Abstract
Motile cilia on the cell surface generate movement and directional fluid flow that is crucial for various biological processes. Dysfunction of these cilia causes human diseases such as sinopulmonary disease and infertility. Here, we show that Ccdc108, a protein linked to male infertility, has an evolutionarily conserved requirement in motile multiciliation. Using Xenopus laevis embryos, Ccdc108 is shown to be required for the migration and docking of basal bodies to the apical membrane in epidermal multiciliated cells (MCCs). We demonstrate that Ccdc108 interacts with the IFT‐B complex, and the ciliation requirement for Ift74 overlaps with Ccdc108 in MCCs. Both Ccdc108 and IFT‐B proteins localize to migrating centrioles, basal bodies, and cilia in MCCs. Importantly, Ccdc108 governs the centriolar recruitment of IFT while IFT licenses the targeting of Ccdc108 to the cilium. Moreover, Ccdc108 is required for the centriolar recruitment of Drg1 and activated RhoA, factors that help establish the apical actin network in MCCs. Together, our studies indicate that Ccdc108 and IFT‐B complex components cooperate in multiciliogenesis.
Keywords: actin cytoskeleton, Ccdc108, ciliogenesis, IFT‐B complex, motile cilia
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Molecular Biology of Disease
Ccdc108, an axonemal central pair apparatus protein, localizes to centrioles and regulates accumulation of IFT‐B complex proteins and PCP‐associated actin cytoskeletal factors required for migration and docking of centrioles to the cell surface during multiciliogenesis.
Introduction
Motile cilia are present on the epithelial cell surface to generate a directional fluid flow that is crucial for various biological processes, such as the establishment of left–right symmetry in the ventral node, mucociliary clearance in airways, and cerebrospinal fluid circulation in the brain (Fliegauf et al, 2007; Brooks & Wallingford, 2014). The axonemal structure of most motile cilia has a “9 + 2” arrangement with nine peripheral outer doublet microtubules surrounding a central microtubule pair, however, motile nodal cilia have an either “9 + 0” or “9 + 4” structure (Feistel & Blum, 2006; Ishikawa & Marshall, 2011; Spassky & Meunier, 2017). The synchronous ciliary motility is governed by protein appendages attached to the axonemal microtubules, which include axonemal dyneins (inner dynein arm [IDA] and outer dynein arm [ODA] complexes), radial spokes, and nexin–dynein regulatory complexes (Teves et al, 2016). Radial spokes interact with IDA and the central pair of microtubules to coordinate the ciliary motility (Yang et al, 2006; Viswanadha et al, 2017). Defects in the assembly and motility of motile cilia can result in various human diseases such as sinopulmonary disease, hearing impairment, hydrocephalus, situs inversus, and male infertility (Nigg & Raff, 2009; Reiter & Leroux, 2017).
In Xenopus laevis epidermis and mammalian airways, the commitment of MCC precursors is transcriptionally controlled by GemC1 and Multicilin (Stubbs et al, 2012; Ma et al, 2014; Kyrousi et al, 2015; Arbi et al, 2016; Terre et al, 2016; Kim et al, 2018), which are necessary and sufficient to induce MCC differentiation. Upon activation of the transcriptional cascade, MCC precursors undergo massive centriole amplification mostly through a deuterosome‐dependent pathway to produce hundreds of centrioles, which sequentially dissociate from deuterosomes, migrate and dock to the apical surface, and convert into basal bodies to initiate axoneme growth (Klos Dehring et al, 2013; Zhao et al, 2013; Brooks & Wallingford, 2014; Spassky & Meunier, 2017). In order for centrioles to dock with the apical membrane they must develop distal appendages following the deuterostome stage (Spassky & Meunier, 2017). MCCs also undergo remodeling of the subapical membrane cytoskeleton, in particular the actin network, to accommodate centriole membrane docking and motile cilium assembly (Antoniades et al, 2014; Sedzinski et al, 2016; Kulkarni et al, 2018). The establishment of the apical actin network involves focal adhesion proteins and components of the planar cell polarity (PCP) pathway including dishevelled (Dvl), Daam1, and the Daam1/Dvl regulator Drg1, which can stimulate the activation of RhoA, a Rho GTPase that regulates the actin polymerization (Park et al, 2008; Antoniades et al, 2014; Yasunaga et al, 2015; Lee et al, 2019).
Assembly and functional maintenance of the cilium require intraflagellar transport (IFT) particles (Davenport et al, 2007; Lechtreck, 2015; Zhu et al, 2017). IFT particles regulate the bidirectional trafficking of ciliary proteins along the axonemal microtubules, and can assemble into linear IFT train structures of varying length (Pigino et al, 2009; Stepanek & Pigino, 2016; Yang & Huang, 2019). The IFT particle is composed of two multiunit subcomplexes, IFT‐A and IFT‐B. The IFT‐A complex functions in retrograde trafficking from the ciliary tip to the base powered by the dynein‐2 motor complex, whereas the IFT‐B complex mediates anterograde protein trafficking from the cell to the ciliary tip through kinesin‐2 motor proteins (Rosenbaum & Witman, 2002; Ishikawa & Marshall, 2011; Sung & Leroux, 2013; Liang et al, 2014). Defects in anterograde trafficking mediated by the IFT‐B complex cause shortened or an absence of cilia (Pazour et al, 2000; Davenport et al, 2007; Lechtreck, 2015; Kubo et al, 2016). IFT‐B subunits IFT20, IFT25, IFT27, and IFT74 are also required for sperm flagella formation in mice (Zhang et al, 2016, 2017; Shi et al, 2019). In the primary cilium, IFT‐B complex proteins accumulate at the mother centriole following removal of the CP110/CEP97 cap that blocks axoneme formation (Goetz et al, 2012; Lu et al, 2015; Kanie et al, 2017). In spite of the important role of IFT proteins in cilium assembly, little is known about how IFT‐B proteins are recruited to centrioles/basal bodies during early ciliogenesis and how this process is scaled/amplified in multiciliogenesis.
CCDC108 (also known as CFAP65) is the ortholog of Akap240, identified as an axonemal protein in Chlamydomonas reinhardtii (Gaillard et al, 2001; Zhao et al, 2019b; Dai et al, 2020). Interestingly, CCDC108 mutations cause abnormal sperm flagellum in patients (Wang et al, 2019; Zhang et al, 2019; Li et al, 2020), and its orthologs in mouse and chicken are essential for sperm motility (Imsland et al, 2012; Li et al, 2020; Wang et al, 2021). These studies suggest that CCDC108 is involved in regulating motile ciliation and/or movement. Here, we investigated the function of Ccdc108 in motile multiciliogenesis. We showed that Ccdc108 is required for ciliation in frog, fish, and mouse multiciliated cells. In Xenopus MCCs, Ccdc108 is needed for basal body migration/docking to the plasma membrane and apical enrichment of F‐actin during multiciliogenesis. We demonstrated that Ccdc108 localizes to centrioles as they migrate to the apical cell surface and in the cilium in Xenopus MCCs. Loss‐of‐function and replacement experiments in vivo demonstrated that the Ccdc108 interacts with the IFT machinery during ciliogenesis and in the mature cilium, and this association is essential for the migration/docking of centrioles to the apical membrane during cilium assembly in MCCs. Finally, we showed that Ccdc108 and its interaction with IFT machinery are crucial for the centriolar distribution of planar cell polarity‐associated actin cytoskeleton regulators Drg1 and RhoA, which contribute to the apical actin polymerization during multiciliation. Together, our findings demonstrate that Ccdc108 and IFT‐B complex components function together in multiciliogenesis.
Results
Ccdc108 is essential for ciliation in multiciliated epidermis of the Xenopus embryo
Previous studies indicate that Ccdc108 is important for proper sperm motility in chicken, mouse, and human (Imsland et al, 2012; Wang et al, 2019, 2021; Zhang et al, 2019; Li et al, 2020). To further investigate Ccdc108 association with motile cilia function, we injected Xenopus laevis embryos with morpholino (MO) oligonucleotides against the ccdc108 mRNA sequence to block the translation of the protein. Morpholino efficiency was confirmed using a GFP reporter RNA containing the MO target site of ccdc108 at the 5’UTR (Fig EV1A) (Romaker et al, 2014). Since the motile cilia of MCCs on the skin of Xenopus embryos generate a directed fluid flow along the posterior ventral axis of the embryo epidermis (Mitchell et al, 2007), fluorescent microbeads were used to visualize the fluid flow. ccdc108 morphants displayed significantly affected bead motility over the epidermis (Fig 1A and B; Movies [Link], [Link], [Link]). Importantly, this defect was rescued by co‐injection of a hemagglutinin (HA)‐tagged ccdc108 mRNA, validating the specificity of the MO (Fig 1A and B; Movies [Link], [Link], [Link]). Defects in fluid flow may be caused by either reduction in cilia number or abnormal cilia motility. To evaluate Ccdc108 requirements in ciliary bead flow in MCCs, we performed immunostaining to examine cilia of epidermal MCCs by confocal and super‐resolution structured illumination microscopy (SIM) and surprisingly found a > 4‐fold reduction in cilia levels upon depletion (Fig 1C and D). We further confirmed that Ccdc108 depletion resulted in fewer and slightly shorter cilia in MCCs using scanning electron microscopy (SEM) (Figs 1E and EV1B). Moreover, we confirmed ciliation requirements for Ccdc108 using CRISPR/Cas9 technology to deplete the protein (Cong et al, 2013; Wang et al, 2015). Consistent with our MO studies, ccdc108 CRISPR mutants displayed a significant reduction in MCC cilia (Figs 1F and EV1C). Because the Chlamydomonas reinhardtii ortholog of Ccdc108, Akap240, is associated with the axoneme microtubule central pair which is important for cilia motility (Rao et al, 2016; Teves et al, 2016), we examined the ciliary beat pattern (CBP) and ciliary beat frequency (CBF) of MCCs by high‐speed video microscopy. MCCs of Ccdc108 morphants displayed an altered CBP and an increased CBF (Fig EV1D; Movies [Link], [Link], [Link]). Importantly, ciliary defects were restored by the expression of wild‐type (WT) Ccdc108 (Figs 1C–E and EV1D). This indicates that cilia present in Ccdc108 morphants have impaired motility. Examination of Ccdc108 expression in Xenopus laevis embryos by in situ hybridization chain reaction (HCR) combined with immunostaining for cilia confirmed that ccdc108 is highly expressed in MCCs but not in the neighboring non‐MCCs (Fig 1G). To further test if Ccdc108 is required for other types of cilia formation, primary cilia of neural progenitors in the neural tube and motile monocilia in the gastrocoel roof plate (GRP) were examined (Schweickert et al, 2007; Toriyama et al, 2017). Notably, neither of these monocilia was affected by Ccdc108 depletion (Fig EV1E and F). Thus, our results indicate that Ccdc108 plays a crucial role in regulating multiciliation in epidermal MCCs.
Figure EV1. Ccdc108 is essential for multiciliogenesis in epidermis of the Xenopus embryo (related to Fig 1).
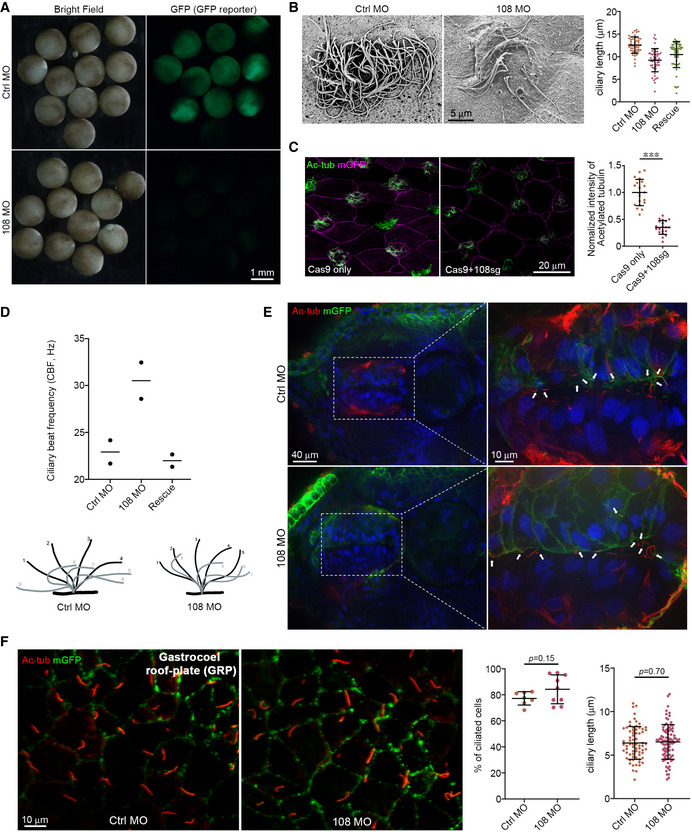
- In vivo validation of morpholino efficiency. ccdc108 morpholino inhibits the expression of the GFP reporter containing the MO target site of ccdc108 at the 5’UTR.
- Scanning electron microscope images and graphical plot show slightly shorter cilia in Ccdc108‐depleted MCCs. Experiment was performed once and 50 cilia from 10 MCCs in different microscopic fields were scored. Mean ± s.d. values are presented.
- Representative confocal images and graphical plot display reduced ciliary Ac‐tub levels in ccdc108 CRISPR mutants. A total of 20 images of 20 embryos for each condition. Cell membranes (mGFP, purple) and cilia (Ac‐tub, green) were labeled with indicated antibodies. Quantitative data from three independent experiments were scored. Unpaired two‐tailed t‐test was performed (***P < 0.001). Mean ± s.d. values are also presented.
- Graphical plot and cartoons show that epidermal MCC cilia beat frequency (CBF) and cilia beat pattern (CBP) are both affected in ccdc108 morpholino‐treated Xenopus embryos from stage 27. The cartoons were generated based on the results of imaging live MCCs by high‐speed video microscopy (Movies [Link], [Link], [Link]). Two biologically independent experiments were performed. Greater than 15 MCCs from four embryos for each condition.
- Transverse section views show that Ccdc108 is dispensable for primary ciliogenesis in the Xenopus neural tube. Embryos at stage 30 were fixed, and stained with the acetylated tubulin antibody (red) and DAPI (blue). mRNA of a membrane‐bound form of GFP (mGFP; green) was co‐injected with each morpholino to indicate targeted cells. Arrows mark primary cilia. Three biologically independent experiments were performed and images from the same experiment were presented.
- Effects of Ccdc108 depletion on motile monocilia formation and length in the gastrocoel roof plate (GRP). Embryos were fixed and stained with the acetylated tubulin antibody (red). mGFP (green) was co‐injected with each morpholino to indicate targeted cells. GRP explants were prepared from embryos at stage 18. Greater than 6 embryos from three biologically independent experiments for each condition were images and 10 cilia from each embryo were scored. Unpaired two‐tailed t‐test was performed. Mean ± s.d. values are also presented.
