Abstract
Purpose:
To study associations across tumor types between genome-wide loss of heterozygosity (gLOH) and alterations in homologous recombination repair (HRR)-associated genes beyond BRCA1 and BRCA2.
Experimental Design:
Genomic profiling using a targeted next-generation sequencing assay examining 324–465 genes (FoundationOne, FoundationOne Heme, and FoundationOne CDx; Foundation Medicine, Inc.) was performed in a cohort of 160,790 samples across different tumor types. Zygosity predictions and gLOH status were calculated and linked with alterations in 18 HRR-associated genes (BRCA1, BRCA2, PALB2, BARD1, ATR, ATRX, ATM, BAP1, RAD51B, RAD51C, RAD51D, BRIP1, NBN, CHEK1, CHEK2, FANCA, FANCC, MRE11) and other genomic features, using Fisher's exact test and Mann–Whitney U tests.
Results:
We identified a strong correlation between elevated gLOH and biallelic alterations in a core set of HRR-associated genes beyond BRCA1 and BRCA2, such as BARD1, PALB2, FANCC, RAD51C, and RAD51D (particularly in breast, ovarian, pancreatic, and prostate cancer). Monoallelic/heterozygous alterations in HRR-associated genes were not associated with elevated gLOH. gLOH was also independently associated with TP53 loss. Co-occurrence of TP53 loss and alterations in HRR-associated genes, and combined loss of TP53-PTEN or TP53-RB1, was associated with a higher gLOH than each of the events separately.
Conclusions:
Biallelic alterations in core HRR-associated genes are frequent, strongly associated with elevated gLOH, and enriched in breast, ovarian, pancreatic, and prostate cancer. This analysis could inform the design of the next generation of clinical trials examining DNA repair–targeting agents, including PARP inhibitors.
Translational Relevance.
Improved understanding of the impact of homologous recombination repair (HRR)-associated gene alterations on tumor genomic signatures and drug sensitivity will enable better design of clinical trials examining DNA repair–targeting agents. Here, we report associations between genome-wide loss of heterozygosity (gLOH) and HRR-associated gene alterations in 160,790 tumors. Known/likely deleterious alterations in HRR-associated genes were found in 18.9% of cases. For both BRCA-associated (breast, ovarian, prostate, pancreatic) and other tumor types, we found strong associations between biallelic alterations and elevated gLOH for a number of core HRR-associated genes, beyond BRCA1 and BRCA2. gLOH differed between tumor types and was also associated with TP53 loss. This analysis suggests that both tumor type and the mutation allelic status may be relevant for clinical interpretation of gLOH. gLOH scores, together with other readouts of HRR deficiency, could enable a more targeted stratification of patients most likely to benefit from DNA repair–targeting agents.
Introduction
The identification and clinical qualification of cancer subtypes based on molecular profiles is central to precision oncology. In this setting, precision oncology aims to tailor therapeutic strategies to each patient based on a comprehensive assessment of individual clinical, genomic, and phenotypic features of the disease (1).
Genomic instability and loss of the physiologic capacity of cells to repair DNA damage are hallmarks of cancer (2). Yet, these features can also represent a tumor's vulnerability; drugs inducing DNA damage and targeting the DNA damage repair systems may be particularly effective in patients with tumors displaying defects in the DNA repair machinery (3–6).
The homologous recombination repair (HRR) pathway is a high-fidelity system involved in repairing double-stranded DNA breaks, and is critical to the resolution of stalled replication forks during cell division (7, 8). BRCA1 and BRCA2 are key components of the HRR pathway (7); inherited mutations in the BRCA1 or BRCA2 genes increase the risk of developing breast, ovarian, prostate, pancreatic, and various other cancers (9).
Therapeutically, inactivation of BRCA1 or BRCA2 typically renders tumors exquisitely sensitive to treatment with PARP inhibitors (4–6). This concept has driven the clinical development of PARP inhibitors in tumor types enriched for inactivation of BRCA1 or BRCA2, related to a germline and/or somatic tumor DNA alteration. Several PARP inhibitors are now approved by the FDA and European Medicines Agency for the treatment of different subtypes of advanced breast, ovarian, prostate, and pancreatic cancer (10). HRR-deficient (HRD) tumors are also more sensitive to other DNA-damaging agents such as platinum-based chemotherapies (3, 11), a feature of particular therapeutic opportunity in cancers with very limited treatment options such as triple-negative breast cancer (3).
In the absence of functional HRR, non-homologous end joining, microhomology-mediated end joining, and single-strand annealing are used to repair double-stranded breaks (12). The preferential use of these errorprone systems in HRD tumors results in accumulation of certain DNA alterations, particularly deletions, and other structural variations (13). Consequently, HRD tumors are characterized by enrichment of characteristic patterns of base substitutions, genome-wide accumulation of loss-of-heterozygosity (LOH) events, large-scale transitions [chromosomal breaks between adjacent regions of ≥10 megabase (Mb)], and subchromosomal regions with allelic imbalance extending to the telomere (14–17). This enrichment results in recognizable patterns, also referred to as “signatures” or “scars,” imprinted on the tumor genome (17). Next-generation sequencing assays have been developed to detect alterations in HRR-associated genes, as well as genomic patterns. This may allow for improved patient stratification for precision medicine approaches, including PARP inhibitors (18).
The total burden of focal genome-wide LOH (gLOH) is enriched in tumors with BRCA1 and/or BRCA2 inactivation (14). In a prior study, in which researchers derived the gLOH score from a clinical grade hybrid capture-based comprehensive genomic profiling (CGP) assay of over 200,000 tumors, a strong association was observed between biallelic BRCA1/2 alterations and gLOH (19).
In a separate study, Jonsson and colleagues demonstrated the lineage-dependence of BRCA1/2 alterations and their impact on tumor evolution (20). In breast, ovarian, prostate, and pancreatic cancer with loss-of-function BRCA1/2 mutations, jointly referred to as “BRCA-associated (BA) tumors,” an enrichment for gLOH was observed (20). In contrast, this was not seen in other tumor types. This observation is suggestive of a selective pressure for biallelic loss preferentially in ovarian, breast, pancreatic, and prostate tumors, whereas in other tumor types, alterations in HRR-associated genes may have a neutral, or at least less relevant, role in cancer progression (20).
In the ARIEL-2 trial of recurrent, platinum-sensitive, high-grade ovarian carcinoma, patients with a high gLOH score but no BRCA1/2 alteration had significantly longer progression-free survival when treated with the PARP inhibitor rucaparib than patients who had lower gLOH scores (6). Indeed, responses to PARP inhibitors have been documented among patients with cancers harboring alterations in HRR-associated genes other than BRCA1/2 [ARIEL-3 (21); PAOLA-1 (22); GALAHAD (23); TOPARP-B (24); TRITON2 (25)]. On a gene-per-gene basis, assessing the contribution to oncogenesis and impact on drug sensitivity of these other HRR-associated gene alterations is challenging due to their low prevalence. Therefore, surrogate biomarkers of HRD could contribute to the clinical qualification of these less common events, drive more effective stratification of patients for clinical trials, and better predict response to platinum-based chemotherapy and PARP inhibitors, while enabling a more precise treatment selection in a significant number of patients with cancer.
