The extensive regulation of HSP70 has received little attention in plant science. Here, we summarize current knowledge gained in large-scale profiling experiments and highlight HSP70s’ role in plant immunity.
Keywords: Biotic interactions, cytokinin, HSP70, immunity, plant defense, phytohormone
Abstract
Heat shock proteins 70 (HSP70s) are steadily gaining more attention in the field of plant biotic interactions. Though their regulation and activity in plants are much less well characterized than are those of their counterparts in mammals, accumulating evidence indicates that the role of HSP70-mediated defense mechanisms in plant cells is indispensable. In this review, we summarize current knowledge of HSP70 post-translational control in plants. We comment on the phytohormonal regulation of HSP70 expression and protein abundance, and identify a prominent role for cytokinin in HSP70 control. We outline HSP70s’ subcellular localizations, chaperone activity, and chaperone-mediated protein degradation. We focus on the role of HSP70s in plant pathogen-associated molecular pattern-triggered immunity and effector-triggered immunity, and discuss the contribution of different HSP70 subfamilies to plant defense against pathogens.
Introduction
Heat shock proteins (HSPs) were discovered by Italian scientist Ferruccio Ritossa who noticed a rapid transcriptional response to heat shock in Drosophila melanogaster and eventually published his results in 1962 (see Ritossa, 1996). The term ‘heat shock proteins’ was adopted 12 years later (Tissiéres et al., 1974). What are HSPs? These ubiquitous and widespread proteins across prokaryotic and eukaryotic organisms were first discovered in response to an increase in temperature, but accumulated evidence indicates that these proteins are involved in much more than that. Foremost, HSPs act as chaperones and are involved in the assembly, stabilization, and maturation of proteins and protein complexes (Wang et al., 2004). Besides their chaperone functions, HSPs play a role in plant development and response to abiotic and biotic stress conditions, including drought, salinity, pathogen infection, and insect attacks (Park and Seo, 2015; Usman et al., 2017; Ul Haq et al., 2019). The HSP family covers a variety of proteins, ranging from 10 kDa to >100 kDa, and the molecular weight is the basis of present-day HSP nomenclature (Ratheesh Kumar et al., 2012). Several recent publications have reviewed the role of plant HSP90 (Tichá et al., 2020), HSP40 (Verma et al., 2019), and small HSPs (Asea et al., 2016; Waters and Vierling, 2020). Here, we will outline the role of HSP70s, one of the most studied HSPs in animals, and discuss their somewhat overlooked roles in plants.
HSP70 structure is evolutionarily conserved
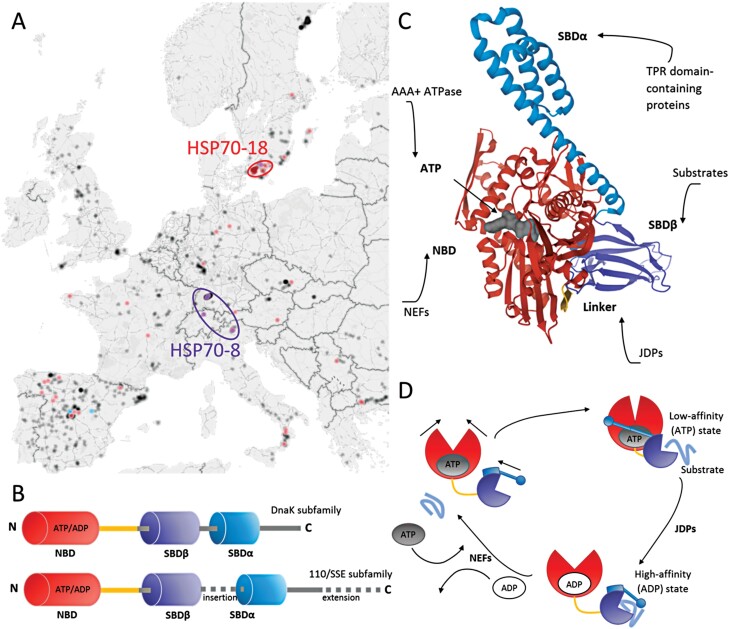
HSP70s are highly conserved ATP-dependent chaperones, and this family shows the highest sequence similarity within a large evolutionary distance (Borchiellini et al., 1998). An example is DnaK protein (prokaryotic HSP70), which shares ~50% amino acid identity with eukaryotic HSP70s. The genetic conservation of the HSP70 sequence is also evident in Arabidopsis accessions. The frequency of mutations is low, seems to correlate with geographical distribution (Fig. 1A), and 10 proteins show zero high-impact mutations (Table 1). Plant HSP70s share at least two of the four structural features of the archetype bacterial HSP70 DnaK: an N-terminal, 45 kDa nucleotide-binding domain (NBD), followed by a 15 kDa substrate-binding domain (SBDβ), a 10 kDa helical lid domain (SBDα), and a disordered C-terminal tail of variable length (Fig. 1C; Sarkar et al., 2013). In eukaryotic cytosolic and nuclear HSP70s, the disordered C-terminal tail frequently ends with a characteristic charged motif (EEVD) that interacts with specific co-chaperones containing TPR repeats, including HSP-organizing protein (HOP; transfer of unfolded HSP70 substrates to foldases; Rosenzweig et al., 2019), HSP-interacting protein (HIP; stabilization of the high-affinity ADP-bound state of HSP70; Zuiderweg et al., 2017), and C-terminus of HSP70-interacting protein (CHIP; ubiquitination mediating degradation of unfolded HSP70 substrates via the proteasome; Fernández-Fernández et al., 2017).
Fig. 1.
HSP70 sequence conservation, domain structure, and chaperone activity. (A) The frequency of mutations in HSP70-8 and HSP70-18 coincides with geographical location. The geographical distribution of all Arabidopsis accessions collected in the 1001 genome database (black) overlaid with the occurrence of high-impact mutations in HSP70-8 (purple), HSP70-13 (blue, no significant clustering), and HSP70-18 (red). Data retrieved from the 1001 genome database (Weigel and Mott, 2009) and visualized using a Microsoft Excel 3D map. (B) Domain organization of DnaK and 110/SSE subfamilies (Lin et al., 2001; Sarkar et al., 2013). (C) Structure of HSP70 with bound ATP. Modeled using the crystal structure of bacterial DnaK (PDB ID: 4B9Q; Kityk et al., 2012). Red, nucleotide-binding domain (NBD); blue, substrate-binding domain SBDα; violet, substrate-binding domain SBDβ; gray, ATP; yellow, linker. Sites of protein–protein interactions with HSP70 co-chaperones are indicated, including tetratricopeptide repeat (TPR) proteins, J-domain proteins (JDP), and nucleotide exchange factors (NEF). (D) Chaperone cycle of HSP70s. The interaction with JDPs stimulates ATP hydrolysis and the transition to the ADP-bound state with a high affinity for protein substrates. NEFs facilitate the HSP70 conversion back to the low-affinity state, the dissociation of ADP and rebinding of ATP, and the release of the protein substrate (Rosenzweig et al., 2019).
Table 1.
Overview of the HSP70 family in the model plant Arabidopsis
| UniProt | Protein names | AGI | SUBA/UniProt (∗) detected in extracellular space | Mutant phenotype | No. of gene models | High-impact mutations in 1001 genomes database |
|---|---|---|---|---|---|---|
| P22953 | HSP70-1 | AT5G02500 | Cytosol, nucleus∗ | A: altered plant growth and development (1), disabled immune response (2, 3), enhanced tolerance to heat shock, hypersensitive to ABA, compromised in the induced stomatal closure (2), γ-ray hypersensitivity and tolerance to salt, cadmium, and arsenic (4) B: did not reveal any difference in the resistance against pathogens Hyaloperonospora parasitica and Pseudomonas syringae pv tomato (3), more susceptible to P. syringae pv. Maculicola ΔhopI (5) |
2 | 1 (SNPs) |
| Q9S7C0 | HSP70-14 | AT1G79930 | Cytosol, nucleus | B: indistinguishable from the wild type (6) | 2 | 4 (SNPs) |
| Q9SAB1 | HSP70-16 | AT1G11660 | Cytosol, nucleus | B: failed flower opening, abnormal floral organ formation, impaired fertilization and seed setting (7), suppressed seed germination under cold stress conditions (8) | 4 | 1 (SNPs) |
| P22954 | HSP70-2 | AT5G02490 | Cytosol, nucleus | B: did not reveal any difference in the resistance against pathogens Hyaloperonospora parasitica and Pseudomonas syringae pv tomato (3), more susceptible to P. syringae pv. Maculicola Δhop (5) | 1 | 0 |
| O65719 | HSP70-3 | AT3G09440 | Cytosol, nucleus∗ | B: did not reveal any difference in the resistance against pathogens Hyaloperonospora parasitica and Pseudomonas syringae pv tomato (3) | 4 | 0 |
| Q9LHA8 | HSP70-4 | AT3G12580 | Cytosol, nucleus | C: abnormal embryogenesis, defective seedlings with high levels of ROS–RNAi in an hsc70-1 mutant background (9) | 1 | 0 |
| Q9S9N1 | HSP70-5 | AT1G16030 | Cytosol∗ | B: delayed growth under normal conditions, reduced survival of both seeds and seedlings after severe heat treatments, decreased growth activity under water deficit (10) | 1 | 0 |
| Q9SKY8 | HSP70-8 | AT2G32120 | Cytosol | 2 | 11 (SNPs) | |
| F4JMJ1 | HSP70-17 | AT4G16660 | Endoplasmic reticulum | 2 | 3 (SNPs) | |
| Q9LKR3 | HSP70-11 (BIP1) | AT5G28540 | Endoplasmic reticulum, nucleus∗ | B: BIP1+BIP2 double mutation has an effect on the fusion of polar nuclei during female gametophyte development (11) | 1 | 0 |
| Q39043 | HSP70-12 (BIP2) | AT5G42020 | endoplasmic reticulum, nucleus ∗ | B: compromised secretion of PR1 (12), enhanced fungal colonization (13), BIP1+ IP2 double mutation has an effect on the fusion of polar nuclei during female gametophyte development (11) | 3 | 0 |
| Q8H1B3 | HSP70-13 (BIP3) | AT1G09080 | Endoplasmic reticulum, nucleus | B: BIP1+BIP2+BIP3 triple mutation is pollen lethal (14) | 2 | 5 (SNPs), 1 (INS), 1 (DEL) |
| F4HQD4 | HSP70-15 | AT1G79920 | Golgi, cytosol, nucleus | B: severe growth retardation, impaired stomatal closure and accelerated wilting, enhanced tolerance to potyvirus infection (6) | 4 | 0 |
| Q9LDZ0 | HSP70-10 | AT5G09590 | Mitochondrion∗ | 1 | 0 | |
| Q8GUM2 | HSP70-9 | AT4G37910 | Mitochondrion, cytosol∗ | B: severe growth defects, abnormal mitochondria and alterations to respiration because of an inhibition of the cytochrome c oxidase (15) | 2 | 0 |
| Q9C7X7 | HSP70-18 | AT1G56410 | Plastid, cytosol∗ | 1 | 10 (SNPs), 7 (INS), 46 (DEL) | |
| Q9STW6 | HSP70-6 | AT4G24280 | Plastid∗ | B: variegated cotyledons, malformed leaves, growth retardation, impaired root growth (16), hsp70-6/hsp70-7 double mutants are lethal (17) | 1 | 0 |
| Q9LTX9 | HSP70-7 | AT5G49910 | Plastid | B: hsp70-6/hsp70-7 double-mutants are lethal (17), defective protein import into chloroplasts during early developmental stages (18) | 1 | 1 (SNPs) |
Based on UniProt (Bateman et al., 2017), SUBA 4.0 (Hooper et al., 2017), the 1001 genome database (Weigel and Mott, 2009), and the previously reported response of HSP70 overexpression (A), loss-of-function mutation (B), and RNAi (C): 1, Sung and Guy (2003); 2, Clément et al. (2011); 3, Noël et al. (2007); 4, Cazalé et al. (2009); 5, Jelenska et al. (2010); 6, Jungkunz et al. (2011); 7, Chen et al. (2019); 8 Ashraf et al. (2021); 9, Lee et al. (2009); 10, Kozeko (2021); 11, Maruyama et al. (2010); 12, Wang et al. (2005); 13, Qiang et al. (2012); 14, Maruyama et al. (2014); 15, Wei et al. (2019); 16, Su and Li (2008); 17, Latijnhouwers et al. (2010); 18, Su and Li (2010). SNP, single nucleotide polymorphism; INS, insertion; DEL, deletion.
