The transcription factor INTERMEDIUM-C is described as regulator involved in tiller formation, integrating environmental conditions such as the shade response in barley.
Keywords: Abscisic acid, barley, bud growth arrest, decapitation, INTERMEDIUM-C, shade avoidance, yield
Abstract
Tiller formation is a key agronomic determinant for grain yield in cereal crops. The modulation of this trait is controlled by transcriptional regulators and plant hormones, tightly regulated by external environmental conditions. While endogenous (genetic) and exogenous (environmental factors) triggers for tiller formation have mostly been investigated separately, it has remained elusive how they are integrated into the developmental program of this trait. The transcription factor gene INTERMEDIUM-C (INT-C), which is the barley ortholog of the maize domestication gene TEOSINTE BRANCHED1 (TB1), has a prominent role in regulating tiller bud outgrowth. Here we show that INT-C is expressed in tiller buds, required for bud growth arrest in response to shade. In contrast to wild-type plants, int-c mutant plants are impaired in their shade response and do not stop tiller production after shading. Gene expression levels of INT-C are up-regulated under light-limiting growth conditions, and down-regulated after decapitation. Transcriptome analysis of wild-type and int-c buds under control and shading conditions identified target genes of INT-C that belong to auxin and gibberellin biosynthesis and signaling pathways. Our study identifies INT-C as an integrator of the shade response into tiller formation, which is prerequisite for implementing shading responses in the breeding of cereal crops.
Introduction
Ensuring yield stability of cereal crops is a major requirement for plant breeding in the face of climate change (Kang et al., 2009). In particular, breeding of new elite varieties with optimized shoot architecture, such as tiller number, will allow the maintenance of high yield potential in unfavorable environments. The effect of genetic factors on this trait was described recently (Haaning et al., 2020). In many countries, particularly at very high latitudes or on shaded slopes, light is a limiting factor affecting crop growth and yield. Moreover, during the past decades, intensive crop management has increased sowing and plant stand densities to improve homogeneity among individual plants that now produce fewer tillers. To increase light capture, plants have evolved refined mechanisms to maximize light harvesting and/or to prevent shade, namely low light intensity and/or a low ratio of red (R) to far-red (FR) light. Suboptimal light triggers a suite of phenotypic changes, defined as the shade avoidance response (SAR), that includes hypocotyl and petiole elongation, an upward orientation of leaves, and early flowering. Additionally, a common characteristic of SAR is the suppression of shoot branching in a wide variety of species (Smith and Jordan, 1994; Tucic et al., 2006; Aguilar-Martinez et al., 2007; González-Grandio et al., 2013).
The extensive work carried out in Arabidopsis has provided a detailed understanding of the SAR (Wang and Wang, 2015). By sensing and responding to R and FR light, the five photoreceptors of the phytochrome family (phyA–phyE) regulate a variety of developmental processes (Franklin and Quail, 2010). Among these, phytochrome B (phyB) appears to be the most important photoreceptor involved in shade detection and avoidance (Reed et al., 1993; Ballaré, 1999), functioning redundantly with other members of its clade (Stamm and Kumar, 2010). Phytochrome proteins act as dimers and exist in two photoconvertible forms: ‘Pr’ (the red-light-absorbing, inactive form) and ‘Pfr’ (the far-red-light-absorbing, active form), with the Pfr:Pr ratio reflecting the R:FR ratio of the environment (Smith, 2000). Upon photoconversion into active Pfr, phytochromes migrate to the nucleus, where they regulate gene expression by interacting with several basic helix–loop–helix (bHLH) transcription factors, including phytochrome-interacting factor (PIF) and PIF-like (PIL) proteins (Chen et al., 2004; Duek and Fankhauser, 2005). In many plant species, genome-wide transcriptional dynamics of the SAR have been studied in petioles or leaf blades at the seedling stage, while SAR is less defined in economically important monocots (Devlin et al., 2003; Tao et al., 2008; Hornitschek et al., 2012; Wang et al., 2016).
In the agricultural production of graminaceous crop species, plant density is a major determinant for crop yield. Shading caused by elevated plant densities reduces not only photosynthetic active radiation (PAR) flux density but also the R/FR ratio of the light reaching the lower strata of the canopy. Generally, increasing plant density results in progressively stronger suppression of tillering due to accelerated apical shoot development and stem elongation. This pattern continues until the beginning of the stem elongation phase (Zadoks et al., 1974). Moreover, early-emerging tillers contribute more to grain yield than do tillers that emerge later. The regulation of barley tiller outgrowth by shade has also been supported by the observation that supplemental FR illumination of elongating leaves or of the main stem base reduced the total number of tillers per plant (Skinner and Simmons, 1993). In spite of its great ecological and economic impact, little is known about the underlying molecular mechanisms linking shade-initiated transcriptional changes with the suppression of tiller outgrowth in barley plants, especially regarding the tiller bud that is one of the most important sites of shade action.
Lateral shoot growth is coordinately controlled by conserved interactions that regulate the biosynthesis and signaling of the hormones auxin, abscisic acid (ABA), strigolactones (SLs), and cytokinins (CKs). Auxin and SLs, synthesized mainly in the shoot apex and root, respectively, inhibit branching, while CKs, synthesized mostly in the root and stem, promote branching (Kebrom et al., 2013). The class II TEOSINTE BRANCHED1, CYCLOIDEA, and PCF (TCP) transcription factors TEOSINTE BRANCHED 1 (TB1)-like, in monocots, and BRANCHED1 (BRC1)-like, in dicots, act locally inside the axillary bud where they are subject to transcriptional regulation by hormonal crosstalk (auxin–SL–CK) and cause bud growth arrest (Doebley et al., 1997; Aguilar-Martinez et al., 2007; Minakuchi et al., 2010; Martin-Trillo et al., 2011; Braun et al., 2012). Moreover, recent studies have implicated the involvement of TB1 orthologs in the shade-induced response of branch suppression. In Sorghum bicolor and maize, active phyB (Pfr) suppresses the expression of the TB1 gene and induces bud outgrowth. On the other hand, light signals that inactivate phyB allow increased expression of TB1 and suppression of bud outgrowth (Kebrom et al., 2006, 2010; Whipple et al., 2011). In Arabidopsis, BRC1 is up-regulated in axillary buds of plants grown at high density and is required for complete branch suppression in these conditions (Aguilar-Martinez et al., 2007). These results suggest that the phytochrome pathway is involved in the control of TB1 and axillary meristem outgrowth, and provides a link between environmental variation and gene action controlling branching (González-Grandio et al., 2013; Reddy et al., 2013). In barley, INT-C is an ortholog of the maize domestication gene TB1, and its mutants exhibit higher tiller numbers and enlarged lateral spikelets (Ramsay et al., 2011).
Here, we investigated the SAR of early formed tiller buds in barley plants before the rapid stem elongation stage, and studied the role of INT-C during the SAR by comparable transcriptome analysis. The analysis revealed that the dynamics of INT-C expression during the SAR are critical for genome-wide reprogramming of gene expression and that the gene categories affected support a central role for INT-C in tiller bud arrest. A comparison of genes responding to shade-induced bud arrest and decapitation-triggered bud activation allowed us to identify key regulators influencing bud dormancy and bud activation genes. Thus, these findings would enable us to better understand the genetic mechanisms controlling the reversible transition of growth to dormancy in barley tiller buds.
