Abstract
Background
Gastric cancer (GC) and gastroesophageal junction adenocarcinomas (GEJ) are molecularly diverse. TP53 is the most frequently altered gene with approximately 50% of patients harboring mutations. This qualitative study describes the distinct genomic alterations in GCs and GEJs stratified by TP53 mutation status.
Patients and Methods
Tumor DNA sequencing results of 324 genes from 3741 patients with GC and GEJ were obtained from Foundation Medicine. Association between gene mutation frequency and TP53 mutation status was examined using Fisher’s exact test. Functional gene groupings representing molecular pathways suggested to be differentially mutated in TP53 wild-type (TP53WT) and TP53 mutant (TP53MUT) tumors were identified. The association of the frequency of tumors containing a gene mutation in the molecular pathways of interest and TP53 mutation status was assessed using Fisher’s exact test with a P-value of <.01 deemed statistically significant for all analyses.
Results
TP53 mutations were noted in 61.6% of 2946 GCs and 81.4% of 795 GEJs (P < .001). Forty-nine genes had statistically different mutation frequencies in TP53WT vs. TP53MUT patients. TP53WT tumors more likely had mutations related to DNA mismatch repair, homologous recombination repair, DNA and histone methylation, Wnt/B-catenin, PI3K/Akt/mTOR, and chromatin remodeling complexes. TP53MUT tumors more likely had mutations related to fibroblast growth factor, epidermal growth factor receptor, other receptor tyrosine kinases, and cyclin and cyclin-dependent kinases.
Conclusion
The mutational profiles of GCs and GEJs varied according to TP53 mutation status. These mutational differences can be used when designing future studies assessing the predictive ability of TP53 mutation status when targeting differentially affected molecular pathways.
Keywords: DNA sequencing, TP53, gastric cancer, gastroesophageal cancer, mutation analysis
Gastric (GC) and gastroesophageal junction adenocarcinomas (GEJ) are molecularly diverse. TP53 is the most frequently altered gene, with approximately 50% of patients harboring mutations. This article describes the distinct genomic alterations in GCs and GEJs stratified by TP53 mutation status.
Implications for Practice.
Despite being mutated in approximately 50% of gastroesophageal adenocarcinomas (GEAs), therapeutic interventions taking TP53 mutation status into account in a predictive capacity have yet to be developed. Additionally, there is limited information available assessing its ability to function as a predictive marker in the era of targeted cancer therapy. The mutational differences of GEAs stratified by TP53 mutation status as described in this manuscript can be used as a tool when conceiving future pre-clinical and clinical studies to assess the predictive ability of TP53 mutational status when using molecularly targeted agents.
Introduction
Gastric and esophageal cancer are the third and sixth leading cause of cancer deaths worldwide.1 In the US alone, 26 250 new cases of gastric cancer and 19 260 new cases of esophageal cancer were diagnosed in 2021, with males representing 61% and 79% of these new diagnoses.2 The western population, in particular, has seen the incidence of gastric cancer (GC) and steady increase of gastroesophageal junction adenocarcinomas (GEJs) due to the increasing prevalence of obesity and gastro-esophageal reflux disease.3-5 GCs and GEJs, collectively known as gastroesophageal adenocarcinomas (GEAs), are molecularly diverse. Attempts have been made to molecularly categorize these tumors to organize future studies and create tailored treatment strategies. In 2014, the Cancer Genome Atlas Research Network (TCGA) analyzed 295 GC samples using array-based somatic copy number analysis, whole-exome sequencing, array-based DNA methylation profiling, messenger RNA (mRNA) sequencing, microRNA (miRNA) sequencing, reverse-phase protein array, and microsatellite instability (MSI) testing to propose separating GC into 4 general molecular subtypes.6 The first 2 molecular subtypes were generated on the basis of CpG island hypermethylation (CIMP): Epstein-Barr virus (EBV)+ CIMP tumors (9%) with distinct hypermethylation patterns not associated with the epigenetic silencing of MLH1, and microsatellite instability (MSI) enriched CIMP EBV-tumors (22%) with hypermethylation patterns consistent with MLH1 epigenetic silencing. The remainder of the GCs were then stratified into one of 2 groups based on their relative burden of somatic copy number aberrations (SCNAs): genomically stable (GS) tumors (20%) had less SCNAs as compared to those deemed to have chromosomal instability (CIN, 50%).6