Source data are available online for this figure.
Figure 1. Ccdc108 is essential for ciliogenesis in multiciliated epidermis of the Xenopus embryo.
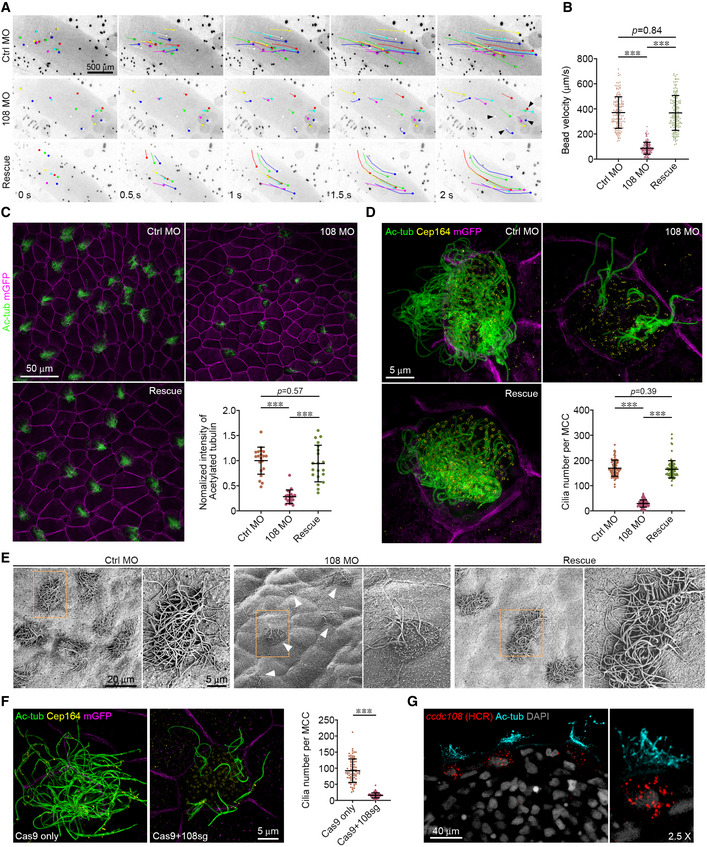
-
A, BDepletion of Ccdc108 markedly affects bead movement by decreasing velocity (Movies [Link], [Link], [Link]). Fluorescent beads were applied to the dorsal surface of Xenopus embryos. Bead tracks every 500 ms over 2 s are shown (A). Plot of average bead velocity shows significantly reduced bead flow in ccdc108 morphants, which can be fully rescued by co‐injecting with Xenopus ccdc108 mRNA (B). A total of 150 beads from 15 embryos for each condition. Arrowheads (A) indicate beads that showed no movement on the embryo surface through the recording.
-
C, DDefective cilia formation in ccdc108 morphants. Embryos at stage 27 were fixed and stained with the acetylated tubulin (Ac‐tub) antibody and/or Cep164 antibody. mRNA of a membrane‐bound form of GFP (mGFP) was co‐injected with each morpholino to indicate targeted cells. Representative confocal images (C) and 3D‐SIM images (D) of the epidermis for each condition and plots show significantly reduced cilia number in Ccdc108‐depleted embryos. (C) Twenty images of 20 embryos and (D) greater than 80 MCCs from six embryos for each condition.
-
EScanning electron microscope images of Xenopus embryos display defective cilia formation in Ccdc108‐depleted MCCs. White arrowheads mark MCCs with reduced ciliation. Boxed areas are magnified to show the details.
-
FDefective cilia formation occurs in ccdc108 CRISPR mutants. Embryos at one‐cell stage were injected with Cas9 protein with or without the sgRNA against ccdc108 (108sg) and fixed at stage 27. Representative 3D‐SIM images and plot show significantly reduced cilia number in ccdc108 CRISPR mutants. Greater than 70 MCCs from six embryos for each condition. Cell membranes (mGFP, purple), cilia (Ac‐tub, green), and basal bodies (Cep164, yellow) were labeled with indicated antibodies.
-
GIn situ hybridization chain reaction (HCR) reveals that ccdc108 mRNA is specially detected in MCCs. Embryos at stage 35 were fixed and subjected to in situ HCR (red). Embryos were then incubated with the Ac‐tub antibody (cyan) and DAPI (gray).
Data information: Quantification data were collected from three independent experiments. Unpaired two‐tailed t‐test was performed (***P < 0.001). Mean ± s.d. values are presented.
Source data are available online for this figure.
Ccdc108 ciliation requirement is evolutionarily conserved in fish and mouse cells
Since Ccdc108 plays an important role in multiciliation in Xenopus, we asked whether this requirement is evolutionarily conserved. We first examined Ccdc108 function in zebrafish. In situ HCR analysis in zebrafish embryos at the long‐pec stage (48 h post‐fertilization) showed that the ccdc108 mRNA was detected in the multiciliated olfactory placode cells and neuromast hair cells, but not in the surrounding cells (Fig 2A), suggesting a potential role of Ccdc108 in these two cell types with 9 + 2 structure cilia (Song et al, 2016). To investigate Ccdc108 ciliary requirements, we injected embryos with a morpholino targeting the translation start site of zebrafish ccdc108. ccdc108 morphants displayed hydrocephalus, a phenotype associated with ciliary motility dysfunction and excess cerebrospinal fluid (CSF) in the brain, which could be rescued by Xenopus Ccdc108 (Fig 2B). Additionally, ccdc108 morphants displayed a reduced number of acetylated tubulin‐positive cilia in both olfactory placode cells and neuromasts, which could be significantly restored by the expression of Xenopus Ccdc108 protein (Fig 2C and D). Using SEM, we confirmed ciliation was affected in olfactory placodes of ccdc108 morphants (Fig 2E). Collectively, these findings demonstrate that Ccdc108 is required for the formation of MCC cilia and the neuromast kinocilia and causes ciliopathy‐like phenotypes upon depletion in zebrafish.
Figure 2. The function of Ccdc108 in regulating multiciliogenesis is evolutionarily conserved across species.
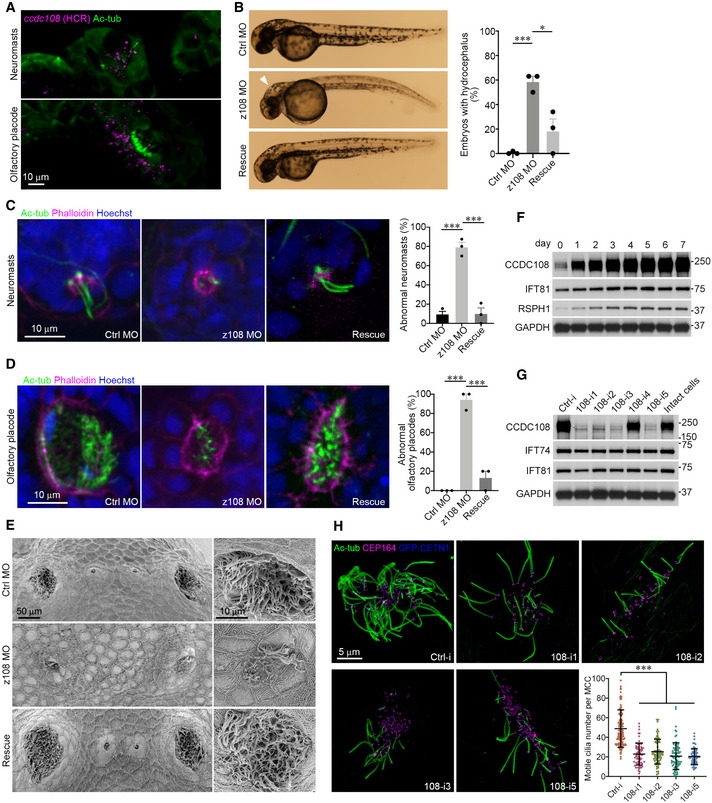
-
AIn situ HCR reveals that ccdc108 mRNA is specially detected in ciliated organs in zebrafish. Zebrafish embryos at the long‐pec stage (48 h post‐fertilization) were fixed and subjected to in situ HCR (purple). Embryos were then incubated with the Ac‐tub antibody (green).
-
BRepresentative images and plots show that ccdc108 morphants display severe hydrocephalus (white arrowhead) in zebrafish, which was rescued by expression of Xenopus Ccdc108.
-
C, DRepresentative confocal images and plots show that ccdc108 morphants displayed ciliogenesis defects in neuromasts (C) and olfactory placodes (D). Zebrafish embryos were fixed and stained with the Ac‐tub antibody (green), phalloidin (purple), and Hoechst (blue).
-
EScanning electron microscope images of zebrafish embryos display a failure in cilia formation in Ccdc108‐depleted olfactory placodes.
-
FCCDC108 shows similar expression pattern to that of indicated cilia‐related proteins in mEPCs. Cells were collected at the indicated day after serum starvation and used for immunoblotting. IFT81 is an IFT complex protein, and RSPH1 is a component of the ciliary radial spoke structure. GAPDH was used as loading control.
-
GFour independent shRNAs (108‐i1, ‐i2, ‐i3, and ‐i5) effectively depleted CCDC108 without affecting the indicated cilia‐related proteins. GAPDH served as loading control. Ctrl‐i, a control shRNA.
-
HRepresentative 3D‐SIM images and plot reveal that depletion of CCDC108 leads to a significant reduction in multiciliogenesis in mEPCs. mEPCs that were infected with lentivirus expressing indicated shRNA were serum starved for 5 days and subjected to immunostaining. GFP‐CETN1 (blue) marked mEPCs infected with lentivirus. Basal bodies (CEP164, purple) and cilia (Ac‐tub, green) were labeled with indicated antibodies. Greater than 60 MCCs from three independent repeats for each condition were counted.
Data information: Quantitative data were from three independent repeats. Unpaired two‐tailed t‐test was performed (***P < 0.001; *P < 0.05). Mean ± s.e.m. (B, C, and D) and mean ± s.d. (H) values are presented.
Source data are available online for this figure.
Next, we evaluated CCDC108 function in motile multiciliated mouse ependymal cells (mEPCs). During the course of ependymal cell differentiation, MCC precursors can be serum starved to induce differentiation and progressively undergo centriole amplification, centriole migration and docking to the plasma membrane, and axoneme extension similar to Xenopus MCCs (Brooks & Wallingford, 2014; Spassky & Meunier, 2017; Zhao et al, 2019a). Proteins functioning in ciliation are upregulated following serum starvation in mEPCs such as the IFT‐B protein IFT81 and the radial spoke protein RSPH1 (Fig 2F) (Pigino et al, 2011; Bhogaraju et al, 2013; Kubo et al, 2016). Thus, we examined the expression pattern of CCDC108 in the differentiating mEPCs. Immunoblotting showed that CCDC108 was gradually upregulated as mEPCs differentiated (Fig 2F), suggesting a potential role in regulating ciliation. To investigate CCDC108 requirements in multiciliation, lentivirus‐mediated stable expression of specific short hairpin RNAs (shRNAs) that decreased CCDC108 protein levels was utilized to deplete CCDC108 in mEPCs (Fig 2G). Strikingly, multiciliation was significantly inhibited by all four CCDC108‐depleting shRNAs without affecting levels of expression of other ciliary proteins (Fig 2G and H). Taken together, our functional analysis in Xenopus, zebrafish, and mouse cells indicates that Ccdc108 has an evolutionarily conserved requirement for multiciliation across species.
Ccdc108 is required for apical trafficking of centrioles and actin assembly in MCCs of the Xenopus epidermis
Since fewer multicilia were observed in frog, fish, and mouse cells following Ccdc108 depletion, we next investigated at what stage Ccdc108 is required during multiciliogenesis. We first examined whether ccdc108 morphants affected centriole amplification and maturation in Xenopus MCCs (Balestra & Gonczy, 2014; Ma et al, 2014; Zhang & Mitchell, 2015; Yan et al, 2016; Spassky & Meunier, 2017; Loncarek & Bettencourt‐Dias, 2018). Although our results revealed a slight decrease in centriole numbers in ccdc108 morphants (162 ± 17 in control morphants vs. 153 ± 17 in Ccdc108 morphants), this reduction does not explain the dramatic reduction in motile cilia in ccdc108 morphants (Figs 1D and 3A). Moreover, the majority of centrioles in Xenopus ccdc108 morphants and CRISPR mutants displayed Cep164 accumulation (Figs 1D and F, and 3A), suggesting that distal appendages form and therefore centriole maturation is unlikely to be affected by Ccdc108 depletion. Likewise, Cep164‐positive structures were observed on the centrioles of mEPCs depleted of Ccdc108 (Fig 2H). Together, these results indicate that Ccdc108 is dispensable for centriole amplification and maturation during ciliogenesis.
Figure 3. Ccdc108 is required for apical trafficking of basal bodies and F‐actin enrichment in MCCs of the Xenopus epidermis.
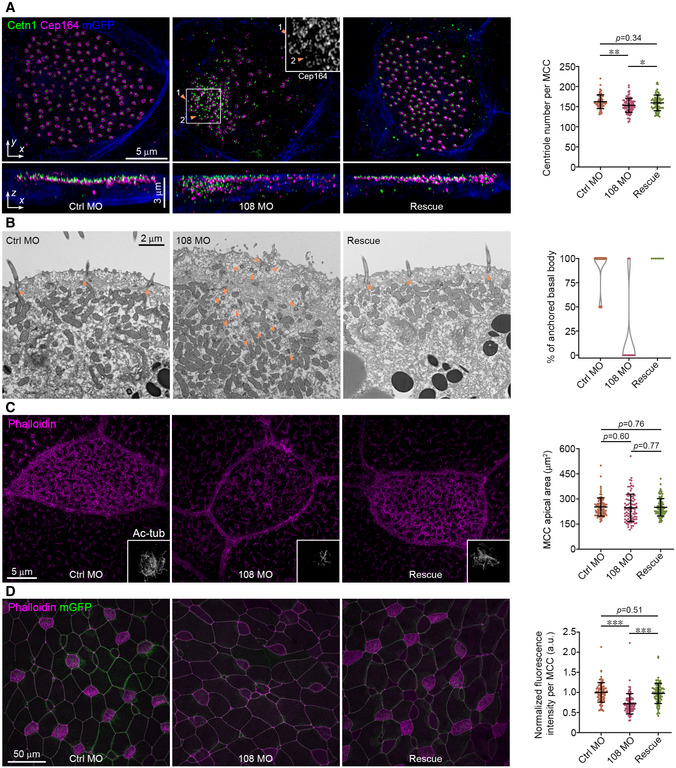
-
ADepletion of Ccdc108 causes disorganized basal body distribution in MCCs. Representative 3D‐SIM images (x–y) and 3D reconstructions (x–z) of MCCs of embryos show a failure of apical trafficking of basal bodies in Ccdc108‐depleted cells. Cell membranes (mGFP, blue), basal bodies (Cetn1, green), and distal appendages (Cep164, purple) were labeled with indicated antibodies. Select area of Cep164 channel was zoomed to show centriolar Cep164 accumulations. Orange arrowheads and numbers mark individual centriole. Greater than 70 MCCs from six embryos for each condition were counted.
