Abstract
Structural investigations of biomolecules are typically confined to in vitro systems under extremely limited conditions. These investigations yield invaluable insights, but such experiments cannot capture important structural features imposed by cellular environments. Structural studies of proteins in their native contexts are not only possible using state-of-the-art sensitivity-enhanced (dynamic nuclear polarization, DNP) solid-state nuclear magnetic resonance (NMR) techniques, but these studies also demonstrate that the cellular context can and does have a dramatic influence on protein structure. In this chapter, we describe methods to prepare samples of isotopically-labeled proteins at endogenous levels in cellular contexts alongside quality control methods to ensure that such samples accurately model important features of the cellular environment.
1. Introduction
For an organism to survive, its proteins must adopt complex conformations in a challenging environment where macromolecular crowding can derail even robust biological pathways. This situation becomes critical when proteins can adopt more than one stable conformation. In these cases, the environment can clearly influence the conformation by favoring one pathway over another. Such decisions can have striking biological consequences, both negative (as is the case for a variety of protein folding diseases(Dobson, 2001)) as well as beneficial(Chakrabortee et al., 2016; Jarosz et al., 2014). In these cases, the “misfolded” form is often, though not always, an amyloid, a tough fibrillar protein polymer. The effect that environment may have on protein conformations becomes even more pronounced when considering the substantial fraction of the proteome that encodes disordered proteins(Dunker et al., 2001). Intrinsically disordered proteins (IDPs) are important components of the cellular signaling machinery, allowing the same polypeptide to undertake different interactions with different consequences(Wright & Dyson, 2015).
Despite the importance of the environment, structural investigations of biomolecules are typically confined to highly purified in vitro systems. Nuclear magnetic resonance (NMR) is a powerful spectroscopic method for studying molecular structure. A key strength NMR is that it can be used to study non-crystalline amorphous samples. Indeed, there have been a handful of high-profile in-cell NMR studies (Banci, Barbieri, Luchinat, & Secci, 2013; Freedberg & Selenko, 2014; Inomata et al., 2009; Sakakibara et al., 2009; Selenko, Serber, Gadea, Ruderman, & Wagner, 2006; Theillet et al., 2016). These studies suggest that while protein structure can be perturbed, it is largely unchanged by the cellular context. However, these studies used solution-state NMR to detect proteins at concentrations two or more orders of magnitude above endogenous levels, radically altering endogenous stoichiometries. Because solution-state NMR is limited by molecular tumbling times, which depends upon molecular size and solvent viscosity, the minority of the protein that might interact with cellular components would likely be undetectable. Moreover, because this population would comprise a small fraction of the total, it would be difficult, if not impossible, to detect the resulting signal loss. Solid-state NMR is not limited by molecular correlation times in this way. Instead, solid-state NMR is limited by its low sensitivity.
Solid-state NMR spectroscopy is currently undergoing a “sensitivity renaissance” with the development of dynamic nuclear polarization (DNP). DNP increases the sensitivity of NMR spectroscopy through the transfer of the large spin polarization that is associated with unpaired electrons to nearby nuclei (Abragam, 1983; Slichter, 1990). Theoretically, DNP can reduce experimental times by more than five orders of magnitude. However, just as for other structural biology techniques, DNP sensitivity enhancements are critically dependent on experimental conditions (Ni et al., 2013) and sample composition (Akbey et al., 2010; Takahashi et al., 2014) and the specificity of NMR is critically dependent upon the choice of isotopic labeling(Wang et al., 2013). In practice, DNP dramatically enhances (~100-fold) the sensitivity of solid state NMR. Thus an experiment that would require decades of experimental time with traditional solid-state NMR methods can be collected in a day with DNP NMR. These sensitivity gains open up the possibility of investigating the structures of macromolecules at concentrations and in environments that more accurately model their complex native environments (Frederick et al., 2015) (Fig. 1).
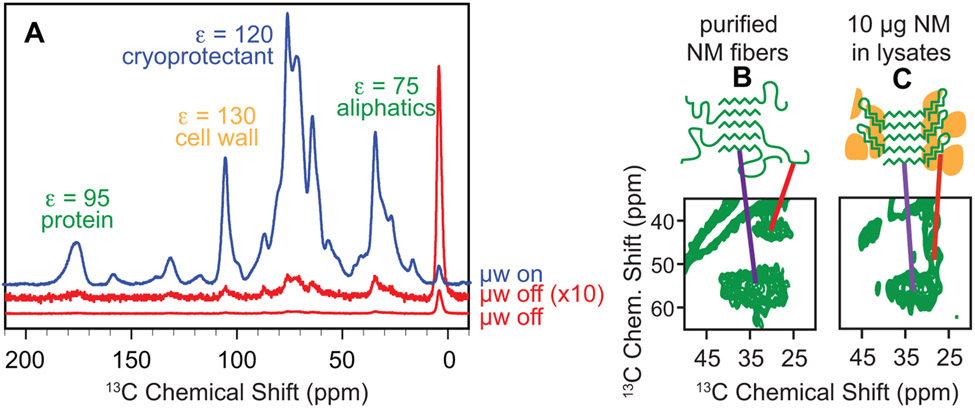
Figure 1:
Dynamic nuclear polarization enhances NMR signals in cellular environments. A) One dimensional 13C{1H} CP spectra both with (blue) and without DNP enhancement (red). DNP NMR results in large signal enhancements (ε) for samples of per-deuterated cellular lysates in a 60:30:10 mixtures of d8-glycerol:D2O:H2O and 10 mM AMUPol at 600 MHz/385 GHz with ω/2π = 12.5 kHz and a sample temperature of 100 K. Spectroscopic selection for the small amount of uniformly isotopically labeled exogenous prepared protein that was added to the cellular lysate mixture reveals that the amyloid cores of this protein have similar conformations (purple line), the intrinsically disordered region (red line) has a very different chemical environment in (B) purified samples than it does in (C) in biological environments.
The application of DNP NMR to complex biological systems has been demonstrated for the yeast prion protein, Sup35. The yeast prion Sup35 has both environmentally-sensitive protein folding landscapes that result in several different amyloid structures as well as intrinsically disordered regions that flank the amyloid fiber. Sup35 is a translation termination factor found in all eukaryotes. In yeast, Sup35 carries an unusual extension with two distinct domains. The most N-terminal region (N) is extremely rich in uncharged polar residues, highly amyloidogenic and responsible for the self-templating activity of the prion. The middle region (M) is highly charged, intrinsically disordered and implicated in chaperone-mediated prion inheritance. Together, N and M constitute the prion-determining domain, NM. The prion state associated with Sup35 amyloid is known as [PSI+]; the non-prion state is designated [psi−]. Biologically, the prion state is easily recognized by a reduction in translation termination, which causes ribosomes to read through stop codons and is readily assayed in the laboratory by the suppression of stop-codon mutations in auxotrophic markers. When a small amount of isotopically labeled NM is added to [PSI+] yeast lysates, the NM protein adopts the amyloid conformation. DNP NMR analysis of NM reveals that NM has a much higher beta sheet content in lysates than in purified samples and that this change in secondary structure is localized to M domain, a region that is intrinsically disordered in purified samples (Frederick et al., 2015).
In this chapter, we describe the approaches our group has developed to determine how different biological environments influence protein structures. This methodology should be useful to groups studying proteins that can take on multiple conformations, have intrinsically disordered regions or form macromolecular complexes. We have developed methods for sample preparation to better represent the structures these proteins can assume in vivo, optimize experimental specificity through careful use of isotopically enriched and depleted carbon, nitrogen and proton sources and improve resolution, through reduction of chemical shift degeneracy via segmental isotopic labeling. These protocols have been optimized for the study of the yeast prion protein, NM, in cellular lysates. However, the approaches here can guide similar studies of proteins with environmentally-sensitivity folding pathways or intrinsically disordered regions at endogenous levels in cellular environments using DNP-assisted NMR spectroscopy.
2. DNP samples of proteins at endogenous levels in native environments.
Most NMR spectroscopy, even DNP-assisted NMR spectroscopy, is performed on highly-concentrated, purified samples. Indeed, much DNP-assisted spectroscopy is focused on the measurement of interactions in samples of purified proteins that were previously sensitivity limited, like long-range distances. In contrast, our focus is on systems previously impossible to study due to very low concentrations. The increase in sample sensitivity enables the study of samples that more accurately capture the native environment of a protein. However the potential pitfalls of low abundance samples are distinct from purified samples and therefore require different controls to assess sample integrity. A major concern is that because the protein we wish to observe is at such low concentrations, signals from natural abundance isotopes that can usually be ignored must be considered. In this section, we describe how to make samples of the NM protein at endogenous levels in native environments. In brief, purified, uniformly-labeled (1H, 13C, 15N) NM protein is added at a concentration of 5 μM to lysates and allowed to polymerize into its amyloid form using the biological template present in [PSI+] cells. In particular, we outline controls to ensure that the sample accurately models important features of the native environment. We monitor cellular phenotypes for yeast grown on isotopically modified media. We monitor protein conformation using a biophysical gel-based aggregation assay to ensure that protein conformation in cellular lysates is maintained through sample manipulation. NMR signal sensitivity is achieved through the use of DNP-enhanced spectroscopy. NMR signal specificity is achieved by pulse sequences that are selective for isotopically enriched sites that are close in space, and therefore over-represented in the exogenously added protein (Figure 2).
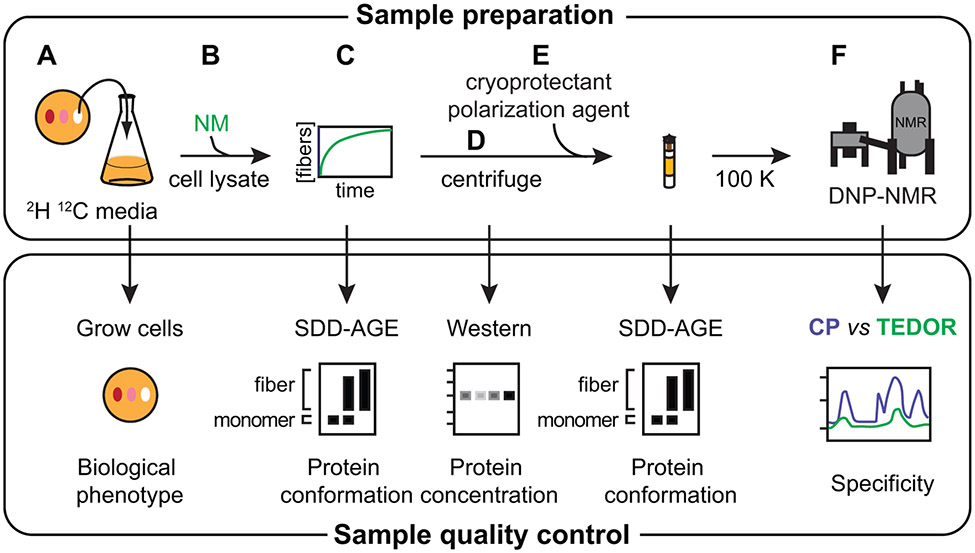
Figure 2:
Sample preparation of proteins at endogenous levels in cellular environments for analysis by DNP NMR requires control of sample integrity. A) Yeast grown in per-deuterated media are phenotyped to ensure that changing the isotopic composition does not affect the biological phenotype of the cells. B) Exogenously-prepared isotopically enriched protein (e.g. NM) is added to lysed cells. C) The exogenously-added protein polymerizes using the cellular material as a template. Assembly of the added protein into the prion form in lysates is monitored by SDD-AGE. D) The insoluble portion of the sample is collected by centrifugation. Western blot analysis using an antibody specific for the protein of interest controls for protein degradation products and allows an estimate of the concentration of the added protein in the insoluble fraction. E) The lysate sample is prepared for DNP with the addition of cryoprotectants and the DNP polarization agent, like AMUPol, and frozen. Maintenance of the protein in the prion form through this process is also monitored by SDD-AGE. F) The specificity of the isotopic labeling schemes, particularly with respect to signals arising from natural abundance isotopes in the cellular lysate (see Table 1), are controlled for by comparison of the spectra of all 13C atoms versus only 13C that are within bonding distance of another isotopically enriched atom.