Chaperone activity of HSP70s
The mediation of protein folding and translocation is the best-described function of HSP70s. The promiscuity of the HSP70 interaction and efficient targeting to substrate proteins are based on a transient interaction of the SBD domain (Fig. 1B–D) with a short degenerative sequence motif that is frequently found in virtually all proteins (Rüdiger et al., 1997; Kityk et al., 2012). The HSP70–substrate interaction depends on a complicated allosteric mechanism that couples ATP hydrolysis in the NBD with substrate capture by the SBD (described in more detail in Qi et al., 2013; Kityk et al., 2015; Rosenzweig et al., 2019). The HSP70 binds to protein substrates with a significantly increased affinity in the ADP-bound state, and ATP hydrolysis is essential for its chaperone function (Kityk et al., 2018). However, the intrinsic ATPase rate is very low (Kityk et al., 2012) and requires co-chaperones, namely J-domain proteins (JDPs; accelerate ATP hydrolysis) and nucleotide exchange factors (NEFs; accelerate ADP–ATP exchange) (Abrams et al., 2014; Bracher and Verghese, 2015; Zuiderweg et al., 2017). The whole chaperone cycle is illustrated in Fig. 1D. In contrast to other chaperones, HSP70s do not require self-interaction for the chaperone functions (Takakuwa et al., 2019).
Besides the described interactions and chaperone complexes that facilitate protein folding, HSP70s are essential for driving the movement of proteins across the membrane and protein translocation into the endoplasmic reticulum (ER), mitochondria, and plastids (Su et al., 2010; Schleiff and Becker, 2011; Nakai, 2018; Sun et al., 2021). Translocases have narrow channels, limiting the transport to a completely unfolded chain or, at most, α-helices (Craig, 2018). Interestingly, organellar HSP70s are more critical for the translocation process than their cytosolic counterparts, and form the core of the so-called import motors. There are different models for the HSP70 motor action mechanism that pulls the protein into an organelle (e.g. Sousa and Lafer, 2019), but the substrate–HSP70 interaction itself is identical to that of the standard chaperone cycle and requires JDP-driven ATP hydrolysis.
HSP70s mediate quality control
HSP70s have an essential role in protein quality control. The fate of misfolded and aggregated proteins is decided by a combination of HSP70 and its interactors, and the defective protein is targeted for degradation through the ubiquitin–proteasome system, as well as through autophagy pathways. The most frequent means of protein degradation in eukaryotic cells requires the ubiquitin–26S proteasome system. Proteins are targeted via ubiquitination through the sequential action of three discrete enzymes: the ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3). The E3 ligase is responsible for substrate recognition, and a highly conserved and chaperone-dependent E3 ligase is CHIP. This enzyme interacts with HSP70 and ubiquitinates protein substrates captured by HSP70. CHIP has been extensively studied in many model organisms, including plants (reviewed in, for example, Zhang et al., 2021), and accumulated knowledge suggests that it is an integral component of both biotic and abiotic stress control. Chaperone-mediated autophagy is mediated by cytosolic HSP70s and targets proteins containing the signal motif KFERQ (Fernández-Fernández et al., 2017). This was the first described chaperone-assisted pathway but requires a receptor that seems to be missing in plant lineages (Lescat et al., 2020). Similarly, the autophagy pathway mediated by endoplasmic HSP70 has not yet been confirmed in plants (Cha-Molstad et al., 2015).
Classification and localization of plant HSP70s
The plant HSP70 protein superfamily consists of two subfamilies: HSP70 (DnaK) and HSP110/SSE (Fig. 1B). The Arabidopsis genome encodes 14 and four members of HSP70 and HSP110/SSE subfamilies, respectively. HSP110/SSE exhibits homology to yeast SSE1 and human HSP110. The HSP110 sequence contains an insertion of acidic residues in the substrate-binding domain, an extension of the C-terminal domain, and the resulting protein is significantly larger (Fig. 1B).
All eukaryotes possess multiple HSP70s. In plants, four distinct sets function in the cytosol, mitochondria, chloroplasts, and endoplasmic reticulum (Table 1). This localization is determined by conserved N- and C-terminal sequences and is often used to divide HSP70s into four major subgroups: cytosolic/nuclear (EEVD motif), ER (HDEL motif), plastidic (PEGDVIDADFTDSK motif), and mitochondrial (PEAEYEEAKK motif) (Guy and Li, 1998). Those targeted to the same compartment share a similar evolutionary history. HSP70s targeted to the ER and cytosol resulted from gene duplication and subsequent divergence, and HSP70s targeted to mitochondria and chloroplasts evolved by gene transfer from endosymbionts and are more related to the prokaryotic DnaK (Waters and Rioflorido, 2007; Huang et al., 2008)
The low correlation between gene expression and protein abundance indicates extensive post-transcriptional or post-translational control of HSP70s
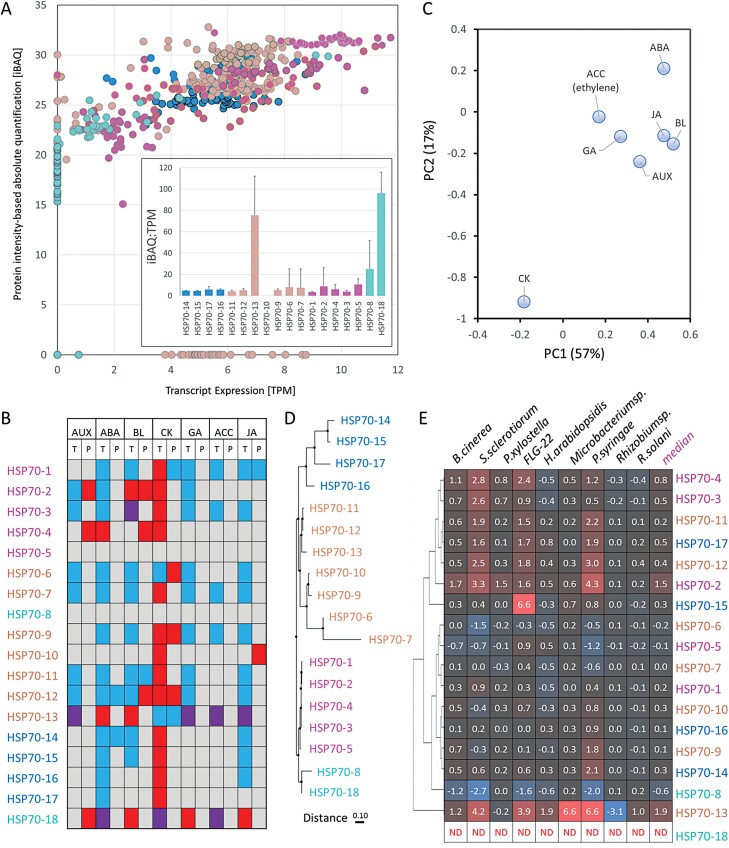
The analysis of available Arabidopsis protein data and expression profiles showed that only a minor fraction of HSP70s has a positive linear correlation between transcript and protein abundance (Fig. 2A). A higher correlation (Spearman correlation coefficient r >0.7) was found only for HSP70-4, HSP70-5, and HSP70-8. The highest deviations were found for HSP70-18 (expression data available only for imbibed seeds) and HSP70-10 (not found in the comparative dataset stored in the Arabidopsis tissue atlas). Translation elongation rates, post-transcriptional regulations, post-translational modifications, and protein life span impact the resulting protein abundances. Protein degradation rates have been estimated for seven Arabidopsis HSP70s (L. Li et al., 2017) and show significant differences. The lowest average degradation rates were found for plastid-localized isoforms HSP70-6 (0.069 d–1) and HSP70-7 (0.082 d–1), indicating an intermediate protein turnover rate. In contrast, cytosolic HSP70s showed a rapid degradation, with an average degradation rate of HSP70-14 reaching 1.878 d–1. Information about the post-transcriptional control of plant HSP70s is missing, but it has been demonstrated that HSP70 pseudogenes, including products of HSP70 alternative splicing, may act as long non-coding RNAs and interfere with the HSP70 translation (Bernabò et al., 2020). Arabidopsis HSP70-18 was considered a pseudogene, but the proteome analyses showed that this is not the case (Fig. 2A). There are 35 predicted HSP70 gene models, and only eight Arabidopsis HSP70 genes are missing putative alternatively spliced variants (Table 1). It is possible that at least some of these variants could participate in transcriptional control, but the sequences are highly similar. Except for AT1G79920.2, the available peptide-based evidence cannot confirm or exclude the existence of the corresponding proteins (van Wijk et al., 2021, Preprint).
Fig. 2.
Transcriptional and translational control of HSP70. (A) Gene expression and estimated protein abundance of HSP70s in Arabidopsis. Comparison of available profiles across a set of 30 matching tissues from the Arabidopsis tissue atlas (Mergner et al., 2020). The inset plot represents the average protein:transcript ratio with the SD. (B) Phytohormones regulate HSP70 biosynthesis. Simplified heat map visualization of reported transcriptional (T; ThaleMine; Krishnakumar et al., 2017) and protein (P; Černý et al., 2016) response to phytohormones. (C) Principal component analysis of the data in (B). Red, up-regulation/increase in protein abundance; blue, down-regulation, decrease in protein abundance; purple, mixed response; AUX, auxin; ABA, abscisic acid; BL, brassinosteroid; CK, cytokinin; GA, gibberellin; ACC, ethylene precursor; JA, jasmonate. (D) Sequence feature similarity visualized by Jalview (Waterhouse et al., 2009) and (E) Arabidopsis HSP70 genes in response to biotic interaction and flagellin peptide fragment (FLG-22; elicits defense response). The ratio represents the median log2 expression compared with mock-treated plants. Expression data were retrieved from the Arabidopsis RNA-Seq Database (Zhang et al., 2020).