Commonly occurring genetic mutations in GEAs include ARID1A, PIK3CA, KRAS, and CDH1.7 However, TP53 is the most frequently altered gene with approximately 50% of GEAs and 70% of CIN GCs harboring mutations.6,8TP53 encodes the tumor suppressor p53 protein that functions as a key member of the G1/S checkpoint to help maintain genetic integrity through the cell cycle.9 In times of cellular stress TP53 expression increases to mediate G1 phase cell cycle arrest to facilitate repair before cellular division or promote apoptosis in cells with an overabundance of molecular derangements that cannot be overcome. When there is a loss of TP53 function, the G1/S checkpoint is effectively lost, and genetically damaged cells have significantly less barriers to unchecked proliferation with negative physiologic consequences. Therefore, it is not surprising that inactivating TP53 mutations have been implicated in the carcinogenesis of a multitude of malignancies, with GEAs being no exception.10 In fact, the Asian Cancer Research Group (ACRG) included TP53 expression as part of their proposed molecular classification of GC.11 Using gene expression panels derived from 300 GCs, the ACRG first separated GCs molecularly into 2 large categories: MSI and microsatellite stable (MSS). MSS tumors were then stratified by the presence of, or lack thereof, an epithelial-to-mesenchymal gene signature (MSS/EMT). The remaining MSS/EMT- tumors were divided into the final 2 categories, MSS/TP53+ and MSS/TP53−, based on levels of TP53 expression with noted differences in patient characteristics between these 2 groups.11
Next-generation sequencing (NGS) is now widely used for most advanced cancers as an aid to elucidate the genomic characteristics of tumors. While therapeutic interventions for GEAs taking TP53 mutation status into account have yet to be developed, its high prevalence makes it a subject of interest to clinical investigators as a potential predictive marker.4,10,12-14 An understanding of commonly co-occurring genetic alterations in TP53 wild-type (TP53WT) and TP53 mutated (TP53MUT) GEAs may help guide clinically practical investigations in the future assessing this potential. This study identified the unique mutational profiles of TP53WT and TP53MUT GEAs by analyzing the DNA sequencing results of 3741 GEA tumor samples.
Patients and Methods
De-identified data were acquired from Foundation Medicine for all available patients with GEA for whom the FoundationOne CDx (F1CDx) assay was performed within the US. F1CDx is a commercially available NGS diagnostic test developed by Foundation Medicine that uses targeted high throughput hybridization-based capture technology for the detection of substitutions, insertion and deletion alterations, and copy number alterations in 324 genes and select gene rearrangements using DNA isolated from formalin-fixed paraffin-embedded tumor tissue specimens.15 The data provided included age at the time of the assay results, gender, tumor mutational burden (TMB), and the distinct genomic alterations noted on DNA sequencing. Information on MSI and PD-L1 status was not available in the dataset. The dataset was sorted by TP53 mutation status. Differences in mutation frequency were detected using Fisher’s exact test of independence with a P-value of <.01 designated as the cutoff value for statistical significance. Statistical significance for continuous data was tested using a 2 samples t-test for mean values and the Mann-Whitney U test for distribution/median values.
Pathway analysis was completed by first isolating the genes with statistically different mutation frequencies between TP53WT and TP53MUT tumors. After studying these results, 12 predetermined functional gene groupings were created to help elucidate molecular pathways suggested to be differentially affected in TP53WT and TP53MUT tumors. The number of tumors containing a mutation in at least one of the prespecified genes in the molecular pathways of interest was counted. Then, the association of the frequency of tumors containing a gene mutation in the molecular pathways of interest and TP53 mutation status was assessed using Fisher’s exact test of independence with a P-value of <.01 deemed statistically significant.
Results
Demographics
The dataset consisted of 3741 patients with GEA: 2946 GCs and 795 GEJs. The entire study population was 65.2% male and 34.8% female. GC patients were 60.6% male and 39.4% female, whereas patients with GEJ were 82.4% male and 17.6% female (P < .001). The median patient age was 63, which was similar between TP53WT and TP53MUT patients.