2.1. Isotopic depletion of yeast for yeast lysate DNP NMR samples.
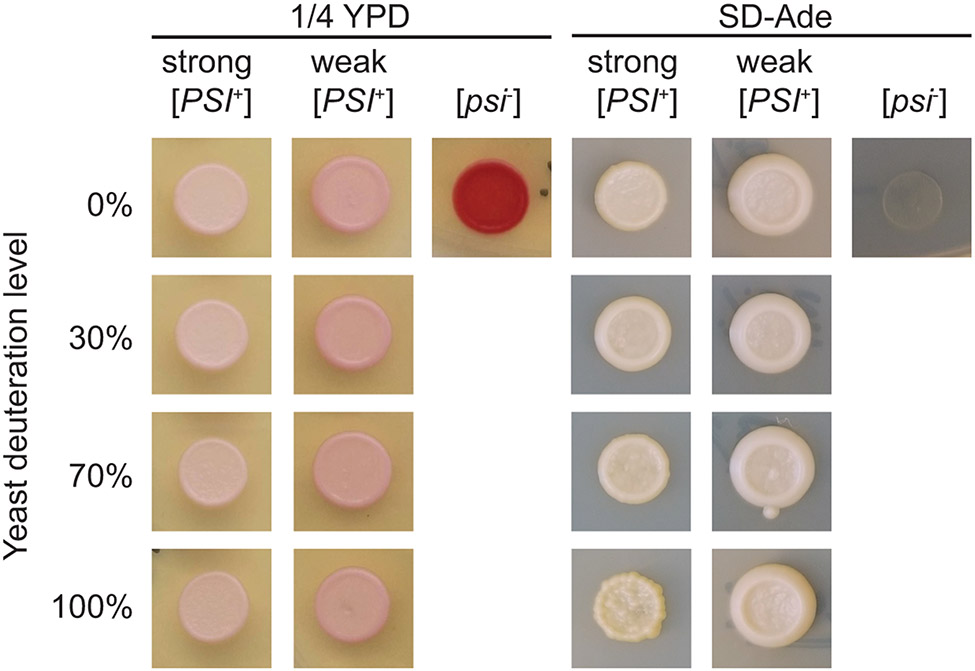
Even very abundant proteins compose a tiny fraction of the cellular biomass of an organism. Thus, the largest contribution of a sample of a protein at endogenous levels is from the biological matrix itself. Because DNP enhancements are highest in deuterated environments, the cellular lysates should be deuterated. However, growth on deuterated media is much slower than growth on protonated media, suggesting that normal cellular processes may be perturbed. Because studying a sample that does not recapitulate the biology under investigation defeats the purpose of including the biological matrix, we control for the ability of the yeast to maintain the phenotype under investigation when cultured in deuterated media. In the case of the yeast prion protein, [psi−] strains have no suppression of the nonsense allele ade1-14. Thus, due to the buildup of an adenine pathway intermediate, they are red on ¼ YPD media and do not grow on SD-ade. Strong [PSI+] strains have higher efficiency ade1-14 suppression and are white on ¼ YPD and grow on SD-ade; weak [PSI+] strains have lower efficiency suppression and are pink on ¼ YPD and grow on SD-ade (Fig. 3). Here we present a generalizable protocol to isotopically deplete yeast of NMR-active isotopes and to check for the maintenance of their prion phenotypes under deuterated growth conditions.
Figure 3:
The yeast prion phenotypes are maintained during growth in media deuterated up to ~100%. A small volume (~ 5 μL) of yeast grown in media with a variety of deuteration levels is spotted on plates for phenotyping immediately before the cells are collected. A) The colony color phenotype on ¼ YPD is maintained for strong [PSI+] (white), weak [PSI+] (pink), and [psi−] (red) yeasts cultured in media with a variety of deuteration levels. B) The growth phenotype on SD-ade is likewise maintained for strong [PSI+] (present), weak [PSI+] (present), and [psi−] (absent) yeasts cultured in media with a variety of deuteration levels, indicating that the protein conformation responsible for this trait is maintained under these conditions.
2.1.1. Equipment
Innova shaker for 30 °C incubation of yeast
Beckmann Coulter Allegra X-14R centrifuge to spin down yeast
Refrigerated Eppendorf centrifuge (e.g. 5427 R)
1.5 mL Eppendorf tubes
−80 °C freezer
2.1.2. Buffers and Reagents
Synthetic defined minimal media (SD-min): Yeast nitrogen base without amino acids (6.7 g/L), D-glucose, 13C-depleted D-glucose or deuterated D-glucose (D-glucose- 1,2,3,4,5,6,-d7 97% atom, ISOTEC) (20 g/L), adenine (0.1 g/L), dissolved in various ratios of water to deuterium oxide (D2O, D 99.8%, Cambridge Isotope Laboratories, Inc.). Prepare 50 mL of media for each DNP NMR sample.
Plates for phenotyping: ¼ Yeast peptone dextrose (YPD): Yeast extract (2.5 g/L), peptone (20 g/L), D-glucose (20 g/L) at pH 7.0, agar (20.0 g/L). SD-ade: Yeast nitrogen base without amino acids (6.7 g/L), SD-ade dropout powder (0.78 g/L, Sunrise Science Products), D-glucose (20 g/L), agar (20.0 g/L).
Prototrophic yeast strain (see notes).
2.1.3. Procedure
Grow yeast to mid-log phase in 50 mL of deuterated SD-min.
Control for maintenance of the biological phenotype. Check that the increasing deuteration conditions do not affect the phenotype of the yeast. For the yeast prion protein, plate 5 μL of the mid-log phase culture on 1/4 YPD to assess colony color and SD-ade plates to assess stop-codon read-through for phenotyping (Fig. 3).
Collect cells by centrifugation at 3,700 x g for 15 minutes and decant supernatant.
Remove residual media by resuspending pellet in 50 mL of sterile water and recollect cells by centrifugation for 3,700 x g for 15 minutes. Decant supernatant.
Resuspend cell pellet in 1 mL of sterile water and transfer mixture to a 1.5 mL tube. Centrifuge for 1 min at 16,000 x g and remove supernatant.
For deuterated samples, re-suspend cell pellet in 1 mL D2O. Centrifuge for 1 min at 16,000 x g, and decant supernatant.
Store cell pellet at −80 °C.
2.1.4. Notes
Most laboratory yeast strains contain mutations in several genetic pathways to allow nutritional selection for ease of genetic manipulations. Control of the isotopic content of yeast biomass requires growth on minimal media, thus all of the amino acid synthesis pathways should be intact.
In the context of the [PSI+] yeast prion strains, a genetic mutation in the adenine synthesis pathway results in a colony color phenotype. Yeast harboring the strong prion strain are thus prototrophic and grow well without additional adenine in the media. Media to grow the yeast harboring the weak prion strains and the prion minus strains are supplemented with adenine.
The deuteration-level of yeast is controlled by the deuteration content of the media. However, yeast preferentially utilizes protons over deuterons. Thus, the deuterium content of the media must be higher than the target deuteration. For target deuteration incorporation levels of ~35%, formulate mediate with 50% (v/v) D2O, for target deuteration incorporation levels of 70%, formulate media with 100% (v/v) D2O and for higher deuteration levels, use deuterated carbon and nitrogen sources in addition to 100% D2O.
Yeast grows slowly in deuterated media. A yeast strain with a doubling time of ~ 2 hours in rich media may take 12 or more hours to double under highly deuterated conditions.
2.2. Addition of isotopically labeled proteins to cellular lysates.
Yeast prion phenotypes are based upon the propagation of the amyloid conformation of NM through the recruitment of monomers into the amyloid form. Making sample for analysis of NM at endogenous levels in the amyloid conformation in cellular lysates for DNP NMR is thus conceptually very simple. We take advantage of the self-templating nature of the amyloid conformation and add a very small amount of denatured exogenously prepared, isotopically-enriched NM protein. The denatured protein adopts the amyloid form using the amyloid template present in the yeast lysates that have been depleted of even natural abundance levels of NMR active isotopes. Following assembly of the isotopically labeled protein into the amyloid conformation, the sample is directly prepared for DNP NMR analysis. Proteins at endogenous levels in native environments make very low sensitivity samples. The success of the experiment therefore relies upon high DNP enhancements, which are very sensitive to sample composition. Empirically, the highest DNP enhancements are attained on cryoprotected samples that have final protonation levels of about 10% with millimolar concentrations of stable bi-nitroxyl radical polarization agents like AMUPol (Sauvée et al., 2013). Because lysate samples are used with minimal downstream manipulation after lysis, the isotopic composition of the buffers is formulated to satisfy these guidelines.
2.2.1. Equipment
QIAGEN Tissulyzer II equipped with QIAGEN TissueLyser Adapter Set.
23-gauge needles
Eppendorf centrifuge (e.g. 5247 R) equipped with a swinging bucket rotor.
Temperature controlled incubators (4 °C - 37 °C)
−80 °C freezer
3.2 mm sapphire rotor, silicon plugs, end cap, and rotor packing tools (Bruker)
2.2.2. Buffers and Reagents
Acid washed glass beads (425-600 μm, Sigma-Aldrich)
Lysis Buffer: 50 mM Tris(hydroxymethyl)aminomethane (Tris) pH 7.4, 200 mM Sodium Chloride (NaCl), 2 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP), 5% d8-glycerol that has been 13C-depleted (glycerol, 12C3, 99.95%, D8, 98%, Cambridge Isotopes, Inc.), 1 mM ethylenediaminetetraacetic acid (EDTA), H2O or D2O. Inhibitors are added right before use: 1x Protease inhibitor cocktail (cOmplete EDTA-free, Roche), 5 μg/mL Aprotinin, 5 μg/mL Leupeptin.
6 M Guanidine hydrochloride (GdHCl)
Purified aggregation-prone protein (e.g. NM, the prion domain of Sup35 (Serio, Cashikar, Moslehi, Kowal, & Lindquist, 1999))
2.2.3. Procedure
Prepare a concentrated (greater than 0.5 mM), denatured solution of purified, isotopically enriched protein (see Note 1).
