Abstract
As more data is generated from medical attendances and as Artificial Neural Networks gain momentum in research and industry, computer-aided medical prognosis has become a promising technology. A common approach to perform automated prognoses relies on textual clinical notes extracted from Electronic Health Records (EHRs). Data from EHRs are fed to neural networks that produce a set with the most probable medical problems to which a patient is subject in her/his clinical future, including clinical conditions, mortality, and readmission. Following this research line, we introduce a methodology that takes advantage of the unstructured text found in clinical notes by applying preprocessing, concepts extraction, and fine-tuned neural networks to predict the most probable medical problems to follow in a patient’s clinical trajectory. Different from former works that focus on word embeddings and raw sets of extracted concepts, we generate a refined set of Unified Medical Language System (UMLS) concepts by applying a similarity threshold filter and a list of acceptable concept types. In our prediction experiments, our method demonstrated AUC-ROC performance of 0.91 for diagnosis codes, 0.93 for mortality, and 0.72 for readmission, determining an efficacy that rivals state-of-the-art works. Our findings contribute to the development of automated prognosis systems in hospitals where text is the main source of clinical history.
Keywords: Clinical notes, MIMIC-III, QuickUMLS, Patient trajectory prediction, Computer-aided prognosis
Introduction
The use of Electronic Health Records (EHRs) [1] to register the patients’ clinical history has increased in medical systems all over the world. EHRs carry many kinds of information including diagnosis, procedures, symptoms, exams, and textual notes written by various professionals involved in the healthcare activity. Such data describes the clinical trajectory of a given patient, that is, her/his sequence of clinical events (admissions) along time, also known as longitudinal medical data [2]. Many works have explored these different kinds of information, but since EHRs are complex, there is room for further investigation. At the same time, the last decade was a turning point for methods based on Artificial Neural Networks whose applicability remarkably advanced with the advent of more processing power, improved algorithms, large data availability, and powerful programming frameworks, which led to the set of techniques widely known as Deep Learning [3]. While supervised Deep Learning leverages the use of data features, such as recurrent combinations of diagnoses or procedures associated with a specific disease, it has become an efficient approach in tasks related to clinical prediction [4].
A promising way of exploring the latent information that lies within EHRs is to use free-text clinical notes [5] written by healthcare professionals. This kind of information is more challenging than the structured information found in EHRs, like those based on standard diagnosis codes: free text is unstructured and highly granular; it carries ambiguities, redundancies, and non-obvious semantics; a set of impreciseness that does not favor computational approaches. The length of clinical notes varies — they can be very long or very short, they can follow a standard structure or be in the form of informal snippets; they can have a number of common sections or result from erratic writing. Furthermore, it is noisy since written text may contain typos and grammatical errors. Yet, previous works have exploited the use of free text in tasks such as predicting the patients’ mortality [6], and readmission [7]. In a similar line of investigation, we use Machine Learning techniques to predict facts related to the patients’ clinical future, including the most probable diagnosis codes, the chances of mortality, and the probability of readmission based on what was annotated in her/his EHR. We tackle the issues of free-text clinical notes by using Natural Language Processing techniques coupled with Deep Learning pattern-recognition capabilities.
Given a sequence of hospital admissions each one comprised of a set of structured diagnosis codes, existing works have proposed computer-aided systems that are able to predict the most probable clinical conditions that a patient is subject to in her/his future. Such systems have achieved good results in terms of anticipating the conditions of patients — refer to Section 3; the drawback, though, is that many hospital systems do not rely on well-curated coding systems, but rather on the free text generated by professionals. This gap asks for computer-aided systems able to digest hundreds, even thousands, of lines of text. This inference has straight applications including preventive medicine, facilitated learning of years of patient history, confirmatory diagnosis, and clinical recommendation.
More specifically, our goal is to use free text to predict the clinical trajectory of a patient, that is, her/his future conditions which could occur following her/his last admission. We work with three modalities of prediction: diagnosis codes prediction, mortality prediction, and readmission prediction. We describe a three-step methodology: (i) we employ Natural Language Processing techniques to preprocess a large set of real-world clinical notes, making them more adequate for a systematic extraction of concepts — explained in Section 4.3; (ii) we extract medical concepts from the notes by using the Unified Medical Language System (UMLS) [8] — details presented in Section 4.4; and (iii) we use the extracted clinical concepts to feed a Neural Network architecture whose output is a set of probabilities indicating the most likely clinical conditions that a given patient will pass through — presented in Section 4.5. We achieve results that compare to the state of the art with respect to metric Area Under the Receiver Operating Characteristic Curve (AUC-ROC) concerning the prediction of diagnosis codes, mortality chances, and readmission likelihood, as we discuss in Sections 3 and 5. Our results demonstrate the feasibility of computer-aided prognosis based on textual notes reaching prediction capabilities that, arguably, approach the needs of real-world applications. At the same time, we elucidate the required steps to use the notes not only for prediction, but for medical-based Machine Learning in general with insights into why our protocol works.