-
BTransmission electron microscopy images of Xenopus embryos displaying aberrant apical trafficking and docking of centrioles in Ccdc108‐depleted MCCs. Orange arrowheads mark centrioles. Percentage of centrioles docked to plasma membrane in each field was scored. At least seven cells for each sample were counted.
-
CRepresentative 3D‐SIM images reveal that depletion of Ccdc108 has no effect on apical expansion in MCCs. Embryos treated as in Fig 1C were fixed and stained with phalloidin (purple). Cilia (Ac‐tub, gray) were also presented. Apical area of MCCs in each condition was measured as described in Methods and plotted. Greater than 80 MCCs from six embryos for each condition.
-
DRepresentative confocal images show a significant reduction in apical actin following depletion of Ccdc108. Cell boundaries and apical actin network were marked with mGFP (green) and phalloidin (purple) and phalloidin intensity levels were measured and plotted. Greater than 80 MCCs from six embryos for each condition.
Data information: Quantitative data in (A, C, and D) were from three independent experiments. Unpaired two‐tailed t‐test was performed (***P < 0.001; **P < 0.01; *P < 0.05). Mean ± s.d. values are presented.
Source data are available online for this figure.
Examination of centriole positioning in Xenopus ccdc108 morphants revealed defects in centriole migration and/or docking to the apical membrane (Fig 3A and B). In comparison to controls, ccdc108 morphants displayed centriole clustering in the cytoplasm by fluorescence microscopy (Fig 3A). This finding was further confirmed by transmission electron microscopy (TEM) (Fig 3B). Only 4 centrioles of 30 from 13 different cells of control morphants were observed in the cytosol, whereas most of the centrioles (71 centrioles of 75 from 12 cells) in ccdc108 morphants remained in the cytoplasm (Fig 3B). As expected, expression of WT Ccdc108 rescued this apical trafficking defect (Fig 3A and B). Together, these results demonstrate that Ccdc108 is important for the apical trafficking and/or docking of basal bodies.
MCC progenitors originate from the basal layer ectoderm and migrate apically before differentiating into MCCs. Once protruded out of the superficial epithelium, the apical surface has to expand to accommodate hundreds of motile cilia (Stubbs et al, 2006; Sedzinski et al, 2016; Kulkarni et al, 2018). Thus, we next examined whether Ccdc108 function in multiciliogenesis is associated with expansion of the apical surface in MCCs. Compared to control MCCs, the area of the apical surface was not affected in Ccdc108‐depleted MCCs at ciliated stage 27 (Fig 3C), indicating that Ccdc108 is dispensable for the apical expansion at this stage in MCCs. However, we observed that the apical enrichment of F‐actin was significantly reduced in MCCs of ccdc108 morphants, while re‐expression of exogenous Ccdc108 rescued the F‐actin levels (Fig 3D). Since there was a noticeable reduction in apical F‐actin but no obvious effect on the apical surface area at this stage, we performed the same analysis with embryos at a later stage (stage 32), and we observed that in MCCs of ccdc108 morphants, centrioles still failed to completely migrate/dock to the apical surface (Fig EV2A). However, unlike MCCs of embryos at stage 27, stage 32 MCCs displayed a significantly reduced apical size with less F‐actin enrichment (Fig 3C vs. Fig EV2B), suggesting that ccdc108 morphants fail to maintain the apical surface due to the reduction in the F‐actin network. Given these results, we conclude that Ccdc108 is required for the apical migration and/or docking of centrioles and apical F‐actin enrichment in multiciliated Xenopus epidermis.
Figure EV2. Ccdc108 is required for apical trafficking of basal bodies and F‐actin enrichment in MCCs of the Xenopus epidermis (related to Fig 3).
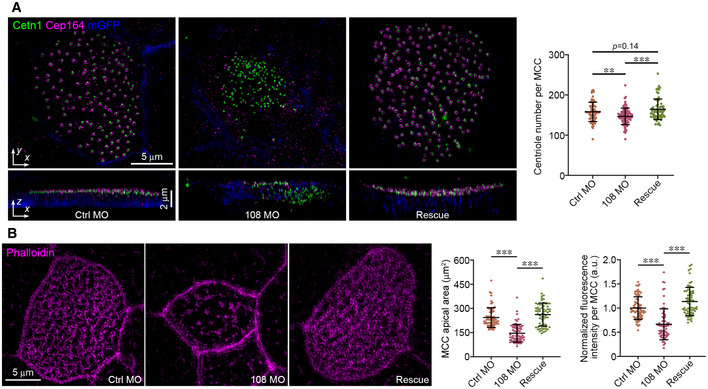
- Ccdc108 depletion affects centriole migration/docking to the apical surface in MCCs. Representative 3D‐SIM images (x–y) and 3D reconstructions (x–z) of MCCs of embryos at stage 32 show a failure of apical trafficking of basal bodies in Ccdc108‐depleted cells. Cell membranes (mGFP, blue), basal bodies (Cetn1, green), and distal appendages (Cep164, purple) were labeled with indicated antibodies. Greater than 60 MCCs from six embryos for each condition were counted.
- Representative 3D‐SIM images show a significant reduction in apical expansion in MCCs of embryos at stage 32. Embryos treated as in Fig 1C were fixed at stage 32 and stained with phalloidin (purple). Greater than 60 MCCs from six embryos for each condition.
Data information: Quantitative data were from three independent experiments. Unpaired two‐tailed t‐test was performed (***P < 0.001; **P < 0.01). Mean ± s.d. values are presented.
Source data are available online for this figure.
Ccdc108 localizes to the ciliary axoneme and basal body in fully ciliated MCCs
To better understand Ccdc108 ciliary function, we investigated its subcellular localization in Xenopus epidermal MCCs. GFP‐Ccdc108 localized to the basal body and along the cilium in fully ciliated MCCs of embryos at stage 27 (Fig 4A). Notably, in MCCs showing higher expression levels, ciliary Ccdc108 was prominently detected in a single punctum in most cilia and could be found along the axoneme anywhere from the ciliary base to the tip, and cytosolic Ccdc108 formed globular structures (Fig EV3A). To further examine the punctate ciliary localization of Ccdc108, we performed immunogold electron microscopy on the Xenopus epidermis using embryos expressing GFP‐Ccdc108, and found that gold particles labeled GFP‐Ccdc108 in electron‐dense regions at the tip of the cilia and at bulges along the axoneme (Fig EV3B), the latter resembling an IFT train‐like structure (Stepanek & Pigino, 2016; Vannuccini et al, 2016). Given that singular Ccdc108 puncta are observed in different areas of cilia, this suggested that Ccdc108 may be trafficked along the axoneme via these IFT train‐like structures. To examine Ccdc108 ciliary trafficking, we performed live cell imaging and found that Ccdc108 puncta display bidirectional movement along the axoneme (Fig EV3C; Movie EV7). Together, these results indicate that Ccdc108 localizes to the basal body and the cilium; localizations consistent with Ccdc108 requirements in multiciliogenesis as well as motile cilia structure and/or function.
Figure 4. Ccdc108 localizes to the ciliary axoneme and basal body in ciliated MCCs, and its ciliary accumulation is dependent on the Ccdc108–IFT interaction.
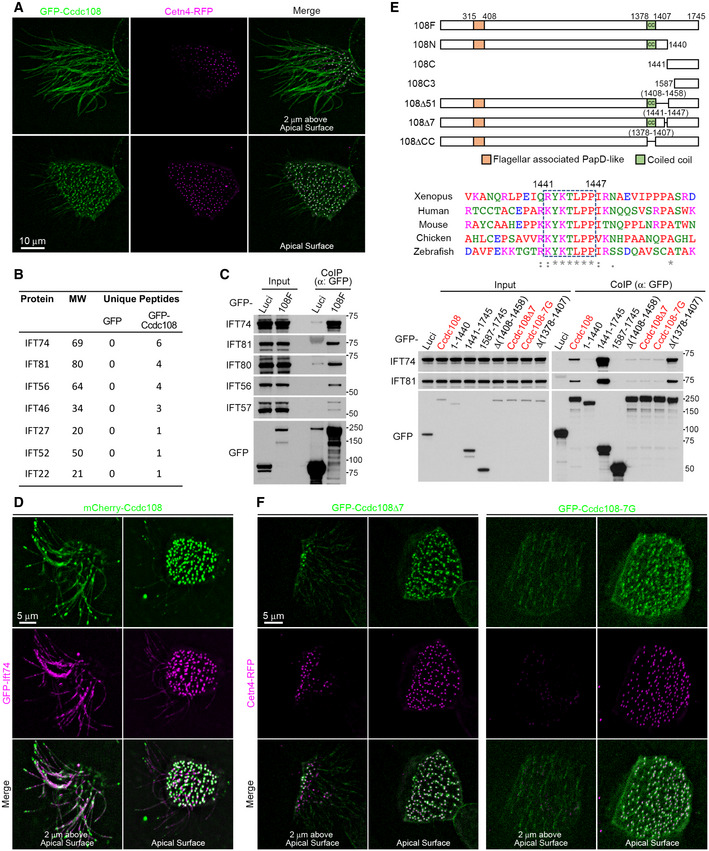
-
ALive imaging of MCCs shows the distribution of Ccdc108 in MCCs cilia and basal bodies. Embryos expressing GFP‐Ccdc108 (green) and Cetn4‐RFP (purple) were imaged at stage 27. To better visualize ciliary structures, maximum intensity projections of images stacks for regions containing cilia and basal bodies are shown separately.
-
B, CCcdc108 interacts with IFT proteins. IFT candidate proteins were identified by Shotgun mass spectrometric analysis (B). Interactions were further determined by co‐immunoprecipitation of endogenous IFT proteins (C) from HEK293T cells with GFP‐Trap agarose beads. The table lists unique peptide numbers of indicated IFT proteins in the GFP and GFP‐Ccdc108 samples.
-
DLive imaging of MCCs shows both Ccdc108 and Ift74 proteins localize to the ciliary axoneme and the basal bodies in MCCs. Embryos expressing mCherry‐Ccdc108 (green) and GFP‐Ift74 (purple) were imaged at stage 27.
-
EIdentification of an evolutionarily conserved seven amino acid motif (dashed box) is essential for the Ccdc108 interaction with IFT‐B proteins. Multiple protein sequences were analyzed with Clustal Omega program (‘*’ indicates fully conserved residue positions; and ‘:’ indicate strongly and weakly conserved residue positions, respectively). Interactions were determined by co‐immunoprecipitation of endogenous IFT74 and IFT81 from HEK 293T cells expressing GFP‐luciferase, GFP‐Ccdc108, and mutant proteins.
-
FLive imaging of MCCs shows the inability of Ccdc108 mutant proteins lacking the seven amino acid IFT interaction domain to localize to cilia. Embryos expressing GFP‐tagged Ccdc108 mutant proteins (green) and Cetn4‐RFP (purple) were imaged at stage 27.
Source data are available online for this figure.
Figure EV3. Overexpression of Ccdc108 forms cytoplasmic granules and ciliary puncta along the axoneme in Xenopus epidermal MCCs (related to Fig 4).
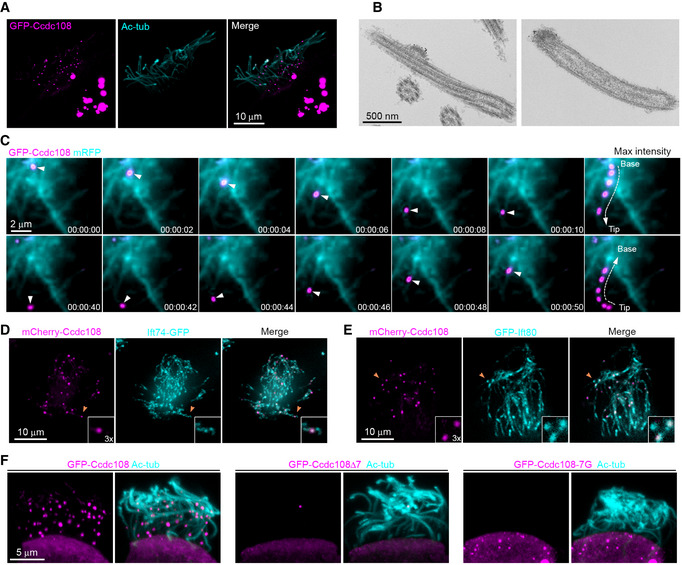
-
ARepresentative confocal images show the distribution of Ccdc108 in MCCs cilia and cytoplasm. Embryos were injected with 500 pg mRNA of GFP‐Ccdc108 (purple) at four‐cell stage, fixed at stage 27 and labeled with the Ac‐tub antibody (cyan).
-
BImmuno‐EM demonstrates that Ccdc108 localizes to the IFT train‐like structures along the axoneme. Embryos expressing GFP‐tagged Ccdc108 were immune labeled with 10 nm gold particles.
-
CLive cell imaging shows that Ccdc108 displays a bidirectional movement along the axoneme (Movie EV7). A live embryo expressing GFP‐Ccdc108 (purple) and membrane‐bound RFP (blue) was imaged at 2 s intervals using a spinning disk confocal microscope.
-
D, ERepresentative confocal images show Ccdc108 puncta co‐localize with IFT proteins at the ciliary axoneme in MCCs. Embryos expressing mCherry‐Ccdc108 (purple) and Ift74‐GFP (D), or GFP‐Ift80 (E) were fixed and labeled with related epitope tag antibodies.
-
FRepresentative confocal images show the inability of Ccdc108 mutant proteins lacking the seven amino acid IFT interaction domain to localize to cilia. Embryos expressing GFP‐Ccdc108 or mutant proteins (purple) were fixed and labeled with the acetylated tubulin antibody (Ac‐tub, cyan).
