ABSTRACT
Anterior segment eye diseases account for a significant proportion of presentations to eye clinics worldwide, including diseases associated with corneal pathologies, anterior chamber abnormalities (e.g. blood or inflammation), and lens diseases. The construction of an automatic tool for segmentation of anterior segment eye lesions would greatly improve the efficiency of clinical care. With research on artificial intelligence progressing in recent years, deep learning models have shown their superiority in image classification and segmentation. The training and evaluation of deep learning models should be based on a large amount of data annotated with expertise; however, such data are relatively scarce in the domain of medicine. Herein, the authors developed a new medical image annotation system, called EyeHealer. It is a large-scale anterior eye segment dataset with both eye structures and lesions annotated at the pixel level. Comprehensive experiments were conducted to verify its performance in disease classification and eye lesion segmentation. The results showed that semantic segmentation models outperformed medical segmentation models. This paper describes the establishment of the system for automated classification and segmentation tasks. The dataset will be made publicly available to encourage future research in this area.
Keywords: anterior segment eye diseases, artificial intelligence, annotation
Introduction
As a vital means of making an initial diagnosis, the slit-lamp provides a convenient way of examining the front part of the eye. Through the diffused light of the microscope, the exposed eye structures can be directly seen, including the eyelid, conjunctiva, cornea, iris, and lens, etc. However, small variances in the normal structures can accumulate to marked differences in the photos. For example, even normal eyes are different from each other as there will be differences in the color or morphology of the iris, or the size of the cornea or the pupil, not to mention eyes that may have various disorders that might occur either alone or concurrently. A series of blinding disorders may develop at the front part of the eye, such as corneal opacity, hyphema, endophthalmitis, and cataract. Compared to fundus images1 or optical coherence tomography (OCT),2 which has been a popular choice in artificial intelligence (AI) research3 in the recent decade, the less uniform modality of slit-lamp images limits its application in AI models to a certain extent, even though this is more common and cost-efficient. To overcome this issue, some researchers chose to make diagnoses within specific diseases that mostly only involve one single structure, such as infectious keratitis,4–6 pterygium,7,8 congenital,9 or senile nuclear cataract.10 However, the requirement of a large-scale dataset for each disease hampers the establishment of deep learning models that cover a variety of ocular diseases. Furthermore, the number of images needed will scale up if there are concurrent disorders or unusual forms of different diseases, both of which are commonly seen during slit-lamp examination.
As the main track in computer vision research, semantic segmentation, that is, the classification of each pixel in an image,11–13 has always been the focus of researchers. For example, in the field of automatic driving, semantic segmentation would be segmentation of common road traffic signs or pedestrians.14 Highly innovative segmentation models, such as DeepLabv3,15 have been proposed. Thus, a medical model falling under the category of semantic segmentation would indicate the segmentation in medical images.
Precise segmentation of the lesion area could be an effective approach to develop an AI model for anterior segment diseases based on imaging. However, manual segmentation is labor-intensive and tedious, and can only be performed by experienced ophthalmologists. With the rapid development of AI, computer-assisted diagnosis (CAD) has drawn great attention over the past few years, with the construction of numerous deep learning models that aim to tackle the lesion segmentation task.16–19 However, sufficient data are essential for the training and validation of these models. Therefore, there is an urgent need for eye lesion datasets that are (1) large, (2) cover a wide range of diseases, (3) open-source, and (4) reliably annotated.
In the current research, EyeHealer was established as a large-scale dataset for anterior eye lesion segmentation, with both eye structures and lesions annotated at the pixel level. To fully evaluate the established dataset, comprehensive experiments were performed to examine its effectiveness in disease classification and eye lesion segmentation. The dataset will be made publicly available to encourage future research in this area.
Methods
Images from human subjects
Anterior eye segment photographs from a slit-lamp camera were retrospectively collected from the Zhongshan Ophthalmic Center, Sun Yat-sen University, and West China Hospital, from January 2019 to March 2020. The dataset consisted of 2192 anterior eye segment photographs. All imaging was performed as part of patients’ routine clinical care. No exclusion criteria were based on race, age, or gender. Images from various brands of slit-lamps (BX-900, Haag-Streit; LS-6 and LS-7, Chongqing SunKingdom) and cameras (Canon 800D, 600D, 80D, and 7D) were collected. Anterior eye segment images were downloaded with a standard JPEG compression format. Ethical reviews were conducted by the Zhongshan Ophthalmic Centre of Sun Yat-sen University and West China Hospital Ethics Review Committee, and approval was obtained.
