Abstract
Cancer cachexia (CC) is a complex metabolic syndrome that accelerates muscle wasting and affects up to 80% of patients with cancer; however, timely diagnostic methods and effective cures are lacking. Although a considerable number of studies have focused on the mechanism of CC-induced muscle atrophy, few novel therapies have been applied in the last decade. In recent years, noncoding RNAs (ncRNAs) have attracted great attention as many differentially expressed ncRNAs in cancer cachectic muscles have been reported to participate in the inhibition of myogenesis and activation of proteolysis. In addition, extracellular vesicles (EVs), which function as ncRNA carriers in intercellular communication, are closely involved in changing ncRNA expression profiles in muscle and promoting the development of muscle wasting; thus, EV-related ncRNAs may represent potential therapeutic targets. This review comprehensively describes the process of ncRNA transmission through EVs and summarizes the pathways and targets of ncRNAs that lead to CC-induced muscle atrophy.
Keywords: cancer cachexia, ncRNA, extracellular vesicles, muscle wasting, endoplasmic reticulum stress
Introduction
Cancer cachexia (CC) is a severe systemic syndrome that is characterized by tissue wasting, decreased energy intake, weight loss, fatigue, metabolic abnormalities, systemic inflammation, and chemotherapy intolerance.1 Furthermore, CC is reported to be responsible for at least 20% of cancer-related deaths and affects 50%–80% of patients in advanced stages.2 Low chemotherapeutic efficacy and chemotherapy tolerance as a result of CC are major factors leading to a poor prognosis, decreased survival, and reduced quality of life for patients.3,4 CC-induced muscle wasting is a severe problem, as it is related to low treatment tolerance and effectiveness, and one in four cancer-related deaths possibly results from muscle atrophy instead of the tumor burden.5 Therefore, a considerable amount of research has been performed to elucidate the molecular mechanisms of muscle atrophy in CC. Despite its clinical relevance, CC is seldom diagnosed and treated early.6,7 Because of the heterogeneous manifestations of CC and the scarcity of diagnostic methods, CC is not detected until the refractory stage, in which therapeutic agents have limited efficacy.8 As CC is a response to host–tumor interactions caused by tumor-derived mediators,5 effective CC biomarkers would contribute to the early identification of patients at risk of CC.9 Nucleic acid biomarkers can be detected more rapidly and less expensively than protein biomarkers, and the techniques are easier to perform.10 In addition, although curing cancer is an effective approach to treat CC, its application is limited.11 Therefore, prolonging survival is the next treatment aim, and this can be achieved by blocking the procachexia signaling pathway.12,13 Thus, noncoding RNAs (ncRNAs) were thought to have potential clinical utility in the treatment of CC-induced muscle wasting.
Extracellular vesicles (EVs) are enriched in numerous bioactive molecules, including nucleic acids and proteins, and can relay signals not only into the tumor microenvironment but also into the circulatory system.14 ncRNAs are defined as heterogeneous RNA transcripts that feature a low protein-coding capability.15 Based on their size, ncRNAs are categorized into short RNAs of < 200 nucleotides, including small interfering RNAs, microRNAs (miRNAs), and piwi-interacting RNAs, and long noncoding RNAs (lncRNAs) of > 200 nucleotides. Over the past decade, a number of ncRNAs have been identified in large-scale analyses. For instance, miRNAs have been identified in endosomal sorting complexes required for transport (ESCRTs) involved in the biogenesis, release, and uptake of exosomes, which can be absorbed by adjacent or distant cells after release. Therefore, miRNAs are thought to participate in tumor immunity, growth, invasion, and metastasis via exosomes.16 In addition, ncRNAs are involved in controlling gene expression and intercellular communication required for skeletal muscle growth by regulating multiple signaling pathways.17
Although numerous ncRNAs are reported to be expressed at abnormal levels in skeletal muscle cells of patients with CC,18,19 few studies have attempted to elucidate their complete mechanisms of action and pathways in CC and muscle wasting. In the present review, we focus on the impact of ncRNAs on the process of muscle wasting induced by CC, briefly describe recent studies of several ncRNAs in CC, and discuss their potential therapeutic applications (Fig. 1).
Figure 1.
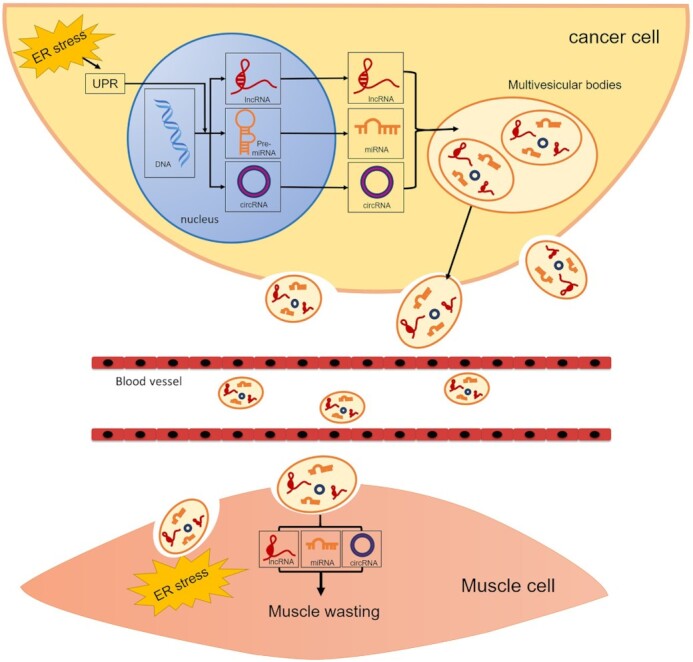
Diagram depicting the mechanism of cancer-induced muscle atrophy by releasing ncRNAs. CC is associated with muscle wasting induced by the release of EVs. DNA is transcribed into ncRNAs, including miRNAs, lncRNAs, and circRNAs, which in turn regulate transcription and translation. Following the biogenesis, release, and uptake of EVs, ncRNAs are transferred into muscle cells and induce muscle wasting through ERS transfer.
