, and Urschler Martin. Instance segmentation and tracking with cosine embeddings and recurrent hourglass networks. In Int. Conf. Medical Image Computing and Computer-Assisted Intervention, pages 3–11. Springer, 2018. [Google Scholar]
- [57].Guerrero Pena Fidel A, Fernandez Pedro D Marrero, Tarr Paul T, Ren Tsang Ing, Meyerowitz Elliot M, and Cunha Alexandre. J-regularization improves imbalanced multiclass segmentation. In IEEE Int. Symp. on Biomedical Imaging (ISBI), pages 1–5, 2020. [Google Scholar]
- [58].PURD-US. http://celltrackingchallenge.net/participants/PURD-US.
- [59].Rapoport Daniel H, Becker Tim, Mamlouk Amir Madany, Schicktanz Simone, and Kruse Charli. A novel validation algorithm allows for automated cell tracking and the extraction of biologically meaningful parameters. PLOS One, 6(11):e27315, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rodriguez-Fraticelli Alejo E, Weinreb Caleb, Wang Shou-Wen, Migueles Rosa P, Jankovic Maja, Usart Marc, Klein Allon M, Lowell Sally, and Camargo Fernando D. Single-cell lineage tracing unveils a role for TCF15 in haematopoiesis. Nature, 583(7817):585–589, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ronneberger Olaf, Fischer Philipp, and Brox Thomas. U-Net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention, pages 234–241. Springer, 2015. [Google Scholar]
- [62].Sarti Alessandro, De Solorzano C Ortiz, Lockett Stephen, and Malladi Ravi. A geometric model for 3-D confocal image analysis. IEEE Transactions on Biomedical Engineering, 47(12):1600–1609, 2000. [DOI] [PubMed] [Google Scholar]
- [63].Scherr Tim, Bartschat Andreas, Reischl Markus, Stegmaier Johannes, and Mikut Ralf. Best Practices in Deep Learning-based Segmentation of Microscopy Images. KIT Scientific Publishing, 2018. [Google Scholar]
- [64].Scherr Tim, Löffler Katharina, Böhland Moritz B, and Mikut Ralf. Cell segmentation and tracking using cnn-based distance predictions and a graph-based matching strategy. PLOS One, 15(12):e0243219, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Schiegg Martin, Hanslovsky Philipp, Haubold Carsten, Koethe Ullrich, Hufnagel Lars, and Fred A Hamprecht. Graphical model for joint segmentation and tracking of multiple dividing cells. Bioinformatics, 31(6):948–956, 2015. [DOI] [PubMed] [Google Scholar]
- [66].Sun Ke, Xiao Bin, Liu Dong, and Wang Jingdong. Deep high-resolution representation learning for human pose estimation. In IEEE Conf. on Computer Vision and Pattern Recognition, pages 5693–5703, 2019. [Google Scholar]
- [67].Todorov Helena and Saeys Yvan. Computational approaches for high-throughput single-cell data analysis. The FEBS Journal, 286(8):1451–1467, 2019. [DOI] [PubMed] [Google Scholar]
- [68].Türetken Engin, Wang Xinchao, Becker Carlos J, Haubold Carsten, and Fua Pascal. Network flow integer programming to track elliptical cells in time-lapse sequences. IEEE Transactions on Medical Imaging, 36(4):942–951, 2016. [DOI] [PubMed] [Google Scholar]
- [69].Tyson Adam L, Rousseau Charly V, Niedworok Christian J, Keshavarzi Sepiedeh, Tsitoura Chryssanthi, Cossell Lee, Strom Molly, and Margrie Troy W. A deep learning algorithm for 3D cell detection in whole mouse brain image datasets. PLOS Computational Biology, 17(5):e1009074, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wählby Carolina, Sintorn I-M, Erlandsson Fredrik, Borgefors Gunilla, and Bengtsson Ewert. Combining intensity, edge and shape information for 2D and 3D segmentation of cell nuclei in tissue sections. J. Microscopy, 215(1):67–76, 2004. [DOI] [PubMed] [Google Scholar]
- [71].Wang Jingdong, Sun Ke, Cheng Tianheng, Jiang Borui, Deng Chaorui, Zhao Yang, Liu Dong, Mu Yadong, Tan Mingkui, Wang Xinggang, et al. Deep high-resolution representation learning for visual recognition. IEEE Transactions on Pattern Analysis and Machine Intelligence, 2020. [DOI] [PubMed] [Google Scholar]
- [72].Wang Meng, Zhou Xiaobo, Li Fuhai, Huckins Jeremy, King Randall W, and Wong Stephen TC. Novel cell segmentation and online SVM for cell cycle phase identification in automated microscopy. Bioinformatics, 24(1):94–101, 2008. [DOI] [PubMed] [Google Scholar]
- [73].Zimmer Christophe, Labruyere Elisabeth, Meas-Yedid Vannary, Guillén Nancy, and Olivo-Marin J-C. Segmentation and tracking of migrating cells in videomicroscopy with parametric active contours: A tool for cell-based drug testing. IEEE Transactions on Medical Imaging, 21(10):1212–1221, 2002. [DOI] [PubMed] [Google Scholar]
- [74].Zimmer Christophe, Zhang Bo, Dufour Alexandre, Thébaud Ayméric, Berlemont Sylvain, Meas-Yedid Vannary, and Marin J-CO. On the digital trail of mobile cells. IEEE Signal Processing Magazine, 23(3):54–62, 2006. [Google Scholar]