Image labeling and annotation
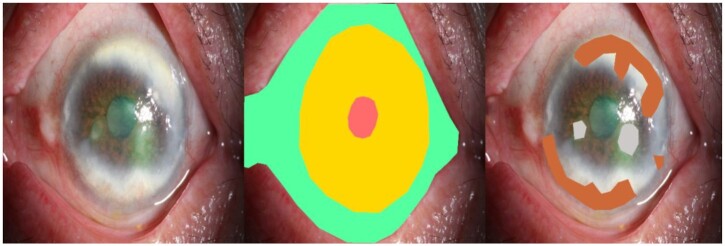
Before training, each image went through a tiered labeling system consisting of multiple layers of ophthalmologists of increasing expertise for verification and correction of image labels. Six ophthalmologists with at least five years of experience in ophthalmology, two of whom had more than eight years of experience, were invited to annotate the photographs. Initial quality control was conducted, in which duplicated, incorrect magnification images, and photographs with a lack of clarity were excluded. Only diffused bright light images with the three specific eye structures, interpalpebral zone, cornea, and pupil, were kept. After the quality control process, the images were separated into six groups and six ophthalmologists independently labeled each group of images. Two types of experiments, classification and segmentation, were conducted. To secure uniformity among different images, the annotation focused on only three eye structures, the eyelid, cornea, and pupil. A total of 23 types of ocular diseases were included in the dataset, with each type corresponding to a certain lesion form. The eye structures and lesions were labeled at the pixel level, where polygons exist and may result in subtle differences from the ground truth. A visual example of the constructed dataset is shown in Fig. 1. The six groups of images with labels were then separated and examined by the two senior ophthalmologists independently. Each of them made sure that no duplicate folders were seen. Images with problematical labels were discussed until the two specialists agreed on the grading. Finally, a retinal specialist with over 20 years of experience was invited to verify the true labels for divergent images.
Figure 1.
Visual illustration of the constructed dataset, including the original photograph, eye structure annotations, and eye lesion annotations.
Classification experiment
The classification experiment was conducted on the top 10 most common diseases (based on the sample size) in the dataset. Three classic pre-trained models in the field of image classification, ResNet-50,20 InceptionV3,21 and DenseNet-121,22 were employed to get a comprehensive evaluation of our dataset. We briefly introduce the features of each model here. ResNet features its strength in handling extremely deep network structures. The key component of ResNet is the residual connection block, which enables the gradient flow to propagate to each layer smoothly. InceptionV3 utilizes a multi-scale filter kernel to extract semantic features at different levels. To reduce computation cost, 'factorization into small convolutions' is applied. DenseNet is based on ResNet. In ResNet, one layer can only get information from the previous layer. However, in DenseNet, with the dense connection design, one layer can get information from an arbitrary layer at the front. For each model, we initialized with weights pre-trained from the ImageNet dataset,21 and we trained all parameters in the model simultaneously.
The data were randomly divided into the training set, validation set, and test set by a ratio of 0.7:0.1:0.2. The size of the hold-out method for testing was 20% of the images for each of the 10 top diseases, resulting in 234, 84, 55, 45, 44, 53, 46, 26, 28, and 20 images. The modeling results were reported only on the held-out test set. An early stopping mechanism was applied, that is, the training of the model would be terminated when the loss of the model on the validation set is significantly higher than the loss on the training set. In this experiment, the terminations were concentrated in the 5th epoch. In such a multi-label classification task, the activation function at the last layer was replaced with the sigmoid function and the weighted binary cross-entropy serving as the loss function for optimization. Each photograph was resized to 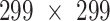 and normalized according to the mean and standard deviation of ImageNet. Adam Optimizer with an initial learning rate at 0.001 was applied and decayed by a factor of 0.8 for every 10 epochs, with a batch size of 8 and a dropout rate of 0.5.
and normalized according to the mean and standard deviation of ImageNet. Adam Optimizer with an initial learning rate at 0.001 was applied and decayed by a factor of 0.8 for every 10 epochs, with a batch size of 8 and a dropout rate of 0.5.