We summarize our contributions as follows:
Method: we introduce a methodology that departs from the preprocessing of clinical notes, evolving to the use of the Unified Medical Language System to transform complex textual snippets into structured sets of encoded concepts — we advance in comparison to the state of the art by using a more elaborated NLP process able to capture a refined set of medical concepts;
Broad experimentation: by using a real-world dataset, we experiment on different concept-extraction settings and neural-network architectures discussing the traits that favored our results — we compare to previous works tracing explanations and suggestions for future work;
Principles: we provide background and formalisms that could assist future researchers in improving data representation based on clinical notes for computer-aided prognosis — for our allegations, we discuss learned lessons about what drove our decisions, raising hypotheses worthy to follow in future works.
Background
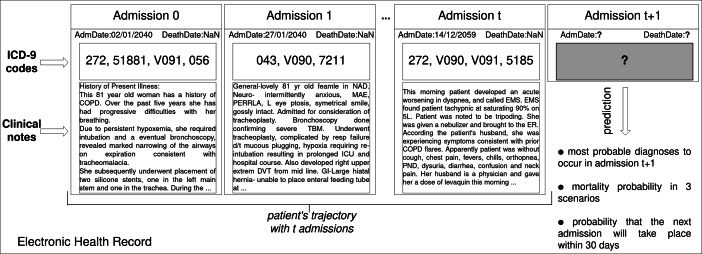
We consider a sequence of admissions (or trajectory) of a patient to a hospital, as stored in her/his EHR. From a structural point of view, the patients’ trajectories refer to sequences of chronologically ordered admissions. Each admission consists of information such as medications, diagnoses, procedures, the three of which represented by means of standard coding systems (such as the International Classification of Diseases (ICD) [9]); they come along with clinical notes that explain or extend the concepts provided in a coded format. In this work, given the clinical notes (text) found in a patient’s trajectory, the goal is to predict what are the most probable clinical conditions in the future, as illustrated in Fig. 1.
Fig. 1.
Illustration of the clinical data prediction problem. Given a set of hospital admissions (Electronic Health Records), each one composed of admission date, death date, ICD-9 codes and textual clinical notes, we use neural network techniques to predict the patient’s clinical conditions including diagnosis, mortality, and readmission
Clinical Notes Representation
To go beyond the traditional Bag-of-Words approach [10], we propose to build a conceptual representation of the patients’ trajectories. The representation is built by annotating the clinical textual notes with medical entities obtained from the knowledge base called Unified Medical Language System (UMLS)[8]. UMLS is a Metathesaurus maintained by the US National Library of Medicine; it contains information about health-related concepts, their different names, and the relationships among them. UMLS was created in 1986 and is organized as a broad compendium of many controlled vocabularies in the biomedical sciences; it also comprises a set of tools whose main purpose is to promote the creation of effective and interoperable biomedical information systems and services, including EHRs [11]. By means of automatic labeling tools and UMLS, a given medical sentence, like the ones found in the clinical annotations, can be associated with canonical medical concepts previously cataloged in the Metathesaurus. Each concept, in turn, is systemically associated with a Concept Unique Identifier (CUI) [12]. For instance, the concept “headache”, whose CUI is C0018681, is related to the strings “headache”, “cranial pain”, and many others that already appear in the medical literature and in the clinical practice. In addition, each concept has a type to indicate its role; the type is encoded as a Type Unique Identifier (TUI). In the case of the concept headache, for example, the type is “Sign or Symptom”, code T184.