The ciliary localization of Ccdc108 relies on the Ccdc108–IFT interaction in MCCs
Given the similarity between IFTs and Ccdc108 in ciliary‐associated localization, we investigated whether overexpressed Xenopus Ccdc108 may interact with IFTs using the non‐motile ciliated mammalian HEK293T cell line. Following co‐immunoprecipitation and subsequent Shotgun mass spectrometry, we identified seven of the sixteen IFT‐B complex components (Fig 4B; Dataset EV1) (Nakayama & Katoh, 2018). To validate these potential IFT‐B complex interactions, we performed co‐immunoprecipitation in HEK293T cells expressing GFP‐Luciferase (Luci) and GFP‐Ccdc108 (108F) and confirmed that IFT‐B complex proteins IFT74, IFT81, and IFT56 interact with Ccdc108 (Fig 4B). We also examined interactions with other IFT‐B complex subunits not identified by mass spectrometry and validated interactions between GFP‐Ccdc108 and IFT80, and IFT57 (Fig 4C). We next examined mCherry–Ccdc108 association with the IFT‐B complex in the cilium of MCCs. Consistent with our biochemical observations, both Ccdc108 and Ift74 were located to the basal body and ciliary axoneme (Fig 4D). In embryos injected with higher levels of ccdc108 mRNA, where single prominent mCherry–Ccdc108 puncta were observed in cilia, the IFT‐B complex proteins consistently co‐localized with these structures (Fig EV3D and E).
To identify the regions of Ccdc108 that are required for IFT‐B complex interactions, Ccdc108 truncation and deletion mutants were generated (Fig 4E). Biochemical pull‐down studies identified a region between Ccdc108 amino acids 1,408–1,459 needed for IFT‐B interactions. Analysis of Ccdc108 ortholog protein sequences in this region identified a conserved seven amino acids motif (IFT‐binding motif—K/RYKTLPP) necessary for IFT binding (Fig 4E). To further confirm the requirement for IFT association, the seven amino acids of this motif were mutated to glycine. The Ccdc108–IFT interactions were dramatically similarly weakened upon deletion or glycine mutation of these seven amino acids (Fig 4E). Shotgun mass spectrometry analysis further confirmed that glycine mutant Ccdc108 protein failed to pull down any IFT components (Dataset EV1). Moreover, these residues were required for Ccdc108 axonemal localization but not for the basal body localization (Figs 4F and EV3F). Together, these results demonstrate that Ccdc108 ciliary localization requires the IFT‐B complex interaction domain.
The Ccdc108–IFT interaction and Ift‐74 are essential for apical trafficking of centrioles and actin assembly in MCCs
We next investigated Ccdc108–IFT interaction requirements in regulating multiciliogenesis using mutations that ablate the interaction of Ccdc108 with the IFT machinery. We tested whether these mutants rescue the loss‐of‐function phenotypes by co‐injecting either WT or IFT‐binding motif mutant ccdc108 mRNA with ccdc108 MO. Strikingly, despite similar expression levels (Fig EV4A), both IFT‐binding motif deletion and glycine substitution constructs failed to rescue the ciliogenesis defects induced by Ccdc108 depletion (Figs 5A and EV4B). Moreover, neither basal body migration/docking defects nor the apical enrichment of F‐actin defects was restored by the expression of mutant Ccdc108 proteins (Fig 5B and C). Thus, these findings demonstrate that the Ccdc108–IFT interaction domain is essential for multiciliogenesis. Furthermore, these findings suggest a previously undescribed role for the IFT‐B complex in regulating multiciliogenesis at the centriole apical migration/docking stage via association with Ccdc108.
Figure EV4. The IFT interaction domain is required for the function of Ccdc108 in multiciliogenesis in Xenopus embryos (related to Fig 5).
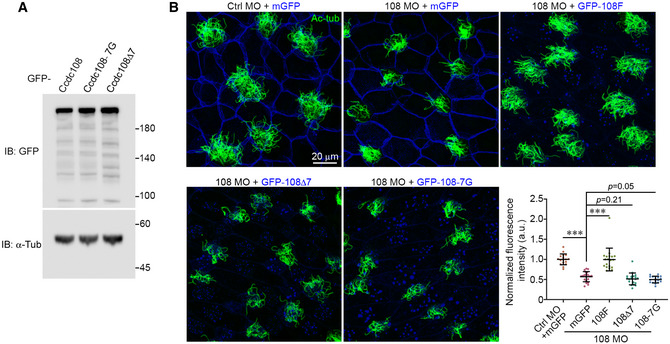
- Immunoblotting reveals that wild‐type and mutant Ccdc108 proteins display similar expression levels. Xenopus embryos expressing GFP‐tagged wild‐type or mutant Ccdc108 proteins were subjected to immunoblotting. α‐tubulin (α‐Tub) served as loading control.
- Representative confocal images show mutant Ccdc108 proteins fail to restore the impaired multiciliogenesis induced by Ccdc108 depletion. Embryos were treated as described in Fig 1C and expressed mGFP (blue) or GFP‐Ccdc108 mutants (blue). Embryos were fixed at stage 27 and labeled with the Ac‐tub antibody (green). Plots show results of greater than 17 images of 17 embryos from three independent experiments. Unpaired two‐tailed t‐test was performed (***P < 0.001). Mean ± s.d. values are presented.
Figure 5. The Ccdc108–IFT interaction is essential for proper multiciliogenesis in MCCs.
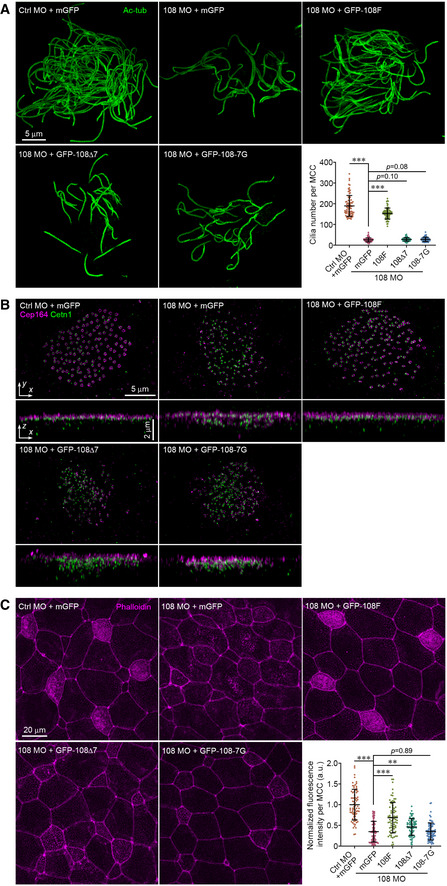
-
A–CThe IFT interaction domain is required for Ccdc108 multiciliogenesis function in Xenopus embryos: (A) impaired multiciliogenesis, (B) defective apical migration/docking of centrioles, and (C) reduced enrichment of apical F‐actin. Embryos at stage 27 were fixed and stained with the acetylated tubulin (A; Ac‐tub; green), Cep164 (B; purple) and Cetn1 (B; green) antibodies, or phalloidin (purple). mRNA of a membrane‐bound form of GFP was co‐injected with each morpholino to indicate targeted cells (A,B). Typical D‐SIM images (A,B) and confocal images (C) are present.
Data information: Embryos were treated as described in Fig 1C and expressed mGFP or GFP‐Ccdc108 mutants. Plots show results from three independent experiments. (A) Greater than 70 MCCs from six embryos, and (C) > 70 MCCs from six embryos for each condition. Unpaired two‐tailed t‐test was performed (***P < 0.001; **P < 0.01). Mean ± s.d. values are presented.
Source data are available online for this figure.
Having established that the IFT interaction domain of Ccdc108 is required for multiciliogenesis, we tested whether the IFT‐B complex protein Ift74, which displayed the highest number of unique peptides in the Ccdc108 mass spectrometry analysis (Fig 4B), is associated with Ccdc108 multiciliogenesis requirements. We were not able to examine the effects of Ift74 knockdown in Xenopus MCCs using morpholinos due to lethality of the two MOs we tested and unrescuable off‐target effects. Moreover, since disruption of IFT function affects primary cilia formation that is crucial for Xenopus embryogenesis (Park et al, 2016), we addressed this issue using CRISPR/Cas9‐mediated gene editing to deplete Ift74, which results in mosaicism (Mehravar et al, 2019) that we anticipated would avert major morphogenetic defects. We tested two different single‐guide RNAs (sgRNAs) that target different coding regions of ift74 and performed functional analysis on embryos in which genome editing by CRISPR/Cas9 was confirmed (Fig EV5). Both sgRNAs caused a significant reduction in multiciliation in regions of the embryos (Fig 6A and B). Further investigations of the centriole distribution revealed that Ift74 depletion affected centriole apical migration and/or docking, but not centriole amplification (Fig 6C). We next examined the effects of Ift74 depletion on expansion of the apical membrane of MCCs and found only a modest but statistically significant effect on the apical surface by one sgRNA against ift74 at stage 27 (Fig 6D). In contrast, the levels of apical F‐actin were reduced by both ift74 sgRNAs (Fig 6E). Overall, these results indicate that Ift74 plays a crucial role in regulating the apical migration and/or docking of centrioles and F‐actin enrichment in multiciliated Xenopus epidermis. Moreover, Ift74 requirements in multiciliogenesis are consistent with a function of Ccdc108 in this process and provide a molecular explanation for why Ccdc108 mutants that are unable to bind the IFT‐B complex fail to restore ciliation in ccdc108‐depleted MCCs.
Figure EV5. In vitro validation of CRISPR‐Cas9‐Mediated Genome Editing (related to Fig 6).
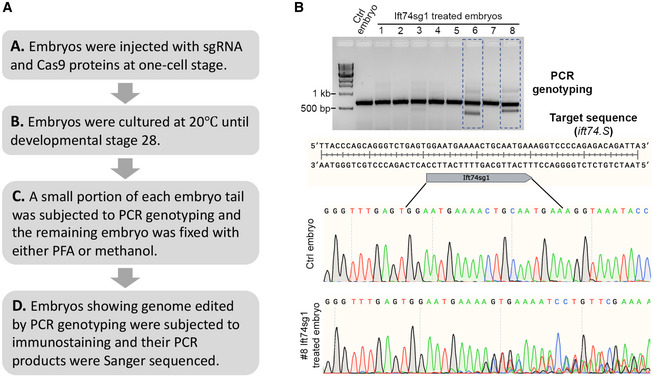
- Experimental design of the in vitro validation.
- In vitro validation results of Xenopus Ift74sg1 used in this study. PCR primers were designed to produce a single‐band PCR product that flanks the genomic edit of interest. The PCR products were resolved in 1.2% agarose gels stained with ethidium bromide. Gel image converted from electropherogram shows that two products (#6 and #8) display multiple bands compared to the control sample. Sequencing chromatograms show DNA editing of #6 PCR product. Underlined region indicates the sgRNA‐binding sequence.
Figure 6. Ift74 regulates apical centriole migration and F‐actin enrichment during multiciliation.
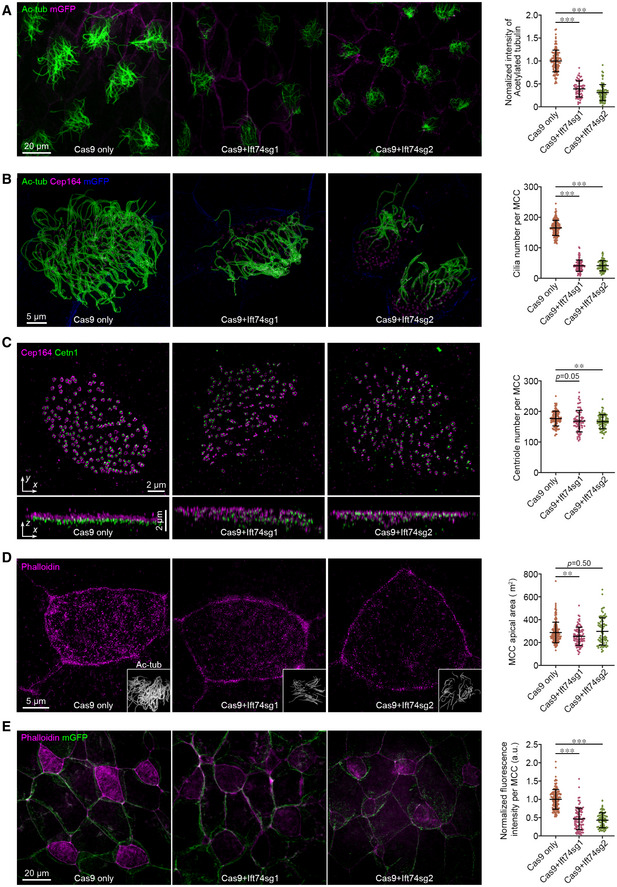
-
A, Bift74 CRISPR mutants display defective cilia formation. Embryos at one‐cell stage were injected with two independent sgRNA‐targeting ift74 and fixed at stage 27. Representative confocal images (A) and 3D‐SIM images (B) of the Xenopus epidermis, and plots display defective ciliation. (A) Greater than 60 MCCs from six embryos and (B) greater than 70 MCCs from six embryos for each condition. Cell membranes (mGFP), cilia (Ac‐tub, green), and basal bodies (Cep164) were labeled with indicated antibodies.
-
CIft74 depletion affects centriole apical migration in MCCs. Representative 3D‐SIM images (x–y) and 3D reconstructions (x–z) of MCCs show failure of apical trafficking of basal bodies. Basal bodies (Cetn1, green) and distal appendages (Cep164, purple) were labeled with indicated antibodies. Quantitative plot shows a slight significant reduction in basal bodies by the depletion of Ift74. Greater than 70 MCCs from six embryos for each condition.
-
DRepresentative 3D‐SIM images and plots reveal that apical expansion of MCCs is slightly affected by Ift74 depletion. Greater than 70 MCCs from six embryos for each condition. Embryos were fixed and stained with phalloidin (purple). Cilia (Ac‐tub, gray) were also presented.
-
ERepresentative confocal images and plot show a significant reduction in apical F‐actin enrichment upon Ift74 depletion. Greater than 70 MCCs from six embryos for each condition. Cell boundaries and apical actin network were marked with mGFP (green) and phalloidin (purple).
Data information: Quantitative data from three independent experiments were scored. Unpaired two‐tailed t‐test was performed (***P < 0.001; **P < 0.01). Mean ± s.d. values are also presented.
Source data are available online for this figure.
Ccdc108 governs the centriolar localization of IFT‐B complex proteins during centriole migration
Since both Ccdc108 and Ift74 function upstream of axoneme formation in centriole migration/docking to the apical surface, which starts from earlier developmental stages (Kulkarni et al, 2021), we set out to investigate the distributions of Ccdc108 and IFT‐B complex proteins in MCCs of embryos at stage 18. Interestingly, both Ccdc108 and IFT‐B complex proteins displayed centriolar localization when centriole migration to the apical surface is expected to be occurring at this stage (Fig 7A and B), supporting involvement of Ccdc108 and IFT‐B proteins in regulating early steps of multiciliation before axoneme elongation. Although Ccdc108 mutants failed to localize to the cilium at later stages (Figs 4F and EV3F), both mutant proteins were able to localize to the centrioles in MCCs of embryos at stage 18 (Fig 7A), similar to what was observed at stage 27 (Fig 4F), confirming the Ccdc108–IFT interactions are dispensable for the centriolar localization of Ccdc108.
