Abstract
Objective
IRX-2 is a homologous cell-derived multi-cytokine biologic with multifaceted immune modulatory effects that has been shown to induce increased lymphocyte infiltration into primary tumors in oral cavity carcinoma. Our objective was to characterize tumor immune gene expression and epigenomic changes after neoadjuvant IRX-2 immunotherapy in patients with squamous cell carcinoma of the oral cavity.
Methods
A randomized phase II trial was conducted of the IRX regimen 3 weeks prior to surgery for previously untreated patients with Stage II-IV oral cavity carcinoma. The treatment regimen consisted of low dose (300mg/m2) cyclophosphamide (day 1) followed by 10 days of regional perilymphatic IRX-2 cytokine injections and daily oral indomethacin, zinc and omeprazole (Regimen 1) compared to the identical regimen without the IRX-2 cytokines (Regimen 2). The NanoString immune panel (730 genes) and Infinium MethylationEPIC BeadChip were performed to assess the gene expression and DNA methylation signatures, respectively, in pre- and post-immunotherapy tumor samples.
Results
A total of 51 and 79 immune-related genes were found upregulated and downregulated, respectively, in the samples from Regimen 1 patients after treatment, while 51 and 56 were found upregulated and downregulated in the samples for Regimen 2. When comparing the changes between the two regimens, we identified 9 genes significantly different, including DMBT1, a potential tumor suppressor, functioning in tumor invasion of head and neck cancer. The exploration of DNA methylation showed slight overall hypermethylation after treatment in both regimens, especially for Regimen 1 immune responders, and methylation-based cell type deconvolution demonstrated high concordance with tumor infiltrating T lymphocyte cell counts.
Conclusion
While a consistent patient response after treatment was observed, most changes were similar between regimens, indicating a subtle, targeted, or patient-specific effect of IRX-2 cytokines. Change in DMBT1 expression was a unique finding that will require further study to better understand its significance.
Keywords: Immunotherapy, Oral cavity carcinoma, Neoadjuvant, NanoString, Tumor infiltrating lymphocytes, Cytokine
Background
Head and neck squamous cell carcinoma (HNSCC) is the sixth most prevalent cancer worldwide, affecting approximately 680,000 patients annually(1,2). Traditional treatment of HNSCC involves a combination of surgery, radiotherapy and/or chemotherapy, and the five-year survival rate ranges from 55% to 66%(3). Immunotherapy with immune checkpoint inhibitors (ICI) has shown revolutionary progress in the treatment of multiple cancers. The PD-1/PD-L1 pathway is a key mechanism targeting T cell regulatory functions to enhance anti-tumor immune response(4), and PD-1 inhibitors pembrolizumab and nivolumab showed clinical advances in recurrent and metastatic HNSCC patients. However, the overall response rate for pembrolizumab is only 16% to 18% regardless of human papillomavirus (HPV) status(5,6), while patients treated with nivolumab have demonstrated significantly longer overall survival than those treated with standard single-drug systemic therapy(7). The challenge remains to discover how to increase these encouraging response rates and clinical results with novel immunotherapy approaches.
IRX-2 is a biologic product that contains multiple cytokines derived from normal donor phytohemagglutinin (PHA) stimulated mononuclear cells. The primary active components in IRX-2 are IL2, IL1β, IFNγ and TNFα. Previous studies showed that IRX-2 can protect T cells from activation-induced cell death and promote the cytolytic functions of natural killer cells(8). In addition to the direct effects of IRX-2 on antitumoral immunity, IRX-2 could potentially prime the tumor for positive response to ICIs. In a completed Phase 2a clinical trial in patients with oral carcinoma, IRX-2 immunotherapy was associated with increased immune infiltration and chemokine receptor expression using multiplex immunohistochemistry (IHC) and transcriptome analysis from 7 matched pre- and post-treatment tumor specimens(9).
Apart from the expression of immune response genes, methylation changes also play an important role in cancer immunotherapy. Hypermethylation at CpG islands and promoters together with global hypomethylation have been documented in HNSCC (10–12). In particular, hypomethylation of human retroelement long interspersed nucleotide element-1 (LINE-1) was associated with higher risk of oral cavity cancer (OCC) relapse, and is a potential predictive biomarker for OCC(13,14). In recent immune-related HNSCC studies, the characterization of DNA methylation profiles predicted a combination of hyper- and hypo-methylation markers in response to ICI, and these methylation signatures could act as surrogate biomarkers to predict responsiveness to PD-1/PD-L1 inhibition(15–18). In addition, accompanied by genomics data, a methylation-based cell type deconvolution approach may distinguish “immune hot” from “immune cold” HNSCCs(19).
Based on promising findings from a prior Phase 2a trial, we studied immune-related gene expression and genome-wide DNA methylation in tumor specimens from a larger scale randomized Phase 2b clinical trial. The trial was designed to determine whether IRX-2 cytokines increase lymphocyte infiltration into primary tumors and how these cytokines are active in previously untreated patients with Stage II-IV OCC undergoing definitive resection (NCT 02609386). The IRX-2 regimen started 3 weeks prior to surgery, consisting of an initial dose of cyclophosphamide (300 mg/m2) followed by 10 days of regional perilymphatic IRX-2 cytokine injections and daily oral indomethacin, zinc and omeprazole (Regimen 1). The control regimen was identical to Regimen 1 without the IRX-2 cytokine injections (Regimen 2). A total of 96 patients were randomized 2:1 to Regimen 1 or Regimen 2 (64:32). To determine the effect of IRX-2 on immune gene expression, a paired transcriptome analysis (NanoString) was conducted on pre- and post-treatment tumor samples from 71 patients. In addition, DNA methylation EPIC BeadChip was conducted on a subset of patients to explore the DNA methylation profiles of patients in both arms before versus after treatment.
Methods
NanoString data analysis
Seventy-one patients out of 96 had usable Nanostring data available both before and after immunotherapy. The expression data were code-count normalized, sample content normalized, and background corrected using the NanoStringNorm R package (20). Paired tests with edgeR-QLF were used to conduct two types of differential analysis: before vs after treatment for each regimen separately, and a contrast between the change in Regimen 1 versus the change in Regimen 2. Significantly differentially expressed genes had false discovery rate (FDR) < 0.05 and fold change > 1.5.
EPIC BeadChip data analysis
MethylationEPIC BeadChip assay data (Illumina, San Diego, CA) were analyzed for 24 patients with both biopsy and resection samples. Raw data were pre-processed with functional normalization funnorm(21). A total of 17611 probes were removed due to detection failurein at least 5% of samples. After dropping cross-reactive probes detected in 450K BeadChip and probes that map to X and Y chromosomes, 777406 probes were used for the remainder of analysis. Linear regression and eBayes function in the limma R package were used to call DMPs (differential methylated probes, with methylation difference > 10% and FDR < 0.05) before versus after treatment in a paired analysis for each regimen, however no probes were significantly different. The R package DMRcate(22) was used to call potential DMRs (differentially methylated regions, with FDR < 0.05 and methylation change > 10%) from DMPs.