Here, we report a genomic analysis in a pan-cancer cohort of 160,790 tumor samples, aiming to describe the distribution of gLOH scores in tumors with these less common HRR-associated gene alterations (e.g., BARD1, RAD51C, RAD51D, and PALB2), assessing other factors that may impact gLOH, and understanding how these associations may differ across tumor types.
Materials and Methods
Genomic profiling of 324–465 genes, including a predefined set of genes directly or indirectly involved in HRR (HRR-associated genes: BRCA1, BRCA2, PALB2, BARD1, ATR, ATRX, ATM, BAP1, RAD51B, RAD51C, RAD51D, BRIP1, NBN, CHEK1, CHEK2, FANCA, FANCC, and MRE11), was performed in a pan-cancer cohort of 160,790 tumor samples for multiple classes of alterations (short variants, insertions, deletions, copy-number events, and rearrangements), using the FoundationOne, FoundationOne Heme, and FoundationOne CDx assays (Foundation Medicine, Inc.; 26, 27). Although the population in this study represents all cases in which profiling data were evaluable, the patient samples sent to Foundation Medicine for sequencing tend to be from later-stage, more advanced disease; thus, the dataset may exhibit bias (e.g., for relapsed disease or more aggressive subtypes).
In this study, we predefined two groups within our population: those patients with breast, ovarian, pancreatic, or prostate cancer (referred to as BA tumor types), and the rest (referred to as “other” or “non–BA” or “NBA tumor types”).
Genomic profiling was carried out in a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists-accredited laboratory (Foundation Medicine, Inc.). At least 50 ng of DNA per specimen was isolated and sequenced to high, uniform coverage (mean, >600×), as described previously (27). The biopsy site was taken from the pathology report for the provided specimen. Variants were interpreted at Foundation Medicine, Inc., based on an analysis of the medical literature and variant databases.
For tumor suppressors, all classes of deletions/truncations were considered pathogenic, including frameshift mutations, core splice site alterations, nonsense mutations, deep deletions, and truncating rearrangement events. Select pathogenic missense mutations were also included on the basis of prior literature and hotspot status (e.g., the inclusion of BRCA1 founder missense mutations). For the HRR-associated genes included in this study, most events were frameshift (36%), nonsense (24%), splice (11%), deletion (8%), and truncating rearrangement (6%) events, with a relatively small proportion of pathogenic missense mutations (12%) and other events (2%).
Zygosity predictions, gLOH, and biallelic status of HRR-associated gene alterations (Supplementary Fig. S1) were calculated as described previously (6, 28). gLOH was assessed as a continuous variable, rather than selecting a cut-off point. When examining gLOH in patients carrying an alteration within an HRR-associated gene (e.g., BRCA1), we excluded the chromosomal arm of the gene to avoid biasing the calculations when examining homozygous versus heterozygous alterations in the gene.
Biomarkers previously associated with benefit from immune checkpoint inhibitors in clinical trials were also evaluated, including microsatellite instability (MSI; 29) and tumor mutational burden (TMB; 30). MSI was calculated on ≥90 loci. TMB was calculated using a range of 0.8–1.2 Mb, excluding driver and germline alterations; high TMB was defined as >10 mutations/Mb (Mut/Mb). Age was captured at specimen collection date, or if not provided, at testing. Ancestry was determined using a SNP-matching approach (31).
All samples that passed sample quality check metrics overall and for gLOH calling were included in the analysis (n = 160,790). Specifically, samples had to have a passed report with an estimated 30% tumor purity and be free of contamination; in addition, samples with noisy copy-number profiles, as assessed by a segment-level signal to noise ratio, were excluded.
For comparisons of categorical variables, we used the Fisher's exact test. For comparisons of continuous variables, the non-parametric Mann–Whitney U test was used.
This study was conducted according to the ethical principles for medical research described in the Declaration of Helsinki. Approval for this analysis, including a waiver of informed consent and a Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (Protocol No. 20152817).
Results
Study population and sample disposition
Genomic profiling data for 160,790 samples sequenced as part of routine clinical care in academic or community centers (outside of clinical trials) were included in this analysis. A summary of the cohort characteristics is shown in Table 1. Median age of the cohort was 63 years (interquartile range: 54–71) and the male:female ratio was 44%:56%. Median TMB was 3.8 Mut/Mb (interquartile range: 2.5–7.5) and 2,431/160,546 (1.5%) samples were classified as MSI-high. The cohort included primary tumor biopsies (n = 66,129; 41%), as well as biopsies acquired from metastatic lesions (n = 50,157; 31%) or lymph nodes (n = 14,002; 9%); in 19% of samples (n = 30,502), the origin of the tissue was ambiguous or not annotated. The three most common cancer types were non–small cell lung cancer (NSCLC; n = 26,511; 16%), colorectal cancer (n = 20,943; 13%), and breast cancer (n = 20,614; 13%; Supplementary Fig. S2). In this population, 44,765/160,790 (28%) cases corresponded to ovarian, breast, pancreatic, or prostate cancers, and were considered together for the analysis as BA tumor types, whereas 116,025 (72%) cases were patients with other tumor types not included in the BA category [“non-BA tumor types” (NBA)]. The overall genomic landscape of the cohort is shown in Supplementary Fig. S3.
Table 1.
Cohort characteristics.
| Characteristic | Patients (N = 160,790) | Median gLOH (interquartile range) |
|---|---|---|
| Overall | 8.22 (3.98–14.04) | |
| Male:Female, % | 44:56 | 7.71 (3.81–12.76); 8.7 (4.13–15.18) |
| TMB, median (interquartile range) | 3.8 (2.5–7.5) | |
| High MSI, % | 1.5 | 2.64 (1.29–5.55) |
| Not MSI-high, % | 98.5 | 8.32 (4.09–14.13) |
| Biopsy type, % | ||
| Local | 41 | 7.25 (3.32–12.82) |
| Metastatic | 31 | 9.11 (4.88–15.06) |
| Lymph node biopsy | 9 | 9.65 (5.19–15.76) |
| Unknown | 19 | 8.11 (3.7–14.14) |
| Tumor type | ||
| Other | 28,208 | 6.42 (2.82–11.4) |
| NSCLC | 26,511 | 10.81 (5.89–16.66) |
| CRC | 20,943 | 6.07 (3.28–9.5) |
| Breast | 20,614 | 12.16 (6.99–19.66) |
| Ovary | 11,427 | 10.9 (4.54–20.65) |
| CUP | 8,533 | 9.62 (5.04–15.42) |
| Glioma | 7,107 | 2.97 (1.27–5.63) |
| Prostate | 6,434 | 8.49 (5.68–11.91) |
| Pancreas | 6,290 | 9.33 (5.91–13.91) |
| Melanoma | 5,141 | 5.99 (3.16–9.81) |
| Endometrial | 4,790 | 6.05 (2.28–12.38) |
| Esophagus | 4,765 | 12.15 (6.62–17.82) |
| Bladder | 3,277 | 8.01 (4.26–12.85) |
| Kidney | 2,482 | 4.51 (1.99–8.32) |
| Cholangiocarcinoma | 2,419 | 10.04 (6.44–14.43) |
| Stomach | 1,849 | 9.18 (3.39–16.94) |
Abbreviations: CRC, colorectal cancer; CUP, carcinoma-of-unknown-primary-origin.
