Abstract
We studied the effect on viral growth of drugs targeting different virus functions using a computer simulation for the intracellular growth of bacteriophage T7. We found that drugs targeting components of negative-feedback loops gain effectiveness against mutant viruses that attenuate the drug-target interaction. The greater inhibition of such mutants than of the wild type suggests a drug design strategy that would hinder the development of drug resistance.
The mutability and resultant adaptability of viruses present a major challenge to the design of antiviral strategies that are effective over the long term. While drug design has gained from advances in the molecular understanding of viral growth processes (13, 22), many initially potent drugs lose efficacy over time because of the emergence of drug-resistant strains (5, 21). When mutations arise that attenuate or compensate for the inhibitory effect of the drug, virus strains that carry such mutations gain a growth advantage and are subsequently selected for in the viral population (2, 9, 11, 14). In some instances, two or more drugs targeting the same or different viral functions have been used to reduce the likelihood that a drug-resistant strain will develop. However, multidrug therapies are often accompanied by adverse side effects (3) and do not prevent the development of multidrug-resistant mutants (12, 15, 17). Here, we present a computational study describing how antiviral strategies might be designed to counter the development of drug-resistant mutants. The central design principle is to choose drug-target interactions that result in selection against likely mutant strains.
Previously, we created a computer simulation that systematized and integrated existing experimental data on the intracellular growth of wild-type phage T7 in a single Escherichia coli cell (6). Our simulation was based on the established mechanisms and rates for the translocation of the phage genome into the host cell, the genome-encoded sequential synthesis of different mRNA species, the synthesis of each T7 gene product, T7 DNA replication, and the intracellular assembly of progeny phage. Lysis of the host cell was not accounted for. Source code, documentation, and an interactive version (T7web) of the simulation are available at http://virus.molsci.org/t7.
Here, we used the simulation to compute the wild-type T7 growth cycles resulting from infection of E. coli BL21 containing a range of antisense RNA concentrations targeting phage mRNA. We chose to simulate the effects of antisense RNA compounds on T7 growth because such compounds can, in principle, be targeted to any viral mRNA (18, 24). Furthermore, the development of drug resistance by the accumulation of point mutations that reduce antisense RNA-mRNA interactions has been observed (2); such laboratory systems may offer a means to test the strategies that we describe here. We computed the effects of different antisense strategies on phage growth by specifying a particular target mRNA, an initial concentration of complementary antisense molecules in the host cell, and an equilibrium binding constant (Keq) for the antisense molecule-target interaction. We used for the interactions between antisense molecules and phage mRNA a range of binding constants consistent with literature values (4, 8, 16, 23). We also performed simulations using kinetic rates for the binding and disassociation of the antisense molecule and the target but did not find qualitative differences from the equilibrium model (data not shown). We assumed that the binding of an antisense molecule to its target prevented the target from being translated. For this work, we defined the degree of inhibition of phage growth as the computed latent time needed to produce 99% the wild-type yield of T7 progeny.
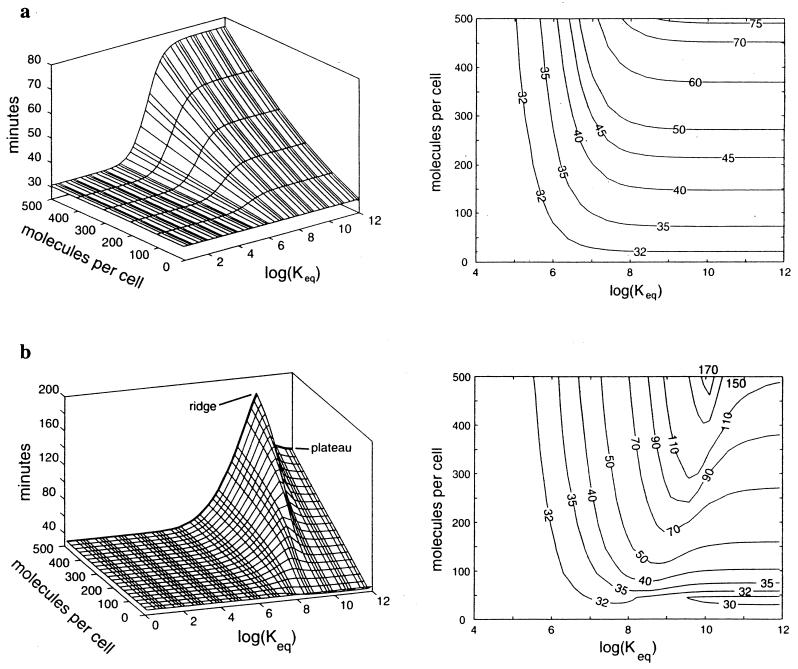
We computed the inhibition of T7 growth by antisense drugs that target mRNA encoding gp10, the T7 capsid protein, and gp1, the T7 RNA polymerase (RNAP). We computed these effects over a range of drug concentrations and affinities for their respective targets (Fig. 1). On these plots, higher elevations correspond to greater inhibition of viral growth. When the gene 10 mRNA was targeted, the degree of inhibition increased as both the concentration and affinity of the drug for its target increased, as we expected (Fig. 1a). Since the capsid protein is an essential component of each virion, depleting its message delays the production of viral progeny. However, when we targeted gene 1 mRNA, the simulation unexpectedly revealed a ridge of maximum inhibition of phage growth in a region of intermediate drug-target binding and a plateau of moderate inhibition in a region of maximum drug-target binding (Fig. 1b).