Materials and methods
Plant material and growth conditions
Hordeum vulgare cv. Bowman, a two-rowed spring barley cultivar, was used as the wild type for comparison with its near isogenic mutant BW421 (int-c.5) (Ramsay et al., 2011). For the experimental analyses except the shading experiment, wild-type and int-c mutant plants were cultivated in a greenhouse. Seeds were sown in either 54- or 96-well trays and germinated in a climate chamber for 10 d at 11 °C day and 7 °C night temperature under 16 h light. After that, seedlings were transferred to pots (diameter 16 cm) filled with two parts of compost, two parts of ‘Substrat 2’ (Klasmann), and one part of quartz sand, and allowed to grow in the greenhouse at 20 °C day/14 °C night under 16 h light. For the density experiments, three barley planting densities (one, five, and 10 plants per pot) were established in pots (size, 21 liter; top diameter, 310 cm; base diameter, 250 cm; height, 214 cm), and the planting density test was repeated four times. For the decapitation experiments, when plants were grown until the early stem elongation stage, the shoot apices were removed, and the apical dominance test was repeated three times. For the shading experiments, seedlings were grown in a climate-controlled growth chamber at a temperature of 12 °C, 70% humidity, and a 12/12 h day/night cycle. Green shading was achieved by using a green plastic filter (122 Fern Green; LEE Filters, Andover, UK) (Kegge et al., 2013).
Sequence retrieval for TCP proteins
In order to identify barley genes putatively encoding TCP transcription factors, the latest barley annotation and genome sequence at http://webblast.ipk-gatersleben.de/barley/viroblast.php was searched using the TBLASTN algorithm with Arabidopsis TCP proteins or TCP domains as query sequence. All redundant sequences were discarded from further analysis based on ClusterW (Thompson et al., 1994). Furthermore, to verify the reliability of the initial results, all non-redundant candidate TCP sequences were analyzed to confirm the presence of the conserved TCP domain using the InterproScan program (Quevillon et al., 2005). The sequences of TCP family members in the genome of Arabidopsis were retrieved from the PlantTFDB plant transcription factor database (http://planttfdb.gao-lab.org/, v3.0). Antirrhinum majus and Oryza sativa TCP sequences, were obtained from NCBI.
Phylogenetic analysis
Multiple sequence alignments were conducted on the amino acid sequences of TCP proteins in Arabidopsis and barley genomes using Cluster X (Thompson et al., 1997) with default settings. Subsequently, MEGA 6.0 software (Tamura et al., 2013) was employed to construct an unrooted phylogenetic tree based on alignments using the Neighbor–Joining (NJ) method with the following parameters: JTT model, pairwise gap deletion, and 1000 bootstraps.
Extraction and quantification of ABA
ABA was extracted from fresh plant materials using ethyl acetate (100%). Isotopically labeled D6-ABA was used as an internal standard and added to each sample during the extraction procedure. Extraction was carried out twice with 1 ml of ethyl acetate at 4 °C. The supernatant collected after centrifugation (13 000 g, 10 min, and 4 °C) was evaporated to dryness at room temperature using a vacuum concentrator. The dried samples were dissolved in acetonitrile:methanol (1:1) and filtered using a 0.8 µm filter (Vivaclear). The filtrate (10 µl) was used for subsequent quantification by LC-MS/MS (Dionex Summit coupled to Varian 1200 L). Chromatographic separation was carried out on a C18 column (4 µm, 100 mm; GENESIS; Vydac/USA). Multiple reaction monitoring (MRM) and quantification were done using the mass traces 263/153 for ABA and 269/159 for D6-ABA. The validity of the extraction and measurement procedure was verified in recovery experiments (~82–95%). Quantification was based on calibration with known ABA standards and individual recovery rates for the samples, as described in Kong et al. (2008).
qRT–PCR and microarray hybridization, and data analyses
RNA was extracted from fresh plant tissues from independent or pooled biological replicates with the same treatment using a plant mini RNA kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol, and its quality and quantity were assessed with a Nano drop device (Peqlab, Erlangen, Germany). A 500 ng aliquot was taken as template for the synthesis of the first cDNA strand, primed by oligo(dT), using a RevertAid cDNA kit (ThermoFisher Scientific, Waltham, MA, USA). The subsequent quantitative reverse transcription–PCR (qRT–PCR) was based on the Power SYBR® Green PCR Master Mix (ThermoFisher Scientific) and conducted in an Applied Biosystems 7900HT Fast Real-Time PCR system (ThermoFisher Scientific) following the manufacturer’s protocol. Evaluation of the data was performed according the MIQE guidelines (Bustin et al., 2009). Relative transcript abundances were obtained using the ΔΔCT method (Livak and Schmittgen, 2001) and were normalized against the abundance of the serine/threonine phosphatase PP2A transcript which was selected as reference earlier (Seiler et al., 2011; Sreenivasulu et al., 2012). The primer sequences employed are given in Supplementary Table S1. The presence of a unique PCR product was verified by dissociation analysis, and each qRT–PCR was repeated at least three times. Each biological replicate was represented by three technical replicates.
For the microarray procedure, the same RNA samples extracted from three biological replicates were used and the quality of the RNA was verified with a Bioanalyzer 2100 device (Agilent Technologies, Santa Clara, CA, USA). The RNA was labeled by using the Low input QuickAmp Labeling kit (Agilent Technologies) and hybridized, following the manufacturer’s protocol, to a custom-synthesized 60 k Barley Microarray (Agilent Technologies) (Koppolu et al., 2013). The resulting data were analyzed using GeneSpring 13.0 GX software (Agilent Technologies). After quantile normalization and baseline transformation to the median of all samples, the probesets (genes) were filtered by coefficient of variation <50%, followed by moderated t-test and Bonferroni–Holm multiple testing corrections. Probesets passing the P-value cut-off of <0.05 with a fold change of ≥2.0 were selected as differentially expressed genes (DEGs). Analyses of functional categories with INT-C-dependent up-regulated and down-regulated genes were performed using MapMan. The fold enrichment was calculated as follows: (number in class input_set/number of total input_set)/(number in class reference_set/ number of total reference_set). The P-value was determined by a hypergeometric distribution test (R Core Team, 2013). The data were sorted by fold enrichment with a cut-off of P<0.05. The specific pathway enrichment analysis by Wilcoxon Rank Sum test was implemented in MapMan. The resulting enrichments for the functional classes (P<0.05, determined by a hypergeometric test; R Core Team, 2013) provided a map of gene modules regulated dependent on INT-C during SAR. To confirm that the common genes of the two identified groups are not random, the test of statistical significance was applied by a web-based tool at http://nemates.org/MA/progs/overlap_stats.html. The co-regulated genes were retrieved from Genevestigator (Zimmermann et al., 2004), and Gene Ontology analysis was performed in agriGO v2.0 with default parameters (Tian et al., 2017).
Statistical analysis
To test the statistical significance of the data, Student’s t-test and one-way ANOVA with Tukey test for significance were used. In the figures, asterisks denote significant differences in Student’s t-tests and different letters denote significant differences in Tukey’s test.
Results
INT-C is a member of the barley TCP gene family
INTERMEDIUM-C (INT-C) is the barley orthologue of TEOSINTE BRANCHED1 (Ramsay et al., 2011), one of the name-giving members of the well-studied TEOSINTE BRANCHED1, CYCLOIDEA, PROLIFERATING CELL NUCLEAR ANTIGEN BINDING FACTOR (TCP) gene family (Li, 2015). TCP transcription factors are defined by the TCP domain, which is a 59 amino acid long bHLH structure which provides the ability to bind GC-rich DNA sequence motifs (Martin-Trillo and Cubas, 2010). In barley, INT-C is encoded by HORVU4Hr1G007040 and was identified as the corresponding gene of the vrs5 locus on chromosome 4 (Ramsay et al., 2011) which is involved in the fertility of lateral spikelets.