Mutations in HRR and other DNA repair genes
We focused our analysis on deleterious or likely deleterious alterations in a broad set of 18 HRR-associated genes (Supplementary Table S1). Deleterious or likely deleterious alterations in HRR-associated genes were found in 30,326/160,790 (18.9%) of cases.
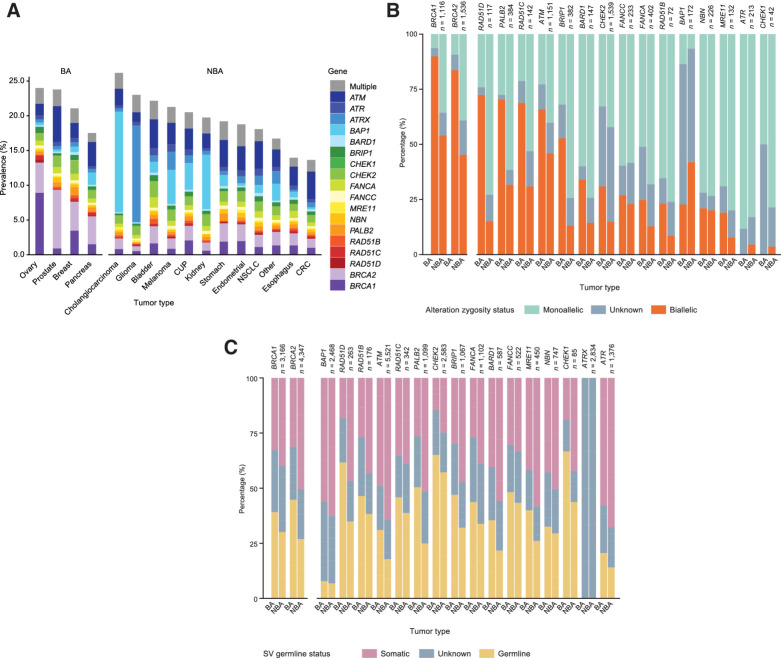
Among BA tumor types, BRCA1/2 were the most commonly altered HRR-associated genes [overall prevalence: n = 2,166/44,765 (5%) BRCA1, 2,517/44,765 (6%) BRCA2; breast cancer: 4% BRCA1, 5% BRCA2; ovarian cancer: 10% BRCA1, 5% BRCA2; prostate cancer: 1% BRCA1, 10% BRCA2; pancreatic cancer: 2% BRCA1, 5% BRCA2; Fig. 1A]. The highest prevalence of non-BRCA1/2 HRR-associated gene alterations was in prostate (14%), breast (13%), pancreatic (12%), and ovarian cancer (10%; Fig. 1A). There were 5,941 cases with ATM alterations; the tumor types with the highest prevalence of ATM alterations were prostate (6%), NSCLC (5%), endometrial (5%), bladder (5%), stomach (5%), and pancreatic cancer (4%). A total of 2,792 cases with CHEK2 alterations (prevalence: 2% breast, 1% ovarian, 2% prostate, and 2% pancreatic cancer) and 1,224 cases with PALB2 mutations (prevalence: 1% breast, 1% ovarian, 1% prostate, and 1% pancreatic cancer) were detected (Fig. 1A).
Figure 1.
Pathogenic variants in HRR-associated genes in the overall cohort (divided by tumor types), including prevalence (A), biallelic/monoallelic status (B), and germline/somatic status (C). CUP, carcinoma of unknown primary origin; Germline/somatic status was limited to short variants.
For other tumor types not included in the BA cohort (NBA), the overall prevalence of HRR-associated gene alterations was 18%; 15% after excluding BRCA1/2 alterations (Fig. 1A). ATM, ATRX, and CHEK2 alterations were most prevalent among the cohort of NBA tumor types (Fig. 1A).
As, in principle, biallelic alterations in HRR-associated genes would be necessary for loss of function, we decided to assess the allelic status of the HRR-associated gene alterations in our cohort (Fig. 1B). Of the samples harboring HRR-associated gene alterations, the percentage that was biallelic was higher among BA tumor types (58% vs. 33% in the cohort of NBA tumor types; P < 1 × 10−100; Fig. 1B). In 7,739/9,086 (85%) of cases, we were able to confidently assess BRCA1/2 allelic status; among these, BRCA1/2 alterations were associated with biallelic loss in BA tumor types (91% BRCA1; 88% BRCA2; Fig. 1B). Among BA tumor types with assessable samples, HRR-associated genes beyond BRCA1/2 in which alterations were most commonly associated with biallelic loss were BAP1 [n = 294/333 (88%)], RAD51D [81/104 (78%)], RAD51B [111/149 (74%)], RAD51C [111/154 (72%)], ATM [823/1,233 (67%)], and PALB2 [259/427 (61%); Fig. 1B]. Beyond ovarian, breast, pancreatic, or prostate cancer, only alterations in BAP1 and RAD51B were commonly associated with biallelic loss. Among cases where biallelic status could be definitively assessed, alterations in ATR, NBN, MRE11, and CHEK1 were only infrequently associated with a biallelic event in both cohorts of BA (8%, 18%, 26%, and 14%, respectively) and NBA (8%, 16%, 11%, and 7%, respectively) tumor types (Fig. 1B; Supplementary Fig. S4).