EVs as ncRNA carriers in intercellular communication
Biogenesis of EVs
EVs can be divided into three types according to their diameter: exosomes (30–100 nm), microvesicles (100–1000 nm), and large oncosomes (1–10 μm).20 Among these types of EVs, exosomes have been studied extensively and found to participate in the intercellular transport of ncRNAs, which have been confirmed to be related to cancer cachexia.16 Therefore, regarding the biogenesis of EVs, we focused on exosomes in the present study. The endosomal system has been demonstrated to be the origin of exosomes.21 First, the inward budding of the membrane of late endosomes generates multivesicular bodies (MVBs), which contain intraluminal vesicles (ILVs). Then, MVBs can fuse with either the lysosome or the cellular plasma membrane. The latter pathway results in release of ILVs. The released ILVs are identified as exosomes.22
RNA sorting mechanism of EVs
The sorting of RNAs is a highly selective and complicated part of the biogenesis of EVs. The ESCRT machinery contains protein complexes that are central for EV packaging. The roles of ESCRT-0, ESCRT-I, and ESCRT-II are to recognize and conceal ubiquitinated membrane proteins in endosomal membranes, while ESCRT-III promotes cleavage and inward budding.23
In addition to the ESCRT machinery, several other factors are involved in the sorting of RNAs into exosomes. A specific sequence (GGAG) in exosomal miRNAs was identified as the EXO motif, a characteristic recognition sequence of heterogeneous ribonucleoprotein A1 (hnRNPA1) and heterogeneous ribonucleoprotein A2B1 (hnRNPA2B1), thus regulating the sorting of miRNAs into EVs.24 However, as numerous miRNAs within EVs lack specific EXO motifs, the specification of RNAs for secretion requires other pathways. Cha et al. found that oncogenic KRAS selectively altered the miRNA profile in exosomes and that ceramide depletion selectively promoted miRNA accumulation in KRAS mutant colorectal cancer cells.25 The RNA-binding protein Y-box protein I (YBX1) was identified to participate in the sorting of miR-223.26 Additionally, the loading of microRNAs into EVs is affected by ectopic changes in the levels of microRNAs or target mRNAs.27
Transport of EVs
The RAB family of small GTPase proteins and soluble NSF-attachment protein receptor (SNARE) complexes play a key role in promoting exosome release. The RAB proteins are involved in vesicle budding, vesicle and organelle migration, and docking to the target. In addition, RAB proteins play a key role in the docking process between the multivesicular body (MVB) and plasma membrane, which allows vesicles to be generated in this compartment.28
The EV content exerts an impact on cell behavior by entering the cytoplasm and possibly even the nucleus. Three main mechanisms for the interaction of EVs with target cells for the delivery of RNAs or other proteins into recipient cells have been proposed: receptor-mediated uptake, direct fusion with the plasma membrane, and endocytosis via phagocytosis.29 Different cell types take up EVs through various mechanisms, leading to functional transfer or degradation of EV contents.29
EVs drive CC via the endoplasmic reticulum stress (ERS) response
Tumor-derived EVs contribute to disease progression, and the cargo transferred by EVs might influence cellular processes in some distant tissues, such as skeletal muscles.30 ERS is characterized by the impairment of physiological functions and the protein-folding environment of the endoplasmic reticulum, and therefore, unfolded proteins accumulate in the endoplasmic reticulum lumen.31 The possibility that ERS-related proteins are overexpressed in cancer cells because of low nutrition, low vascularization, and hypoxia has been suggested.32 The unfolded protein response (UPR) signaling cascade is initiated in cells to restore homeostasis. Some components of the UPR induced by ERS are activated in the skeletal muscle of patients with CC.33 However, the role of ERS in muscle atrophy is poorly studied. Recently, our research group found that exosomal miR-181a-3p regulates the transmissible endoplasmic reticulum stress (TERS) pathway and induces the activation of the UPR, which subsequently accelerates muscle cell atrophy.34 This finding indicates that UPR activation may participate in the regulation of skeletal muscle quality along with other signaling pathways. The revealed role for EVs in the ERS response in CC strongly indicates the urgency of analyzing the mechanism and application potential of their contents.
The role of ncRNAs in muscle wasting induced by CC
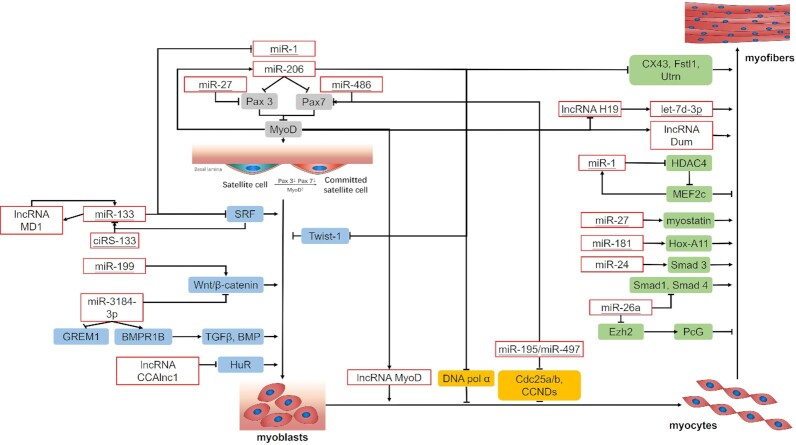
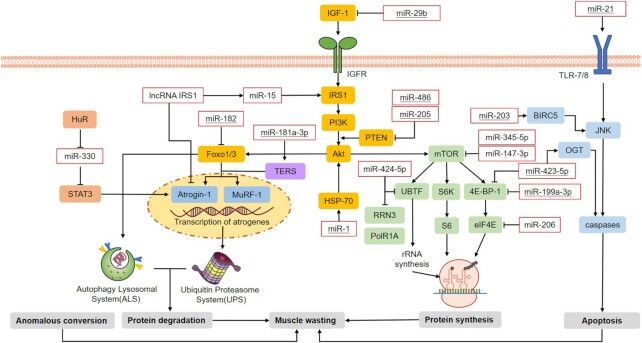
Muscle mass loss induced by CC is a combined result of the inhibition of myogenesis and activation of muscle protein degradation.35 Because the effect of EVs on the progression of CC is a topic of special concern, ncRNAs are categorized into EV-related and other ncRNAs, and both types are differentially expressed in cancer cachectic muscle. Through the investigation of the upregulated or downregulated ncRNAs, clear evidence has been obtained that these ncRNAs regulate pathways implicated in muscle wasting in patients with cancer (Figs. 2 and 3).
Figure 2.
Roles of ncRNAs in pathways and processes regulating CC-induced inhibition of myogenesis. Myogenesis is a result of the combination of several factors, including the activation of satellite cells, myoblast proliferation, cell cycle exit, and terminal differentiation. Evidence clearly indicates that the upregulation or downregulation of ncRNAs regulates the inhibition of myogenesis in patients with cancer. The role of ncRNAs in the myogenic process is visually illustrated. The underlined ncRNAs are related to EVs.
Figure 3.
Roles of ncRNAs in pathways regulating CC-induced muscle atrophy. Four pathways regulating the process of CC-induced muscle wasting were identified: protein degradation, anomalous conversion, protein synthesis inhibition, and muscle cell apoptosis. Most studies on the relevant topic have focused on the roles of ncRNAs that regulate the expression of Atrogin-1 and MuRF-1, thereby promoting the activation of the UPS and autophagy-lysosome system (ALS). In particular, ncRNAs related to EVs are underlined.
The role of EV-related ncRNAs during muscle wasting in CC
Recently, many studies have revealed a number of EVs containing ncRNAs with a potential impact on distant tissues that regulate the progression of cachexia36 by performing in vitro experiments, mouse models, and human studies.18,37 Exosomal ncRNAs are presumed to regulate gene expression in recipient cells by targeting mRNAs and directly binding to proteins, thereby modulating inflammatory pathways and degradation pathways in skeletal muscle (Table 1).
Table 1.