Lyse yeast by adding 200 μL of lysis buffer and 200 μL acid-washed glass beads to cell pellet from 50 mL of culture and bead beating (Section 2.1) in the TissueLyser II for 8 minutes at 30 Hz.
Remove glass beads by placing the bottom of the 1.5 mL tube containing the lysate on top of a new 1.5 mL tube, puncturing the bottom of tube with a 23-gauge needle, and centrifuging the lysate into the new 1.5 mL tube for 2 minutes at 3,700 x g.
Add isotopically-enriched, denatured purified protein to the yeast lysates. In the case of the NM, add protein from a stock solution at a concentration of 0.5 mM or greater to a final concentration of 5 μM (Fig. 2B). Vortex briefly and incubate, quiescent, at prion strain appropriate temperature for 24 hours; strong prion strains are propagated at 4 °C and weak prion strains are propagated at 25 °C. Assess polymerization by semi-denaturing detergent agarose electrophoresis (SDD-AGE) (Section 2.3. Fig. 2C, 4B).
Collect the insoluble fraction of the polymerization reaction by centrifugation at 16,000 x g for 1 hour at 4 °C (Fig. 2D). Remove the supernatant (see note 4).
Prepare the insoluble cell pellet for DNP analysis. Estimate the volume of the pellet. Add deuterated, 13C-depleted d8-glycerol and polarization agent (e.g. AMUPol) (Fig. 2E) to desired final concentrations (see note 3).
Pack the sample into 3.2 mm sapphire rotor by transferring the cryoprotected cell pellet into the rotor with a pipette. Centrifuge at 16,000 x g for 30 seconds in a swinging-bucket centrifuge for even packing. Seal the sample with a silicon plug using the tool kit and instructions provided by Bruker. Store at −80 °C.
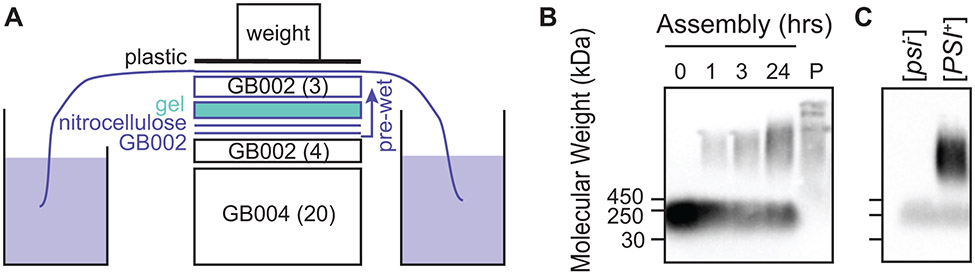
Figure 4:
Semi-Denaturing Detergent-Agarose Gel Electrophoresis (SDD-AGE) is used to visualize the aggregation state of recombinantly-expressed, exogenously-added proteins. After separation of large protein aggregates on agarose gels, the proteins are transferred to a nitrocellulose membrane using capillary transfer. Because the read-out is antibody based, SDD-AGE is appropriate for analysis of proteins at low concentrations in complex mixtures. A) Transfer of proteins to the nitrocellulose membrane is accomplished by capillary transfer. The agarose gel is placed in direct contact with a wet nitrocellulose membrane. The gel and membrane are placed on a stack of dry blotting paper, covered by wet blotting paper kept moist through the action of a wick in a reservoir of buffer. B) The kinetics of assembly of exogenously-added NM (visualized in this case by an antibody against a his6 tag) into the amyloid form in yeast lysates (e.g. Figure 2C). Monomeric NM is removed by centrifugation as evidenced by analysis of the pellet (P) as described in Figure 2D. C) Amyloid aggregates remain intact after the addition of cryoprotectants, polarization agents and freezing (e.g. Figure 2E). Moreover, these manipulations do not induce the formation of amyloid as evidenced by the lack of high molecular weight aggregates in samples prepared using [psi−] yeast.
2.2.4. Notes
In the case of the NM, purified protein stocks(Serio et al., 1999) are stored at −80 °C as 1:5 solutions of concentrated purified protein:methanol. Remove protein from methanol by centrifugation for 10 minutes at 16,000 x g. Decant the supernatant, solubilize the pellet in a small volume of 6 M GdHCl and boil for 10 minutes to denature the protein before use.
Estimate the volume of the pellet by putting a known volume of water in a same size tube. In our hands, 50 mL of yeast culture at mid-log phase gives a ~30 μL volume of cell pellet.
The cryoprotectant composes a large proportion of the sample volume. Thus, because the amount of 13C from natural abundance is greater than the 13C from the exogenously added protein (see Table 1), deuterated, 13C-depleted, glycerol is used to simplify the spectroscopy.
Empirically, DNP samples with proton content of ~10% (v/v) have the highest DNP enhancements. Lysis buffers for cellular lysate DNP samples are therefore formulated to provide 10% protonation upon the final sample formulation. Deuterated cryoprotectant and D2O-containing buffer comprise the remainder of the DNP sample volume. Thus, for a 60:30:10 (d8-glycerol:D2O:H2O) DNP sample containing 10 mM AMUPol, use lysis buffer that is 75% (v/v) D2O so that the sample will contain 10% (v/v) H2O upon addition of 60% (v/v) d8-glycerol.
At step 6, the soluble portion of the lysates can be analyzed by 1H solution state NMR to determine the level of incorporation of deuterium into the cellular biomass (Perkins, 1981).
Table 1.
Spectroscopic Filtering for Adjacent Isotopic Sites Enables Specific Detection of the Uniformly Isotopically Enriched Protein in the Background of Yeast Cellular Lysates Grown Using Natural Abundance or Isotopically Depleted Media
| Protein (uniformly isotopically labeled) | Yeast (natural abundance media) | Yeast (isotopically depleted media) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Quantity (µg) | Total | Carbon | Nitrogen | Total | Carbon | Nitrogen | Total | Carbon | Nitrogen |
| All isotopes | 20.0 | 10.8 | 3.3 | 30,000.0 | 13800.0 | 2700.0 | 30,000.0 | 13800.0 | 2700.0 |
| NMR-visible | 20.0 | 10.8 | 3.3 | 151.8 | 10.0 | 69.0 | 0.3 | ||
| 13C-adjacent isotopes | 10.8 | 3.3 | 1.7 | 0.1 | 0.3 | 0.0 | |||
2.3. Control of protein conformation with SDD-AGE
Unlike samples of purified proteins, most biophysical methods to verify sample integrity cannot be applied to samples at endogenous levels in cellular contexts because the cellular contexts limit the specificity for the protein of interest and the very low sample quantity limit the sensitivity of most experiments. Antibody-based methods are specific for the protein of interest (or an attached epitope, like a his6 tag) and are sensitive enough to detect proteins at their endogenous levels. Thus, a critical control of DNP sample integrity is a simple Western blot. Compare the exogenously added protein signal in lysates without exogenously added protein to lysates containing exogenously added protein alongside several samples of purified exogenous protein at known concentrations. Western analysis of the NMR sample ensures that the exogenously added protein has not been degraded and allows estimation of the concentration of the protein of interest in the NMR sample (Figure 2D).
For aggregation-prone proteins, additional biophysical information about the protein conformation can be determined. The amyloid conformation is a very stable high molecular weight polymer. Indeed, one of the hallmarks of this conformation is its stability at room temperature in 2% SDS and instability under the same conditions when boiled. SDD-AGE separates proteins based on molecular weight, but uses larger pore sizes of agarose gels (“DNA gels”) to separate higher molecular weight protein aggregates. This allows for detection of SDS-stable high molecular weight aggregates using an antibody specific for the protein of interest (Halfmann & Lindquist, 2008). Because the technique is somewhat non-standard due to the agarose gel, which requires capillary transfer of the proteins from the gel to the nitrocellulose membrane rather than the more familiar semi-dry electrotransfer technique, we provide a protocol for SDD-AGE here. The methodology can be used to monitor amyloid assembly kinetics in minimally diluted cellular lysates as well as to determine if cryoprotection, freezing, the addition of the polarization agent, as well as any other experimental, environmental, or genetic perturbations affect amyloid formation.
2.3.1. Equipment
Standard DNA gel electrophoresis equipment
Turboblotter Kit including Whatman paper (GE Healthcare)
Imager for visualization of Western blots
2.3.2. Buffers and Reagents
1X Tris-acetate-EDTA (TAE): 40 mM Tris, 40 mM Glacial Acetic Acid, 1 mM EDTA
1.5% agarose gel (Medium to high gel strength, low EEO agarose (Research Products International) gel with composition 1x TAE + 0.1% Sodium Dodecyl Sulfate (SDS)
2x protein loading dye: 100 mM Tris pH 6.8, 4% beta-mercaptol ethanol, 4% SDS, 10% glycerol, bromphenol blue to preference.
Running buffer: 1X TAE, 0.1% SDS
HiMark pre-stained protein standard (Thermo Fisher Scientific)
1x Tris-buffered saline (TBS): 50 mM Tris pH 7.5, 150 mM NaCl
Nitrocellulose membrane (Amersham™ Hybond ECL, 0.45 μm, 200x 200 mm, GE Healthcare)
Standard Western blotting materials
2.2.3. Procedure
Prepare gel for SDD-AGE by boiling 1.5% agarose in 1X TAE. Once dissolved let this mixture cool, then add 0.1% SDS. Mix and pour gel.
Prepare samples for SDD-AGE analysis by the addition of a 2X protein loading dye at room temperature; boiling will destroy amyloid aggregates.
Load samples and HiMark ladder and run gel at a low voltage (less than or equal to 3 V/cm) in running buffer until samples are 1 cm from the end of the gel.
Assemble capillary transfer stack. Cut all materials to the size of the agarose gel. Stack 20 sheets of dry Whatman paper GB004 under 4 sheets of dry Whatman paper GB002. Wet the wick, four sheets of Whatman paper GB002, and the nitrocellulose membrane in TBS. Ensuring there are no bubbles, place one piece of GB002 on the stack of dry Whatman paper, followed by the nitrocellulose membrane, agarose gel, four more pieces of Whatman paper, the wick, a piece of plastic and an ~ 0.25 kg weight (Fig. 4A).
Transfer protein to nitrocellulose membrane using an overnight capillary transfer.
Visualize protein of interest with an antibody that is specific for the protein of interest using standard Western protocol.
2.3.4. Notes
Add the concentrated SDS solution to the gel just before pouring to avoid boiling or bubble formation.
Because the protein of interest may be distributed over a large range of molecular weights, the sensitivity of SDD-AGE membranes may be lower than that of a typical Western blot. Higher sensitivity electrochemiluminescence (ECL) reagents for detection of horseradish peroxidase (HRP) enzyme, such as Supersignal West Femto maximal sensitivity reagent (Thermo Scientific) may be required.