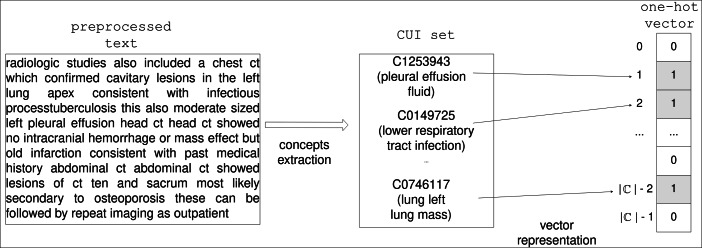
Given a medical textual sentence, software tools based on UMLS and on string-matching algorithms — such as MetaMap or QuickUMLS (our choice) — can retrieve a set of CUIs, each one categorized as a specific TUI. This possibility addresses the problem of using unstructured medical text in computing systems. By converting the notes of a patient’s hospital admission to sets of codes, the information becomes discretely numerical making it ideal for use by Machine Learning methods. In our work, given a corpus of free textual notes originated from a database of EHRs related to a number of patients’ admissions, the set union of all the corresponding CUIs produces an ordered set with elements. From this set, we are able to represent one given admission as a binary vector of length , in which the i-th entry has value 1 if the i-th CUI is present in the textual note, or 0 otherwise.
Formally, let be the set of all the detected CUI codes. Each admission is represented as a one-hot encoding vector of size . That is, by having ci as the ith element of the vector, we have:
| 1 |
As illustrated in Fig. 2.
Fig. 2.
Hot vector CUI-based representation of the textual notes found in an EHR
Diagnoses Representation
The International Classification of Diseases (ICD) is a coding system used by medical institutions; it is held by the World Health Organization. Although ICD is highly structured and precise, its 10th version, for instance, comprises over 70,000 codes, which poses challenges for statistical and computing purposes — many times the encoded information is not registered to reflect the actual details, being specific just as much as to maximize reimbursement purposes; in other situations, a specific code might carry details way too specific for an analytical purpose; not to mention the numerical issues that arise from such a large cardinality. For Machine Learning, the problem comes from the fact that the statistical distribution of the codes’ usage might be sparse or imbalanced, which does not favor a robust learning. As these problems are recurrent, they motivated the creation of initiative Clinical Classification Software (CCS) [13], which maps the ICD codes to a less granular set. If we take breast cancer as an example, there are more than a hundred different ICD-9 codes referring to the several manifestations of this disease, each one with just a few different details; on the contrary, there is only one CCS code to describe it. As a result, the ICD-9 standard, originally with nearly 15,000 codes is mapped to only 283 unique CCS codes. Despite the loss of details, the diagnosis information provided by the CCS standard has been used with good results in several works [14–16].
Related Works
Previous works have demonstrated efficacy in using free-text clinical notes to perform Machine Learning tasks. Sushil et al. [17] created unsupervised dense patient representations from clinical notes over dataset MIMIC-III [18]. They focus on different techniques to learn dense patient representations using only textual data, exploring the usage of two neural representation-learning architectures — a Stacked Denoising Autoencoder (SDAE), and the paragraph vector architecture doc2vec. They also employ two independent representations; the first one is based on the doc2vec architecture to get an embedding dense vector; the other one uses the UMLS concept recognizer CLAMP toolkit [19], which maps a given note to a set of UMLS CUI codes (refer to Section 2.1). Next, they transferred the representations from the complete patient space to different supervised tasks with the aim to generalize on the tasks for which they had limited labeled data. By using either one of the representations, or the concatenation of the two, the authors managed to perform four tasks: mortality prediction, primary diagnostic, procedural category, and gender classification. They did so by using a simple fully connected architecture. The work of Sushil et al. provides valuable experimentation on the usage of different combinations of dense and sparse data representations to which we compare and discuss demonstrating superior results with respect to mortality prediction, and primary diagnostic.
Grnarova et al. [6] use a document embedding layer to encode the clinical notes, obtaining a vector that is semantically machine-readable. Their goal is, given a patient record, to predict the mortality probability in three scenarios: (1) during the hospital stay, (2) within 30 days after discharge, or, (3) within 1 year after discharge. They use a two-layer convolutional neural network (CNN) architecture; the first layer creates vector representations of the sentences in the clinical note, and the second layer combines the sentence vectors into one single vector that represents the entire note. For optimization, they replicate the classification loss at the sentence level, improving the regularization of the first layer. For this task, we adapted our codes prediction method to a classification problem concerning the three possible mortality scenarios; our results significantly outperformed those of Grnarova et al.
Dubois et al. [16] map clinical notes to CUI concepts that are reduced to a Bag-of-Words (BoW) vector. The vectors are used as input to create patient representations by employing two different techniques. The first one, called embed-and-aggregate, embeds the BoW vectors using method GloVe [20] and then aggregates the resulting representations using operators min, max, or mean. The other one uses a Recurrent Neural Network (RNN) to process the BoW vectors in supervised fashion using CCS diagnosis codes; the final hidden state of the network is used as a representation of the patients. The two representations are used with L2 logistic regression for predicting mortality, inpatient admission, and emergency room visits. These tasks are simpler than predicting the diagnosis codes for a patient; yet, we obtained results significantly superior for similar metrics and, specifically, for the task of mortality prediction.
