Abstract
Silica passivating agents have shown great success in minimizing nonspecific protein binding to glass surfaces for imaging and microscopy applications. Amine-derivatized surfaces are commonly used in conjugation with amide coupling agents to immobilize peptides/proteins through C-terminal or side-chain carboxylic acids. In the case of the single-molecule fluorosequencing of peptides, attachment occurs via the C-terminus and nonspecific surface binding has previously been a source of error in peptide identification. Here, we employ fluorosequencing as a high-throughput, single-molecule sensitivity assay to identify and quantify the extent of nonspecific binding of peptides to amine-derivatized surfaces. We show that there is little improvement when using common passivating agents in combination with the surface derivatizing agent 3-aminopropyl-triethoxysilane (APTES) to couple the peptides to the modified surface. Furthermore, many xanthene fluorophores have carboxylic acids in the appended phenyl ring at positions ortho and meta or ortho and para, and the literature shows that conjugation through the ortho position is not favored. Because xanthene-derived fluorophores are commonly used for single-molecule applications, we devised a novel assay to probe the conjugation of peptides via their fluorophores relative to their C-termini on silane-derivatized surfaces. We find significant attachment to the ortho position, which is a warning to those attempting to immobilize fluorophore-labeled peptides to silica surfaces via amide coupling agents. However, eliminating all amines on the surface by switching to 3-azidopropyl-triethoxysilane (AzTES) for coupling via copper-catalyzed azide–alkyne cycloaddition (CuAAC) and omitting additional passivation agents allowed us to achieve a high level of C-terminally bound peptides relative to nonspecifically or ortho-phenyl-bound, fluorophore-labeled peptides. This strategy substantially improves the specificity of peptide immobilization for single-molecule fluorosequencing experiments.
Graphical Abstract
INTRODUCTION
Amine-functionalized silanes (H2N-R-SiX3) are among the most widely used agents for modifying silicate surfaces (glass) in the life sciences, such as beads, biochips, and microscope slides,1–6 due to the ease of biomolecule attachment of the surface-exposed amines to carboxylic acids via the use of amide coupling agents. Various coupling reactions with the amine moiety allow for covalent attachment at specific sites to biomolecules and other chemicals, allowing for a variety of analytical protocols. However, the resulting surfaces are prone to nonspecific binding, and considerable research has gone into understanding how to passivate these surfaces to reduce undesirable binding and chemical reactions without reducing the efficiency of the desired amine-coupling reactions.7–9
Minimizing the nonspecific binding of biomolecules is particularly important for single-molecule imaging applications, which are powerful techniques for imaging intracellular targets10 and quantitative biological assays,11 and is essential to the high-throughput peptide sequencing technology known as fluorosequencing.12 This technology relies upon single-molecule imaging of peptides derivatized with fluorescent dyes on specific amino acids as they are subjected to cycles of Edman degradation to reveal the sequence positions of the labeled amino acids. However, nonspecific binding can occur between the surface and the hydrophobic fluorophores used as positional reporters of amino acids. Furthermore, the nonspecific attachment of free fluorophores can lead to increased nonpeptide background signals.9 Both forms of nonspecific binding led to increased errors and lowered the throughput and accuracy of fluorosequencing, just as analogous nonspecific binding events can similarly interfere with many other types of single-molecule assays.
Generally, nonspecific binding is the result of interactions between the biomolecule targets and/or their fluorophore labels with the imaging surface.13,14 This can be due to hydrophobic interactions of the fluorophore with the surface as well as hydrogen bonding, van der Waals forces, and charge attractions between the surface and the target. Often passivating agents are used to block the surface in an attempt to minimize the unwanted interactions.13,15,16
A common method of passivating silicate surfaces is to attach poly(ethylene glycol) (PEG) to the surface.7–9 Full coverage with PEG requires several rounds of time-consuming treatments and can block the attachment sites of the biomolecule of interest as well as limit diffusion to the biomolecule by reactants in the solution.15,17,18 One possible way to minimize these adverse effects is to mix an amino silane with the PEG silane, controlling the ratio of the passivation agent to the specific biological attachment site.19 In addition to PEG, other siloxane functional groups have been shown to provide improved passivation of silica surfaces against fluorescent molecules.20
Here, we evaluate surface preparation cocktails using the six different functionalized silanes in Figure 1. The shorter-chain alkanes were selected to prevent entanglement of the peptides with the passivating agents. Hydrophobic hexyl, n-propyl, and t-butyl were selected to compare with hydrophilic PEG and examine the effects of length and branching of the alkane on passivating the surface. We use single-molecule microscopy to determine the optimal surface preparation to minimize the off-target binding of both peptide and fluorophore entities. Importantly, the use of total internal reflection fluorescence (TIRF) microscopy provides the sensitivity required for the counting of low signal levels.
Figure 1.
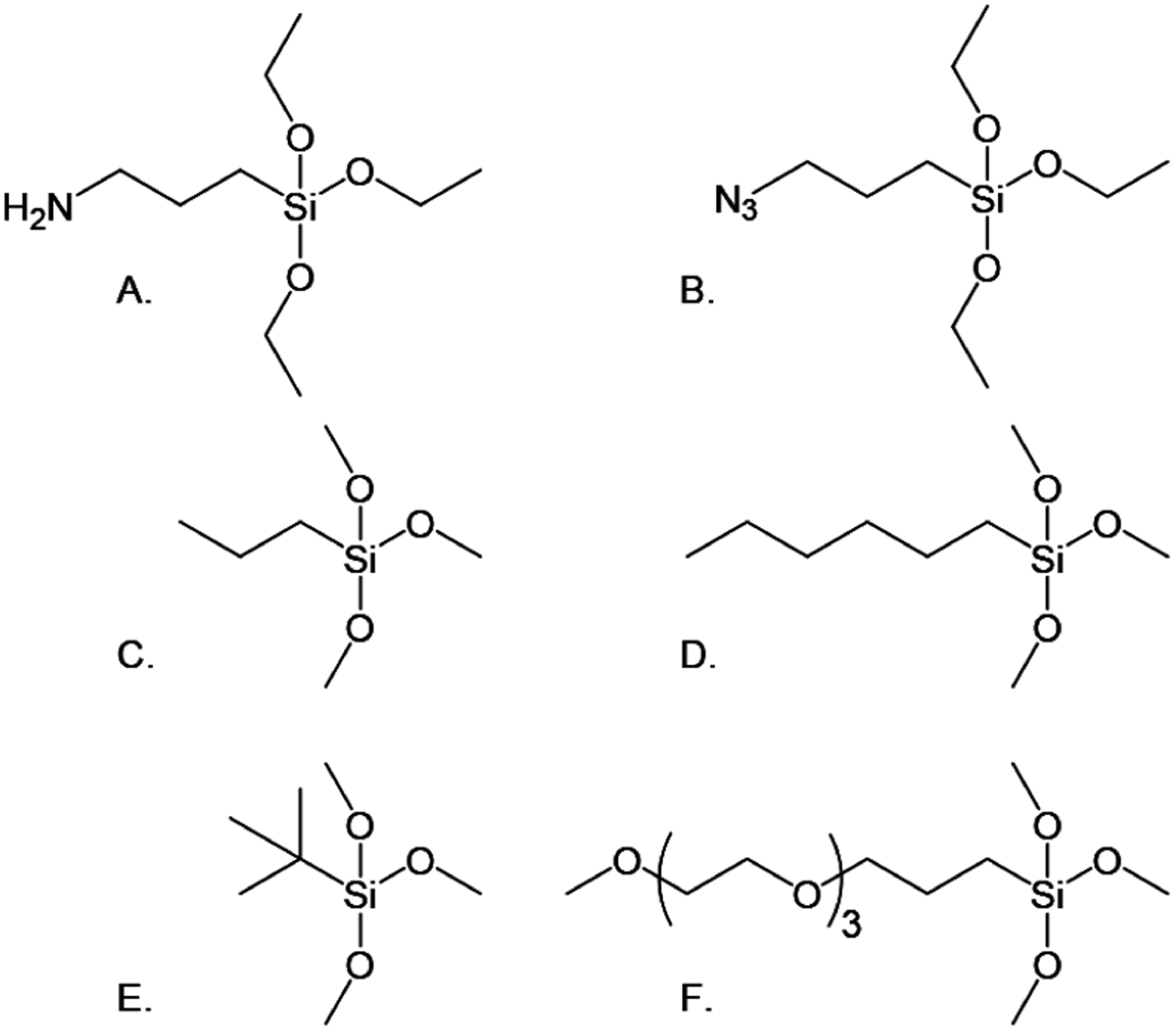
Silylating agents. (A) 3-Aminopropyl-triethoxysilane (APTES). (B) 3-Azidopropyltriethoxysilane, used to covalently attach peptides via their C-termini, and passivating agents (C–F). (C) n-Propyltrimethoxysilane (n-propyl). (D) Hexyltrimethoxysilane (hexyl). (E) t-Butyltri-methoxysilane (t-butyl). (F) Methoxytri-ethyleneoxypropyl-trimethoxysilane (PEG-3).