Mutations Stratified by TP53 Mutation Status
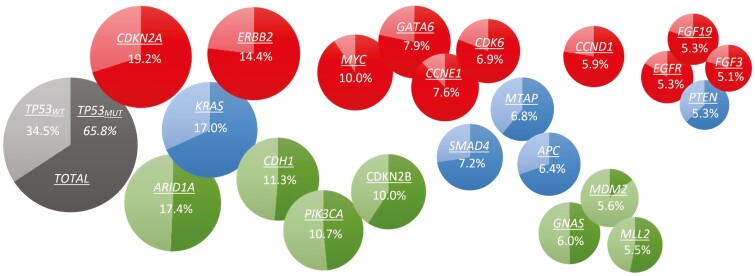
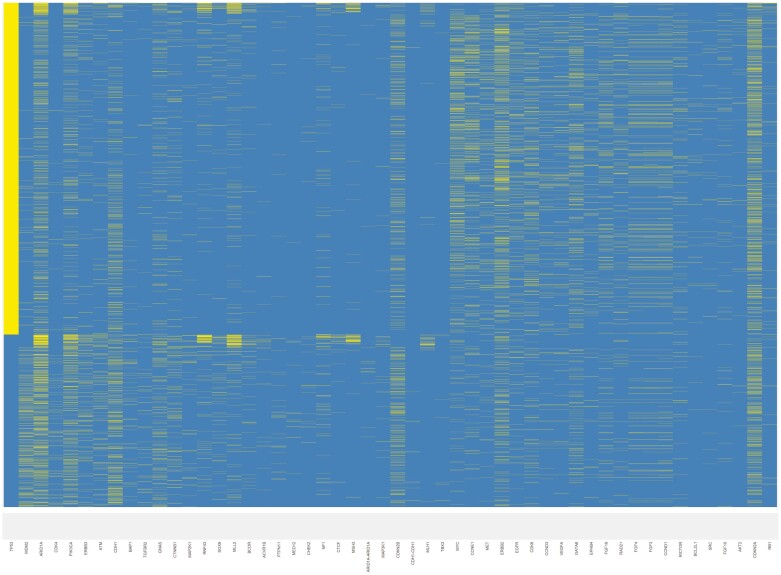
TP53 mutations were present in 65.8% of specimens. TP53 mutations were noted in 61.6% of GCs and 81.4% of GEJs (P < .001). The median TMB score was lower in TP53WT as compared to TP53MUT GEAs (2.5 vs. 3.8, P < .001). The frequency of tumors with a TMB score >10 mutations/megabase (11.1% vs. 9.4%; P = .11) were similar in TP53WT and TP53MUT groups, respectively. The most commonly mutated genes in the entire population other than TP53 were CDKN2A (19.1%), ARID1A (17.4%), KRAS (17.0%), ERBB2 (14.4%), and CDH1 (11.3%) (Fig. 1). Forty-nine of the 324 genes in the panel (15.1%) had statistically different mutation frequencies in TP53WT vs. TP53MUT patients (Fig. 2). The genetic alterations with the greatest over-representation in TP53WT patients included amplification mutations of MDM2 and CDK4, truncating mutations of ARID1A, point mutations of PIK3CA, and amplification/point mutations of ERBB3 (Table 1). Amplification mutations of MYC, CCNE1, and MET, along with amplification/point mutations of EGRF and ERBB2, were the most over-represented genetic alterations in TP53MUT patients (Table 2).
Figure 1.
Compound bubble pie chart of the 22 genes with >5% gene mutation frequency within the entire dataset (GEAs) in order of decreasing frequency. Each pie chart represents an individual gene broken down by the relative frequency in which patients harbor a mutation stratified by TP53 mutation status: the lighter color represents TP53WT patients, and the darker color represents TP53MUT patients. The numbers within the pie charts describe the frequency of the gene mutation within the entire dataset. Statistical significance of mutation frequency between TP53WT and TP53MUT tumors is designated using a color-coded scheme. Green: TP53MUT < TP53WT. Red: TP53MUT > TP53WT. Blue: TP53MUT = TP53WT.
Figure 2.
Distribution of gene mutations on an individual basis. Each row represents an individual. Each column represents one of the 49 genes noted to be differentially mutated stratified by TP53 mutation status. The yellow tick mark designates that the patient contains a mutation in the gene of interest. The background blue color denotes wild-type status. The 49 gene names, in order by column, are as follows: TP53, MDM2, ARID1A, CDK4, PIK3CA, ERBB3, ATM, CDH1, BAP1, TGFBR2, GNAS, CTNNB1, MAP2K1, RNF43, SOX9, MLL2, BCOR, ACVR1B, PTPN11, MED12, CHEK2, NF1, CTCF, MSH3, ARID1A-ARID1A, MAP3K1, CDKN2B, CDH1-CDH1, MLH1, TBX3, MYC, CCNE1, MET, ERBB2, EGFR, CDK6, CCND3, VEGFA, GATA6, EPHB4, FGF19, RAD21, FGF4, FGF3, CCND1, RICTOR, BCL2L1, SRC, FGF10, AKT2, CDKN2A, RB1.
Table 1.