ClinicalBERT by Huang et al. [7] uses bidirectional encoder representations from transformers (BERT) [21]. BERT achieved state-of-the-art performance for a wide range of tasks in Natural Language Processing such as Question Answering, or Named Entity Recognition. ClinicalBERT was experimented over the MIMIC-III clinical notes with improvements in transposing BERT to the clinical domain. This model, which is remarkably more complex than ours, uses contextualized word embeddings based on tokens; such embeddings are input to the attention mechanism of BERT, which is trained to predict the readmission probability of a given patient. The method outperformed models such as the bidirectional Long Short-Term Memory (BiLSTM), and Logistic Regression using a Bag-of-Words representation in the task of 30-day patient readmission prediction. We achieved results comparable to the work of Huang et al. with a more versatile, yet simpler, architecture.
In Section 6, we compare our method to these related works considering metrics Precision@, Recall@ and, particularly, metric AUC-ROC — for details on the metrics, refer to Section 5.1. In Table 1, for quick reference, we present the characteristics of each work considered hereafter. In comparison to previous works, we further elaborate on the concepts-extraction step, which produced a set of medical concepts arguably with higher potential for trajectory prediction.
Table 1.
Related works summary
| Work | Prediction Task(s) | Methodology | AUC-ROC |
|---|---|---|---|
| Sushil et al. [17] | Primary | SDAE + | 0.94 |
| diagnostic | Doc2Vec then FFN | (Hospital) | |
| Mortality | 0.81 | ||
| (30 days) | |||
| 0.83 | |||
| (1 year) | |||
| Grnarova et al. [6] | Mortality | CNN (supervised) | 0.963 |
| (Hospital) | |||
| 0.858 | |||
| (30 days) | |||
| 0.853 | |||
| (1 year) | |||
| Dubois et al. [16] | Emergency room | Embed-and-aggregate | ER Visit |
| (GloVe;min/max/mean) | GloVe: 0.775 | ||
| Inpatient admission | or RNN | RNN: 0.76 | |
| Mortality | L2 regularized | Inpatient | |
| logistic regression | admission | ||
| GloVe: 0.80 | |||
| RNN: 0.81 | |||
| Mortality | |||
| GloVe: 0.9 | |||
| RNN: 0.86 | |||
| Huang et al. [7] | Readmission | Fine-tune pre-trained | 0.768 |
| BERT using | (30-day readmission) | ||
| clinical data | |||
| (ClinicalBERT) | 0.673 | ||
| (24h-48h readmission) | |||
| 0.674 | |||
| (48h-72h | |||
| readmission) |
Materials and Methods
Methodology Overview
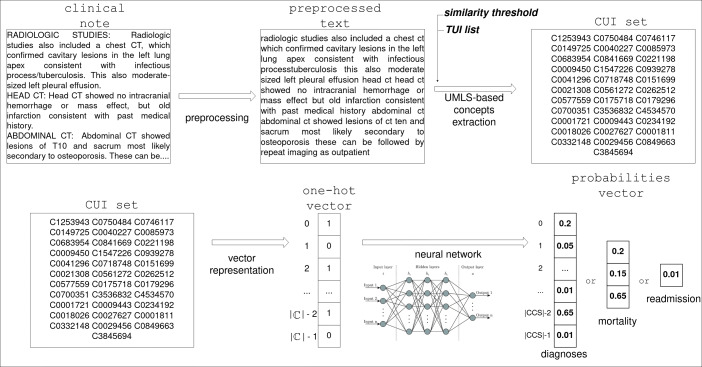
In this section, we introduce our methodology which consists of the following steps, illustrated in Fig. 3:
First, we perform text preprocessing on the clinical notes to clean the textual information, making it denser in terms of cardinality and meaning of words, as detailed in Section 4.3;
Then, we extract clinical concepts from the notes using a UMLS concept recognizer — presented in Section 4.4;
Finally, we use the detected concepts, represented as one-hot vectors, to experiment on different neural network architectures for patient trajectory prediction, as discussed in Section 4.5.
Fig. 3.
