Abstract
Telomerase plays a pivotal role in tumorigenesis by both telomere‐dependent and telomere‐independent activities, although the underlying mechanisms are not completely understood. Using single‐sample gene set enrichment analysis (ssGSEA) across 9,264 tumour samples, we observe that expression of telomerase reverse transcriptase (TERT) is closely associated with immunosuppressive signatures. We demonstrate that TERT can activate a subclass of endogenous retroviruses (ERVs) independent of its telomerase activity to form double‐stranded RNAs (dsRNAs), which are sensed by the RIG‐1/MDA5‐MAVS signalling pathway and trigger interferon signalling in cancer cells. Furthermore, we show that TERT‐induced ERV/interferon signalling stimulates the expression of chemokines, including CXCL10, which induces the infiltration of suppressive T–cell populations with increased percentage of CD4+ and FOXP3+ cells. These data reveal an unanticipated role for telomerase as a transcriptional activator of ERVs and provide strong evidence that TERT‐mediated ERV/interferon signalling contributes to immune suppression in tumours.
Keywords: dsRNA, ERVs, immune suppression, interferon signalling, TERT
Subject Categories: Cancer; Chromatin, Transcription & Genomics; Immunology
Expression of telomerase reverse transcriptase is closely associated with immunosuppressive signatures in tumours. TERT induces dsRNAs by bidirectional transcription of ERVs, which triggers interferon response and attracts suppressive T cell populations.
Introduction
Telomerase reverse transcriptase (TERT), the catalytic subunit of telomerase, plays a critical role in cancer formation by maintaining telomere homeostasis and the cellular proliferation potential (Blasco, 2005). Most human somatic cells have undetectable telomerase activity due to transcriptional repression of TERT during embryonic development. However, TERT is universally reactivated during tumorigenesis and has been considered a hallmark of cancer cells (Cong et al, 2002). Recent studies have shown that in addition to telomere maintenance, TERT is also involved in several cellular processes independent of its telomerase activity, including the WNT/β‐catenin signalling pathway (Park et al, 2009), NF‐κB signalling pathway (Ghosh et al, 2012; Ding et al, 2013), and epithelial–mesenchymal transition (Liu et al, 2013). However, the underlying mechanisms of TERT in tumorigenesis are not completely understood.
ERVs, which integrated into the genome of mammals millions of years ago via exogenous retroviral infections of germ cells (Feschotte & Gilbert, 2012), represent ~8% of the human genome (Lander et al, 2001). Although it is generally believed that most of ERVs do not encode proteins due to drastic genetic alterations (Vargiu et al, 2016), recent studies have shown that some coding‐competent ERVs are implicated in embryonic development (Mi et al, 2000) and diseases, including neurological diseases (Küry et al, 2018) and cancer (Matteucci et al, 2018). However, the relationship between TERT and ERVs in tumorigenesis is not known.
In the current study, we found that expression of TERT is closely associated with immunosuppressive signatures. We then demonstrated that TERT can trigger interferon signalling by activating a subclass of ERVs independent of its telomerase activity in cancer cells. The TERT‐induced ERV/interferon signalling stimulated the expression of chemokines, such as CXCL10, which induced suppressive T‐cell infiltration, contributing to the establishment of immunosuppressive tumour microenvironment.
Results and Discussion
TERT expression is associated with immunosuppressive signatures
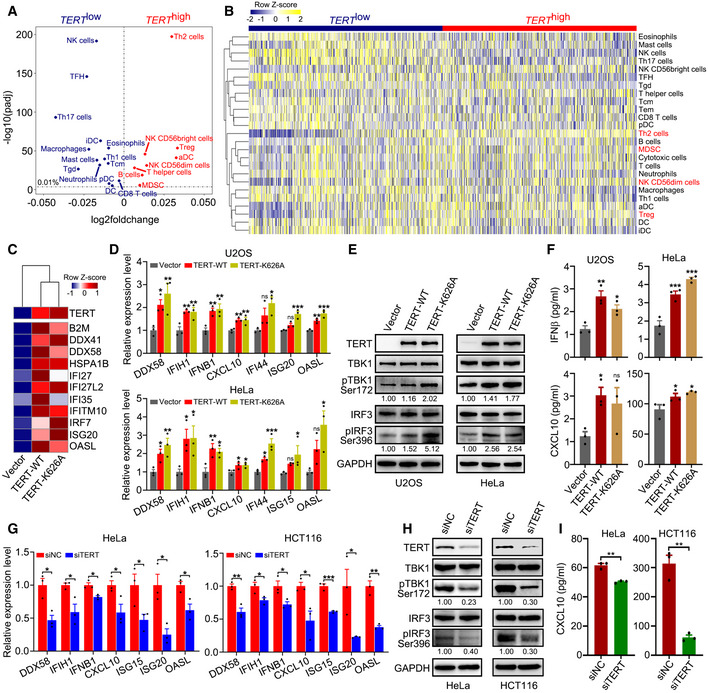
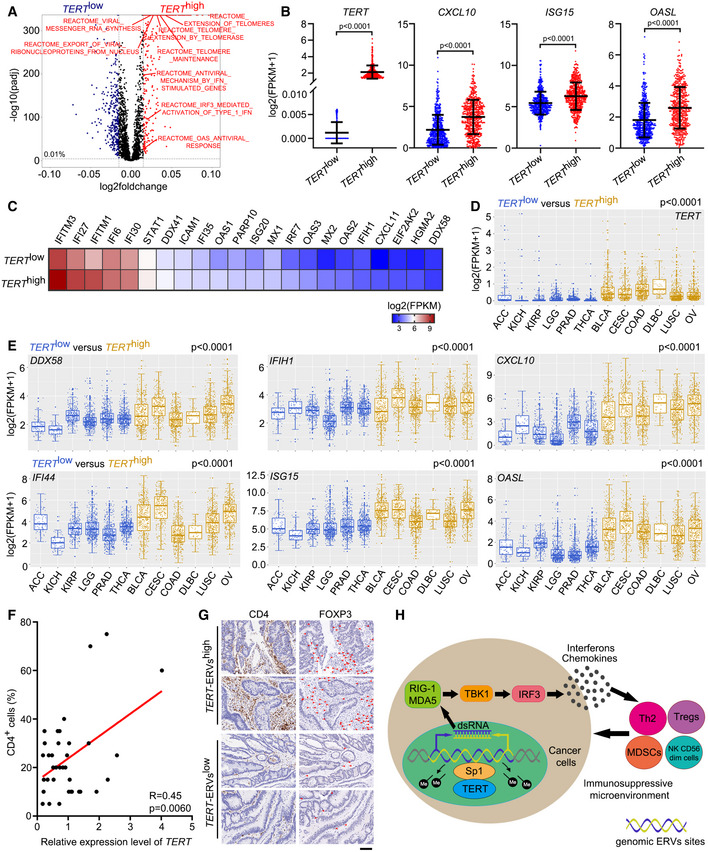
To comprehensively characterize genome‐wide expression signatures associated with TERT expression in cancer cells, we divided 9,264 tumour samples across 24 cancer types derived from The Cancer Genome Atlas (TCGA) (Rahman et al, 2015) into two groups (TERT high and TERT low) based on their TERT expression levels. Using single‐sample gene set enrichment analysis (ssGSEA) for immune cell infiltration (Bindea et al, 2013; Charoentong et al, 2017), we observed that TERT expression was closely associated with immunosuppressive signatures, at the top of which was type 2 T helper cells (Th2 cells), followed by regulatory T cells (Treg cells), CD56dim natural killer cells (NK CD56dim cells), and myeloid‐derived suppressor cells (MDSCs) (Jia et al, 2018) (Fig 1A). The heat map of ssGSEA for TCGA tumour samples grouped into TERT high/low categories confirmed robust and significant association of TERT high tumours with these immunosuppressive signatures (Fig 1B). This indicated that TERT may participate in immune suppressive function.
Figure 1. TERT expression is associated with immunosuppressive signatures and triggers interferon response.
-
AScatterplot representing differences in ssGSEA immune signatures in TERT high versus TERT low tumours (n = 4,632 tumours in each group) across TCGA. −log10(padj) for enrichment of ssGSEA immune signatures in TERT high versus TERT low tumours is shown on the y‐axis. Signatures more highly represented in TERT high tumours are shown on the right, while those in TERT low tumours are shown on the left.
-
BHeat map for ssGSEA of immune signatures in TERT high and TERT low tumours (n = 4,632 tumours in each group) across TCGA. Th1, Th2, Th17: T helper 1, 2, 17 cells, Tcm: central memory T cells, Tem: effector memory T cells, Tgd: gamma delta T cells, TFH: follicular helper T cells, Treg: regulatory T cells, MDSC: myeloid‐derived suppressor cells, DC: dendritic cells, aDC: activated DCs, iDC: immature DCs, pDC: plasmacytoid DCs, NK cells: natural killer cells.
-
CHeat map of interferon‐related genes in U2OS cells with TERT‐WT or TERT‐K626A ectopic expression.
-
D–FRT–qPCR analysis of interferon‐related genes (D), western blot analysis of TERT, TBK1, pTBK1, IRF3, pIRF3, and GAPDH levels (E), and ELISA of IFNβ and CXCL10 in culture supernatant (F) in U2OS and HeLa cells ectopically expressing TERT‐WT or TERT‐K626A.