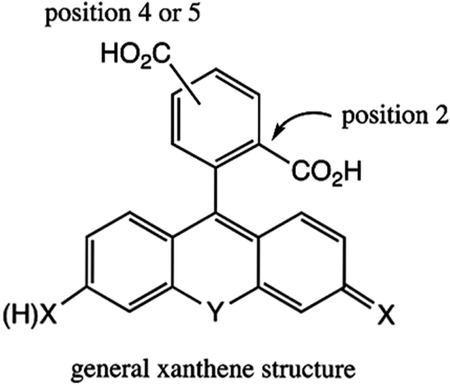
Additionally, many fluorophores used to label biomolecules, such as peptides and proteins, are xanthene dyes. These dyes commonly have two carboxylic acids in the appended phenyl ring, in the ortho position and either the meta or para position (either 2,4- or 2,5-constitutional isomers), and the literature shows several examples where conjugation to a biomolecule is to the less sterically hindered meta or para positions.21,22 However, this leaves the 2-position open for coupling to an amine-derivatized surface via amide coupling, and we set out to explore the extent to which this coupling occurs during surface attachment relative to a C-terminal carboxylic acid.
Because bright and stable fluorophores are required for most single-molecule applications, xanthene dyes are often used. This is particularly true for fluorosequencing because only a limited number of fluorophores survive the reagents and sequencing conditions. Two suitable fluorophores are Atto647N and Janelia Fluor 549 (JF549).12 Fluorophore JF549 is among the class of structures that contains a second ortho carboxylic acid that can potentially form a covalent bond with free amines on surfaces when aqueous coupling conditions are used. One source of error in previous work has been attributed to nonspecific binding of peptides, which may potentially occur due to coupling via this carboxylate moiety.12 The direct differentiation of different binding modes, e.g., whether mediated via the dye or the C-terminal carboxylate, of individual peptide molecules has not been possible in the past. However, the sensitivity of fluorosequencing allowed us to determine the extent of both coupling modalities, as described herein. We use this assay to compare the degree of off-target coupling to a glass surface observed for different dyes and silanes (amino- and azido-), and we observe a marked improvement by using azido couplings.
EXPERIMENTAL SECTION
Materials.
3-Aminopropyl-triethoxysilane (APTES, SIA0610.1), 3-azidopropyltriethoxysilane (AzTES, SIA0777.0), methoxy(trietheneoxy)propyltrichloro silane) (PEG-3, SIM6493.4), n-propyltrimethoxysilane (n-propyl, SIP6918.0, hexyltrimethoxysilane (hexyl, SIH6168.5), t-butyltrimethoxysilane (t-butyl, SIB1989.0), and methoxytriethyleneoxypropyltrimethoxysilane (PEG-3, SIM6493.4) were purchased from Gelest Inc. Fmoc-azidolysine was purchased from Novabiochem. Lipoic acid, N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC), N-hydroxysuccinimide (NHS), 6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox), methanol, triethylamine, pyridine, phenylisothiocyanate (PITC), acetonitrile, copper(II) sulfate, sodium ascorbate, 4-(dimethylamino)pyridine (DMAP), 4-ethynylbenzealdehyde, copper-(I) iodide, sodium ascorbate, tris(3-hydroxypropyltriazolylmethyl) amine (THPTA) trifluoroacetic acid (TFA), tris(2-carboxyethyl)-phosphine (TCEP), N,N-diisopropylethylamine (DIPEA), and all other Fmoc amino acids were purchased from Sigma-Aldrich. (2-(N-Morpholino) ethanesulfonic acid (MES) was purchased from Thermo Scientific. All other solid-phase peptide synthesis reagents were purchased from Chem-Impex. Janelia Fluor 549, SE was purchased from Tocris. Atto647N iodoacetamide and NHS-ester were purchased from Atto-Tec. Tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA) was synthesized using standard methods.23
Peptide Synthesis.
Three peptides were synthesized and labeled, (P1) Fmoc-GK*ASRG, (P2) Fmoc-GK+AGC*AGAY[Pra]R, and (P3) Fmoc- GKAGC*AGAY[Pra]R, along with (PS1) Fmoc-GC*AGK+AGAGAYG, where + is the JF549 label and * is the A647N label (Figure 2 and Figure S1A). Peptides were synthesized using standard solid-phase peptide synthesis methods24,25 using a Liberty Blue automated microwave peptide synthesizer (CEM Corp.). To simplify labeling, δ-azido lysine was used in place of standard lysine for peptide P1 (Figure 2). Fluorophore labeling of peptides P2 and P3 was accomplished using standard methods and commercial fluorophores.26
Figure 2.
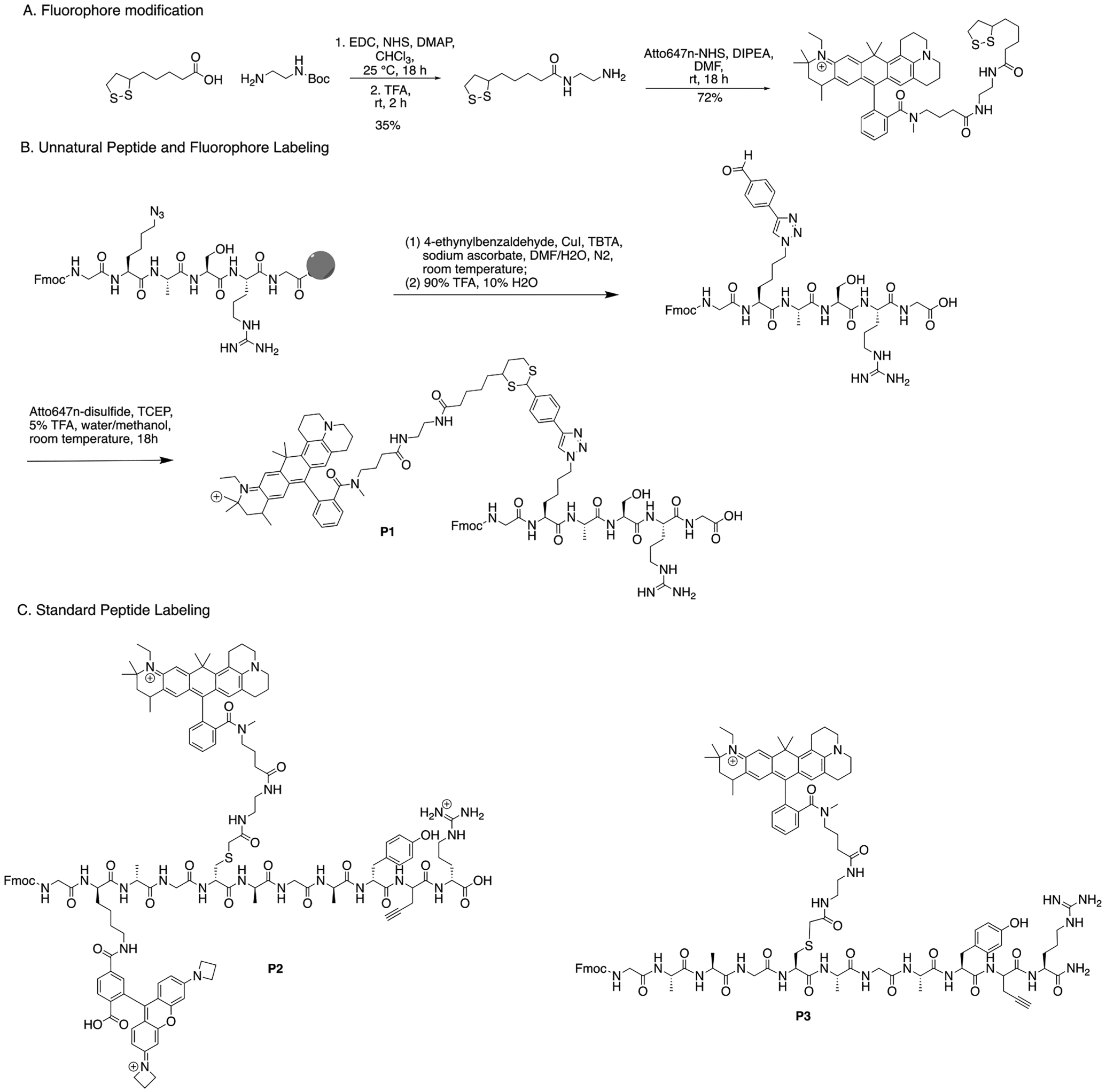
(A) Modified Atto647N fluorophore synthesis. (B) Unnatural peptide, P1, modification and labeling with Atto647N. (C) Standard peptides with conventional fluorophore labeling. Peptide P2 has JF549 at the second amino acid position and Atto647N at the fifth. Peptide P3 has Atto647N at the fifth position.
Dithiol-Functionalized Atto647N (A647N) Fluorophore.