Process flow of our methodology
As illustrated in Fig. 3, the output of our neural network is a vector of probabilities. The cardinality depends on the target task — for instance, in the case of diagnoses prediction, it is equal to the number of possible CCS codes found in the database. For each code, the vector holds a probability value indicating the likelihood of the corresponding clinical condition to manifest in the patient’s future.
In turn, predicting mortality corresponds to computing the likelihood of the patient dying in one of three circumstances: (i) during the hospital stay; (ii) within up to 30 days after discharge; and (iii) within 1 year after discharge. This computation is useful to estimate the severity of a patient’s condition, and to decide the amount of attention required [17]. To achieve this task, the output of our prediction architecture was set to a vector with three values, each one corresponding to one of the three mortality circumstances.
Predicting the readmission corresponds to computing the single probability of the patient returning to the hospital within 30 days after discharge, having an output set to one single value.
Following, we explain the architectures tested for our problems. We used the same architecture for all the three tasks, adapting the output accordingly and carrying out the network re-training for each task. The complete code for reproducing our work is available at https://github.com/JamilProg/patient_trajectory_prediction.
Dataset
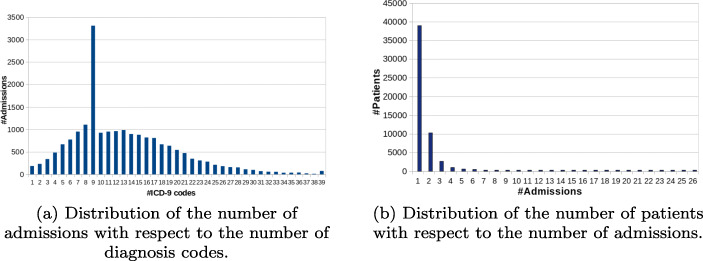
We work with the open-access dataset Medical Information Mart for Intensive Care III (MIMIC-III) [22], a large critical care database released by the Massachusetts Institute of Technology. This dataset integrates deidentified, comprehensive clinical data of patients admitted to the critical care unit of the Beth Israel Deaconess Medical Center in Boston, MA, USA. It contains data from approximately 48,520 patients collected from 2001 through 2012. For each patient, there is a set of admissions, each one consisting of textual clinical notes, and a sequence of diagnoses given in ICD-9 codes (on average 13 diagnoses per admission) — refer to Fig. 4a. In this work, the clinical notes and diagnosis codes are employed to predict the patients’ trajectories. Figure 1 illustrates this structure. We use only the admissions that carry clinical notes and the patients with at least two admissions; as a result, we worked with 7,314 patients — refer to Fig. 4b. Because of privacy, the data is anonymized — dates are fictitious and names are tossed off. We chose MIMIC-III because it is a high-quality public available dataset, allowing for direct comparison to other works.
Fig. 4.
Basic distributions related to dataset MIMIC-III
Notes Preprocessing
Medical notes are often unstructured and noisy, demanding the use of Natural Language Processing techniques to reduce the unusable information. The first preprocessing was to remove the anonymized information that, in MIMIC, is unreadable but kept in the notes. We continued by converting the text to lowercase, tossing off special characters, stop-words, rare words (which are often erroneous, such as typos), multiple spaces in a row, and duplicated, special or noisy characters, segmenting free-text to paragraphs, converting numbers to text and so on. The paragraph segmentation is important to the concepts extraction stage; this is because most UMLS concepts-extraction tools work on a paragraph-by-paragraph basis and the concepts extraction depends on the input quality.
Concepts Extraction
We use the software QuickUMLS, by Soldaini et al.’s [23], for concepts extraction, available at software repository GitHub [24]. We chose this extractor because it offers computation speed and high accuracy based on algorithm SimString to evaluate the clinical concepts from a given text. Given two strings to compare, it computes a score between 1 (perfect match) and 0 (no match at all). We retrieve the concepts by defining a threshold value with respect to the similarity of the sentences in the text and the cataloged concepts. We reduced the number of candidate concepts by increasing the threshold and restricting concepts to a selected list of semantic Type Unique Identifiers (TUIs).
For experimentation, we defined two TUI lists named α and β — the lists are available in the GitHub repository of this project. The TUI identifiers were selected considering types more related to the clinical domain in contrast to those related to, for example, non-medical concepts, or non-chemical substances.
Semantic type list α consists of 47 types related to chemicals, disorders, clinical phenomena, and physiology — for example, we kept TUI T109 for “Organic Chemical”;
Semantic type list β consists of 85 types; an extension of α, with additional types related to living beings, temporal concepts, anatomy, and procedures; for example, we added TUI T030 for “Body Space or Junction”.