Next, we used a previously validated algorithm based on variant allele frequency, locus copy number, and minor allele fraction to infer the HRR-associated gene alterations that were likely to originate in germline DNA (Fig. 1C). Overall, 28% of HRR-associated gene alterations were predicted to be a germline alteration, compared with 42% predicted to be somatic alteration (in the remaining 30%, we were not able to predict a somatic vs. germline origin based on tumor next-generation sequencing data; Fig. 1C). CHEK2 (60%), CHEK1 (49%), FANCC (45%), RAD51D (44%), RAD51C (42%), RAD51B (41%), BRIP1 (36%), BRCA1 (35%), and BRCA2 (35%) alterations were commonly predicted to be of germline origin, whereas alterations in ATM (21%), ATR (15%), and BAP1 (7%) were less commonly predicted to be germline events (Fig. 1C). For patients with predicted germline alterations, we observed a modestly higher rate of biallelic alterations for most genes (Supplementary Fig. S5).
gLOH score distribution among tumor types and correlation with HRR-associated genes
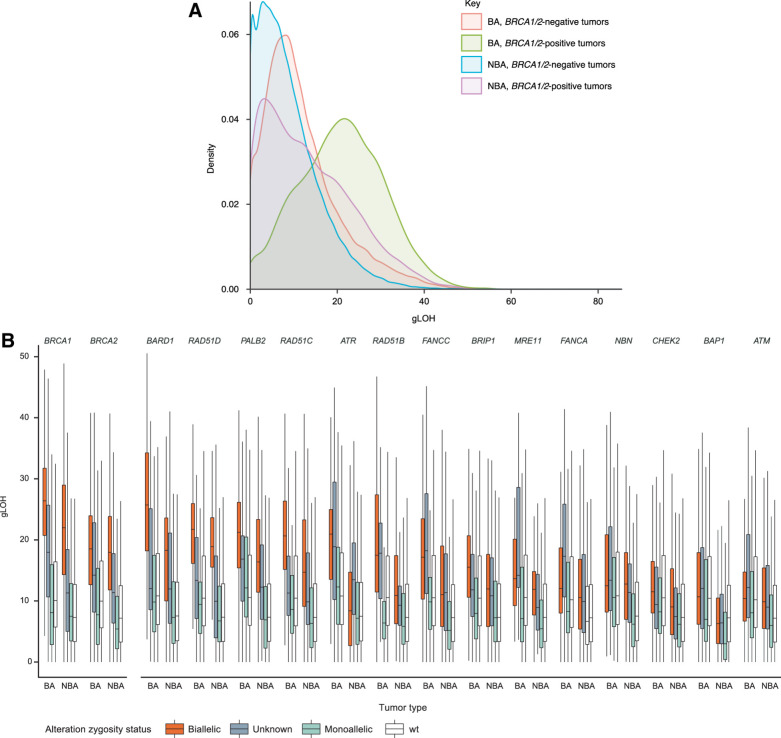
As gLOH has been proposed as a clinically relevant marker of HRD, we next assessed gLOH in this cohort, focusing on tumor-type differences and the genomic correlates of gLOH scores (Fig. 2A). Overall, gLOH showed a multimodal distribution and had a median of 8% (interquartile range: 4%–14%); median gLOH was higher in BA tumor types [11% vs. 7% in the cohort of other (NBA) tumor types], even after removing BRCA1/2-altered cases (10% vs. 7%). The tumor types with the highest median gLOH scores were breast (12%), esophageal (12%), and ovarian (11%).
Figure 2.
Overall distribution of gLOH across BA and NBA tumor types (A) and association of HRR-associated gene alterations (B) with gLOH across all cancers. wt, wild type; Only genes with at least five assessable alterations in each category were considered; wt refers to the gene being analyzed.
There was an association between age and gLOH only in the BA cohort, with younger individuals having more elevated gLOH (Supplementary Fig. S6A). Within each tumor type, gLOH scores were relatively similar across different ancestries (African, Central and South American, East Asian, European, and South Asian), except for melanoma, where Europeans had a lower gLOH (P < 0.01), and ovarian cancer, where gLOH was higher in South Asian and East Asian populations (Supplementary Fig. S6B; P < 0.01). Of note, in ovarian cancer, patients with South or East Asian ancestry had a median age 7 years younger than other patients (56 vs. 63 years; P = 4 × 10−34).
In both cohorts (BA and NBA tumor types), strong positive associations between biallelic HRR-associated gene alterations and gLOH (50% increase in gLOH relative to wild type; P < 1 × 10−5) were observed for BRCA1 (BA tumor types: 162% increase; NBA tumor types: 201% increase), BRCA2 (85%; 149%), BARD1 (137%; 142%), PALB2 (100%; 123%), FANCC (62%; 51%), RAD51C (97%; 100%), and RAD51D (107%; 157%; Fig. 2B; Supplementary Table S2). Strong positive associations were also observed for RAD51B in the cohort of BA tumor types (65% increase) and for NBN (69%) and BRIP1 (63%) in the cohort of NBA tumor types. In both BA and NBA tumor types, monoallelic alterations were not associated with elevated gLOH for any gene examined.
The association between biallelic loss of these genes and elevated gLOH was consistent across tumor types (Supplementary Fig. S7A–S7F; Supplementary Tables S2 and S3). In particular, based on statistical significance, strong positive associations with gLOH were observed for alterations in BRCA1, BRCA2, BARD1, PALB2, RAD51C, and RAD51D across breast, colorectal cancer, NSCLC, ovarian, pancreatic, and prostate cancer (effect sizes >50%; P < 1 × 10−5; Supplementary Fig. S7A–S7F). Some tissue-specific effects were observed. For example, in prostate cancer samples with BRCA1/2 biallelic alterations, median gLOH was lower than in the other BA tumor types (Supplementary Fig. S7C). Biallelic ATM and CHEK2 alterations were associated with elevated gLOH only in stomach and esophageal cancer (Supplementary Fig. S8A and S8B; Supplementary Tables S4 and S5); biallelic BAP1 associations were not associated with strongly elevated gLOH in any tumor type examined (all effect sizes <50%; Supplementary Fig. S8C; Supplementary Table S6).
Impact of other genomic features upon gLOH scores
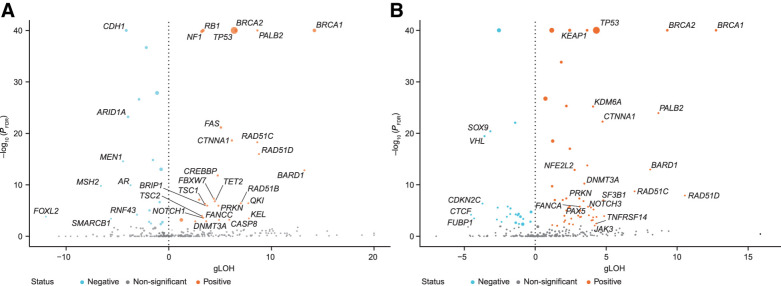
Next, we investigated correlations between gLOH scores and alterations in all baited genes, including genes not in our curated HRR-associated pathway list (Fig. 3; Supplementary Fig. S8; Supplementary Tables S4–S8).
Figure 3.
Association of biallelic gene alterations and gLOH in BA (A) and NBA (B) tumor types. FDR, false discovery rate; volcano plots show the median gLOH difference and P value between biallelic and wild-type samples using a non-parametric Mann–Whitney U test of distributions.