To identify the genes closest to INT-C, the barley TCP gene family was analyzed using the latest barley annotation and genome sequence at http://webblast.ipk-gatersleben.de/barley/viroblast.php and the TBLASTN algorithm with Arabidopsis TCP proteins or TCP domains as query sequence (Fig. 1A; Supplementary Table S2). This family consists of 19 genes which contain a putative TCP-HLH-type domain at the N-terminus (Fig. 1B). The phylogenetic tree which was built based on multiple alignments of the TCP domain in TCP proteins showed that barley TCP proteins could be divided into two groups, as for all species so far (Fig. 1A, B). The class I group was formed by nine predicted proteins related to the PCF rice factors (Kosugi and Ohashi, 1997), while class II was comprised of 10 predicted proteins related to the Antirrhinum CYC and CIN genes and to OsTB1 (Luo et al., 1996; Doebley et al., 1997; Nath et al., 2003; Takeda et al., 2003). In addition, the class II group could be further divided into two subclades: the CIN group formed by seven members and the CYC/TB1 group formed by three members. HvTC16 and HvTC17 from the class II CYC/TB1 contain an R domain that is also found in HvTC12 from the class II CIN group (Fig. 1C), as previously described in Arabidopsis (Yao et al., 2007). Although in eudicots several CYC/TB1 sequences are found, and phylogenetic analyses have suggested that duplications within this clade occurred at the base of eudicots, in monocots only one type of CYC/TB1 has been identified (e.g. OsTB1) (Howarth and Donoghue, 2006). Our phylogenetic analysis revealed that, based on the absence of the R domain, none of the newly identified barley TCP genes can be considered as a paralog of INT-C.
Fig. 1.
Phylogenetic tree and alignments of Arabidopsis and barley TCP proteins. (A) The phylogenetic tree was built based on multiple alignment of the TCP domain in the TCP proteins using the Neighbor–Joining method with 1000 bootstrap replicates. Blue, light blue, and orange lines indicate the PCF, CYC/TB1, and CIN clades, respectively. Each Arabidopsis protein is indicated by a square, each barley protein is indicated by a triangle. (B) Alignment of the TCP domain and adjoining sequence for the predicted barley TCP proteins. Overall conserved amino acids are shaded in black. Amino acids 80% or 100% conserved in Class II or Class I are shaded in light gray and dark gray, respectively. The basic, helix I, loop, and helix II regions are indicated. (C) Alignment of the R-domain of Class II subfamily members. Amino acids are expressed in the standard single letter code. Sequences were aligned with ClustalW and represented with Genedoc. (D) Alignment of putative target areas for miR319a (aligned in reverse). Asterisks indicate INT-C (HvTCP16).
As described for the model plant Arabidopsis, five of the CIN subclade members are post-transcriptionally regulated by miRNA319 (AtTCP2, 3, 4, 10, and 24) (Palatnik et al., 2003, 2007; Ori et al., 2007). In barley, hvu-miR319a (UUGGACUGAAGGGAGCUCCC) is encoded by CL16998_Contig1 (Ozhuner et al., 2013). The closest barley homologs of these Arabidopsis genes are the five genes, HvTCP3, HvTCP6, HvTCP9, HvTCP12, and HvTCP15. These barley CIN subclade members contain sequences with putative binding sites for hvu-miR319 (Supplementary Table S2). Figure 1D shows the alignment of the target sites of these genes with the miR319 sequence. This suggests that regulation of leaf development by a redundant set of miRNA-regulated homologous TCP genes could occur in barley, while INT-C does not represent a miR319 target.
INT-C loss of function leads to an early promotion of tiller bud outgrowth in barley
Previous studies noticed that int-c mutants in various cultivars produce more tillers than the respective wild-type plants during early vegetative stages (Ramsay et al., 2011; Liller et al., 2015). However, this increased tiller formation did not translate into more productive tillers. In contrast, the number of tillers was significantly lower at the point of harvest. The growth experiment described here performed with the near isogenic mutant BW421 (int-c.5) and the corresponding wild type confirmed these earlier observations. An increase in tiller number in int-c was only detectable at the early developmental stages (Fig. 2A) between 2 and 5 weeks after germination. At later developmental stages (6–8 weeks after germination), the pattern of tiller number was reversed. This observation correlated with an earlier anthesis of int-c compared with the wild type cv. Bowman (Supplementary Fig. S1), leading to an earlier arrest of tiller bud production in int-c. The tillers of barley are formed in a sequential order, starting with the first tiller bud under the coleoptile. The development of the tiller buds in the axils of successive leaves was studied by dissecting the plants at different developmental stages (Fig. 2B). To investigate the involvement of INT-C in bud initiation and bud outgrowth, the primary tiller buds were classified (Fig. 2C, D) as dormant bud (800–1200 μm), outgrowing bud (1.5–100 mm), or tiller emergence (10–35 cm) in each leaf axil at an early developmental stage (2–3 weeks after germination). Figure 2D indicates the enhanced bud outgrowth in int-c mutants compared with the wild type. This result suggests that the outgrowth of tiller buds is accelerated in int-c mutants at early developmental stages but is slowed down at later stages (>5 weeks).
Fig. 2.
INT-C is involved in barley plant architecture by tiller bud outgrowth. Analysis of tiller development in wild-type and int-c plants. (A) Tiller number of int-c and wild-type plants 1–8 weeks after germination (n=25–30 plants). Asterisks indicate significant differences (Student’s t-test, P<0.001) between wild-type and int-c mutant plants. (B) Dissected tillers from successive leaf axils in ~2- to 3-week-old seedlings. Ct, coleoptile tiller; 1–4, order of leaves; scale bars=5 cm; arrows, tiller buds. (C) Exemplary tiller bud formation stage in the third leaf axil. The area of the close-up view is outlined with a white box in the left image. Dissection of a tiller bud at this stage will reveal a shoot apex with leaf primordia and a meristematic dome. Scale bars represent 200 μm. (D) Schematic representations of tiller bud production in each leaf axil of the wild type and int-c in 2- to 3-week-old seedlings. Each column stands for a single plant, and each row stands for a leaf axil in order from bottom to top, starting with the coleoptile tiller. Different colored squares denote different tiller bud lengths. (E) INT-C (HvTCP16) mRNA levels in different tissues and (F) during spike development as analyzed by real-time qRT–PCR. Bars represent means ±SD; n=3 biological replicates. Serine/threonine protein phosphatase HvPP2A-4 mRNA was used as a reference. TM, triple mound; GP, glume primordium; SP, stamen primordium; AP, awn primordium.
INT-C mRNA levels were analyzed by real-time qRT–PCR in different tissues and during spike development (Fig. 2E, F). INT-C mRNA was detectable at the highest levels in tiller buds, supporting its role in the control of tiller bud development. It was expressed at lower levels in other tissues such as root, stem, and leaf. During spike meristem development, mRNA levels of INT-C peaked at the glume primordium stage. In the later stages (stamen primordium and awn primordium), relatively high levels of INT-C mRNA persisted. This peak of expression correlated with the observation that after the awn primordium stage, profound differences in the development of the lateral spikelet in int-c occurred compared with the wild type (Ramsay et al., 2011).