-
G–IRT–qPCR analysis of interferon‐related genes (G), western blot analysis of TERT, TBK1, pTBK1, IRF3, pIRF3, and GAPDH levels (H), and ELISA analysis of CXCL10 in culture supernatant (I) in HeLa and HCT116 cells with TERT knockdown.
Data information: Data represent mean ± SEM of three biological replicates (D, F, G and I). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant. One‐way ANOVA with Fisher's LSD tests was used for (D) and (F), unpaired two‐tailed Student’s t‐tests were used for (G) and (I). Relative levels of pTBK1 and pIRF3 were quantified with ImageJ software and normalized to GAPDH, as indicated at the bottom of the blot (E and H).
Source data are available online for this figure.
Expression of TERT triggers interferon response
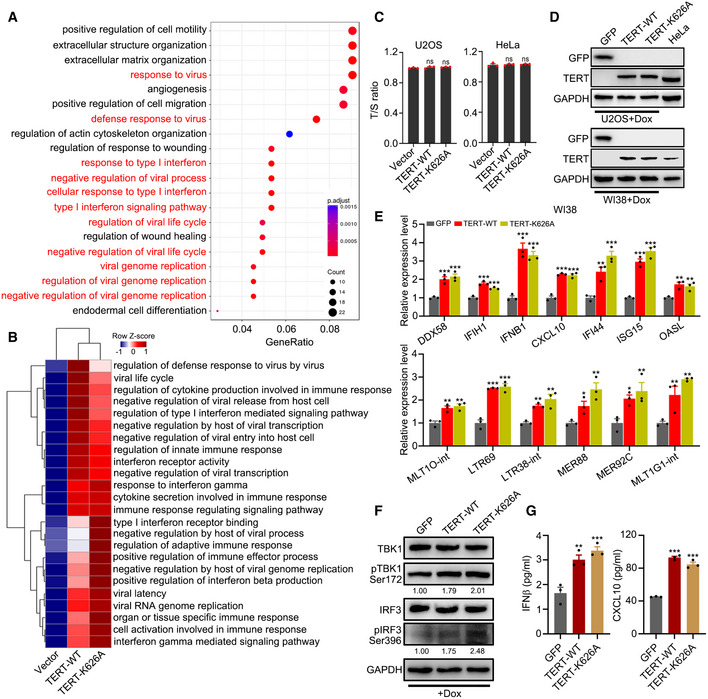
To assess whether TERT is involved in immune suppressive function independent of telomere maintenance, we ectopically expressed TERT‐WT or catalytically inactive mutant TERT‐K626A (Ding et al, 2013) in U2OS (telomerase‐negative) and HeLa (telomerase‐positive) cells. RNA‐seq and Gene Ontology (GO) analysis of the U2OS cells revealed that the expression of TERT induced significant enrichments of GO terms associated with viral defence and type I interferon response (Fig EV1A). Furthermore, ssGSEA analysis indicated that both TERT‐WT and TERT‐K626A induced interferon response (Fig EV1B). Consistently, expression of a panel of genes involved in interferon response, including IFNB1, ISG15, ISG20, IFI27, IFI35, IFI44, OASL, and CXCL10, were activated by both TERT‐WT and TERT‐K626A (Fig 1C and D). The expression of these genes has been reported to be tightly associated with immunosuppressive microenvironment (Cañadas et al, 2018; Guldner et al, 2020). Similar results were also obtained in HeLa cells (Fig 1D). Interestingly, we observed that RNA cytosolic sensor genes (DDX41, DDX58/RIG‐1, and IFIH1/MDA5) were also co‐ordinately upregulated (Fig 1C and D). Considering that DDX58/RIG‐1 and IFIH1/MDA5 are the key viral RNA sensing proteins in the cytoplasm and that their activation induces downstream signalling via MAVS, TBK1, and IRF3 (Hur, 2019), we speculated that TERT may trigger the RIG‐1/MDA5‐MAVS signalling pathway. Using western blot analysis, we confirmed the activation of TBK1 (pTBK1 Ser172) and IRF3 (pIRF3 Ser396) signalling in U2OS and HeLa cells, which act downstream of the MAVS signalling pathway, upon ectopic expression of TERT‐WT or TERT‐K626A (Fig 1E). Furthermore, enzyme‐linked immunosorbent assays (ELISA) revealed that levels of IFNβ and CXCL10 increased in the culture supernatant of U2OS and HeLa cells ectopically expressing TERT‐WT or TERT‐K626A (Fig 1F). In addition, knockdown of TERT by siRNAs in HeLa and HCT116 cells resulted in downregulated expression of interferon‐related genes (Fig 1G), reduced phosphorylation of TBK1 and IRF3 (Fig 1H), and significantly decreased level of CXCL10 in culture supernatant (Fig 1I). These data suggest that TERT is involved in interferon response.
Figure EV1. Ectopic expression of TERT triggers interferon response.
-
AEnriched biological processes (BP) with TERT ectopic expression. GO was analysed for upregulated genes by TERT compared to those in the empty vector control. GO terms in red are biological processes related to interferon response.
-
BssGSEA of GO terms related to interferon response in U2OS cells ectopically expressing TERT‐WT or TERT‐K626A.
-
CT/S ratio of U2OS and HeLa cells with transient transfection of TERT‐WT or TERT‐K626A.
-
DWestern blot analysis of GFP, TERT and GAPDH levels in U2OS (upper) and WI38 (lower) cells ectopically expressing TERT‐WT or TERT‐K626A using Dox‐inducible system to induce expression of TERT at the levels comparable to that of HeLa cells. GFP as negative control.
-
E–GRT–qPCR analysis of interferon‐related genes and TA‐ERVs (E), western blot analysis of TERT, TBK1, pTBK1, IRF3, pIRF3, and GAPDH levels (F), and ELISA of IFNβ and CXCL10 in culture supernatant (G) in WI38 cells ectopically expressing TERT‐WT or TERT‐K626A using Dox‐inducible system. GFP as negative control. Relative pTBK1 and pIRF3 protein levels were quantified with ImageJ software and normalized to GAPDH, as indicated at the bottom of the blot (F).
Data information: Data represent mean ± SEM of three biological replicates (C, E and G). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, using one‐way ANOVA with Fisher's LSD tests (C, E and G).
Source data are available online for this figure.
TERT triggers interferon response via induction of dsRNAs transcribed from ERVs
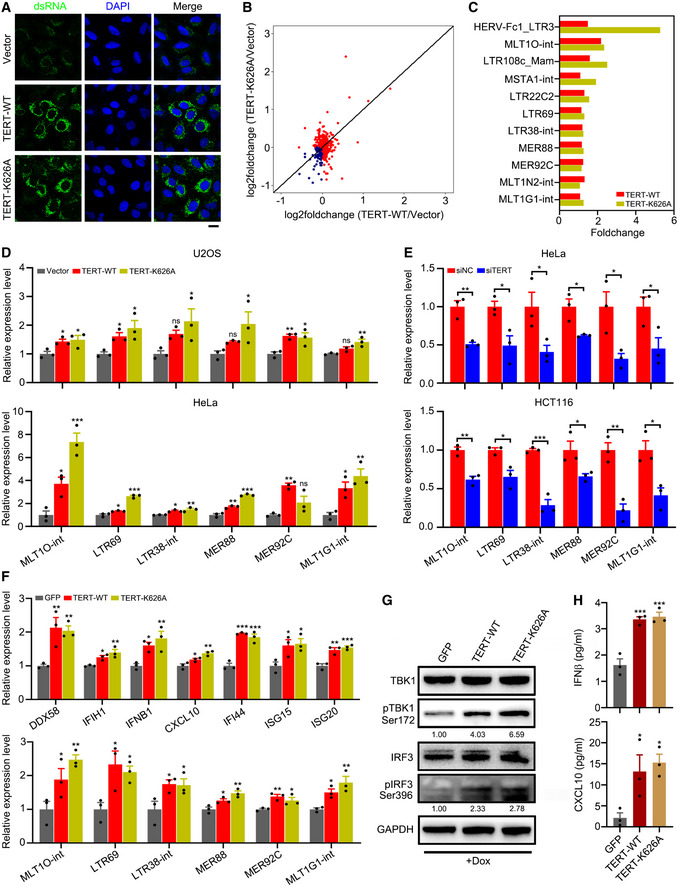
Studies have shown that cellular double‐stranded RNA (dsRNA) can activate viral cytosolic RNA sensors (Hur, 2019). We reasoned that activation of viral cytosolic RNA sensors by TERT might be due to increase in dsRNA levels in cells. By immunofluorescence analysis with dsRNA‐specific antibody (J2), we observed that both TERT‐WT and TERT‐K626A induced dsRNA expression (Fig 2A). As active ERVs can fold into dsRNA secondary structures that can trigger interferon response (Chiappinelli et al, 2015; Roulois et al, 2015), we analysed the expression of ERVs in U2OS cells. Results revealed that a large number of ERVs were upregulated by TERT‐WT or TERT‐K626A expression (Fig 2B). We named these TERT‐activated ERVs TA‐ERVs. The top upregulated TA‐ERVs are listed in Fig 2C, which included MLT1O‐int, LTR69, LTR38‐int, MER88, MER92C, and MLT1G1‐int. The activation of these ERVs by TERT‐WT or TERT‐K626A were further confirmed using quantitative reverse transcription polymerase chain reaction (RT–qPCR) in U2OS and HeLa cells (Fig 2D). Conversely, knockdown of TERT led to reduced expression of TA‐ERVs in HeLa and HCT116 cells (Fig 2E). To exclude the effects of telomere elongation associated with TERT ectopic expression, we determined the telomere lengths in U2OS and HeLa cells ectopically expressing TERT‐WT or TERT‐K626A using qPCR analysis (T/S ratio) (Gil & Coetzer, 2004). The results showed that transient transfection of TERT‐WT or TERT‐K626A did not significantly affect telomere length (Fig EV1C), confirming that the activation of ERVs by TERT is independent of telomere lengths.