2.4. Detection of proteins at endogenous levels in native environments using DNP spectroscopy
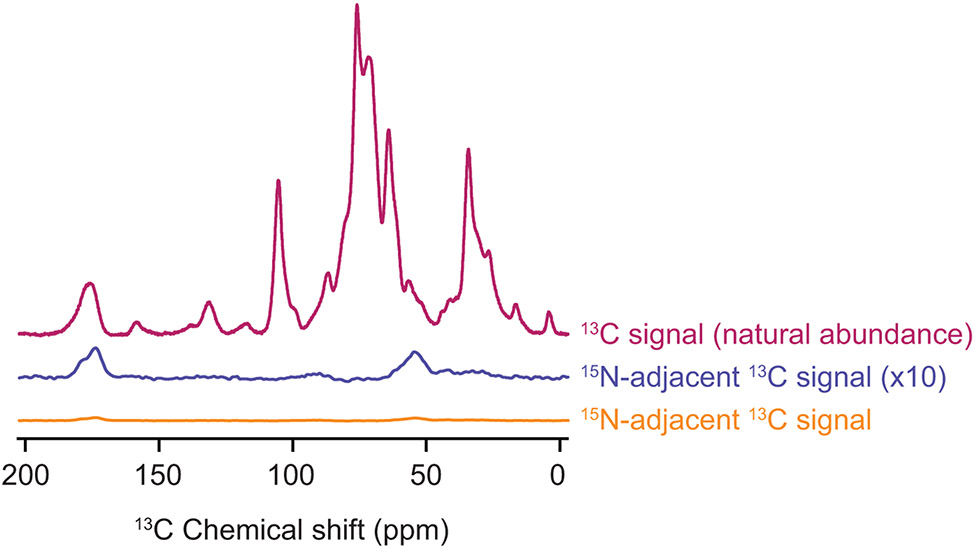
The cell lysates compose the vast majority of the sample volume for study of proteins at endogenous levels in native environments. Even when the cells are grown on NMR active isotope depleted proton, carbon and nitrogen sources, the NMR active isotopic content of the cellular lysates is still greater than that of the added uniformly isotopically enriched protein. However, because the natural abundance of 13C is 1.1%, only 0.01% of the 13C sites in the cell lysates are adjacent to another 13C site. Conversely, all the 13C sites in the exogenously added uniformly 13C,15N-labeled NM have 13C sites adjacent to another isotopically labeled site. To isolate 13C signals from NM and filter out background 13C signals from the cell lysates, we use spectroscopic filtering for adjacent isotopically labeled sites (Fig. 2F, Fig. 5). The transfer echo double resonance (TEDOR) pulse sequence specifically reports on 13C sites that dipolar couple to a nearby 15N site (Jaroniec, Filip, & Griffin, 2002). Because TEDOR achieves heteronuclear coupling through an repetitive INEPT-based π-pulse trains, despite having somewhat lower experimental efficiencies than other heteronuclear pulse schemes, this pulse sequence is preferred due to the challenges involved in optimization of other pulse sequences like double cross polarization (CP) (Schaefer, McKay, & Stejskal, 1979), on low sensitivity samples. By selectively observing only signals from adjacent 13C and 15N sites, the signals from the exogenously added uniformly isotopically labeled protein are an order of magnitude larger than those from the cellular lysate (Table 1).
Figure 5:
The combination of isotopic depletion of the cellular lysates with spectroscopic selection for adjacent isotopically labeled sites enables highly specific detection of proteins at endogenous concentrations. One dimensional 13C{1H} CP spectra report on all of the 13C in the sample (magenta). The TEDOR experiment with a 1.6 ms mixing time (orange) is selective for 13C sites with adjacent 15N atoms. Such sites are much less common (see Table 1) than 13C sites overall. The TEDOR spectra (orange) is multiplied by a factor of 4 to account for differences in experimental efficiency between the 13C CP experiment (magenta) and the TEDOR experiment (orange). It is further multiplied by a factor of 10 (blue) to allow comparison of the difference in features of the peak that reports on the carbonyl carbon chemical shift around 175 ppm. The peak near 175 ppm in the TEDOR spectra is shifted towards values that are consistent with beta sheet conformations and has a shoulder near 180 ppm, in line with the amino acid composition of the protein. Specificity for the 1 μM concentration of added protein from the cellular background can be obtained for samples of per-deuterated cellular lysates in a 60:30:10 mixtures of d8-glycerol:D2O:H2O and 10 mM AMUPol at 600 MHz/385 GHz with ω/2π = 12.5 kHz and a sample temperature of 100 K.
2.4.1. Equipment
Dynamic nuclear polarization magic angle spinning nuclear magnetic resonance (DNP MAS NMR) experiments were performed on a 600 MHz Bruker Ascend DNP NMR spectrometer/7.2 T Cryogenfree gyrotron magnet (Bruker), equipped with a 1H, 13C, 15N triple-resonace, 3.2 mm low temperature (LT) DNP MAS NMR Bruker probe (600 MHz).
3.2 mm sapphire rotor, silicon plugs, end cap, and rotor packing tools (Bruker)
2.4.2. Buffers and Reagents
Standard Proline sample: 60% d8-glycerol that is 13C-depleted (glycerol, 12C3, 99.95%, D8, 98%, Cambridge Isotopes, Inc.), 30% D2O (D2O, D 99.8%, Cambridge Isotope Laboratories, Inc.), 10% H2O, 1.5 mg uniformly 13C-15N labeled L-Proline (98 atom % 13C, 98 atom % 15N, 95% (CP), ISOTEC) with 10 mM AMUPol packed in a 3.2 mm sapphire rotor with silicon plug and ceramic end cap.
Sample containing your protein at endogenous concentrations in cellular lysates made in Section 2.2 capped with ceramic end cap.
2.4.3. Procedure
Determine the maximum instrumental enhancement by measuring the enhancement of the standard proline sample. Record 1D 13C CP spectra with and without microwaves irradiation. Multiply the spectrum recorded without microwave irradiation to match the intensity of spectrum recorded with microwave irradiation; this number is your DNP enhancement. Spectra of the standard proline sample with good signal to noise can usually be collected with 8 scans. DNP enhancements for proline at 600 MHz with ~8 kHz MAS at 100 K are typically between 120 and 140.
Determine the DNP enhancement your protein at endogenous concentrations in cellular lysates. Record 1D 13C CP spectra with and without microwaves irradiation. These samples have very low sensitivity, therefore spectra with good signal to noise typically require 2048 or more scans.
Selectively observe your labeled protein of interest by recording a 1D TEDOR spectra. The TEDOR is selective for 13C-15N pairs, which are less abundant (Table 1) and heteronuclear transfers are less sensitive than homonuclear transfers therefore spectra colleted with microwave irradiation typically require 2048 or more scans (Figure 5).
2.4.4. Notes
DNP enhancement of proteins at endogenous concentrations in cellular lysates are typically ~ 80% of the standard proline sample.
Typically, the 90 degree pulse for carbon is at 4.5 μs (55.5 kHz) and nitrogen is at 6 μs (41.7 kHz) for MAS NMR experiments. γB1/2π are 83 to 100 kHz for 1H (π/2 = 2.5 to 3 μs), 50 to 83 kHz for 13C (π/2 = 3 to 5 μs), and 40 to 50 kHz for 15N.
When optimizing TEDOR use a tangential shape pulse at the carbon or nitrogen channel, while the 1H RF amplitude was kept constant at ωS/2π + ωr/2π) = ωH/2π kHz. Keep 1H decoupling at 90 kHz. Recycle delays for these samples are typically under 5 s.
The total time of TEDOR mixing is the duration of the 13C and 15N π-pulse trains in the sequence and should be ~ 1.8 ms (one-bond distance) for this experiment. Rotor synchronization is required for accuracy of dipolar coupling measurement.
3. Purified amyloid samples
An essential prerequisite to determining whether cellular environments have an effect on protein structure is structural information about the protein studied in isolation. Comparison of MAS NMR spectra of a purified protein acquired under traditional conditions to DNP NMR spectra of a protein in cellular lysates could be informative about regions of high molecular order. However, differences in the experimental conditions between traditional NMR samples and DNP NMR samples introduce confounding factors. DNP NMR samples contain cryoprotectants and are collected at low experimental temperatures. While these differences might not have large effects on the structure of the rigid core of amyloid fibers, this region is often flanked by highly dynamic or intrinsically disordered regions(Fitzpatrick et al., 2017; Heise et al., 2005; Helmus, Surewicz, Nadaud, Surewicz, & Jaroniec, 2008a). To isolate structural differences between purified fibers and amyloid fibers formed in cellular lysates to differences imposed by the cellular lysates rather than simply the freezing of dynamic motions or interactions with the cyroprotectant, the spectra of fibers formed in cellular lysates must be compared with purified fibers under the same experimental conditions. We provide protocols to prepare such samples in the following section.
3.1. Cellular lysate-derived amyloid seeds
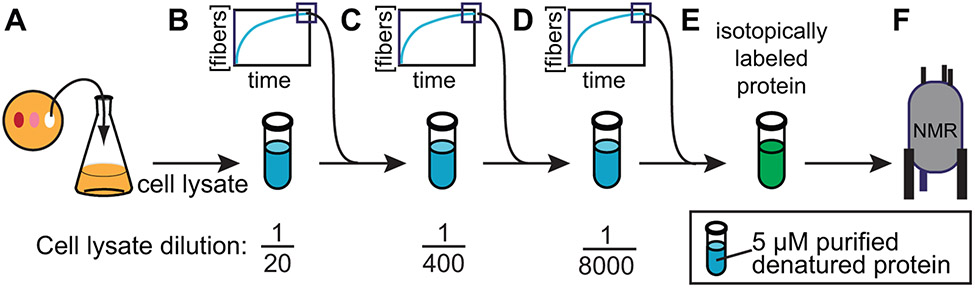
Dilute solutions of amyloid-forming proteins will stochastically assemble into amyloid fibers. However, the de novo assembled fibers may not all have the same conformation and differences in buffer conditions or temperature can alter the dominant fibril form. Variability between preparations of amyloid fibers can be eliminated by using a template, or seed, to start the assembly reaction. While the source of the template is important consideration for amyloid fibers in general, it is of heightened interest in this context. To delineate the effect of the cellular environment from differences in the in vivo versus de novo amyloid initiation events, using the same template for both the purified sample and the cellular lysate-containing sample is critical. To faithfully produce isotopically labeled NM samples of a pure fiber type, we have developed protocols to template recombinant denatured NM proteins into the amyloid conformation using cellular lysates as the template. We lyse phenotypically strong or weak [PSI+] yeasts by bead beating, and then add purified denatured NM to the lysates and allow the protein to template from the bona fide biological prion. The assembled material is then diluted and used as the template in three subsequent assembly reactions, simultaneously amplifying the quantity of amyloid and diluting away cellular lysates (Fig. 6).
Figure 6:
Fiber formation can be templated by yeast cell lysates. A) Cultured yeast cells harboring the desired prion phenotype are grown to mid-log phase and then lysed by bead beating. B) Lysates are diluted into buffer containing low concentrations of purified recombinant denatured protein. C & D) Subsequent rounds of polymerization are seeded by addition of a small volume of the previous reactions, effectively removing cellular material by dilution. E) These lysate templated seeds are used to polymerize a large volume of purified recombinant isotopically enriched protein into the amyloid form. F) This sample serves as a control for the effects of cellular environment on protein structure to complement study of the same protein in cellular lysates.