Gene mutations overrepresented in TP53WT GEAs.
| Gene | TP53MUT (%) | TP53WT (%) | −log10(P-value) |
|---|---|---|---|
| MDM2 | 1.2 | 14.1 | 56.2 |
| ARID1A | 13.4 | 25.2 | 17.8 |
| CDK4 | 0.4 | 3.9 | 14.4 |
| PIK3CA | 7.9 | 16.2 | 13.3 |
| ERBB3 | 2.1 | 7 | 12.5 |
| ATM | 1.7 | 6 | 11 |
| CDH1 | 8.8 | 16 | 10 |
| BAP1 | 0.7 | 3.6 | 9.5 |
| TGFBR2 | 0.6 | 2.9 | 7.5 |
| GNAS | 4.5 | 8.8 | 6.5 |
| CTNNB1 | 2.8 | 6.1 | 5.5 |
| MAP2K1 | 0.8 | 2.7 | 5.4 |
| RNF43 | 3 | 6.3 | 5.2 |
| SOX9 | 1.5 | 3.8 | 5.1 |
| MLL2 | 4.4 | 7.6 | 4 |
| BCOR | 1.1 | 3 | 4 |
| ACVR1B | 0.3 | 1.4 | 3.5 |
| PTPN11 | 0.5 | 1.7 | 3.4 |
| MED12 | 0.1 | 0.8 | 3.2 |
| CHEK2 | 0.3 | 1.2 | 2.5 |
| NF1 | 2.2 | 3.8 | 2.4 |
| CTCF | 0.5 | 1.5 | 2.4 |
| MSH3 | 1.4 | 2.7 | 2.3 |
| MAP3K1 | 0.5 | 1.4 | 2.3 |
| CDKN2B | 9 | 11.9 | 2.2 |
| MLH1 | 0.8 | 1.8 | 2.1 |
| TBX3 | 0.1 | 0.5 | 2 |
Table 2.
Gene mutations overrepresented in TP53MUT GEAs.
| Gene | TP53MUT (%) | TP53WT (%) | −log10(P-value) |
|---|---|---|---|
| MYC | 13.8 | 2.8 | 30.4 |
| CCNE1 | 10.2 | 2.7 | 18.2 |
| MET | 6.1 | 1.8 | 9.6 |
| ERBB2 | 16.8 | 9.7 | 8.6 |
| EGFR | 6.8 | 2.5 | 8.3 |
| CDK6 | 8.5 | 3.9 | 7.1 |
| CCND3 | 4.8 | 1.6 | 6.7 |
| VEGFA | 4.8 | 1.6 | 6.3 |
| GATA6 | 9.3 | 5 | 5.8 |
| EPHB4 | 3.7 | 1.2 | 5.6 |
| FGF19 | 6.5 | 3 | 5.3 |
| RAD21 | 4.8 | 2 | 5.2 |
| FGF4 | 6.1 | 2.8 | 5.1 |
| FGF3 | 6.2 | 3 | 4.7 |
| CCND1 | 6.9 | 3.8 | 4 |
| RICTOR | 3.9 | 1.6 | 3.9 |
| BCL2L1 | 1.4 | 0.2 | 3.4 |
| SRC | 1.3 | 0.3 | 2.8 |
| FGF10 | 2.3 | 0.9 | 2.5 |
| AKT2 | 1.1 | 0.2 | 2.5 |
| CDKN2A | 20.5 | 16.8 | 2.2 |
| RB1 | 1.9 | 0.8 | 2 |
Pathway Analysis
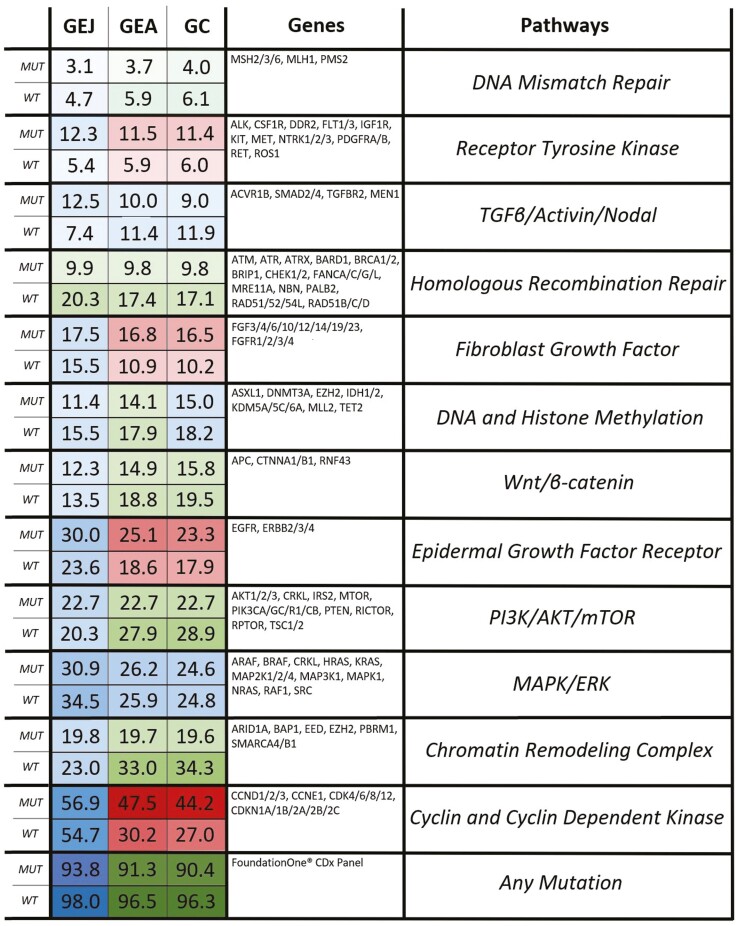
The pathway analysis results are represented in Fig. 3. TP53WT GEAs were more likely to contain a mutation in a gene related to DNA mismatch repair, homologous recombination repair, DNA and histone methylation, Wnt/B-catenin, PI3K/Akt/mTOR, and chromatin remodeling complexes. Furthermore, TP53WT tumors were more likely to contain any mutation in the gene panel as compared to TP53MUT tumors. TP53MUT GEAs were more likely to contain a mutation in a gene related to fibroblast growth factor, epidermal growth factor receptor, other receptor tyrosine kinases, and cyclin and cyclin-dependent kinases. When broken down by tumor type, GCs had pathway analyses results that closely mirrored those of the entire dataset with the only difference being that the frequency of mutations in DNA and histone methylation genes was no longer statistically different between TP53WT and TP53MUT GCs (P = .026). On the other hand, the lone pathway that was differentially affected in TP53WT and TP53MUT GEJs was homologous recombination repair (20.3% vs. 9.9%, P = .001).
Figure 3.
The final pathway analysis results. The column on the right describes the 12 functional gene groupings that were created to represent molecular pathways suggested to be differentially affected in TP53WT and TP53MUT GEAs. The purpose of this figure is to illustrate whether the mutational involvement of these pathways/gene groupings varied between TP53WT and TP53MUT GEAs (entire dataset), GCs, and GEJs. The column in the middle contains the pre-specified genes within the pathways of interest. The numbers in the left column describe the percentage of patients containing a mutation in at least one of the genes in the pathway of interest. The left column is broken down into 3 sub-columns by tumor type. Each row is broken down into 2 sub-rows denoting TP53 mutation status. Statistical significance of mutation frequency between TP53WT and TP53MUT tumors is designated using a color-coded scheme. Green: TP53MUT < TP53WT. Red: TP53MUT > TP53WT. Blue: TP53MUT = TP53WT.