Figure 2. TERT triggers interferon response via induction of dsRNA transcribed from ERVs.
-
AConfocal microscopy of U2OS cells ectopically expressing TERT‐WT or TERT‐K626A. Total dsRNA is stained green, and nuclei are stained blue (DAPI). Scale bar = 20 µm.
-
BGenome‐wide upregulation of ERVs in U2OS cells ectopically expressing TERT‐WT or TERT‐K626A. RNA‐seq data for TERT‐WT/TERT‐K626A and empty vector control were used to calculate the log2 fold change values. Red dots indicate elements with increased expression, while blue dots indicate the elements with decreased expression.
-
CRepresentative TA‐ERVs activated by TERT‐WT or TERT‐K626A.
-
DRT–qPCR analysis of TA‐ERVs in U2OS and HeLa cells ectopically expressing TERT‐WT or TERT‐K626A.
-
ERT–qPCR analysis of TA‐ERVs in HeLa and HCT116 cells with TERT knockdown.
-
F–HRT–qPCR analysis of interferon‐related genes and TA‐ERVs (F), western blot analysis of TERT, TBK1, pTBK1, IRF3, pIRF3, and GAPDH levels (G), and ELISA of IFNβ and CXCL10 in culture supernatant (H) in U2OS cells ectopically expressing TERT‐WT or TERT‐K626A using Dox‐inducible system. GFP was used as negative control. Relative levels of pTBK1 and pIRF3 were quantified with ImageJ software and normalized to GAPDH, as indicated at the bottom of the blot (G).
Data information: Data represent mean ± SEM of three biological replicates (D, E, F and H). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant. One‐way ANOVA with Fisher's LSD tests was used for (D), (F) and (H), unpaired two‐tailed Student’s t‐tests were used for (E).
Source data are available online for this figure.
To ascertain the interferon response and TA‐ERVs activation are related to TERT specifically, we have ectopically expressed TERT in U2OS and human fibroblasts WI38 cells (both telomerase‐negative) at the levels comparable to that of HeLa cells by using doxycycline (Dox)‐inducible system (Fig EV1D). We found that both interferon‐related genes and TA‐ERVs were upregulated (Figs 2F and EV1E), TBK1 and IRF3 were activated (Figs 2G and EV1F), and levels of IFNβ and CXCL10 increased in culture supernatant (Figs 2H and EV1G), upon induction of TERT expression. These results support that TERT regulates interferon response and TA‐ERVs expression.
To further verify that dsRNAs are formed from TA‐ERVs, we precipitated dsRNA from U2OS cells ectopically expressing TERT‐WT or TERT‐K626A using dsRNA‐specific antibody (J2) and performed strand‐specific RNA‐seq (dsRIP‐seq). The results of dsRIP‐seq showed that both sense and anti‐sense transcripts of these TA‐ERVs were expressed at comparable levels (Fig 3A). In addition, using strand‐specific RT–qPCR analysis, we detected bidirectional transcription producing sense and anti‐sense transcripts for MLT1O‐int, LTR69, LTR38‐int, MER88, MER92C, and MLT1G1‐int, but not for β‐actin, in U2OS and HeLa cells ectopically expressing TERT‐WT or TERT‐K626A (Fig 3B). These data suggest that TERT can activate ERVs to form dsRNA via bidirectional transcription, thereby triggering interferon response.
Figure 3. TERT activates ERVs by interacting with Sp1.
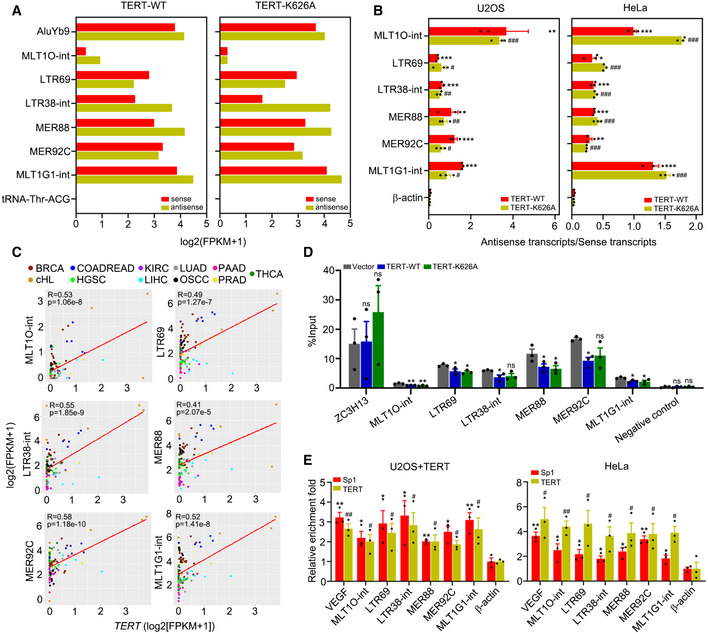
-
AIdentification of dsRNAs activated by TERT‐WT or TERT‐K626A using dsRIP‐seq in U2OS cells. AluYb9 and tRNA‐Thr‐ACG as positive and negative controls, respectively.
-
BQuantification of antisense/sense transcripts of ERVs activated by TERT‐WT or TERT‐K626A in U2OS and HeLa cells using strand‐specific RT–qPCR.
-
CRegression analysis of the correlation between TERT and TA‐ERVs expression in 103 tumour samples across 11 cancer types. BRCA (breast invasive carcinoma), n = 17. cHL (classical Hodgkin lymphoma), n = 5. COADREAD (colon adenocarcinoma/rectum adenocarcinoma), n = 6. HGSC (high‐grade serous ovarian cancer), n = 6. KIRC (kidney renal clear cell carcinoma), n = 12. LIHC (liver hepatocellular carcinoma), n = 9. LUAD (lung adenocarcinoma), n = 6. OSCC (oral squamous cell carcinoma), n = 8. PAAD (pancreatic adenocarcinoma), n = 10. PRAD (prostate adenocarcinoma), n = 6. THCA (thyroid carcinoma), n = 18.
-
DMeDIP‐qPCR analysis of TA‐ERVs sites in HeLa cells ectopically expressing TERT‐WT or TERT‐K626A. Primer sets recognizing exon 8 of the human ZC3H13 gene and a gene desert on chromosome 12 were used for positive and negative controls, respectively.
-
EChIP‐qPCR for TERT and Sp1 targets on TA‐ERVs in U2OS cells with TERT ectopic expression and in HeLa cells. β‐actin promoter was used as a negative control, and VEGF promoter was used as a positive control.
Data information: Data represent mean ± SEM of three biological replicates (B, D and E). */# P < 0.05, **/## P < 0.01, ***/### P < 0.001, ns, not significant. Unpaired two‐tailed Student’s t‐tests were used, and asterisks indicating significance for each ERV versus β‐actin in TERT‐WT while hashtags indicating significance for each ERV versus β‐actin in TERT‐K626A in (B) and (E), one‐way ANOVA with Fisher's LSD tests was used for (D).
TERT expression is significant positive correlation with TA‐ERVs in tumours
To further validate our data, we analysed the expression of TERT and ERVs in 103 tumour samples across 11 cancer types available from the Gene Expression Omnibus (GEO) and observed significant positive correlation between TERT and TA‐ERVs (Fig 3C). In addition, we analysed the expression of TERT and TA‐ERVs in different human tissues from the GEO. Results indicated that TERT is expressed more in immune tissues, including the appendix, bone marrow, lymph node, and thymus (Fig EV2A). Concomitantly, the TA‐ERVs are also highly expressed in these tissues (Fig EV2A), suggesting that the TERT‐ERVs axis is involved in immune function.
Figure EV2. TERT activates ERVs via interaction with Sp1.
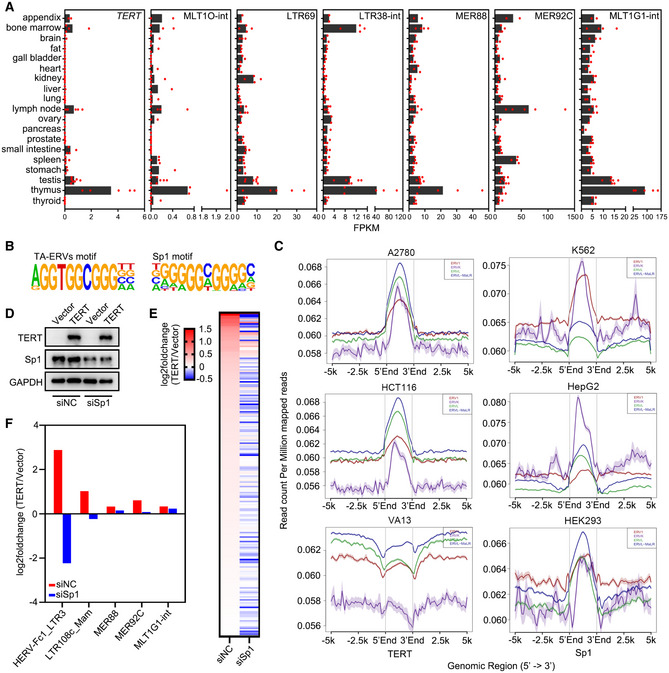
-
AExpression of TERT and ERVs in human tissues using RNA‐seq data from the GEO database. Data represent average expression levels of TERT and ERVs. Appendix, n = 2. Bone marrow, n = 4. Brain, n = 3. Fat, n = 3. Gall bladder, n = 3. Heart, n = 3. Kidney, n = 4. Liver, n = 3. Lung, n = 4. Lymph node, n = 5. Ovary, n = 2. Pancreas, n = 2. Prostate, n = 4. Small intestine, n = 4. Spleen, n = 4. Stomach, n = 3. Testis, n = 7. Thymus, n = 5. Thyroid, n = 4.