EV-related ncRNAs mentioned above and their targets in skeletal muscle wasting induced by CC.
| Organism)reference( | ncRNA | Expression | Target(s) | Function |
|---|---|---|---|---|
| Glioblastoma38 | miR-1 | downregulated | Pax3/7,YY1, EIF4E3 | activation of quiescent satellite cells |
| HDAC4 | myoblast proliferation and differentiation | |||
| HSP70, Akt, Foxo3 | protein anabolism, activation of UPS and ALS | |||
| Oral squamous cell carcinoma39 | miR-27 | downregulated | Pax3 | activation of quiescent satellite cells |
| myostatin | myoblast proliferation and differentiation | |||
| Lung adenocarcinoma40 | miR-486 | upregulated | PTEN | suppress skeletal muscle proteolysis |
| Pax7 | activation of quiescent satellite cells | |||
| Gastric cancer41 | ciRS-133/miR-133 | upregulated | SRF | myoblast proliferation and differentiation |
| Cholangiocarcinoma42 | miR-199a-3p | upregulated | Wnt/β-catenin signaling proteins | myoblast proliferation and differentiation |
| Pancreatic and colorectal cancer37 | miR-423-5p | upregulated | OGT | cell apoptosis |
| EIF4EBP1 | protein synthesis | |||
| Lung cancer19 | miR-424-5p | upregulated | UBTF, PolR1A, RRN3 | protein synthesis |
| ribosomal RNA | maturation of muscle fibers | |||
| Osteosarcoma and bladder cancer43 | miR-195/miR-497 | upregulated | Cdc25a/b, CCNDs | process of myoblasts exiting cell cycle |
| Nasopharyngeal carcinoma44 | let-7d-3p | upregulated | TFRC | myoblast proliferation and differentiation |
| Breast cancer45; lung cancer18,46 | miR-205 | PTEN, PHLPP2 | protein anabolism | |
| Bladder cancer47 | lncH19 | upregulated | IGF2, let-7 family, miR-675 | myoblast proliferation and differentiation |
| Cervical cancer48 | miR-203 | upregulated | BIRC5 | myoblast proliferation and differentiation |
| Lung and pancreatic cancer49 | miR-21 | upregulated | TLR 7/8, YY1, eIF4E3 | cell apoptotic response |
| Lung cancer50 | miR-181/miR-181a-3p | downregulated | Hox-A11 | myoblast differentiation |
| MuRF1, Atrogin-1 and TRAF6 | muscle atrophy | |||
| Location undetermined51 | miR-182 | undetermined | Foxo3 | attenuation of UPS and ALS |
| Lung cancer52 | miR-29b | upregulated | IGF-1,PI3K(p85a),Foxo3 | activation of UPS and ALS |
| Gastric cancer53,54 | miR-26 | downregulated | PcG complex, Ezh2, Smad 1, Smad 4 | myoblast proliferation and differentiation |
miR-1
The presence of miR-1 was reported in glioblastoma-derived EVs, and miRNA is downregulated in CC.38 In addition, miR-1 can inhibit Pax3/7, which can induce the formation of muscle fibers when muscles are damaged but remains in an inactive state under normal physiological conditions.55,56 Moreover, miR-1 can bind to YY1 (a transcription factor) and eIF4E3 (a translation initiation factor), thereby interfering with muscle formation.53 Furthermore, miR-1 is thought to downregulate histone deacetylase 4 (HDAC4), which represses the expression of MEF2C, a key muscle-related transcription factor, thereby inhibiting the expression of muscle-related genes.57
Additionally, increased miR-1 expression was shown to promote a reduction in the activity of heat shock protein 70 (HSP70), which is critical for Akt phosphorylation (p-Akt). The decrease in HSP70 expression was related to decreased levels of p-Akt. The reduction in p-Akt levels promoted a decrease in the level of FoxO3 phosphorylation, thereby promoting the upregulation of Atrogin-1 and MurF-1, eventually contributing to muscle atrophy.58
miR-27
Oncogenic miR-27 was detected in large quantities in the EVs of patients with oral squamous cell carcinoma (OSCC) and was downregulated in muscle cells.39,53 Additionally, miR-27 was found to participate in the regulation of Pax3 expression in embryonic myotomes and stimulated myogenic robustness by activating satellite cells.59 Furthermore, miR-27a/b participates in promoting myogenic differentiation by targeting the 3’UTR of myostatin and increases its levels, but it decreases myoblast proliferation.60
miR-486
PTEN phosphatase, a negative regulator of p-Akt, is downregulated by miR-486, thereby repressing FoxO1 protein translation and increasing FoxO1 phosphorylation.61 These reactions prevent the translocation of FoxO1 into the nucleus, thereby suppressing skeletal muscle proteolysis by the ubiquitin-proteasome system (UPS).61 Additionally, miR-486 exerts its inhibitory effect on differentiation by suppressing the activity of MyoD, which targets the Pax7 3’ UTR in satellite cells to maintain the muscle stem cell status.62 A recent study further supported a relationship among miR-486, EVs, and cancer cachexia, although the mechanisms are not completely understood. Reports have indicated that miR-486 expression is significantly increased in EVs from patients with lung adenocarcinoma.40
ciRS-133 and miR-133
A previous study showed that circulating exosomal ciRS-133 is overexpressed in gastric cancer and acts as a miR-133 sponge.41 The upregulation of miR-133 during CC inhibits myoblast differentiation and maintains myoblast proliferation via serum response factor (SRF).57 As the transcription of miR-133a and miR-1 is directly regulated by SRF, a negative feedback loop is formed by miR-133 and SRF that balances myoblast proliferation and myoblast differentiation.63
miR-199a-3p
Although reports have indicated that miR-199 family members are downregulated in exosomes isolated from cholangiocarcinoma cell lines, miR-199a-3p is upregulated in cancer cachectic muscle.37,42 Upregulated miR-199a-3p in cancer cachectic muscle likely affects myogenesis and muscle regeneration by regulating the levels of proteins involved in the Wnt signaling pathway, collectively regulating myoblast proliferation and myoblast differentiation.64
miR-423-5p
The level of miR-423-5p was reported to be increased in serum exosomes and cancer cachectic muscle, indicating that miR-423-5p likely induces apoptosis via direct targeting of O-GlcNAc transferase (OGT), thereby causing muscle wasting.37,65 EIF4EBP1, a target of miR-199a-3p and miR-423-5p, has been demonstrated to participate in the mTOR pathway, which is related to muscle protein synthesis.37
miR-424-5p
Although miR-424-5p was found to be downregulated in exosomes derived from cholangiocarcinoma cells,66 it was found to be overexpressed in cancer cachectic muscle.19In vitro, the synthesis of ribosomal RNA was found to be inhibited by miR-424-5p via effects on UBTF, PolR1A, and RRN3. Overexpression of miR-424-5p, therefore, may lead to a decrease in protein synthesis. In vivo, a significant decrease in fiber diameter was observed when miR-424-5p was overexpressed.67
miR-203
The upregulation of miR-203 has been investigated in the humoral circulation of patients with cervical cancer.48 Similarly, Okugawa et al.68 revealed that overexpressed miR-203 markedly accelerates apoptosis in human skeletal muscle cells, possibly through downregulation of BIRC5, which inhibits the initiator caspase 9 and executor caspases 3 and 7, thus suppressing apoptosis.69
miR-195/miR-497
High-throughput sequencing identified the upregulation of miR-195 in exosomes from patients with osteosarcoma and the upregulation of miR-497 in exosomes from patients with bladder cancer.43 When adult skeletal muscle stem cells were treated with miR-195/497 inhibitors, the mRNA expression of the cell cycle regulators Cdc25 and Ccnd was induced, and the expression of Pax7 was decreased, subsequently decreasing the regenerative myogenesis of adult skeletal muscle stem cells.70
let-7d-3p
let-7d-3p was reported to be upregulated in serum from patients with nasopharyngeal carcinoma and in the cancer cachectic muscle.37,44 Myogenic differentiation might be impaired by downregulation of the transferrin receptor (TFRC), which is a target of let-7d-3p. Thus, let-7d-3p was proposed to participate in muscle cell proliferation via downregulation of the target gene TFRC, thereby affecting muscle cell myogenic differentiation and proliferation and eventually reducing the ability of muscle to regenerate.37
miR-205
Reports have indicated that miR-205 from the EV cargo plays a role in suppressing the metastasis of breast cancer cells.45 In CC samples, upregulated miR-205 directly caused a decrease in the levels of PTEN and PHLPP2 and simultaneously activated the Akt/FoxO3a and Akt/mTOR signaling pathways, inducing a negative protein balance and skeletal muscle loss by disrupting muscle protein anabolism.18,46
lncRNA H19
Wang et al. validated that exosomal lncRNA H19 is stable in exosomes derived from the serum of patients with bladder cancer.47 The reduced expression of MyoD in muscles of CC directly activates lncRNA H19 transcription, while the lncRNA H19 downregulates expression of IGF2 during muscle differentiation.71 Additionally, lncRNA H19 contains multiple binding sites for the let-7 family of miRNAs, and thus acts as a molecular sponge for most members of the let-7 family of miRNAs.72
miR-203
Notably, miR-203 was found to be upregulated in EVs in the humoral circulation of patients with cervical cancer.73 Similarly, Okugawa et al.recently revealed that upregulated miR-203 in skeletal muscle cells from patients with colorectal cancer promoted apoptosis, possibly through downregulation of BIRC5, which may inhibit apoptosis by inhibiting the initiator caspase-9 and executor caspases 3 and 7.68 These results demonstrate that serum miR-203 has potential utility in predicting cancer cachexia.