The dithiol handle was synthesized by stirring 1 equiv of lipoic acid and 1.1 equiv of N-Boc-ethylenediamine with EDC, NHS (both 1.1 equiv), and DMAP (0.1 equiv). The crude product was isolated by washing with water (50 mL ×3) and brine (50 mL ×3) before treating with excess TFA to remove the Boc group. The final product was purified by flash chromatography using C18 with a 5–95% methanol gradient over 20 min. The lipoic-amine was then reacted (1 equiv) with Atto647N-NHS (1 equiv) in 100 μL of 62 mM DIPEA in DMF and nutated for 18 h. The final product was isolated by HPLC using the same method above.
Peptide Functionalization.
Solid-phase copper-catalyzed azide–alkyne cycloaddition (CuAAC) with TBTA as the ligand was used to functionalize the azide-containing peptide with 4-ethynylbenzyl-aldehyde. Established methods to synthesize and cleave the final aldehyde peptide from the resin were used.27 The aldehyde peptides (1.2 equiv) were then labeled with lipoic-acid-functionalized Atto647N (1 equiv) in 200 μL of 50/50 water/methanol with 5% TFA and TCEP (0.4 equiv) and nutated for 18 h. Labeled peptides were purified via HPLC and lyophilized to afford A647N-labeled peptides (P1).
Cysteines in peptides P2, P3, and PS1 and lysines in peptides P2 and PS1 were functionalized by mixing 200 μg of peptide with 1 equiv of DIPEA, 1.1 equiv of Atto47N-iodoacetamide for cysteines and/or 1.1 equiv of JF549, and SE for lysines in 100 μL of DMF for 16 h before HPLC purification.
Slide Preparation.
Surfaces were prepared with 40 mm of a round no. 1.5 (1.5 mm) glass cover glass (Bioptiechs), first cleaned with UV/ozone (Jelight Company) for 20 min on each side and functionalized through amino-silanization with APTES or AzTES and PEG-3, n-propyl, hexyl, or t-butyl silylating agents (Figure 1) using a modified version of the vendor-supplied protocol.19 The clean slides were then submerged in silylation solutions consisting of 0.0–0.43 mM APTES and/or 0.0–0.86 mM passivating agent in methanol for 30 min. The slides were washed with methanol and water and then dried with nitrogen. The slides were cured at 120 °C for 20 min under vacuum (−20 in Hg) and then allowed to cool to room temperature overnight under vacuum.
Total Internal Reflection Fluorescence (TIRF) Microscopy.
Single-molecule TIRF microscopy experiments were performed with a Nikon Ti-E inverted microscope equipped with a CFI Apo 60X/1.49NA oil-immersion objective lens and a 1.5× tube lens, a motorized stage (ProScan II, Prior Scientific), an iXon3 DU-897E 512×512 EMCCD detector (Andor) operated at −70 °C, and a MLC400B (Keysight) laser combiner with 561 and 647 nm lasers. Atto647N (A647) was excited using the 647 nm laser at 6.0 mW (surface passivation) and 2.8 mW (fluorosequencing) via the 647LP dichroic and collected through 665LP and 705/72BP emission filters and a Janelia Fluor 549 (JF549) 2.7 mW 561 nm laser power via the 561LP dichroic and collected through 575LP and 600/50BP emission filters. Each image represents a 92 μm × 92 μm square region of the sample.
Surface Passivation Screening.
Passivation of the cover glass surface with aminosilanes was measured by attaching fluorophore-labeled peptides to the surface on two halves of the same slide surface. The peptide concentration was chosen to provide a density that ensured that each diffraction-limited spot in the image contained only one peptide. Half of the slide surface was incubated in a solution of 800 pM synthetic peptide and 1.88 mM EDC in 0.1 M MES buffer (EDC side). The other half was incubated in a solution of 800 pM peptide in 0.1 M MES buffer (MES side). The peptide solutions were incubated on the slides at room temperature for 60 min before washing with methanol and water and drying with nitrogen. The prepared cover glass was then placed in a flow cell (FCS2, Bioptechs) with the sample facing toward the interior of the flow chamber. The flow cell was placed on a TIRF microscope as shown in Figure 3D and washed with 1 mM Trolox in methanol (imaging solution), and Atto647N was imaged as described above by collecting a 3 × 3 array of images. The surfaces were then washed and incubated in 100% TFA for 300 s and washed with the imaging solution, and then images were acquired in the same 3 × 3 grid locations. This process was repeated three times. The images were analyzed using Image-J software by setting the threshold mask to between 0.1 and 0.2% of the maximum signal to distinguished individual peptide signals. Peaks were counted for each image using Image-J’s Analyze Particles module, filtering for peak sizes greater than 4 pixels.2 The average count for the nine images acquired after each wash was recorded.
Figure 3.
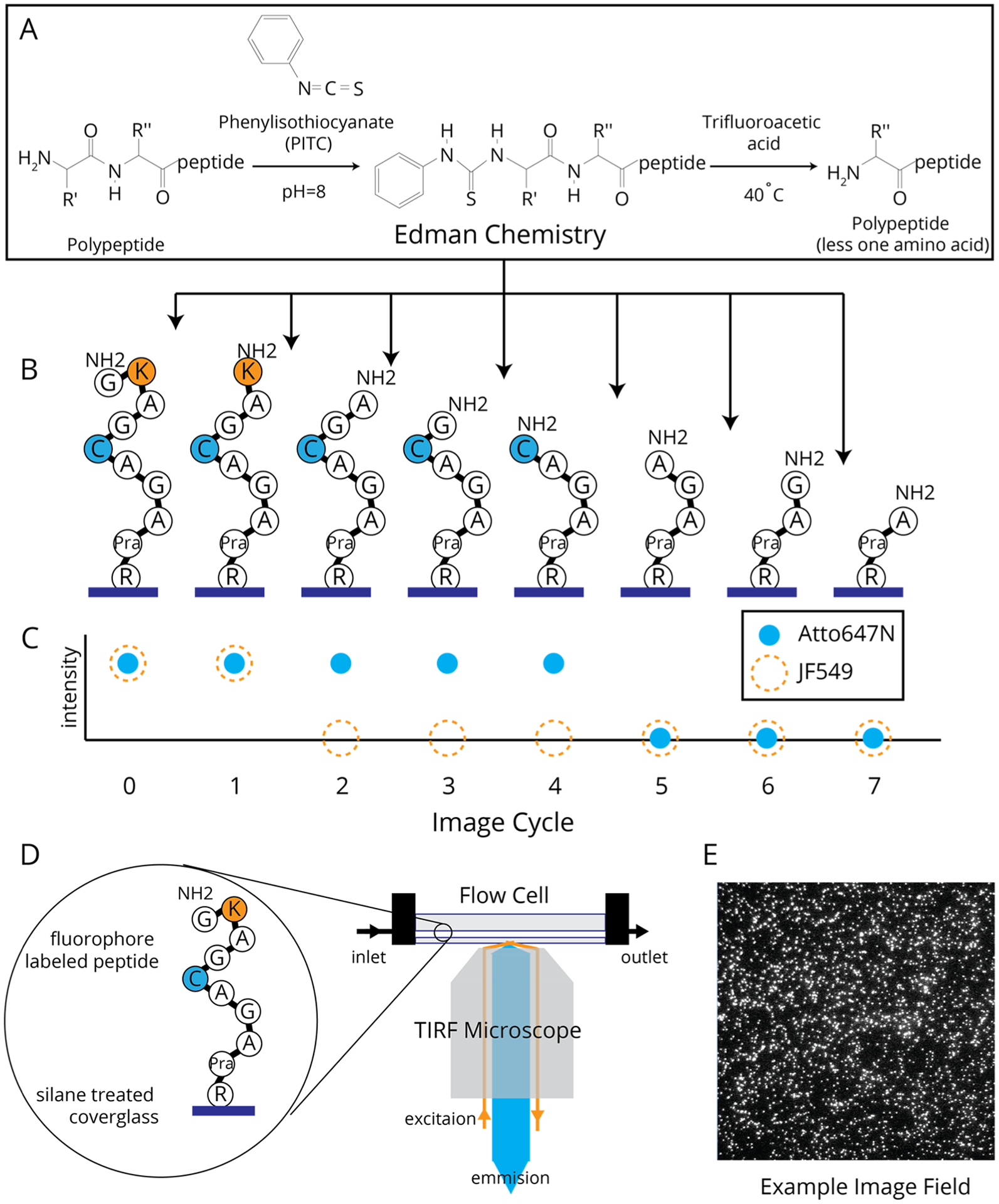
(A) Edman degradation pathway. (B) Expected sequence changes of a single P2 peptide attached through the C-terminus after cycles of Edman degradation. (C) Relative fluorescence changes for Atto647N and JF549 on peptide P2 where signal loss is correlated to the liberation of a labeled amino acid after the corresponding cycles of Edman degradation. (D) Diagram of the imaging setup for both passivation screening and single-molecule fluorosequecning experiments.
Single-Molecule Fluorosequencing.