Discussion
The most common predictive biomarkers currently used in GEAs are microsatellite instability or mismatch repair deficiency status and expression of human epidermal growth factor receptor 2 (HER2) and programmed death-ligand 1 (PD-L1).16,17 There are other biomarkers that will soon have therapeutic implications, such as Claudin 18.2 and FGFR2b.18,19 The use of these predictive biomarkers has helped us tailor treatments for patients with GEA, yet there is still an unmet need to identify additional targets for novel treatment strategies. In our study, we were able to demonstrate that the mutational profiles of GEAs differed when stratified according to TP53 mutation status. TP53 mutations were identified at higher rates in our sample of GEAs as compared to those described by TCGA and other manuscripts reporting such data.4,8,20,21 Genes directly involved in the DNA damage response and epigenetic regulation of DNA were more frequently mutated in TP53WT tumors. Meanwhile, growth factor and receptor tyrosine kinase genes were more frequently mutated in TP53MUT tumors.
GEJ tumors were more likely to be TP53MUT than their GC counterparts (81.4% vs. 61.6%, P-value = <.001) in this dataset, which is consistent with DNA sequencing results previously described.22,23 Given this variability, there was interest in comparing GEJ and GC mutation profiles against one another to assess any further genetic differences stratified by TP53 mutational status that may distinguish the 2 GEA malignancies. Homologous recombination repair genes were more frequently mutated in both TP53WT GEJs and GCs. Additionally, the ratio of mutational involvement of DNA mismatch repair, DNA and histone methylation, epidermal growth factor, and receptor tyrosine kinase genes appeared to be consistent in GEJs and GCs when broken down by TP53 mutation status. The remaining functional gene groupings, however, did not seem to have as much mutational variation between TP53WT and TP53MUT tumors in GEJs as compared to GCs. This can be most readily seen when looking at the involvement of cyclin and cyclin-dependent kinase genes, which play an integral role in the maintenance of the cell cycle in concert with TP53. Twenty-seven percent vs. 44.2% of TP53WT and TP53MUT GCs had cyclin and cyclin-dependent kinase gene mutations. Meanwhile, 54.7% vs. 56.9% of TP53WT and TP53MUT GEJs had cyclin and cyclin-dependent kinase gene mutations. On a gene-specific level, ZNF217 (15.5%) and AURKA (8.1%) amplification mutations were disproportionately seen in TP53WT GEJs as compared to the rest of the dataset (4.4% and 2.1%, respectively). As both ZNF217 and AURKA genes are located on chromosome 20q13, it stands to reason that 20q13 amplification may play a prominent role in the carcinogenesis of TP53WT GEJs.20,24 These findings suggest that despite their anatomic proximity the genetic makeup of GEJs and GCs are distinct from one another.