FIG. 1.
Computed effect of antisense drugs on the intracellular growth of phage T7 in E. coli. Antisense targets are mRNAs that encode the viral capsid protein (gp10) (a) and the viral RNAP (gp1) (b). Surface elevations and contour plots represent the time required to achieve 99% the wild-type growth yield. Log(Keq), the base-10 log of Keq for the antisense drug and its target mRNA, is a measure of the potency of the drug. Molecules per cell, number of antisense molecules per host cell at the start of the T7 infection. In both panels, the horizontal surface for minimum log(Keq) represents the wild-type growth region (31 ± 1 min).
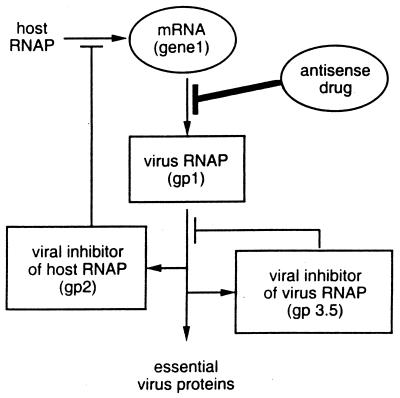
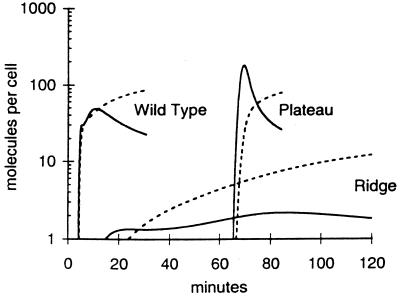
Differences between the inhibition landscapes reflect differences in the function and regulation of the targets. The gene 1 landscape is more complex than the gene 10 landscape because, unlike gp10, gp1 regulates both its own activity and the activity of other essential viral gene products (Fig. 2). To understand the gene 1 landscape, we computed the concentrations of gp1 and the gene 2 mRNA, encoding a host RNAP inhibitor (gp2), for regions on the gene 1 inhibition landscape corresponding to no drug (wild type), high concentrations of a high-affinity drug (plateau), and high concentrations of an intermediate-affinity drug (ridge) (Fig. 3). We found that high concentrations of a high-affinity drug, after a long delay, allowed bursts in the concentrations of active gp1 and gene 2 mRNA that were qualitatively similar to bursts observed during wild-type growth. The long delay in the initiation of gp1 and gene 2 mRNA syntheses reflected the time required by the virus to titrate away the drug. In contrast, we found that high concentrations of an intermediate-affinity drug resulted in incomplete inactivation of gene 1 mRNA, thereby allowing a small amount of gp1 synthesis and the subsequent gp1-mediated activation of the gp2 and gp3.5 negative-feedback loops. Notably, the activation of these loops occurred without the burst in the syntheses of gp1 and gene 2 mRNA seen in the wild-type or plateau cases. The combination of the negative-feedback loops embedded in T7 growth and the antisense drug-based inhibition of an intermediate of one of these negative-feedback loops resulted in the maximum inhibition of viral growth. Locally, gp2 inactivated host RNAP, thereby inhibiting its ability to titrate away the effect of the antisense drug. Globally, the pattern of continuous low-level viral RNAP expression reduced the expression of essential virus proteins.
FIG. 2.
Schematic of the T7 RNAP (gp1) regulatory circuit that includes the interaction of gene 1 mRNA with an antisense drug. Transcription by host RNAP creates mRNA for gene 1, which is translated to produce gp1. gp1 then transcribes other essential viral genes while regulating its own activity indirectly and directly by transcribing an inhibitor of host RNAP (gene 2) and an inhibitor of itself (gene 3.5), respectively.