Plant hormones control HSP70 expression
The phytohormones are small-molecule regulators that collectively regulate plant growth and development, and facilitate the integration of environmental cues, including biotic and abiotic stressors. It is thus not surprising to find Arabidopsis HSP70 among the list of phytohormone-responsive genes and proteins (Fig. 2B, C). Only two cytosolic HSP70s appear to be missing in response to phytohormones, namely HSP70-5 and HSP70-8. Somewhat counterintuitive is the reported response to hormones traditionally associated with a stress response. Abscisic acid, jasmonates, and ethylene showed a predominantly negative effect on HSP70 expression and the abundances of the corresponding proteins. Arabidopsis data for salicylic acid treatment were not available in the selected datasets, but previous proteomic analyses showed a decrease in HSP70 after salicylic acid treatment of Cucumis sativus and Cunninghaimia lanceolata (Černý et al., 2016). In contrast, cytokinin up-regulates most HSP70s and positively impacts HSP70 abundances (Fig. 2B). That could coincide with the predicted role of cytokinin in response to heat shock (Černý et al., 2014). Interestingly, abscisic acid and cytokinin antagonistically regulate many developmental processes (Huang et al., 2018), and the observed antagonistic effect on HSP70s (Fig. 2B, C) indicates that its regulation could be a part of the crosstalk between these two phytohormones.
HSP70s are subject to extensive post-translational control
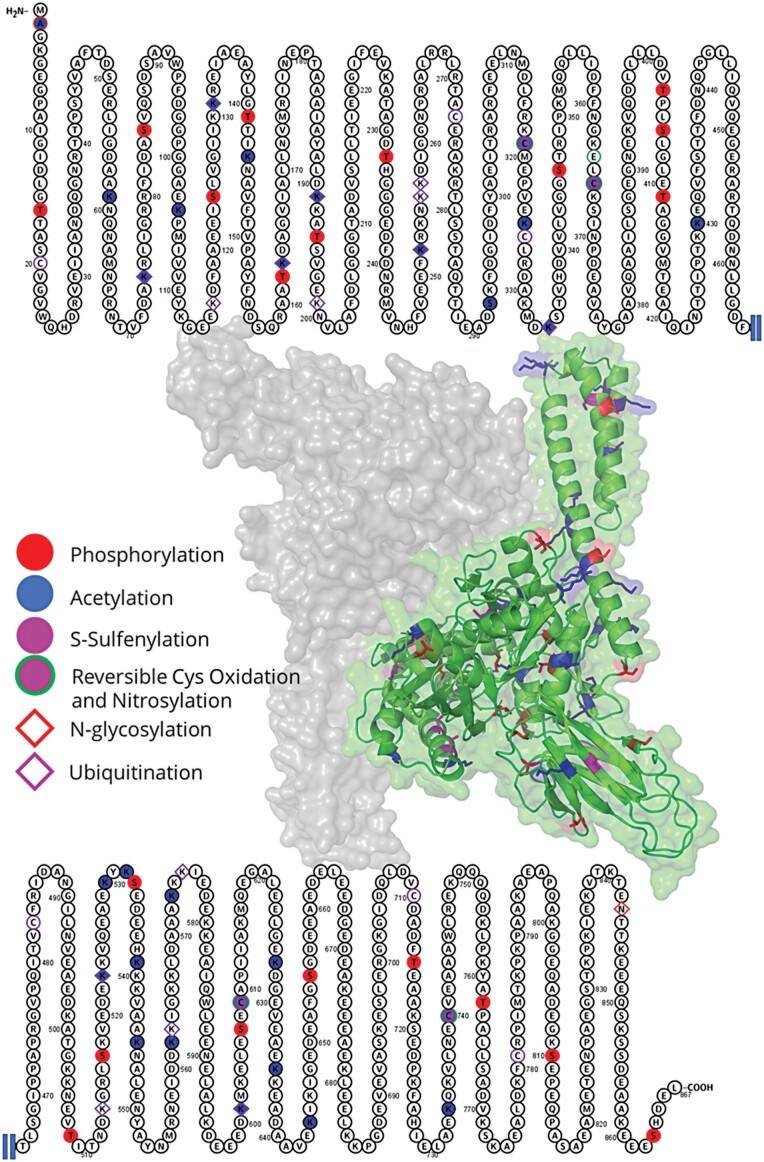
The Plant PTM viewer lists almost 300 HSP70 peptides with a post-translational modification (Willems et al., 2019). The most frequent modifications are phosphorylation (65 peptides), acetylation (53 peptides), sulfenylation (33 peptides), and ubiquitination (27 peptides). However, these are the most intensively studied plant protein modifications, and the apparent enrichment probably does not reflect the true proportion of different modifications. As illustrated in Fig. 3, the sites of these modifications are highly conserved and most can be mapped onto the consensus of HSP70 family sequences. Experimental evidence in plant protein science is largely missing, but HSP70 modifications have been established as the so-called ‘chaperone code’ and can determine protein localization, selectivity, activity, and protein–protein interactions (Cloutier and Coulombe, 2013; Nitika et al., 2020). These modifications represent both the fine-tuning of HSP70 biology and a means of rapid response to stress (Xu et al., 2019). The activation of HSP70 expression is rapid, but corresponding protein biosynthesis is significantly slower. For instance, the delay in heat-induced accumulation of Arabidopsis HSP70-4 and HSP70-5 is >2 h compared with the corresponding transcripts (Palmblad et al., 2008). Post-translational modifications are thus the first line of defense. Phosphorylation and acetylation have been shown to regulate the oligomerization of HSP70s (Morgner et al., 2015). The dimerization of HSP70s effectively buries its binding domains and inhibits protein–protein interactions (Takakuwa et al., 2019), and thus the regulation of the monomer/oligomer pool represents a rapid control mechanism to regulate the activity of HSP70s.
Fig. 3.
Reported post-translational modifications of Arabidopsis HSP70s mapped onto the consensus sequence. The consensus of the HSP70 sequence generated by Jalview (Waterhouse et al., 2009), visualized by Protter (Omasits et al., 2014), and projected into the 3D dimeric structure by SWISS-MODEL (Waterhouse et al., 2018). Data were retrieved from the Plant PTM Viewer database (Willems et al., 2019).
The Arabidopsis HSP70 sequences contain on average 1.2% cysteines, and sulfenylation and nitrosylation have been reported for these residues (Fig. 3). Cysteine is the most reactive proteinogenic amino acid, and thiols often act as a redox molecular switch regulating protein activity (e.g. Černý et al., 2013). In fact, sulfenylation of HSP70 has been demonstrated to activate heat shock transcription and induce thermotolerance in yeast (Wang et al., 2012). Interestingly, HSP70 may also directly participate in reactive oxygen metabolism by regulating the transport of superoxide dismutase into mitochondria (Zemanovic et al., 2018).
Role of HSP70s in plant immunity
Analysis of publicly available data stored in the Arabidopsis RNA-Seq Database (Zhang et al., 2020) showed that most HSP70 genes are up-regulated in response to biotic stressors (Fig. 2E). Only cytosolic HSP70-8 and chloroplastic HSP70-6 and HSP70-7 were predominantly repressed, and expression profiles for HSP70-18 were unavailable. Comparison of the expression profile and sequence feature similarity (Fig. 2D, E) did not show any significant similarity in the clusterings (adjusted Rand index 0.16), indicating that the response to biotic stress is not correlated with the HSP70 sequence. However, the contrasting response of chloroplastic HSP70 genes and the available literature indicate that HSP70 localization is the key factor.
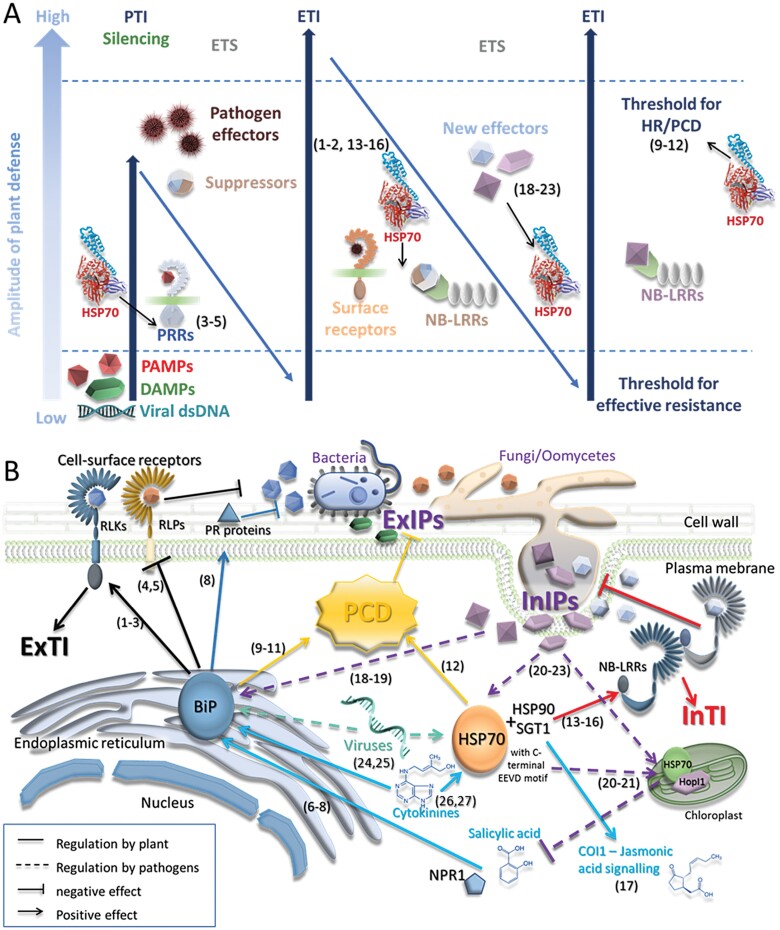
Plants respond to pathogen invasion using a two-branched innate immune system consisting of PAMP (pathogen-associated molecular pattern)-triggered immunity (PTI) and effector-triggered immunity (ETI; Abdul Malik et al., 2020). PTI represents an early response mechanism mediated by pattern recognition receptors (PRRs) that recognize and bind PAMPs (Nürnberger and Kemmerling, 2009). ETI is a reaction to pathogen-secreted effectors interfering with PTI. This second defensive barrier recruits resistance (R) proteins that specifically recognize pathogen effectors and usually triggers a hypersensitive response and cell death (Abdul Malik et al., 2020). HSP70s are an integral part of plant immunity and participate in both PTI and ETI responses (Fig. 4).
Fig. 4.