Studies assessing the prognostic impact of TP53 mutations on the outcomes of GEAs have demonstrated conflicting results.25-29 The molecular characterization of GEAs in relation to TCGA subtype, co-occurring mutations, and the type of TP53 mutation itself all seem to influence TP53’s prognostic significance.11,12,30 Equally, the ability of TP53 mutation status to function as a predictive marker of treatment response is also unknown. A meta-analysis of 13 studies including 564 cases concluded that GCs with high levels of p53 expression on immunohistochemistry (IHC) have a decreased response to chemotherapy.31 Unfortunately, the correlation of p53 expression on IHC with TP53 mutational status is not consistent, making it difficult to extrapolate the findings of this analysis toward tumor DNA sequencing results.32,33 More recent studies have attempted to delineate the predictive potential of TP53 mutations in the era of molecularly targeted therapies and immunotherapies. One manuscript supporting its predictive ability in targeting the vascular endothelial growth factor (VEGF) pathway was recently published.34 In 48 patients with GC who received second-line ramicurimab/paclitaxel combination therapy the median OS was 9.5 months for carriers of TP53Inactive mutations, 8.6 months for carriers of other TP53 mutations, 6.0 months for carriers of TP53Active missense mutations, and 4.5 months for TP53WT patients (P = .01). VEGFA was accordingly more mutated in our group of TP53MUT GEAs as compared to TP53WT GEAs (4.8% vs. 1.6%, −log10(P-value) = 6.3). In another review of 356 GCs from the TCGA database, it was found that TP53MUT tumors had lower levels of anti-tumor immunity and a decreased response to immune checkpoint inhibitor therapy as compared to TP53WT tumors.35 On the other hand, sub-group analysis of the phase III GOLD trial investigating olaparib and paclitaxel combination therapy in recurrent and metastatic GC found no difference in therapeutic response based on TP53 mutation status.36 More studies are needed to assess the predictive ability of TP53 mutation in isolation and in conjunction with other molecular alterations.
The limitations of this study include missing data regarding clinical outcomes, microsatellite instability status, expression of other protein biomarkers, and additional patient characteristics necessary to organize these tumors into one of the 4 molecular subtypes as proposed by the Cancer Genome Atlas Research Network. In addition, while tissue-based assays offer a comprehensive genomic profile, there can be intra- and intertumoral (primary vs. metastatic site) heterogeneity which may not be fully encompassed on tissue NGS.37 This potentially limits the clinical utility of tissue-based mutation analysis as the full mutational landscape of the patient’s tumors may not be elucidated due to the sampling limitations inherent with this method of genetic profiling. This notion, along with the invasiveness of obtaining tumor samples for molecular analysis, has led to the increased use of liquid biopsies assessing circulating tumor DNA (ctDNA) NGS results in conjunction with tissue NGS to guide clinical decision-making. Nonetheless, as tissue-based DNA/RNA sequencing results yet remain standard in clinical practice, the observations of this study could serve as a foundation for practical future preclinical and clinical investigations that can help elucidate the clinical correlations of our findings. For example, genes in the PI3K/AKT/mTOR pathway were significantly more mutated in TP53WT tumors, which was largely driven by the greater presence of PIK3CA mutations in TP53WT as compared to TP53MUT tumors (16.2% vs. 7.9%). Trials are currently underway assessing the role of PI3K inhibitors for PIK3CA mutated tumors in GEAs. It would be useful to perform a subgroup analysis of the results of these trials stratified by TP53 mutation status to determine if these findings signal a pathophysiologic difference that carries predictive relevance in the real-world setting. Similar analyses could be completed for additional studies investigating therapeutic interventions targeting genes and cellular products related to the DNA damage response, epigenetic regulation of DNA, growth factors, receptor tyrosine kinases, cyclins and cyclin-dependent kinases, and the PI3K/AKT/mTOR pathway.