Figure 7. Ccdc108 governs the centriolar localization of IFT‐B complex proteins during centriole migration.
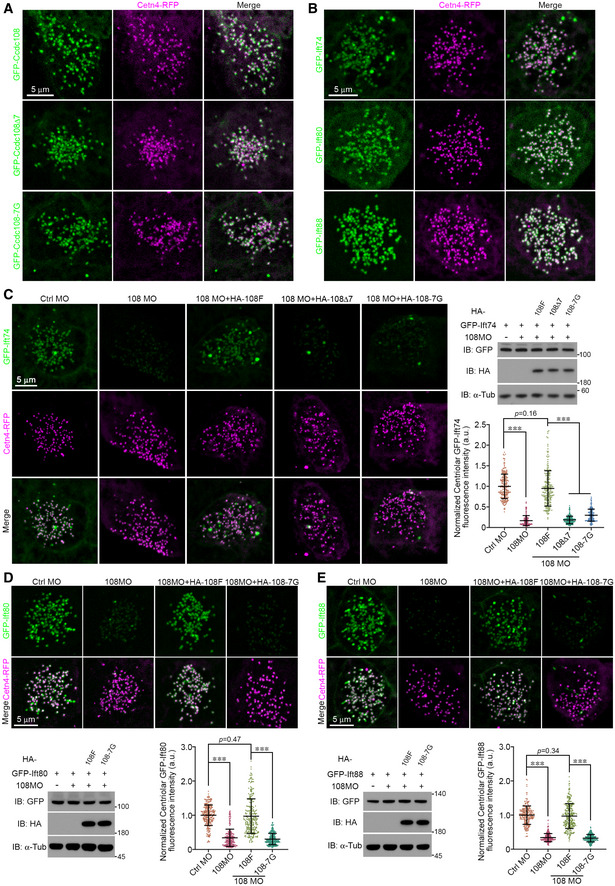
-
ALive imaging of MCCs shows the ability of wild‐type Ccdc108 and mutant proteins lacking the seven amino acid IFT interaction domain to localize to migrating centrioles. Embryos expressing GFP‐tagged Ccdc108 proteins (green) and Cetn4‐RFP (purple) were imaged at stage 18.
-
BLive imaging of MCCs shows the ability of IFT‐B complex proteins to localize to migrating centrioles. Embryos expressing GFP‐tagged IFT‐B proteins (green) and Cetn4‐RFP (purple) were imaged at stage 18.
-
C–ECcdc108 depletion affects centriolar distribution of IFT‐B proteins during centriole migration in MCCs. Embryos were co‐injected with GFP‐IFTs (C: Ift74; D: Ift80; E: Ift88; green), Cetn4‐RFP (purple), and each morpholino, and imaged at stage 18. HA‐tagged Ccdc108 proteins were co‐expressed to test for the rescue of phenotypic defects. Xenopus embryos were subjected to immunoblotting to evaluate the protein expression levels. α‐tubulin (α‐Tub) served as loading control. Quantitative plots show that Ccdc108‐depleted MCCs showed a significant decrease in centriolar distribution of IFT‐B proteins (Ift74, Ift80, and Ift88), and Ccdc108–IFT interaction deficient mutants are unable to rescue the decreased centriolar accumulation of IFT‐B proteins. Greater than 40 MCCs from six embryos for each condition.
Data information: Quantitative data from three independent experiments were scored. Unpaired two‐tailed t‐test was performed (***P < 0.001). Mean ± s.d. values are also presented.
Source data are available online for this figure.
Next, we investigated if Ccdc108 depletion affects IFT‐B centriole localization during centriole migration at stages 18–20. Strikingly, Ccdc108‐depleted MCCs showed a significant decrease in centriolar distribution of IFT‐B proteins (Ift74, Ift80, and Ift88) relative to control MCCs despite comparable protein expression levels between control and morphant embryos (Fig 7C–E). Strikingly, in contrast to WT Ccdc108 that can fully rescue the centriolar IFT‐B protein levels, neither of Ccdc108–IFT interaction‐deficient mutants can rescue centriole accumulation of IFT‐B proteins (Fig 7C–E). Together, these findings indicate that IFT‐B accumulation at the centrioles is dependent on Ccdc108, but the centriolar localization of Ccdc108 is not dependent on the IFT‐B proteins (as evidenced by the Δ7 and 7G mutants).
Ccdc108 regulates centriolar planar cell polarity effectors during centriole migration
In MCCs, centrioles are nucleated in the cytoplasm and transported to the apical surface through an actin cytoskeleton‐dependent mechanism, which involves components of the planar cell polarity (PCP) pathway including Dvl and Dvl‐associated cytoskeletal regulators Drg1 and RhoA (Park et al, 2006, 2008; Yasunaga et al, 2015; Lee et al, 2019), as well as ciliary adhesion complex‐related proteins such as Fak and Cp110 (Antoniades et al, 2014; Walentek et al, 2016). Since Ccdc108 is required for centriole migration/docking and establishing the apical actin network (Fig 3), we considered if Ccdc108 would be associated with the actin cytoskeletal regulators linked to the PCP pathway and/or the ciliary adhesion complex during multiciliogenesis. Thus, we examined the distributions of Drg1 and RBD (the RhoA‐binding domain of rhotekin; an indicator of active RhoA (Benink & Bement, 2005; Lee et al, 2019)), and ciliary adhesion complex‐related proteins, Fak and Cp110, in MCCs of embryos at stage 18 (Fig 8). Our results showed that all GFP‐tagged proteins localized to the centrioles at this stage (Fig 8), which is consistent with their functions in regulating actin network establishment needed for centriole migration/docking. Next, we determined whether Ccdc108 depletion affected the centriolar localization of these factors. Interestingly, we observed that levels of centriolar Drg1 and active RhoA were significantly reduced in Ccdc108‐depleted MCCs (Fig 8C and D), while neither Fak nor Cp110 displayed a decrease in centriolar distribution (Fig 8A and B). Together these results indicate a role for Ccdc108 in regulating the accumulation of cytoskeletal regulators linked to the PCP pathway but not ciliary adhesion complex proteins during centriole migration. Unlike WT Ccdc108, both Ccdc108–IFT‐binding mutant proteins failed to rescue the reduction in centriolar Drg1 and RBD in Ccdc108‐depleted cells (Fig 8C and D). Given the requirements for Ccdc108–IFT interaction for IFT‐B complex protein centriolar localizations at the same stage (Fig 7), we speculate that Ccdc108 recruits IFT‐B proteins to the centrioles. This in turn mediates the centriolar accumulations of PCP‐associated cytoskeleton regulators to induce subsequent actin polymerization for the establishment of the apical actin network and the proper apical migration and docking of centrioles in MCCs.
Figure 8. Ccdc108 and its interaction with the IFT‐B complex is required for actin cytoskeleton regulator recruitment to centrioles for their apical membrane migration in MCCs.
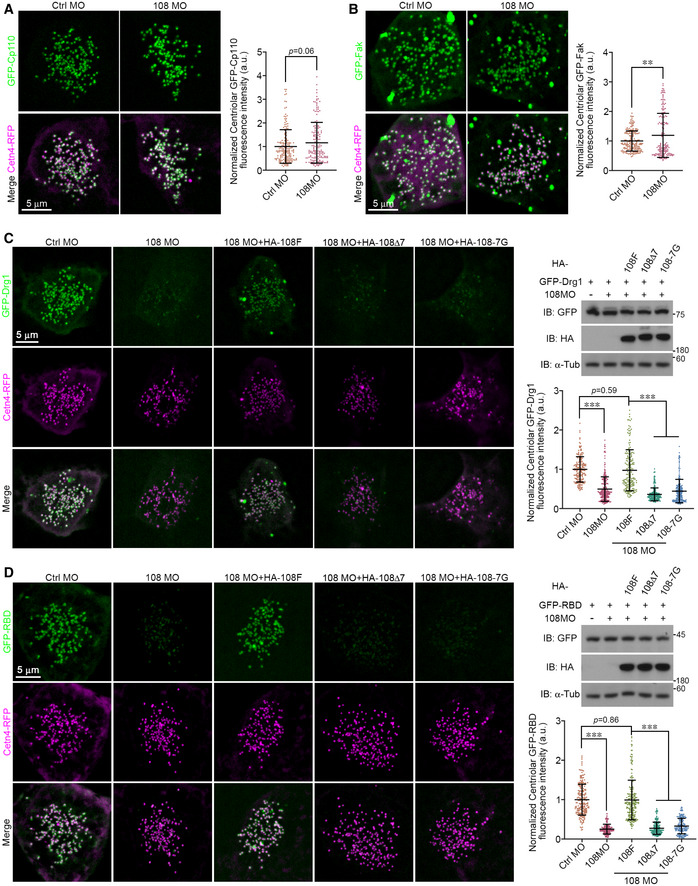
-
A, BCentriolar distribution of ciliary adhesion complex‐related proteins Fak and Cp110 is not reduced following Ccdc108 depletion. Embryos were co‐injected with GFP‐Cp110 (A), GFP‐Fak (B), Cetn4‐RFP (purple), and each morpholino, and imaged at stage 18. Greater than 30 MCCs from six embryos for each condition.
-
C, DCcdc108 depletion affects centriolar distribution of PCP‐associated actin cytoskeleton regulators important for centriole migration in MCCs. Embryos were co‐injected with GFP‐Drg1 (C) GFP‐RBD (D), Cetn4‐RFP (purple), and each morpholino, and imaged at stage 18. HA‐tagged Ccdc108 proteins were co‐expressed to test for the rescue of phenotypic defects. Xenopus embryos were subjected to immunoblotting to evaluate the protein expression levels. α‐tubulin (α‐Tub) served as loading control. Quantitative plots show that Ccdc108‐depleted MCCs showed a significant decrease in centriolar distribution of Drg1 and RBD, and neither of Ccdc108–IFT interaction‐deficient mutants can rescue the centriolar phenotypic defects. Greater than 30 MCCs from six embryos for each condition.
Data information: Quantitative data from three independent experiments were scored. Unpaired two‐tailed t‐test was performed (***P < 0.001; **P < 0.01). Mean ± s.d. values are also presented.
Source data are available online for this figure.
Discussion
In this study, we utilize motile cilia models to assess Ccdc108 ciliation function. Unlike later roles reported for CCCDC108 in the central apparatus for motility, we discover novel requirements specifically during earlier multiciliation, where it localizes to migrating centrioles, basal bodies, and cilia in ciliated Xenopus MCCs. Functional analysis reveals that Ccdc108 is required for multiciliation in Xenopus epidermis, zebrafish embryos, and mouse ependymal cells. Our investigations in Xenopus MCCs demonstrate a requirement of Ccdc108 for the docking of centrioles to the apical membrane during ciliogenesis, but is dispensable for centriole biogenesis and maturation. Moreover, Ccdc108 depletion prevents apical enrichment of F‐actin below the membrane surface, which could explain the observed impairment in multiciliogenesis. Remarkably, we demonstrate that Ccdc108 biochemically interacts with components of the IFT‐B complex via a conserved IFT‐B interaction domain which is required for Ccdc108 ciliary localization and ciliogenesis function, and depletion of the IFT‐B complex protein Ift74 causes similar multiciliogenesis defects. IFT‐B complex proteins also localize to the centrioles during the migration phase in MCCs, and we demonstrate that IFT‐B protein localization at this migration stage requires the Ccdc108‐IFTB interaction motif, but this region is dispensable for Ccdc108 centriole localization. Importantly, we find that Ccdc108 is required for the centriolar accumulations of actin cytoskeletal regulators, which require the Ccdc108–IFT‐B interaction motif as well. Thus, based on these results, we conclude that Ccdc108 regulates recruitment of centriolar IFT‐B and PCP‐associated cytoskeletal proteins necessary for migration and docking of centrioles to the cell surface during multiciliogenesis.
TEM studies of MCC morphants demonstrate requirements of Ccdc108 for centriole targeting to the apical surface during ciliogenesis. This function is further supported by immunofluorescence‐based studies of centriole position in Ccdc108 and Ift74‐depleted MCCs, which in the former case cannot be rescued by Ccdc108 mutant proteins that are incapable of interacting with IFT‐B proteins. Another striking feature of this study is that IFT‐B complex proteins are observed to localize to the centrioles during the apical migration of centrioles to the cell surface, and their centriolar localizations require the Ccdc108–IFT‐B interaction. To our knowledge this is the first report implicating the IFT‐B complex in the migration and/or docking of centrioles during MCC multiciliogenesis. IFT proteins have long been described as adapters between cargoes and motors needed for ciliary transport, and also been linked to organizing centriolar function, outside of ciliary roles, in mitotic spindle formation (Vitre et al, 2020). However, unlike these IFT‐associated cellular transport functions, centriole migration to the apical surface is thought to occur independently of microtubules (Boisvieux‐Ulrich et al, 1990; Brooks & Wallingford, 2014). Interestingly, actin has been shown to regulate IFT accumulation in Chlamydomonas reinhardtii flagella (Avasthi et al, 2014), and our results showing both IFT‐binding deficiency mutant proteins failed to restore the centriolar localization of PCP‐associated cytoskeletal regulators in Ccdc108‐depleted MCCs suggest a potential involvement of IFT‐B complex proteins in the regulation of actin dynamics during multiciliogenesis. However, due to mosaic effects of CRISPR/Cas9 system used in embryos (Mehravar et al, 2019), we could not evaluate if IFT‐B complex proteins are directly involved in regulating the centriolar recruitments of Drg1 and RBD as was done for Ccdc108. Taken together, our findings support a model whereby a Ccdc108–IFT‐B complex regulates actin‐dependent centriole migration and/or apical F‐actin accumulation needed for ciliogenesis in MCCs.
There is little known about how the apical F‐actin network is established in multiciliogenesis. During MCC differentiation, progenitor cells migrate apically and protrude out of the superficial epithelial, and the apical surface has to expand sequentially to accommodate hundreds of basal bodies/motile cilia (Stubbs et al, 2006; Sedzinski et al, 2016; Kulkarni et al, 2018). Actin‐generated pushing forces have been reported to be important for driving emergence of the apical membrane surface (Sedzinski et al, 2016). Another study demonstrated that basal bodies are connected to the actin cytoskeleton through ciliary adhesions (Antoniades et al, 2014). Like the chicken or the egg causality question, it is still not clear which comes first: the centriole apical membrane docking or the apical F‐actin enrichment. One possibility is there may be two sub‐types or layers of the F‐actin network associated with multiciliogenesis (Werner et al, 2011; Antoniades et al, 2014; Mahuzier et al, 2018). The first generates pushing forces needed for apical surface expansion, while the other is established later and contributes to further enrichment of F‐actin at the apical surface after basal body docking. Our observations that Ccdc108 and the IFT‐B complex are dispensable for apical expansion and are implicated in regulating the actin assembly through affecting cytoskeletal regulators linked to the PCP‐signaling pathway provide new clues into this poorly understood ciliogenesis stage in MCCs.