INT-C mRNA abundance decreased after decapitation
Apical dominance is the inhibitory control exerted by the shoot apex over the outgrowth of the lateral buds (Cline, 1997). Decapitation stimulates bud reactivation after breaking the apical dominance (Hall and Hillman, 1975; Napoli et al., 1999; Cline, 2000; Tatematsu et al., 2005; Aguilar-Martinez et al., 2007). To analyze the involvement of INT-C in integrating the decapitation response into bud outgrowth, barley int-c mutant plants were compared with the respective wild-type plants. For this approach, 3-week-old plants that had undergone early stem elongation were decapitated. Two weeks later, two tiller buds of decapitated Bowman plants had elongated prematurely. Thus, total tiller number in decapitated wild-type plants reached the same level as in int-c mutants, in which decapitation had no effect (Fig. 3A). To investigate whether this response was related to a transcriptional down-regulation of INT-C, mRNA levels in axillary buds were analyzed by real-time qRT–PCR after decapitation, before any visible sign of bud outgrowth (Fig. 3B). INT-C mRNA decreased significantly 6 h after decapitation. DRM1/ARP (DORMANCY-ASSOCIATED GENE/AUXIN-REPRESSED PROTEIN) mRNA, an early marker for bud dormancy (Stafstrom et al., 1998; Tatematsu et al., 2005), also showed reduction and reached its minimum 24 h after decapitation. These results support the idea that INT-C is involved in the early response to bud release from apical dominance and required for bud activation.
Fig. 3.
INT-C expression in response to decapitation. (A) Tiller number of cv. Bowman (wild-type) and int-c plants 2 weeks after decapitation. Bars represent means ±SD; n=3 replicates with ≥16 plants. Different letters indicate significant differences according to Tukey’s test (P<0.05). (B) Ratio of mRNA levels of INT-C and DRM1 in tiller buds between decapitated and non-decapitated plants. Relative mRNA abundance of INT-C mRNA was analyzed by real-time qRT–PCR. Bars represent means ±SD; n=4 biological replicates. Serine/threonine protein phosphatase HvPP2A-4 was used as a reference gene. Analyzed is the early transcriptional response within 24 h after decapitation.
Transcriptome analysis of buds after decapitation
To investigate the transcriptome response of dormant versus activated buds, Agilent 8 × 60K customized barley microarray expression analysis (Thirulogachandar et al., 2017) using total mRNA prepared from tiller buds at the early stem elongation stage was performed. The time point 24 h after decapitation was chosen for the following reasons: (i) down-regulation of DRM1 expression was evident at this time point (Fig. 3B); (ii) this time point was 1 d before the first visible effects of decapitation on the growth were detectable—2 d later, the vegetative buds of decapitated plants had activated and begun to elongate; and (iii) the time point enabled us to exclude genes affected by circadian rhythm.
A total of 1704 DEGs were detected in tiller buds of decapitated versus non-decapitated plants (Supplementary Table S3), among those 1011 were down-regulated and 693 up-regulated. Microarray results were confirmed by qRT–PCR for selected genes (Supplementary Fig. S2). Besides DRM1, which was found among the down-regulated genes, several genes encoding transcription regulators were detected. A total of 81 out of 494 DEGs regulated in the opposite direction under shading conditions (see below) could be mapped to the term ‘transcription regulator associated with ABA’. Among them are the auxin-binding protein ABP44 and a putative ripening-related bZIP protein (CAB85632). The up-regulated genes included a large number of ribosomal proteins, cell organization, and cell cycle-related genes.
Plant density affects INT-C expression
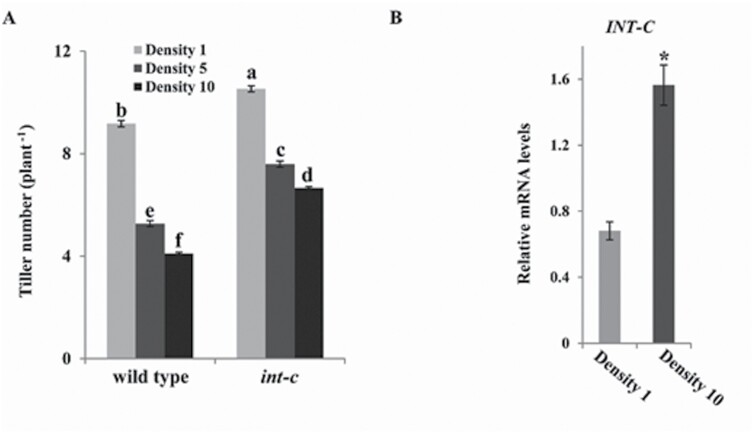
Planting density affects shoot branching in many plant species. Low plant density results in more branches, compared with growth in dense plant stands as a result of a neighbor-sensing response (Casal et al., 1986). To test the impact of plant density on INT-C, wild-type and int-c plants were grown in three different densities under greenhouse conditions (one, five, and 10 plants per pot). Tiller numbers were counted until the early stem elongation stage (Fig. 4).
Fig. 4.
INT-C expression responds to planting density. (A) Tiller number of wild-type and int-c plants grown at planting densities of one, five, or 10 plants per pot. Plants were analyzed 5 weeks after sowing. Bars represent means ±SD; n=3 replicates with ≥20 plants. Different letters indicate significant differences according to Tukey’s test (P<0.01). (B) Transcript levels of INT-C in the tiller bud tissue analyzed by real-time PCR at a density of one or 10 plants per pot. Bars represent means ±SD; n=3 biological replicates. Serine/threonine protein phosphatase HvPP2A-4 was used as a reference gene. The asterisk indicates a significant difference according to Student’s t-test at ∗P<0.001.
Wild-type plants responded to increased planting density with reduced tillering. At a density of 10 plants per pot, tiller bud suppression was more than half, as tiller number was 56% lower than in plants grown at one plant per pot. However, int-c mutants showed reduced sensitivity to this condition (33% reduction compared with plants at one plant per pot). The mRNA levels of INT-C were then analyzed by qRT–PCR in wild-type plants grown at low (one plant per pot) and high (10 plants per pot) density. At high density, INT-C mRNA levels showed a 2-fold increase compared with those at low density (Fig. 4B). These results support the involvement of transcriptional regulation of INT-C in bud dormancy.
INT-C is up-regulated as part of the shade avoidance response
To further examine whether INT-C is participating in the SAR, plants were grown under two different light conditions: control light (PAR=840 μmol m−2 s−1; R:FR ratio=2.2) and low R:FR light mimicking shade imposition by neighboring plants (green shade, PAR=260 μmol m−2 s−1; R:FR ratio=0.2) (Kegge et al., 2013). To minimize a putative bias resulting from variations in leaf number and int-c-mediated earlier flowering, plants were grown under conditions attenuating the int-c early flowering phenotype (Fig. 5A).
Fig. 5.
Effect of shading on tiller bud outgrowth and INT-C expression. (A) Tillering phenotype of wild-type and int-c plants grown under control conditions or shading. Red dots indicate the primary tillers. (B) Tiller number of wild-type and int-c plants grown under control or shade conditions. Bars represent means ±SD; three independent experiments with n≥35 plants each. Different letters indicate significant differences according to Tukey’s test (P<0.01). (C) Transcript levels of INT-C analyzed by qPCR, in buds of shaded plants, relative to levels in control plants. Bars represent means ±SD; n=3 biological replicates. Serine/threonine protein phosphatase PP2A-4 was used as a reference gene. Asterisks indicate significant differences according to Student’s t-test at ∗P<0.05. (D) ABA concentrations in tiller buds of wild-type and int-c plants 6 h after exposure to shade. Bars represent means ±SD of six independent biological replicates. Different letters indicate significant differences according to Tukey’s test (P<0.05).