-
BMotif analysis using the sequences of representative TA‐ERVs.
-
CChIP‐seq analysis for TERT and Sp1 from the GEO database. Average binding profiles of TERT on ERV1, ERVK, ERVL, and ERVL‐MaLR in A2780 and HCT116 cells, VA13 cells as the negative control (left); and Sp1 binding profiles in K562, HepG2, and HEK293 cells (right).
-
D–FWestern blot analysis of TERT and Sp1 expression (D), log2 fold change of ERVs plotted as heat map (E), and log2 fold change of representative ERVs (F) in siNC or siSp1 transfected U2OS cells expressing TERT versus control vector.
Source data are available online for this figure.
TERT activates ERVs by interacting with Sp1
In general, ERVs are silenced by the host, and DNA methylation plays an important role in preserving the silent state of ERVs (Groh & Schotta, 2017). However, ERVs can be derepressed due to reduced DNA methylation in cancer cells (Strissel et al, 2012; Espinet et al, 2021). Using methylated DNA immunoprecipitation (MeDIP)‐qPCR, we observed that the methylation levels of MLT1O‐int, LTR69, LTR38‐int, MER88, MER92C, and MLT1G1‐int indeed decreased in HeLa cells ectopically expressing TERT‐WT or TERT‐K626A (Fig 3D), which is consistent with the transcriptionally active status of these ERVs. Therefore, we analysed the sequences of the significantly upregulated TA‐ERVs and identified the motif of the transcription factor Sp1 in the TA‐ERV sequences (Fig EV2B). Sp1 has been reported to bind to ERVs and protect them from methylation, thereby activating the ERVs (Manghera & Douville, 2013). In line with these observations, we have previously demonstrated that TERT can interact with Sp1 to activate vascular endothelial growth factor (VEGF) expression (Liu et al, 2016). Our analyses of ChIP‐seq data from the GEO indicated that both TERT and Sp1 can bind to the genomic sites of ERV families, including ERV1, ERVK, ERVL, and ERVL‐MaLR (Fig EV2C). Therefore, we performed ChIP‐qPCR analysis using antibodies against TERT and Sp1 in U2OS cells ectopically expressing TERT (U2OS+TERT) and in HeLa cells, and the results showed that TERT and Sp1 share a common binding sites on TA‐ERVs (Fig 3E). Furthermore, Sp1 knockdown compromised TERT‐mediated ERV expression, as revealed by RNA‐seq analysis (Fig EV2D–F). These data indicate that TERT can activate ERV expression by interacting with transcription factor Sp1.
Deletion of Tert leads to decreased ERV expression and compromised interferon response in mice
As demonstrated above, TA‐ERVs can form dsRNA to trigger interferon response. To determine whether TERT participated in interferon response in vivo, we compared WT mice with G1 Tert −/− mice that have no apparent telomere dysfunction (Liu et al, 2000). We first analysed the expression levels of murine ERVs in the blood of these mice using RT–qPCR and observed that expression of MuERVL, IAP, MusD (also known as ETnERV), and MMERGLN was reduced in G1 Tert −/− mice compared to WT mice (Fig 4A). We then analysed the expression levels of Tert and ERVs in different tissues of mice from the GEO and found a positive correlation between Tert and MuLV‐int, MMETn‐int, ETnERV‐int and MMERGLN expression in different tissues of mice (Fig EV3A). Interestingly, Tert and ERVs were synchronously expressed at higher levels in immune tissues, such as the spleen and thymus, in agreement with the observation in human tissues (Figs EV2A and EV3A). Furthermore, we analysed the expression of ERVs in the livers of WT and G1 Tert −/− mice treated with ENU (a reagent that can activate ERVs in mouse (Wu et al, 2012)) or saline (control) using RT–qPCR. The results showed lower expression of MuERVL, MuLV, IAP, MusD, and MMERGLN in the livers of G1 Tert −/− mice compared to WT mice (Fig EV3B). Notably, ENU significantly upregulated MMERGLN in the livers of WT mice, but not in those of G1 Tert −/− mice (Fig 4B). We then performed GO analyses between WT and G1 Tert −/− mice both under control (saline) and ENU treatment and found significant upregulation for GO terms associated with viral defence and interferon response in WT mice, especially under ENU treatment, compared to G1 Tert −/− mice (Fig 4C). In addition, RNA‐seq analysis revealed that the expression of interferon‐related genes was significantly upregulated by ENU in WT mice but not in G1 Tert −/− mice (Fig 4D). Furthermore, we performed cytokine/chemokine array analysis and observed that ENU significantly upregulated cytokine/chemokine expression in WT mice, but not in G1 Tert −/− mice (Fig 4E). It has previously reported that there is a positive feedback loop between ERVs and interferon response (Cañadas et al, 2018). To assess the relationship between TA‐ERVs and interferon response in vivo, we treated WT and G1 Tert −/− mice using dsRNA mimics poly(I:C) and measured CXCL10 levels in their serum using ELISA. Results showed that poly(I:C) significantly induced CXCL10 expression in WT mice, but not in G1 Tert −/− mice (Fig 4F). On the other hand, we measured the populations of CD4+/FOXP3+ cells in blood of WT and G1 Tert −/− mice at baseline and upon stimulation with poly(I:C) by flow cytometry analysis. The results showed that the population of CD4+ and CD4+FOXP3+ cells increased under poly(I:C) treatment in both WT and G1 Tert −/− mice, but there is no significant difference observed in population of CD4+ and CD4+FOXP3+ cells between WT and G1 Tert −/− mice at baseline or upon stimulation with poly(I:C) (Fig EV3C and D). Thus, differences observed in expression of ERVs and cytokine/chemokine are due to the activation of ERVs by TERT, but not differences in population of immune cells in blood. Together, these results demonstrate that the absence of Tert compromised ERV‐mediated interferon response in vivo.
Figure 4. Deletion of Tert leads to decreased ERV expression and compromised interferon response in mice.
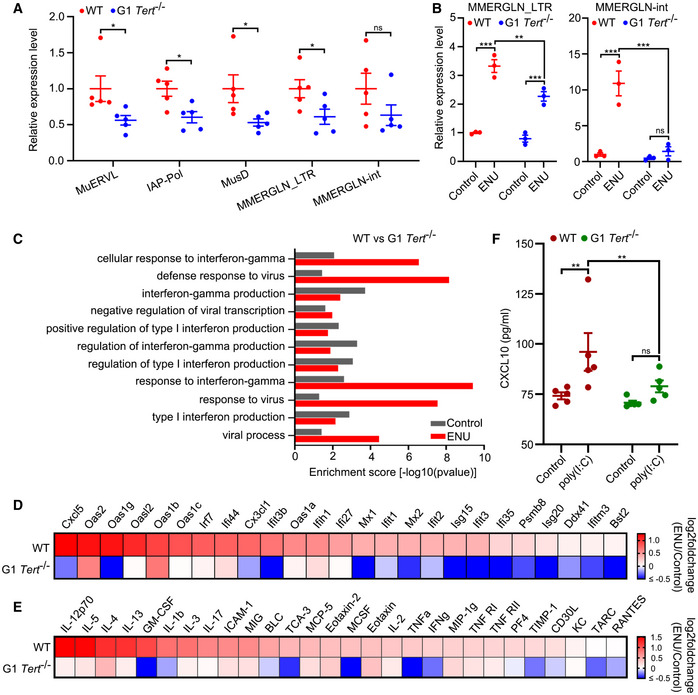
-
AExpression of murine ERVs in blood of WT and G1 Tert −/− mice. n = 5 mice per group.
-
BExpression of MMERGLN_LTR and MMERGLN‐int in livers of WT and G1 Tert −/− mice treated with ENU or saline (control). n = 3 mice per group.
-
CEnriched biological processes (BP) for WT versus G1 Tert −/− mice. GO was analysed for upregulated genes in WT mice compared to G1 Tert −/− mice under control or ENU treatment.
-
DHeat map showing log2 fold change differences of interferon‐related genes expression between ENU treatment and control (saline) in WT and G1 Tert −/− mice by RNA‐seq. n = 3 mice per group.
-
EHeat map showing log2 fold change differences of cytokine/chemokine between ENU treatment and control (saline) in WT and G1 Tert −/− mice by cytokine/chemokine array. n = 5 mice per group.
-
FELISA of CXCL10 in WT and G1 Tert −/− mice treated with poly(I:C) or saline (control). n = 5 mice per group.
Data information: Data represent mean ± SEM (A, B and F). *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant. Unpaired two‐tailed Student’s t‐tests were used for A, and two‐way ANOVA with Fisher's LSD tests was used for (B) and (F).