Interestingly, we show that Ccdc108, IFT‐B complex proteins, and actin cytoskeleton regulators all display centriolar localization during centriole migration to the apical surface, and these proteins are required for proper centriole migration during multiciliogenesis, suggesting a complex protein network associated with centrioles that regulates this process. Although Ift80 is not considered as a IFT‐B core complex protein (Taschner et al, 2012), this peripheral IFT‐B subunit was still located at the centriole and its centriole accumulation was reduced by Ccdc108 depletion as well (Fig 7B and D). These data suggest that both core and peripheral subunits of the IFT‐B complex might be implicated in centriole migration distinct from their critical roles in formation and maintenance of ciliary structures. Notably, we did not observe IFT‐A complex proteins in our Ccdc108 mass spectrometry results, however, this does not rule out an involvement of IFT‐A complex in this process. Consistent with this idea, peripheral IFT‐A complex proteins are recruited to basal bodies by the CPLANE complex which is required for MCC ciliogenesis (Toriyama et al, 2016). Moreover, in contrast to Ccdc108, the CPLANE complex associates with the IFT‐A complex but not the IFT‐B complex (Toriyama et al, 2016). Although it is still not clear how Ccdc108 itself is recruited to the centrioles, our findings demonstrate that Ccdc108 governs the centriolar accumulations of IFT‐B complex proteins and Drg1 and activated Rho, and the Ccdc108–IFT interaction is also crucial for these factors' centriolar localization.
Consistent with the finding that the Chlamydomonas reinhardtii ortholog of Ccdc108, Akap240, was reported to be required for ciliary motility through affecting the structure of the axoneme microtubule central pair (Rao et al, 2016; Teves et al, 2016; Zhao et al, 2019b), the remaining motile cilia in Ccdc108‐depleted MCCs displayed an altered CBP and an increased CBF as well (Fig EV1D), indicating a similar requirement of Ccdc108 in regulating the ciliary motility as other axoneme microtubule central pair‐related proteins such as Hydin, Spef1, and Spef2 (Lechtreck & Witman, 2007; Olbrich et al, 2012; Zheng et al, 2019; Cindric et al, 2020). Here, we show Ccdc108 localizes to centrioles ahead of the docking of centrioles to the apical surface in MCCs, but whether other central pair apparatus proteins such as Cfap70 are also present at this early stage remains unknown. Nevertheless, this novel function of Ccdc108 in regulating centriolar accumulation of IFT‐B and cytoskeletal regulator proteins appears to be specific to multiciliogenesis, since formation of either primary cilia or motile monocilia was not affected by Ccdc108 depletion in Xenopus embryos, although we note that kinocilium formation in zebrafish plane‐polarized hair cells was affected by Ccdc108 depletion.
Our findings provide evidence that mutations in CCDC108 may be associated with human ciliopathy. Studies examining patients with primary infertility reveal that mutations in CCDC108 cause abnormal sperm flagella and lead to male infertility (Wang et al, 2019; Zhang et al, 2019; Li et al, 2020). Recently, Ccdc108 knockout mice were shown to affect sperm acrosome biogenesis and mitochondrial sheath assembly which prevented flagellar elongation (Wang et al, 2021). Our results are consistent with their findings that Ccdc108 localizes to the axoneme and has additional specific functions upstream of axonemal formation. Although other cilia‐type functions were not investigated in this report, Ccdc108 knockout mice were noted to have developmental delays and reduced body size and weight, which are phenotypes also observed in knockout mouse models of genes involved in human primary ciliary dyskinesia (PCD) (Lechtreck et al, 2008; Chiani et al, 2019). Considering our results in this context, Ccdc108 is likely to have downstream roles in regulating cilia motility in addition to these novel and earlier functions in multiciliogenesis required for axoneme assembly.
Materials and Methods
Plasmid constructs
Full‐length Xenopus ccdc108 (NM_001123490; MXT1765‐202715229) and fak (NM_001091540; MXL1736‐202787211) were obtained from Horizon Discovery. Xenopus ift74 (NM_001095800) was amplified from a Xenopus cDNA library and human IFT88 (NM_175605) from a HEK293T cDNA library by polymerase chain reaction (PCR). Xenopus cp110, drg1, and RBD constructs were previously described (Lee et al, 2019). Either full‐length or relative fragments were PCR‐amplified and subcloned into the recombinational donor vector pDONR221 to generate entry clones (ThermoFisher, 11789100). LR recombination reactions between entry clones and desired pCS2 gateway destination vectors (Addgene, Kit #1000000107) were performed to generate the expression constructs (ThermoFisher, 11791100).
To generate the bacterial expression constructs, mouse Ccdc108 fragment (CCDC108M: 601–1,200 aa; NM_001039495) was PCR amplified from a mouse testis cDNA library and subcloned into pDONR221. pDEST‐15 (ThermoFisher, 11802014) and pDEST‐HisMBP (Addgene, Plasmid #11085) destination vectors were used to create the His‐MBP‐ and GST‐ Ccdc108M constructs.
To generate the lentiviral shRNA constructs, oligonucleotides were synthesized, annealed, and inserted into a modified pLKO.1 vector in which the puromycin resistance gene was replaced with GFP‐Cetn1 (Zhao et al, 2013). The four shRNA sequences used for the functional studies have the following sequences: 108‐i1 5′‐CGGCAACTGCACCCTCTATTA‐3′; 108‐i2 5′‐CAGGATCTGAAGACCATAATA‐3′; 108‐i3 5′‐CAGCATAGACTTGATATGCAA‐3′; 108‐i5 5′‐GGAGACTTGGTTCTGTCTGAA‐3′.
All constructs were verified via Sanger sequencing analysis.
Xenopus and zebrafish embryo microinjections
Xenopus and zebrafish experimental procedures were performed in accordance with the protocols (ASP #20‐433 and ASP #20‐416) approved by the Animal Care and Use Committee of the National Cancer Institute at Frederick in compliance with the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines.
For Xenopus experiments, embryos were obtained by standard methods (Sun et al, 2020). Capped mRNAs for microinjection were in vitro transcribed with the mMessage mMachine SP6 Transcription Kit (ThermoFisher, AM1340). sgRNA sequences were designed by the ZiFiT Targeter program website (http://zifit.partners.org/ZiFiT/ChoiceMenu.aspx), and evaluated for the specificity by the website (https://www.crisprscan.org) (Nakayama et al, 2013). The 5′ oligonucleotide sequence (5′‐GCAGCTAATACGACTCACTATA~20 nt~GTTTTAGAGCTAGAAATA‐3′) containing each specific sgRNA sequence (20 nucleotides) and the 3′ common oligonucleotide sequence (5′‐AAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC‐3′) were synthesized, annealed, and PCR amplified. sgRNAs were in vitro transcribed using the MEGAscript T7 Transcription Kit (ThermoFisher, AM1334). Embryos were injected with morpholinos into one ventral blastomere at four‐cell stage, and sgRNAs with the Cas9 protein (PNA Bio, CP01) at one‐cell stage. Small portions of embryos injected with sgRNAs were PCR genotyped and embryos showing genome edited by CRISPR/Cas9 were further subjected to functional analysis. The morpholinos were obtained from Gene Tools with the following sequences: Ctrl MO 5′‐CTAAACTTGTGGTTCTGGCGGATA‐3′ and 108 MO 5′‐CTAGCATCATACTGTCTTTTCTAAG‐3′. The sgRNA target sequences used are as follows: 108sg 5′‐GGCATTACATTGCGGGCCAG‐3′; Ift74sg1 5′‐GGAATGAAAACTGCAATGAA‐3′; and Ift74sg2 5′‐GTGGTCGCATTCCACTTGG‐3′.
For zebrafish experiments, one‐cell stage TAB‐5 embryos were micro‐injected with morpholinos obtained from Gene Tools (Ctrl MO 5′‐CCTCTTACCTCAGTTACAATTTAT‐3′ and z108 MO 5′‐ACCCTCTCAGTTCAGTTGTGTATTC‐3′).
Neither randomization nor blinding was applied for animal studies.
Whole‐mount in situ hybridization chain reaction (HCR)
HCR probe sets targeting zebrafish ccdc108 (XM_002663322) and Xenopus ccdc108 (XM_018235717) were designed and synthesized by Molecular Instruments. Whole‐mount in situ HCRs were performed following the HCR v3.0 protocol for whole‐mount zebrafish embryos and larvae provided by Molecular Instruments. Once RNA signals are amplified by HCR, embryo samples are subjected to further immunostaining treatment.
Antibodies
His‐MBP‐ and GST‐Ccdc108M proteins were expressed in BL21 CodonPlus (DE3) RIPL bacteria strain (Agilent, #230280), and purified with amylose resin (NEB, E8021S) or glutathione agarose beads (Sigma, G4510). To generate the chicken polyclonal Ccdc108 antibody, His‐MBP‐Ccdc108 proteins were used to immobilize two chickens and immobilization was carried out by Pocono Rabbit Farm & Laboratory. Subsequent affinity purification was performed with GST‐Ccdc108M protein.
The following primary antibodies were used for immunostaining: rabbit anti‐CEP164 (Proteintech; 22227‐1‐AP; 1:300), mouse anti‐CETN1 (Sigma; 04‐1624; 1:300), mouse anti‐acetylated Tubulin (Sigma; T6793; 1:500), rat anti‐GFP (Biolegend; 338002; 1:300), rat anti‐HA (Sigma; 11867423001; 1:300), rat anti‐mCherry (ThermoFisher; M11217; 1:300), and chicken anti‐GFP (Abcam; ab13970; 1:1,000).
The following primary antibodies were used for western blot experiments: Rabbit antibodies against IFT81 (Proteintech; 11744‐1‐AP; 1:500), IFT80 (Proteintech; 25230‐1‐AP; 1:1,000), IFT74 (Proteintech; 27334‐1‐AP; 1:1,000), IFT57 (Proteintech; 11083‐1‐AP; 1:1,000), IFT56 (Novus; NBP1‐84035; 1:1,000), GAPDH (Proteintech; 10494‐1‐AP; 1:5,000), Rsph1 (generous gift from Xueliang Zhu; 1:4,000), mouse anti‐α‐Tubulin (Sigma; T9026; 1:5,000), chicken anti‐Ccdc108 (homemade; 1:1,000), and horseradish peroxidase (HRP)‐conjugated mouse anti‐GFP (Miltenyi Biotec; 130‐091‐833; 1:3,000) and rat anti‐HA (Sigma; 12013819001; 1:3,000).
The following secondary antibodies were used: DyLight 405 donkey anti‐rabbit (Jackson ImmunoResearch; 711‐475‐152; 1:200), DyLight 405 donkey anti‐mouse (Jackson ImmunoResearch; 715‐475‐151; 1:200), Alexa Fluor 488 goat anti‐rat (ThermoFisher; A‐11006; 1:1,000), Alexa Fluor 488 goat anti‐chicken IgY (ThermoFisher; A‐11039; 1:1,000), Alexa Fluor 568 goat anti‐rat (ThermoFisher; A‐11077; 1:1,000), Alexa Fluor 568 donkey anti‐mouse (ThermoFisher; A‐10037; 1:1,000), Alexa Fluor 488 donkey anti‐mouse (ThermoFisher; A‐32766; 1:500), Alexa Fluor 647 goat anti‐rabbit (ThermoFisher; A‐21245; 1:1,000), and HRP‐conjugated donkey anti‐rabbit (GE; NA934V; 1:5,000).
Immunofluorescent microscopy
Xenopus embryos or mEPCs grown on glass‐bottomed dishes (Cellvis, D29‐14‐1.5‐N) were fixed with 4% fresh paraformaldehyde in PBS for 15 min at room temperature (RT), followed by extraction and penetration with 0.5% Triton X‐100 in PBS for 15 min and blocked with blocking buffer (4% BSA in TBST) for 1 h at RT. For staining of endogenous Cetn1 and Cep164, Xenopus embryos were fixed with Dent’s fixative (80% methanol and 20% dimethyl sulphoxide) overnight at 4°C and were incubated in blocking buffer (4% BSA in TBST) for 1 h at RT. For imaging motile monocilia, the gastrocoel roof plate (GRP) was dissected and fixed with MEMFA (0.1 M MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde) at stages 18–19. For staining primary cilia in the neural tube, embryos were fixed at stage 30 with MEMFA, followed by methanol dehydration. Embryos were then embedded in 4% low melting agarose gel and were sectioned at a thickness of 50 μm with the vibratome (LEICA VT 1200S). Primary and secondary antibodies were prepared in blocking buffer and applied to samples at 4°C overnight, respectively. Anti‐GFP antibody and Alexa Fluor‐488‐conjugated secondary antibody were used to enhance GFP fluorescent signals. Alexa Fluor 647‐conjugated phalloidin (ThermoFisher; A22287; 1:400) was co‐incubated with secondary antibodies. Super‐resolution images were obtained with a 3D‐structured illumination microscope at 125 nm intervals (GE DeltaVision OMX SR imaging system), and further processed for maximum intensity projection with the SoftWoRx software and 3D reconstruction with the Imaris (Bitplane). The confocal images were acquired using a Zeiss LSM710 laser scanning confocal microscope with a 63×/1.4 NA oil immersion objective. Optical sections were captured at 0.5 μm intervals and z‐stack images were obtained with maximum intensity projections (Carl Zeiss Microimaging, Germany).
To normalize the intensity of acetylated tubulin and phalloidin, the intensity values of control embryos from the same repeat were averaged, and the intensity values of this repeat were normalized to the average intensity of control morphants (Arnold et al, 2019). The numbers of cilia and centrioles, and the apical area of individual MCC were measured from 3D‐SIM images. To count the number of cilia, individual cilium was identified by observing the axoneme staining (Ac‐tub) and the distal appendage staining (Cep164) at the cilia base. The measurements of fluorescence intensity and the apical area of individual MCC were carried out using the image analysis module of Adobe Photoshop software. Due to the mosaic effects when CRISPR/Cas9 system was used in embryo studies (Mehravar et al, 2019), fields displaying phenotypic defects, gauged by reduced cilia numbers or affected centriole positioning, in the crispant samples were imaged, while similar defects were rarely observed in control samples.