miR-21
He et al. demonstrated that miR-21, a miRNA present in EVs, promotes cell death by activating Toll-like 7/8 receptors (TLR 7/8) on myoblasts. Furthermore, c-Jun N-terminal kinase activity is required to modulate this apoptotic response.49 YY1 (a transcription factor) and eIF4E3 (a translation initiation factor) were demonstrated to be the downstream targets of miR-21.53
miR-181/miR-181a-3p
Downregulated miRNAs include miR-181,50 which has been previously studied as an exosomal diagnostic biomarker for nonsmall-cell lung cancer.74 Moreover, miR-181 downregulates the homeobox protein Hox-A11 during cell differentiation, thus participating in the establishment of the muscle phenotype.75 Recently, exosomes containing miR-181a-3p, which are secreted by OSCC cells, were shown to regulate TERS signaling in C2C12 myotubes and upregulate the expression of genes related to muscle atrophy, including MuRF-1 and Atrogin-1, thus eventually inducing muscle wasting.34
miR-182
MiR-182 in isolated exosomes was found to decrease the expression of FoxO3 mRNA by 30% and FoxO3 protein by 67% and to prevent glucocorticoid-induced upregulation of FoxO3 gene targets, such as Atrogin-1, thereby probably attenuating protein degradation.51
miR-29b
miR-29b was found to be overexpressed in CC-induced muscle atrophy models.50,52 The levels of IGF-1, PI3K (p85a), and downstream targets were decreased in CC models in vitro and were reduced by miR-29b in vivo. Moreover, miR-29b reduced the levels of IGF-1 and PI3K (p85a) to decrease the phosphorylation of FoxO3a, thus inducing the expression of Atrogin-1 and Murf-1 and eventually promoting muscle loss.52
miR-26
The epidermal growth factor receptor (EGFR) is transferred to liver cells by exosomes derived from gastric cancer cells and then regulates the microenvironment of liver cells via downregulation of miR-26a/b.54 Intriguingly, the level of miR-26 is also decreased in muscle cells from patients with CC.53 Downregulation of miR-26 promotes the expression and function of the PCG complex in muscle cells, thereby inhibiting the expression and differentiation of muscle-specific genes in CC.76 Additionally, miR-26a mediates myoblasts by targeting and suppressing enhancer of zeste homolog 2 (Ezh2).77 Similarly, downregulation of miR-26 inhibits myogenic differentiation by inhibiting the expression of Smad 1 and Smad 4, both of which are repressed in CC.53,76
The roles of other ncRNAs in CC-induced muscle wasting
Although direct evidence that ncRNAs are transported by EVs is lacking, ncRNAs are differentially expressed in cancer cachectic muscles and regulate the process of muscle formation, and their inherent biological functions remain stable (Table 2).
Table 2.
ncRNAs mentioned above and their targets in skeletal muscle wasting induced by CC.
| Organism (reference) | ncRNA | Expression | Target(s) | Function |
|---|---|---|---|---|
| Pancreatic ductal adenocarcinoma78,79 | miR-206 | upregulated | Pax3/7, YY1, EIF4E3 | activation of quiescent satellite cells |
| MyoD, Twist-1 | myoblast proliferation and differentiation | |||
| CX43, Fstl1, Utrn | maturation of skeletal muscle | |||
| Pancreatic and colorectal cancer37 | miR-3184-3p/miR-1296-5p | upregulated | Wnt/β-catenin signaling, BMPR1B, GREM1, TGF-β | myoblast proliferation and differentiation |
| Pancreatic and colorectal cancer37 | miR-345-5p | upregulated | NOV, COL1A1, CYR61 | activation of UPS and ALS |
| Pancreatic and colorectal cancer37 | miR-423-3p | upregulated | Cox6a2 | ATP regulation and myogenic differentiation |
| Breast cancer80 | miR-147-3p | upregulated | Akt/mTOR | protein anabolism |
| Liver cancer81 | lncMyoD | downregulated | IMP2-Mediated mRNA | myoblast proliferation and differentiation |
| Liver cancer81,82 | lncDum | downregulated | Dppa2 | myoblast proliferation and differentiation |
| Location undetermined83 | lncCAA1/miR-330 | downregulated | HuR/STAT3 | myoblast differentiation |
| Colon carcinoma53 | miR-15 | downregulated | IRS1, IGF1/PI3K/AKT signal | myogenesis and atrophy |
| Colon carcinoma53,84 | lncIRS1 | upregulated | miR-15, Atrogin-1, MuRF1 | myogenesis and atrophy |
miR-206
In rats with CC, MyoD expression was found to be decreased in the diaphragm and gastrocnemius.81 Low expression of MyoD upregulates miR-206 expression.78 Similar to miR-1, miR-206 also inhibits Pax3/7 and binds to the transcription factor YY1 and translation promoter eIF4E3 to regulate muscle wasting.55,56 Additionally, miR-206 overexpression downregulates the level of Twist-1, promotes the differentiation of skeletal muscle cells, and induces myogenesis.78 Notably, in immortalized myoblastic cell systems, the overexpression of Twist-1 inhibits MyoD activity and further suppresses myoblast differentiation.85 Intriguingly, in another study, the level of Twist-1 was decreased in skeletal muscle, and Twist-1 was found to regulate muscle cachexia via activin/myostatin signaling.79 The overexpression of miR-206 activates the p180 subunit of DNA polymerase alpha and the subsequent differentiation of muscle cells.86 Moreover, neuromuscular junctions (NMJs) are enriched in miR-206,87 which regulates various other muscle-specific genes, such as Connexin 43 (CX43), Follistatin-like 1 (Fstl1), and Utrophin (Utrn). These genes are key to the maturation of skeletal muscle and promote the formation of NMJ endplates.86,88,89
miR-3184-3p/miR-1296-5p
Both miR-3184-3p and miR-1296-5p are upregulated in human cachectic muscle tissue and participate in Wnt/β-catenin signaling to impair myogenic differentiation.37,90 BMPR1B and GREM1 are the targets of miR-3184-3p, which may contribute to CC and participate in TGF-β and BMP signaling.37 Additionally, HTR2A, one target of miR-1296-5p, plays a role in serotonin signaling. Serotonin was found to promote the longitudinal growth of muscle fibers, thus participating in myogenesis.91