3.1.1. Equipment
Sonicator (e.g. Branson Digital Sonifier 450).
Plate reading fluorimeter with filters for ThT fluorescence (ex. 460 nm, em. 480 nm).
3.1.2. Buffers and reagents
YPD: 10 g/L of yeast extract, 20 g/L of peptone, and 20 g/L of dextrose at pH 7.0.
Seeding lysis buffer: 50 mM Tris pH 7.4, 200 mM NaCl, 2 mM TCEP, 5% glycerol, 1 mM EDTA, 4 mM phenylmethylsulfonyl fluoride protease inhibitor (PMSF), 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 0.1 μg/mL cOmplete™ Protease Inhibitor Cocktail pellet (Roche).
Congo red binding buffer (CRBB): 5 mM potassium phosphate pH 7.5 and 150 mM NaCl.
Assembly buffer: 50 mM potassium phosphate pH 7.5, 1.5 M NaCl, 5 μg/ml aprotinin, 5 μg/ml leupeptin
Glass beads, acid-washed, 425-600 μm (Sigma Aldrich)
5 mM Thioflavin T
Purified, denatured, aggregation-prone protein (e.g. NM, the prion domain of Sup35 (Serio et al., 1999))
3.1.3. Procedure
Grow 50 mL of yeast culture with appropriate phenotype in YPD, instead of SD-min, following protocol in section 2.1.3.
Obtain yeast lysate by following steps 2-3 in section 2.2.3 using seeding lysis buffer instead of lysis buffer.
Dilute lysate 5x with assembly buffer containing purified, denatured NM (or your protein of interest) at a concentration of 5 μM (Fig. 6B).
Incubate reactions quiescently for 24 hours at 4°C for lysates of cells carrying strong [PSI+] and at 25 °C for lysates of cells carrying weak [PSI+].
Sonicate the reactions on ice (30% 450 W, 50% duty cycle, 30 s).
Make a fresh 5 μM solution of denatured NM in CRBB, and start a second round of polymerization by the addition of 5% of reaction volume (Fig. 6C). Repeat, diluting away cellular constituents (Fig. 6D). The resulting fibers can be now be used to polymerize purified, denatured uniformly isotopically enriched protein at 1 to 2% w/w (Fig. 6E) for NMR analysis (Fig. 6F)
3.1.4. Notes
Seeds may be stored for later use at −80 °C in CRBB.
Template polymerization reactions with cells that do not contain a prion template like [psi−] as a control for spontaneous fibril formation in lysate-templated reactions.
Include 20 μM of ThT in the reaction mixtures and monitor fiber growth by ThT fluorescence (420 nm excitation and 480 nm emission) after the second round of lysate seeding; strong and weak prion fibers have different polymerization kinetics (Frederick et al., 2014)
The structural homogeneity of prion fibers can be determined by fiber transformation of prion fibers into [psi−] yeasts. Robust protocols for fiber transformation are described elsewhere (Tanaka and Weissman, 2006). Transformation efficiencies using lysate seeded fibers are lower than efficiencies with in vitro prepared fibers (under 2%)(Frederick et al., 2014), but the lysate-seeded fiber produce only one, rather than a mix, of phenotypes in transformed yeasts(Hess, Lindquist, & Scheibel, 2007).
3.2. Production of segmentally isotopically labeled proteins to reduce chemical shift degeneracy
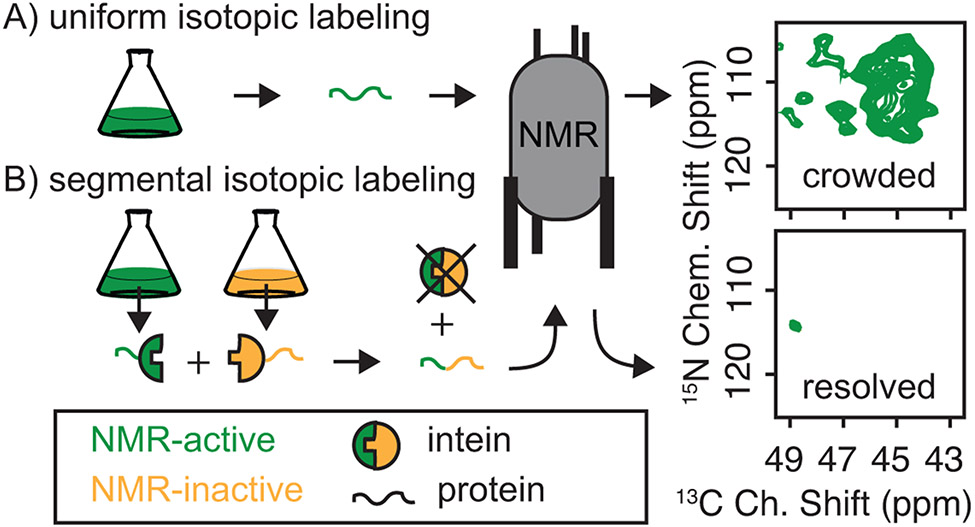
Technically, bio-molecular magic angle spinning (MAS) solid state NMR has no size limit, making it particularly suited for structural investigations of polymers like amyloids with molecular weights in the megadalton range. Practically however, MAS NMR is limited by spectral resolution (Fig 1). Segmental isotopic labeling opens up the possibility of investigating short segments of very large and complicated systems in atomic detail by NMR. Segmental isotopic labeling is particularly advantageous for DNP studies. Samples for DNP NMR are frozen. This tends to increase spectral line widths due to the freezing out of small motions and eliminates the possibility of using dynamic filtering to simplify spectra of large molecules. While in some cases this is advantageous (Bayro et al., 2011), it can complicate analyses.
To create segmentally labeled proteins we have taken advantage of split intein technology. Inteins are amino acid sequences that excise themselves from a polypeptide chain, ligating the flanking sequences via a native peptide bond. Inteins can be cut into (or exist naturally as) two pieces that associate non-covalently upon reconstitution and ligate their flanking sequences. This technology has long been suggested to solve problems such as those discussed here (Volkmann & Iwaï, 2010) but it has been extremely difficult to implement, largely due to issues with purity and yield. We and others have recently implemented segmental isotopic labeling of proteins for structural analysis by solid state NMR (Frederick et al., 2017; Ghosh, Dong, Wall, & Frederick, 2018; Gupta & Tycko, 2018; Murray et al., 2017; Schubeis et al., 2015).
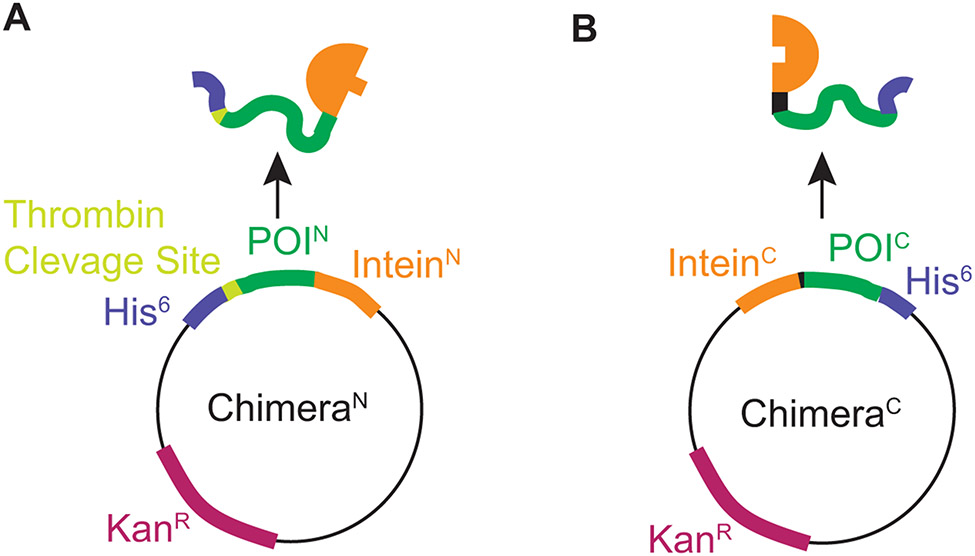
Although using split inteins to accomplish this is intellectually simple, much of the protein chemistry for high yields and purity is defined by the extein – the protein to be segmentally labeled – rather than the inteins. Protein biochemistry on both amyloid proteins and split inteins present different, somewhat orthogonal challenges. Amyloids tend to self-assemble, so much of the purification must be done under denaturing conditions. Split inteins must be able to come together, an interaction driven by electrostatic complementarity, and then be competent to carry out the protein trans-splicing reaction that has variable yields and requires semi-native reducing conditions. NM presents an additional challenge because the highly-charged M domain interferes with the inteinN-inteinC interaction, lowers affinity for Ni-NTA resin, and defines the physico-chemical properties of the molecules that contain it. These issues make downstream purification of off-pathway products of the intein reaction from the trans-spliced product complicated. We overcame these challenges for NM and have created full-length NM molecules that are uniformly labeled with NMR active stable isotopes at the first 14 or first 32 amino acids. (Frederick et al., 2017; Ghosh et al., 2018). To do so we prepared his6-NM(1-14)-inteinN and his6-NM(1-32)-inteinN ChimeriaN, expressed in minimal media with 13C glucose and 15N ammonium chloride as the sole carbon and nitrogen sources. These were ligated to the matching construct, inteinC-NM(15-253)-his6 or inteinC-NM(33-253)-his6 ChimeraC expressed in natural abundance media (Fig. 8). After the ligation reaction we separated the product from the reactants taking advantage of the fact that the correct product has two his6-tags and therefore increased affinity for Ni-NTA (Fig. 9). After eluting the product with an imidazole gradient the N-terminal his6 tags were cleaved off with thrombin. This approach allows examination of several consecutive labeled residues in the context of the full-length unlabeled protein, greatly simplifying the spectroscopy (Fig. 7). Because the ligation conditions and purification approaches required to create and isolate segmentally isotopically labeled proteins are largely defined by the chemistry of the exteins, we include a protocol to produce segmentally isotopically labeled versions of NM to complement protocols using a similar intein to segmentally isotopically label proteins with different biochemical and biophysical properties (Gupta & Tycko, 2018; Schubeis, Nagaraj, & Ritter, 2017).
Figure 8:
Plasmid constructs designed to express chimeric proteins designed to produce segmentally isotopically labeled yeast prion protein. A) The plasmid for ChimeraN. The N terminal segment of the protein of interest is expressed as a chimera with a cleavable his6 tag for purification adjacent to the segment to be isotopically labeled and followed by the N terminal piece of the CfaGEP intein. B) The plasmid for ChimeraC. The C terminal segment of the protein of interest is expressed as a chimera that starts with the C terminal piece of the CfaGEP intein followed by the C terminal region of the protein of interest with a non-cleavable his6 tag. The intein has sequence requirements at the amino acids directly adjacent to the intein thus for optimal ligation, the intein introduces a cysteine mutation at the junction between the labeled and unlabeled region and the ligation site was chosen to be at a position the fulfilled the preference for a native bulky hydrophobic amino acid (Y) in the second position from the C-terminal intein ligation site.