HSP70s and plant immunity. (A) Simplified diagram representing the classical view of plant–pathogen interaction, with the highlighted role for HSP70s in pattern-triggered immunity (PTI), effector-triggered immunity (ETI), programmed cell death, and in interaction with pathogen effector proteins. PAMPs, pathogen-associated molecular patterns; DAMPs, damage-associated molecular patterns; PPRs, pattern recognition receptors; NB-LRRs, nucleotide-binding site-leucine-rich repeats; HR, hypersensitive response; PCD, programmed cell death; ETS, effector-triggered susceptibility. (B) Subcellular localization and interactions of HSP70 in the spatial immunity model. BiPs are components of extracellularly triggered immunity (ExTI). These HSP70s are regulated by salicylic acid and NPR1, and play a role in the secretion of pathogenesis-related (PR) proteins. The cytosolic HSP70s participate in intracellularly triggered immunity (InTI) and influence perception of jasmonic acid. Cytokinin up-regulates most HSP70 genes and positively impacts HSP70 abundance. Moreover, HSP70s interact with pathogen effector proteins and are involved in virus replication and translocation. COI1, coronatine-insensitive protein 1; ExIPs, extracellular immunogenic patterns; InIPs, intracellular immunogenic patterns; NB-LRRs, nucleotide-binding site-leucine-rich repeats; NPR1, non-expresser of PR genes 1; PCD, programmed cell death; RLKs, receptor-like kinases; RLPs, receptor-like proteins. References: 1–2, Liebrand et al. (2012, 2014); 3, Nekrasov et al. (2009); 4–5, Park et al. (2010, 2014); 6, Nagashima et al. (2014); 7, Poór et al. (2019); 8, Wang et al. (2005); 9, Carvalho et al. (2014); 10, Qiang et al. (2012); 11, Xu et al. (2012); 12, Kanzaki et al. (2003); 13, Noël et al. (2007); 14, Thao et al. (2007); 15, Nakashima et al. (2008); 16, Shirasu (2009); 17, X.-C. Zhang et al. (2015); 18, Jing et al. (2016); 19, Zhou et al. (2021); 20–21, Jelenska et al. (2007, 2010); 22, Lee et al. (2018); 23, Kim and Hwang (2015); 24, Hýsková et al. (2021b); 25, Ye et al. (2011); 26, Černý et al. (2016); 27, Krishnakumar et al. (2017). For details about the spatial immunity model, see, for example, van der Burgh et al. (2019).
Cytosolic HSP70s regulate ETI
The HSP70 subfamily (DnaK) members with a C-terminal EEVD motif (sometimes referred to as heat shock cognate 70 chaperones) regulate immune responses in plants by cooperating with cytosolic chaperones such as SGT1, HSP90, and RAR1. This interaction is an integral part of the processing and folding of NB-LRR (nucleotide-binding domain-leucine-rich repeat) immune sensors (Shirasu, 2009). There are four cognate HSP70 genes in Arabidopsis (isoforms 1–4), and three of these are strongly up-regulated in response to different pathogens (Fig. 2E). Limited evidence indicates that these HSP70 genes are at least partially redundant, and experiments with a loss-of-function mutation of individual HSP70 isoforms 1, 2, and 3 did not reveal any difference in the resistance against pathogens (Noël et al., 2007; Table 1). Surprisingly, HSP70-1 is mostly unresponsive in biotic stress experiments (Fig. 2E; Table 2). A transgenic line overexpressing HSP70-1 has a reduced NB-LRR-dependent immunity response (Noël et al., 2007), but that could coincide with an attenuated expression of five different HSP70 genes (Sung and Guy, 2003). The HSP70 interactor SGT1 is a co-chaperone of HSP90 and, in addition to ETI, also regulates auxin and jasmonic acid signaling by maintaining steady-state levels of the corresponding receptors TIR1 and COI1 (X.-C. Zhang et al., 2015; Wang et al., 2016). It has been proven that the COI1 stabilization is facilitated by SGT1b–HSP70–HSP90 chaperone complex (X.-C. Zhang et al., 2015), and HSP70s thus directly influence hormonal perception.
Table 2.
Summary of recent reports supporting the role of HSP70 in biotic interactions
| Host | Pathogen | Arabidopsis ortholog | Regulation | Reference |
|---|---|---|---|---|
| Arabidopsis thaliana | Phytophthora capsici | HSP70-4 | ↑ | (1) |
| HSP70-5 | ↑ | (1) | ||
| Pseudomonas syringae pv tomato | HSP70-2 | ↑ | (2) | |
| HSP70-4 | ↑ | (2) | ||
| Brassica rapa | Turnip mosaic virus | HSP70-4 | ↑ | (3) |
| HSP70-1 | ↑ | (3) | ||
| Capsicum annum | Bemisia tabaci | HSP70-2 | ↑ | (4) |
| HSP70-1 | ↑ | (4) | ||
| Pepper golden mosaic virus | HSP70-4 | ↑ | (5) | |
| Castanea sativa | Phytophthora cinnamomi | HSP70-1 | ↑ | (6) |
| HSP70-4 | ↓ | (6) | ||
| HSP70-12 | ↑ | (6) | ||
| HSP70-10 | ↑↓ | (6) | ||
| HSP70-16 | ↑ | (6) | ||
| Cicer arietinum | Fusarium oxysporum | HSP70-1 | ↑ | (7) |
| Glycine max | Heterodera glycines | HSP70-2 | ↑ | (8) |
| HSP70-4 | ↑ | (8) | ||
| Hordeum vulgare | Phytophthora palmivora | HSP70-4 | ↓ | (9) |
| Blumeria graminis | HSP70-4 | ↑ | (10) | |
| Puccinia graminis | HSP70-1 | ↑ | (11) | |
| Malus domestica | Alternaria alternata | HSP70-4 | ↑↓ | (12) |
| HSP70-5 | ↑ | (12) | ||
| Nicotiana benthamiana | Potato virus X | HSP70-12 | ↑ | (13) |
| Potato virus Y | HSP70-4 | ↑ | (14) | |
| Tobacco mosaic virus | N/A | ↑ | (15) | |
| Nicotiana tabacum | Potato virus Y | N/A | ↑ | (16) |
| Pennisetum glaucum | Puccinia substriata | HSP70-4 | ↓ | (17) |
| Pisum sativum | Pea seed-borne mosaic virus | HSP70-3 | ↑ | (18) |
| Quercus suber | Phytophthora cinnamomi | HSP70-11 | ↑ | (19) |
| Saccharum spp. | Sporisorium scitamineum | HSP70-2 | ↓ | (20) |
| Solanum lycopersicum | Fusarium oxysporum f. sp. radicis-lycopersici | HSP70-10 | ↑ | (21) |
| Phytophthora parasitica | HSP70-4 | ↑ | (22) | |
| Tomato yellow leaf curl virus | N/A | ↓ | (23) | |
| HSP70-4 | ↓ | (24) | ||
| HSP70-2 | ↓ | (24) | ||
| Solanum lycopersicum | Rhizopus nigricans | HSP70-7 | ↓ | (25) |
| Solanum tuberosum L. | Synchytrium endobioticum | HSP70-4 | ↓∗ | (26) |
| HSP70-10 | ↓∗ | (26) | ||
| HSP70-2 | ↑∗ | (26) | ||
| Triticum aestivum | Blumeria graminis | HSP70-2 | ↑ | (27) |
| Puccinia striiformis | Hsp70-2 | ↑ | (27) | |
| Chinese wheat mosaic furovirus | HSP70-4 | ↑ | (28) | |
| Vigna unguiculata | Cowpea severe mosaic virus | HSP70-2 | ↓ | (29) |
| HSP70-3 | ↓ | (30) | ||
| HSP70-4 | ↓ | (30) | ||
| HSP70-5 | ↓ | (30) | ||
| HSP70-9 | ↓ | (30) | ||
| HSP70-18 | ↓ | (30) | ||
| Vitis vinifera | Plasmopara viticola | HSP70-4 | ↑ | (31) |
| Zea mays | Sugarcane mosaic virus | HSP70-12 | ↑ | (32) |
For the sake of simplicity, diverse non-canonical nomenclature has been replaced with the corresponding Arabidopsis orthologs. Asterisks indicate differences between susceptible and resistant cultivars. References: 1, Song et al. (2015); 2, Noël et al. (2007); 3, Lyu et al. (2020); 4, Wu et al. (2019); 5, Góngora-Castillo et al. (2012); 6, Saiz-Fernández et al. (2020); 7, Gupta et al. (2017); 8, Klink et al. (2007); 9, Berka et al. (2020); 10, Molitor et al. (2011); 11, Sharma Poudel et al. (2019); 12, C. Zhang et al. (2015); 13, Ye et al. (2011); 14, Hafrén et al. (2010); 15, Caplan et al. (2009); 16, Hýsková et al. (2021b); 17, Crampton et al. (2009); 18, Cerna et al. (2017); 19, Coelho et al. (2021); 20, Singh et al. (2019); 21, Vitale et al. (2014); 22, Naveed and Ali (2018); 23, Gorovits et al. (2013); 24, Chen et al. (2013); 25, Pan et al. (2013); 26, Szajko et al. (2020); 27, Guo et al. (2021); 28, Yang et al. (2017); 29, Paiva et al. (2016); 30, Varela et al. (2017); 31, Liu et al. (2021); 32, Chen et al. (2017).
The critical role of cytosolic HSP70s in plant defense is supported by the fact that these proteins are targeted by pathogen effector proteins. HopI1, a virulence effector of pathogenic Pseudomonas syringae, binds directly to the host HSP70 and recruits it to chloroplasts, the site of HopI1 localization (Jelenska et al., 2007, 2010). The interaction was confirmed by co-immunoprecipitation for all four Arabidopsis isoforms (1–4) and for chloroplastic HSP70-6. Further, plants with depleted cytosolic HSP70-1 were more susceptible to P. syringae (Jelenska et al., 2010), and silencing of cytosolic HSP70 in pepper (Capsicum annuum) increased the plant’s susceptibility to Xanthomonas campestris (Kim and Hwang, 2015). Finally, the effector Pi23226 of Phytophthora infestans was co-immunoprecipitated with HSP70s, and transient overexpression of these HSP70s in Nicotiana benthamiana inhibited Phytophthora growth (Lee et al., 2018).
HSP70s resident in the endoplasmic reticulum are related to endoplasmic reticulum-mediated plant immunity
Recent studies have pointed out the ability of plant pathogens to utilize their effectors to bind to the host ER stress pathway and manipulate it to their advantage (reviewed, for example, in Jing and Wang, 2020). ER stress occurs when the ER’s ability to fold proteins becomes saturated and can induce programmed cell death (PCD; Eichmann and Schäfer, 2012). Cells have evolved an unfolded protein response mechanism that induces the expression of genes that encode ER chaperones, including HSP70 genes. ER-resident HSP70s (also called binding protein, BiPs) participate in plant immunity as a central regulator and are the link to ER-supported immunity functions. BiP genes are induced by salicylic acid (Nagashima et al., 2014; Poór et al., 2019) and multiple biotic stressors (Figs 2E, 4B), and the expression is reportedly regulated by NPR1, the master regulator of systemic acquired resistance (Wang et al., 2005). BiPs stabilize folding intermediates, prevent aggregation, and aid in the subsequent protein folding and assembly (Padmanabhan and Dinesh-Kumar, 2010). In contrast to mammals and yeast, flowering plants contain several BiP proteins (HSP11–13 in Arabidopsis; Noh et al., 2003). BiP1 and BiP2 are highly similar based on their amino acid sequence, and they are ubiquitously expressed under non-stressed conditions, whereas BiP3 is more distantly related, highly induced upon ER stress, and therefore classified as a marker gene of the Arabidopsis unfolded protein response (Strasser, 2018). Several studies have shown that a process called ER quality control is essential for the proper function of the pattern recognition receptors, and part of this pathway relies on the BiP chaperone complex (Anelli and Sitia, 2008; Strasser, 2018). Nekrasov et al. (2009) reported that a subset of LRR-receptor-like kinases (RLKs), including EF-Tu (elongation factor thermounstable) receptor (EFR), require one or several ER complexes comprising SDF2, ERdj3B (Hsp40 co-chaperone), and BiP for their accumulation and subcellular localization. This chaperone complex regulates the biogenesis of multiple components of the plant defense system, including Cf proteins (resistance to the foliar pathogen Cladosporium fulvum; Liebrand et al., 2012) and Ve1 (resistance to the vascular fungal pathogen Verticillium dahliae; Liebrand et al., 2014). In contrast to the positive effect of cytosolic HSP70 overexpression, BiP3 overexpression compromised immunity against Xanthomonas oryzae (Park et al., 2010, 2014). OsBiP3 can bind to the critical XA21 receptor that facilitates resistance, and the presence of high levels of BiP reduces its stability and function (Park et al., 2010). The silencing of the individual BiPs did not affect EFR or Cf function, but a knockdown of the different BiPs in tomatoes resulted in a compromised Ve1-mediated resistance to V. dahliae (Nekrasov et al., 2009; Liebrand et al., 2012, 2014). The bip2 mutant showed compromised secretion of PR1, leading to an impaired resistance against the bacterial pathogen Pseudomonas syringae (Wang et al., 2005).