miR-345-5p
miR-345-5p is upregulated in patients with CC and subsequently downregulates the expression of NOV and COL1A1 and upregulates the expression of CYR61 in muscle cells.37 NOV and CYR61 are postulated to mediate protein synthesis via the IGF-1, Akt, and mTOR pathways. In animal models, downregulation of COL1A1 is related to muscle wasting. Both let-7d-3p and miR-345-5p are proposed to mediate the expression of genes associated with CC-related muscle atrophy, such as TFRC and COL1A1.37,92
miR-423-3p
MiR-423-3p was found to be upregulated in skeletal muscle from individuals with CC and to be involved in myogenic differentiation. During myogenic differentiation in vitro, the level of miR-423-3p was increased and thus dramatically reduced the levels of Cox6a2 and ATP, thereby negatively influencing myogenic differentiation.37,93
miR-147-3p
In breast cancer cells, ectopic expression of miR-147 was found to decrease the levels of components of the Akt/mTOR protein synthesis cascade, indicating a potential approach by which overexpression of miR-147-3p induced by CC may repress protein anabolism in cachectic muscle.18,80
lncRNA MyoD/Dum/CAAlnc1
lncMyoD is a downstream target identified for MyoD, which is downregulated in CC,81 regulates cell cycle exit and participates in myogenesis. MyoD also reduces the expression of the lncRNA Dum and is reported to act as a promyogenic factor in the process of muscle regeneration.82
lncCAA1/miR-330
Shen et al. found that the knockdown of CAAlnc1 in mice with CC enhances the binding of Hu antigen R (HuR) to RNAs, thereby stabilizing the mRNAs encoding some myogenic factors in the process of muscle differentiation.83 Another study indicated that HuR prevents the translation inhibition regulated by miR-330, thus promoting the translation of the signal transducer and activator of transcription 3 (STAT3) mRNA and inducing muscle loss.94
lncRNA IRS1/miR-15
lncIRS is a novel lncRNA that functions as a sponge for members of the miR-15 family (miR-15a, miR-15b-5p, and miR-15c-5p), activates IRS1, and regulates the IGF-1 signaling pathway, thereby participating in skeletal muscle myogenesis.53,84 Moreover, the overexpression of lncIRS1 decreases the levels of Atrogin-1 and MuRF-1, thus attenuating muscle wasting.84
Clinical applications of ncRNAs in CC
Because of the differences in the clinical manifestations of CC and the lack of effective testing criteria, muscle wasting cannot be diagnosed in a timely manner. Recently, the level of miRNA-130a was shown to contribute to improving the accuracy of Subjective Global Assessment (SGA) in predicting the risk of CC in patients with head and neck cancer.95 Notably, miRNAs identified 59.5% of the patients who lost more than 10% of their body mass, while the SGA score identified 33.3% of these patients (P = 0.049). By promoting inflammation mediated by TNF-α, patients with HNC presenting low miRNA-130a expression are more prone to cachexia.
Liang et al. silenced the SpHK2 gene in HCC cells to inhibit the sorting of miR-21 into exosomes through nanoparticle transport, thus inhibiting tumor cell migration and the tumorigenic function of exosomes.96 Additionally, the antitumor miRNA let-7a was able to be delivered to breast cancer tissue in Rag2 (-/-) mice via exosomes, revealing the therapeutic potential of exosomes carrying nucleic acids.97
Conclusions and perspectives
Recent studies have explored the molecular mechanisms related to muscle wasting in CC.98–100 This review summarizes recent research advances in determining the roles of ncRNAs in promoting muscle atrophy and inhibiting myogenesis during CC and highlights the involvement of EVs, thereby providing a new opportunity for the recognition of CC-induced muscle loss. Although the understanding of CC is increasing, the role of ncRNAs in muscle wasting in CC remains a promising field of research because the roles of most differentially expressed ncRNAs in the muscle of patients with CC are still unknown. An integrative meta-analysis may be a promising approach for further research. For instance, miR-145a was identified as a novel potential FoxO1 regulator during muscle wasting in CC by an integrative meta-analysis.98 Additionally, ncRNAs appear to be targets for the design of specific therapeutic treatments to attenuate the negative impact of muscle wasting on the quality of life of patients with various cancers. In clinical practice, a further challenge is to exploit the critical roles of EV-related ncRNAs. Specific EV-related ncRNAs may become not only markers for clinical diagnosis but also targets for anticancer drugs. By blocking the transmission of key ncRNAs through EVs, desirable alleviation of CC progression would be achieved.
ACKNOWLEDGEMENTS
We gratefully acknowledge the financial support from the National Natural Science Foundation of China (Grant No. 81972546) and the National Training Programs of Innovation and Entrepreneurship for Undergraduates of China (Grant No. 201910611653).
Contributor Information
Xueer Zhou, State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Dept. of Head and Neck Oncology, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Shoushan Hu, State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Dept. of Head and Neck Oncology, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Yunan Zhang, State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Dept. of Head and Neck Oncology, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Guannan Du, State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Dept. of Head and Neck Oncology, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Yi Li, State Key Laboratory of Oral Diseases & National Clinical Research Center for Oral Diseases & Dept. of Head and Neck Oncology, West China Hospital of Stomatology, Sichuan University, Chengdu 610041, China.
Conflict of interest
None declared.