Determination of response to neoadjuvant immunotherapy
Two types of response to treatment were calculated for each patient having the relevant data: clinical tumor response and immune response based on the change in T cell counts in the tumor microenvironment (Table 1). Clinical response was categorized as partial response (PR), stable (SD), or progressive disease (PD) based on RECIST (Response Evaluation Criteria in Solid Tumors) criteria, evaluated from unbiased centralized independent interpretation of CT and/or MRI scans (Parexel Informatics). Immune responders (IR) were defined as having an increase in CD8+ TIL (tumor infiltrating lymphocytes) infiltrate score of at least 10 cells/mm2, as previously published(23).
Table 1:
The number of patients with immune response and clinical response status available in Regimen 1 and Regimen 2 respectively.
| Regimen 1 | Regimen 2 | ||
|---|---|---|---|
| Immune response (n=33) |
Responder | 15 | 4 |
| Non-responder | 5 | 9 | |
| Clinical response (n=77) |
PR | 6 | 2 |
| SD | 33 | 17 | |
| PD | 10 | 9 |
Additional analyses
Detailed analytic methods for the (1) Phase 2b clinical trial; (2) tumor sample preparation; (3) DNA extraction and EPIC BeadChip protocol; (4) RNA extraction and NanoString protocol; (5) DNA methylation-based cell type deconvolution are available in Supplementary Methods.
Results
Both regimens resulted in a differential pattern of immune gene expression
From 2016 to 2018, we collected clinical data and tumor specimens from a total of 96 patients with newly diagnosed OCC who participated in the neoadjuvant Phase 2b clinical trial with IRX-2 regimen. To determine the effect of IRX-2 on immune response, we captured the expression level of 730 immune response genes on 71 patients for paired biopsy (3 weeks prior to surgery) and surgical resection (after 21 days of immunotherapy) specimens (Table S1).. First, to understand the degree of heterogeneity based on signatures in our cohort, principal component analysis (PCA) was used. Although the samples did not separate by regimen, a major source of heterogeneity was the distinction between biopsy and resection, suggesting treatment induced immune response change in both regimens (Figure S1A). By using the log2 fold change (FC) of resection versus biopsy samples, we did not observe any separation based on regimen, immune response, or clinical response (Figure S1B).
By comparing the baseline (biopsy) to resection, we elucidated changes in key immune response genes. A total of 51 and 79 immune response genes were found upregulated and downregulated, respectively, in the resection samples of Regimen 1 patients, while 51 and 56 were found upregulated and downregulated, respectively, in the resection samples of Regimen 2 patients. Among these differentially expressed genes, 69 overlapped between the two regimens, with 33 upregulated and 36 downregulated in resection (Figure 1A). Some of these overlapping upregulated genes have been previously linked with mutations in cancer, including CD63, BCL2, MME1, DMBT1, and CXCL12, while the overlapping downregulated cancer genes include S100A7, HRAS, MST1R, CDH1, and IL1B. A complete list of differentially expressed genes in each regimen are listed in Table S2. Noticeably, CD8A, a gene marker for CD8 T cells, was only upregulated in Regimen 1.
Figure 1:
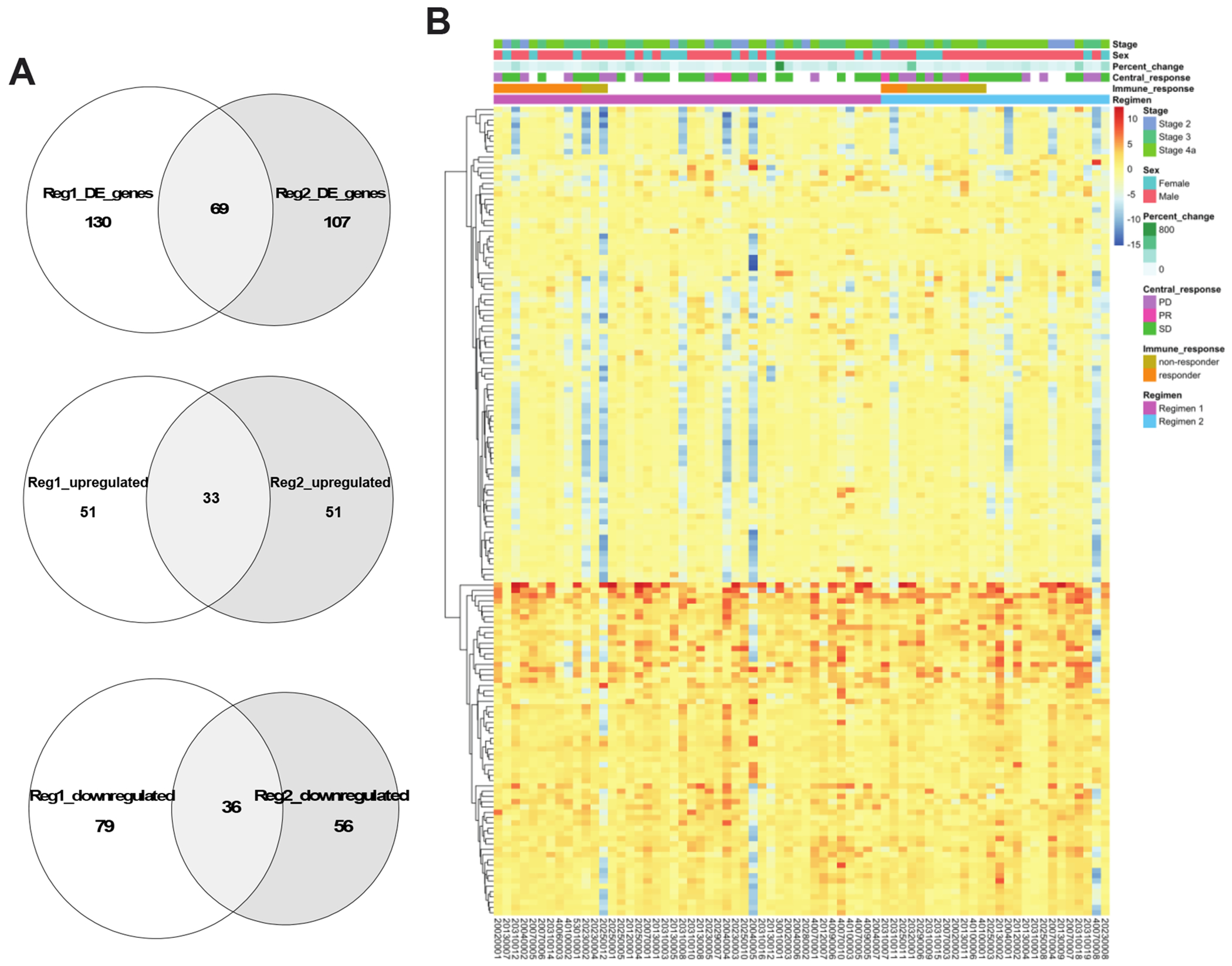
Immune response genes changed similarly in both Regimen 1 and Regimen 2. (A) Venn diagrams showing the total number of differentially expressed genes, the number of upregulated genes, and the number of downregulated genes after treatment that are overlapped between Regimen 1 and Regimen 2, respectively. (B) Heatmap showing the log2 fold change of gene expression before versus after treatment (log2FC, resection/biopsy) of all 153 differentially expressed immune genes in either regimen. Samples are ordered in columns by regimen and immune responder status, and hierarchical clustering was performed on genes only, which are displayed as rows.