As expected, in the pan-gene analysis, the strongest associations between elevated gLOH and biallelic gene alterations were observed in BRCA1, BRCA2, PALB2, RAD51C, RAD51D, and BARD1 (Fig. 3A and B). Beyond these HRR-associated genes, CTNNA1 and TP53 alterations were highly associated with elevated gLOH scores, in both BA and NBA tumor type cohorts (Supplementary Fig. S8D and S8E; Supplementary Tables S7 and S8).
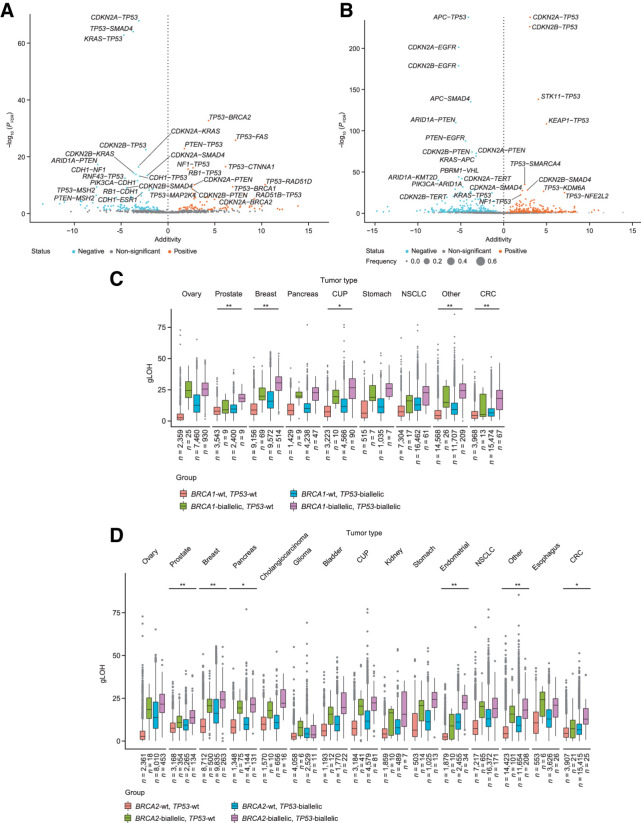
Because many of these alterations are known to co-occur (e.g., BRCA1 and TP53), we explored how these alterations interacted, for example, whether co-occurrence of certain alterations associated with a higher gLOH than each of the alterations separately (Fig. 4). In the cohort of BA tumor types, we observed significant additivity for TP53 alterations with several other genes, including RAD51D, RAD51B, BRCA1, BRCA2, CTNNA1, and FAS (Fig. 4A). In contrast, in the cohort of NBA tumor types, these associations were largely absent with modest additivity observed for KEAP1–TP53, STK11–TP53, NFE2L2–TP53, and KDM6A–TP53 (Fig. 4B). Co-occurrent loss of several tumor suppressors, such as TP53–RB1, TP53–PTEN, or TP53–NF1 resulted in a mild but statistically significant additivity effect toward elevation of gLOH (Fig. 4B). Contrarily, the co-occurrence of TP53 with either MSH2 or KRAS alterations was associated with tumors having a significantly lower-than-expected gLOH score (Fig. 4B).
Figure 4.
Association of co-occurring gene alterations and gLOH in BA (A) and NBA (B) tumor types. Additivity of TP53 alterations with BRCA1 (C) and BRCA2 (D) across tumor types. CRC, colorectal cancer; CUP, carcinoma of unknown primary origin; FDR, false discovery rate; wt, wild type; asterisks denote level of significance after false discovery correction; *, P < 0.05; **, P < 0.01.
Next, we focused on the impact of TP53 alterations on the gLOH scores of BRCA1/2-altered tumors, the population in which PARP inhibitors have been more widely introduced into clinical practice (Fig. 4C and D). The additivity was significant in prostate and breast cancer; beyond BA tumor types, the impact of TP53 alterations was most relevant in bladder, endometrial, and colorectal cancer (Fig. 4C and D).
Correlation of gLOH with TMB and MSI
We examined associations between HRR-associated gene alterations and biomarkers that have been associated with benefit from immune checkpoint inhibitors in clinical trials, including MSI and TMB (Supplementary Fig. S9).
Across the cohort, median TMB was 3.8 Mut/Mb (interquartile range: 2.5–7.5) and was 3.5 and 4.3 in the cohorts of BA and NBA tumor types, respectively. Among tumor types with the highest number of samples available (melanoma, NSCLC, bladder, esophagus, stomach, carcinoma of unknown primary origin, breast, endometrial, colorectal cancer, kidney, ovary, glioma, cholangiocarcinoma, prostate, pancreas), the median TMB was the highest in melanoma (10.0 Mut/Mb), NSCLC (7.8 Mut/Mb), and bladder cancer (7.5 Mut/Mb). Overall, 2,431/160,546 (1.5%) samples had genomic evidence of high MSI, with lower rates of MSI in BA versus NBA tumor type cohorts (1% vs. 2%, P < 1 × 10−50). MSI was most prevalent in endometrial (n = 690/4,771; 14%), stomach (86/1,848; 5%), and prostate cancer (179/6,413; 3%).
In our cohort, there was an inverse relationship between gLOH and MSI status (Supplementary Fig. S9A); gLOH was significantly lower in MSI-high groups, compared with microsatellite-stable tumors (median 2.6 vs. 8.3; P < 1 × 10−100).
For most tumor types, there was little association between gLOH and TMB (Supplementary Figs. S9B and S10). However, gLOH was significantly lower in TMB-high tumors in endometrial cancer (median 7.5 vs. 2.8; P = 2 × 10−126), and significantly higher in TMB-high tumors in ovarian cancer (median 10.7 vs. 19.1; P = 6 × 10−21; Supplementary Fig. S9B).
Discussion
In this study, we identified a strong correlation between elevated gLOH and biallelic likely/known deleterious tumor alterations in HRR-associated genes beyond BRCA1 and BRCA2, namely BARD1, PALB2, FANCC, RAD51C, and RAD51D; contrarily, other HRR-associated genes presented weak or absent associations with gLOH. We also demonstrated that gLOH distribution differs between tumor types and is associated with genomic events beyond HRR-associated genes, particularly loss of TP53. Finally, we identified genomic interactions that were associated with significant increases in gLOH.
Recent clinical trials of PARP inhibitors in prostate, ovarian, and pancreatic cancers have broadened the focus on HRD as a potential common denominator for this therapeutic strategy [ARIEL-3 (21); PAOLA-1 (22); GALAHAD (23); TOPARP-B (24); TRITON2 (25)]. It is necessary to better understand the functional consequences of alterations in different HRR-associated genes to inform clinical trial design in the future, assess impact on clinical practice, and enable personalized therapeutic approaches. This may be relevant to clinical trials such as LODESTAR (NCT04171700), a phase II open-label study assessing the efficacy of rucaparib in patients with solid tumors and deleterious mutations in HRR-associated genes, or KEYLYNK-007 (NCT04123366), a phase II trial investigating olaparib combined with pembrolizumab in the treatment of cancers with mutations in HRR-associated genes and/or HRD.