FIG. 3.
Response of virus growth cycle components to antisense molecules targeting T7 gene 1 mRNA. Computed concentrations of active gp1 (solid lines) and gene 2 mRNA, a viral inhibitor of host RNAP (broken lines), are shown corresponding to landscape features (plateau and ridge) from Fig. 1b at an antisense drug concentration of 500 molecules per cell. The wild-type profiles computed in the absence of any drug are shown for comparison.
Examination of the gene 10 landscape (Fig. 1a) revealed how mutant viruses might easily escape antisense drugs targeting mRNA. From any dose and drug affinity corresponding to a point on the landscape, gene 10 mRNA mutants with reduced Keq values for the antisense molecule-mRNA interaction will always be inhibited less. Such mutants have a growth advantage relative to parent strains and should be selected for, other factors being equal. However, targeting of T7 gene 1 produced an entirely different landscape (Fig. 1b). When gene 1 mRNA was targeted, a ridge formed on the landscape due to the involvement of gp1 in a negative-feedback loop (Fig. 2). The ridge provides an example of how evolutionary robust drug strategies might work. Starting from the right of the ridge, at log(Keq) values above 10, the ridge serves as an evolutionary barrier that selects against potential escape mutants. Virus strains carrying mutations that attenuate the drug-target affinity must initially move “uphill” on the landscape, putting them at a selective disadvantage relative to their parent strains. Thus, the regulatory circuitry of the virus is exploited to block viral escape. Obviously, for the ridge to be effective, the decrease in binding affinity caused by a point mutation must not exceed the ridge width. Changes in the antisense molecule-target binding constant can be as high as 400-fold per nucleotide mismatch for binding between 18-mers (8), a value which is somewhat smaller than the width of the ridge in our example.
In this work, we showed how inhibiting components of negative-feedback loops under a specific set of conditions might be used to create barriers against the development of drug resistance. While we examined antisense mRNAs as inhibitor molecules, the results should extend to small-molecule drugs that inhibit specific virus proteins. For simplicity, we neglected the effects of RNA secondary structure or folding associated with the specific antisense molecule-target interaction. Such effects have been studied elsewhere (2, 10, 19) and will need to be taken into account as appropriate target sequences for antisense inhibition are developed. We also neglected issues associated with the abundance, availability, and localization of antisense molecules, important factors that need to be addressed for their in vivo application. Moreover, our simulation considered only viral intracellular growth and ignored any dynamics associated with the cell-to-cell spread of viral mutants, which have been studied in other systems (1, 20). Finally, while we have proposed a means to design evolutionary barriers against obvious escape routes, we recognize that new antiviral strategies merely create new selection environments.
Acknowledgments
This work was supported by the National Science Foundation (grant BES-9896067) and the Office of Naval Research (grant N00014-98-1-0226).
Lingchong You assisted with Fig. 1.
REFERENCES
- 1.Bonhoeffer S, May R M, Shaw G M, Nowak M A. Virus dynamics and drug therapy. Proc Natl Acad Sci USA. 1997;94:6971–6976. doi: 10.1073/pnas.94.13.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bull J, Jacobson A, Badgett M, Molineux I. Viral escape from antisense RNA. Mol Microbiol. 1998;28:835–846. doi: 10.1046/j.1365-2958.1998.00847.x. [DOI] [PubMed] [Google Scholar]
- 3.Cameron D W, Japour A J, Xu Y, Hsu A, Mellors J, Farthing C, Cohen C, Poretz D, Markowitz M, Follansbee S, Angel J B, McMahon D, Ho D, Devanarayan V, Rode R, Salgo M, Kempf D J, Granneman R, Leonard J M, Sun E. Ritonavir and saquinavir combination therapy for the treatment of HIV infection. AIDS. 1999;13:213–224. doi: 10.1097/00002030-199902040-00009. [DOI] [PubMed] [Google Scholar]
- 4.Daly T J, Doten R C, Rusche J R, Auer M. The amino terminal domain of HIV-1 Rev is required for discrimination of the RRE from nonspecific RNA. J Mol Biol. 1995;253:243–258. doi: 10.1006/jmbi.1995.0549. [DOI] [PubMed] [Google Scholar]