To visualize the expression patterns of upregulated and downregulated genes in both regimens, we performed hierarchical clustering on the union of differentially expression genes in Regimen 1 and Regimen 2 (Figure 1B). The resulting heatmap separated up and downregulated genes, except for a few patients whose gene expression consistently decreased after treatment for nearly all genes. Interestingly, while the upregulated genes demonstrated a consistent pattern across patients in both regimens, the downregulated genes were due to large decreases in a subset of patients irrespective of regimen. No clear distinctions were observed between the two regimens or between the two immune response groups.
Higher DMBT1 upregulation after treatment in Regimen 1 than Regimen 2
Comparing the changes between the regimens, we identified 9 genes which displayed significantly different changes between the regimens (Figure 2A). Of these genes, six (DMBT1, SEMG1, CD160, CCR9, CCL16, IL2) were associated with a larger change between biopsy and resection samples in Regimen 1, while the changes in Regimen 2 were larger for KIR3DL2, IFNA7 and CTAG1B. Only DMBT1 was upregulated in both regimens. The only gene that showed previous mutations in HNSCC was DMBT1, a potential tumor suppressor gene functioning in interaction between tumor cells and immune system, as well as invasion in HNSCC(24,25). The average increase in DMBT1 was 59.3-fold in Regimen 1 versus 19.43 in Regimen 2 (Figure 2B). Visualizing individuals’ DMBT1 expression across response groups, we did not observe a strong association between change in DMBT1 expression level and either clinical tumor response (Figure S2) or immune response (Figure 2B) in either regimen. However, this conclusion was limited because few patients with NanoString data were characterized as clinical responders (n=4), and only 26 patients had immune response information available, with only three immune responders in Regimen 2.
Figure 2:
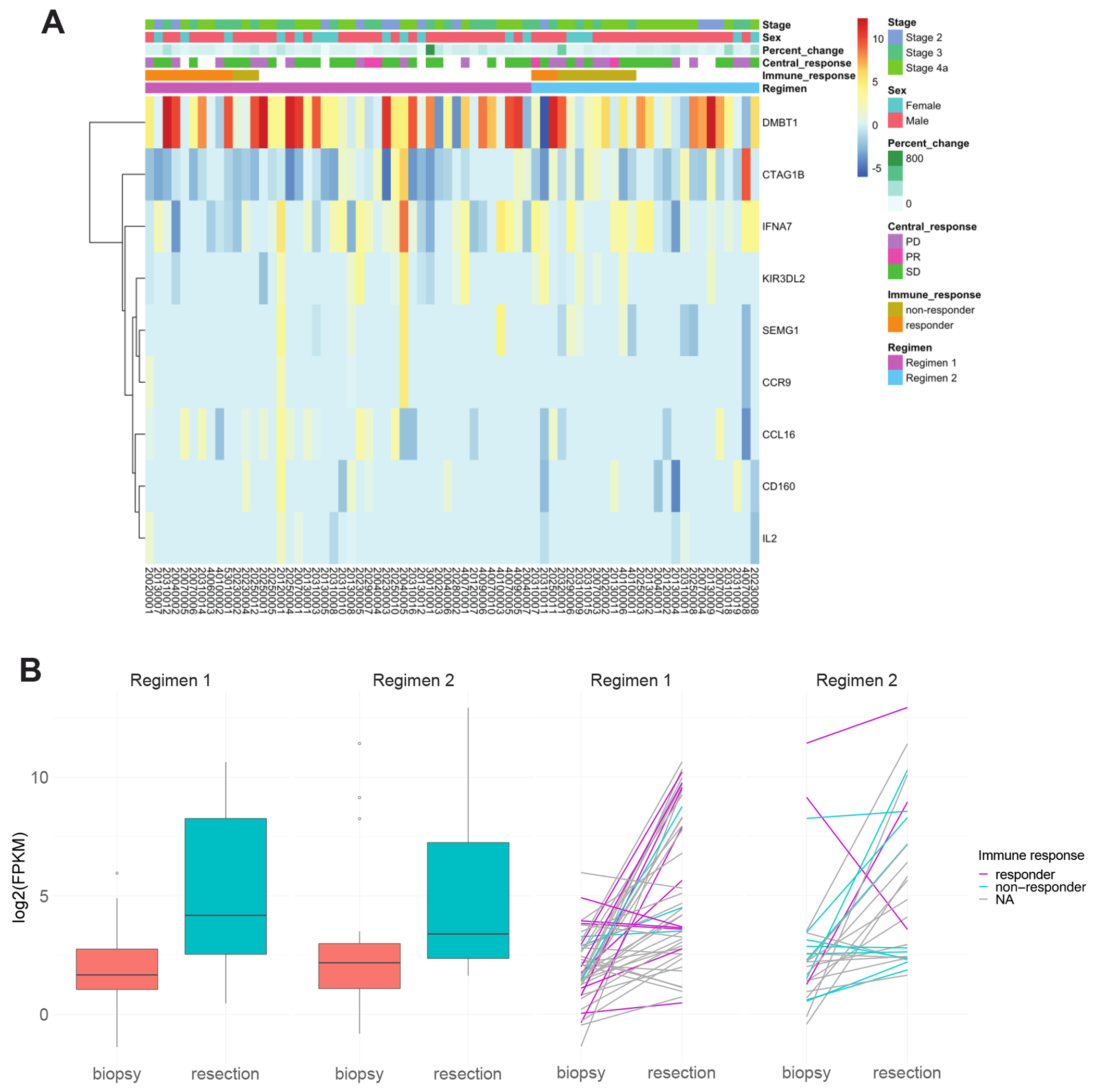
A total of 9 genes showed significantly different changes after treatment between the two regimens. (A) Heatmap showing the log2 fold change (resection/biopsy) of the 9 genes whose difference before vs after treatment is significantly different between Regimen 1 and Regimen 2. (B) Box plot of the DMBT1 gene expression at biopsy and resection in the two regimens separately. Spaghetti plot showed the change of DMBT1 expression in patients of different immune response groups.