References
- 1. Argilés JM, Stemmler B, López-Soriano FJ, et al. Inter-tissue communication in cancer cachexia. Nat Rev Endocrinol. 2018;15(1):9–20.. doi: https://doi.org/10.1038/s41574-018-0123-0. [DOI] [PubMed] [Google Scholar]
- 2. Ryan AM, Power DG, Daly L, et al. . Cancer-associated malnutrition, cachexia and sarcopenia: the skeleton in the hospital closet 40 years later. Proc Nutr Soc. 2016;75:199–211.. doi: https://doi.org/10.1017/S002966511500419X. [DOI] [PubMed] [Google Scholar]
- 3. da Rocha IMG, Marcadenti A, de Medeiros GOCet al. . Is cachexia associated with chemotherapy toxicities in gastrointestinal cancer patients? A prospective study. J Cachexia Sarcopenia Muscle. 2019;10:445–54.. doi: https://doi.org/10.1002/jcsm.12391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caillet P, Liuu E, Raynaud Simon A, et al. . Association between cachexia, chemotherapy and outcomes in older cancer patients: A systematic review. Clin Nutr. 2017;36:1473–82. doi: https://doi.org/10.1016/j.clnu.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 5. Talbert EE, Guttridge DC. Impaired regeneration: A role for the muscle microenvironment in cancer cachexia. Semin Cell Dev Biol. 2016;54:82–91.. doi: https://doi.org/10.1016/j.semcdb.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aversa Z, Costelli P, Muscaritoli M. Cancer-induced muscle wasting: latest findings in prevention and treatment. Ther Adv Med Oncol. 2017;9:369–82. doi: https://doi.org/10.1177/1758834017698643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Crawford J. What are the criteria for response to cachexia treatment?. Ann Palliat Med. 2019;8:43–9. doi: https://doi.org/10.21037/apm.2018.12.08. [DOI] [PubMed] [Google Scholar]
- 8. van der Meij B, Teleni L, McCarthy A, et al. . Cancer cachexia: an overview of diagnostic criteria and therapeutic approaches for the accredited practicing dietitian. J Hum Nutr Diet. 2021;34:243–54. doi: https://doi.org/10.1111/jhn.12811. [DOI] [PubMed] [Google Scholar]
- 9. Loumaye A, Thissen JP. Biomarkers of cancer cachexia. Clin Biochem. 2017;50:1281. doi: https://doi.org/10.1016/j.clinbiochem.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 10. Condrat C, Thompson D, Barbu M, et al. . miRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells. 2020;9(2):276. doi: https://doi.org/10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Inui A. Cancer anorexia-cachexia syndrome: current issues in research and management. CA Cancer J Clin. 2002;52:72–91.. doi: https://doi.org/10.3322/canjclin.52.2.72. [DOI] [PubMed] [Google Scholar]
- 12. Baracos V, Martin L, Korc Met al. . Cancer-associated cachexia. Nat Rev Dis Primers. 2018;4:17105. doi: https://doi.org/10.1038/nrdp.2017.105. [DOI] [PubMed] [Google Scholar]
- 13. Tseng Y, Kulp S, Lai I, et al. . Preclinical investigation of the novel histone deacetylase inhibitor AR-42 in the treatment of cancer-induced cachexia. J Natl Cancer Inst. 2015;107:djv274. doi: https://doi.org/10.1093/jnci/djv274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–83. doi: https://doi.org/10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diederichs S, Bartsch L, Berkmann Jet al. . The dark matter of the cancer genome: aberrations in regulatory elements, untranslated regions, splice sites, non-coding RNA and synonymous mutations. EMBO Mol Med. 2016;8:442–57. doi: https://doi.org/10.15252/emmm.201506055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun Z, Shi K, Yang Set al. . Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17:147. doi: https://doi.org/10.1186/s12943-018-0897-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van de Worp WRPH, Schols AMWJ, Dingemans A-MCet al. . Identification of microRNAs in skeletal muscle associated with lung cancer cachexia. J Cachexia Sarcopenia Muscle. 2020;11(2):452-63. doi: https://doi.org/10.1002/jcsm.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee DE, Brown JL, Rosa-Caldwell ME, et al. . Cancer cachexia-induced muscle atrophy: evidence for alterations in microRNAs important for muscle size. Physiol Genomics. 2017;49:253–60. doi: https://doi.org/10.1152/physiolgenomics.00006.2017. [DOI] [PubMed] [Google Scholar]
- 19. Van De Worp WRPH, Schols AMWJ, Harel-Bellan Aet al. . Identification of miRNAs in skeletal muscle associated with lung cancer cachexia. J Cachexia Sarcopenia Muscle. 2018;9:1153–4. doi: https://doi.org/10.1002/jcsm.12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Minciacchi VR, Freeman MR, Di Vizio D. Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol. 2015;40:41–51.. doi: https://doi.org/10.1016/j.semcdb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36:301–12. doi: https://doi.org/10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao H, Im H, Castro CM, et al. . New technologies for analysis of extracellular vesicles. Chem Rev. 2018;118:1917–50. doi: https://doi.org/ 10.1021/acs.chemrev.7b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–52. doi: https://doi.org/10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 24. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. . Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: https://doi.org/10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cha DJ, Franklin JL, Dou Yet al. . KRAS-dependent sorting of miRNA to exosomes. eLife. 2015;4:e07197. doi: https://doi.org/10.7554/eLife.07197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, et al. . Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. eLife. 2016;5:e19276. doi: https://doi.org/10.7554/eLife.19276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Squadrito ML, Baer C, Burdet F,et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–46. doi: https://doi.org/10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 28. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–25. doi: https://doi.org/10.1016/j.ceb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 29. Svensson KJ, Christianson HC, Wittrup Aet al. . Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288:17713–24. doi: https://doi.org/10.1074/jbc.M112.445403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chitti S, Fonseka P, Mathivanan S. Emerging role of extracellular vesicles in mediating cancer cachexia. Biochem Soc Trans. 2018;46:1129–36. doi: https://doi.org/10.1042/bst20180213. [DOI] [PubMed] [Google Scholar]
- 31. Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7:1013–30.. doi: https://doi.org/10.1038/nrd2755. [DOI] [PubMed] [Google Scholar]
- 32. Liu CY, Hsu CC, Huang TTet al. . ER stress-related ATF6 upregulates CIP2A and contributes to poor prognosis of colon cancer. Mol Oncol. 2018;12:1706–17. doi: https://doi.org/10.1002/1878-0261.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Afroze D, Kumar A. ER stress in skeletal muscle remodeling and myopathies. FEBS J. 2019;286:379–98. doi: https://doi.org/10.1111/febs.14358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu L, Chen W, Wu Cet al. . Exosomes of oral squamous cell carcinoma cells containing miR-181a-3p induce muscle cell atrophy and apoptosis by transmissible endoplasmic reticulum stress signaling. Biochem Biophys Res Commun. 2020;533(4):831-37. doi: https://doi.org/10.1016/j.bbrc.2020.09.066. [DOI] [PubMed] [Google Scholar]
- 35. Dodson S, Baracos VE, Jatoi Aet al. . Muscle wasting in cancer cachexia: clinical implications, diagnosis, and emerging treatment strategies. Annu Rev Med. 2011;62:265–79. doi: https://doi.org/10.1146/annurev-med-061509-131248. [DOI] [PubMed] [Google Scholar]