Our lists were approved by two medical professionals (radiology and visceral surgery) affiliated with the Centre Hospitalier Emile Roux, in the city of Le Puy-en-Velay, France. They provided opinions on the set of TUIs with respect to their adequacy in summarizing the clinical past of the patients; their feedback allowed us to formulate our two sets.
Clinical Future Prediction
After running QuickUMLS, each admission is represented as a set of concepts represented in the form of CUI codes. As explained in Section 2, each set of codes becomes a one-hot encoding vector, ideal for Machine Learning — after our concepts extraction, we ended up with vectors whose length could go from 16,000 to 39,000, that is, the number of unique concepts detected. The next step is to build artificial neural network models to predict the most probable future clinical conditions of a given patient, including the most probable diagnoses, mortality risk, and 30-day readmission expectation. These problems can be treated in the form of a multi-label classification to which we propose two different strategies, discussed next. We report on strategies that passed through many rounds of experimentation for fine-tuning of hyper-parameters, including number of epochs (50, 100, 500, 1500, 3000, 5000, 8000, and 10 iterations without improvement), learning rate (10− 1, 10− 2, and 10− 3), batch size (10, 50, 100, and 200), optimizers (Adam, Adadelta and Stochastic Gradient Descent) [25, 26], number of neurons in the hidden layer (10, 50, 100, 200, 1000, 5000, 10000, and 15000), dropout (0, 0.2, 0.5), and including or not Xavier initialization for weights [27]. We found out that the simpler configuration, detailed in the next sections, yielded the best results; probably because, given the limited size of the dataset, we achieved the balance between size, information richness, and model complexity. We also tried configurations with 1, 2, and 3 hidden layers, but the more layers the worse the performance. Empirically, we verified that more layers increased the number of weights, which demanded more iterations to converge, but always with a performance decay in comparison to configurations with fewer layers.
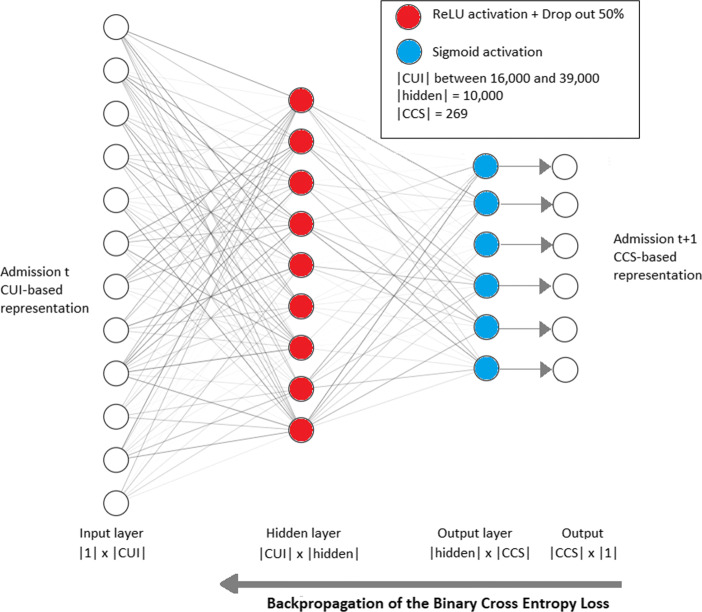
The first solution we employed was a fully connected Feed-Forward architecture; Fig. 5 illustrates this architecture for the task of diagnoses prediction, for the other tasks, only the output layer is altered, which demands re-training. This solution answers for a clinical prediction that takes into account only the patient’s last admission, instead of her/his entire history of admissions. The corresponding network contains an input layer, one single fully connected hidden layer, and one output layer whose cardinality of neurons is 269 (number of CCS codes) for diagnoses, 3 for mortality, and 1 for readmission. The output logits pass through a sigmoid activation (squeezing) to produce numbers between 0 and 1, as for setting a probability distribution. During training, the network evaluation comes from the supervised comparison of the predicted probabilities and the actual values at time t + 1; the evaluation is expressed by a Binary Cross-Entropy loss function that guides the backpropagation feedback to the network. The loss function is given by:
| 2 |
where N is the number of output probabilities, yi is 1 if the i-th condition is true for a specific training sample, and p(yi) is the output probability computed with the sigmoid operation. Notice that the number of output probabilities depends on the task, either diagnoses, mortality, or readmission, as explained in Section 4.1.
Fig. 5.