Figure EV3. Expression of Tert and ERVs in mice.
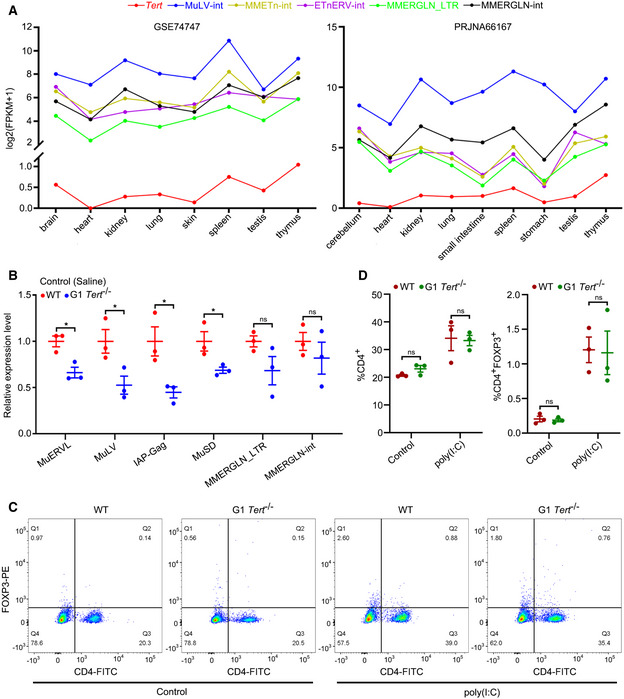
-
AExpression analysis of Tert and ERVs in mouse tissues using the GEO.
-
BRT–qPCR analysis of ERV expression in livers of WT and G1 Tert −/− mice treated with saline (control). n = 3 mice per group.
-
C, DRepresentative flow cytometry images (C) and statistical analyses (D) of CD4+ and CD4+FOXP3+ cells among mouse peripheral blood mononuclear cells (PBMCs). n = 3 mice per group.
Data information: Data represent mean ± SEM. *P < 0.05, ns, not significant. Unpaired two‐tailed Student’s t‐tests were used for (B), and two‐way ANOVA with Fisher's LSD tests was used for (D).
TERT expression is associated with interferon response in tumours
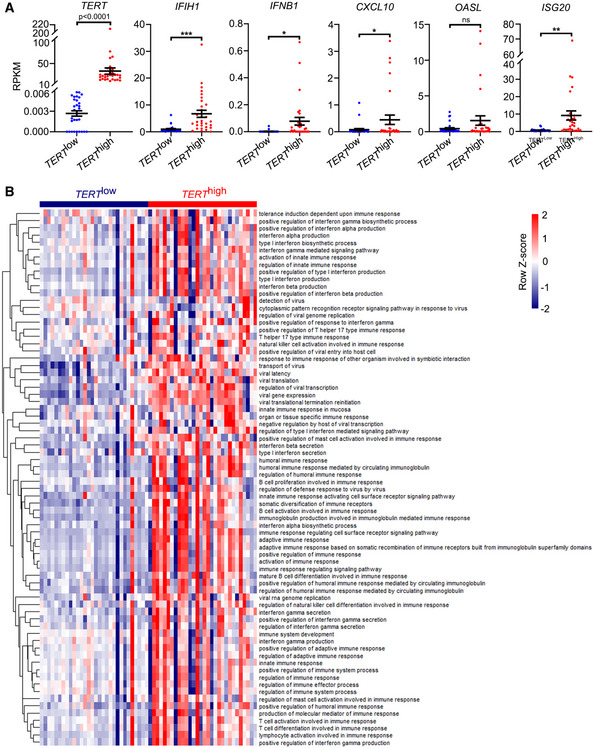
By performing ssGSEA of cellular pathways in TERT high and TERT low tumours across TCGA, we found that in addition to “Extension of telomeres” and “Telomere maintenance”, interferon‐related pathways such as “Antiviral mechanism by IFN stimulated genes”, “IRF3 mediated activation of type 1 IFN” and “OAS antiviral response” were significantly enriched in TERT high tumours compared to TERT low tumours (Fig 5A). Consistent with the results of ssGSEA, a number of interferon‐related genes, including CXCL10, ISG15 and OASL, were highly expressed in TERT high tumours (Fig 5B and C). Furthermore, by an analysis of six TERT low cancer types (ACC, KICH, KIRP, LGG, PRAD and THCA) and six TERT high cancer types (BLCA, CESC, COAD, DLBC, LUSC and OV) across TCGA (Fig 5D), we found that TERT high cancer types showed higher expression of genes related to interferon signalling, including DDX58, IFIH1, CXCL10, IFI44, OASL and ISG15, compared to the TERT low cancer types (Fig 5E). In addition, we analysed TERT and interferon‐related gene expression using the Cancer Cell Line Encyclopaedia (CCLE) (Barretina et al, 2012) and found that levels of TERT expression were closely associated with those of interferon‐related genes IFIH1, IFNB1, CXCL10, OASL and ISG20 (Fig EV4A). Consistently, results of our ssGSEA have implicated TERT in interferon response (Fig EV4B).
Figure 5. TERT promotes tumour immune suppression.
-
AScatterplot representing differences in ssGSEA of cellular pathways in TERT high versus TERT low tumours (n = 4,632 tumours in each group) across TCGA.
-
BTCGA fragments per kilobase million (FPKM) values of TERT, CXCL10, ISG15, and OASL in tumours grouped in TERT high (n = 500) and TERT low (n = 500) groups. Data represent mean ± SD. P‐values were analysed using unpaired two‐tailed Student’s t‐tests.
-
CTCGA FPKM values of interferon‐related genes in tumours grouped in TERT high (n = 500) and TERT low (n = 500) groups. Heat map showing mean FPKM values of each gene.
-
D, EExpression of TERT (D) and DDX58, IFIH1, CXCL10, IFI44, ISG15, and OASL (E) in TERT high and TERT low cancer types (6 types in each). ACC (adrenocortical carcinoma), n = 79. KICH (kidney chromophobe), n = 66. KIRP (kidney renal papillary cell carcinoma), n = 291. LGG (brain lower grade glioma), n = 532. PRAD, n = 502. THCA, n = 513. BLCA (bladder urothelial carcinoma), n = 414. CESC (cervical squamous cell carcinoma and endocervical adenocarcinoma), n = 306. DLBC (lymphoid neoplasm diffuse large B‐cell lymphoma), n = 48. LUSC (lung squamous cell carcinoma), n = 502. OV (ovarian serous cystadenocarcinoma), n = 430. All boxplots include the median line, box indicates the interquartile range (IQR) and whiskers denote the 1.5 × IQR. P‐values were analysed using two‐tailed Mann–Whitney U test.
-
FRegression analysis of the correlation between TERT expression and percentage of CD4+ cells in colon tumours. n = 36.
-
GRepresentative sections of immunohistochemical staining of CD4 and FOXP3 in TERT‐ERVshigh and TERT‐ERVslow colon tumours. Red arrowheads indicate FOXP3+ cells. Scale bar = 100 μm.
-
HSchematic model showing TERT contributing to immunosuppressive tumour microenvironment by activating ERVs.
Figure EV4. Analysis of gene expression and ssGSEA in the Cancer Cell Line Encyclopaedia (CCLE).
-
ACCLE reads per kilobase million (RPKM) values of TERT, IFIH1, IFNB1, CXCL10, ISG20, and OASL in cancer cell lines grouped in TERT high and TERT low groups (n = 30 each). Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ns, not significant, using unpaired two‐tailed Student’s t test.
-
BssGSEA of GO terms related to interferon response in TERT high and TERT low cancer cell lines (n = 30, respectively) across CCLE.
TERT promotes tumour immunouppression
To place our observations in a clinical context, we assessed relationship between TERT, ERVs and T‐cell infiltration in colon tumours. Towards this, we analysed the expression of TERT, and LTR69, LTR38‐int and MER88 in colon tumours using RT–qPCR and observed significantly positive correlation between TERT, and LTR69, LTR38‐int and MER88 expression (Fig EV5). Notably, the expression level of TERT is significantly correlated with percentage of CD4+ cells in colon tumours determined by immunohistochemistry (Fig 5F). Furthermore, we observed increased FOXP3+ cells among CD4+ cells in TERT‐ERVshigh tumours, compared to TERT‐ERVslow tumours (Fig 5G). These results indicated that TERT and ERV expression are associated with infiltration of CD4 T cells, especially Treg cells, in colon tumours, consistent with the results of ssGSEA for immune cell infiltration across TCGA (Fig 1A and B). In infectious diseases, the continual accumulation of Treg cells at sites of infection can upset the homeostasis of the infected organ and cause local immunosuppression and such an effect might be particularly substantial in the context of chronic systemic infection, such as with human immunodeficiency virus (HIV) or hepatitis C virus (HCV) (Belkaid & Rouse, 2005). Our data imply that the infiltration of these CD4 cells, including Th2 cells and Treg cells, contributed to an immunosuppressive microenvironment in TERT high tumours, similar to a chronic virus‐infected state.
Figure EV5. Regression analysis of the correlation between TERT and LTR69, LTR38‐int and MER88 expression in colon tumours.