In the past, pan-cancer trials in precision oncology assessed alterations in known HRR-associated genes without considering allelic status or the impact on genomic signatures, such as gLOH; thus, potentially confounding the interpretation of the effect of biomarker-matched drugs in this setting. The need to better understand the functional relevance of each of these alterations is further reflected in recent studies showing little activity of PARP inhibitors in ATM- or CHEK2-altered breast cancer (32), or seemingly differential activity of PARP inhibitors in prostate cancer with alterations in BRCA1 versus BRCA2 (33).
Although recent clinical studies have demonstrated elevated gLOH to predict a higher magnitude of response to PARP inhibitor treatment in ovarian cancer (6), analysis of the ARIEL-3 trial showed that there is still clinical benefit in patients responsive to platinum-based chemotherapy with a low gLOH and treated with rucaparib versus placebo (21). Clinical trials calculating an HRD score, using gLOH, large-scale transitions, and subchromosomal regions with allelic imbalance extending to the telomere, have demonstrated HRD positivity based on such a combined score to be significantly associated with improved clinical response to treatment with DNA-damaging agents in the breast cancer setting, with minimal benefit in the HRD-negative population (34, 35).
Using a CGP assay to analyze a cohort of 160,790 samples from a broad range of different malignancies allowed us to demonstrate that both germline and somatic alterations in HRR-associated genes, including genes beyond BRCA1 and BRCA2, are common across cancer types, but particularly frequent in breast, ovarian, prostate, and pancreatic cancer (referred to in these analysis as “BA tumor types”). In these tumor types, biallelic alterations of core HRR-associated genes were associated with an elevation in gLOH. However, in other tumor types, and although alterations in these HRR-associated genes were still prevalent, they were less commonly linked with biallelic gene loss, suggesting that HRR-associated gene alterations may less frequently be a driving force of disease progression in these other tumor types. This is consistent with findings from previous studies (20). Our data are relevant when planning basket trials of DNA damage repair–targeting agents, or when attempting to interpret tumor-agnostic biomarker studies. At the therapeutic level, the data presented here can inform precision oncology efforts, because dedicated molecular tumor boards have become available at many referral centers. These interdisciplinary expert meetings interpret tumor profiling results to inform clinical practice and explore targeted treatment options (36). With the knowledge that most likely deleterious alterations in key HRR-associated genes detected in tumor types beyond breast, ovarian, pancreatic, or prostate cancers may be heterozygous and thus potentially passenger mutations, therapeutic interventions that are most likely to be ineffective can be avoided. The presence of biallelic alterations in core HRR-associated genes, in association with elevated gLOH, may more accurately identify tumors more likely to benefit from PARP inhibitors and DNA-damaging agents in these other tumor types. Although, individually, these are not common, in sum they represent a significant number of patients who may benefit from these therapeutic approaches. We acknowledge, however, that some mechanisms leading to biallelic HRR-associated gene loss may not be captured by this next-generation sequencing CGP panel, for example, BRCA1 methylation in ovarian cancer, which has been shown to occur in up to 19.3% of cases and is predictive of response to PARP inhibitors (37, 38). This may explain, at least in part, recent studies in breast and prostate cancer reporting PARP inhibitor efficacy in patients harboring BRCA1/2 mutations, but with no evidence of second allele loss by next-generation sequencing (39–41).
Whole-genome or whole-exome studies previously identified genomic signatures of HRD (42); however, these assays are not easy to implement in clinical practice, due to their cost and high burden of bioinformatics requirements. Instead, deriving biomarkers of HRD using a clinical-grade CGP panel test can facilitate identification of HRD tumors in clinical practice.
The lack of elevated gLOH in the setting of biallelic ATM, BAP1, and CHEK2 alterations add to the current evidence with regards to the lower responsiveness of these tumors to PARP inhibitors alone (43–45). Still, these tumors may be targeted with other approaches leveraging their DNA repair impairment, and ATR (46) or DNA-dependent protein kinase inhibitors (47) are being tested in clinical trials. CTNNA1 encodes for alpha-catenin, which has been reported to be present in the nucleus and potentially play a role in DNA repair (48). In the current study, the association of biallelic CTNNA1 loss with elevated gLOH suggests a novel role for CTNNA1 in maintenance of genomic stability and possibly HRR; this requires further investigation (48).
Prospective validation of gLOH as a pan-cancer predictive biomarker for PARP inhibitor treatment has not yet been pursued. On the basis of our results, such an effort should consider tumor-type specificities in gLOH distribution. Moreover, we identified how TP53 alterations or concomitant loss of tumor suppressor genes may result in tumors with an elevated gLOH despite not presenting HRR-associated gene alterations, or how co-occurrence between TP53 loss and HRR-associated gene alterations may significantly elevate gLOH. The relationship between TP53 alterations, HRD, and genomic instability is complex; loss of TP53 may lead to chromosomal instability and aneuploidy (49), which could elevate gLOH scores. Yet, loss of TP53 may allow some cells to tolerate acute loss (biallelic alterations) better in key HRR-associated genes; thus, TP53 alterations may collaborate with HRD in some settings. Nevertheless, there is no clinical evidence demonstrating loss of TP53 alone to result in response to PARP inhibition. Therefore, the impact of TP53 loss in gLOH should be considered when evaluating the performance of gLOH as a predictive biomarker in clinical trials of PARP inhibitors.
We also observed an inverse correlation between gLOH and MSI status, and generally no associations between gLOH and TMB, except for ovarian cancer. This may be relevant to defining the optimal population for combination trials of PARP inhibitors and immune checkpoint inhibitors in different tumor types (50).
Our study is among the largest published so far examining putative surrogate biomarkers of HRD in clinical samples and could contribute to the development of precision oncology. However, we acknowledge several limitations. First, as a pan-cancer study, our cohort is inherently heterogeneous, so other variables intrinsic to different tumor types that we have not analyzed may confound some of the results. Second, as our cohort includes samples collected at different disease stages, we did not account for the potential effect of treatment-induced selective pressure on an enrichment of HRD features in advanced cancers. Third, cutoffs (effect sizes >50%) were chosen to explore associations between gLOH and gene alterations; it has yet to be seen whether more modest effects on gLOH are associated with potential clinical actionability. Finally, as we lack treatment outcome data, we cannot confirm the clinical value of our findings, but we are confident that this dataset will inform the design of the next generation of clinical trials examining DNA repair–targeting agents.
In conclusion, we show that alterations in HRR-associated genes are very common across cancer types and that biallelic alterations in core HRR genes associate with elevated gLOH scores, which can be assessed from clinical samples using a CGP assay.