The shading treatment was started at the two-leaf stage. Four weeks later, primary tiller number was quantified. Bowman plants grown under shade conditions responded strongly and had four times fewer tillers than plants grown in control conditions (Fig. 5B), indicating that exposure of plants with young vegetative buds to a low R:FR ratio promotes bud arrest in barley. In contrast, the response of int-c mutants to shading was much weaker. int-c plants grown under shade had only 1.4 times fewer tillers than plants grown under normal light conditions (Fig. 5A, B). Other phenotypic SARs, such as hyponasty and stem elongation, were indistinguishable between the wild-type and mutant plants (Fig. 5A). The short-term response of INT-C to shade treatment was analyzed by transferring plants at the five-leaf stage (when plants had small vegetative buds) to low R:FR light. The mRNA levels of INT-C in tiller buds increased after 4, 6, or 8 h exposure to shade, indicating a transcriptional activation of INT-C in response to shade (Fig. 5C). This result is in agreement with the increase of tillers in plants grown in dense stands. The higher tiller number in int-c coincided with a lower concentration of ABA in tiller buds (Fig. 5B, D), supporting the role of ABA as a mediator of INT-C-dependent tiller bud suppression in the shade.
Transcriptome analysis of tiller buds after shading identifies INT-C-dependent genes
To obtain further insight into the molecular mechanisms of INT-C-mediated growth responses, the transcriptome of tiller buds in wild-type and int-c plants was analyzed. The plants were exposed to control conditions or shade (Fig. 5) and the transcriptome was analyzed 6 h after exposure to shade using an Agilent 8 × 60K customized barley microarray. As INT-C expression was highest in tiller buds (Fig. 2), we selected this tissue for detailed analysis of RNA expression levels.
While under control conditions a minor influence of int-c-dependent transcript changes was found, 305 DEGs were identified between the wild type and int-c, out of which 185 were up-regulated and 120 were down-regulated. Under shading, a stronger effect of int-c on the transcriptome was noted. In wild-type buds, 2726 shade-responsive DEGs (1803 DEG up-regulated and 923 DEGs down-regulated) were detected. In the int-c mutant, a total of 906 DEGs were identified in response to shade, with 226 up-regulated genes and 680 down-regulated genes (Fig. 6; Supplementary Table S3). DEGs detected by microarray were validated and confirmed for five up-regulated and five down-regulated genes by qRT–PCR (Supplementary Fig. S3). The number of DEGs in response to shading decreased by ~67% in int-c (Fig. 6; Supplementary Table S3). This drop supports the hypothesis of the involvement of INT-C in the shade response. The overlapping DEGs detectable in the wild type and int-c are related to a common SAR independent of INT-C, while DEGs detectable in the wild type could directly or indirectly depend on INT-C function. These 2154 DEGs were termed INT-C-dependent genes of the shade response (Fig. 6; Supplementary Table S3).
Fig. 6.
Venn diagram of differentially expressed genes (DEGs) detected after shade treatment in cv. Bowman wild type (Wt shade) and int-c (int-c shade). These blocks were compared with DEGs detectable after decapitation (decap) in the int-c mutant and wild type (int-c versus WT). Numbers indicate transcript fold changes ≥2 at an FDR of P<0.05. (shade, INT-C mRNA induced; int-c, no functional INT-C; decap, INT-C mRNA reduced).
In Bowman buds, the up-regulation of INT-C after shading [2.56-fold increase at a false discovery rate (FDR) of 4.78E-04] could be confirmed. In the BW421 (int-c.5) deletion mutant, a similar induction of INT-C (2.10-fold increase, FDR=0.0086) was detectable (Supplementary Table S3). However, in the int-c.5 mutant, the deletion in the INT-C gene leads to a frameshift in the C-terminus downstream of the R-domain. This mutation results in a non-functional gene product. The inspection of the respective marker genes HAT4/ATHB2, PIF3, and PIF4 (Leivar and Monte, 2014) by their transcriptional activation validated the applied experimental shade conditions (Supplementary Table S3). Among the significantly up-regulated genes after shade in the wild type, transcript levels of the marker gene DRM1 that is associated with tiller bud dormancy (Stafstrom et al., 1998; Tatematsu et al., 2005) were found to be >2-fold higher (Supplementary Table S3).
As shown in Fig. 5A, the shade response led to a transcriptional activation of INT-C and a repression of tiller outgrowth. In contrast, the decapitation of barley plants resulted in a temporal reduced expression of INT-C and the opposite phenotype. As INT-C expression was reduced under this simulated condition, the generated dataset was used to validate the list of INT-C-dependent shade response genes (Supplementary Table S3).
The Venn diagram (Fig. 6) illustrates 753 overlapping DEGs after decapitation and after shading (27% of 2726 DEGs after shading and 44% of 1704 DEGs after decapitation). A total of 495 (29%) of the decapitation-induced DEGs were also detectable in response to shading. Almost all of these DEGs were oppositely regulated after decapitation and shading, respectively (Supplementary Table S4). This list was defined as INT-C dependent genes.
MapMan software was used to define gene functional categories (Thimm et al., 2004; Usadel et al., 2005). For the INT-C-dependent genes, we identified hormone- and stress-related genes among the up-regulated genes, and cell division- and protein synthesis-related genes among the down-regulated genes as the most prominent functional categories.
As the majority of INT-C-dependent genes showed opposite responses after decapitation and under shade, these gene sets infer the causal mechanisms associated with bud activation and bud arrest, respectively. A total of 123 genes (25%) found to be up-regulated after decapitation were down-regulated under shade, while 372 (75%) genes which were down-regulated after decapitation were up-regulated under shade (Fig. 6; Supplementary Table S4). During decapitation-induced bud activation, INT-C was rapidly down-regulated (Fig. 3B). Further, a strong overlap of DEGs responding to decapitation and shading was observed. Theoretically, the 372 down-regulated (after decapitation) genes might be directly involved in the promotion of axillary bud arrest. This group included a number of genes related to ethylene, auxin, and gibberellin signaling (AP2/ERBP, ACC, ERF1, IAA17, and GID1L2), as well as protein degradation (SKIP1, SKIP5, SKP2A, UBQ3, and UBQ4). Moreover, genes related to sugar metabolism and transport were also identified within that group (TPS6, sucrose transporter). Trehalose-6-phosphate is known to be involved in sugar signaling (Figueroa and Lunn, 2016). On the other hand, the 123 up-regulated genes (after decapitation) could be involved in promoting axillary bud growth. This group included many genes associated with chloroplast function and chlorophyll synthesis (chlorophyll-binding protein, ATP synthase), protein synthesis (EF-Ts, ribosomal protein), and chromatin structure (HISTONE H3.2, HISTONE 2B.3). This set of genes was co-regulated with a subset of genes related to functional categories involving the thylakoid and photosynthesis (Supplementary Fig. S4).
Discussion
The present study shows that INT-C is transcriptionally regulated by apical dominance and by light perception, and that INT-C expression in tiller buds regulates tillering in response to these signals. This highlights the role of INT-C as a major transcriptional regulator integrating endogenous with environmental signals to determine the outgrowth of tiller buds. Based on the microarray hybridization experiment, INT-C-dependent genes are defined.