Whole‐mount zebrafish immunostaining was performed as described previously (Cuenca et al, 2019; Insinna et al, 2019). Briefly, embryos at 48 h post‐fertilization (hpf) were fixed with 4% paraformaldehyde in PBS following a brief pre‐permeation incubation into a solution of 0.5% Triton X‐100 and incubation with blocking solution (PBS with 0.1% Triton X‐100 and 5% goat serum) for 30 min at RT. Primary and secondary antibodies were applied to samples for 2 h at RT. Fluorophore‐conjugated phalloidin (ThermoFisher; A22287; 1:400) and Hoechst (Immunochemistry; #639; 1:200) were co‐incubated with secondary antibodies. Imaging was carried out on a spinning disk confocal microscope with a Zeiss EC Plan‐Neofluar 40×/1.30 (Intelligent Imaging Innovations). Bright‐field images of whole embryos were performed using a 4× water immersion objective (0.13 NA) and a Nikon Eclipse Ni‐E upright microscope equipped with a DS‐Ri2 camera.
Live cell fluorescence imaging
Live imaging of Xenopus epidermal MCCs was carried out on a spinning disk confocal microscope with a Zeiss Plan‐Apochromat 63×/1.4 or 100×/1.46 oil immersion objective (Intelligent Imaging Innovations) at RT. Xenopus embryos were mounted on a glass‐bottom dish in agarose. For imaging the IFT transport, single‐layer image acquisition was recorded every 2 s using a HAMAMATSU digital CMOS camera (HAMAMATSU Photonics, C11440). Processing of images was carried out using either ImageJ or SlideBook software.
Electron microscopy
For transmission EM, skins of Xenopus embryos injected with morpholinos were dissected and fixed in 100 mM cacodylate buffer containing 2.5% glutaraldehyde and 4% fresh paraformaldehyde for 15 min at RT and overnight at 4°C. Samples were then post‐fixed in 1% aqueous osmium tetroxide for 1 h on ice, and incubated in 1% uranyl acetate overnight at 4°C. Dehydration was performed through an ethanol series. Samples were gradually infiltrated through an ascending series of low viscosity ethanol–resin mixture (25, 50, 75, and 100% Epon 812 resin) and polymerized at 60°C for 48 h. For immuno‐EM, Xenopus embryos expressing GFP‐tagged Ccdc108 were fixed in PBS containing 4% paraformaldehyde and 0.25% glutaraldehyde for 15 min, and extracted with 0.5% Triton X‐100 in PBS for 15 min at RT. Embryos were then incubated with the rat anti‐GFP antibody (Biolegend; 338002; 1:300) for 16 h at 4°C and 4 h at RT. The incubation with a goat anti‐rat IgG–gold antibody (Sigma; G7035; 1:50) was carried out for 1 h at RT. The samples were then subjected to TEM sample preparation. Ultrathin sections of approximately 70 nm thickness were sectioned with a Leica Ultracut UCT ultramicrotome and collected on copper mesh grids for transmission EM. EM images were captured with a Hitachi 7600 transmission electron microscope.
For SEM, Xenopus and zebrafish embryos were fixed in 100 mM cacodylate buffer containing 2.5% glutaraldehyde and 4% fresh paraformaldehyde overnight at 4°C, post‐fixed with 1% aqueous osmium tetroxide in H2O on ice for 1 h, and dehydrated in an ethanol series. Dehydrated samples were dried using tetramethylsilane before sputter coating. SEM images were collected on a scanning electron microscope (Hitachi S4500).
Fluorescent bead assay and cilia beat analysis
The fluorescent bead assay was performed as described previously (Lee et al, 2019). In brief, fluorescent beads (ThermoFisher, F8836) were washed twice with 0.1× Modified Barth’s Saline (MBS), resuspended with 0.1× MBS containing 1.5% glycerol and loaded onto anesthetized embryos at stages 26–28. The bead movement was recorded using a dissecting scope equipped with a digital camera (Canon PowerShot G10) at 30× magnification. The distance travelled by each individual fluorescent bead moved in 500 ms was measured to calculate the velocity. The average velocity of five continuous tracks over 500 ms was determined for each individual bead. Ten beads per embryo and 5 embryos per treatment were scored in each independent repeat.
For cilia beat pattern and beat frequency analysis, Xenopus embryos at stage 27 were imaged at room temperature in bright field with a Plan Apo λ 20× objective on a Nikon ECLIPSE Ti2 microscope using a Hammamtsu ORCA‐Fusion CMOS camera. Images were captured at 2633 frames per sec for 1.5 s. The first 500 images (0.1875 s) were analyzed using Fiji Kymograph‐Reslice‐Wide plugin as described (Rachev et al, 2020). Cilia beat frequencies (Hz) were determined by measuring time between cilia beat repeats (2 per MCC) and plotted using the Prism 8 software (GraphPad Software).
Cell culture and transfection
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (R&D Systems, S11150) and 1% penicillin/streptomycin (ThermoFisher, 15140122). For lentiviral productions, HEK 293T cells were transfected with lentiviral plasmid, pCMV‐D8.9, and pCMV‐VSVG at the ratio of 5:3:2 using the PolyJet In Vitro DNA Transfection Reagent (SignaGen Laboratories, SL100688). mEPC precursors isolated from P0 neonates were cultured and infected with lentivirus particles as described previously (Zhao et al, 2019a). Briefly, telencephala were digested with the dissection solution containing 10 U/ml papain (Worthington, LS003126), 0.2 mg/ml L‐Cysteine, 0.5 mM EDTA, 1 mM CaCl2, 1.5 mM NaOH, and 0.15% DNase I (Sigma, D5025) for 30 min at 37°C. Released cells were collected by centrifugation, resuspended with DMEM medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin, and inoculated into the laminin‐coated flask (Sigma, L2020). After removing differentiated oligodendrocytes and neurons mechanically by striking the flask roughly, cells were trypsinized and inoculated into laminin‐coated glass‐bottom dishes. FBS was removed from the medium to initiate differentiation when cells had grown to full confluence.
Experiments involving mouse tissues were performed in accordance with the protocol (ASP #17‐041) approved by the animal care and use committee of the National Cancer Institute at Frederick in compliance with the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC) guidelines.
Immunoprecipitation and mass spectrometry
HEK 293T cells expressing indicated proteins were harvested and lysed with lysis buffer (20 mM Tris‐HCl, pH 7.5, 150 mM KCl, 1 mM EDTA, 0.5% NP‐40, 10% glycerol, 10 mM sodium pyrophosphate, 3 mM dithiothreitol, and 0.5 mM phenylmethyl sulphonyl fluoride [PMSF, ThermoFisher, 36978] and protease inhibitor cocktail [Sigma, 539134]). The cell lysates were cleared by centrifugation at 14,000 g for 10 min at 4°C. GFP‐Trap affinity resin (Chromotek, gta‐20) was used to immunoprecipitate GFP fusion proteins. After washing four times with the lysis buffer, beads were either incubated with 65 μl 2× SDS loading buffer or subjected to mass spectrometry analysis (Protein Characterization Laboratory [PCL] Mass Spectrometry Center, NCI; Walia et al, 2019).
Statistical analysis
Quantification results are presented as mean ± s.d. unless specified in the figure legend. The unpaired two‐tailed t‐test was performed to determine statistical significance using the Prism 8 software (GraphPad Software). Differences were considered significant when P was < 0.05.
Author contributions
Huijie Zhao: Conceptualization; Data curation; Investigation; Writing—original draft. Jian Sun: Data curation; Investigation; Methodology; Writing—review & editing. Christine Insinna: Investigation; Writing—review & editing. Quanlong Lu: Methodology. Ziqiu Wang: Investigation. Kunio Nagashima: Investigation. Jimmy Stauffer: Investigation. Thorkell Andresson: Formal analysis. Suzanne Specht: Resources. Sumeth Perera: Writing—review & editing. Ira O Daar: Conceptualization; Supervision; Project administration; Writing—review & editing. Christopher J Westlake: Conceptualization; Supervision; Investigation; Project administration; Writing—wrote paper, review and editing.
In addition to the CRediT author contributions listed above, the contributions in detail are:
HZ and JS performed major experiments; CI and JS carried out zebrafish‐related analysis; QL contributed to collect live cell imaging data; ZW and KN helped with TEM and SEM data collection; TA performed Shotgun mass spectrometric analysis; SS maintained cell lines used in the laboratory; SP contributed to the draft editing; CW, ID, HZ, and JS designed experiments, interpreted data, and wrote the paper.
Supporting information
Expanded View Figures PDF
Dataset EV1
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Source Data for Expanded View
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
We thank John Wallingford (University of Texas at Austin) for providing the Cetn4‐RFP and GFP‐Ift80 expression plasmids; Jaeho Yoon, Moonsup Lee, Yoo‐Seok Hwang, and Hyun‐Kyung Lee (members of Dr. Ira O. Daar’s laboratory) for assistance in Xenopus embryo preparation; Xufeng Wu (Light Microscopy Core of National Heart, Lung, and Blood Institute) for providing super‐resolution microscope imaging assistance; Stanley Midy (Nikon Instruments Inc.) for assistance with high‐speed live cell imaging, Gerard Duncan (Protein Characterization Laboratory of National Cancer Institute); Justin Smith, Christine Perella, and Jacui Clem (Frederick National Laboratory of National Cancer Institute) for instrumental and technical supports. This research was supported by the National Cancer Institute, National Institutes of Health, Intramural Research Program, and was funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health under contract HHSN26120080001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
EMBO reports (2022) 23: e52775.
Contributor Information
Ira O Daar, Email: daari@mail.nih.gov.
Christopher J Westlake, Email: westlakecj@mail.nih.gov.
Data availability
No data were deposited in a public database.
References
- Antoniades I, Stylianou P, Skourides PA (2014) Making the connection: ciliary adhesion complexes anchor basal bodies to the actin cytoskeleton. Dev Cell 28: 70–80 [DOI] [PubMed] [Google Scholar]
- Arbi M, Pefani DE, Kyrousi C, Lalioti ME, Kalogeropoulou A, Papanastasiou AD, Taraviras S, Lygerou Z (2016) GemC1 controls multiciliogenesis in the airway epithelium. EMBO Rep 17: 400–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold TR, Shawky JH, Stephenson RE, Dinshaw KM, Higashi T, Huq F, Davidson LA, Miller AL (2019) Anillin regulates epithelial cell mechanics by structuring the medial‐apical actomyosin network. eLife 8: e39065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avasthi P, Onishi M, Karpiak J, Yamamoto R, Mackinder L, Jonikas MC, Sale WS, Shoichet B, Pringle JR, Marshall WF (2014) Actin is required for IFT regulation in Chlamydomonas reinhardtii. Curr Biol 24: 2025–2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balestra FR, Gonczy P (2014) Multiciliogenesis: multicilin directs transcriptional activation of centriole formation. Curr Biol 24: R746–749 [DOI] [PubMed] [Google Scholar]
- Benink HA, Bement WM (2005) Concentric zones of active RhoA and Cdc42 around single cell wounds. J Cell Biol 168: 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S, Cajanek L, Fort C, Blisnick T, Weber K, Taschner M, Mizuno N, Lamla S, Bastin P, Nigg EA et al (2013) Molecular basis of tubulin transport within the cilium by IFT74 and IFT81. Science 341: 1009–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvieux‐Ulrich E, Laine MC, Sandoz D (1990) Cytochalasin D inhibits basal body migration and ciliary elongation in quail oviduct epithelium. Cell Tissue Res 259: 443–454 [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB (2014) Multiciliated cells. Curr Biol 24: R973–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiani F, Orsini T, Gambadoro A, Pasquini M, Putti S, Cirilli M, Ermakova O, Tocchini‐Valentini GP (2019) Functional loss of Ccdc1 51 leads to hydrocephalus in a mouse model of primary ciliary dyskinesia. Dis Model Mech 12: dmm038489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cindric S, Dougherty GW, Olbrich H, Hjeij R, Loges NT, Amirav I, Philipsen MC, Marthin JK, Nielsen KG, Sutharsan S et al (2020) SPEF2‐ and HYDIN‐Mutant Cilia Lack the Central Pair‐associated Protein SPEF2, Aiding Primary Ciliary Dyskinesia Diagnostics. Am J Respir Cell Mol Biol 62: 382–396 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca A, Insinna C, Zhao H, John P, Weiss MA, Lu Q, Walia V, Specht S, Manivannan S, Stauffer J et al (2019) The C7orf43/TRAPPC14 component links the TRAPPII complex to Rabin8 for preciliary vesicle tethering at the mother centriole during ciliogenesis. J Biol Chem 294: 15418–15434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai D, Ichikawa M, Peri K, Rebinsky R, Huy Bui K (2020) Identification and mapping of central pair proteins by proteomic analysis. Biophys Physicobiol 17: 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport JR, Watts AJ, Roper VC, Croyle MJ, van Groen T, Wyss JM, Nagy TR, Kesterson RA, Yoder BK (2007) Disruption of intraflagellar transport in adult mice leads to obesity and slow‐onset cystic kidney disease. Curr Biol 17: 1586–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feistel K, Blum M (2006) Three types of cilia including a novel 9+4 axoneme on the notochordal plate of the rabbit embryo. Dev Dyn 235: 3348–3358 [DOI] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H (2007) When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol 8: 880–893 [DOI] [PubMed] [Google Scholar]
- Gaillard AR, Diener DR, Rosenbaum JL, Sale WS (2001) Flagellar radial spoke protein 3 is an A‐kinase anchoring protein (AKAP). J Cell Biol 153: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Liem KF Jr, Anderson KV (2012) The spinocerebellar ataxia‐associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell 151: 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imsland F, Feng C, Boije H, Bed'hom B, Fillon V, Dorshorst B, Rubin C‐J, Liu R, Gao YU, Gu X et al (2012) The Rose‐comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet 8: e1002775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insinna C, Lu Q, Teixeira I, Harned A, Semler EM, Stauffer J, Magidson V, Tiwari A, Kenworthy AK, Narayan K et al (2019) Investigation of F‐BAR domain PACSIN proteins uncovers membrane tubulation function in cilia assembly and transport. Nat Commun 10: 428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF (2011) Ciliogenesis: building the cell's antenna. Nat Rev Mol Cell Biol 12: 222–234 [DOI] [PubMed] [Google Scholar]