Segmentation workflow
In this part, segmentation experiments were performed on all images. First, the ophthalmologists labeled the three eye structures, the interpalpebral zone, cornea, and pupil, using polygons serving as location marks. Then, eye lesions were segmented. Two medical image segmentation models, DRUNet23 and SegNet,24 and two state-of-the-art semantic segmentation models, PspNet25 and DeepLabv3,15 were applied. The final convolutional map was activated by a sigmoid function, with weighted binary cross-entropy as the loss function. Again, the data were randomly classified into the training set, validation set, and test set by the same ratio mentioned above.
Only one type of lesion was segmented at a time. Each photograph was resized to 256 × 256 and rescaled to [0,1] through the dividing procedure by 255. Also, an SGD optimizer with an initial learning rate of 0.01 and momentum of 0.9 was employed. The data were classified as mentioned above.
Evaluation metrics
To evaluate the performance and stability of the model, the two experiments adopted different quantitative metrics. In the classification model, precision (PRE), recall (REC) and F1 Score were calculated, as follows.
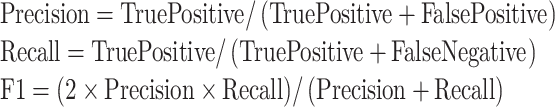 |
Precision and recall are the common evaluation indicators for classification. F1 Score measures the balance of the positive and negative samples at the same time.26
In the segmentation model, Intersection over Union (IoU), Dice coefficient (Dice), and Pixel Accuracy (PA) were applied to measure the performance.
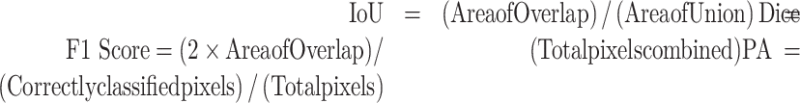 |
The IoU and Dice are the most commonly used metrics for segmentation experiments, while PA is considered to be the simplest indicator.
Results
Fundamental concept of EyeHealer
Anterior segment images serve as a routine and standard of care in the ophthalmic clinic. These can be easily acquired and give clinicians a thorough impression of the eye at first sight. The initial diagnosis is made, then decisions on whether to perform further examinations or start treatments. Unfortunately, even though this appears to be a simple procedure of taking photographs, some lesions could be ambiguous, making it hard to make correct diagnoses. Misdiagnosis can happen, which might lead to irreversible loss of vision and decreased quality of life. Hence, the EyeHealer system was established and trained on a professional level with annotated anterior segment photos to provide patients with accurate diagnoses.
EyeHealer divides ocular images into basic anatomical structures and provides annotations of the lesions. The system is highly formalized to ensure the accuracy and precision of the annotation. Specifically, three ocular structures will be strictly segmented first: the interpalpebral zone, cornea, and pupil. Subsequently, the pathological area will be marked out based on the training datasets. Hence, the system provides both ocular disease diagnosis and accurate lesion labeling. With the help of the system, visual and reliable ocular disease diagnoses can be made for patients, and shared with clinicians from other departments.
Imaging datasets
A total of 3813 slit-lamp images that met the criteria for inclusion during the quality control process were annotated by six experienced ophthalmologists. A statistical overview of the constructed dataset is shown in Table 1. In total, 23 types of diseases were included, sorted by the numbers of samples. The top 10 most common diseases, with sample sizes >100 cases, were used for the classification labeling. The remaining diseases were excluded to ensure the model's performance and accuracy rate. As a result, a total of 3194 images were included.
Table 1.