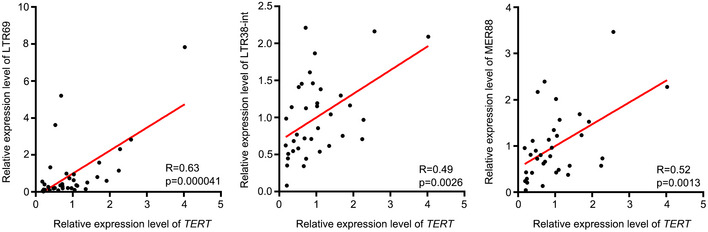
The expression of TERT, and LTR69, LTR38‐int and MER88 were determined using RT–qPCR. n = 36.
The unique feature of telomerase is its low or undetectable expression in normal human somatic cells owing to transcriptional repression of the TERT gene during embryonic differentiation, but is reactivated in over 90% of cancerous cells by upregulation of TERT gene expression during cellular transformation (Cong et al, 2002), and high TERT expression is often associated with poor patient outcome (Bertorelle et al, 2013; Mocellin et al, 2013; Borah et al, 2015). Recent progress from our and other groups suggests a critical role for TERT in tumorigenesis, apart from its function in telomere maintenance (Park et al, 2009; Ghosh et al, 2012; Ding et al, 2013; Liu et al, 2013, 2016). Telomerase is considered as an attractive anti‐cancer target (Mocellin et al, 2013). Therefore, understanding the mechanisms of action of telomerase in cancer development and progression would provide novel insights into the development of better therapeutic strategies.
In this study, we demonstrated that TERT‐mediated ERV activation contributed to tumour immunosuppression. We found that TERT induced dsRNA expression derived from bidirectional transcription of ERVs by interacting with Sp1. Importantly, these TERT‐mediated dsRNAs triggered interferon response and increased chemokine secretion in tumours. Subsequently, these chemokines attracted suppressive T‐cell populations, including Th2 cells, Treg cells, NK CD56dim cells and MDSCs and established an immunosuppressive microenvironment to promote tumour progression (Fig 5H). Our findings reveal an unanticipated role for TERT as a transcriptional activator of ERVs and provide strong evidence that TA‐ERV/interferon signalling contributes to immune suppression in tumours, offering insights to combinational cancer therapeutic strategy with telomerase inhibitors, which not only restrict cancer cell proliferation but also compromise tumour immunosuppressive microenvironments, especially for tumours highly expressing TERT.
Given that ERVs represent ~8% of the human genome (Lander et al, 2001) and their activation is a common feature in human cancers (Matteucci et al, 2018), future work on the regulation and function of ERVs in physiological and pathological contexts would provide important insights into the role of ERVs in cancer and opportunities to develop novel cancer therapeutic interventions.
Materials and Methods
Cell culture
U2OS, HeLa, WI38 and HEK293T cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% foetal bovine serum (FBS), HCT116 cells were cultured in McCoy’s 5A medium (Biological Industries) with 10% FBS, at 37°C and 5% CO2.
Plasmids and siRNAs transfection
Cells were transfected with pcDNA3.1‐TERT‐WT, pcDNA3.1‐TERT‐K626A or empty vector using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions for 48 h.
Control siRNA (siNC), TERT and Sp1 siRNAs were purchased from GenePharma (Shanghai, China). Transfection of siRNAs was performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instructions. The sequences of siRNAs are listed in Appendix Table S1.
Stable cell lines generation and inducing expression
GFP, TERT‐WT and TERT‐K626A were cloned into the pLVX‐TetOne‐Puro vector with the EcoRI/AgeI sites and confirmed by DNA sequencing. HEK293T cells were co‐transfected with the resultant vectors, together with psPAX2 and pMD2.G. Lentiviruses were harvested, filtered and concentrated. U2OS and WI38 cells were infected with lentiviruses containing inducible GFP, TERT‐WT or TERT‐K626A, and stable cell lines were generated by puromycin selection. Then, the concentration of doxycycline was tested to induce the ectopic expression of TERT in U2OS and WI38 at the levels similar to that of HeLa cells. Doxycycline (Selleck) (for U2OS, 0.02 μg ml−1; for WI38, 0.05 μg ml−1) was added into the medium of inducible U2OS or WI38 cell lines, GFP was used as negative control. After 48 h of induction (doxycycline added every 24 h), the samples were harvested and tested.
Quantitative real‐time PCR
Total RNA was extracted using TRIzol (for cell and tissue samples) or TRIzol LS (for blood samples) Reagent (Invitrogen), and the cDNA was generated using FastKing RT Kit (with gDNase) (TIANGEN) according to the manufacturer’s protocol. Quantitative real‐time PCR (qPCR) were performed with iTaqTM Universal SYBR Green Supermix (Bio‐Rad), and GAPDH was measured as an internal control. Primers used for qPCR were confirmed their specificity with dissociation curves, and the sequences are listed in Appendix Table S2.
Immunofluorescence microscopy
U2OS cells transfected by TERT (WT or K626A) or vector were fixed with 3.7% paraformaldehyde and permeabilized in 0.1% Triton X‐100. After blocking of nonspecific antigens with 3% goat serum, cells were incubated with primary antibody against dsRNA (J2, English and Scientific Consulting Kft., 10010200) overnight at 4°C. Then, cells were washed and subsequently incubated with secondary antibody Alexa Fluor 488 donkey anti‐mouse IgG (H + L) (Invitrogen). Samples were washed and counterstained with DAPI in Vectashield mounting medium (Vectorlabs). Fluorescence was detected and imaged using a Zeiss confocal fluorescence microscope.
Telomere measurement by quantitative real‐time PCR
Quantitative real‐time PCR was used to measure relative telomere length based on previously described (Zhang et al, 2020). Genome DNA was extracted from cells using DNeasy Blood & Tissue Kit (Qiagen). Average telomere length was measured from total genomic DNA using a qPCR assay, modified for measurement of human telomeres. The qPCRs were performed using telomeric primers and primers for the reference control gene (human 36B4 single copy gene) (Appendix Table S3). For each qPCR, a standard curve was made by serial dilutions of known amounts of human genomic DNA. The telomere signal was normalized to the signal from the single copy gene to generate a T/S ratio indicative of relative telomere length.
Western blot
Protein samples were subjected to polyacrylamide gel electrophoresis using the 10% SurePAGE™ gels (GenScript), transferred to PVDF (Millipore) membranes, and immunoblotted using antibodies that specifically recognize TERT (Abcam, ab32020), TBK1 (CST, 3013), pTBK1 Ser172 (CST, 5483), IRF3 (CST, 4302), pIRF3 Ser396 (CST, 4947), Sp1 (CST, 9389), GFP (Santa Cruz, sc‐9996) and GAPDH (HuaAn Biotechnology, EM1101). Secondary antibodies were Goat anti‐Rabbit IgG (whole molecule) (Sigma‐Aldrich, A9169) and Rabbit anti‐Mouse IgG (whole molecule) (Sigma‐Aldrich, A9044). Imaging of blots was performed using the Bio‐Rad ChemiDoc™ MP Imaging System.
dsRNA enrichment
For dsRNA enrichment, total RNA was immunoprecipitated using the J2 dsRNA‐specific antibody (English and Scientific Consulting Kft., 10010200). In brief, the product of 9 μg RNA was incubated in binding buffer (150 mM NaCl, 50 mM Tris pH 8.0, 1 mM EDTA, 1% NP‐40) with 5 μg J2 antibody and 20 μl Magna ChIP™ Protein A + G Magnetic Beads (Millipore, 16‐663), rotating overnight at 4°C, followed by five washes in cold binding buffer. RNA was then extracted with TRIzol Reagent and expression levels of indicated ERVs were analysed by strand‐specific RNA‐seq.
First‐strand cDNA synthesis and strand‐specific qPCR for detection of sense and antisense ERV transcripts
FastKing RT Kit (with gDNase) (TIANGEN) was adapted to perform strand‐specific reverse transcription with RNA from U2OS or HeLa cells transfected by TERT (WT or K626A). For first‐strand cDNA synthesis, 1 μg RNA was used for MLT1O‐int, LTR69, LTR38‐int, MER88, MER92C, MLT1G1‐int and β‐actin. A strand‐specific primer (0.1 μM) for each ERV or β‐actin (Appendix Table S4) was implemented in the reaction. First‐strand cDNA was generated according to the manufacturer’s protocol. Afterwards, strand‐specific qPCR was performed with iTaqTM Universal SYBR Green Supermix (Bio‐Rad). β‐actin was used as internal negative control for that was previously demonstrated to have no antisense transcript (Chen et al, 2004).