HSP70s were found to play a role in developmentally regulated PCD (Rowarth et al., 2020), and several studies suggest that BiPs are responsible for that (see, for example, Zhang et al., 2014). Furthermore, this mechanism is utilized by both mutualistic fungi and pathogens. For example, the Arabidopsis mutant bip2 showed enhanced fungal colonization rates compared with the wild type, and BiP protein levels were reduced in colonized wild-type roots compared with non-colonized roots, suggesting that impaired BiP accumulation supports fungal development during cell death-associated colonization (Qiang et al., 2012). Similarly, silencing of BiP resulted in a delay of the hypersensitive response and PCD induced by Xanthomonas oryzae pv. oryzae (Xu et al., 2012), and BiP-overexpressing lines displayed an accelerated hypersensitive response triggered by Pseudomonas syringae pv. tomato in soybean and tobacco (Carvalho et al., 2014). It has also been found that the effector PsAvh262 secreted by Phytophthora sojae stabilizes BiPs and suppresses ER stress-triggered cell death (Jing et al., 2016). Viruses can also exploit this pathway (Gaguancela et al., 2016), and up-regulation of BiP was found in many previous studies, including response to Potato virus X, Turnip crinkle virus, Garlic virus X, and Sugarcane mosaic virus (Ye et al., 2011; Lu et al., 2016; Moon et al., 2016; Chen et al., 2017). A recent study showed that the BiP accumulation inhibits the ER to nucleus translocation of Bcl-2-associated athanogene (BAG7; Zhou et al., 2021). This protein is a member of a multifunctional group of co-chaperones, and its role in heat tolerance, unfolded protein response, and basal immunity is well established (Williams et al., 2010; Li et al., 2016; Y. Li et al., 2017). The accumulated evidence indicates that the BAG7 translocation to the nucleus is critical for its protective function and that its BiP-induced retention in the ER promotes susceptibility to pathogens (Zhou et al., 2021).
HSP70s mediate virus replication and cell to cell transport
HSP70 genes have been found to be up-regulated in response to many viruses, and this effect is not limited to BiPs. For example, up-regulation or an increase in HSP70 was reported in response to Tobacco mosaic virus (N. benthamiana; cytosolic HSP70 genes; Caplan et al., 2009), Turnip mosaic virus (Brassica rapa; two cytosolic HSP70 genes; Lyu et al., 2020), and Pea seed-borne mosaic virus (Pisum sativum; two cytosolic HSP70 genes; Cerna et al., 2017). The role of these regulations is far from fully understood. The induction of HSP70 may represent a host reaction to the synthesis of a large number of exogenous proteins (Alfenas-Zerbini et al., 2009), and some resistance genes reportedly encode regulators of HSP70 expression (Hayano-Saito and Hayashi, 2020). However, accumulated evidence indicates that the HSP70 expression is, in fact, triggered by pathogens. Up-regulation of host HSP70 genes has not been found in response to Grapevine leafroll virus 3 closterovirus infection, but this virus has its own HSP70 gene (Espinoza et al., 2007; Prator et al., 2020). Aside from closteroviruses, plant viral genomes do not encode HSP70 genes, but many viruses seem to recruit host HSP70s to assist in virion assembly, replication, and cell to cell movement (e.g. Mine et al., 2012).
The cell to cell transport of plant viruses is mediated by specific virally encoded factors termed movement proteins and coat proteins which can create complexes with HSP70s (Nelson and Citovsky, 2005). It is believed that these complexes mediate virus interaction with the cytoskeleton and facilitate its transfer to and through the plasmodesmata (Aoki et al., 2002; Krenz et al., 2010). The interaction of HSP70s with coat proteins also facilitated the intracellular movement of Tomato yellow leaf curl virus into the nucleus, and the inactivation of HSP70 resulted in a decrease in viral replication (Gorovits et al., 2013). Similarly, a positive effect of HSP70s on viral replication was reported for Chinese wheat mosaic furovirus (Yang et al., 2017) and Cowpea severe mosaic virus (Paiva et al., 2016; Varela et al., 2017), and indicated for Tomato bushy stunt virus (Pogany et al., 2008). Further, Nicotiana tabacum plants exposed to heat shock and inoculated with Potato virus Y showed higher amounts of HSP70 genes and higher virus content than the corresponding controls inoculated at standard temperature (Hýsková et al., 2021b).
HSP70 accumulation and plant resistance to pathogens
The reported changes in HSP70 expression and protein accumulation show a predominantly positive response to biotic stimuli (Fig. 5; Table 2), and many reports have indicated that an increase in the plant pathogen resistance positively correlates with the HSP70 accumulation. For instance, the comparison of tomato accessions showed that the accession resistant to Phytophthora parasitica had a significantly higher HSP70 expression (Naveed et al., 2018). Similarly, faster induction of HSP70 genes was correlated with enhanced resistance to Blumeria graminis in barley (Molitor et al., 2011). In contrast, at least some pathogens exploit the host’s HSP70 machinery, and a loss-of-function mutation or silencing of HSP70 reportedly stimulate the resistance of the plant to viruses (reviewed in, for example, Hýsková et al., 2021a). However, the effect of mutation is not that simple in a partially redundant HSP70 gene family, and the interpretation should be carefully evaluated. The analysis of gene expression of Arabidopsis hsp70-15 knockout plants showed that the mutation significantly increased HSP70-4 (14-fold change) and HSP70-2 (6-fold change) (Jungkunz et al., 2011). These members of the DnaK subfamily are the most frequently found HSP70s in the biotic response, and the observed mutant resistance to the Turnip mosaic virus could coincide with that.
Fig. 5.
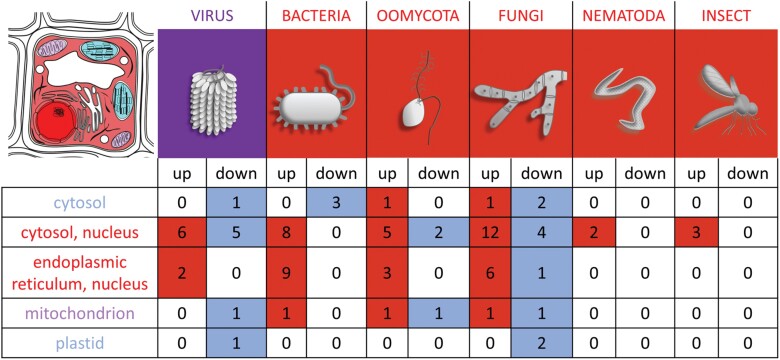
Comparison of the HSP70 response to biotic stress and its localization. Summary of reported transcriptomic and protein analyses outlined in Fig. 4B and Table 2. Localization is based on the expected localization of Arabidopsis HSP70s and corresponding putative Arabidopsis orthologs. The color of the cellular compartments represents the predominant response of the resident HSP70s to biotic stimuli. Red, blue, and purple represent a positive, negative, and mixed response to biotic stimuli, respectively.
Conclusion and future perspectives
Plant HSP70 research is lagging behind its animal counterparts, and our knowledge about post-transcriptional and post-translational control is only rudimentary. It is obvious that this should be the next objective in plant HSP70 research. Animal HSP70s play a significant signaling role in the extracellular space, particularly in the inflammatory and immune responses (see, for example, Mambula et al., 2007). According to available data and similar to its animal counterparts, none of the Arabidopsis HSP70s contains a targeting signal for secretion. Despite the presence of cell walls, plants can secrete proteins without the consensus N-terminal peptide sequence via alternative, unconventional protein secretory pathways into the extracellular space. The exact extent of these alternative pathways in plants has not yet been fully elucidated, and the role of plant HSP70s in the extracellular space (if any) is unknown. However, at least nine Arabidopsis HSP70s have been detected in the extracellular space (Table 1), and it has been confirmed that the extracellular vesicles in the plant secretome contribute to the plant defense system (Regente et al., 2017). The role of extracellular HSP70 could be evolutionarily conserved and could provide an additional explanation for the observed accumulation in response to pathogens. Finally, our review highlighted a prominent role for the plant hormone cytokinin in the regulation of HSP70 genes. Cytokinin promotes plant resistance against pathogens such as bacteria, fungi, and pest insects (Akhtar et al., 2020), and the up-regulation of HSP70 genes is probably an overlooked part of that effect.
Author contributions
MB, RK, VB, BB, and MC: performing the literature review; MB, RK, VB, and MC: writing – original draft; MC, BB, VB, and RK: preparation of figures; MC: writing – review & editing. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding
Funding support for this work was provided by the Ministry of Education, Youth and Sports of the Czech Republic (grant no. CZ.02.1.01/0.0/0.0/16_019/0000738) with support from the European Regional Development Fund - Project ‘Centre for Experimental Plant Biology’ (MC, RK, and BB), and by the Ministry of Education, Youth and Sports of the Czech Republic (grant no. CZ.02.2.69/0.0/0.0/19_073/0016670) with support from the European Social Fund - Project ‘Internal grants of Mendel University in Brno’ (MB, RK, VB, and MC).