Change in immune signature after treatment
To study the effect of IRX-2 treatment on key immune signatures, the change in expression of PD1 and PDL1 in both regimens was assessed. Expression levels of both PDL1 (CD274) and PD1 (PDCD1) revealed large individual variability (Figure 3). The differential expression analysis between biopsy and resection revealed downregulation of PDL1 after treatment in Regimen 2; this drop in expression was concentrated in non-immune responders. Patients characterized as immune responders showed little to no change in either gene in either regimen.
Figure 3:
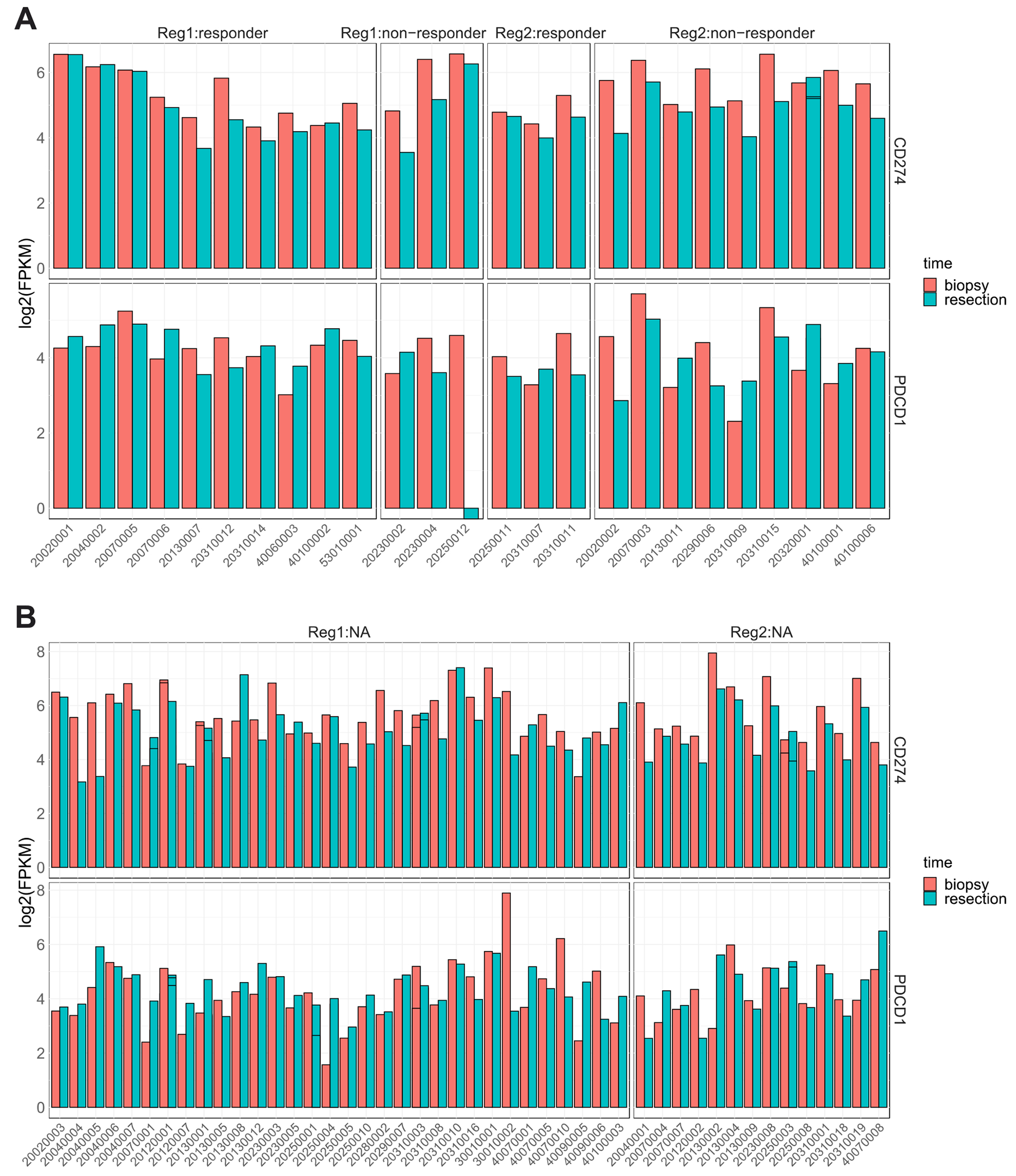
The expression levels of CD274 (PDL1) and PDCD1 (PD1) in patients (A) with and (B) without immune response groups of the two regimens.
Beyond PD1 and PDL1, we examined per patient changes in interferon, cytokine, antigen and inflammation gene groups, which were defined in Li et al.(26) and identified as being important in immune response of epithelial cancers. Overall, we observed a high level of variability among patients, yet 25% of the genes were differential expressed between biopsy and resection in both Regimen 1 and Regimen 2 (Table S3). After separating patients by respective regimen and immune response, we discovered that the changes in interferon and antigen genes were more subtle in the immune responders of Regimen 1 than Regimen 2 and non-responders, but this trend was not as prominent for cytokine or inflammation genes (Figure S3A-D). Although a definitive conclusion is difficult due to the existence of a few outlier patients with extremely large downregulation, we found that 62% and 75% of Regimen 1 immune responders had increased cytokine and inflammation gene expression; only 1 of 3 Regimen 2 immune responders had a similar increase.
Higher methylation level after treatment in both regimens
DNA methylation was assessed using the MethylationEPIC BeadChip assay on 24 patients (14 in Regimen 1 and 10 in Regimen 2), to explore the changes in DNA methylation caused by IRX-2. The overall global methylation distribution results showed a slight overall increase in methylation after treatment in both regimens, and this increase was consistent across CpG islands, shores and shelves (Figure S4). The same trend was especially prominent on the LINE-1 and LINE-2 elements of Regimen 1 compared to Regimen 2 and was mainly due to the immune responders in both regimens (Figure 4A). Since the focus of this study was on the effect of IRX-2 on immune response, we also looked at the methylation level of the CpGs that fall on the promoter regions of immune response genes. In Regimen 1, the methylation level increase after treatment was more prominent in immune responders, which was not observed in Regimen 2 (Figure 4B).