Fully connected Feed-Forward network to predict the future diagnoses of a patient considering her/his last admission only. The same architecture is used for the tasks of mortality and readmission, but each one with its specific output layer, and training/testing sessions
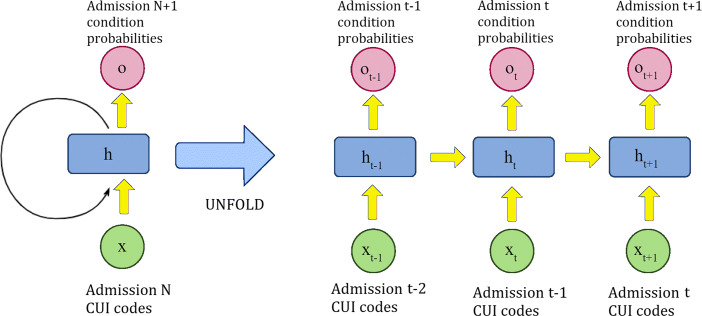
The second solution was to use a Recurrent Neural Network (RNN), whose underlying mechanism is illustrated in Fig. 6. It uses a recurrent neuron capable of benefiting from the whole history of admissions. This strategy is interesting because it allowed us to evaluate the importance of the history of the patient with respect to her/his clinical future. The corresponding network comprises one input layer, one hidden RNN layer, and one output layer. For a given patient, we use the first admission to predict the clinical condition of the second admission. Then, given the first and second admissions, we predict the condition of the third one, and so on; so, for n admissions, we perform n − 1 experimental predictions — thus, admission n, the last one, is never used for training. Obviously, each task (diagnoses, mortality, or readmission) demanded a specific training/testing. The strength of this architecture comes from the fact that if there is any temporal dependency between admissions, then it will learn it. However, since MIMIC is an intensive care database, the temporal dependencies are not strong, which impacted our results for RNNs.
Fig. 6.
Recurrent neuron unfolding mechanism to predict the future clinical condition of a patient considering her/his entire admissions history
Experiments and Results
As explained in Section 4.4, two factors define our material for experimentation: the threshold similarity and the TUI list. Accordingly, using QuickUMLS, we produced four datasets by combining TUI lists α and β and thresholds 0.7 and 0.9 (the higher the threshold, the more restrictive it is), as presented in Table 2. The similarity threshold of QuickUMLS has a default value, which is 0.7. When the threshold is smaller than this value, we capture too many UMLS concepts per admission, which leads to memory issues. On the other hand, increasing the threshold reduces the number of captured concepts per admission. Hence, we made experiments with 0.7, 0.8, and 0.9 as threshold values. Using a threshold of 1 (perfect match) does not make sense because we would miss too many concepts.
Table 2.
The dataset configurations used in our experiments
| Dataset | Threshold | TUI list | Number of CUI codes |
|---|---|---|---|
| A | 0.7 | α (47 types) | 33,752 |
| B | 0.7 | β (85 types) | 39,049 |
| C | 0.9 | α (47 types) | 16,723 |
| D | 0.9 | β (85 types) | 22,820 |
We experimented with other parameters, like thresholds smaller than 0.7 and more comprehensive TUI lists. The resulting datasets were excessively large without, necessarily, providing prediction improvements. Accordingly, we report results related only to datasets A, B, C, and D. Furthermore, in Sections 5.2, 5.3, and 5.4, we experiment only over the task of diagnoses prediction, the more complex of the three tasks, whose results are, arguably, generalizable to the other tasks regarding architectural definitions. In Section 5.5, we report on the experiments of the three tasks diagnoses, mortality, and readmission.
Our networks were implemented over framework PyTorch (https://pytorch.org/). For each task, diagnoses, mortality, or readmission, the training step occurred during 5,000 epochs for the FFN, and 1,500 epochs for the RNN; we had batches of size 100 for the FFN, and 10 for the RNN. We used hardware NVIDIA Quadro P6000 GPU. In every case, we split the dataset using 80% for training and 20% for testing, averaging the results after a 5-fold cross-validation. For the network based on the Feed-Forward architecture, we used optimizer Adam for mortality prediction, and optimizer Stochastic Gradient Descent (SGD) [26] for diagnoses and readmission. When experimenting with Recurrent Neural Networks, we used optimizer Adam.