Statistical overview of the proposed dataset sorted by the number of samples.
| Disease name | Sample number | Avg size ratio | Disease name | Sample number | Avg size ratio |
|---|---|---|---|---|---|
| Cataract | 1160 | 8.49% | Corneal edema | 80 | 34.18% |
| Pterygium | 482 | 13.62% | Subconjunctival hemorrhage | 58 | 23.87% |
| Cornea arcus senillis | 320 | 32.82% | Nevus of iris | 58 | 1.53% |
| Pinguecula | 238 | 4.64% | Corneal degeneration | 43 | 11.13% |
| Conjunctival hyperemia | 221 | 4.46% | Synechia | 43 | 3.47% |
| Corneal neovascularization | 214 | 12.35% | Hyphema | 42 | 9.70% |
| Corneal opacity | 209 | 11.55% | Hypopyon | 27 | 4.25% |
| Corneal leucoma | 130 | 10.92% | Iris astrophy | 27 | 7.25% |
| Corneal ulcer | 112 | 8.83% | Eyelid tumor | 25 | 4.80% |
| Discoria | 108 | 10.14% | Pigment adhesion to lens capsule | 25 | 5.01% |
| Ciliary congestion | 85 | 69.43% | Lens dislocation | 24 | 20.53% |
| Conjunctival nevus | 82 | 10.78% |
Sorted by sample number.
Avg size ratio, average size ratio = (lesion area)/(total image area) × 100%.
Anterior ocular disease classification with three pre-trained models
The results of the classification experiment are shown in Table 2. Precision, recall, and F1 Score were used to evaluate the performance of models in each of the 10 diseases. Comparison of the model classification performance over different diseases revealed the best results in cataract because of the ample amount of images. Both the precision rate and recall rate were >80%. The F1 Score is used to indicate the balance between the two indices and was >85% in all three pre-trained models. For pterygium, the precision and recall of DenseNet were >90%, with a percentage of >70 on InceptionV3 and 50 on ResNet. However, when we compare cataract and pterygium with other diseases, the performance is relatively low. The reason for this could be the large differences in numbers of sample images across different diseases. The problem persisted even after adjustment of label weights.
Table 2.
Experimental results of classification over the top 10 most common diseases.
| Disease name | ResNet | InceptionV3 | DenseNet | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Pre | Rec | F1 | Pre | Rec | F1 | Pre | Rec | F1 | |
| Cataract | 0.858 | 0.862 | 0.860 | 0.821 | 0.909 | 0.863 | 0.827 | 0.931 | 0.876 |
| Pterygium | 0.716 | 0.578 | 0.640 | 0.784 | 0.746 | 0.765 | 0.903 | 0.903 | 0.903 |
| Cornea arcus senillis | 0.422 | 0.545 | 0.476 | 0.387 | 0.654 | 0.486 | 0.391 | 0.69 | 0.499 |
| Pinguecula | 0.273 | 0.511 | 0.356 | 0.232 | 0.577 | 0.331 | 0.382 | 0.577 | 0.460 |
| Conjunctival hyperemia | 0.370 | 0.522 | 0.433 | 0.394 | 0.636 | 0.487 | 0.491 | 0.509 | 0.499 |
| Corneal neovascularization | 0.431 | 0.716 | 0.538 | 0.439 | 0.754 | 0.555 | 0.631 | 0.679 | 0.654 |
| Corneal opacity | 0.329 | 0.673 | 0.442 | 0.433 | 0.5 | 0.464 | 0.56 | 0.608 | 0.583 |
| Corneal leucoma | 0.288 | 0.576 | 0.384 | 0.25 | 0.807 | 0.382 | 0.472 | 0.653 | 0.548 |
| Corneal ulcer | 0.514 | 0.642 | 0.571 | 0.375 | 0.75 | 0.500 | 0.5 | 0.607 | 0.548 |
| Discoria | 0.281 | 0.45 | 0.346 | 0.23 | 0.45 | 0.304 | 0.5 | 0.35 | 0.412 |
Pre, precision; Rec, recall.
Lesion segmentation performance of the four models
As above, the top 10 most common diseases were included, and the area of each lesion was outlined using polygons as segmentation labels. Attempts to segment 10 different lesions at the same time in one model brought no satisfying results. Consequently, only one type of lesion was segmented at a time. The segmentation performance was evaluated with three evaluation metrics: IoU, Dice, and PA. The results are separated into Table 3 and Table 4, respectively. Comparison of the two medical image segmentation models, DRUNet and SegNet, revealed similar performance, so did comparison of the semantic segmentation models, PspNet and DeepLabv3. The models performed significantly well in cataract and pterygium, the two most common blinding anterior ocular diseases. Moreover, semantic segmentation models were found to outperform medical segmentation models. PA for cataract in medical image segmentation models and semantic segmentation models were all around 0.90, but for pterygium, medical image segmentation models were around 0.75, and for semantic segmentation models around 0.85.