INT-C integrates environmental signals to regulate tiller bud outgrowth
In crop plants, tillering is an important agronomic trait for yield formation. However, in barley, little is known about the genetic mechanisms acting inside tiller buds to cause growth arrest. Although a quantitative trait locus (QTL) study revealed several genes associated with tiller number (Haaning et al., 2020), the fact that some tillering genes do not overlap with identified QTLs point toward the complex involvement of environmental influence. Here, we show that the bHLH transcription factor gene INT-C is involved in the integration of different branching signals mediating a suppressive effect on bud outgrowth (Fig. 3). INT-C itself is regulated on the transcriptional level as a response of the investigated environmental conditions. This modulation of INT-C transcription appears to be under tight regulation of environmental and developmental stimuli that are correlated with bud outgrowth and activation of tillering. INT-C up-regulation was observed after shading or under high planting densities (Figs 3, 4), linked to reduced tiller numbers; in contrast, suppression of INT-C expression (i.e. after decapitation) triggered bud outgrowth (Fig. 5). This emphasizes a role for INT-C as a regulator integrating signals within the axillary bud to determine tiller number. This finding agrees with the results in rice where TB1 was also reported to mediate a negative function in tillering (Takeda et al., 2003; Choi et al., 2012). A comparable approach using transgenic overexpression and antisense-mediated repression of TB1 resulted in the modulation of tiller and panicle development in rice. In rice, the duplication of Tb1 and neofunctionalization of Tb2 resulted in a novel regulatory hub for tiller development (Lyu et al., 2020). Such a hub was not detectable in barley.
The influence and integration of environmental stimuli in rice were only addressed in response to greenhouse and paddy field conditions. In maize, the involvement of the SPL gene family as direct regulators of TB1 in plant architectural traits was described, and optimized plants were engineered for high-density planting (Wei et al., 2018). Whether directed modification of INT-C can help to improve the yield of barley plants is very difficult to predict. As the association of tiller formation and yield or fertile tillers is also dependent on the developmental stage and environmental condition, only a modulation of INT-C function might be helpful. As INT-C was not detected among the genes in tiller QTLs (Haaning et al., 2020), an easy association of the presence or absence of the gene with yield or fertile tillers is not possible.
As the shade-induced reduction of tiller number was observed in both wild-type and mutant plants (Fig. 5), it can be concluded that the underlying regulation is not solely mediated by INT-C. Also, other factors might contribute to the integration of the environmental signals.
Definition of INT-C-dependent genes
The analysis of the shading response at the transcriptome level led to the identification of important DEGs that were directly or indirectly dependent on INT-C. Our data suggest that the number of INT-C-dependent genes (372 up-regulated and 123 down-regulated) was closely associated with tiller bud activation, confirmed through int-c mutant study and INT-C function inferred from the stimulus experiments (either shade or decapitation). These target genes are promising candidates to play an important role in tiller bud transition between repression and promotion of growth. The MAPMAN-based categorization of DEGs resulted in a significant over-representation of the term thylakoid and genes associated with photosynthesis. In addition to this, several target genes of sink/source properties of a tissue are found (e.g. TPS6, sucrose transporters). This supports the idea of bud activation, accompanied by generation of a new initial sink tissue. Also, photosynthesis will be activated in this new tiller. The scheme in Fig. 7 shows the integration of the respective categories and important genes involved in INT-C-mediated bud growth.
Fig. 7.
Working model for the dynamic balance of INT-C-dependent transcriptional programming to regulate tiller bud outgrowth in barley. Shade perception occurring in tiller buds activates the ‘master-switch’ transcription factor INT-C, thus altering INT-C-dependent target genes; among them is a set of up-regulated hormone and stress response genes and down-regulated cell division- and ribosome-related genes. The output of this transcriptional regulation mediates bud transition from outgrowth to arrest.
ABA signaling and the promotion of bud dormancy
ABA has been related to the maintenance and promotion of bud dormancy in many plant species: elevated ABA levels in buds are associated with the inhibition of branching during plant development (Tamas et al., 1979; Knox and Wareing, 1984; Gocal et al., 1991; Mader et al., 2003; Destefano-Beltran et al., 2006; Ruttink et al., 2007), as well as in the context of responses to low R:FR ratios (Tucker and Mansfield, 1971; Reddy et al., 2013; González-Grandio et al., 2017). Moreover, a correlation has been found between the up-regulation of ABA response genes in axillary buds and bud dormancy (Ruttink et al., 2007; González-Grandio et al., 2013; Kebrom and Mullet, 2016). So far, ABA measurements have not been reported in tiller buds, and neither have ABA-inducible genes been studied in the tiller bud development of barley (Hussien et al., 2014). Nonetheless, it has been presumed that in barley a correlation exists between ABA signaling and bud arrest. Although the precise role of ABA in the promotion of bud dormancy in Arabidopsis is still not clear, a strong overlap of common regulated genes can be found (González-Grandio et al., 2013). Recently, Luo et al. (2019) have shown that application of ABA to hydroponic cultures of rice SL mutants and wild-type plants suppressed axillary bud outgrowth. SL and ABA biosynthesis share the same precursor and are closely related in regulating the tiller number in barley (Wang et al., 2018). In the present study, we demonstrate that INT-C as a regulatory hub controls various ABA hormone pathway genes as well decapitation and shade responses. The large number of transcription factors found in the list of defined INT-C-dependent genes also points toward a function of INT-C as a regulatory hub controlling a complex downstream network. Our transcript data provide evidence that wild-type tiller buds display a strong increase in the global response of ABA-related genes, while int-c buds, which exhibit less dormancy and continue to grow, show reduced ABA-related responses. Consistently, the number of differentially expressed ABA-related genes in the tiller buds was higher after shading. These findings are subsumed in the model (Fig. 7), building the hypothesis that INT-C employs the ABA signaling pathway in conjunction with other hormones to mediate bud growth arrest.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Flowering in wild-type and int-c mutant plants at 15 weeks after germination.
Fig. S2. Validation of microarray data by qRT–PCR. mRNA levels of a subset of genes identified as responding to the decapitation treatment, and Pearson’s correlations between gene expression levels determined by qRT–PCR and microarray expression profiling for the same genes.
Fig. S3. Validation of microarray data by qRT–PCR. mRNA levels of a subset of genes identified as responding to the shade treatment in both wild-type and int-c samples, and Pearson’s correlations between gene expression levels determined by qRT–PCR and microarray expression profiling for the same genes.
Fig. S4. MAPMAN analysis of DEGs for regulation overview, biotic stress, chloroplast, and metabolism overview.
Table S1. List of the primer sequences used in this study.
Table S2. List of TCP genes and associated gene expression derived from public databases.
Table S3. List of differentially expressed genes (DEGs) for the wild-type versus int-c in control, shading, and decapitation condition.
Table S4. List of INT-C-dependent genes, including comparison of direction after shading and decapitation.
Acknowledgements
We thank Barbara Kettig, IPK Gatersleben, for excellent technical assistance.
Author contributions
HW: planning and design; HW, CS, and KE: performing experiments and data analysis; HW, MK, and NvW: conceptualization; HW and MK: writing.
Conflict of interest
The authors declare no competing interests.
Funding
This work was supported by IZN (Interdisciplinary Centre for Crop Plant Research), Halle (Saale), Saxony-Anhalt, Germany and the Leibniz Graduate School ‘Yield Formation in cereals—overcoming yield-limiting factors’ IPK.