Supplementary Material
Acknowledgments
All authors acknowledge support for third-party medical writing assistance (including copyediting, editorial assistance, and production assistance), furnished by Stephen Salem, BSc, of Health Interactions, provided by F. Hoffmann-La Roche Ltd, Basel, Switzerland. S. Ganesan acknowledges support from NCI (P30 CA072720, P01 CA250957). J. Mateo acknowledges support from the CRIS Cancer Foundation (PR_TCL_2020-10) and “la Caixa” Foundation (CaixaResearch Advanced Oncology Research Program).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
C.B. Westphalen reports other support from Roche during the conduct of the study; grants from Roche and personal fees from Roche outside the submitted work; and Faculty European Society of Medical Oncology, Translational Research and Precision Medicine Working Group. A.D. Fine reports personal fees from Foundation Medicine and other support from Roche during the conduct of the study. F. André reports grants from Roche, Daiichi, Lilly, AstraZeneca, Pfizer, and Novartis outside the submitted work. S. Ganesan reports grants from NCI during the conduct of the study; personal fees from Merck, EQRX, Foundation Medicine, Roche, SilaGene, KayoThera, and EMD Serano outside the submitted work. V. Heinemann reports grants, personal fees, and non-financial support from Merck, Roche, Amgen, and SIRTEX; grants and personal fees from Celgene and Boehringer; personal fees from Sanofi, Lilly, Taiho, and Halozyme; personal fees and non-financial support from Servier, MSD, and BMS; grants and non-financial support from Shire outside the submitted work. E. Rouleau reports grants and other support from AstraZeneca; other support from Clovis and GSK during the conduct of the study; grants and other support from Roche and other support from BMS outside the submitted work. C. Turnbull reports personal fees from AstraZeneca and Roche outside the submitted work. L. Garcia Palacios reports other support from Roche Farma S.A during the conduct of the study; other support from Roche Farma S.A outside the submitted work. J.-A. Lopez is a Roche full-time employee of F. Hoffmann-La Roche. J.-A. Lopez has access to company shares. E.S. Sokol reports other support from Foundation Medicine and Roche during the conduct of the study; in addition, E.S. Sokol has a patent for HRD Biomarker pending to Foundation Medicine. J. Mateo reports personal fees from Roche outside the submitted work; grants and personal fees from AstraZeneca and Pfizer Oncology; personal fees from Merck/MSD, Guardant Health, Janssen, Monterosa, and Clovis Oncology outside the submitted work. No other disclosures were reported.
Authors' Contributions
C.B. Westphalen: study design, data interpretation, preparation of manuscript, review of manuscript and approval of decision to submit. A.D. Fine: data collection, analysis and interpretation, preparation of manuscript, review of manuscript and approval of decision to submit. F. Andre: data interpretation, review of manuscript and approval of decision to submit. S. Ganesan: data interpretation, review of manuscript and approval of decision to submit. V. Heinemann: data interpretation, review of manuscript and approval of decision to submit. E. Rouleau: data interpretation, review of manuscript and approval of decision to submit. C. Turnbull: data interpretation, review of manuscript and approval of decision to submit. L. Garcia-Palacios: obtaining funding, review of manuscript and approval of decision to submit. J.A. Lopez: obtaining funding, review of manuscript and approval of decision to submit. E.S. Sokol: study design, study supervision, data collection, analysis and interpretation, preparation of manuscript, review of manuscript and approval of decision to submit. J. Mateo: study design, study supervision, data analysis and interpretation, preparation of manuscript, review of manuscript and approval of decision to submit.
References
- 1. Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med 2020;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100:57–70. [DOI] [PubMed] [Google Scholar]
- 3. Isakoff SJ, Mayer EL, He L, Traina TA, Carey LA, Krag KJ, et al. TBCRC009: a multicenter phase II clinical trial of platinum monotherapy with biomarker assessment in metastatic triple-negative breast cancer. J Clin Oncol 2015;33:1902–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moore K, Colombo N, Scambia G, Kim BG, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018;379:2495–505. [DOI] [PubMed] [Google Scholar]
- 5. Diéras V, Han HS, Kaufman B, Wildiers H, Friedlander M, Ayoub JP, et al. Veliparib with carboplatin and paclitaxel in BRCA-mutated advanced breast cancer (BROCADE3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 2020;21:1269–82. [DOI] [PubMed] [Google Scholar]
- 6. Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- 7. Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer 2016;16:110–20. [DOI] [PubMed] [Google Scholar]
- 8. Nguyen L, WMM J, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun 2020;11:5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mersch J, Jackson MA, Park M, Nebgen D, Peterson SK, Singletary C, et al. Cancers associated with BRCA1 and BRCA2 mutations other than breast and ovarian. Cancer 2015;121:269–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rose M, Burgess JT, O'Byrne K, Richard DJ, Bolderson E. PARP inhibitors: clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol 2020;8:564601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O'Reilly EM, Lee JW, Zalupski M, Capanu M, Park J, Golan T, et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol 2020;38:1378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cannan WJ, Pederson DS. Mechanisms and consequences of double-strand DNA break formation in chromatin. J Cell Physiol 2016;231:3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 2016;534:47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer 2012;107:1776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Birkbak NJ, Wang ZC, Kim JY, Eklund AC, Li Q, Tian R, et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov 2012;2:366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Popova T, Manié E, Rieunier G, Caux-Moncoutier V, Tirapo C, Dubois T, et al. Ploidy and large-scale genomic instability consistently identify basal-like breast carcinomas with BRCA1/2 inactivation. Cancer Res 2012;72:5454–62. [DOI] [PubMed] [Google Scholar]
- 17. Takaya H, Nakai H, Takamatsu S, Mandai M, Matsumura N. Homologous recombination deficiency status-based classification of high-grade serous ovarian carcinoma. Sci Rep 2020;10:2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pellegrino B, Musolino A, Llop-Guevara A, Serra V, De Silva P, Hlavata Z, et al. Homologous recombination repair deficiency and the immune response in breast cancer: a literature review. Transl Oncol 2020;13:410–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sokol ES, Pavlick D, Khiabanian H, Frampton GM, Ross JS, Gregg JP, et al. Pan-cancer analysis of BRCA1 and BRCA2 genomic alterations and their association with genomic instability as measured by genome-wide loss of heterozygosity. JCO Precis Oncol 2020;4:442–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jonsson P, Bandlamudi C, Cheng ML, Srinivasan P, Chavan SS, Friedman ND, et al. Tumour lineage shapes BRCA-mediated phenotypes. Nature 2019;571:576–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coleman RL, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. [DOI] [PubMed] [Google Scholar]
- 23. Smith MR, Sandhu SK, Kelly WK, Scher HI, Efstathiou E, Lara PN, et al. Pre-specified interim analysis of GALAHAD: a phase II study of niraparib in patients (pts) with metastatic castration-resistant prostate cancer (mCRPC) and biallelic DNA-repair gene defects (DRD). Ann Oncol 2019;30:LBA50. [Google Scholar]
- 24. Mateo J, Porta N, Bianchini D, McGovern U, Elliott T, Jones R, et al. Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): a multicentre, open-label, randomised, phase 2 trial. Lancet Oncol 2020;21:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abida W, Patnaik A, Campbell D, Shapiro J, Bryce AH, McDermott R, et al. Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol 2020;38:3763–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Food and Drug Administration (FDA). FoundationOne®CDx: Summary of safety and effectiveness data (SSED); 2019. Available from: https://www.accessdata.fda.gov/cdrh_docs/pdf17/P170019S006B.pdf.