Table 3.
Experimental results of eye lesion segmentation with DRUNet and SegNet.
| Disease name | DRUNet | SegNet | ||||
|---|---|---|---|---|---|---|
| IoU | Dice | PA | IoU | Dice | PA | |
| Cataract | 0.735 | 0.837 | 0.91 | 0.728 | 0.832 | 0.883 |
| Pterygium | 0.608 | 0.732 | 0.743 | 0.595 | 0.723 | 0.759 |
| Cornea arcus senillis | 0.312 | 0.421 | 0.599 | 0.261 | 0.381 | 0.583 |
| Pinguecula | 0.289 | 0.397 | 0.497 | 0.207 | 0.315 | 0.598 |
| Conjunctival hyperemia | 0.237 | 0.326 | 0.607 | 0.213 | 0.306 | 0.444 |
| Corneal neovascularization | 0.199 | 0.301 | 0.534 | 0.108 | 0.169 | 0.303 |
| Corneal opacity | 0.195 | 0.281 | 0.368 | 0.168 | 0.253 | 0.493 |
| Corneal leucoma | 0.29 | 0.393 | 0.504 | 0.252 | 0.367 | 0.622 |
| Corneal ulcer | 0.302 | 0.402 | 0.663 | 0.232 | 0.347 | 0.716 |
| Discoria | 0.252 | 0.357 | 0.638 | 0.206 | 0.313 | 0.742 |
IoU, intersection over union; Dice, dice coefficient; PA, pixel accuracy.
Table 4.
Experimental results of eye lesion segmentation with PspNet and DeepLabv3.
| Disease name | PspNet | DeepLabv3 | ||||
|---|---|---|---|---|---|---|
| IoU | Dice | PA | IoU | Dice | PA | |
| Cataract | 0.784 | 0.869 | 0.905 | 0.777 | 0.87 | 0.895 |
| Pterygium | 0.684 | 0.797 | 0.858 | 0.699 | 0.81 | 0.854 |
| Cornea arcus senillis | 0.36 | 0.46 | 0.6 | 0.355 | 0.456 | 0.595 |
| Pinguecula | 0.312 | 0.44 | 0.65 | 0.277 | 0.395 | 0.631 |
| Conjunctival hyperemia | 0.236 | 0.322 | 0.421 | 0.267 | 0.353 | 0.542 |
| Corneal neovascularization | 0.202 | 0.297 | 0.489 | 0.223 | 0.32 | 0.386 |
| Corneal opacity | 0.21 | 0.309 | 0.553 | 0.205 | 0.299 | 0.497 |
| Corneal leucoma | 0.333 | 0.457 | 0.61 | 0.289 | 0.396 | 0.514 |
| Corneal ulcer | 0.263 | 0.353 | 0.487 | 0.299 | 0.402 | 0.538 |
| Discoria | 0.332 | 0.429 | 0.55 | 0.307 | 0.377 | 0.437 |
Discussion
This study describes establishment of an AI system called EyeHealer with the potential of diagnosing and annotating visible ocular lesions. This technique was combined with deep learning, and at the current stage, applied to several common anterior segment ocular diseases. Even with the help of modern machine learning algorithms, such as transfer learning, deep learning classifiers require tens of thousands of images of every disease category to acquire convincing accuracy. Moreover, it is difficult to obtain such large numbers of medical images, as even for the commonly seen diseases, different features might present in different patients. Therefore, the current system adopted clear images with highly accurate annotations by ophthalmic experts using polygons, sharply reducing the number of images in the training dataset to hundreds of images per class to obtain a relatively precise diagnosis.