Despite the inherent limitations of this qualitative study, compelling trends within a large database of tumor DNA sequencing results derived from clinically accessible information were identified. A clinical trial assessing the efficacy of berzosertib and irinotecan in TP53 mutant GEA cancers is currently ongoing (NCT03641313) which is the first clinical trial selected for patients based on TP53 status. The mutational differences of GEAs stratified by TP53 mutation status as described in this manuscript can be used as a tool when designing and analyzing future pre-clinical and clinical trials to assess the predictive ability of TP53 mutational status when targeting differentially affected molecular pathways.
Funding
This work was supported in part by the Biostatistics and Bioinformatics Shared Resource at the H. Lee Moffitt Cancer Center & Research Institute, an NCI designated Comprehensive Cancer Center (P30-CA076292).
Conflict of Interest
Sarah Hoffe: ViewRay and Varian (RF); Merck, MyCareGorithm (C/A); Beyond the White Coat LLC (OI); Vit INC (SAB); H: UptoDATE (H). Christine Walko: Jackson Genetic Laboratories Molecular Tumor Board (SAB); Rutika Mehta: Eli Lilly, BMS, Astellas (SAB); Eli Lilly, Natera, Daiichi Sankyo (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board.
Author Contributions
Conception/Design: A.C.W., Y.Z., Q.M., L.C., C.M.W., R.M. Provision of study material/patients: A.C.W., R.M. Collection and/or assembly of data: A.C.W., Y.Z., Q.M., L.C., R.M. Data analysis and interpretation: A.C.W., Y.Z., Q.M., L.C., R.M. Manuscript writing: A.C. W., Y.Z., Q.M., L.C., J.F., S.E.H., J.F., S.P.D., J.M.P., C.M.W., R.M., Final approval of manuscript: All authors.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394-424. [DOI] [PubMed] [Google Scholar]
- 2. https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html.
- 3. Thrift AP, Whiteman DC.. The incidence of esophageal adenocarcinoma continues to rise: analysis of period and birth cohort effects on recent trends. Ann Oncol. 2012;23(12):3155-3162. [DOI] [PubMed] [Google Scholar]
- 4. GBD 2017 Gastro-oesophageal Reflux Disease Collaborators. The global, regional, and national burden of gastro-oesophageal reflux disease in 195 countries and territories, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5(6):561-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390(10113):2627-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tan P, Yeoh KG.. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149(5):1153-1162.e3. [DOI] [PubMed] [Google Scholar]
- 8. Hanazono K, Natsugoe S, Stein HJ, Aikou T, Hoefler H, Siewert JR.. Distribution of p53 mutations in esophageal and gastric carcinomas and the relationship with p53 expression. Oncol Rep. 2006;15(4):821-824. [PubMed] [Google Scholar]
- 9. Mantovani F, Collavin L, Del Sal G.. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 2019;26(2):199-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Busuttil RA, Zapparoli GV, Haupt S, et al. Role of p53 in the progression of gastric cancer. Oncotarget. 2014;5(23):12016-12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015;21(5):449-456. [DOI] [PubMed] [Google Scholar]
- 12. Park S, Lee J, Kim YH, Park J, Shin JW, Nam S.. Clinical relevance and molecular phenotypes in gastric cancer, of TP53 mutations and gene expressions, in combination with other gene mutations. Sci Rep. 2016;6:34822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yildirim M, Kaya V, Demirpence O, Gunduz S, Bozcuk H.. Prognostic significance of p53 in gastric cancer: a meta-analysis. Asian Pac J Cancer Prev. 2015;16(1):327-332. [DOI] [PubMed] [Google Scholar]
- 14. Xu HY, Xu WL, Wang LQ, Chen MB, Shen HL.. Relationship between p53 status and response to chemotherapy in patients with gastric cancer: a meta-analysis. PLoS One. 2014;9(4):e95371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.https://www.foundationmedicine.com/test/foundationone-cdx
- 16. Bang YJ, Van Cutsem E, Feyereislova A, et al. ; ToGA Trial Investigators . Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet. 2010;376(9742):687-697. [DOI] [PubMed] [Google Scholar]
- 17. Chao J, Fuchs CS, Shitara K, et al. Assessment of pembrolizumab therapy for the treatment of microsatellite instability-high gastric or gastroesophageal junction cancer among patients in the KEYNOTE-059, KEYNOTE-061, and KEYNOTE-062 clinical trials. JAMA Oncol. 2021;7(6):895-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sahin U, Türeci Ö, Manikhas G, et al. FAST: a randomised phase II study of zolbetuximab (IMAB362) plus EOX versus EOX alone for first-line treatment of advanced CLDN18.2-positive gastric and gastro-oesophageal adenocarcinoma. Ann Oncol. 2021;32(5):609-619. [DOI] [PubMed] [Google Scholar]