Figure 9:
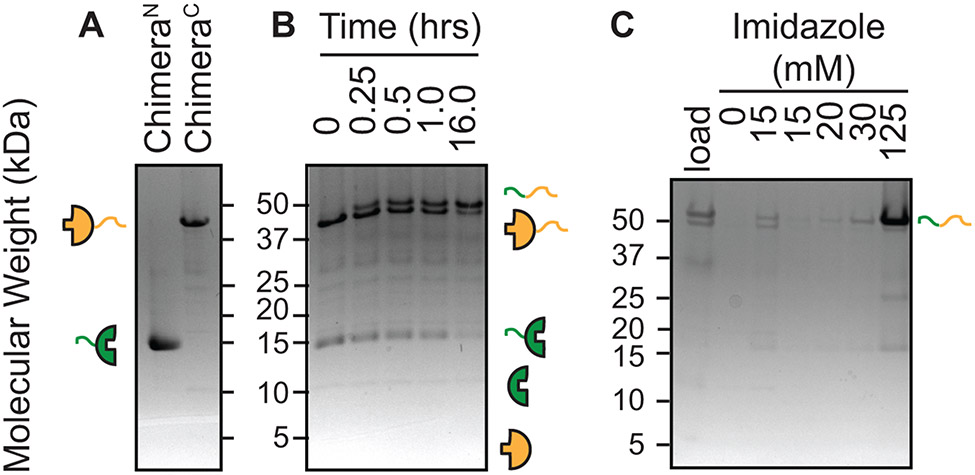
Chimeric protein purification, chimeric protein ligation and spliced product purification can by analyzed be SDS-PAGE. Inteins are represented by half circles, exteins are represented by curved lines and isotopic enrichment and depletion is represented by green and yellow color, respectively. A) Chimeric proteins were purified to homogeneity using Ni-NTA resin. B) Ligation of ChimeraN and ChimeraC to form the full-length protein of interest was followed over 16 hours. Appearance of the full-length protein of interest and the N-terminal fragment of the intein serves as a criteria to assess the progression of the ligation reaction. The smaller (4 kDa) C-terminal intein fragment is often difficult to visualize by SDS-PAGE. C) The full-length protein, which contains two his6 tags, is purified away from the precursors by taking advantage of its increases affinity for Ni-NTA using a step gradient of increasing imidazole concentrations.
Figure 7:
Segmental isotopic labeling simplifies NMR spectra. A) The glycine region of a 13C-15N correlation spectrum for uniformly 13C and 15N labeled NM has many peaks, consistent with a protein that contains 23 glycine resides. B) The same region for a full-length NM protein that is segmentally isotopically labeled has one peak, consistent with a single glycine in the isotopically labeled region. NMR was segmentally isotopically labeled by intein-mediated ligation of NM-intein chimeras purified from bacteria grown in media containing either NMR active (13C & 15N, green) or NMR inactive (12C & 14N, yellow) carbon and nitrogen sources.
3.2.1. Equipment
Innova temperature controlled shaker for 37 °C incubation of bacteria
Centrifuge equipped to spin to 35,000 x g
Beckmann Coulter Allegra X-14R centrifuge to concentrate protein
Gravity flow columns with frets (e.g. PD-10 empty columns, GE Healthcare)
Sonicator (e.g. Branson Digital Sonifier 450).
37 °C incubator
Standard SDS-PAGE equipment
10 kDa molecular weight cut off (MWCO) centrifuge filters (10 MWCO, amicon)
3.2.2. Buffers and Reagents
M9 Minimal media for growth of isotopically labeled chimeric protein: 48 mM sodium phosphate dibasic (Na2HPO4), 22 mM potassium phosphate monobasic (KH2PO4), 9 mM NaCl, adjusted to pH 7.4 by the addition of sodium hydroxide (NaOH) 10 mg/L iron (II) suflate (FeSO4), 2 mM magnesium sulfate (MgSO4), 100 μM calcium chloride (CaCl2), 10 mg/L of thiamine hydrochloride, then sterile filtered. The media is typically supplemented with 1 g/L of Ammonium-15N Chloride (15NH4Cl, 15N, 98%, ISOTEC) and 2 g/L of D-glucose (U-13C6, 99%, Cambridge Isotope Laboratories, Inc.) for expression of isotopically enriched proteins.
Rich media for growth of unlabeled chimeric protein: Per Liter: Tryptone 16.0 g, Yeast extract 10.0 g, NaCl 5.0 g, sterile filtered.
1M Isopropyl-Beta-D-Thiogalactoside (IPTG) – final concentration 1 mM in a liter of culture
Ni-NTA gravity flow resin (Qiagen)
Binding buffer: 8 M urea, 100 mM NaH2PO4, 500 mM NaCl, 10 mM Tris base pH 8.0, Adjust pH to 8 with NaOH.
Elution solution: 8 M urea, 100 mM NH2PO4, 500 mM NaCl, 10 mM Tris base pH 4.0. Adjust pH to 4 with hydrocholic acid (HCl).
Ligation buffer: 10% glycerol, 300 mM NaCl, 50 mM Tris base pH 7.0, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP), 0.5 mM EDTA.
Binding buffer with increasing amounts of imidazole, typically 15 – 125 mM with 0.5 mM TCEP.
Cleavage buffer: 20 mM Tris base pH 8.4, 150 mM NaCl
Thrombin cleavage capture kit (Millipore)
3.3.3. Procedure
Overexpress the chimeric protein that contains the region to be isotopically enriched in the M9 media. Overexpress the chimeric protein that contains the other region in 2xYT media. Collect cells by centrifugation and store at −80 °C.
Lyse cell pellets by suspension of bacterial growth in 20 mL/L of binding buffer.
Remove insoluble cellular debris by centrifugation at 35,000 x g for 20 minutes at 4 °C.
Bind chimeras to Ni-NTA resin by incubation of cleared lysates with Ni-NTA for at ~ 1 hour at room temperature with gentle rocking.
Transfer resin slurry to an empty gravity flow column.
Wash the resin with ~5 column volumes of binding buffer.
Elute the isotopically labeled and unlabeled chimeras with elution solution (Fig. 9A).
Initiate protein trans-splicing by mixing the chimeric proteins in a ratio of 1:1.2 molar ratio of the isotopically labeled chimera to unlabeled chimera out of the elution solution to a final urea concentration of 1 M and final protein concentrations between 5 – 20 μM into ligation buffer. Incubate overnight at 37 °C. To monitor the progress of the ligation reactions, remove 50 μL aliquots immediately after mixing the chimeric proteins and throughout the incubation period and stop the reactions by addition of protein loading dye and freezing at −20 °C for SDS-PAGE analysis (Fig. 9B).
To purify the segmentally isotopically labeled protein from the ligation mixture, concentrate the cleared ligation reaction to < 10 mL. dilute the concentrated ligation reaction 10-fold with binding buffer to reduce the concentration of EDTA. Pour diluted ligation reaction over Ni-NTA column.
Wash the column with a step gradient of imidazole to separate the doubly his6-tagged segmental isotopically labeled protein from the singly his6-tagged chimeric precursors (Fig. 9C).
Remove N-terminal his6-tag by diluting the elution fraction containing the segmental isotopically labeled protein 8-fold to 1 M urea concentration with cleavage buffer and add 1.2 μL of thrombin per mg of protein, incubate at room temperature overnight.
Remove biotinylated thrombin by concentrating reaction to <1 mL with Amicon spin filter and then following the manufactures directions from the thrombin capture kit.
3.2.4. Notes:
For constructs containing chimeric constructs that are codon optimized for bacterial expression, protein expression levels are typically high, and we routinely obtain yields of more than 20 mg of purified chimeric protein per liter of bacterial culture, even in M9 media for growth at 37 °C and induction times of ~ 5 hours. Expression will be largely determined by the chemistry and toxicity of the exteins and expression conditions should be optimized with this in mind.
Addition of trace vitamins and metals can increases protein yields of bacterial growths in M9 media. Make trace vitamin supplement by dissolving 1 Centrum A-Z complete vitamin in 40 mL of water. Remove the insoluble fraction by centrifuging at 35,000 x g for 20 min. Sterile filter the supernatant. Add at 4 mL/L to M9 media. Store vitamin supplement stock at −20 °C.
Sonication of the denaturing improves lysis can increase the yield of the purification.
After trans-splicing, there is often some protein precipitate. Remove the precipitated protein by centrifugation in 50 mL conicals at 2100 x g for 10 minutes. Proceed with the purification using the cleared ligation reaction mix.
The protocol uses the CfaGEP intein (Stevens et al., 2017), though conditions and yields for NM using the NpuDnaE intein are similar(Frederick et al., 2017). Ligation conditions and optimal stoichiometries for highest yields will depend upon both the intein and the extein(Schubeis et al., 2017).
For inteins such as CfaGEP that require a cysteine, all reactions must be carries out in the presence of a reducing agent like 1 mM TCEP.
For some constructs, mass shifts are hard to resolve by SDS-PAGE. MALDI-TOF is helpful to confirm ligation (Frederick et al., 2017).
3.3. Preparation of purified proteins for DNP NMR analysis
Purified amyloid fibers are typically studied by MAS NMR by data collection on pelleted samples of amyloid fibers at experimental temperatures that are at or near room temperature. Amyloid fibers are usually allowed to polymerize in dilute solution in buffer and collected by ultracentrifugation. The amyloid fibers are then either lyophilized to further concentrate them or simply packed as a wet protein pellet into an NMR rotor and analyzed by MAS NMR at or near room temperature. However, because the amyloid conformation is typically very biophysically stable, yet is often flanked by regions of disorder, the sample is sometimes frozen, without damage, to quench dynamic motions that prevent detection of mobile regions of the protein.
DNP NMR is most efficient at low temperatures because the electron spin relaxation rates of the DNP polarization agent are temperature dependent. However, we cannot simply add AMUPol and freeze the sample of purified amyloid fibers. The highest DNP enhancements are obtained at a concentration of AMUPol that is the best compromise between increased enhancement due to higher concentrations of the polarization agent and the bleaching of the NMR signal caused by the proximity to the unpaired electron. Uneven distribution of the polarization agent will therefore result in low overall DNP enhancements. Non-vitreous ice formation causes uneven distribution of the DNP polarization agent. Thus DNP samples must be cryo-protected to achieve high DNP enhancements. Empirically, the best condition for analysis of purified proteins by DNP NMR are in mixtures of d8-glycerol:D2O:H2O at 60:30:10 ratio in the presence of 10 mM AMUPol. Because the both the d8-glycerol and DNP polarization reagents are expensive, we provide a protocol to prepare homogenous samples of purified amyloid proteins that minimizes reagent use for analysis of purified samples of amyloid proteins.
3.3.1. Equipment
Ultracentrifuge for deciliter scale volumes (e.g. Beckman Coulter Optima L-90k, equipped with a Ti-70 rotor)
Ultracentrifuge for milliliter scale volumes (e.g. Beckman Coulter Optima MAX-xp equipped with a TLA-110 Fixed Angle Centrifuge Rotor)
3.2 mL polycarbonate thick wall tubes (Beckman Coulter)
Eppendorf centrifuge (e.g. 5427 R) equipped with a swinging bucket rotor.