Three main approaches were applied to standardize the training datasets. First, in contrast to other anterior segmentation systems, the current system focuses on only three eye structures to eliminate unnecessary and misleading information, with these three structures serving as location labels. The interpalpebral zone, with the eyeball area inside and the eyelid with eyelashes outside the boundaries, functions as the outermost part of the eye; the cornea, as the intermediate area, covers the middle part of the eye; and the pupil represents the center of the entire eye. Therefore, in the initial quality control step, it is vital to make sure that all three structures are visualized in one single image. This method of localization can help establish unity out of the difference, which greatly improves the performance of the system.
Only bright light images were used to avoid redundant and inaccurate labeling from blue light or slit-light images. The slit-lamp is a multi-function examination that helps with the diagnoses of different diseases. Blue light focuses on the integrity of the cornea and is also used to diagnose tear film related disease. With the difference in the image color, even a small spot of fluorescein staining could constitute confusing information for the machine. As for the slit-light examination, even in one eye of a patient, difference in the width or angles of the light can lead to differences in images, thereby causing deviation in the process of machine learning. In the future, it will be necessary to add photos taken with different means of examination and generalize the system for its use in a wider disease spectrum.
In addition, unlike other systems that use quadrate to label, the current system applies irregular polygons to segment the normal eye structure as well as the lesion area. The quadrate is simpler and has been widely used in a diversity of image labeling systems. By comparison, polygon segmentations require more calculations in the training process but can achieve a more precise diagnosis, especially with medical images.
During the training of the system, different pre-trained models were compared to apply the model with the best performance. DenseNet demonstrated the best results among the three models. In the segmentation exam, different commonly used models were also compared. Semantic segmentation models were found to outperform medical segmentation models. DeepLabv3 and PspNet exhibited similar performance. The semantic segmentation network obtained smooth and clear lesion segmentation boundaries, achieving a high accuracy in common anterior diseases.
This paper describes the preliminary establishment of the system. More work needs to be done to improve the system performance. The large differences in the numbers of sample cases across different diseases and limited numbers of images led to relatively low classification performance. Corneal opacity could obscure the view of the anterior chamber or even lens abnormalities, which could affect the accuracy of AI-based diagnoses of diseases of the posterior structures. In future studies, we will apply our system to more images of corneal opacity, especially with posterior abnormalities. More clearly annotated images need to be applied. Moreover, people from more diverse ethnic groups should be included for the generalization of the system. Further progression in the field of neural networks could also improve the performance of the current approach.
Furthermore, few-shot or low-shot learning could be applied when images of less common diseases are relatively limited. Other than bright-field images, the current system has the potential to be applied to a wide range of slit-lamp images, including the ones mentioned above, namely blue light or slit-light photos. With the additional information of different types of images, the diagnostic results could be more convincing. Anterior images are simple to acquire and even handheld devices like mobile phones could be used to obtain images, so in future it might be possible to develop a mobile application based on the current system, which could be used for diagnosis or provide referral or treatment suggestions.
Conclusions
This paper describes EyeHealer, a large-scale anterior eye segment dataset with eye structure and lesion annotated. To our knowledge, this publicly available dataset is by far the largest and most extensive in this field. Experiments on this dataset demonstrated that currently both disease classification and lesion segmentation are challenging in model design. The EyeHealer dataset still requires great improvements in future research.
Data and code availability
De-identified and anonymized data are stored in a secured patient confidentiality compliant cloud in China, and codes are available on reasonable request to the corresponding authors. All data or codes access requests will be carefully reviewed and granted by the Data Access Committee.
Supplementary Material
ACKNOWLEDGEMENTS
This study was funded by the National Key Research and Development Program of China (Grant No. 2017YFC1104600), and Recruitment Program of Leading Talents of Guangdong Province (Grant No. 2016LJ06Y375).
Contributor Information
Wenjia Cai, State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University, Guangzhou 510060, China.
Jie Xu, Beijing Institute of Ophthalmology, Capital Medical University, Beijing Tongren Hospital, Beijing 100730, China.
Ke Wang, Department of Computer Science and Technology & BNRist, Tsinghua University, Beijing 100084, China.
Xiaohong Liu, Department of Computer Science and Technology & BNRist, Tsinghua University, Beijing 100084, China.