Single-molecule fluorosequencing has been described in detail in Swaminathan et al.12 and is outlined in Figure 3. Briefly, for the experiments presented, synthetic peptides were labeled with JF549 and/or A647N, with each reporter labeling all occurrences of a specific amino acid type (e.g., cysteine or lysine; Figure 2). After being labeled, the peptides were purified and attached via their C-termini to a functionalized cover glass, as described above for APTES. For AzTES, the cover glasses were incubated with a mixture of 0.02 mM copper sulfate, 0.01 mM THPTA, 0.5 mM sodium ascorbate, and 20 pM alkyne-modified peptide in HEPES buffer for 30 min.
The prepared cover glass was then clamped in a flow cell with the sample facing toward the interior of the flow chamber. The flow cell was mounted on a TIRF microscope as shown in Figure 3D and heated to 40 °C. We first removed the N-terminal Fmoc by incubating the flow chamber with 20% piperidine in DMF for 30 min. The samples were then subjected to 10 cycles of fluorosequencing (Figure 3A–C). Each cycle consists of (1) imaging to determine the sequence state of the peptide and (2) Edman degradation to remove the current N-terminal amino acid. Each imaging step consisted of acquiring an array of 200 fields of view separated by 150 μm. Individual images were collected for both the A647N and JF549 emissions for each field before moving to the next field. For the Edman degradation step, the flow cell was exposed to (1) a mixture of acetonitrile, pyridine, triethylamine, and water (10:3:2:1 v/v) for 5 min, (2) a mixture of acetonitrile and phenylisothiocyanate (PITC) (9:1 v/v) for 20 min, (3) 100% trifluoroacetic acid for 15 min, and (4) 1 mM Trolox in methanol before performing the next round of imaging. For the first two cycles, the PITC solution was replaced with acetonitrile to obtain a background measure of signal loss not associated with amino acid liberation though Edman degradation.
After sequencing, the images from each field of view were aligned across Edman cycles and the intensity values (“tracks”) associated with each peptide were extracted with using the SigProc software tool, available as part of the Plaster package at https://github.com/erisyon/plaster. Each peptide’s intensity track was then analyzed using the sequencing-fitter software of Swaminathan et al.12 to compute a frequency histogram showing the counts of peptide molecules exhibiting signal loss following each Edman cycle. The background loss due to chemical and photodestruction was determined by fitting all cycles, except those where sequencing is expected, to an exponential decay (Scipy, Python). This background was removed by simple subtraction of the fitted values from the measured counts.
RESULTS
Testing the Impact of Surface Passivation.
To assess nonspecific binding and the effectiveness of passivating silanes, peptide P1 was designed using Atto647N, a nonxanthene dye, in order to test if it would exhibit specific binding through the C-terminus or nonspecifically through the fluorophore. An initial screening of the four passivation agents PEG-3, hexyl, n-propyl, and t-butyl, prepared in 1:2 (mol/mol) APTES/passivating agent, was performed to determine the relative passivation due to each agent. Figure 4 shows the count of fluorescently labeled P1 peptide molecules measured for preparations with (black) and without (gray) the EDC coupling agent. The downward trend after each TFA wash from the initial count (“Pre”) is seen for all slide preparations and indicates a high degree of peptides bound nonspecifically to the surface on both halves of the slide, i.e., with and without the EDC coupling reagent. As expected for the buffer-only samples, the peak count returned to background levels after washing, indicating that nearly all noncovalently bound peptide was removed. For the samples with EDC coupling, the peptides bound to the surface remained after the TFA washes due to proper immobilization, thus showing significantly higher counts compared to the buffer-only preparations. It is significant to note that with EDC present there are far more nonspecifically bound peptides in all cases relative to the buffer-only samples (Table S1). Thus, EDC treatment itself leads to an increase in the initial nonspecific binding.
Figure 4.
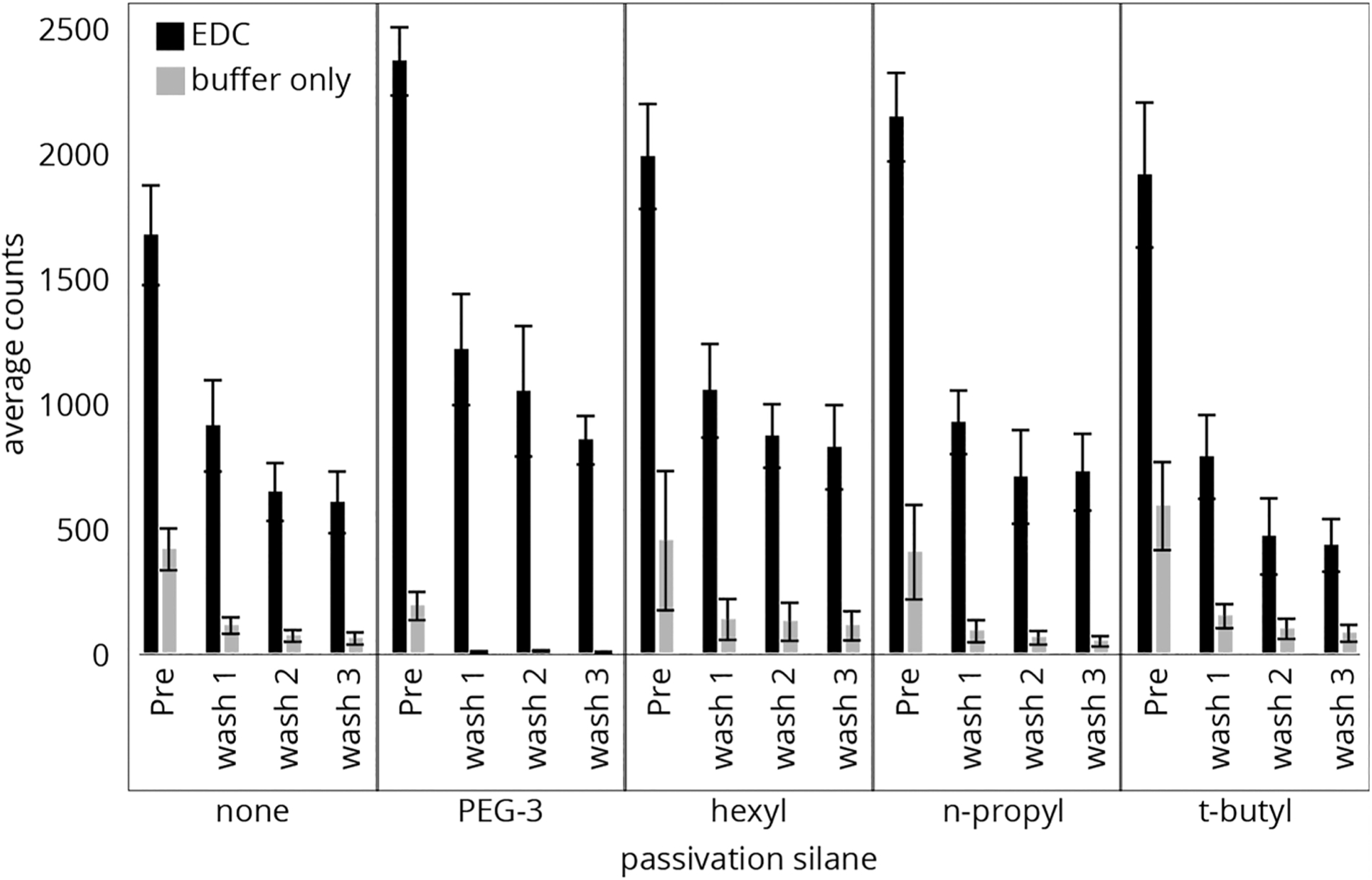
Counts of individual P1 peptide molecules from slide surfaces made with APTES (0.43 mM) and PEG-3, hexyl, n-propyl, and t-butyl (2 equiv). Atto647N-labeled peptides were bound to the surfaces before imaging and washing with TFA three times, with imaging after each cycle.
The surface preparation with the lowest proportions of nonspecifically bound peptide, as indicated by the final counts for the buffer-only preparations, were the surfaces passivated with PEG-3. In comparison, n-propyl had a similar performance to APTES alone. The t-butyl had low buffer-only counts; however, it also showed the lowest proportion of properly bound peptide on the EDC treated portion of the slides. It is notable that PEG-3 also retained peptides at a higher average in comparison to the APTES-only preparation, indicating some overall improvement in properly bound to nonspecifically bound peptides. These results suggest that PEG-3 performs the best of the four surface treatments at preventing nonspecifically bound peptide from adhering to the surface while still allowing labeled peptide to covalently bond to the surface amines. For this reason, PEG-3 was the only passivation agent chosen for further studies.
Next, the optimal concentration of PEG-3 was determined. Here, slides were prepared with a 0, 0.75, 1.5, 2, and 2.5 molar ratios of PEG-3/APTES, and then peptides were attached and analyzed in the same manner as above. Figure 5 shows the count of labeled peptides on the surface for preparations with (black) and without (gray) the EDC coupling agent. As expected, increasing the passivation agent decreased both the nonspecifically bound and the specifically coupled peptides, but with little improvement above a molar ratio of 1.5, as indicated by the counts remaining after wash 3 for samples with and without EDC. A ratio of 2:1 PEG-3/APTES was chosen for the subsequent passivation studies using fluorosequencing.