3.2 mm sapphire rotors, silicon caps, end cap, and rotor packing tool (Bruker)
Ball-peen hammer and an 18-gauge needle.
3.4.2. Buffers and reagents
Glycerol-AMUpol stock solution: AMUPol (F.W. 726 g/mol) dissolved in deuterated, 13C-depleted glycerol (glycerol, 12C3, 99.95%, D8, 98%, Cambridge Isotopes, Inc.), at 16.7 mM. Store at −80 °C.
CRBB (Section 2.1.2) made with 75% D2O, (D, 99.8%) (Cambridge Isotope Laboratories, Inc)
3.3.3. Procedure
Polymerize ~ 15 mg of purified isotopically-labeled protein into the amyloid form (3.1.3 step 6). Collect the fibers by centrifugation for 1 hour at 60,000 x g at 4 °C using a floor model ultracentrifuge. Discard the supernatant.
Resuspend pelleted fibers in 1 mL of CRBB 75% D2O.
Transfer the resuspended fibers into a pre-weighed thick-wall polycarbonate ultra-centrifuge tube. Ensure the tube is rated for the sample volume and g-force.
Pellet the amyloid fibers by centrifugation for 1 hour at 100,000 x g and 4 °C. Remove all of the supernatant using a pipette.
Weigh the thick-wall tubes containing fiber pellets to determine weight of the pellet in milligrams.
Add glycerol-AMUPol stock solution to the tube containing the pellet. Add 1.2 μL of glycerol-AMUPol stock per 1 mg of pellet for a final sample composition of 60:30:10 glycerol:D2O:H2O and 10 mM of AMUPol.
After addition of the glycerol-AMUpol, place the centrifuge tube back into ultracentrifuge rotor, rotated by 180 degrees so that the pellet is near the rotor axis and will have to travel through the glycerol to pellet against the opposite side of the tube.
Spin for 5 min at 30,000 x g at 4 °C.
Rotate the tube 180 degrees and spin at 30,000 x g. Repeat until the mixture appears homogenous, usually three times.
Pellet the amyloid fibers a final time by spinning at 100,000 x g for 1 hour at 4 °C. Remove any supernatant using a pipette.
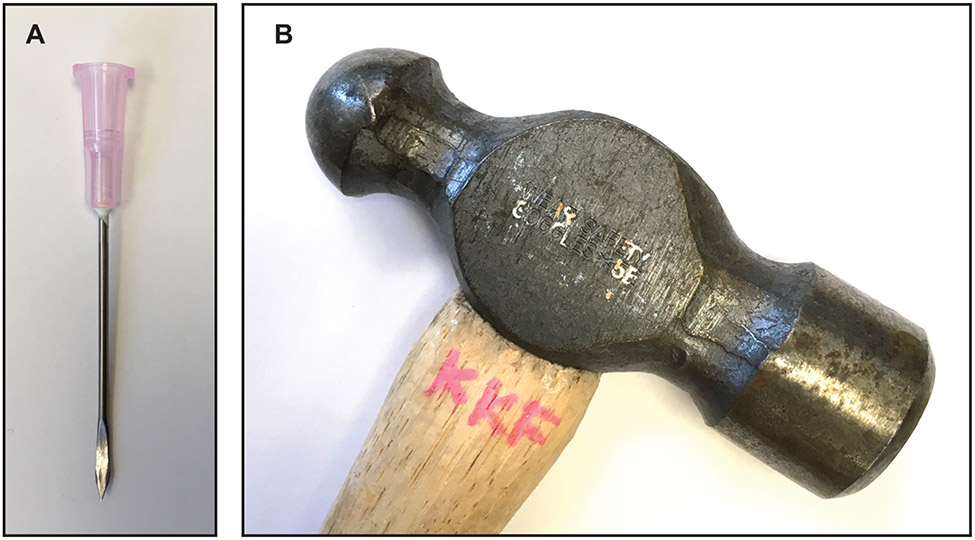
Flatten an 18-gauge needle with a ball-peen hammer (Figure 10). Clean and dry the flattened needle.
Transfer the pelleted fibers to a 3.2 mm Sapphire NMR rotor using the flattened needle.
Distribute the fibril pellet evenly in the 3.2 mm Sapphire rotor. During transfer of the pelleted fibers to the rotor, periodically place the rotor in an Eppendorf tube and centrifuge in a swinging bucket equipped rotor for two minutes at ~ 4,000 x g and 4 °C.
Seal the rotor with silicon plug using the tool kit and instructions provided by the manufacturer and store the sample at −80 °C.
Figure 10:
Custom tool for packing samples for DNP NMR analysis. A) An 18 gauge needle is flattened on one end to create a scoop to transfer ultracentrifuged cell pellets from the centrifuge tube to the NMR rotor using a B) ball-peen hammer.
3.3.4. Notes
Glycerol is viscous and difficult to pipette. Using weight (1.26 g/cm3) as a proxy for the volume transferred.
We routinely achieve enhancements that range between 40-60 for samples of purified amyloid fibers in 60:30:10 glycerol:D2O:H2O mixtures containing 10 mM AMUPol measured at 600 MHz and 100 K.
4. Summary and conclusions
Most proteins have the capacity to form amyloids under the right conditions and this fold has enormous importance in many realms of biology. For example, accumulation of large amyloid aggregates is a shared feature of neurodegenerative disorders. However, the study of these aggregated forms is notoriously difficult, in part because amyloid formation is an environmentally sensitive process; different conditions can bias formation of one amyloid form over another. Yet such differences are critical. Different amyloid forms can result in different biological phenotypes in yeast(Toyama, Kelly, Gross, & Weissman, 2007) and different disease etiologies in human (Qiang, Yau, Lu, Collinge, & Tycko, 2017). A significant shortcoming with structural studies of such metastable proteins is that the conformations that are amplified in vitro for structural work are not necessarily those that are most prevalent in vivo. For example, recent work with lysate-seeded amyloid fibers of NM found that each strain of the this protein is a mixture of two distinct amyloid conformations (Ghosh et al., 2018). It is formally possible that a single yeast prion strain is a result of a propogated mixture of two distinct amyloid conformations; these preparations faithfully confer a single prion phenotype when transformed into [psi−] yeast. Yet, the transformation efficiency is significantly lower than obtained from preparations of de novo prepared fibers, raising the possibility that the lower transformation efficiency is a result of only one of the two conformations being the one that is propagated in vivo. This is reminiscent of some of the barriers encountered when attempting to amplify amyloid aggregates of the proteins that are involved in neurodegenerative diseases. For example, structural information from solid-state NMR on fibrils prepared by seeded growth from extracts of Alzheimer's disease brain cortex reveal structural variation in fibril morphology that correlates with clinical subtype (Qiang et al., 2017). Yet, even when seeding in vitro filament growth with extracts from diseased brains (Lu et al., 2013; Qiang et al., 2017; Tuttle et al., 2016) structures may not be amplified according to their relative abundance in the brain. Comparison of amyloid fibers amplified in vitro from lysate-derived seed with fibers in cellular lysates may eliminate questions of preferential amplification of certain amyloid forms that may not reflect those that are most common in vivo. The ability to observe proteins at endogenous level in cellular environments using DNP NMR may enable identification and study of the most biologically important conformations.
The ability to observe proteins at endogenous level in cellular environments using DNP NMR may likewise prove to be invaluable to our efforts to structurally characterize intrinsically disordered regions. The amyloid core of NM, like most amyloids – at least when studied in isolation – is flanked by intrinsically disordered regions that are not visualizable using traditional structural biology approaches (Fitzpatrick et al., 2017; Heise et al., 2005; Helmus, Surewicz, Nadaud, Surewicz, & Jaroniec, 2008b; Wasmer et al., 2008). Yet, many interactions with cellular factors (Allen et al., 2005; Helsen & Glover, 2012), mostly chaperones, are mediated through the intrinsically disordered region (Frederick et al., 2014; Toyama et al., 2007). Deletion of this disordered region, which is structured in native environments(Frederick et al., 2015), results in a loss of inheritance of the prion form to the daughter cells(Liu, Sondheimer, & Lindquist, 2002). This raises the possibility that interactions of cellular factors with the regions that are disordered in purified samples are key to prion inheritance. Intriguingly, many of the proteins that misfold in neurodegenerative diseases also have prion-like self-templating dispersion properties in vivo (Jucker & Walker, 2013; Polymenidou & Cleveland, 2012; Sanders et al., 2014; Watts et al., 2013). Moreover, amyloid fibers interact with many cellular factors, especially chaperone proteins (Cox et al., 2018; Shelton et al., 2017). This raises questions about whether regions of intrinsic disorder mediate such interactions and if so, whether and how these regions are structured when they interact with their cellular binding partners.
In this chapter, we describe methods to prepare samples of isotopically-labeled proteins at endogenous levels in cellular contexts alongside quality control methods to ensure that such samples accurately model important features of the cellular environment. The methodologies outlined here for DNP NMR of proteins at endogenous levels in cellular environments provide a strategy to understand how cellular environment influence the structures of proteins that have environmentally sensitive folding pathways and regions of intrinsic disorder.
Acknowledgements:
W.N.C. was supported by a graduate research fellowship from the NSF. This work was supported by grants from the National Science Foundation [1751174]; the Welch Foundation [1-1923-20170325]; the Lupe Murchison Foundation, the Ted Nash Long Life Foundation and the Kinship Foundation (Searle Scholars Program) to K.K.F.