References
- Abdul Malik NA, Kumar IS, Nadarajah K, Malik NAA, Kumar IS, Nadarajah K.. 2020. Elicitor and receptor molecules: orchestrators of plant defense and immunity. International Journal of Molecular Sciences 21, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrams JL, Verghese J, Gibney PA, Morano KA.. 2014. Hierarchical functional specificity of cytosolic heat shock protein 70 (Hsp70) nucleotide exchange factors in yeast. Journal of Biological Chemistry 289, 13155–13167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar SS, Mekureyaw MF, Pandey C, Roitsch T.. 2020. Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Frontiers in Plant Science 10, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfenas-Zerbini P, Maia IG, Fávaro RD, Cascardo JCM, Brommonschenkel SH, Zerbini FM.. 2009. Genome-wide analysis of differentially expressed genes during the early stages of tomato infection by a potyvirus. Molecular Plant-Microbe Interactions 22, 352–361. [DOI] [PubMed] [Google Scholar]
- Anelli T, Sitia R.. 2008. Protein quality control in the early secretory pathway. The EMBO Journal 27, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki K, Kragler F, Xoconostle-Cazares B, Lucas WJ, Xoconostle-Cázares B, Lucas WJ.. 2002. A subclass of plant heat shock cognate 70 chaperones carries a motif that facilitates trafficking through plasmodesmata. Proceedings of the National Academy of Sciences, USA 99, 16342–16347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asea AAA,Calderwood SK, Kaur P., eds.2016. Heat shock proteins and plants. Cham: Springer International Publishing. [Google Scholar]
- Ashraf M, Mao Q, Hong J, Shi L, Ran X, Liaquat F, Uzair M, Liang W, Fernie AR, Shi J.. 2021. HSP70-16 and VDAC3 jointly inhibit seed germination under cold stress in Arabidopsis. Plant, Cell & Environment 44, 3616–3627. [DOI] [PubMed] [Google Scholar]
- Bateman A, Martin MJ, O’Donovan C, et al. . 2017. UniProt: the universal protein knowledgebase. Nucleic Acids Research 45, D158–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka M, Greplová M, Saiz-Fernández I, Novák J, Luklová M, Zelená P, Tomšovský M, Brzobohatý B, Černý M.. 2020. Peptide-based identification of Phytophthora isolates and Phytophthora detection in planta. International Journal of Molecular Sciences 21, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernabò P, Viero G, Lencioni V.. 2020. A long noncoding RNA acts as a posttranscriptional regulator of heat shock protein (HSP70) synthesis in the cold hardy Diamesa tonsa under heat shock. PLoS One 15, e0227172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchiellini C, Boury-Esnault N, Vacelet J, Le Parco Y.. 1998. Phylogenetic analysis of the Hsp70 sequences reveals the monophyly of Metazoa and specific phylogenetic relationships between animals and fungi. Molecular Biology and Evolution 15, 647–655. [DOI] [PubMed] [Google Scholar]
- Bracher A, Verghese J.. 2015. The nucleotide exchange factors of Hsp70 molecular chaperones. Frontiers in Molecular Biosciences 2, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan JL, Zhu X, Mamillapalli P, Marathe R, Anandalakshmi R, Dinesh-Kumar SPP.. 2009. Induced ER chaperones regulate a receptor-like kinase to mediate antiviral innate immune response in plants. Cell Host & Microbe 6, 457–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho HH, Silva PA, Mendes GC, Brustolini OJB, Pimenta MR, Gouveia BC, Valente MAS, Ramos HJO, Soares-Ramos JRL, Fontes EPB.. 2014. The endoplasmic reticulum binding protein BiP displays dual function in modulating cell death events. Plant Physiology 164, 654–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalé A-C, Clément M, Chiarenza S, Roncato M-A, Pochon N, Creff A, Marin E, Leonhardt N, Noël LD.. 2009. Altered expression of cytosolic/nuclear HSC70-1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. Journal of Experimental Botany 60, 2653–2664. [DOI] [PubMed] [Google Scholar]
- Cerna H, Černý M, Habánová H, Šafářová D, Abushamsiya K, Navrátil M, Brzobohatý B.. 2017. Proteomics offers insight to the mechanism behind Pisum sativum L. response to Pea seed-borne mosaic virus (PSbMV). Journal of Proteomics 153, 78–88. [DOI] [PubMed] [Google Scholar]
- Černý M, Jedelský PL, Novák J, Schlosser A, Brzobohatý B.. 2014. Cytokinin modulates proteomic, transcriptomic and growth responses to temperature shocks in Arabidopsis. Plant, Cell & Environment 37, 1641–1655. [DOI] [PubMed] [Google Scholar]
- Černý M, Novák J, Habánová H, Cerna H, Brzobohatý B.. 2016. Role of the proteome in phytohormonal signaling. Biochimica et Biophysica Acta 1864, 1003–1015. [DOI] [PubMed] [Google Scholar]
- Černý M, Skalák J, Cerna H, Brzobohatý B.. 2013. Advances in purification and separation of posttranslationally modified proteins. Journal of Proteomics 92, 2–27. [DOI] [PubMed] [Google Scholar]
- Cha-Molstad H, Sung KS, Hwang J, et al. . 2015. Amino-terminal arginylation targets endoplasmic reticulum chaperone BiP for autophagy through p62 binding. Nature Cell Biology 17, 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Cao Y, Li Y, Xia Z, Xie J, Carr JP, Wu B, Fan Z, Zhou T.. 2017. Identification of differentially regulated maize proteins conditioning Sugarcane mosaic virus systemic infection. New Phytologist 215, 1156–1172. [DOI] [PubMed] [Google Scholar]
- Chen T, Lv Y, Zhao T, Li N, Yang Y, Yu W, He X, Liu T, Zhang B.. 2013. Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PLoS One 8, e80816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shi L, Chen Y, Zhu L, Zhang D, Xiao S, Aharoni A, Shi J, Xu J.. 2019. Arabidopsis HSP70-16 is required for flower opening under normal or mild heat stress temperatures. Plant, Cell & Environment 42, 1190–1204. [DOI] [PubMed] [Google Scholar]
- Clément M, Leonhardt N, Droillard M-JJ, et al. . 2011. The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant Physiology 156, 1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier P, Coulombe B.. 2013. Regulation of molecular chaperones through post-translational modifications: decrypting the chaperone code. Biochimica et Biophysica Acta 1829, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho AC, Pires R, Schütz G, Santa CC, Manadas B, Pinto PP, Schutz G, Santa CC, Manadas B, Pinto PP.. 2021. Disclosing proteins in the leaves of cork oak plants associated with the immune response to Phytophthora cinnamomi inoculation in the roots: a long-term proteomics approach. PLoS One 16, e0245148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA. 2018. Hsp70 at the membrane: driving protein translocation. BMC Biology 16, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampton BG, Hein I, Berger DK.. 2009. Salicylic acid confers resistance to a biotrophic rust pathogen, Puccinia substriata, in pearl millet (Pennisetum glaucum). Molecular Plant Pathology 10, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichmann R, Schäfer P.. 2012. The endoplasmic reticulum in plant immunity and cell death. Frontiers in Plant Science 3, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza C, Vega A, Medina C, Schlauch K, Cramer G, Arce-Johnson P.. 2007. Gene expression associated with compatible viral diseases in grapevine cultivars. Functional & Integrative Genomics 7, 95–110. [DOI] [PubMed] [Google Scholar]
- Fernández-Fernández MR, Gragera M, Ochoa-Ibarrola L, Quintana-Gallardo L, Valpuesta JM.. 2017. Hsp70—a master regulator in protein degradation. FEBS Letters 591, 2648–2660. [DOI] [PubMed] [Google Scholar]
- Gaguancela OA, Źuñiga LP, Arias AV, Halterman D, Flores FJ, Johansen IE, Wang A, Yamaji Y, Verchot J.. 2016. The IRE1/bZIP60 pathway and bax inhibitor 1 suppress systemic accumulation of potyviruses and potexviruses in Arabidopsis and Nicotiana benthamiana plants. Molecular Plant-Microbe Interactions 29, 750–766. [DOI] [PubMed] [Google Scholar]
- Góngora-Castillo E, Ibarra-Laclette E, Trejo-Saavedra DL, Rivera-Bustamante RF.. 2012. Transcriptome analysis of symptomatic and recovered leaves of geminivirus-infected pepper (Capsicum annuum). Virology Journal 9, 295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorovits R, Moshe A, Ghanim M, Czosnek H.. 2013. Recruitment of the host plant heat shock protein 70 by Tomato yellow leaf curl virus coat protein is required for virus infection. PLoS One 8, e70280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Zhang H, Wang G, Wang C, Wang Y, Liu X, Ji W.. 2021. Identification and expression analysis of heat-shock proteins in wheat infected with powdery mildew and stripe rust. The Plant Genome 14, e20092. [DOI] [PubMed] [Google Scholar]
- Gupta S, Bhar A, Chatterjee M, Ghosh A, Das S, Gupta V.. 2017. Transcriptomic dissection reveals wide spread differential expression in chickpea during early time points of Fusarium oxysporum f. sp. ciceri Race 1 attack. PLoS One 12, e0178164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL, Li Q-B.. 1998. The organization and evolution of the spinach stress 70 molecular chaperone gene family. The Plant Cell 10, 539–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K.. 2010. HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. The Plant Cell 22, 523–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayano-Saito Y, Hayashi K.. 2020. Stvb-i, a rice gene conferring durable resistance to Rice stripe virus, protects plant growth from heat stress. Frontiers in Plant Science 11, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper CM, Castleden IR, Tanz SK, Aryamanesh N, Millar AH.. 2017. SUBA4: the interactive data analysis centre for Arabidopsis subcellular protein locations. Nucleic Acids Research 45, D1064–D1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L-H, Wang H-S, Kang L.. 2008. Different evolutionary lineages of large and small heat shock proteins in eukaryotes. Cell Research 18, 1074–1076. [DOI] [PubMed] [Google Scholar]
- Huang X, Hou L, Meng J, You H, Li Z, Gong Z, Yang S, Shi Y.. 2018. The antagonistic action of abscisic acid and cytokinin signaling mediates drought stress response in Arabidopsis. Molecular Plant 11, 970–982. [DOI] [PubMed] [Google Scholar]
- Hýsková V, Bělonožníková K, Čeřovská N, Ryšlavá H.. 2021a. HSP70 plays an ambiguous role during viral infections in plants. Biologia Plantarum65, 68–79. [Google Scholar]
- Hýsková V, Bělonožníková K, Doričová V, Kavan D, Gillarová S, Henke S, Synková H, Ryšlavá H, Čeřovská N.. 2021b. Effects of heat treatment on metabolism of tobacco plants infected with Potato virus Y. Plant Biology 23, 131–141. [DOI] [PubMed] [Google Scholar]
- Jelenska J, van Hal JA, Greenberg JT.. 2010. Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proceedings of the National Academy of Sciences, USA 107, 13177–13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Yao N, Vinatzer BA, Wright CM, Brodsky JL, Greenberg JT.. 2007. A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Current Biology 17, 499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Guo B, Li H, et al. . 2016. A Phytophthora sojae effector suppresses endoplasmic reticulum stress-mediated immunity by stabilizing plant binding immunoglobulin proteins. Nature Communications 7, 11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing M, Wang Y.. 2020. Plant pathogens utilize effectors to hijack the host endoplasmic reticulum as part of their infection strategy. Engineering 6, 500–504. [Google Scholar]
- Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, Sonnewald U.. 2011. AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. The Plant Journal 66, 983–995. [DOI] [PubMed] [Google Scholar]