Figure 4:
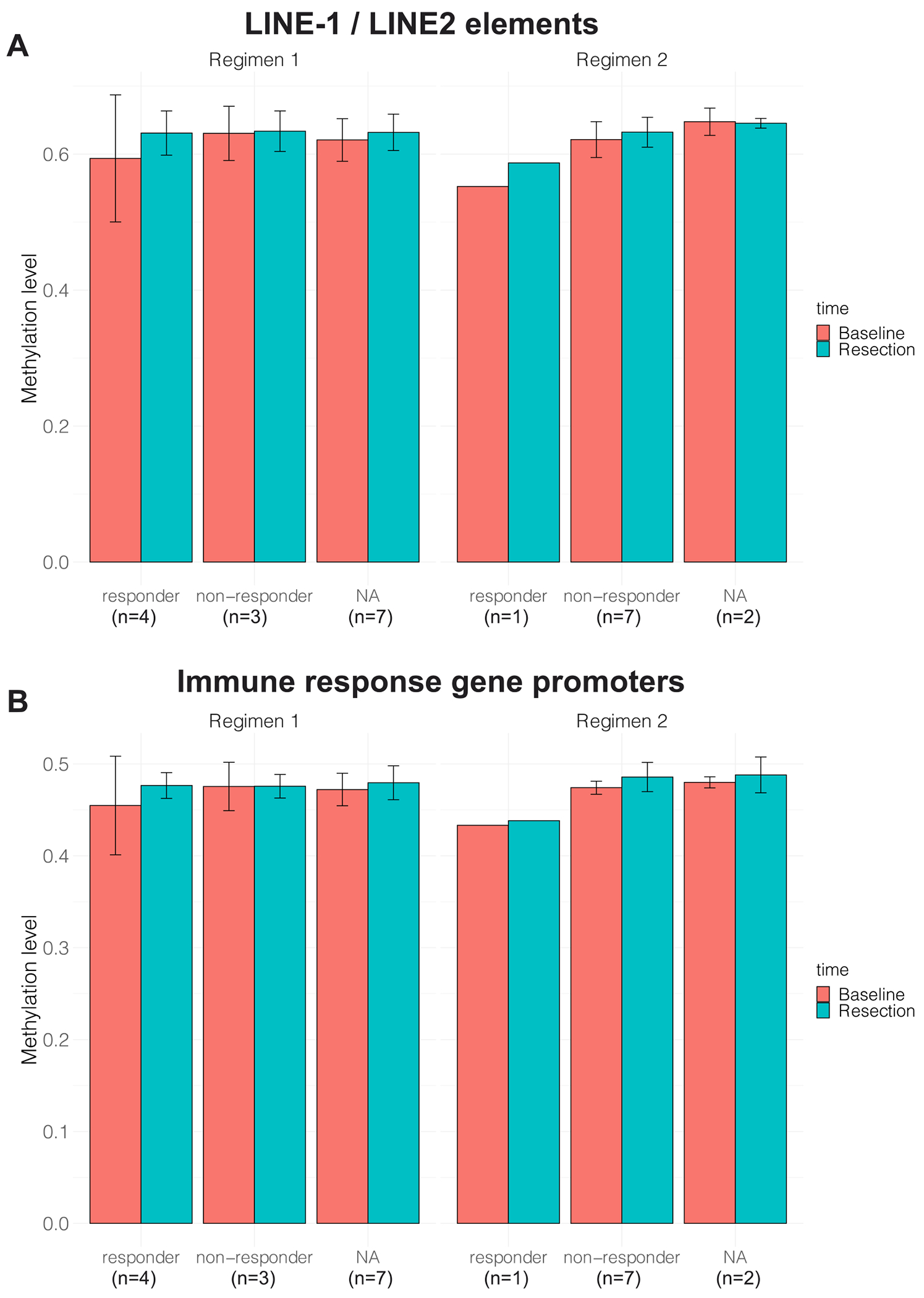
The average DNA methylation levels in both regimens at (A) LINE-1 and LINE-2 elements and (B) immune response gene promoters.
Altered methylation of immune response and keratinization pathways
By comparing methylation profiles before versus after treatment, we did not identify any differentially methylated probes (DMPs) in either regimen. However, testing differentially methylated regions (DMRs), we identified 523 and 1 potential hyper and hypo-methylated regions in the resection samples of Regimen 1 respectively, including those mapped to immune response genes IFI27 and SPINK5 which were downregulated after treatment based on NanoString data (Table 2). Conversely, a total of 721 and 112 potential regions were found to be hyper- and hypo-methylated respectively in the resection samples of Regimen 2; of genes with potential hypermethylation, CXCL14, CLEC5A, LAMP3, TOLLIP and IFITM1 displayed downregulation. A total of 21 potential DMRs were hyper-methylated in both regimens, including keratinocyte differentiation gene RIPK4, but none of the overlapped genes were among the immune response genes selected for NanoString. Pathway enrichment of the DMRs suggested cornification, keratinization, and epidermic development and differentiation were enriched with hypermethylation in Regimen 1 (Figure S5A). For Regimen 2, enrichment pathways including organ morphogenesis, epithelium development and embryonic development were most significant, but not keratinization or cornification (Figure S5B).
Table 2:
The number of potential differentially methylated regions (DMRs) derived from EPIC BeadChip data that are hyper- or hypo-methylated in Regimen 1 and Regimen 2, separately.
| Direction in resection | Regimen 1 | Regimen 2 |
|---|---|---|
| Hypermethylated | 523 | 721 |
| Hypomethylated | 1 | 112 |
| Total | 524 | 833 |
Lower percentage of cancer cells and higher percentage of T cells after treatment for immune responders
Cell type deconvolution analysis using bulk gene expression or DNA methylation data has become an important tool for interpreting changes in light of potential shifts in cell type proportions. Recently, it was shown that cell type deconvolution is more accurate using DNA methylation data than gene expression data (19). Due to the uncertainty in defining immune responders based on any one criterion, cell type deconvolution provides a complementary perspective. To determine the effect of IRX-2 treatment on cell type proportions in each regimen, we performed deconvolution with MethylCIBERSORT on 48 samples (28 in Regimen 1 and 20 in Regimen 2) with DNA methylation data, estimating the proportions of 11 cell types (see Supplementary Methods). None of the cell types were significantly different either between the two regimens or between biopsy and resection samples. However, we observed decreased cancer cell proportions as well as increased CD8+ T cell proportions in immune responders of Regimen 1, which was not replicated in non-immune responders or Regimen 2 patients (Figure 5A). The cell type deconvolution results for CD8+ T cells were confirmed by the high correlation between the estimated CD8+ T cell proportion from MethylCIBERSORT and the CD8+ T cell density counts performed previously on tissue microarrays(23) (Figure 5B).