- 5.Domingo E, Menéndez-Arias L, Quiñones-Mateu M, Holguín A, Gutiérrez-Rivas M, Martínez M, Novella I, Holland J. Viral quasispecies and the problem of vaccine-escape and drug-resistant mutants. Prog Drug Res. 1997;48:99–128. doi: 10.1007/978-3-0348-8861-5_4. [DOI] [PubMed] [Google Scholar]
- 6.Endy A, Kong D, Yin J. Intracellular kinetics of a growing virus: a genetically structured simulation for bacteriophage T7. Biotechnol Bioeng. 1997;55:375–389. doi: 10.1002/(SICI)1097-0290(19970720)55:2<375::AID-BIT15>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 7.Endy D. Development and application of a genetically-structured simulation for bacteriophage T7. Ph.D. thesis. Hanover, N.H.: Dartmouth College; 1997. [Google Scholar]
- 8.Freier S M, Lima W F, Yogesh S S, Vickers T, Zounes M, Cook P D, Ecker D J. Thermodynamics of antisense oligonucleotide hybridization. In: Erickson R P, Izant J G, editors. Gene regulation: biology of antisense RNA and DNA. New York, N.Y: Raven Press, Ltd.; 1992. pp. 95–107. [Google Scholar]
- 9.Heinz B A, Rueckert R R, Shepard D A, Dutko F J, McKinlay M A, Fancher M, Rossmann M G, Badger J, Smith T J. Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J Virol. 1989;63:2476–2485. doi: 10.1128/jvi.63.6.2476-2485.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjalt T A, Wagner E G. Bulged-out nucleotides in an antisense RNA are required for rapid target RNA binding in vitro and inhibition in vivo. Nucleic Acids Res. 1995;23:580–587. doi: 10.1093/nar/23.4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho D D, Toyoshima T, Mo H, Kempf D J, Norbeck D, Chen C M, Wideburg N E, Burt S K, Erickson J W, Singh M K. Characterization of human immunodeficiency virus type 1 variants with increased resistance to a C2-symmetric protease inhibitor. J Virol. 1994;68:2016–2020. doi: 10.1128/jvi.68.3.2016-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iversen A K, Shafer R W, Wehrly K, Winters M A, Mullins J I, Chesebro B, Merigan T C. Multidrug-resistant human immunodeficiency virus type 1 strains resulting from combination antiretroviral therapy. J Virol. 1996;70:1086–1090. doi: 10.1128/jvi.70.2.1086-1090.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffries D J, De Clercq E, editors. Antiviral chemotherapy. Chichester, England: John Wiley & Sons; 1995. [Google Scholar]
- 14.Larder B, Coates K, Kemp S. Zidovudine-resistant human immunodeficiency virus selected by passage in cell culture. J Virol. 1991;65:5232–5236. doi: 10.1128/jvi.65.10.5232-5236.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larder B A, Kellam P, Kemp S D. Convergent combination therapy can select viable multidrug-resistant HIV-1 in vitro. Nature. 1993;365:451–453. doi: 10.1038/365451a0. [DOI] [PubMed] [Google Scholar]
- 16.Lima W F, Monia B P, Ecker D J, Freier S M. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry. 1992;31:12055–12061. doi: 10.1021/bi00163a013. [DOI] [PubMed] [Google Scholar]
- 17.Moutouh L, Corbeil J, Richman D D. Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci USA. 1996;93:6106–6111. doi: 10.1073/pnas.93.12.6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray J A H. Antisense RNA and DNA. New York, N.Y: Wiley-Liss, Inc.; 1992. [Google Scholar]
- 19.Patzel V, Sczakiel G. Theoretical design of antisense RNA structures substantially improves annealing kinetics and efficacy in human cells. Nat Biotechnol. 1998;16:64–68. doi: 10.1038/nbt0198-64. [DOI] [PubMed] [Google Scholar]
- 20.Perelson A, Neumann A, Markowitz M, Leonard J, Ho D. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 21.Richman D D, editor. Antiviral drug resistance. Chichester, England: John Wiley & Sons; 1996. [Google Scholar]
- 22.Saunders J, Cameron J M. Recent development in the design of antiviral agents. Med Res Rev. 1995;15:497–531. doi: 10.1002/med.2610150604. [DOI] [PubMed] [Google Scholar]
- 23.Tomizawa J. Control of ColE1 plasmid replication: the process of binding of RNA I to the primer transcript. Cell. 1984;38:861–870. doi: 10.1016/0092-8674(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 24.Uhlmann E, Peyman A. Antisense oligonucleotides: a new therapeutic principle. Chem Rev. 1990;90:543–584. [Google Scholar]