Wenqin Xu, Center for Biomedicine and Innovations, Faculty of Medicine, Macau University of Science and Technology and University Hospital, Macau 999078, China.
Huimin Cai, Center for Biomedicine and Innovations, Faculty of Medicine, Macau University of Science and Technology and University Hospital, Macau 999078, China.
Yuanxu Gao, Center for Biomedicine and Innovations, Faculty of Medicine, Macau University of Science and Technology and University Hospital, Macau 999078, China.
Yuandong Su, Center for Translational Innovations, West China Hospital and Sichuan University, Chengdu 610041, China.
Meixia Zhang, Center for Translational Innovations, West China Hospital and Sichuan University, Chengdu 610041, China.
Jie Zhu, Guangzhou Women and Children's Medical Center, Guangzhou Medical University, Guangzhou 510623, China.
Charlotte L Zhang, Bioland Laboratory, Guangzhou 510005, China.
Edward E Zhang, Bioland Laboratory, Guangzhou 510005, China.
Fangfei Wang, Bioland Laboratory, Guangzhou 510005, China.
Yun Yin, School of Business, Macau University of Science and Technology, Macau 999078, China.
Iat Fan Lai, Ophthalmic Center, Kiang Wu Hospital, Macau, China.
Guangyu Wang, School of Information and Communication Engineering, Beijing University of Posts and Telecommunications, Beijing 100876, China.
Kang Zhang, Center for Biomedicine and Innovations, Faculty of Medicine, Macau University of Science and Technology and University Hospital, Macau 999078, China; Center for Translational Innovations, West China Hospital and Sichuan University, Chengdu 610041, China.
Yingfeng Zheng, State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University, Guangzhou 510060, China.
Author contributions
W.C., J.X., K.W., X.L., W.X., H.C., Y.G., Y.S., M.Z., J.Z., C.L.Z., E.Z., F.W., Y.Y., IF.L., GY.W., and K.Z. collected and analyzed the data. Y.Z., K.Z. and GY.W. conceived and supervised the project, and wrote the manuscript. All authors discussed the results, and reviewed and approved the manuscript.
Conflict of interest
As a Co-EIC of Precision Clinical Medicine, the corresponding author Dr. Kang Zhang was blinded from reviewing or making decisions on this manuscript.
References
- 1. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–10.. doi:10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 2. Kermany DS, Goldbaum M, Cai Wet al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172:1122–31..doi:10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 3. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521:436–44.. doi:10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 4. Gu H, Guo Y, Gu L, et al. Deep learning for identifying corneal diseases from ocular surface slit-lamp photographs. Sci Rep. 2020;10:17851. doi:10.1038/s41598-020-75027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vupparaboina KK, Vedula SN, Aithu S, et al. Artificial intelligence based detection of infectious keratitis using slit-lamp images. In: ARVO Annual Meeting, July 2019. Invest Ophthalmol Vis Sci. 2019; 60: 4236. [Google Scholar]
- 6. Xu Y, Kong M, Xie Wet al. Deep sequential feature learning in clinical image classification of infectious keratitis. Engineering. 2020. doi:10.1016/j.eng.2020.04.012. [Google Scholar]
- 7. Wan Zaki WMD, Mat Daud M, Abdani SR, et al. Automated pterygium detection method of anterior segment photographed images. Comput Methods Programs Biomed. 2018;154:71–8.. doi:10.1016/j.cmpb.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 8. Zamani NSM, Zaki WMDW, Huddin AB, et al. Automated pterygium detection using deep neural network. IEEE Access. 2020;8:191659–72.. doi:10.1109/ACCESS.2020.3030787. [Google Scholar]
- 9. Long E, Lin H, Liu Z, et al. An artificial intelligence platform for the multihospital collaborative management of congenital cataracts. Nature Biomedical Engineering. 2017;1:0024. doi:10.1038/s41551-016-0024. [Google Scholar]
- 10. Gao X, Lin S, Wong TY. Automatic feature learning to grade nuclear cataracts based on deep learning. IEEE Trans Biomed Eng. 2015;62:2693–701.. doi:10.1109/TBME.2015.2444389. [DOI] [PubMed] [Google Scholar]
- 11. Noh H, Hong S, Han B. Learning deconvolution network for semantic segmentation. In:Proceedings of the IEEE international conference on computer vision. 2015, pp. 1520–8.. doi:10.1109/ICCV.2015.178. [Google Scholar]
- 12. Garcia-Garcia A, Orts-Escolano S, Oprea S, et al. A review on deep learning techniques applied to semantic segmentation. arXiv [cs.CV]. 2017, arXiv:1704.06857.