Figure 5:
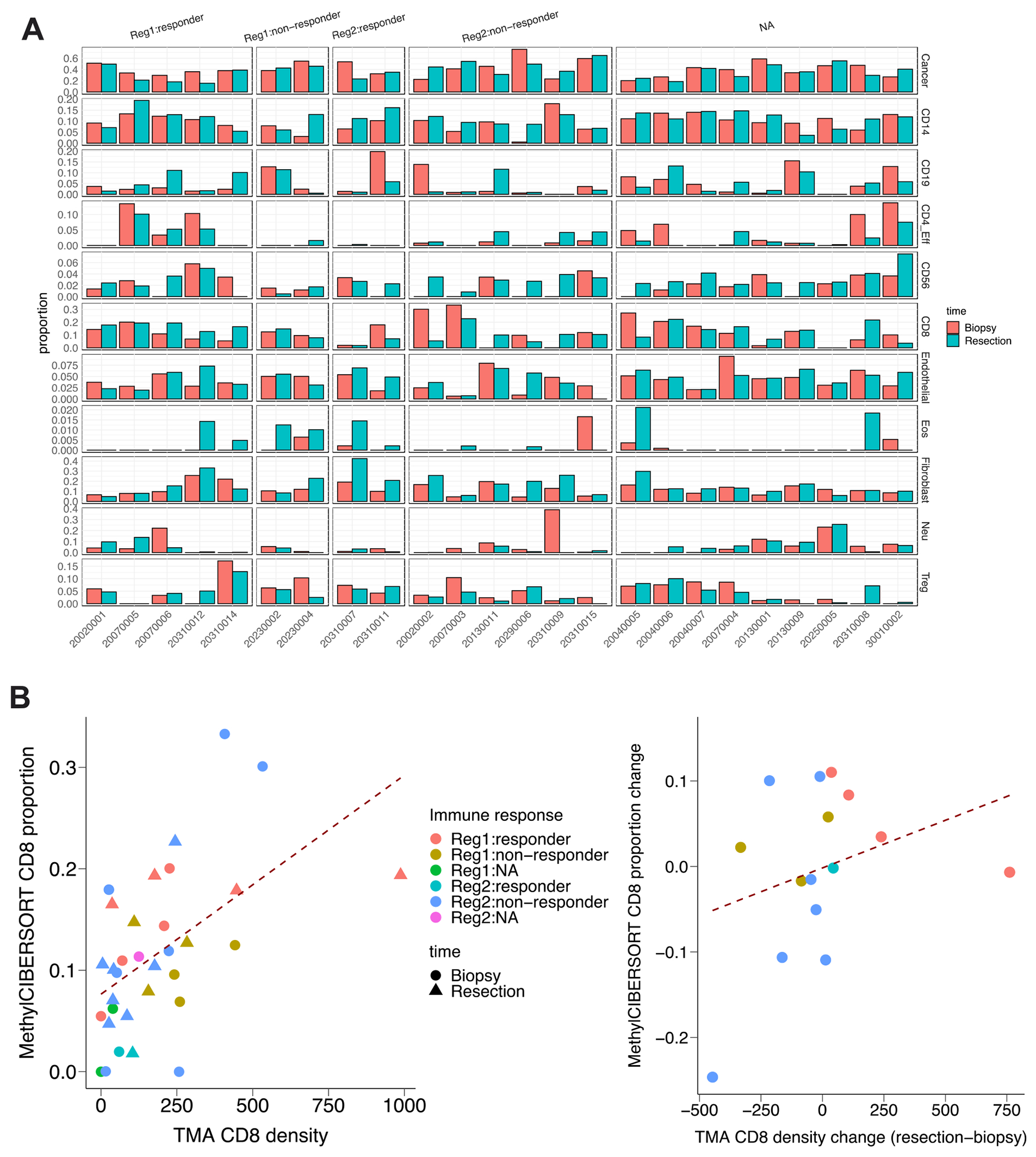
Cell type deconvolution using DNA methylation data revealed higher CD8 in immune responders of Regimen 1. (A) Bar plot showing the proportion of different cell types from 48 patients in different immune response groups. (B) Dot plot indicating the correlation between MethylCIBERSORT CD8 proportion and TMA density (r = 0.51), and the correlation between MethylCIBERSORT CD8 proportion change (resection - biopsy) andTMA CD8 density change (r = 0.31), respectively.
Discussion
Our study is the first to characterize immune-related gene expression and DNA methylation profiles after neoadjuvant immunotherapy with the IRX-2 regimen in a randomized clinical trial. We identified expression changes in immune response genes after treatment with both regimens. Surprisingly, the differential expression pattern was similar in Regimen 1 and Regimen 2, with more than half of the differentially expressed genes in common between the two regimens, suggesting much of the action in the tumor microenvironment (TME) of the complete IRX-2 treatment protocol is due to elements other than the cocktail of cytokines directed at the regional lymph nodes. For example, one cytokine component in IRX-2, IL1B, was downregulated after both regimens. However, the IL2 cytokine component, showed more significant change between biopsy and resection samples for Regimen 1 patients (Figure 2A), indicating a potential linkage between IRX-2 treatment and higher activated CD4+ T cells and activated CD8+ T cells, major sources of IL2 in the TME(27). DMBT1 (deleted in malignant brain tumor 1) is a potential tumor suppressor gene encoding a pattern recognition molecule that plays a key role within the innate immune system(28,29). Uniquely, DMBT1 was found to have more extensive upregulation in Regimen 1 than Regimen 2. Previous studies confirmed the critical role DMBT1 plays in the interaction of tumor cells and the immune system, and downregulation of DMBT1 is thought to promote invasion in head and neck cancer(24,25), which could suggest modulation of tumor invasiveness after IRX-2 treatment in Regimen 1 in our study. However, follow-up studies are needed to further understand the downstream effects of DMBT1 upregulation in patients with OCC and the results reported here are in a limited number of patients.