- Kanzaki H, Saitoh H, Ito A, Fujisawa S, Kamoun S, Katou S, Yoshioka H, Terauchi R.. 2003. Cytosolic HSP90 and HSP70 are essential components of INF1-mediated hypersensitive response and non-host resistance to Pseudomonas cichorii in Nicotiana benthamiana. Molecular Plant Pathology 4, 383–391. [DOI] [PubMed] [Google Scholar]
- Kim NH, Hwang BK.. 2015. Pepper heat shock protein 70a interacts with the type III effector AvrBsT and triggers plant cell death and immunity. Plant Physiology 167, 307–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kityk R, Kopp J, Mayer MP.. 2018. Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Molecular Cell 69, 227-237.e4. [DOI] [PubMed] [Google Scholar]
- Kityk R, Kopp J, Sinning I, Mayer MP.. 2012. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Molecular Cell 48, 863–874. [DOI] [PubMed] [Google Scholar]
- Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP.. 2015. Pathways of allosteric regulation in Hsp70 chaperones. Nature Communications 6, 8308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink VP, Overall CC, Alkharouf NW, MacDonald MH, Matthews BF.. 2007. Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 226, 1389–1409. [DOI] [PubMed] [Google Scholar]
- Kozeko L. 2021. Different roles of inducible and constitutive HSP70 and HSP90 in tolerance of Arabidopsis thaliana to high temperature and water deficit. Acta Physiologiae Plantarum 43, 58. [Google Scholar]
- Krenz B, Windeisen V, Wege C, Jeske H, Kleinow T.. 2010. A plastid-targeted heat shock cognate 70 kDa protein interacts with the Abutilon mosaic virus movement protein. Virology 401, 6–17. [DOI] [PubMed] [Google Scholar]
- Krishnakumar V, Contrino S, Cheng CY, Belyaeva I, Ferlanti ES, Miller JR, Vaughn MW, Micklem G, Town CD, Chan AP.. 2017. Thalemine: a warehouse for Arabidopsis data integration and discovery. Plant & Cell Physiology 58, e4. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M, Xu X-MM, Møller SG.. 2010. Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta 232, 567–578. [DOI] [PubMed] [Google Scholar]
- Lee JH, Lee SE, Oh S, Seo E, Choi D.. 2018. HSP70s enhance a Phytophthora infestans effector-induced cell death via an MAPK cascade in Nicotiana benthamiana. Molecular Plant-Microbe Interactions 31, 356–362. [DOI] [PubMed] [Google Scholar]
- Lee SS, Lee DW, Lee Y, Mayer U, Stierhof Y-DD, Lee SS, Jürgens G, Hwang I, Jürgens G, Hwang I.. 2009. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin–26S proteasome system in Arabidopsis. The Plant Cell 21, 3984–4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescat L, Véron V, Mourot B, et al. . 2020. Chaperone-mediated autophagy in the light of evolution: insight from fish. Molecular Biology and Evolution 37, 2887–2899. [DOI] [PubMed] [Google Scholar]
- Li L, Nelson CJ, Trösch J, Castleden I, Huang S, Millar AH.. 2017. Protein degradation rate in Arabidopsis thaliana leaf growth and development. The Plant Cell 29, 207–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Kabbage M, Liu W, Dickman MB.. 2016. Aspartyl protease-mediated cleavage of BAG6 is necessary for autophagy and fungal resistance in plants. The Plant Cell 28, 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Williams B, Dickman M.. 2017. Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytologist 214, 695–705. [DOI] [PubMed] [Google Scholar]
- Liebrand TWH, Kombrink A, Zhang Z, Sklenar J, Jones AME, Robatzek S, Thomma BPHJ, Joosten MHAJ.. 2014. Chaperones of the endoplasmic reticulum are required for Ve1-mediated resistance to Verticillium. Molecular Plant Pathology 15, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebrand TWH, Smit P, Abd-El-Haliem A, et al. . 2012. Endoplasmic reticulum-quality control chaperones facilitate the biogenesis of Cf receptor-like proteins involved in pathogen resistance of tomato. Plant Physiology 159, 1819–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M.. 2001. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress and Chaperones 6, 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G-TT, Wang B-BB, Lecourieux D, et al. . 2021. Proteomic analysis of early-stage incompatible and compatible interactions between grapevine and P. viticola. Horticulture Research 8, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Yin M, Wang X, et al. . 2016. The unfolded protein response and programmed cell death are induced by expression of Garlic virus X p11 in Nicotiana benthamiana. Journal of General Virology 97, 1462–1468. [DOI] [PubMed] [Google Scholar]
- Lyu S, Gao L, Zhang R, Zhang C, Hou X.. 2020. Correlation analysis of expression profile and quantitative iTRAQ-LC-MS/MS proteomics reveals resistance mechanism against TuMV in Chinese cabbage (Brassica rapa ssp. pekinensis). Frontiers in Genetics 11, 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mambula SS, Stevenson MA, Ogawa K, Calderwood SK.. 2007. Mechanisms for Hsp70 secretion: crossing membranes without a leader. Methods 43, 168–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Endo T, Nishikawa S-i.. 2010. BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 107, 1684–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama D, Sugiyama T, Endo T, Nishikawa SI.. 2014. Multiple BiP genes of Arabidopsis thaliana are required for male gametogenesis and pollen competitiveness. Plant & Cell Physiology 55, 801–810. [DOI] [PubMed] [Google Scholar]
- Mergner J, Frejno M, List M, et al. . 2020. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 579, 409–414. [DOI] [PubMed] [Google Scholar]
- Mine A, Hyodo K, Tajima Y, Kusumanegara K, Taniguchi T, Kaido M, Mise K, Taniguchi H, Okuno T.. 2012. Differential roles of Hsp70 and Hsp90 in the assembly of the replicase complex of a positive-strand RNA plant virus. Journal of Virology 86, 12091–12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molitor A, Zajic D, Voll LM, Pons-Kühnemann J, Samans B, Kogel KH, Waller F.. 2011. Barley leaf transcriptome and metabolite analysis reveals new aspects of compatibility and Piriformospora indica-mediated systemic induced resistance to powdery mildew. Molecular Plant-Microbe Interactions 24, 1427–1439. [DOI] [PubMed] [Google Scholar]
- Moon JY, Lee JH, Oh C-SS, Kang H-GG, Park JM.. 2016. Endoplasmic reticulum stress responses function in the HRT-mediated hypersensitive response in Nicotiana benthamiana. Molecular Plant Pathology 17, 1382–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgner N, Schmidt C, Beilsten-Edmands V, et al. . 2015. Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Reports 11, 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y, Iwata Y, Ashida M, Mishiba KI, Koizumi N.. 2014. Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant & Cell Physiology 55, 1772–1778. [DOI] [PubMed] [Google Scholar]
- Nakai M. 2018. New perspectives on chloroplast protein import. Plant & Cell Physiology 59, 1111–1119. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Chen L, Thao NP, et al. . 2008. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. The Plant Cell 20, 2265–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveed ZA, Ali GS.. 2018. Comparative transcriptome analysis between a resistant and a susceptible wild tomato accession in response to Phytophthora parasitica. International Journal of Molecular Sciences 19, 3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, et al. . 2009. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. The EMBO Journal 28, 3428–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RS, Citovsky V.. 2005. Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiology 138, 1809–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitika, Porter CM, Truman AW, Truttmann MC.. 2020. Post-translational modifications of Hsp70 family proteins: expanding the chaperone code. Journal of Biological Chemistry 295, 10689–10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël LD, Cagna G, Stuttmann J, et al. . 2007. Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. The Plant Cell 19, 4061–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh S-JJ, Kwon CS, Oh D-HH, Moon JS, Chung W-I.. 2003. Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311, 81–91. [DOI] [PubMed] [Google Scholar]
- Nürnberger T, Kemmerling B.. 2009. Pathogen-associated molecular patterns (PAMP) and PAMP-triggered immunity. Annual Plant Reviews 34, 16–47. [Google Scholar]
- Omasits U, Ahrens CH, Müller S, Wollscheid B.. 2014. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Dinesh-Kumar SP.. 2010. All hands on deck—the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Molecular Plant-Microbe Interactions 23, 1368–1380. [DOI] [PubMed] [Google Scholar]
- Paiva ALSS, Oliveira JTAA, de Souza GA, Vasconcelos IM.. 2016. Label-free proteomic reveals that Cowpea severe mosaic virus transiently suppresses the host leaf protein accumulation during the compatible interaction with cowpea (Vigna unguiculata [L.] Walp.). Journal of Proteome Research 15, 4208–4220. [DOI] [PubMed] [Google Scholar]
- Palmblad M, Mills DJ, Bindschedler LV.. 2008. Heat-shock response in Arabidopsis thaliana explored by multiplexed quantitative proteomics using differential metabolic labeling. Journal of Proteome Research 7, 780–785. [DOI] [PubMed] [Google Scholar]
- Pan X, Zhu B, Luo Y, Fu D.. 2013. Unraveling the protein network of tomato fruit in response to necrotrophic phytopathogenic Rhizopus nigricans. PLoS One 8, e73034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C-JJ, Bart R, Chern M, Canlas PE, Bai W, Ronald PC.. 2010. Overexpression of the endoplasmic reticulum chaperone BiP3 regulates XA21-mediated innate immunity in rice. PLoS One 5, e9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C-JJ, Seo Y-SS.. 2015. Heat shock proteins: a review of the molecular chaperones for plant immunity. Plant Pathology Journal 31, 323–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CJ, Song MY, Kim CY, Jeon JS, Ronald PC.. 2014. Rice BiP3 regulates immunity mediated by the PRRs XA3 and XA21 but not immunity mediated by the NB-LRR protein, Pi5. Biochemical and Biophysical Research Communications 448, 70–75. [DOI] [PubMed] [Google Scholar]
- Pogany J, Stork J, Li Z, Nagy PD.. 2008. In vitro assembly of the Tomato bushy stunt virus replicase requires the host heat shock protein 70. Proceedings of the National Academy of Sciences, USA 105, 19956–19961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poór P, Czékus Z, Tari I, Ördög A.. 2019. The multifaceted roles of plant hormone salicylic acid in endoplasmic reticulum stress and unfolded protein response. International Journal of Molecular Sciences 20, 5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prator CA, Chooi KM, Jones D, Davy MW, MacDiarmid RM, Almeida RPPP.. 2020. Comparison of two different host plant genera responding to grapevine leafroll-associated virus 3 infection. Scientific Reports 10, 8505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi R, Sarbeng EB, Liu QQ, et al. . 2013. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nature Structural & Molecular Biology 20, 900–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang X, Zechmann B, Reitz MU, Kogel KH, Schäfer P.. 2012. The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress-triggered caspase-dependent cell death. The Plant Cell 24, 794–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh Kumar R, Nagarajan NS, Arunraj SP, Sinha D, Veedin Rajan VB, Esthaki VK, D’Silva P.. 2012. HSPIR: a manually annotated heat shock protein information resource. Bioinformatics 28, 2853–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, de la Canal L.. 2017. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. Journal of Experimental Botany 68, 5485–5495. [DOI] [PubMed] [Google Scholar]