Data availability
The datasets generated during and/or analysed during the current study are available in the e!DAL repository: https://doi.ipk-gatersleben.de/DOI/73ddd8d1-046c-4934-80fa-189a9cf0bdc8/dc282984-1535-465a-a7e0-9b61fa831fcf/2/1847940088
References
- Aguilar-Martínez JA, Poza-Carrión C, Cubas P.. 2007. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. The Plant Cell 19, 458–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL. 1999. Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends in Plant Science 4, 201. [DOI] [PubMed] [Google Scholar]
- Braun N, de Saint Germain A, Pillot JP, et al. 2012. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiology 158, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, et al. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55, 611–622. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Sanchez RA, Deregibus VA.. 1986. The effect of plant-density on tillering—the involvement of R/Fr ratio and the proportion of radiation intercepted per plant. Environmental and Experimental Botany 26, 365–371. [Google Scholar]
- Chen M, Chory J, Fankhauser C.. 2004. Light signal transduction in higher plants. Annual Review of Genetics 38, 87–117. [DOI] [PubMed] [Google Scholar]
- Choi MS, Woo MO, Koh EB, Lee J, Ham TH, Seo HS, Koh HJ.. 2012. Teosinte Branched 1 modulates tillering in rice plants. Plant Cell Reports 31, 57–65. [DOI] [PubMed] [Google Scholar]
- Cline M. 1997. Concepts and terminology of apical dominance. American Journal of Botany 84, 1064. [PubMed] [Google Scholar]
- Cline MG. 2000. Execution of the auxin replacement apical dominance experiment in temperate woody species. American Journal of Botany 87, 182–190. [PubMed] [Google Scholar]
- Destefano-Beltrán L, Knauber D, Huckle L, Suttle J.. 2006. Chemically forced dormancy termination mimics natural dormancy progression in potato tuber meristems by reducing ABA content and modifying expression of genes involved in regulating ABA synthesis and metabolism. Journal of Experimental Botany 57, 2879–2886. [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA.. 2003. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiology 133, 1617–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley J, Stec A, Hubbard L.. 1997. The evolution of apical dominance in maize. Nature 386, 485–488. [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C.. 2005. bHLH class transcription factors take centre stage in phytochrome signalling. Trends in Plant Science 10, 51–54. [DOI] [PubMed] [Google Scholar]
- Figueroa CM, Lunn JE.. 2016. A tale of two sugars: trehalose 6-phosphate and sucrose. Plant Physiology 172, 7–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH.. 2010. Phytochrome functions in Arabidopsis development. Journal of Experimental Botany 61, 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gocal GF, Pharis RP, Yeung EC, Pearce D.. 1991. Changes after decapitation in concentrations of indole-3-acetic acid and abscisic acid in the larger axillary bud of Phaseolus vulgaris L. cv tender green. Plant Physiology 95, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E, Pajoro A, Franco-Zorrilla JM, Tarancón C, Immink RG, Cubas P.. 2017. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proceedings of the National Academy of Sciences, USA 114, E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Grandío E, Poza-Carrión C, Sorzano CO, Cubas P.. 2013. BRANCHED1 promotes axillary bud dormancy in response to shade in Arabidopsis. The Plant Cell 25, 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaning AM, Smith KP, Brown-Guedira GL, Chao S, Tyagi P, Muehlbauer GJ.. 2020. Natural genetic variation underlying tiller development in barley (Hordeum vulgare L). G3 10, 1197–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall SM, Hillman JR.. 1975. Correlative inhibition of lateral bud growth in Phaseolus vulgaris L. Timing of bud growth following decapitation. Planta 123, 137–143. [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, et al. 2012. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. The Plant Journal 71, 699–711. [DOI] [PubMed] [Google Scholar]
- Howarth DG, Donoghue MJ.. 2006. Phylogenetic analysis of the ‘ECE’ (CYC/TB1) clade reveals duplications predating the core eudicots. Proceedings of the National Academy of Sciences, USA 103, 9101–9106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussien A, Tavakol E, Horner DS, Munoz-Amatriain M, Muehlbauer GJ, Rossini L.. 2014. Genetics of tillering in rice and barley. Plant Genome 7, 226–256. [Google Scholar]
- Kang YH, Khan S, Ma XY.. 2009. Climate change impacts on crop yield, crop water productivity and food security—a review. Progress in Natural Science-Materials International 19, 1665–1674. [Google Scholar]
- Kebrom TH, Brutnell TP, Finlayson SA.. 2010. Suppression of sorghum axillary bud outgrowth by shade, phyB and defoliation signalling pathways. Plant, Cell & Environment 33, 48–58. [DOI] [PubMed] [Google Scholar]
- Kebrom TH, Burson BL, Finlayson SA.. 2006. Phytochrome B represses Teosinte Branched1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiology 140, 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Mullet JE.. 2016. Transcriptome profiling of tiller buds provides new insights into PhyB regulation of tillering and indeterminate growth in Sorghum. Plant Physiology 170, 2232–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kebrom TH, Spielmeyer W, Finnegan EJ.. 2013. Grasses provide new insights into regulation of shoot branching. Trends in Plant Science 18, 41–48. [DOI] [PubMed] [Google Scholar]
- Kegge W, Weldegergis BT, Soler R, Eijk MV, Dicke M, Voesenek LACJ, Pierik R.. 2013. Canopy light cues affect emission of constitutive and methyl jasmonate-induced volatile organic compounds in Arabidopsis thaliana. New Phytologist 200, 861–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox JP, Wareing PF.. 1984. Apical dominance in Phaseolus vulgaris L—the possible roles of abscisic and indole-3-acetic-acid. Journal of Experimental Botany 35, 239–244. [Google Scholar]
- Kong L, Abrams SR, Owen SJ, Graham H, von Aderkas P.. 2008. Phytohormones and their metabolites during long shoot development in Douglas-fir following cone induction by gibberellin injection. Tree Physiology 28, 1357–1364. [DOI] [PubMed] [Google Scholar]
- Koppolu R, Anwar N, Sakuma S, et al. 2013. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proceedings of the National Academy of Sciences, USA 110, 13198–13203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y.. 1997. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. The Plant Cell 9, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E.. 2014. PIFs: systems integrators in plant development. The Plant Cell 26, 56–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S. 2015. The Arabidopsis thaliana TCP transcription factors: a broadening horizon beyond development. Plant Signaling & Behavior 10, e1044192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liller CB, Neuhaus R, von Korff M, Koornneef M, van Esse W.. 2015. Mutations in barley row type genes have pleiotropic effects on shoot branching. PLoS One 10, e0140246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E.. 1996. Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799. [DOI] [PubMed] [Google Scholar]
- Luo L, Takahashi M, Kameoka H, Qin R, Shiga T, Kanno Y, Seo M, Ito M, Xu G, Kyozuka J.. 2019. Developmental analysis of the early steps in strigolactone-mediated axillary bud dormancy in rice. The Plant Journal 97, 1006–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J, Huang L, Zhang S, et al. 2020. Neo-functionalization of a Teosinte branched 1 homologue mediates adaptations of upland rice. Nature Communications 11, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader JC, Emery RJN, Turnbull CGN.. 2003. Spatial and temporal changes in multiple hormone groups during lateral bud release shortly following apex decapitation of chickpea (Cicer arietinum) seedlings. Physiologia Plantarum 119, 295–308. [Google Scholar]