Cyclophosphamide, a component of both regimens, slows cancer growth by inhibiting protein synthesis and can suppress regulatory T cells with a single low dose(30). Indomethacin and zinc are also known immunomodulators. Given the complexity of the complete IRX-2 regimen and unknown interactions among its components, it is difficult to predict a priori the effects it has on PD1 and PDL1 expression. In our study with only limited numbers, we did not observe a clear distinction based on PD1 and PDL1 changes in the TME between the two regimens, and in fact the PD1 and PDL1 levels remained relatively stable for immune responders in Regimen 1 suggesting that the effect of IRX-2 cytokines likely differs from traditional checkpoint inhibitors and might help reconstitute the immune response by stimulating T cell infiltration. Further, our measures did not specifically differentiate between tumor and immune cell expression of these biomarkers. However, both PD1 and PDL1 expression dropped in Regimen 2, particularly for non-responders and for non-responders in Regimen 1, which is consistent with an effect on PD-1 blockade(31) suggesting that such effects might not be clinically beneficial. In prior work we noted increases in CD68 tumor associated macrophages associated with IRX Regimen 2 which would be consistent with such a negative effect(23). Interacting factors makes clear interpretations difficult, however the lack of toxicity also makes IRX-2 an attractive approach combined with other immune modulators.
Utilizing the DNA methylation data, we were able to identify an increase in global methylation levels after treatment in both regimens, with a more prominent trend in responders of Regimen 1 and in LINE repetitive elements. Since global and repetitive element hypomethylation is common across cancer types, this increase is shifting the tumor’s methylation status closer to “normal”. Other publications also report the potential importance of PD1/PDL1 methylation in the prognosis of HNSCC patients(15,32), however we did not observe any significant methylation change in or near PD1 or PDL1. Pathway enrichment analysis with the DNA methylation data revealed pathways linked with hyper-methylation after treatment in Regimen 1, including cornification, keratinization and epidermic development, which could point to a change in the differentiation status of tumors(33,34). Although our immune-focused NanoString data did not allow us to assess keratinization gene changes, this could be a counterpoint to the benefit of IRX-2 worth studying, since low keratinization has been associated with higher recurrence rates and lower 5-year survival rates in oral cancer patients(35).
Previously, our group published the preliminary findings of this phase2b clinical trial that Regimen 1 resulted in higher levels of CD8+ T cell density post-treatment than did Regimen 2(23). Our study reconfirms this finding based on significant CD8 markers’ gene expression (CD8A and IL2) in Regimen 1 only and on methylation-based cell type deconvolution. However, our analysis also suggested that most of the observed immune pattern changes beyond this and the PD1/PDL1 findings, did not significantly differ by regimen. The main potential benefit of IRX-2 has been thought to be increased activation and migration of T cells from the regional lymph nodes into the tumor microenvironment. Direct effects of the cytokine preparation on the primary tumor would not necessarily be expected unless they are mediated by changes in TILs. Our current study focused on changes in the tumor microenvironment rather than T cells, therefore the lack of major changes in gene expression or methylation is not surprising. We found general changes associated with both immune modulating treatment regimens that probably reflect the non-cytokine components of the regimens but also encouraging findings that confirmed the T cell density counts previously reported with Regimen 1.
A limitation of our study is the inconsistent availability of different data types. Ninety-six patients were enrolled in the Phase 2b clinical trial, whose clinical characteristics and survival information were collected in a prospective manner. Correlations of our findings with patient survival are currently awaiting further follow-up. However, only 71 patients had data sufficient for gene expression analysis, and only a subset of those (24) had DNA methylation analysis. Only 26 patients had CD8+ T cell density measured for determining immune responder status, and clinical tumor response was only available for 77 patients, resulting in missing response data in a large percent of patients with gene expression or DNA methylation data. With gene expression analysis limited to NanoString instead of full RNA-seq, our study also lacked the ability to study expression of other important pathways such as keratinization and differentiation status.
Conclusion
Future studies with larger sample size and more complete data will provide a more comprehensive picture and a better understanding of the effects of IRX-2 treatment and help in selecting patients most suitable for immunotherapy. As one of the first studies to capture the genetic and epigenetic effects of neoadjuvant immunotherapy with the IRX-2 regimen, we conclude that the cytokine components of IRX-2 have definite but subtle effects on patient immune response, and that some of the effects of the complete IRX-2 treatment are due to the non-cytokine components. Specific benefits in the tumor microenvironment of the cytokine cocktail in IRX-2 remain undefined, but our findings in this very limited study are consistent with increases in CD8+ T cell density, modest global and repetitive element re-methylation of the genome, upregulation of the tumor suppressor DMBT1, and unchanged PD1/PDL1 for the subset of patients showing immune cell infiltration.
Supplementary Material
Highlights:
First study to capture transcriptomic and epigenomic effects of IRX-2 immunotherapy
Both neoadjuvant regimens resulted in significant immune expression profile changes
Methylation-based cell type deconvolution captured CD8+ T cell density changes
Acknowledgement
We thank the Advanced Genomics Core at the University of Michigan for their careful sample and library preparations.
Funding
Research reported in this publication was supported in part by grant funding from Brooklyn ImmunoTherapeutics, Inc., Brooklyn, New York and the National Cancer Institute of the National Institutes of Health under Award Number P30CA046592 by the use of the following Cancer Center Shared Resource(s): Tissue and Molecular Pathology. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Clinical trial was sponsored by Brooklyn ImmunoTherapeutics, Inc. and the Rogel Cancer Center at University of Michigan.
Conflict of Interest
Dr. Wolf was a recipient of grant funding from the clinical trial sponsor, Brooklyn ImmunoTherapeutics, for central laboratory tissue collection and correlative immune studies. Dr. Wolf also serves on the Scientific Advisory Board for Brooklyn ImmunoTherapeutics, Inc.
Members of the INSPIRE Trial Clinical Investigators include: 1Jeff Moyer, 2Mihir Patel, 3Audrey Erman, 4Wanessa A. Martins, 5Jason Newman, 6Michael Kaplan, 7Frabicio Oliveira, 8Ana Paula Victorina, 9R. Bryan Bell, 10Gustavo C. Girotto, 11Jorge Nieva, 12Joseph Valentino, 13Greg Krempl, 14Claudio R. Cernea, 15Dennis Kraus, 16Kevin Higgins, 17Felipe JSM Cruz, 18Aru Panwar, 19Clodoaldo Z Campos, 20Jim McCaul
1University of Michigan, Ann Arbor, MI 2Emory University Winship Cancer Institute, Atlanta, GA; 3University of Arizona Medical Center, Tucson, AZ; 4Instituo Goiano de Oncologia e Hematologia, Brazil; 5Hospital of the University of Pennsylvania, Philadelphia, PA; 6Stanford University Medical Center, Stanford, CA; 7Hospital Erasto Gaertner/Liga Paranaense de Combate Cancer, Brazil; 8Instituto Nacional do Cancer, Brazil; 9Providence Cancer Center, Providence, RI; 10Hospital de Base de Sao Jose do Rio Preto, Brazil; 11University of Southern California, Los Angeles, CA; 12University of Kentucky Medical Center, Lexington, KY; 13University of Oklahoma Health Sciences Center, Oklahoma City, OK; 14Instituto do Cancer do Estado de Sao Paulo; 15Northwell Health Lenox Hill Hospital, New York, NY; 16Sunnybrook Research Institute, Toronto, CA; 17Instituto Brasileiro de Controle ao Cancer; 18University of Nebraska Medical Center, Omaha, NE; 19Hospital do Cancer de Londrina, Brazil; 20Queen Elizabeth University Hospital, Glasgow, UK.