- 36. Chen X, Liang H, Zhang J,et al. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–32. doi: https://doi.org/10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 37. Narasimhan A, Ghosh S, Stretch C, et al. Small RNAome profiling from human skeletal muscle: novel miRNAs and their targets associated with cancer cachexia. J Cachexia Sarcopenia Muscle. 2017;8:405–16. doi: https://doi.org/10.1002/jcsm.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bronisz A, Wang Y, Nowicki MOet al. . Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res. 2014;74:738–50. doi: https://doi.org/10.1158/0008-5472.CAN-13-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Z, Yanfang W, Li J, et al. . Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:237–50. doi: https://doi.org/10.1016/j.canlet.2018.04.035. [DOI] [PubMed] [Google Scholar]
- 40. Yao B, Qu S, Hu R, et al. . A panel of miRNAs derived from plasma extracellular vesicles as novel diagnostic biomarkers of lung adenocarcinoma. FEBS Open Bio. 2019;9:2149–58. doi: https://doi.org/10.1002/2211-5463.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H, Zhu L, Bai M, et al. Exosomal circRNA derived from gastric tumor promotes white adipose browning by targeting the miR-133/PRDM16 pathway. Int J Cancer. 2019;144:2501–15. doi: https://doi.org/10.1002/ijc.31977. [DOI] [PubMed] [Google Scholar]
- 42. Kitdumrongthum S, Metheetrairut C, Charoensawan Vet al. . Dysregulated microRNA expression profiles in cholangiocarcinoma cell-derived exosomes. Life Sci. 2018a;210:65–75.. doi: https://doi.org/10.1016/j.lfs.2018.08.058. [DOI] [PubMed] [Google Scholar]
- 43. Ye Z, Zheng Z, Peng L. MicroRNA profiling of serum exosomes in patients with osteosarcoma by high-throughput sequencing. J Investig Med. 2020;68:893–901.. doi: https://doi.org/10.1136/jim-2019-001196. [DOI] [PubMed] [Google Scholar]
- 44. Zou X, Zhu D, Zhang Het al. . MicroRNA expression profiling analysis in serum for nasopharyngeal carcinoma diagnosis. Gene. 2020;727:144243. doi: https://doi.org/10.1016/j.gene.2019.144243. [DOI] [PubMed] [Google Scholar]
- 45. Vallabhaneni KC, Penfornis P, Xing F, et al. . Stromal cell extracellular vesicular cargo mediated regulation of breast cancer cell metastasis via ubiquitin conjugating enzyme E2 N pathway. Oncotarget. 2017;8:109861–76. doi: https://doi.org/10.18632/oncotarget.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cai J, Fang L, Huang Y, et al. . miR-205 targets PTEN and PHLPP2 to augment AKT signaling and drive malignant phenotypes in non-small cell lung cancer. Cancer Res. 2013;73:5402. doi: https://doi.org/10.1158/0008-5472.CAN-13-0297. [DOI] [PubMed] [Google Scholar]
- 47. Wang J, Yang K, Yuan Wet al. . Determination of serum exosomal H19 as a noninvasive biomarker for bladder cancer diagnosis and prognosis. Med Sci Monit. 2018;24:9307–16. doi: https://doi.org/10.12659/msm.912018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hasanzadeh M, Movahedi M, Rejali M. The potential prognostic and therapeutic application of tissue and circulating microRNAs in cervical cancer. J Cell Physiol. 2019;234:1289–94. doi: https://doi.org/10.1002/jcp.27160. [DOI] [PubMed] [Google Scholar]
- 49. He WA, Calore F, Londhe P,et al. Microvesicles containing miRNAs promote muscle cell death in cancer cachexia via TLR7. PNAS. 2014;111:4525–9. doi: https://doi.org/10.1073/pnas.1402714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fernandez G, Ferreira J, Vechetti I, et al. . MicroRNA-mRNA Co-sequencing identifies transcriptional and post-transcriptional regulatory networks underlying muscle wasting in cancer cachexia. Front Genet. 2020;11:541. doi: https://doi.org/10.3389/fgene.2020.00541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hudson MB, Rahnert JA, Zheng B, et al. miR-182 attenuates atrophy-related gene expression by targeting FoxO3 in skeletal muscle. Am J Physiol Cell Physiol. 2014;307:C314–9. doi: https://doi.org/10.1152/ajpcell.00395.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li J, Chan MC, Yu Y, et al. . miR-29b contributes to multiple types of muscle atrophy. Nat Commun. 2017;8:15201. doi: https://doi.org/10.1038/ncomms15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Soares RJ, Cagnin S, Chemello F, et al. . Involvement of MicroRNAs in the regulation of muscle wasting during catabolic conditions. J Biol Chem. 2014;289:21909–25. doi: https://doi.org/10.1074/jbc.M114.561845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang H, Deng T, Liu R, et al. . Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat Commun. 2017;8:15016. doi: https://doi.org/10.1038/ncomms15016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chen JF, Tao Y, Li Jet al. . microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7. J Cell Biol. 2010;190:867–79. doi: https://doi.org/10.1083/jcb.200911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Goljanek-Whysall K, Sweetman D, Abu-Elmagd M, et al. MicroRNA regulation of the paired-box transcription factor Pax3 confers robustness to developmental timing of myogenesis. PNAS. 2011;108:11936–41. doi: https://doi.org/10.1073/pnas.1105362108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chen JF, Mandel EM, Thomson JM, et al. . The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–33. doi: https://doi.org/10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kukreti H, Amuthavalli K, Harikumar A, et al. . Muscle-specific microRNA1 (miR1) targets heat shock protein 70 (HSP70) during dexamethasone-mediated atrophy. J Biol Chem. 2013;288:6663–78. doi: https://doi.org/10.1074/jbc.M112.390369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Crist CG, Montarras D, Pallafacchina G, et al. . Muscle stem cell behavior is modified by microRNA-27 regulation of Pax3 expression. PNAS. 2009;106:13383–7. doi: https://doi.org/10.1073/pnas.0900210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mcfarlane C, Vajjala A, Arigela H, et al. . Negative auto-regulation of myostatin expression is mediated by Smad3 and microRNA-27. PLoS One, 2014;9(1):e87687. doi: https://doi.org/10.1371/journal.pone.0087687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu J, Li R, Workeneh Bet al. . Transcription factor FoxO1, the dominant mediator of muscle wasting in chronic kidney disease, is inhibited by microRNA-486. Kidney Int. 2012;82:401–11. doi: https://doi.org/10.1038/ki.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dey B, Gagan J, Dutta A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol Cell Biol. 2011;31:203–14. doi: https://doi.org/10.1128/MCB.05013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Richardson AJ, Liu N, Basselduby R, et al. . microRNA-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. 2008;22:3242–54. doi: https://doi.org/10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Alexander MS, Kawahara G, Motohashi N, et al. . MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. 2013;20:1194–208. doi: https://doi.org/10.1038/cdd.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Luo P, He T, Jiang R, et al. . MicroRNA-423-5p targets O-GlcNAc transferase to induce apoptosis in cardiomyocytes. Mol Med Rep. 2015;12:1163–8. doi: https://doi.org/10.3892/mmr.2015.3491. [DOI] [PubMed] [Google Scholar]
- 66. Kitdumrongthum S, Metheetrairut C, Charoensawan V, et al. . Dysregulated microRNA expression profiles in cholangiocarcinoma cell-derived exosomes. Life Sci. 2018;210:65–75.. doi: https://doi.org/10.1016/j.lfs.2018.08.058. [DOI] [PubMed] [Google Scholar]
- 67. Connolly M, Paul R, Farre-Garros Ret al. . miR-424-5p reduces ribosomal RNA and protein synthesis in muscle wasting. J Cachexia Sarcopenia Muscle. 2018;9:400–16. doi: https://doi.org/10.1002/jcsm.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Okugawa Y, Toiyama Y, Hur K, et al. . Circulating miR-203 derived from metastatic tissues promotes myopenia in colorectal cancer patients. J Cachexia Sarcopenia Muscle. 2019;10:536–48. doi: https://doi.org/10.1002/jcsm.12403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kotipatruni RR, Nalla AK, Asuthkar S, et al. . Apoptosis induced by knockdown of uPAR and MMP-9 is mediated by inactivation of EGFR/STAT3 signaling in medulloblastoma. PLoS One. 2012;7:e44798. doi: https://doi.org/10.1371/journal.pone.0044798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sato T, Yamamoto T, Sehara-Fujisawa A. miR-195/497 induce postnatal quiescence of skeletal muscle stem cells. Nat Commun. 2014;5:4597. doi: https://doi.org/10.1038/ncomms5597. [DOI] [PubMed] [Google Scholar]