Figure 5.
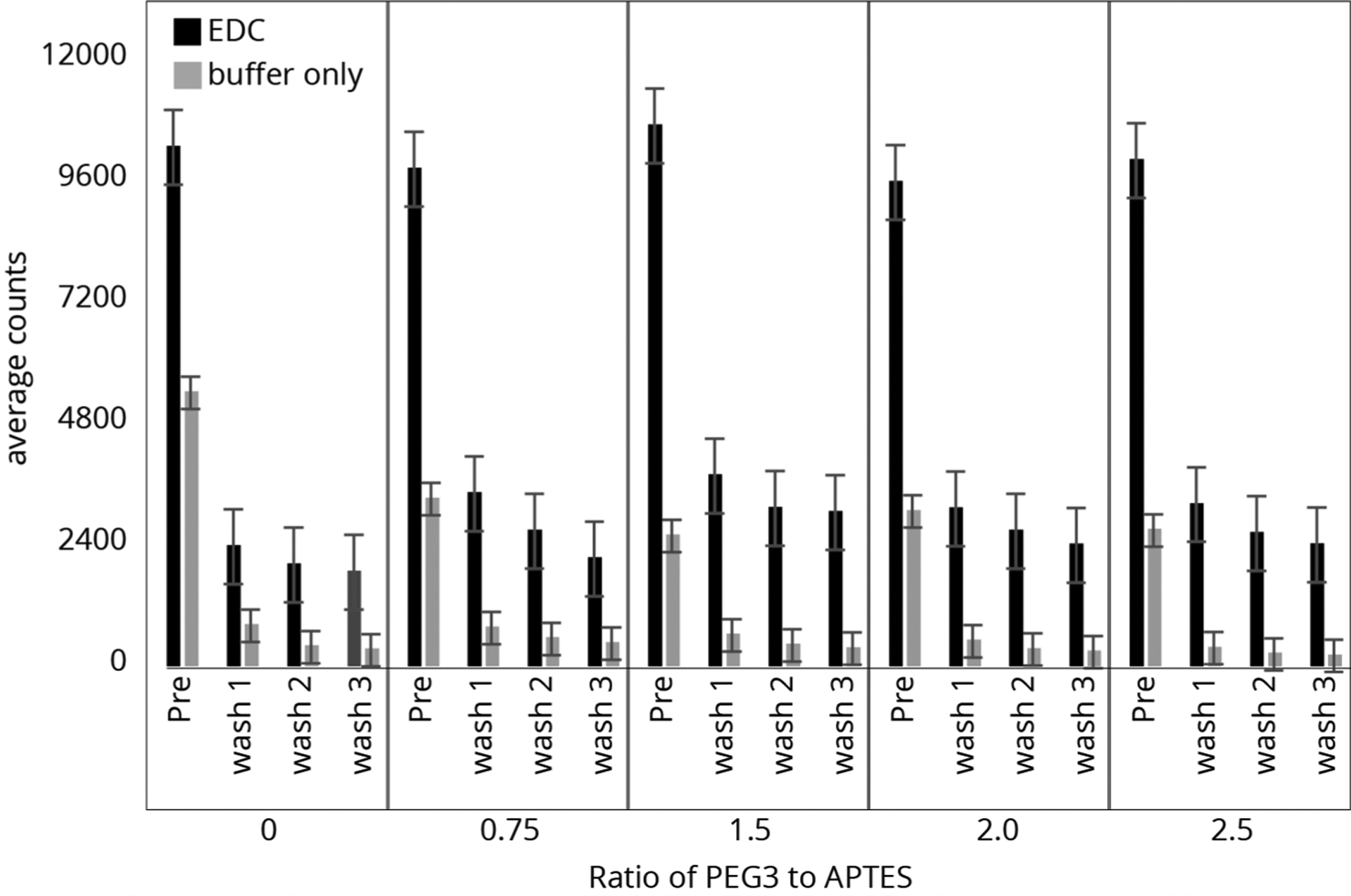
Counts of individual P1 peptide molecules from slide surfaces of APTES (0.43 mM) and PEG-3 (0–2.5 equiv). Atto647N-labeled peptides were bound to the surfaces before imaging and were washed with TFA three times, imaging after each cycle.
Finally, to determine the reproducibility of the surfaces from the same batch, three replicate slides for each of four types of preparations were prepared: a slide with no silane (0P, 0A), no PEG-3 with 1 equiv of APTES (0P, 1A), 2 equiv of PEG-3 and no APTES (2P, 0A), and 2 equiv of PEG-3 and 1 equiv of APTES (2P, 1A). As above, peptide P1 was incubated on the surface, imaged, and analyzed. The counts before (black) and after three TFA washes (gray) were recorded and are shown in Figure 6. From this study, it appears that there is no statistical difference in the performance of the surfaces with or without PEG-3 passivation. In fact, the slides with APTES (1A) (where the peptide should bind to the surface under the EDC coupling conditions) and without PEG-3 (0P, 1A) retained a slightly higher fluorophore count. This could be due to either competition between the PEG-3 silane and APTES to bind to the surface or difficulty in attaching the peptide to the APTES on the surface when PEG is present.
Figure 6.
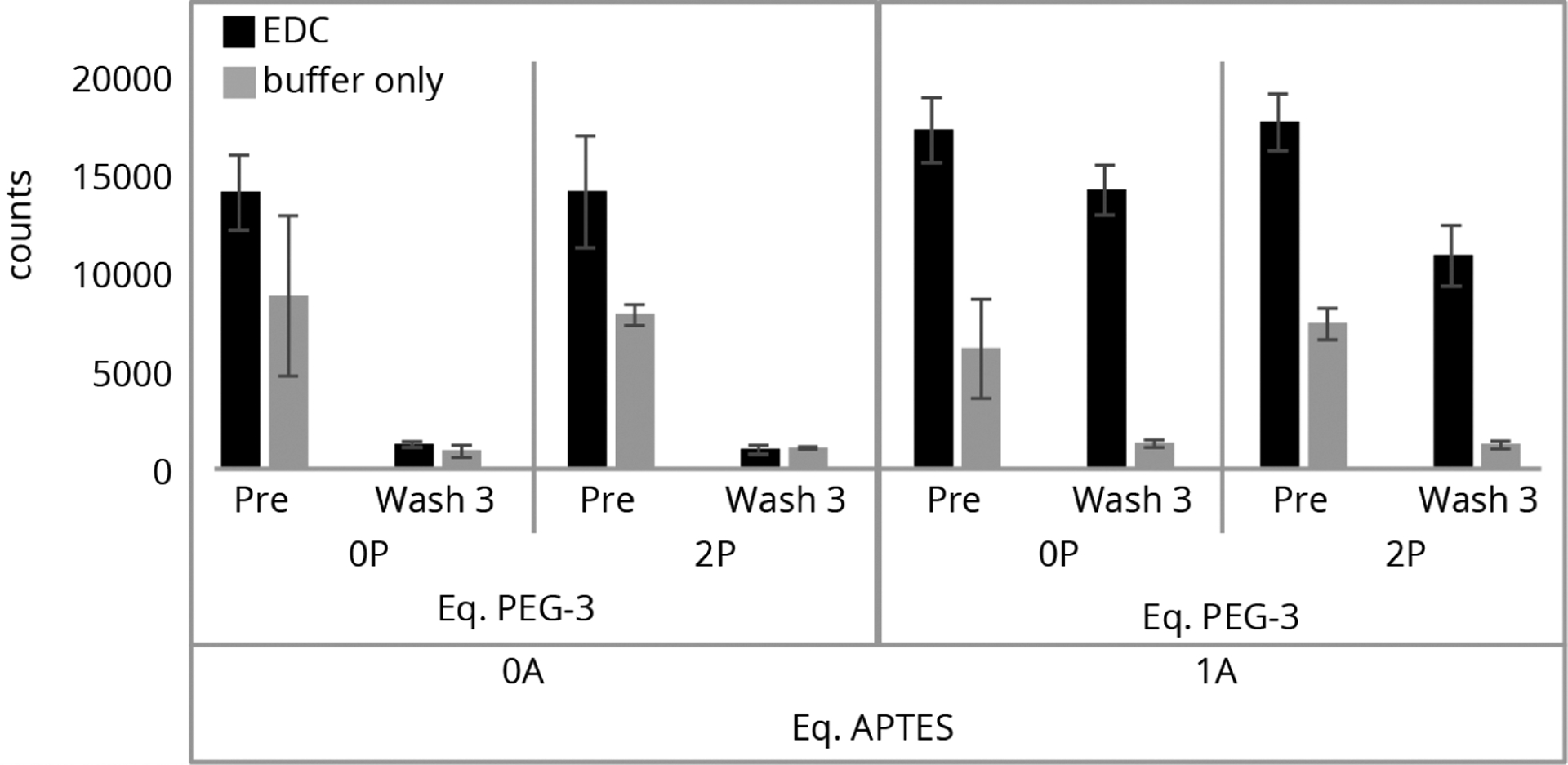
0P and 2P ratio of passivation with PEG-3 relative to the 1A level of APTES. 0A surfaces were made without APTES. Each type was analyzed in triplicate, labeled, and imaged on the same day.