- Martín-Trillo M, Cubas P.. 2010. TCP genes: a family snapshot ten years later. Trends in Plant Science 15, 31–39. [DOI] [PubMed] [Google Scholar]
- Martín-Trillo M, Grandío EG, Serra F, Marcel F, Rodríguez-Buey ML, Schmitz G, Theres K, Bendahmane A, Dopazo H, Cubas P.. 2011. Role of tomato BRANCHED1-like genes in the control of shoot branching. The Plant Journal 67, 701–714. [DOI] [PubMed] [Google Scholar]
- Minakuchi K, Kameoka H, Yasuno N, et al. 2010. FINE CULM1 (FC1) works downstream of strigolactones to inhibit the outgrowth of axillary buds in rice. Plant & Cell Physiology 51, 1127–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli CA, Beveridge CA, Snowden KC.. 1999. Reevaluating concepts of apical dominance and the control of axillary bud outgrowth. Current Topics in Developmental Biology 44, 127–169. [DOI] [PubMed] [Google Scholar]
- Nath U, Crawford BC, Carpenter R, Coen E.. 2003. Genetic control of surface curvature. Science 299, 1404–1407. [DOI] [PubMed] [Google Scholar]
- Ori N, Cohen AR, Etzioni A, et al. 2007. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nature Genetics 39, 787–791. [DOI] [PubMed] [Google Scholar]
- Ozhuner E, Eldem V, Ipek A, Okay S, Sakcali S, Zhang B, Boke H, Unver T.. 2013. Boron stress responsive microRNAs and their targets in barley. PLoS One 8, e59543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatnik JF, Allen E, Wu X, Schommer C, Schwab R, Carrington JC, Weigel D.. 2003. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. [DOI] [PubMed] [Google Scholar]
- Palatnik JF, Wollmann H, Schommer C, et al. 2007. Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Developmental Cell 13, 115–125. [DOI] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R.. 2005. InterProScan: protein domains identifier. Nucleic Acids Research 33, W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay L, Comadran J, Druka A, et al. 2011. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nature Genetics 43, 169–172. [DOI] [PubMed] [Google Scholar]
- R Core Team. 2013. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Reddy SK, Holalu SV, Casal JJ, Finlayson SA.. 2013. Abscisic acid regulates axillary bud outgrowth responses to the ratio of red to far-red light. Plant Physiology 163, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J.. 1993. Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. The Plant Cell 5, 147–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttink T, Arend M, Morreel K, Storme V, Rombauts S, Fromm J, Bhalerao RP, Boerjan W, Rohde A.. 2007. A molecular timetable for apical bud formation and dormancy induction in poplar. The Plant cell 19, 2370–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler C, Harshavardhan VT, Rajesh K, Reddy PS, Strickert M, Rolletschek H, Scholz U, Wobus U, Sreenivasulu N.. 2011. ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought-stress conditions. Journal of Experimental Botany 62, 2615–2632. [DOI] [PubMed] [Google Scholar]
- Skinner RH, Simmons SR.. 1993. Modulation of leaf elongation, tiller appearance and tiller senescence in spring barley by far-red light. Plant, Cell & Environment 16, 555–562. [Google Scholar]
- Smith H. 2000. Phytochromes and light signal perception by plants—an emerging synthesis. Nature 407, 585–591. [DOI] [PubMed] [Google Scholar]
- Smith JE, Jordan PW.. 1994. Stand density effects on branching in an annual legume (Senna obtusifolia). Annals of Botany 74, 17–25. [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N, Harshavardhan VT, Govind G, Seiler C, Kohli A.. 2012. Contrapuntal role of ABA: does it mediate stress tolerance or plant growth retardation under long-term drought stress? Gene 506, 265–273. [DOI] [PubMed] [Google Scholar]
- Stafstrom JP, Ripley BD, Devitt ML, Drake B.. 1998. Dormancy-associated gene expression in pea axillary buds. Cloning and expression of PsDRM1 and PsDRM2. Planta 205, 547–552. [DOI] [PubMed] [Google Scholar]
- Stamm P, Kumar PP.. 2010. The phytohormone signal network regulating elongation growth during shade avoidance. Journal of Experimental Botany 61, 2889–2903. [DOI] [PubMed] [Google Scholar]
- Takeda T, Suwa Y, Suzuki M, Kitano H, Ueguchi-Tanaka M, Ashikari M, Matsuoka M, Ueguchi C.. 2003. The OsTB1 gene negatively regulates lateral branching in rice. The Plant Journal 33, 513–520. [DOI] [PubMed] [Google Scholar]
- Tamas IA, Ozbun JL, Wallace DH.. 1979. Effect of fruits on dormancy and abscisic acid concentration in the axillary buds of Phaseolus vulgaris L. Plant Physiology 64, 615–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S.. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30, 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133, 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Ward S, Leyser O, Kamiya Y, Nambara E.. 2005. Identification of cis-elements that regulate gene expression during initiation of axillary bud outgrowth in Arabidopsis. Plant Physiology 138, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M.. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Thirulogachandar V, Alqudah AM, Koppolu R, et al. 2017. Leaf primordium size specifies leaf width and vein number among row-type classes in barley. The Plant Journal 91, 601–612. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG.. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ.. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Liu Y, Yan H, You Q, Yi X, Du Z, Xu W, Su Z.. 2017. agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Research 45, W122–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucic B, Ducic J, Pemac D.. 2006. Phenotypic responses to early signals of neighbour proximity in Picea omorika, a pioneer conifer tree. Basic and Applied Ecology 7, 443–454. [Google Scholar]
- Tucker DJ, Mansfield TA.. 1971. Effects of light quality on apical dominance in Xanthium strumarium and the associated changes in endogenous levels of abscisic acid and cytokinins. Planta 102, 140–151. [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, et al. 2005. Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiology 138, 1195–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang H.. 2015. Phytochrome signaling: time to tighten up the loose ends. Molecular Plant 8, 540–551. [DOI] [PubMed] [Google Scholar]
- Wang H, Wu GX, Zhao BB, Wang BB, Lang ZH, Zhang CY, Wang HY.. 2016. Regulatory modules controlling early shade avoidance response in maize seedlings. BMC Genomics 17, 269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Chen W, Eggert K, et al. 2018. Abscisic acid influences tillering by modulation of strigolactones in barley. Journal of Experimental Botany 69, 3883–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei H, Zhao Y, Xie Y, Wang H.. 2018. Exploiting SPL genes to improve maize plant architecture tailored for high-density planting. Journal of Experimental Botany 69, 4675–4688. [DOI] [PubMed] [Google Scholar]
- Whipple CJ, Kebrom TH, Weber AL, Yang F, Hall D, Meeley R, Schmidt R, Doebley J, Brutnell TP, Jackson DP.. 2011. grassy tillers1 promotes apical dominance in maize and responds to shade signals in the grasses. Proceedings of the National Academy of Sciences, USA 108, E506–E512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Ma H, Wang J, Zhang DB.. 2007. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. Journal of Integrative Plant Biology 49, 885–897. [Google Scholar]
- Zadoks JC, Chang TT, Konzak CF.. 1974. A decimal code for the growth-stages of cereals. Weed Research 14, 415–421. [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W.. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136, 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the e!DAL repository: https://doi.ipk-gatersleben.de/DOI/73ddd8d1-046c-4934-80fa-189a9cf0bdc8/dc282984-1535-465a-a7e0-9b61fa831fcf/2/1847940088