Reference
- 1.Argiris A, Karamouzis MV, Raben D, Ferris RL. Head and neck cancer. Lancet (London, England) [Internet]. 2008;371:1695–709. Available from: http://www.sciencedirect.com/science/article/pii/S014067360860728X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019. [DOI] [PubMed] [Google Scholar]
- 3.Pulte D, Brenner H. Changes in Survival in Head and Neck Cancers in the Late 20th and Early 21st Century: A Period Analysis. Oncologist. 2010; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cramer JD, Burtness B, Ferris RL. Immunotherapy for head and neck cancer: Recent advances and future directions. Oral Oncol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, et al. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016; [DOI] [PubMed] [Google Scholar]
- 6.Bauml J, Seiwert TY, Pfister DG, Worden F, Liu SV., Gilbert J, et al. Pembrolizumab for platinum- and cetuximab-refractory head and neck cancer: Results from a single-arm, phase II study. J Clin Oncol. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferris RL, Blumenschein G, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N Engl J Med. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolf GT, Moyer JS, Kaplan MJ, Newman JG, Egan JE, Berinstein NL, et al. IRX-2 natural cytokine biologic for immunotherapy in patients with head and neck cancers. Onco. Targets. Ther 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berinstein NL, McNamara M, Nguyen A, Egan J, Wolf GT. Increased immune infiltration and chemokine receptor expression in head and neck epithelial tumors after neoadjuvant immunotherapy with the IRX-2 regimen. Oncoimmunology. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007; [DOI] [PubMed] [Google Scholar]
- 11.Castilho RM, Squarize CH, Almeida LO. Epigenetic modifications and head and neck cancer: Implications for tumor progression and resistance to therapy. Int J Mol Sci. 2017;18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaździcka J, Gołąbek K, Strzelczyk JK, Ostrowska Z. Epigenetic Modifications in Head and Neck Cancer. Biochem. Genet 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung H, Kim HS, Kim JY, Sun JM, Ahn JS, Ahn MJ, et al. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat Commun. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Misawa K, Yamada S, Mima M, Nakagawa T, Kurokawa T, Imai A, et al. Long interspersed nuclear element 1 hypomethylation has novel prognostic value and potential utility in liquid biopsy for oral cavity cancer. Biomark Res. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goltz D, Gevensleben H, Dietrich J, Schroeck F, de Vos L, Droege F, et al. PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Darvin P, Toor SM, Sasidharan Nair V, Elkord E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue G, Cui Z-J, Zhou X-H, Zhu Y-X, Chen Y, Liang F-J, et al. DNA Methylation Biomarkers Predict Objective Responses to PD-1/PD-L1 Inhibition Blockade. Front Genet. Frontiers Media S.A; 2019;10:724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Starzer AM, Heller G, Tomasich E, Feldmann K, Hatziioannou T, Traint S, et al. DNA methylation profiling in patients with head and neck squamous cell carcinoma treated with immune checkpoint inhibitors. J Clin Oncol. American Society of Clinical Oncology (ASCO); 2020;38:e18527–e18527. [Google Scholar]
- 19.Chakravarthy A, Furness A, Joshi K, Ghorani E, Ford K, Ward MJ, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat Commun. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: An extensible R package for the pre-processing of nanostring mRNA and miRNA data. Bioinformatics. 2012; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peters TJ, Buckley MJ, Statham AL, Pidsley R, Samaras K, V Lord R, et al. De novo identification of differentially methylated regions in the human genome. Epigenetics and Chromatin. 2015; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wolf GT, Liu S, Bellile E, Sartor M, Rozek L, Thomas D, et al. Tumor infiltrating lymphocytes after neoadjuvant IRX-2 immunotherapy in oral squamous cell carcinoma: Interim findings from the INSPIRE trial. Oral Oncol. 2020; [DOI] [PubMed] [Google Scholar]
- 24.PIAO S, PIAO S, BANERJEE R, LIU M, INGLEHART RC, D’SILVA N. DOWNREGULATION OF DMBT1 PROMOTES INVASION IN HEAD AND NECK CANCER. Oral Surg Oral Med Oral Pathol Oral Radiol. 2017; [DOI] [PubMed] [Google Scholar]
- 25.Singh P, Banerjee R, Piao S, Costa de Medeiros M, Bellile E, Liu M, et al. Squamous cell carcinoma subverts adjacent histologically normal epithelium to promote lateral invasion. J Exp Med. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RWC, Vatapalli R, et al. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao W, Lin JX, Leonard WJ. IL-2 family cytokines: New insights into the complex roles of IL-2 as a broad regulator of T helper cell differentiation. Curr. Opin. Immunol 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.End C, Bikker F, Renner M, Bergmann G, Lyer S, Blaich S, et al. DMBT1 functions as pattern-recognition molecule for poly-sulfated and poly-phosphorylated ligands. Eur J Immunol. 2009; [DOI] [PubMed] [Google Scholar]
- 29.Tuttolomondo M, Casella C, Hansen PL, Polo E, Herda LM, Dawson KA, et al. Human DMBT1-Derived Cell-Penetrating Peptides for Intracellular siRNA Delivery. Mol Ther - Nucleic Acids. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahlmann M, Hempel G. The effect of cyclophosphamide on the immune system: implications for clinical cancer therapy. Cancer Chemother. Pharmacol 2016. [DOI] [PubMed] [Google Scholar]
- 31.Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, et al. Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bais MV Impact of Epigenetic Regulation on Head and Neck Squamous Cell Carcinoma. J. Dent. Res 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Outh-Gauer S, Alt M, Le Tourneau C, Augustin J, Broudin C, Gasne C, et al. Immunotherapy in head and neck cancers: A new challenge for immunologists, pathologists and clinicians. Cancer Treat. Rev 2018. [DOI] [PubMed] [Google Scholar]
- 34.Canning M, Guo G, Yu M, Myint C, Groves MW, Byrd JK, et al. Heterogeneity of the head and neck squamous cell carcinoma immune landscape and its impact on immunotherapy. Front. Cell Dev. Biol 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfer S, Elstner S, Schultze-Mosgau S. Degree of Keratinization Is an Independent Prognostic Factor in Oral Squamous Cell Carcinoma. J Oral Maxillofac Surg. 2018; [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