- Ritossa F. 1996. Discovery of the heat shock response. Cell Stress & Chaperones 1, 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R, Nillegoda NB, Mayer MP, Bukau B.. 2019. The Hsp70 chaperone network. Nature Reviews. Molecular Cell Biology 20, 665–680. [DOI] [PubMed] [Google Scholar]
- Rowarth NM, Dauphinee AN, Denbigh GL, Gunawardena AH.. 2020. Hsp70 plays a role in programmed cell death during the remodelling of leaves of the lace plant (Aponogeton madagascariensis). Journal of Experimental Botany 71, 907–918. [DOI] [PubMed] [Google Scholar]
- Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B, Rudiger S.. 1997. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. The EMBO Journal 16, 1501–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz-Fernández I, Milenković I, Berka M, Černý M, Tomšovský M, Brzobohatý B, Kerchev P.. 2020. Integrated proteomic and metabolomic profiling of Phytophthora cinnamomi attack on sweet chestnut (Castanea sativa) reveals distinct molecular reprogramming proximal to the infection site and away from it. International Journal of Molecular Sciences 21, 8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar NK, Kundnani P, Grover A.. 2013. Functional analysis of Hsp70 superfamily proteins of rice (Oryza sativa). Cell Stress and Chaperones 18, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleiff E, Becker T.. 2011. Common ground for protein translocation: access control for mitochondria and chloroplasts. Nature Reviews. Molecular Cell Biology 12, 48–59. [DOI] [PubMed] [Google Scholar]
- Sharma Poudel R, Richards J, Shrestha S, Solanki S, Brueggeman R.. 2019. Transcriptome-wide association study identifies putative elicitors/suppressor of Puccinia graminis f. sp. tritici that modulate barley rpg4-mediated stem rust resistance. BMC Genomics 20, 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu K. 2009. The HSP90–SGT1 chaperone complex for NLR immune sensors. Annual Review of Plant Biology 60, 139–164. [DOI] [PubMed] [Google Scholar]
- Singh P, Song Q-Q, Singh R, Li H-B, Solanki M, Malviya M, Verma K, Yang L-T, Li Y-R.. 2019. Proteomic analysis of the resistance mechanisms in sugarcane during Sporisorium scitamineum infection. International Journal of Molecular Sciences 20, 569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song T, Ma Z, Shen D, Li Q, Li W, Su L, Ye T, Zhang M, Wang Y, Dou D.. 2015. An oomycete CRN effector reprograms expression of plant HSP genes by targeting their promoters. PLoS Pathogens 11, e1005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa R, Lafer EM.. 2019. The physics of entropic pulling: a novel model for the Hsp70 motor mechanism. International Journal of Molecular Sciences 20, 2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R. 2018. Protein quality control in the endoplasmic reticulum of plants. Annual Review of Plant Biology 69, 147–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM.. 2008. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermotolerance of germinating seeds. Plant Physiology 146, 1231–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P-HH, Li HM.. 2010. Stromal hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. The Plant Cell 22, 1516–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J-LL, Li J-YY, Wang M-JJ, Song Z-TT, Liu J-XX.. 2021. Protein quality control in plant organelles: current progress and future perspectives. Molecular Plant 14, 95–114. [DOI] [PubMed] [Google Scholar]
- Sung DY, Guy CL.. 2003. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiology 132, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szajko K, Plich J, Przetakiewicz J, Sołtys-Kalina D, Marczewski W.. 2020. Comparative proteomic analysis of resistant and susceptible potato cultivars during Synchytrium endobioticum infestation. Planta 251, 4. [DOI] [PubMed] [Google Scholar]
- Takakuwa JE, Nitika Knighton LE, Truman AW.. 2019. Oligomerization of Hsp70: current perspectives on regulation and function. Frontiers in Molecular Biosciences 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thao NP, Chen L, Nakashima A, Hara SI, Umemura K, Takahashi A, Shirasu K, Kawasaki T, Shimamoto K.. 2007. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. The Plant Cell 19, 4035–4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichá T, Samakovli D, Kuchařová A, Vavrdová T, Šamaj J.. 2020. Multifaceted roles of HEAT SHOCK PROTEIN 90 molecular chaperones in plant development. Journal of Experimental Botany 71, 3966–3985. [DOI] [PubMed] [Google Scholar]
- Tissiéres A, Mitchell HK, Tracy UM.. 1974. Protein synthesis in salivary glands of Drosophila melanogaster: relation to chromosome puffs. Journal of Molecular Biology 84, 389–398. [DOI] [PubMed] [Google Scholar]
- Ul Haq S, Khan A, Ali M, Khattak AM, Gai WX, Zhang HX, Wei AM, Gong ZH.. 2019. Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. International Journal of Molecular Sciences 20, 5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman MG, Rafii MY, Martini MY, Yusuff OA, Ismail MR, Miah G.. 2017. Molecular analysis of Hsp70 mechanisms in plants and their function in response to stress. Biotechnology and Genetic Engineering Reviews 33, 26–39. [DOI] [PubMed] [Google Scholar]
- van der Burgh A M, Joosten MHAJ.. 2019. Plant immunity: thinking outside and inside the box. Trends in Plant Science 24, 587–601. [DOI] [PubMed] [Google Scholar]
- van Wijk KJ, LeppertT, Sun Q, Boguraev SS, Sun Z, Mendoza L, Deutsch EW.. 2021. The Arabidopsis thaliana PeptideAtlas; harnessing world-wide proteomics data for a comprehensive community proteomics resource. bioRxiv 2021, 05.03.442425. [Preprint]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varela ALN, Komatsu S, Wang X, Silva RGG, Souza PFN, Lobo AKM, Vasconcelos IM, Silveira JAG, Oliveira JTA.. 2017. Gel-free/label-free proteomic, photosynthetic, and biochemical analysis of cowpea (Vigna unguiculata [L.] Walp.) resistance against Cowpea severe mosaic virus (CPSMV). Journal of Proteomics 163, 76–91. [DOI] [PubMed] [Google Scholar]
- Verma AK, Tamadaddi C, Tak Y, Lal SS, Cole SJ, Hines JK, Sahi C.. 2019. The expanding world of plant J-domain proteins. Critical Reviews in Plant Sciences 38, 382–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Rocco M, Arena S, et al. . 2014. Tomato susceptibility to Fusarium crown and root rot: effect of grafting combination and proteomic analysis of tolerance expression in the rootstock. Plant Physiology and Biochemistry 83, 207–216. [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, Dong X.. 2005. Induction of protein secretory pathway is required for systemic acquired resistance. Science 308, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M.. 2016. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nature Communications 7, 10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Vinocur B, Shoseyov O, Altman A.. 2004. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends in Plant Science 9, 244–252. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gibney PA, West JD, Morano KA.. 2012. The yeast Hsp70 Ssa1 is a sensor for activation of the heat shock response by thiol-reactive compounds. Molecular Biology of the Cell 23, 3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse A, Bertoni M, Bienert S, et al. . 2018. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Research 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMAA, Clamp M, Barton GJ.. 2009. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER, Rioflorido I.. 2007. Evolutionary analysis of the small heat shock proteins in five complete algal genomes. Journal of Molecular Evolution 65, 162–174. [DOI] [PubMed] [Google Scholar]
- Waters ER, Vierling E.. 2020. Plant small heat shock proteins—evolutionary and functional diversity. New Phytologist 227, 24–37. [DOI] [PubMed] [Google Scholar]
- Wei S-SS, Niu W-TT, Zhai X-TT, et al. . 2019. Arabidopsis mtHSC70-1 plays important roles in the establishment of COX-dependent respiration and redox homeostasis. Journal of Experimental Botany 70, 5575–5590. [DOI] [PubMed] [Google Scholar]
- Weigel D, Mott R.. 2009. The 1001 Genomes Project for Arabidopsis thaliana. Genome Biology 10, 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems P, Horne A, Van Parys T, Goormachtig S, De Smet I, Botzki A, Van Breusegem F, Gevaert K.. 2019. The Plant PTM Viewer, a central resource for exploring plant protein modifications. The Plant Journal 99, 752–762. [DOI] [PubMed] [Google Scholar]
- Williams B, Kabbage M, Britt R, Dickman MB.. 2010. AtBAG7, an Arabidopsis Bcl-2-associated athanogene, resides in the endoplasmic reticulum and is involved in the unfolded protein response. Proceedings of the National Academy of Sciences, USA 107, 6088–6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yan J, Wu Y, Zhang H, Mo S, Xu X, Zhou F, Ding H.. 2019. Proteomic analysis by iTRAQ-PRM provides integrated insight into mechanisms of resistance in pepper to Bemisia tabaci (Gennadius). BMC Plant Biology 19, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Li S, Xie K, Zhang Q, Wang Y, Tang Y, Liu D, Hong Y, He C, Liu Y.. 2012. Plant ERD2-like proteins function as endoplasmic reticulum luminal protein receptors and participate in programmed cell death during innate immunity. The Plant Journal 72, 57–69. [DOI] [PubMed] [Google Scholar]
- Xu L , Nitika Hasin N, Cuskelly DD, Wolfgeher D, Doyle S, Moynagh P, Perrett S, Jones GW, Truman AW.. 2019. Rapid deacetylation of yeast Hsp70 mediates the cellular response to heat stress. Scientific Reports 9, 16260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang F, Cai N-JJ, Wu N, Chen X, Li J, Meng X-FF, Zhu T-QQ, Chen J-PP, Zhang H-MM.. 2017. A furoviral replicase recruits host HSP70 to membranes for viral RNA replication. Scientific Reports 7, 45590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye C, Dickman MB, Whitham SA, Payton M, Verchot J.. 2011. The unfolded protein response is triggered by a plant viral movement protein. Plant Physiology 156, 741–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemanovic S, Ivanov MV., Ivanova LV., et al. . 2018. Dynamic phosphorylation of the C terminus of Hsp70 regulates the mitochondrial import of SOD2 and redox balance. Cell Reports 25, 2605–2616.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Tian Y, Cong P.. 2015. Proteome analysis of pathogen-responsive proteins from apple leaves induced by the alternaria blotch Alternaria alternata. PLoS One 10, e0122233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang F, Yu Y, Feng L, Jia J, Liu B, Li B, Guo H, Zhai J.. 2020. A comprehensive online database for exploring ~20,000 public Arabidopsis RNA-Seq libraries. Molecular Plant 13, 1231–1233. [DOI] [PubMed] [Google Scholar]
- Zhang X-C, Millet YA, Cheng Z, Bush J, Ausubel FM.. 2015. Jasmonate signalling in Arabidopsis involves SGT1b–HSP70–HSP90 chaperone complexes. Nature Plants 1, 15049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xia G, Zhu Q.. 2021. Conserved and unique roles of chaperone-dependent E3 ubiquitin ligase CHIP in plants. Frontiers in Plant Science 12, 699756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Shrestha J, Tateda C, Greenberg JT.. 2014. Salicylic acid signaling controls the maturation and localization of the Arabidopsis defense protein ACCELERATED CELL DEATH6. Molecular Plant 7, 1365–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Yang K, Cheng M, et al. . 2021. Double-faced role of Bcl-2-associated athanogene 7 in plant–Phytophthora interaction. Journal of Experimental Botany 72, 5751–5765. [DOI] [PubMed] [Google Scholar]
- Zuiderweg ERP, Hightower LE, Gestwicki JE.. 2017. The remarkable multivalency of the Hsp70 chaperones. Cell Stress and Chaperones 22, 173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]