Here again, the unexpected influence of EDC is interesting. The surfaces without APTES but with the coupling reagent show higher nonspecific binding, calculated by the difference between the pre- and postwash counts, as compared to those without EDC (Table S2). This result is seen both with and without PEG-3 passivation and suggests that the coupling conditions play a significant role in controlling the initial nonspecific binding.
The role of EDC in increasing the nonspecific binding is not clear, but we speculate that it can react with the APTES amines on the slide surface to generate guanidine species.28 Because of the structure of EDC, this would introduce short alkyl chains and a secondary ammonium ion which may interact with peptides and/or Atto647N.
Single-Molecule Fluorosequencing Assay Determines Modes of Peptide Coupling.
With surface passivation using coupling agents showing minimal success in lowering nonspecific binding, our next challenge was to determine if passivation would protect against the off-target binding of peptides in fluorosequencing experiments with xanthene-type fluorophores such as JF549, which possess ortho-phenyl carboxylic acids. We also wanted to explore whether APTES may be contributing to this off-target coupling.
To study these effects, we took advantage of the single-molecule sensitivity of fluorosequencing12 to distinguish specific from nonspecific binding on a molecule-by-molecule basis. This method provides direct information on which amino acid positions are participating in surface attachment and to what degree.
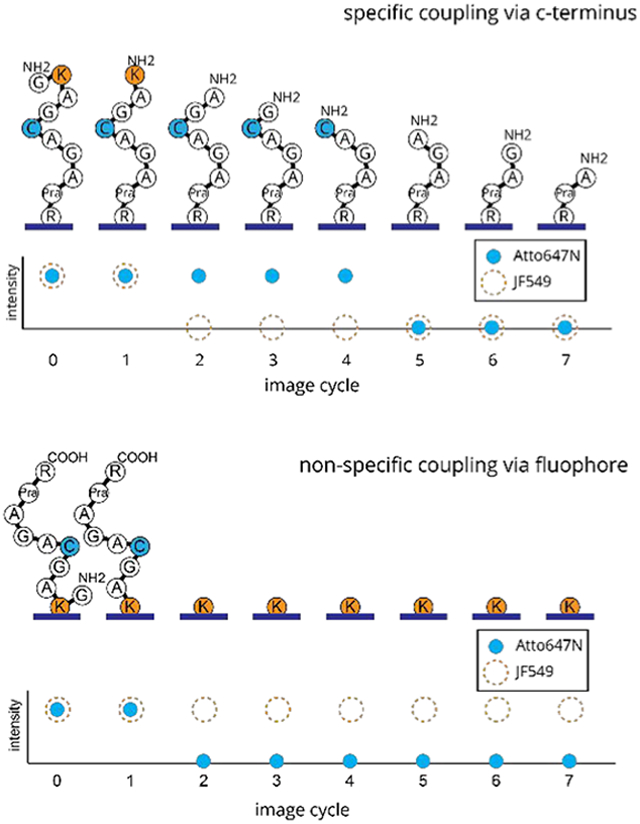
We designed peptide P2 with two different fluorophores selected to indicate alternate modes of surface attachment. The label attached at the second amino acid (lysine), xanthene dye JF549, was used to test for binding via the 2-position carboxylic acid on the appended phenyl ring, and the label attached at the fifth amino acid (cysteine), A647N, was incorporated as a reporter of the peptide’s orientation relative to the surface in the fluorescence-based assay. Furthermore, P2 was designed such that it could be attached either by its C-terminus or via a CuAAC to an azide-derivatized surface though an alkyne on the penultimate amino acid.
In this assay, for P2 peptides that couple to the silane surface via their C-termini (Figure 3B), the fluorosequence for the A647N fluorescence is expected to exhibit signal loss after the fifth Edman cycle (Figure 3C). However, for peptides that couple to the surface though the JF549 (Figure 7A), the fluorosequence will shift, showing the loss of the A647N signal after the second Edman cycle (Figure 7B). This shift occurs because once the second amino acid is removed though Edman degradation, JF549 remains attached to the surface, and the peptide with the A647N reporter is instead liberated. From this assay, we consider the ratio of the number of peptides showing fifth-cycle drops to those showing second-cycle drops (c5/c2) as a metric for surface passivation against nonspecific binding and/or position 2-carboxylate binding. Note that peptides with off-target coupling though the reporter dye, i.e., Atto647N, would show no Edman-related signal drop and thus would be ignored or contribute to the photobleaching background.
Figure 7.
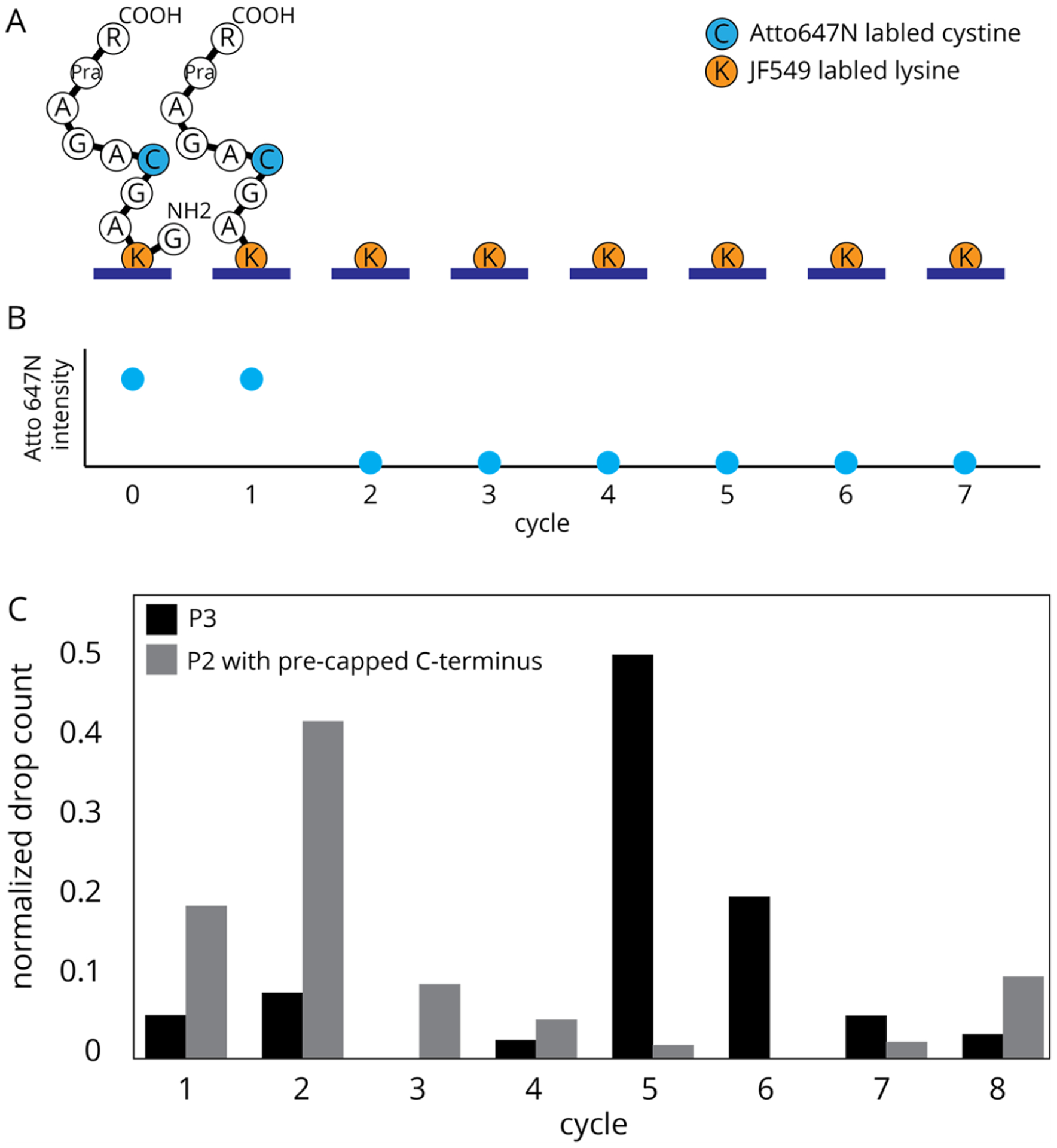
(A) Expected sequence changes of a single P2 peptide misattached though the JF549 dye after rounds of Edman degradation. (B) Relative fluorescence changes expected for Atto647N on peptide P2 misattached via JF549, where signal loss is correlated to the liberation of the remaining peptide after Edman degradation at the labeled lysine. (C) Fluorosequencing of control peptide P3, with no JF549 label, results in mostly C-terminal attachment and a peak drop count after the fifth Edman cycle (n = 2304). Fluorosequencing results for P2 in which the C-terminus was capped prior to attachment to the surface, thus resulting in mostly attachment though the JF549, shows a prominent peak drop count after the second Edman cycle (n = 1380).