MeDIP DNA preparation and qPCR
MeDIP was performed based on previously described (Morris et al, 2011). RNA‐free genomic DNA was extracted from transfected HeLa cells using DNeasy Blood & Tissue Kit (Qiagen). A measure of 6 μg of each DNA sample was digested overnight at 37°C with 24 U of MseI (NEB). The reactions were stopped by heating at 65°C for 20 min, and the success of the reactions was verified by running a small aliquot of the digested DNA on an agarose gel. DNA was purified by phenol/chloroform extraction and precipitation and quantified using a NanoDrop spectrophotometer (Thermo Scientific), and 1 μg of DNA was diluted in TE buffer to a final volume of 450 μl. The DNA was denatured at 95°C for 10 min. A volume of 50 μl of 10 × IP buffer (100 mM Na‐Phosphate pH 7.0, 1.4 M NaCl, 0.5% Triton X‐100) and 1 μg of 5mC antibody (Epigentek, A‐1014) or IgG control (CST, 5415) with 20 μl Magna ChIP™ Protein A + G Magnetic Beads (Millipore, 16‐663) were added to the digested DNA, and the mixture was incubated overnight at 4°C with gentle rotation. The beads were washed three times with 1× IP buffer and resuspended in 250 μl digestion buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS). A volume of 3.5 μl of proteinase K was added to the above mixture, and incubation was carried out 3 h at 50°C. DNA was purified by phenol/chloroform extraction and ethanol precipitation (the latter with the addition of 25 μg glycogen to facilitate visualization of the DNA pellet). 10 ng input DNA and 1 μl IP DNA were used for qPCR assay using primers listed in Appendix Table S2. Commercial primer sets recognizing exon 8 of the human ZC3H13 gene (Human Positive Control Primer Set ZC3H13, Active Motif, 71009) and a gene desert on chromosome 12 (Human Negative Control Primer Set 1, Active Motif, 71001) were used for positive and negative controls, respectively.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed with a Pierce Magnetic ChIP Kit (Thermo Scientific, 26157) according to the manufacturer’s instruction. Briefly, cells were crosslinked, lysed, digested with MNase and sonicated to generate DNA fragments. ChIP was performed using antibodies against TERT (Abcam, ab32020), Sp1 (CST, 9389), or normal rabbit IgG (provided by the ChIP kit) as a negative control. The immunoprecipitates were then washed and eluted. The eluates were de‐crosslinked, and ChIP‐enriched DNA was purified for qPCR analysis using primers listed in Appendix Table S2.
Animal studies
Tert +/− mice on the C57BL/6/129/SvJ genetic background were kindly provided by Dr Yie Liu (NIA, Baltimore, MD, USA). Heterozygous (Tert +/−) pairs were intercrossed to generate first‐generation Tert +/+ (WT) and Tert −/− (KO) strains. All animal experiments were performed according to the guidelines of the Animal Care and Use Committee of Hangzhou Normal University. This study was compliant with all of the relevant ethical regulations regarding animal research.
For ENU treatment, age‐matched WT and G1 Tert −/− mouse cohorts were treated with 150 μg g‐1 ENU (Sigma‐Aldrich) or saline by intraperitoneal, serum or livers were extracted after 24 h of injection.
For poly(I:C) treatment, age‐matched WT and G1 Tert −/− mouse cohorts were treated with 50 μg poly(I:C) HMW (InvivoGen) or saline by intraperitoneal, serum was extracted after 24 h of injection.
ELISA and cytokine array
WT and G1 Tert −/− mice were treated with poly(I:C) or saline and serum was extracted as described above, Mouse CRG‐2 (CXCL10) ELISA Kit (RayBiotech, ELM‐CRG2) was used according to the manufacturer’s instruction. For the ELISA of cell culture supernatant, cell culture supernatant was collected after 48 h of treatment. Human IP‐10 (CXCL10) ELISA Kit (RayBiotech, ELH‐IP10/IQH‐IP10) and IFNβ ELISA Kit (R&D, DIFNB0) were used according to the manufacturer's instruction.
For cytokine/chemokine array, WT and G1 Tert −/− mice were treated with ENU or saline and serum was extracted as described above. Quantibody® Mouse Inflammation Array 1 (RayBiotech, QAM‐INF‐1) was used according to manufacturer’s instructions.
Flow cytometry analysis
The peripheral blood mononuclear cells (PBMCs) from mouse blood were isolated using Mouse Peripheral Blood Mononuclear Cells Separation Kit (Solarbio) according to the manufacturer’s instruction. Mouse PBMCs were stained with FITC Rat Anti‐Mouse CD4 (BD Pharmingen, 553650), PE Rat anti‐Mouse FOXP3 (BD Pharmingen, 563101), or isotype IgG control antibodies (BD Pharmingen, 554688 and 554689) for flow cytometry analysis. Briefly, cells were firstly stained with CD4‐FITC or FITC isotype control. After washed, the cells were fixed and permeabilized, then stained with FOXP3‐PE or PE isotype control. After washed, the cells were resuspended in PBS and analysed on a BD LSRFortessa. Fluorescence intensity of CD4 and FOXP3 was normalized to isotype controls. The data analyses were performed with FlowJo software.
Library preparation for transcriptome sequencing
A total amount of 3 µg RNA per sample was used as input material for the RNA sample preparations. Sequencing libraries were generated using NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, mRNA was purified from total RNA using poly‐T oligo‐attached magnetic beads. Fragmentation was carried out using divalent cations under elevated temperature in NEBNext First Strand Synthesis Reaction Buffer (5×). First‐strand cDNA was synthesized using random hexamer primer and M‐MuLV Reverse Transcriptase (RNase H‐). Second strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure were ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 250–300 bp in length, the library fragments were purified with AMPure XP system (Beckman Coulter). Then 3 µl USER Enzyme (NEB) was used with size‐selected, adaptor‐ligated cDNA at 37°C for 15 min followed by 5 min at 95°C before PCR. Then, PCR was performed with Phusion High‐Fidelity DNA polymerase, Universal PCR primers and Index (X) Primer. At last, PCR products were purified (AMPure XP system) and library quality was assessed on the Agilent Bioanalyzer 2100 system.
Clustering and sequencing
The clustering of the index‐coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3‐cBot‐HS (Illumia) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina Hiseq platform and 125 bp/150 bp paired‐end reads were generated.
RNA‐seq analysis for expression of genes
Raw data (raw reads) of fastq format were firstly processed through in‐house perl scripts. In this step, clean data (clean reads) were obtained by removing reads containing adapter, reads containing poly‐N and low quality reads from raw data. At the same time, Q20, Q30 and GC content of the clean data were calculated. All the downstream analyses were based on the clean data with high quality.
Reference genome and gene model annotation files were downloaded from genome website directly. Index of the reference genome was built using Hisat2 and paired‐end clean reads were aligned to the reference genome using Hisat2 (Kim et al, 2015). HTSeq (Anders et al, 2015) was used to count the reads numbers mapped to each gene. And then FPKM of each gene was calculated based on the length of the gene and reads count mapped to this gene. Differential expression analyses of two conditions/groups were performed using the DESeq2 R package (Love et al, 2014). The resulting P‐values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P‐value < 0.05 found by DESeq2 were assigned as differentially expressed.
GO enrichment analysis of differentially expressed genes
Gene Ontology (GO) enrichment analysis of differentially expressed genes was implemented by the clusterProfiler R package (Yu et al, 2012), in which gene length bias was corrected. GO terms with corrected P‐value less than 0.05 were considered significantly enriched by differential expressed genes.
RNA‐seq analysis for expression of ERVs
To assess the expression levels of ERVs, RNA‐seq clean reads were mapped to the hg38 or mm10 genome using the STAR aligner software (Dobin et al, 2013), allowing up to three mismatches and filtering out reads that mapped to > 500 positions in the genome. Mapped files were then processed using the makeTagDirectory script of HOMER with the ‐keepOne option (Heinz et al, 2010). The tag directories of the mapped files were analysed using the analyzeRepeats.pl script of HOMER with the option ‘repeat’ and ‐noadj or ‐fpkm. This script adds the reads that map to multiple loci to the expression of the repeat class they represent regardless of where they are specifically placed. Differential repeats (ERVs included) expression analyses were performed using DESeq2.
Single‐sample gene set enrichment analysis (ssGSEA)
The ssGSEA (Barbie et al, 2009) was introduced to quantify the relative infiltration of 25 immune cell types in the tumour microenvironment for 9,264 tumour samples from 24 cancer types in TCGA database (Rahman et al, 2015). Feature gene panels for each immune cell type were obtained from recent publications (Bindea et al, 2013; Charoentong et al, 2017). The relative abundance of each immune cell type was represented by an enrichment score in ssGSEA analysis using GSVA R package (Hänzelmann et al, 2013).
For ssGSEA of GO or cellular pathways, GMT files (GO: Gene Ontology gene sets and CP: Canonical pathways) were downloaded from Molecular Signatures Database on GSEA website and ssGSEA analyses were performed by GSVA R package (Hänzelmann et al, 2013).
RNA‐seq analysis for expression of genes and ERVs from the GEO database
Published raw data of RNA‐seq were downloaded from the GEO database (Accession numbers are listed in Appendix Table S5). Analyses for expression of genes and ERVs were performed as described above.
ChIP‐seq analysis
Published ChIP‐seq raw data for TERT (A2780: GSM2044817; HCT116: GSM2044819; VA13: GSM2154981, GSM2154982) and Sp1 (K562: GSM803505; HEK293: GSM4141459, GSM4141460; HepG2: GSM2797629) were downloaded from the GEO database. The raw data of the ChIP‐seq libraries were mapped to hg38 genome using Bowtie2 with parameter ‐‐local ‐k 1 (Langmead & Salzberg, 2012). Average binding plots were generated using ngs.plot (Shen et al, 2014).
Colon tumour samples and tissue microarrays
The cDNA and tissue microarrays of human colon tumour samples were purchased from Shanghai Outdo Biotechnology. Immunohistochemical staining was performed with primary antibodies against CD4 (Abcarta, PA285) and FOXP3 (CST, 12653).