References:
- Abragam A (1983). The principles of nuclear magnetism. Clarendon Press. [Google Scholar]
- Akbey Ü, Franks WT, Linden A, Lange S, Griffin RG, van Rossum B-J, & Oschkinat H (2010). Dynamic nuclear polarization of deuterated proteins. Angewandte Chemie International Edition, 49(42), 7803–7806. 10.1002/anie.201002044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen KD, Wegrzyn RD, Chernova TA, Müller S, Newnam GP, Winslett PA, et al. (2005). Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics, 169(3), 1227–1242. 10.1534/genetics.104.037168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Barbieri L, Luchinat E, & Secci E (2013). Visualization of redox-controlled protein fold in living cells, 20(6), 747–752. 10.1016/j.chembiol.2013.05.007 [DOI] [PubMed] [Google Scholar]
- Bayro MJ, Debelouchina GT, Eddy MT, Birkett NR, MacPhee CE, Rosay M, et al. (2011). Intermolecular Structure Determination of Amyloid Fibrils with Magic-Angle Spinning and Dynamic Nuclear Polarization NMR. Journal of the American Chemical Society, 133(35), 13967–13974. 10.1021/ja203756x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, Byers JS, Jones S, Garcia DM, Bhullar B, Chang A, et al. (2016). Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell, 167(2), 369–381.e12. 10.1016/j.cell.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D, Whiten DR, Brown JWP, Horrocks MH, San Gil R, Dobson CM, et al. (2018). The small heat shock protein Hsp27 binds α-synuclein fibrils, preventing elongation and cytotoxicity. Journal of Biological Chemistry, 293(12), 4486–4497. 10.1074/jbc.M117.813865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM (2001). The structural basis of protein folding and its links with human disease. Philosophical Transactions of the Royal Society B: Biological Sciences, 356(1406), 133–145. 10.1098/rstb.2000.0758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, et al. (2001). Intrinsically disordered protein. Journal of Molecular Graphics & Modelling, 19(1), 26–59. Retrieved from http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=11381529&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, et al. (2017). Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature, 547(7662), 185–190. 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick KK, Debelouchina GT, Kayatekin C, Dorminy T, Jacavone AC, Griffin RG, & Lindquist S (2014). Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics., 21(2), 295–305. 10.1016/j.chembiol.2013.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick KK, Michaelis VK, Caporini MA, Andreas LB, Debelouchina GT, Griffin RG, & Lindquist S (2017). Combining DNP NMR with segmental and specific labeling to study a yeast prion protein strain that is not parallel in-register. Proceedings of the National Academy of Sciences of the United States of America, 114(14), 3642–3647. 10.1073/pnas.1619051114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick KK, Michaelis VK, Corzilius B, Ong T-C, Jacavone AC, Griffin RG, & Lindquist S (2015). Sensitivity-Enhanced NMR Reveals Alterations in Protein Structure by Cellular Milieus. Cell, 163(3), 620–628. 10.1016/j.cell.2015.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg DI, & Selenko P (2014). Live cell NMR. Annual Review of Biophysics, 43, 171–192. 10.1146/annurev-biophys-051013-023136 [DOI] [PubMed] [Google Scholar]
- Ghosh R, Dong J, Wall J, & Frederick KK (2018). Amyloid fibrils embodying distinctive yeast prion phenotypes exhibit diverse morphologies. FEMS Yeast Research. 10.1093/femsyr/foy059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, & Tycko R (2018). Segmental isotopic labeling of HIV-1 capsid protein assemblies for solid state NMR. Journal of Biomolecular NMR, 70(2), 103–114. 10.1007/s10858-017-0162-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann R, & Lindquist S (2008). Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. Journal of Visualized Experiments : JoVE, (17). 10.3791/838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, & Baldus M (2005). Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America, 102(44), 15871–15876. 10.1073/pnas.0506109102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, & Jaroniec CP (2008a). Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America, 105(17), 6284–6289. 10.1073/pnas.0711716105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, & Jaroniec CP (2008b). Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proceedings of the National Academy of Sciences of the United States of America, 105(17), 6284–6289. 10.1073/pnas.0711716105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsen CW, & Glover JR (2012). Insight into molecular basis of curing of [PSI+] prion by overexpression of 104-kDa heat shock protein (Hsp104). Journal of Biological Chemistry, 287(1), 542–556. 10.1074/jbc.M111.302869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S, Lindquist SL, & Scheibel T (2007). Alternative assembly pathways of the amyloidogenic yeast prion determinant Sup35–NM. EMBO Reports, 8(12), 1196–1201. 10.1038/sj.embor.7401096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K, Ohno A, Tochio H, Isogai S, Tenno T, Nakase I, et al. (2009). High-resolution multi-dimensional NMR spectroscopy of proteins in human cells. Nature, 458(7234), 106–109. 10.1038/nature07839 [DOI] [PubMed] [Google Scholar]
- Jaroniec CP, Filip C, & Griffin RG (2002). 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly (13)C,(15)N-labeled solids. Journal of the American Chemical Society, 124(36), 10728–10742. 10.1021/ja026385y [DOI] [PubMed] [Google Scholar]
- Jarosz DF, Brown JCS, Walker GA, Datta MS, Ung WL, Lancaster AK, et al. (2014). Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell, 158(5), 1083–1093. 10.1016/j.cell.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M, & Walker LC (2013). Self-propagation of pathogenic protein aggregates in neurodegenerative diseases. Nature, 501(7465), 45–51. 10.1038/nature12481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J-J, Sondheimer N, & Lindquist SL (2002). Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. Proceedings of the National Academy of Sciences of the United States of America, 99 Suppl 4, 16446–16453. 10.1073/pnas.252652099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J-X, Qiang W, Yau W-M, Schwieters CD, Meredith SC, & Tycko R (2013). Molecular Structure of b-Amyloid Fibrils in Alzheimer’s Disease Brain Tissue. Cell, 154(6), 1257–1268. 10.1016/j.cell.2013.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, & Tycko R (2017). Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell, 171(3), 615–627.e16. 10.1016/j.cell.2017.08.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni QZ, Daviso E, Can TV, Markhasin E, Jawla SK, Swager TM, et al. (2013). High Frequency Dynamic Nuclear Polarization. Accounts of Chemical Research, 46(9), 1933–1941. 10.1021/ar300348n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins SJ (1981). Estimation of deuteration levels in whole cells and cellular proteins by1H n.m.r. spectroscopy and neutron scattering. Biochemical Journal, 199(1), 163–170. 10.1042/bj1990163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymenidou M, & Cleveland DW (2012). Prion-like spread of protein aggregates in neurodegeneration. The Journal of Experimental Medicine, 209(5), 889–893. 10.1084/jem.20120741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang W, Yau W-M, Lu J-X, Collinge J, & Tycko R (2017). Structural variation in amyloid-β fibrils from Alzheimer's disease clinical subtypes. Nature, 541(7636), 217–221. 10.1038/nature20814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakakibara D, Sasaki A, Ikeya T, Hamatsu J, Hanashima T, Mishima M, et al. (2009). Protein structure determination in living cells by in-cell NMR spectroscopy. Nature, 457(7234), 102–105. 10.1038/nature07814 [DOI] [PubMed] [Google Scholar]
- Sanders DW, Kaufman SK, DeVos SL, Sharma AM, Mirbaha H, Li A, et al. (2014). Distinct Tau Prion Strains Propagate in Cells and Mice and Define Different Tauopathies. Neuron, 82(6), 1271–1288. 10.1016/j.neuron.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvee C, Rosay M, Casano G, Aussenac F, Weber RT, Ouari O, & Tordo P (2013). Highly efficient, water-soluble polarizing agents for dynamic nuclear polarization at high frequency. Angewandte Chemie International Edition, 52(41), 10858–10861. 10.1002/anie.201304657 [DOI] [PubMed] [Google Scholar]
- Schaefer J, McKay RA, & Stejskal EO (1979). Double-cross-polarization NMR of solids. Journal of Magnetic Resonance (1969), 34(2), 443–447. 10.1016/0022-2364(79)90022-2 [DOI] [Google Scholar]
- Schubeis T, Nagaraj M, & Ritter C (2017). Segmental Isotope Labeling of Insoluble Proteins for Solid-State NMR by Protein Trans-Splicing. Methods in Molecular Biology (Clifton, NJ), 1495, 147–160. 10.1007/978-1-4939-6451-2_10 [DOI] [PubMed] [Google Scholar]
- Schubeis T, Yuan P, Ahmed M, Nagaraj M, van Rossum B-J, & Ritter C (2015). Untangling a Repetitive Amyloid Sequence: Correlating Biofilm-Derived and Segmentally Labeled Curli Fimbriae by Solid-State NMR Spectroscopy. Angewandte Chemie International Edition, 54(49), 14669–14672. 10.1002/anie.201506772 [DOI] [PubMed] [Google Scholar]
- Selenko P, Serber Z, Gadea B, Ruderman J, & Wagner G (2006). Quantitative NMR analysis of the protein G B1 domain in Xenopus laevis egg extracts and intact oocytes. Proceedings of the National Academy of Sciences of the United States of America, 103(32), 11904–11909. 10.1073/pnas.0604667103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Cashikar AG, Moslehi JJ, Kowal AS, & Lindquist SL (1999). Yeast prion [psi +] and its determinant, Sup35p. Methods in Enzymology, 309, 649–673. [DOI] [PubMed] [Google Scholar]
- Shelton LB, Baker JD, Zheng D, Sullivan LE, Solanki PK, Webster JM, et al. (2017). Hsp90 activator Aha1 drives production of pathological tau aggregates. Proceedings of the National Academy of Sciences of the United States of America, 114(36), 9707–9712. 10.1073/pnas.1707039114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slichter CP (1990). Principles of Magnetic Resonance. Springer Science & Business Media. [Google Scholar]
- Stevens AJ, Sekar G, Shah NH, Mostafavi AZ, Cowburn D, & Muir TW (2017). A promiscuous split intein with expanded protein engineering applications. Proceedings of the National Academy of Sciences of the United States of America, 114(32), 8538–8543. 10.1073/pnas.1701083114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Fernández-de-Alba C, Lee D, Maurel V, Gambarelli S, Bardet M, et al. (2014). Optimization of an absolute sensitivity in a glassy matrix during DNP-enhanced multidimensional solid-state NMR experiments. Journal of Magnetic Resonance (San Diego, Calif: 1997), 239, 91–99. 10.1016/j.jmr.2013.12.005 [DOI] [PubMed] [Google Scholar]
- Theillet F-X, Binolfi A, Bekei B, Martorana A, Rose HM, Stuiver M, et al. (2016). Structural disorder of monomeric α-synuclein persists in mammalian cells. Nature, 530(7588), 45–50. 10.1038/nature16531 [DOI] [PubMed] [Google Scholar]
- Toyama BH, Kelly MJS, Gross JD, & Weissman JS (2007). The structural basis of yeast prion strain variants. Nature, 449(7159), 233–237. 10.1038/nature06108 [DOI] [PubMed] [Google Scholar]
- Tuttle MD, Comellas G, Nieuwkoop AJ, Covell DJ, Berthold DA, Kloepper KD, et al. (2016). Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein. Nature Structural & Molecular Biology, 23(5), 409–415. 10.1038/nsmb.3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkmann G, & Iwaï H (2010). Protein trans-splicing and its use in structural biology: opportunities and limitations. Molecular bioSystems, 6(11), 2110–2121. 10.1039/c0mb00034e [DOI] [PubMed] [Google Scholar]
- Wang T, Park YB, Caporini MA, Rosay M, Zhong L, Cosgrove DJ, & Hong M (2013). Sensitivity-enhanced solid-state NMR detection of expansin's target in plant cell walls. Proceedings of the National Academy of Sciences of the United States of America, 110(41), 16444–16449. 10.1073/pnas.1316290110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, & Meier BH (2008). Amyloid fibrils of the HET-s(218-289) prion form a beta solenoid with a triangular hydrophobic core. Science, 319(5869), 1523–1526. 10.1126/science.1151839 [DOI] [PubMed] [Google Scholar]
- Watts JC, Giles K, Oehler A, Middleton L, Dexter DT, Gentleman SM, et al. (2013). Transmission of multiple system atrophy prions to transgenic mice. Proceedings of the National Academy of Sciences of the United States of America, 110(48), 19555–19560. 10.1073/pnas.1318268110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright PE, & Dyson HJ (2015). Intrinsically disordered proteins incellular signalling and regulation. Nature Reviews Molecular Cell Biology, 16(1), 18–29. 10.1038/nrm3920 [DOI] [PMC free article] [PubMed] [Google Scholar]