- 19. Wainberg ZA, Enzinger PC, Kang YK, et al. Randomized double-blind placebo-controlled phase 2 study of bemarituzumab combined with modified FOLFOX6 (mFOLFOX6) in first-line (1L) treatment of advanced gastric/gastroesophageal junction adenocarcinoma (FIGHT). J Clin Oncol. 2021;39(suppl 3):abstr 160. [Google Scholar]
- 20. Poremba C, Yandell DW, Huang Q, et al. Frequency and spectrum of p53 mutations in gastric cancer—a molecular genetic and immunohistochemical study. Virchows Arch. 1995;426(5):447-455. [DOI] [PubMed] [Google Scholar]
- 21. Li-Chang HH, Kasaian K, Ng Y, et al. Retrospective review using targeted deep sequencing reveals mutational differences between gastroesophageal junction and gastric carcinomas. BMC Cancer. 2015;15:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fléjou JF, Gratio V, Muzeau F, Hamelin R.. p53 abnormalities in adenocarcinoma of the gastric cardia and antrum. Mol Pathol. 1999;52(5):263-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ireland AP, Shibata DK, Chandrasoma P, Lord RV, Peters JH, DeMeester TR.. Clinical significance of p53 mutations in adenocarcinoma of the esophagus and cardia. Ann Surg. 2000;231(2):179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cohen PA, Donini CF, Nguyen NT, Lincet H, Vendrell JA.. The dark side of ZNF217, a key regulator of tumorigenesis with powerful biomarker value. Oncotarget. 2015;6(39):41566-41581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pasello G, Agata S, Bonaldi L, et al. DNA copy number alterations correlate with survival of esophageal adenocarcinoma patients. Mod Pathol. 2009;22(1):58-65. [DOI] [PubMed] [Google Scholar]
- 26. Ikari N, Serizawa A, Mitani S, Yamamoto M, Furukawa T.. Near-comprehensive resequencing of cancer-associated genes in surgically resected metastatic liver tumors of gastric cancer. Am J Pathol. 2019;189(4):784-796. [DOI] [PubMed] [Google Scholar]
- 27. Blanchet A., et al. Isoforms of the p53 family and gastric cancer: a ménage à trois for an unfinished affair. Cancers (Basel). 2021;13(4):916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lim BH, Soong R, Grieu F, Robbins PD, House AK, Iacopetta BJ.. p53 accumulation and mutation are prognostic indicators of poor survival in human gastric carcinoma. Int J Cancer. 1996;69(3):200-204. [DOI] [PubMed] [Google Scholar]
- 29. Deng W, Hao Q, Vadgama J, Wu Y.. Wild-type TP53 predicts poor prognosis in patients with gastric cancer. J Cancer Sci Clin Ther. 2021;5(1):134-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tahara T, Shibata T, Okamoto Y, et al. Mutation spectrum of TP53 gene predicts clinicopathological features and survival of gastric cancer. Oncotarget. 2016;7(27):42252-42260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xu HY, Xu WL, Wang LQ, Chen MB, Shen HL.. Relationship between p53 status and response to chemotherapy in patients with gastric cancer: a meta-analysis. PLoS One. 2014;9(4):e95371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hwang HJ, Nam SK, Park H, et al. Prediction of TP53 mutations by p53 immunohistochemistry and their prognostic significance in gastric cancer. J Pathol Transl Med. 2020;54(5):378-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schoop I, Maleki SS, Behrens HM, Krüger S, Haag J, Röcken C.. p53 immunostaining cannot be used to predict TP53 mutations in gastric cancer: results from a large Central European cohort. Hum Pathol. 2020;105:53-66. [DOI] [PubMed] [Google Scholar]
- 34. Graziano F., et al. TP53 mutation analysis in gastric cancer and clinical outcomes of patients with metastatic disease treated with Ramucirumab/Paclitaxel or standard chemotherapy. Cancers (Basel). 2020;12(8):2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li L, Li M, Wang X.. Cancer type-dependent correlations between TP53 mutations and antitumor immunity. DNA Repair (Amst). 2020;88:102785. [DOI] [PubMed] [Google Scholar]
- 36. Liu YZ, et al. Olaparib plus paclitaxel sensitivity in biomarker subgroups of gastric cancer. Ann Oncol. 2018;29(suppl_8):814-857. [Google Scholar]
- 37. Gao JP, Xu W, Liu WT, Yan M, Zhu ZG.. Tumor heterogeneity of gastric cancer: from the perspective of tumor-initiating cell. World J Gastroenterol. 2018;24(24):2567-2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.