- 13. Wang P, Chen P, Yuan Y, et al. Understanding convolution for semantic segmentation. In:2018 IEEE Winter Conference on Applications of Computer Vision (WACV). 2018, pp. 1451–60.. doi:10.1109/WACV.2018.00163. [Google Scholar]
- 14. Chen B-K, Gong C, Yang J. Importance-aware semantic segmentation for autonomous driving system. In: Proceedings of the Twenty-Sixth International Joint Conference on Artificial Intelligence (IJCAI-17). 2017, pp.1504–10. [Google Scholar]
- 15. Chen L-C, Papandreou G, Kokkinos I, et al. DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40(4):834–48.. doi:10.1109/tpami.2017.2699184. [DOI] [PubMed] [Google Scholar]
- 16. Puech P, Betrouni N, Makni Net al. Computer-assisted diagnosis of prostate cancer using DCE-MRI data: design, implementation and preliminary results. Int J Comput Assist Radiol Surg. 2009;4:1–10.. doi:10.1007/s11548-008-0261-2. [DOI] [PubMed] [Google Scholar]
- 17. Eadie LH, Taylor P, Gibson AP. A systematic review of computer-assisted diagnosis in diagnostic cancer imaging. Eur J Radiol. 2012;81:e70–6.. doi:10.1016/j.ejrad.2011.01.098. [DOI] [PubMed] [Google Scholar]
- 18. Ozawa T, Ishihara S, Fujishiro Met al. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc. 2019;89:416–21.. e1, doi:10.1016/j.gie.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 19. di Ruffano LF, Takwoingi Y, Dinnes J. Computer-assisted diagnosis techniques (dermoscopy and spectroscopy-based) for diagnosing skin cancer in adults. Cochrane Database Syst Rev. 2018;12(12):CD013186. doi:10.1002/14651858.CD013186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. He K, Zhang X, Ren Set al. Deep residual learning for image recognition. In:Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016, pp. 770–778.. doi:10.1109/CVPR.2016.90.
- 21. Szegedy C, Vanhoucke V, Ioffe S. Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR). 2016, pp. 2818–26.. Available: https://www.cv-foundation.org/openaccess/content_cvpr_2016/html/Szegedy_Rethinking_the_Inception_CVPR_2016_paper.html. [Google Scholar]
- 22. Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. In:Proceedings of the IEEE conference on computer vision and pattern recognition. 2017, pp.2261–2269.. doi:10.1109/CVPR.2017.243. [Google Scholar]
- 23. Devalla SK, Renukanand PK, Sreedhar BKet al. DRUNET: a dilated-residual U-Net deep learning network to segment optic nerve head tissues in optical coherence tomography images. Biomed Opt Express. 2018;9:3244–65.. doi:10.1364/BOE.9.003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Badrinarayanan V, Kendall A, Cipolla R. SegNet: A Deep Convolutional Encoder-Decoder Architecture for Image Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39:2481–95.. doi:10.1109/TPAMI.2016.2644615. [DOI] [PubMed] [Google Scholar]
- 25. Zhao H, Shi J, Qi X, et al. Pyramid scene parsing network. In: Proceedings of the IEEE conference on computer vision and pattern recognition. 2017, pp. 6230–39.. doi:10.1109/CVPR.2017.660. [Google Scholar]
- 26. Sokolova M, Lapalme G. A systematic analysis of performance measures for classification tasks. Inf Process Manag. 2009;45:427–37.. doi:10.1016/j.ipm.2009.03.002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
De-identified and anonymized data are stored in a secured patient confidentiality compliant cloud in China, and codes are available on reasonable request to the corresponding authors. All data or codes access requests will be carefully reviewed and granted by the Data Access Committee.