- 27. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sun JX, He Y, Sanford E, Montesion M, Frampton GM, Vignot S, et al. ( 2018) A computational approach to distinguish somatic vs. germline origin of genomic alterations from deep sequencing of cancer specimens without a matched normal. PLoS Comput Biol 14(2): e1005965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Trabucco SE, Gowen K, Maund SL, Sanford E, Fabrizio DA, Hall MJ, et al. A novel next-generation sequencing approach to detecting microsatellite instability and pan-tumor characterization of 1000 microsatellite instability-high cases in 67,000 patient samples. J Mol Diagn 2019;21:1053–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chalmers ZR, Connelly CF, Fabrizio D, Gay L, Ali SM, Ennis R, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 2017;9:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Connelly CF, Carrot-Zhang J, Stephens PJ, Frampton GM. Abstract 1227:Somatic genome alterations in cancer as compared to inferred patient ancestry. Cancer Res 2018. (78) (13 Supplement) 1227. [Google Scholar]
- 32. Tung NM, Robson ME, Ventz S, Santa-Maria CA, Nanda R, Marcom PK, et al. TBCRC 048: phase II study of olaparib for metastatic breast cancer and mutations in homologous recombination-related genes. J Clin Oncol 2020;38:4274–82. [DOI] [PubMed] [Google Scholar]
- 33. Taza F, Holler AE, Fu W, Wang H, Adra N, Albany C, et al. Differential Activity of PARP Inhibitors in BRCA1- Versus BRCA2-Altered Metastatic Castration-Resistant Prostate Cancer. JCO Precision Oncology 2021;5:1200–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Loibl S, Weber KE, Timms KM, Elkin EP, Hahnen E, Fasching PA, et al. Survival analysis of carboplatin added to an anthracycline/taxane-based neoadjuvant chemotherapy and HRD score as predictor of response-final results from GeparSixto. Ann Oncol 2018;29:2341–7. [DOI] [PubMed] [Google Scholar]
- 35. Sharma P, Barlow WE, Godwin AK, Pathak H, Isakova K, Williams D, et al. Impact of homologous recombination deficiency biomarkers on outcomes in patients with triple-negative breast cancer treated with adjuvant doxorubicin and cyclophosphamide (SWOG S9313). Ann Oncol 2018;29:654–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luchini C, Lawlor RT, Milella M, Scarpa A. Molecular tumor boards in clinical practice. Trends Cancer 2020;6:738–44. [DOI] [PubMed] [Google Scholar]
- 37. Kondrashova O, Topp M, Nesic K, Lieschke E, Ho GY, Harrell MI, et al. Methylation of all BRCA1 copies predicts response to the PARP inhibitor rucaparib in ovarian carcinoma. Nat Commun 2018;9:3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sahnane N, Carnevali I, Formenti G, Casarin J, Facchi S, Bombelli R, et al. BRCA methylation testing identifies a subset of ovarian carcinomas without germline variants that can benefit from PARP inhibitor. Int J Mol Sci 2020;21:9708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robson M, Lai Z, Dearden S, Barrett JC, Harrington EA, Timms KM, et al. 1936P - Analysis of BRCA genes and homologous recombination deficiency (HRD) scores in tumours from patients (pts) with metastatic breast cancer (mBC) in the OlympiAD trial. Ann Oncol 2019;30:v780–1. [DOI] [PubMed] [Google Scholar]
- 40. Litton JK, Laird AD, Rugo HS, Ettl J, Hurvitz SA, Martin M, et al. Abstract CT072: Exploration of impact of tumor BRCA zygosity and genomic loss-of-heterozygosity (gLOH) on efficacy in Phase 3 EMBRACA study of talazoparib in patients (pts) with HER2-negative (HER2−) advanced breast cancer (ABC) and a germline BRCA1/2 (gBRCA1/2) mutation. Cancer Res 2020. (80) (16 Supplement) CT072. [Google Scholar]
- 41. Carreira S, Porta N, Arce-Gallego S, Seed G, Llop-Guevara A, Bianchini D, et al. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B trial. Cancer Discov 2021;11:2812–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, et al. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med 2017;23:517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 2020;382:2091–102. [DOI] [PubMed] [Google Scholar]
- 44. Jette NR, Radhamani S, Ye R, Yu Y, Arthur G, Goutam S, et al. ATM-deficient lung, prostate and pancreatic cancer cells are acutely sensitive to the combination of olaparib and the ATR inhibitor AZD6738. Genome Instab Dis 2020;1:197–205. [Google Scholar]
- 45. Hassan R, Mian I, Wagner C, Mallory Y, Agra M, Padiernos E, et al. Phase II study of olaparib in malignant mesothelioma (MM) to correlate efficacy with germline and somatic mutations in DNA repair genes. J Clin Oncol 38:15s, 2020. (suppl; abstr 9054). [Google Scholar]
- 46. Yap TA, Tan DSP, Terbuch A, Caldwell R, Guo C, Goh BC, et al. First-in-human trial of the oral ataxia telangiectasia and RAD3-related (ATR) inhibitor BAY 1895344 in patients with advanced solid tumors. Cancer Discov 2021;11:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fok JHL, Ramos-Montoya A, Vazquez-Chantada M, Wijnhoven PWG, Follia V, James N, et al. AZD7648 is a potent and selective DNA-PK inhibitor that enhances radiation, chemotherapy and olaparib activity. Nat Commun 2019;10:5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Serebryannyy LA, Yemelyanov A, Gottardi CJ, de Lanerolle P. Nuclear α-catenin mediates the DNA damage response via β-catenin and nuclear actin. J Cell Sci 2017;130:1717–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bronder D, Tighe A, Wangsa D, Zong D, Meyer TJ, Wardenaar R, et al. TP53 loss initiates chromosomal instability in fallopian tube epithelial cells. Dis Model Mech 2021; dmm.049001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vikas P, Borcherding N, Chennamadhavuni A, Garje R. Therapeutic potential of combining PARP inhibitor and immunotherapy in solid tumors. Front Oncol 2020;10:570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.