To explore this sensitivity of the assay, several peptides were fluorosequenced. First, as a positive control, a purified sample of peptide P3 (labeled only on position 5) was prepared, immobilized with EDC, and sequenced. Having no JF549, these peptides attach to the surface primarily though their C-termini and show a peak signal drop after the fifth Edman cycle, with a c5/c2 of 42 (Figure 7C, gray bars). Next, as a negative control, P2 was allowed to react with 10:1 molar mono-t-boc ethylenediamine in 0.2 mM sodium bicarbonate for 1 h, thus blocking the C-terminus, and possibly the 2-position carboxylate (if reactive during this coupling). Thus, the surface attachment should occur only though off-target binding, i.e., not the C-terminus. When sequenced, the peak drop for the A647N reporter occurs after the second Edman cycle, with a c5/c2 of 0.04 (Figure 7C, black bars). These results show that the c5/c2 ratio provides a sensitive method for measuring if the peptides are predominantly immobilized by their C-termini or the JF549 dyes.
We then used this assay to compare the passivation of APTES surfaces with and without PEG-3. A sample of peptide P2 with a free C-terminus was attached using EDC coupling. Sequencing was performed in triplicate for these two surface treatments. Both samples (Figure 8A) showed signal drops after the second and fifth Edman cycles, indicating that some peptides attached via the C-terminus while others were attached though the JF549. The APTES-only slides resulted in a c5/c2 value of 1.3 ± 0.2, and the APTES with PEG-3 slides resulted in a c5/c2 value of 1.4 ± 0.3. These results show no signification improvement in off-target binding by adding the PEG-3 to an APTES silane preparation.
Figure 8.
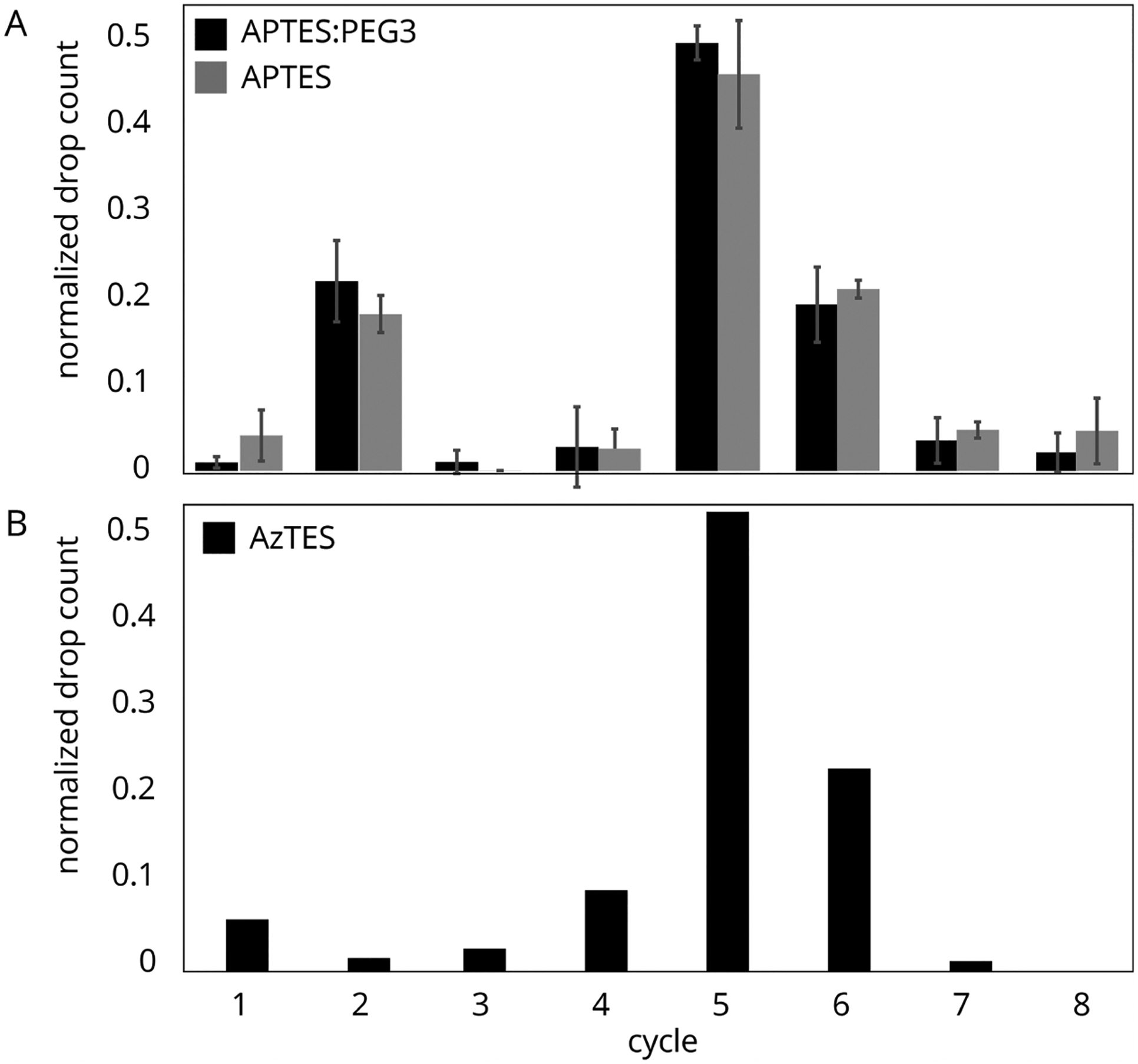
(A) Fluorosequencing of peptide P2, attached using EDC coupling to APTES/PEG3 (black) and APTES-derivatized cover glass (gray). Results consist of five technical replicates (n = 1070, 245, 1786, 4826, and 1878 and n = 4307, 5364, 4791, 8343, and 4812 molecules imaged, respectively). The drop peaks after both the second and fifth Edman cycles indicate both modes of attachment with a significant portion coupling though the JF549. (B) Fluorosequencing of peptide P2, attached using CuAAC coupling to AzTES-derivatized cover glass (n = 4643). Here, the large drop peak after the fifth Edman cycle indicates that primary coupling is through the C-terminal propargylglycine.
To complete this study, a peptide with the dye positions reversed (PS1) was prepared and sequenced (Figure S1). Here, the JF549 acts as the reporter molecule for the off-target coupling of Atto647N. The results indicate some nonspecific binding of Atto647N (c5/c2 = 0.35), consistent with the passivation study. However, the generally poor quality of these sequencing experiments suggests that the attachment is partially via the 2-carboxylate of the JF549, making this peptide a poor reporter for this assay.
Markedly Reduced Nonspecific Binding When Coupling via CuAAC.
The screening experiments indicated that passivation did not significantly decrease nonspecific binding to amines on the slides or block attachment of the xanthene-fluorophores via their 2-carboxylic acid. Thus, as an alternative solution, we considered a silane for which the peptides could be attached by a different coupling. We treated the cover glass with AzTES, a silane presenting azide instead of amine functionality, prepared at the same concentration as the APTES-only surfaces. Peptide P2 was attached to the slides via a propargylglycine using CuAAC. These samples were sequenced as above, with the results shown in Figure 8B. In contrast to APTES, we observed a strong preference for the (correct) fifth Edman cycle and a c5/c2 ratio of 32.4, indicating a dominant mode of attachment via the propargylglycine and little to no nonspecific binding of the JF549 or attachment via the ortho-carboxyphenyl ring.
CONCLUSIONS
Preparing surfaces for specific attachment while minimizing nonspecific binding is an ongoing challenge for single-molecule or other microscopy-based assays. Our results show that simply relying on secondary passivation is not sufficient for a significant reduction in nonspecific binding in single-molecule imaging using fluorophores.
We have shown that APTES and EDC used to form the amide bond between fluorophore-labeled peptides and the surface result in increased levels of nonspecifically bound peptide. Although PEG-3 showed slightly improved results when compared to the other passivating agents in screening experiments, we saw no significant improvement to the nonspecific surface binding when we attempted to couple peptides directly to the surface using EDC.
Using a single-molecule sensitivity assay based on fluorosequencing that can determine peptide binding orientations on a molecule-by-molecule basis, we measured the extent of nonspecific and 2-phenyl coupling of xanthene (JF549) fluorophore-derivatized peptides. We observed a large proportion of covalent attachment via the fluorophore’s 2-phenyl group. In principle, this method could be extended to read out interactions involving other synthetic labels or naturally occurring side chains in order to better understand the interaction of biological molecules with surface materials.