Statistical analysis
All graphs depict mean ± SEM unless otherwise indicated. Tests for differences between two groups were performed using two‐tailed unpaired Student’s t‐test or two‐tailed Mann–Whitney U test, as specified in the figure legends. One‐way or two‐way analysis of variance (ANOVA) was performed where applicable using the Fisher's LSD test. P values were considered significant if less than 0.05. Statistical significance was defined as */#P < 0.05, **/##P < 0.01, ***/###P < 0.001, or ns, not significant. GraphPad Prism 8 or R were used for statistical analysis of experiments, data processing, and presentation.
Author contributions
Jian Mao: Conceptualization; Investigation; Writing—original draft. Qian Zhang: Conceptualization; Investigation. Yaxiang Wang: Investigation. Yang Zhuang: Investigation. Lu Xu: Investigation. Xiaohe Ma: Investigation. Di Guan: Investigation. Junzhi Zhou: Investigation. Jiang Liu: Investigation. Xiaoying Wu: Project administration. Qian Liang: Investigation. Miao Wang: Investigation. Yu‐Sheng Cong: Conceptualization; Supervision; Funding acquisition; Investigation; Writing—review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
JM, QZ, and Y‐SC conceived of and designed the experiments. JM and QZ performed most of the experiments and data analyses. YW, YZ, LX, XM, DG, JZ, JL, XW, and QL, MW helped with experiments. JM, QZ, and Y‐SC wrote the paper.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
Source Data for Figure 1
Source Data for Figure 2
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31730020, 32000512, 31801155), and the Hangzhou Science and Technology Bureau (20182014B01).
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
EMBO reports (2022) 23: e52984.
Data availability
RNA‐seq data are available in the GEO database with GSE169715 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE169715).
References
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high‐throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C et al (2009) Systematic RNA interference reveals that oncogenic KRAS‐driven cancers require TBK1. Nature 462: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D et al (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Rouse BT (2005) Natural regulatory T cells in infectious disease. Nat Immunol 6: 353–360 [DOI] [PubMed] [Google Scholar]
- Bertorelle R, Briarava M, Rampazzo E, Biasini L, Agostini M, Maretto I, Lonardi S, Friso ML, Mescoli C, Zagonel V et al (2013) Telomerase is an independent prognostic marker of overall survival in patients with colorectal cancer. Br J Cancer 108: 278–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindea G, Mlecnik B, Tosolini M, Kirilovsky A, Waldner M, Obenauf A, Angell H, Fredriksen T, Lafontaine L, Berger A et al (2013) Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39: 782–795 [DOI] [PubMed] [Google Scholar]
- Blasco MA (2005) Telomeres and human disease: ageing, cancer and beyond. Nat Rev Genet 6: 611–622 [DOI] [PubMed] [Google Scholar]
- Borah S, Xi L, Zaug AJ, Powell NM, Dancik GM, Cohen SB, Costello JC, Theodorescu D, Cech TR (2015) Cancer. TERT promoter mutations and telomerase reactivation in urothelial cancer. Science 347: 1006–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, Campisi M, Kuang Y, Zhang Y, Gjini E et al (2018) Tumor innate immunity primed by specific interferon‐stimulated endogenous retroviruses. Nat Med 24: 1143–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoentong P, Finotello F, Angelova M, Mayer C, Efremova M, Rieder D, Hackl H, Trajanoski Z (2017) Pan‐cancer immunogenomic analyses reveal genotype‐immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 18: 248–262 [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD (2004) Over 20% of human transcripts might form sense‐antisense pairs. Nucleic Acids Res 32: 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli K, Strissel P, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote N, Cope L, Snyder A et al (2015) Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 162: 974–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y‐S, Wright WE, Shay JW (2002) Human telomerase and its regulation. Microbiol Mol Biol Rev 66: 407–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding D, Xi P, Zhou J, Wang M, Cong Y‐S (2013) Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF‐κB‐dependent transcription. FASEB J 27: 4375–4383 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA‐seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinet E, Gu Z, Imbusch CD, Giese NA, Büscher M, Safavi M, Weisenburger S, Klein C, Vogel V, Falcone M et al (2021) Aggressive PDACs show hypomethylation of repetitive elements and the execution of an intrinsic IFN program linked to a ductal cell of origin. Cancer Discov 11: 638–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Gilbert C (2012) Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13: 283–296 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Saginc G, Leow SC, Khattar E, Shin EM, Yan TD, Wong M, Zhang Z, Li G, Sung W‐K et al (2012) Telomerase directly regulates NF‐κB‐dependent transcription. Nat Cell Biol 14: 1270–1281 [DOI] [PubMed] [Google Scholar]
- Gil ME, Coetzer TL (2004) Real‐time quantitative PCR of telomere length. Mol Biotechnol 27: 169–172 [DOI] [PubMed] [Google Scholar]
- Groh S, Schotta G (2017) Silencing of endogenous retroviruses by heterochromatin. Cell Mol Life Sci 74: 2055–2065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guldner IH, Wang Q, Yang L, Golomb SM, Zhao Z, Lopez JA, Brunory A, Howe EN, Zhang Y, Palakurthi B et al (2020) CNS‐native myeloid cells drive immune suppression in the brain metastatic niche through Cxcl10. Cell 183: 1234–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänzelmann S, Castelo R, Guinney J (2013) GSVA: gene set variation analysis for microarray and RNA‐seq data. BMC Bioinformatics 14: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK (2010) Simple combinations of lineage‐determining transcription factors prime cis‐regulatory elements required for macrophage and B cell identities. Mol Cell 38: 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hur S (2019) Double‐stranded RNA sensors and modulators in innate immunity. Annu Rev Immunol 37: 349–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Q, Wu W, Wang Y, Alexander PB, Sun C, Gong Z, Cheng J‐N, Sun H, Guan Y, Xia X et al (2018) Local mutational diversity drives intratumoral immune heterogeneity in non‐small cell lung cancer. Nat Commun 9: 5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küry P, Nath A, Créange A, Dolei A, Marche P, Gold J, Giovannoni G, Hartung H‐P, Perron H (2018) Human endogenous retroviruses in neurological diseases. Trends Mol Med 24: 379–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped‐read alignment with Bowtie 2. Nat Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Ding D, Hao W, Yang F, Wu X, Wang M, Xu X, Ju Z, Liu J‐P, Song Z et al (2016) hTERT promotes tumor angiogenesis by activating VEGF via interactions with the Sp1 transcription factor. Nucleic Acids Res 44: 8693–8703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO et al (2000) The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo . Curr Biol 10: 1459–1462 [DOI] [PubMed] [Google Scholar]
- Liu Z, Li Q, Li K, Chen L, Li W, Hou M, Liu T, Yang J, Lindvall C, Björkholm M et al (2013) Telomerase reverse transcriptase promotes epithelial‐mesenchymal transition and stem cell‐like traits in cancer cells. Oncogene 32: 4203–4213 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15: 550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manghera M, Douville RN (2013) Endogenous retrovirus‐K promoter: a landing strip for inflammatory transcription factors? Retrovirology 10: 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matteucci C, Balestrieri E, Argaw‐Denboba A, Sinibaldi‐Vallebona P (2018) Human endogenous retroviruses role in cancer cell stemness. Semin Cancer Biol 53: 17–30 [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X‐P, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang X‐Y, Edouard P, Howes S et al (2000) Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403: 785–789 [DOI] [PubMed] [Google Scholar]
- Mocellin S, Pooley KA, Nitti D (2013) Telomerase and the search for the end of cancer. Trends Mol Med 19: 125–133 [DOI] [PubMed] [Google Scholar]
- Morris MR, Ricketts CJ, Gentle D, McRonald F, Carli N, Khalili H, Brown M, Kishida T, Yao M, Banks RE et al (2011) Genome‐wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 30: 1390–1401 [DOI] [PubMed] [Google Scholar]
- Park J‐I, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H et al (2009) Telomerase modulates Wnt signalling by association with target gene chromatin. Nature 460: 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Jackson LK, Johnson WE, Li DY, Bild AH, Piccolo SR (2015) Alternative preprocessing of RNA‐Sequencing data in The Cancer Genome Atlas leads to improved analysis results. Bioinformatics 31: 3666–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen S, Han H, Liang G, Jones P, Pugh T et al (2015) DNA‐demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162: 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao N, Liu X, Nestler E (2014) ngs.plot: Quick mining and visualization of next‐generation sequencing data by integrating genomic databases. BMC Genom 15: 284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strissel PL, Ruebner M, Thiel F, Wachter D, Ekici AB, Wolf F, Thieme F, Ruprecht K, Beckmann MW, Strick R (2012) Reactivation of codogenic endogenous retroviral (ERV) envelope genes in human endometrial carcinoma and prestages: Emergence of new molecular targets. Oncotarget 3: 1204–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargiu L, Rodriguez‐Tomé P, Sperber GO, Cadeddu M, Grandi N, Blikstad V, Tramontano E, Blomberg J (2016) Classification and characterization of human endogenous retroviruses; mosaic forms are common. Retrovirology 13: 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Qi X, Gong L, Xing G, Chen M, Miao L, Yao J, Suzuki T, Furihata C, Luan Y et al (2012) Identification of BC005512 as a DNA damage responsive murine endogenous retrovirus of GLN family involved in cell growth regulation. PLoS One 7: e35010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L‐G, Han Y, He Q‐Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16: 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Liu N, Bai J, Zhou QI, Mao J, Xu LU, Liu J, Wei H, Ren C, Wu X et al (2020) Human telomerase reverse transcriptase is a novel target of Hippo‐YAP pathway. FASEB J 34: 4178–4188 [DOI] [PubMed] [Google Scholar]