- 71. Runge S, Nielsen FC, Nielsen J, et al. . H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem. 2000;275:29562–9. doi: https://doi.org/10.1074/jbc.M001156200. [DOI] [PubMed] [Google Scholar]
- 72. Kallen AN, Zhou XB, Xu Jet al. . The imprinted H19 LncRNA antagonizes Let-7 MicroRNAs. Mol Cell. 2013;52:101–12. doi: https://doi.org/10.1016/j.molcel.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hasanzadeh M, Movahedi M, Rejali Met al. . The potential prognostic and therapeutic application of tissue and circulating microRNAs in cervical cancer. J Cell Physiol. 2019;234:1289–94. doi: https://doi.org/10.1002/jcp.27160 . [DOI] [PubMed] [Google Scholar]
- 74. Jin X, Chen Y, Chen H, et al. . Evaluation of tumor-derived exosomal miRNA as potential diagnostic biomarkers for early-stage non-small cell lung cancer using next-generation sequencing. Clin Cancer Res. 2017;23:5311–9. doi: https://doi.org/10.1158/1078-0432.Ccr-17-0577. [DOI] [PubMed] [Google Scholar]
- 75. Naguibneva I, Ameyar-Zazoua M, Polesskaya A, et al. . The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–84. doi: https://doi.org/10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 76. Dey BK, Gagan J, Yan Zet al. . miR-26a is required for skeletal muscle differentiation and regeneration in mice. Genes Dev. 2012;26:2180–91. doi: https://doi.org/10.1101/gad.198085.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase Enhancer of Zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–43. doi: https://doi.org/10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 78. Koutalianos D, Koutsoulidou A, Mastroyiannopoulos NPet al. . MyoD transcription factor induces myogenesis by inhibiting Twist-1 through miR-206.J Cell Sci2015;128:3631–45. doi: https://doi.org/10.1242/jcs.172288. [DOI] [PubMed]
- 79. Parajuli P, Kumar S, Loumaye A. et al. Twist1 activation in muscle progenitor cells causes muscle loss akin to cancer cachexia. Dev Cell. 2018;45:712–25.e6. doi: https://doi.org/10.1016/j.devcel.2018.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhang Y, Zhang H, Liu Z. MicroRNA-147 suppresses proliferation, invasion and migration through the AKT/mTOR signaling pathway in breast cancer. Oncol Lett. 2016;11:405–10. doi: https://doi.org/10.3892/ol.2015.3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Salazar-Degracia A, Busquets S, Argilés Jet al. . Effects of the beta agonist formoterol on atrophy signaling, autophagy, and muscle phenotype in respiratory and limb muscles of rats with cancer-induced cachexia. Biochimie. 2018;149:79–91.. doi: https://doi.org/10.1016/j.biochi.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 82. Wang L, Zhao Y, Bao X, et al. . LncRNA Dum interacts with Dnmts to regulate Dppa2 expression during myogenic differentiation and muscle regeneration. Cell Res. 2015;25:335–50.. doi: https://doi.org/10.1038/cr.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Shen L, Han J, Wang H, et al. . Cachexia-related long noncoding RNA, CAAlnc1, suppresses adipogenesis by blocking the binding of HuR to adipogenic transcription factor mRNAs. Int J Cancer. 2019;145:1809–21. doi: https://doi.org/10.1002/ijc.32236. [DOI] [PubMed] [Google Scholar]
- 84. Li Z, Cai B, Abdalla BA, et al. . LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J Cachexia Sarcopenia Muscle. 2019;10:391–410.. doi: https://doi.org/10.1002/jcsm.12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ghouzzi VE, Merrer ML, Perrin-Schmitt F, et al. . Mutations of the TWIST gene in Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–6. doi: https://doi.org/10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- 86. Kim HK, Yong SL, Sivaprasad Uet al. . Muscle-specific microRNA MIR206 promotes muscle differentiation. J Cell Biol. 2006;174:677–87. doi: https://www.jcb.org/cgi/doi/10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Williams AH, Valdez G, Moresi V, et al. . MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;326:1549–54. doi: https://doi.org/10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Curtis A, Heath C, Rudolf W. MIR-206 regulates connexin43 expression during skeletal muscle development. Nucleic Acids Res. 2006;34::5863–71.. doi: https://doi.org/10.1093/nar/gkl743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Rosenberg MI, Georges SA, Asawachaicharn A, et al. . MyoD inhibits Fstl1 and Utrn expression by inducing transcription of miR-206. J Cell Biol. 2006;175:77–85.. doi: https://doi.org/10.1083/jcb.200603039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Suzuki A, Scruggs A, Iwata J. The temporal specific role of WNT/β-catenin signaling during myogenesis. J Nat Sci2015;1:e143. PMCID: PMC4499510 [PMC free article] [PubMed]
- 91. Chandran S, Guo T, Tolliver Tet al. . Effects of serotonin on skeletal muscle growth. BMC Proceedings. 2012;6:O3. doi: https://doi.org/10.1186/1753-6561-6-S3-O3. [Google Scholar]
- 92. Rommel C, Bodine SC, Clarke BA, et al. . Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat Cell Biol. 2001;3(11):1009-13. doi: https://doi.org/10.1038/ncb1101-1009. [DOI] [PubMed] [Google Scholar]
- 93. Puntita S, Nares T, Eduard Met al. . MicroRNAs regulate cellular ATP levels by targeting mitochondrial energy metabolism genes during C2C12 myoblast differentiation. PLoS One. 2015;10:e0127850. doi: https://doi.org/10.1371/journal.pone.0127850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mubaid S, Ma J, Omer A, et al. . HuR counteracts miR-330 to promote STAT3 translation during inflammation-induced muscle wasting. PNAS. 2019;116:17261–70. doi: https://doi.org/10.1073/pnas.1905172116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Powrozek T, Mlak R, Brzozowska Aet al. . MiRNA-130a significantly improves accuracy of SGA nutritional assessment tool in prediction of malnutrition and cachexia in radiotherapy treated head and neck cancer patients. J Cachexia Sarcopenia Muscle, 2018;9:1180. doi: https://doi.org/10.3390/cancers10090294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Liang J, Zhang X, He S, et al. . Sphk2 RNAi nanoparticles suppress tumor growth via downregulating cancer cell derived exosomal microRNA. J Control Release. 2018;286:348–57. doi: https://doi.org/10.1016/j.jconrel.2018.07.039. [DOI] [PubMed] [Google Scholar]
- 97. Ohno S-i, Takanashi M, Sudo K, et al. . Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–91. doi: https://doi.org/10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Freire PP, Fernandez GJ, Cury SS, et al. . The pathway to cancer cachexia: MicroRNA-Regulated networks in muscle wasting based on integrative Meta-Analysis. Int J Mol Sci. 2019;20::1962doi: https://doi.org/10.3390/ijms20081962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Pillai VB, Gupta MP. Cellular mechanisms promoting cachexia and how they are opposed by sirtuins. Can J Physiol Pharmacol. 2019;97:235–45. doi: https://doi.org/10.1139/cjpp-2018-0479. [DOI] [PubMed] [Google Scholar]
- 100. Zhao R, Li FQ, Tian LLet al. . Comprehensive analysis of the whole coding and non-coding RNA transcriptome expression profiles and construction of the circRNA-lncRNA co-regulated ceRNA network in laryngeal squamous cell carcinoma. Funct Integr Genomics. 2019;19:109–21. doi: https://doi.org/10.1007/s10142-018-0631-y. [DOI] [PubMed] [Google Scholar]