Taken together, our results support transitioning away from amine functionalization (and avoiding EDC) and instead coupling peptides to azide surfaces, such as through the use of CuAAC to covalently bond the peptides via an alkyne, which resulted in far fewer nonspecifically bound peptides on the surfaces. The utility of CuAAC has been widely demonstrated as a useful tool for efficient covalent attachment, with many reactions reaching completion in minutes.29,30 In applications where Cu(II) may interfere with the underlying biology, the copper catalyst can be omitted by utilizing strained alkynes for surface attachment.31 We expect this strategy to improve single-molecule fluorescence assays in which the peptide orientation and mode of coupling are important, most notably, in single-molecule protein-sequencing experiments.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Welch Regents Chair (F-0046) to E.V.A. and support from Erisyon, Inc., to E.M.M. and E.V.A. E.M.M. acknowledges additional support from the Welch Foundation (F-1515), NIH (R35 GM122480, R01 HD085901, and R01 DK110520), and an NSF STTR grant (1938726) with Erisyon Inc. More data can be found at 10.5281/zenodo.5514438.
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.langmuir.1c02644
The authors declare the following competing financial interest(s): A.M.B., J.S., E.M.M., and E.V.A. are co-founders and shareholders of Erisyon, Inc., and E.M.M. and E.V.A. serve on the scientific advisory board. C.M.H., A.M.B., J.S, E.M.M, and E.V.A. are co-inventors on pending and granted patents relevant to this work.
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.langmuir.1c02644.
Differences between the exposure of various surfaces to EDC and buffer; sequencing data and all peptide MS data (PDF)
Contributor Information
Caroline M. Hinson, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States.
Angela M. Bardo, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, United States
Cassie E. Shannon, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States.
Sebastian Rivera, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States.
Jagannath Swaminathan, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, United States.
Edward M. Marcotte, Department of Molecular Biosciences, The University of Texas at Austin, Austin, Texas 78712, United States.
Eric V. Anslyn, Department of Chemistry, The University of Texas at Austin, Austin, Texas 78712, United States.
REFERENCES
- (1).Taylor AP; Webb RI; Barry JC; et al. Adhesion of microbes using 3-aminopropyl triethoxy silane and specimen stabilisation techniques for analytical transmission electron microscopy. J. Microsc 2000, 199 (1), 56–67. [DOI] [PubMed] [Google Scholar]
- (2).Trévisiol E; Berre-Anton VL; Leclaire J; et al. Dendrislides, dendrichips: a simple chemical functionalization of glass slides with phosphorus dendrimers as an effective means for the preparation of biochips. New J. Chem 2003, 27 (12), 1713–1719. [Google Scholar]
- (3).Kratz SRA; Bachmann B; Spitz S; et al. A compression transmission device for the evaluation of bonding strength of biocompatible microfluidic and biochip materials and systems. Sci. Rep 2020, 10 (1), 1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Ma M; Zhang Y; Yu W; et al. Preparation and characterization of magnetite nanoparticles coated by amino silane. Colloids Surf., A 2003, 212 (2), 219–226. [Google Scholar]
- (5).Jedlicka SS; Rickus JL; Zemlyanov DY Surface Analysis by X-ray Photoelectron Spectroscopy of Sol–Gel Silica Modified with Covalently Bound Peptides. J. Phys. Chem. B 2007, 111 (40), 11850–11857. [DOI] [PubMed] [Google Scholar]
- (6).Kirkness MWH; Korosec CS; Forde NR Modified Pluronic F127 Surface for Bioconjugation and Blocking Nonspecific Adsorption of Microspheres and Biomacromolecules. Langmuir 2018, 34 (45), 13550–13557. [DOI] [PubMed] [Google Scholar]
- (7).Stutz H Protein attachment onto silica surfaces – a survey of molecular fundamentals, resulting effects and novel preventive strategies in CE. Electrophoresis 2009, 30 (12), 2032–2061. [DOI] [PubMed] [Google Scholar]
- (8).Mikhail AS; Ranger JJ; Liu L; et al. Rapid and Efficient Assembly of Functional Silicone Surfaces Protected by PEG: Cell Adhesion to Peptide-Modified PDMS. J. Biomater. Sci., Polym. Ed 2010, 21 (6–7), 821–842. [DOI] [PubMed] [Google Scholar]
- (9).Maurice V; Slostowski C; Herlin-Boime N; Carrot G Polymer-Grafted Silicon Nanoparticles Obtained Either via Peptide Bonding or Click Chemistry. Macromol. Chem. Phys 2012, 213 (23), 2498–2503. [Google Scholar]
- (10).Fish KN Total Internal Reflection Fluorescence (TIRF) Microscopy. Curr. Protoc. Cytom 2009, 50 (1), 12.18.1–12.18.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Salehi-Reyhani A Evaluating single molecule detection methods for microarrays with high dynamic range for quantitative single cell analysis. Sci. Rep 2017, 7 (1), 17957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Swaminathan J; Boulgakov AA; Hernandez ET; et al. Highly parallel single-molecule identification of proteins in zeptomole-scale mixtures. Nat. Biotechnol 2018, 36 (11), 1076–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ren C; Schlapak R; Hager R; Szleifer I; Howorka S Molecular and Thermodynamic Factors Explain the Passivation Properties of Poly(ethylene glycol)-Coated Substrate Surfaces against Fluorophore-Labeled DNA Oligonucleotides. Langmuir 2015, 31 (42), 11491–11501. [DOI] [PubMed] [Google Scholar]
- (14).Zanetti-Domingues LC; Tynan CJ; Rolfe DJ; Clarke DT; Martin-Fernandez M Hydrophobic Fluorescent Probes Introduce Artifacts into Single Molecule Tracking Experiments Due to Non-Specific Binding. PLoS One 2013, 8 (9), e74200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Heyes CD; Groll J; Möller M; Nienhaus GU Synthesis, patterning and applications of star-shaped poly(ethylene glycol) biofunctionalized surfaces. Mol. BioSyst 2007, 3 (6), 419–430. [DOI] [PubMed] [Google Scholar]
- (16).Feldman K; Hähner G; Spencer ND; Harder P; Grunze M Probing Resistance to Protein Adsorption of Oligo(ethylene glycol)-Terminated Self-Assembled Monolayers by Scanning Force Microscopy. J. Am. Chem. Soc 1999, 121 (43), 10134–10141. [Google Scholar]
- (17).Ha T; Rasnik I; Cheng W; et al. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 2002, 419 (6907), 638–641. [DOI] [PubMed] [Google Scholar]
- (18).Lamichhane R; Solem A; Black W; Rueda D Single Molecule FRET of Protein-Nucleic Acid and Protein-Protein complexes: Surface Passivation and Immobilization. Methods 2010, 52 (2), 192–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Silane, Silicone & Metal-Organic Materials Innovation|Gelest; https://www.gelest.com/ (accessed Feb 24, 2021). [Google Scholar]
- (20).Crowe LL; Tolbert LM Silica Passivation Efficiency Monitored By a Surface-Bound Fluorescent Dye. Langmuir 2008, 24 (16), 8541–8546. [DOI] [PubMed] [Google Scholar]
- (21).Grimm JB; English BP; Chen J; et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 2015, 12 (3), 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Grimm JB; Klein T; Kopek BG; et al. Synthesis of a Far-Red Photoactivatable Silicon-Containing Rhodamine for Super-Resolution Microscopy. Angew. Chem., Int. Ed 2016, 55 (5), 1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lee B-Y; Park SR; Jeon HB; Kim KS A new solvent system for efficient synthesis of 1,2,3-triazoles. Tetrahedron Lett 2006, 47 (29), 5105–5109. [Google Scholar]
- (24).CEM Corporation https://cem.com/en/ (accessed Mar 5, 2021).
- (25).Vanier GS Microwave-Assisted Solid-Phase Peptide Synthesis Based on the Fmoc Protecting Group Strategy (CEM). In Peptide Synthesis and Applications; Jensen KJ, Tofteng Shelton P, Pedersen SL, Eds.; Methods in Molecular Biology; Humana Press, Totowa, NJ, 2013; pp 235–249. [DOI] [PubMed] [Google Scholar]
- (26).Hernandez ET; Swaminathan J; Marcotte EM; Anslyn EV Solution-phase and solid-phase sequential, selective modification of side chains in KDYWEC and KDYWE as models for usage in single-molecule protein sequencing. New J. Chem 2017, 41 (2), 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Reuther JF; Dees JL; Kolesnichenko IV; et al. Dynamic covalent chemistry enables formation of antimicrobial peptide quaternary assemblies in a completely abiotic manner. Nat. Chem 2018, 10 (1), 45–50. [DOI] [PubMed] [Google Scholar]
- (28).Hopkins TP; Dener JM; Boldi AM Solid-Phase Synthesis of Trisubsituted Guanidines. J. Comb. Chem 2002, 4 (2), 167–174. [DOI] [PubMed] [Google Scholar]
- (29).Castro V; Rodríguez H; Albericio F CuAAC: An Efficient Click Chemistry Reaction on Solid Phase. ACS Comb. Sci 2016, 18 (1), 1–14. [DOI] [PubMed] [Google Scholar]
- (30).Hong V; Steinmetz NF; Manchester M; Finn MG Labeling Live Cells by Copper-Catalyzed Alkyne–Azide Click Chemistry. Bioconjugate Chem 2010, 21 (10), 1912–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kaur J; Saxena M; Rishi N An Overview of Recent Advances in Biomedical Applications of Click Chemistry. Bioconjugate Chem 2021, 32 (8), 1455–1471. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