- Kanie T, Abbott KL, Mooney NA, Plowey ED, Demeter J, Jackson PK (2017) The CEP19‐RABL2 GTPase complex binds IFT‐B to initiate intraflagellar transport at the ciliary base. Dev Cell 42: 22–36.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Ma L, Shokhirev MN, Quigley I, Kintner C (2018) Multicilin and activated E2f4 induce multiciliated cell differentiation in primary fibroblasts. Sci Rep 8: 12369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klos Dehring DA, Vladar EK, Werner ME, Mitchell JW, Hwang P, Mitchell BJ (2013) Deuterosome‐mediated centriole biogenesis. Dev Cell 27: 103–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T, Brown JM, Bellve K, Craige B, Craft JM, Fogarty K, Lechtreck KF, Witman GB (2016) Together, the IFT81 and IFT74 N‐termini form the main module for intraflagellar transport of tubulin. J Cell Sci 129: 2106–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni SS, Griffin JN, Date PP, Liem KF Jr, Khokha MK (2018) WDR5 stabilizes actin architecture to promote multiciliated cell formation. Dev Cell 46: 595–610.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Marquez J, Date P, Ventrella R, Mitchell BJ, Khokha MK (2021) Mechanical stretch scales centriole number to apical area via Piezo1 in multiciliated cells. eLife 10: e66076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrousi C, Arbi M, Pilz GA, Pefani DE, Lalioti ME, Ninkovic J, Gotz M, Lygerou Z, Taraviras S (2015) Mcidas and GemC1 are key regulators for the generation of multiciliated ependymal cells in the adult neurogenic niche. Development 142: 3661–3674 [DOI] [PubMed] [Google Scholar]
- Lechtreck KF (2015) IFT‐cargo interactions and protein transport in cilia. Trends Biochem Sci 40: 765–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Delmotte P, Robinson ML, Sanderson MJ, Witman GB (2008) Mutations in Hydin impair ciliary motility in mice. J Cell Biol 180: 633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck KF, Witman GB (2007) Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J Cell Biol 176: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Hwang YS, Yoon J, Sun J, Harned A, Nagashima K, Daar IO (2019) Developmentally regulated GTP‐binding protein 1 modulates ciliogenesis via an interaction with Dishevelled. J Cell Biol 218: 2659–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wu H, Li F, Tian S, Kherraf ZE, Zhang J, Ni X, Lv M, Liu C, Tan Q et al (2020) Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J Med Genet 57: 89–95 [DOI] [PubMed] [Google Scholar]
- Liang Y, Pang Y, Wu Q, Hu Z, Han X, Xu Y, Deng H, Pan J (2014) FLA8/KIF3B phosphorylation regulates kinesin‐II interaction with IFT‐B to control IFT entry and turnaround. Dev Cell 30: 585–597 [DOI] [PubMed] [Google Scholar]
- Loncarek J, Bettencourt‐Dias M (2018) Building the right centriole for each cell type. J Cell Biol 217: 823–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Insinna C, Ott C, Stauffer J, Pintado PA, Rahajeng J, Baxa U, Walia V, Cuenca A, Hwang YS et al (2015) Early steps in primary cilium assembly require EHD1/EHD3‐dependent ciliary vesicle formation. Nat Cell Biol 17: 531 [DOI] [PubMed] [Google Scholar]
- Ma L, Quigley I, Omran H, Kintner C (2014) Multicilin drives centriole biogenesis via E2f proteins. Genes Dev 28: 1461–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahuzier A, Shihavuddin A, Fournier C, Lansade P, Faucourt M, Menezes N, Meunier A, Garfa‐Traoré M, Carlier M‐F, Voituriez R et al (2018) Ependymal cilia beating induces an actin network to protect centrioles against shear stress. Nat Commun 9: 2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehravar M, Shirazi A, Nazari M, Banan M (2019) Mosaicism in CRISPR/Cas9‐mediated genome editing. Dev Biol 445: 156–162 [DOI] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, Kintner C (2007) A positive feedback mechanism governs the polarity and motion of motile cilia. Nature 447: 97–101 [DOI] [PubMed] [Google Scholar]
- Nakayama K, Katoh Y (2018) Ciliary protein trafficking mediated by IFT and BBSome complexes with the aid of kinesin‐2 and dynein‐2 motors. J Biochem 163: 155–164 [DOI] [PubMed] [Google Scholar]
- Nakayama T, Fish MB, Fisher M, Oomen‐Hajagos J, Thomsen GH, Grainger RM (2013) Simple and efficient CRISPR/Cas9‐mediated targeted mutagenesis in Xenopus tropicalis. Genesis 51: 835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678 [DOI] [PubMed] [Google Scholar]
- Olbrich H, Schmidts M, Werner C, Onoufriadis A, Loges N, Raidt J, Banki N, Shoemark A, Burgoyne T, Al Turki S et al (2012) Recessive HYDIN mutations cause primary ciliary dyskinesia without randomization of left‐right body asymmetry. Am J Hum Genet 91: 672–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I, Lee H‐K, Kim C, Ismail T, Kim Y‐K, Park J‐W, Kwon O‐S, Kang BS, Lee D‐S, Park T‐J et al (2016) IFT46 plays crucial roles in craniofacial and cilia development. Biochem Biophys Res Commun 477: 419–425 [DOI] [PubMed] [Google Scholar]
- Park TJ, Haigo SL, Wallingford JB (2006) Ciliogenesis defects in embryos lacking inturned or fuzzy function are associated with failure of planar cell polarity and Hedgehog signaling. Nat Genet 38: 303–311 [DOI] [PubMed] [Google Scholar]
- Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB (2008) Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet 40: 871–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazour GJ, Dickert BL, Vucica Y, Seeley ES, Rosenbaum JL, Witman GB, Cole DG (2000) Chlamydomonas IFT88 and its mouse homologue, polycystic kidney disease gene tg737, are required for assembly of cilia and flagella. J Cell Biol 151: 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Bui KH, Maheshwari A, Lupetti P, Diener D, Ishikawa T (2011) Cryoelectron tomography of radial spokes in cilia and flagella. J Cell Biol 195: 673–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pigino G, Geimer S, Lanzavecchia S, Paccagnini E, Cantele F, Diener DR, Rosenbaum JL, Lupetti P (2009) Electron‐tomographic analysis of intraflagellar transport particle trains in situ. J Cell Biol 187: 135–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachev EV, Schuster‐Gossler K, Fuhl F, Ott T, Tveriakhina L, Beckers A, Hegermann J, Boldt K, Mai M, Kremmer E et al (2020) CFAP43 modulates ciliary beating in mouse and Xenopus. Dev Biol 459: 109–125 [DOI] [PubMed] [Google Scholar]
- Rao VG, Sarafdar RB, Chowdhury TS, Sivadas P, Yang P, Dongre PM, D'Souza JS (2016) Myc‐binding protein orthologue interacts with AKAP240 in the central pair apparatus of the Chlamydomonas flagella. BMC Cell Biol 17: 24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter JF, Leroux MR (2017) Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol 18: 533–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romaker D, Kumar V, Cerqueira DM, Cox RM, Wessely O (2014) MicroRNAs are critical regulators of tuberous sclerosis complex and mTORC1 activity in the size control of the Xenopus kidney. Proc Natl Acad Sci U S A 111: 6335–6340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum JL, Witman GB (2002) Intraflagellar transport. Nat Rev Mol Cell Biol 3: 813–825 [DOI] [PubMed] [Google Scholar]
- Schweickert A, Weber T, Beyer T, Vick P, Bogusch S, Feistel K, Blum M (2007) Cilia‐driven leftward flow determines laterality in Xenopus. Curr Biol 17: 60–66 [DOI] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB (2016) Emergence of an apical epithelial cell surface in vivo . Dev Cell 36: 24–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Zhou T, Huang Q, Zhang S, Li W, Zhang L, Hess RA, Pazour GJ, Zhang Z (2019) Intraflagellar transport protein 74 is essential for spermatogenesis and male fertility in micedagger. Biol Reprod 101: 188–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Zhang X, Jia S, Yelick PC, Zhao C (2016) Zebrafish as a model for human ciliopathies. J Genet Genomics 43: 107–120 [DOI] [PubMed] [Google Scholar]
- Spassky N, Meunier A (2017) The development and functions of multiciliated epithelia. Nat Rev Mol Cell Biol 18: 423–436 [DOI] [PubMed] [Google Scholar]
- Stepanek L, Pigino G (2016) Microtubule doublets are double‐track railways for intraflagellar transport trains. Science 352: 721–724 [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Davidson L, Keller R, Kintner C (2006) Radial intercalation of ciliated cells during Xenopus skin development. Development 133: 2507–2515 [DOI] [PubMed] [Google Scholar]
- Stubbs JL, Vladar EK, Axelrod JD, Kintner C (2012) Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat Cell Biol 14: 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Yoon J, Lee M, Hwang YS, Daar IO (2020) Sprouty2 regulates positioning of retinal progenitors through suppressing the Ras/Raf/MAPK pathway. Sci Rep 10: 13752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CH, Leroux MR (2013) The roles of evolutionarily conserved functional modules in cilia‐related trafficking. Nat Cell Biol 15: 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner M, Bhogaraju S, Lorentzen E (2012) Architecture and function of IFT complex proteins in ciliogenesis. Differentiation 83: S12–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terre B, Piergiovanni G, Segura‐Bayona S, Gil‐Gomez G, Youssef SA, Attolini CS, Wilsch‐Brauninger M, Jung C, Rojas AM, Marjanovic M et al (2016) GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J 35: 942–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves ME, Nagarkatti‐Gude DR, Zhang Z, Strauss JF 3rd (2016) Mammalian axoneme central pair complex proteins: Broader roles revealed by gene knockout phenotypes. Cytoskeleton 73: 3–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M, Lee C, Taylor SP, Duran I, Cohn DH, Bruel A‐L, Tabler JM, Drew K, Kelly MR, Kim S et al (2016) The ciliopathy‐associated CPLANE proteins direct basal body recruitment of intraflagellar transport machinery. Nat Genet 48: 648–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama M, Toriyama M, Wallingford JB, Finnell RH (2017) Folate‐dependent methylation of septins governs ciliogenesis during neural tube closure. FASEB J 31: 3622–3635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannuccini E, Paccagnini E, Cantele F, Gentile M, Dini D, Fino F, Diener D, Mencarelli C, Lupetti P (2016) Two classes of short intraflagellar transport train with different 3D structures are present in Chlamydomonas flagella. J Cell Sci 129: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanadha R, Sale WS, Porter ME (2017) Ciliary Motility: Regulation of Axonemal Dynein Motors. Cold Spring Harb Perspect Biol 9: a018325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitre B, Taulet N, Guesdon A, Douanier A, Dosdane A, Cisneros M, Maurin J, Hettinger S, Anguille C, Taschner M et al (2020) IFT proteins interact with HSET to promote supernumerary centrosome clustering in mitosis. EMBO Rep 21: e49234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentek P, Quigley IK, Sun DI, Sajjan UK, Kintner C, Harland RM (2016) Ciliary transcription factors and miRNAs precisely regulate Cp110 levels required for ciliary adhesions and ciliogenesis. eLife 5: e17557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walia V, Cuenca A, Vetter M, Insinna C, Perera S, Lu Q, Ritt DA, Semler E, Specht S, Stauffer J et al (2019) Akt regulates a Rab11‐effector switch required for ciliogenesis. Dev Cell 50: 229–246.e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Shi Z, Cui Y, Guo X, Shi YB, Chen Y (2015) Targeted gene disruption in Xenopus laevis using CRISPR/Cas9. Cell Biosci 5: 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Tian S, Nie H, Tu C, Liu C, Li Y, Li D, Yang X, Meng L, Hu T et al (2021) CFAP65 is required in the acrosome biogenesis and mitochondrial sheath assembly during spermiogenesis. Hum Mol Genet 30: 2240–2254 [DOI] [PubMed] [Google Scholar]
- Wang W, Tu C, Nie H, Meng L, Li Y, Yuan S, Zhang Q, Du J, Wang J, Gong F et al (2019) Biallelic mutations in CFAP65 lead to severe asthenoteratospermia due to acrosome hypoplasia and flagellum malformations. J Med Genet 56: 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ (2011) Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J Cell Biol 195: 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhao H, Zhu X (2016) Production of Basal Bodies in bulk for dense multicilia formation. F1000Research 5: 1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Huang K (2019) Dissecting the vesicular trafficking function of IFT subunits. Front Cell Dev Biol 7: 352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Diener DR, Yang C, Kohno T, Pazour GJ, Dienes JM, Agrin NS, King SM, Sale WS, Kamiya R et al (2006) Radial spoke proteins of Chlamydomonas flagella. J Cell Sci 119: 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasunaga T, Hoff S, Schell C, Helmstädter M, Kretz O, Kuechlin S, Yakulov TA, Engel C, Müller B, Bensch R et al (2015) The polarity protein Inturned links NPHP4 to Daam1 to control the subapical actin network in multiciliated cells. J Cell Biol 211: 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Mitchell BJ (2015) Centriole biogenesis and function in multiciliated cells. Methods Cell Biol 129: 103–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shen Y, Wang X, Yuan G, Zhang C, Yang Y (2019) A novel homozygous CFAP65 mutation in humans causes male infertility with multiple morphological abnormalities of the sperm flagella. Clin Genet 96: 541–548 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu H, Li W, Zhang Z, Shang X, Zhang D, Li Y, Zhang S, Liu J, Hess RA et al (2017) Intraflagellar transporter protein (IFT27), an IFT25 binding partner, is essential for male fertility and spermiogenesis in mice. Dev Biol 432: 125–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li W, Zhang Y, Zhang L, Teves ME, Liu H, Strauss JF, Pazour GJ, Foster JA, Hess RA et al (2016) Intraflagellar transport protein IFT20 is essential for male fertility and spermiogenesis in mice. Mol Biol Cell 27: 3705–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Chen Q, Fang C, Huang Q, Zhou J, Yan X, Zhu X (2019a) Parental centrioles are dispensable for deuterosome formation and function during basal body amplification. EMBO Rep 20: e46735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Zhu L, Zhu Y, Cao J, Li S, Huang Q, Xu T, Huang X, Yan X, Zhu X (2013) The Cep63 paralogue Deup1 enables massive de novo centriole biogenesis for vertebrate multiciliogenesis. Nat Cell Biol 15: 1434–1444 [DOI] [PubMed] [Google Scholar]
- Zhao L, Hou Y, Picariello T, Craige B, Witman GB (2019b) Proteome of the central apparatus of a ciliary axoneme. J Cell Biol 218: 2051–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Liu H, Zhu L, Chen Y, Zhao H, Zhang W, Li F, Xie L, Yan X, Zhu X (2019) Microtubule‐bundling protein Spef1 enables mammalian ciliary central apparatus formation. J Mol Cell Biol 11: 67–77 [DOI] [PubMed] [Google Scholar]
- Zhu B, Zhu X, Wang L, Liang Y, Feng Q, Pan J (2017) Functional exploration of the IFT‐A complex in intraflagellar transport and ciliogenesis. PLoS Genet 13: e1006627 [DOI] [PMC free article] [PubMed] [Google Scholar]