Evaluation Metrics
Due to the characteristics of the problem, two metrics commonly used for recommendation systems are employed: Precision and Recall at the top-k recommendations. In our case, the top k recommendations refer to the k diagnosis codes that received the highest probabilities of appearing in the next admission. Precision@k refers to the percentage of all the actual codes that appear in the top-k recommendations, expressed by:
| 3 |
Recall@k refers to the percentage of recommended codes that are correct with respect to the entire set of correct codes, expressed by:
| 4 |
Notice from the very equations (3) and (4) that when the value of k increases, the value of Precision decreases, while the value of Recall increases. So, a good performance is indicated by a slowly decaying Precision, and by a steadily increasing Recall.
The Area Under the Receiver Operating Characteristic Curve (AUC-ROC) is also computed; it refers to a metric based on the concepts of Sensitivity and Specificity designed to measure the ability of a binary classifier along the spectrum of its discriminative threshold. We consider a given prediction as truly positive if it belongs to the actual answer set; and truly negative otherwise. This procedure allows the construction of a confusion matrix for different thresholds over the probability scores. For non-binary classification, we compute the AUC-ROC in one-versus-the-rest fashion, then average.
Experiments on the Parameters for CUI Selection
Our first round of experiments aimed at elucidating the impact of the CUI selection in the prediction performance. For this initial experiment, we relied on the task of diagnoses prediction only, the aim was to identify the most appropriate set of CUI codes to be used in the other tasks of mortality and readmission. Accordingly, the input was the set of clinical text notes converted to a TUI-filtered set of CUI codes; and the output was a set of predicted CCS codes — as discussed in Section 2. This course of action summarizes our problem setting, which holds for all the experiments.
For these first experiments, we used the Feed-Forward neural network architecture described in Section 4.5. The protocol was to compute the average of 5-fold cross-validation experiments having the dataset shuffled for each run.
Table 3 presents the metrics computed over the four dataset configurations — the higher the better for all of them. From the table, one can see that dataset D had the best performance, followed by dataset B, dataset C, and dataset A, in order of higher performance. The results indicate that the TUI list had the highest impact — the more comprehensive list β produced the best results (datasets D and B). Next, the threshold made a significant difference as the highest threshold of 0.9 produced the best results for both lists β and α.
Table 3.
Diagnoses prediction metrics Precision@, Recall@, and AUC-ROC computed over the four dataset configurations using a Feed-Forward network
| Dataset | P@1 | P@2 | P@3 | R@10 | R@20 | R@30 | AUC-ROC |
|---|---|---|---|---|---|---|---|
| A (0.7 & α) | 0.732 | 0.671 | 0.624 | 0.382 | 0.563 | 0.677 | 0.901 |
| B (0.7 & β) | 0.742 | 0.679 | 0.631 | 0.387 | 0.570 | 0.683 | 0.905 |
| C (0.9 & α) | 0.729 | 0.672 | 0.627 | 0.385 | 0.567 | 0.681 | 0.903 |
| D (0.9 & β) | 0.750 | 0.688 | 0.638 | 0.392 | 0.576 | 0.689 | 0.911 |
Highest marks in boldface
Although our first results are significant, with metric marks ranging from 40% to more than 70%, in the next section, we explore the possibility of using additional information to aid in the prediction task, as suggested by Pham et. al. [28].
Experiments on Combining Text and Diagnosis Codes
Since neural networks learn patterns from data, they can benefit from additional information, provided this aid is non-redundant and non-noisy. Accordingly, in this round of experiments, we used the diagnosis codes that constitute the EHRs together with the textual notes to perform the diagnoses prediction task. The 269 CCS codes, in the form of one-hot vectors, were concatenated to the CUI-coded vectors derived from the textual notes. Now, the input to the problem was a set of clinical text notes (CUI codes) and of diagnosis codes (CCS codes), and the output was a set of CCS codes. We used the same Feed-Forward network as of Section 5.2, altering only the cardinality of the first two layers. The CUI codes were generated using the best parameters (threshold 0.9 and TUI list β) detected in the first round of experiments.
Table 4 presents the results of using CUI-only, CUI+CCS, and CCS-only as input. By comparison, one can see that the use of the diagnosis CCS codes combined with the CUI codes significantly improved the prediction performance. The Precision@ increased by over 4%, while the Recall@ increased by over 3%. The improvement is even higher when compared to the use of CCS codes only — the worst result, discarding the hypothesis that the structured CCS data would be enough for prediction.
Table 4.
Diagnoses prediction performance comparison regarding inputs CUI codes only, CUI codes combined to CCS codes, and CCS codes only, using the best QuickUMLS settings threshold 0.9 and TUI list β
| Input | P@1 | P@2 | P@3 | R@10 | R@20 | R@30 | AUC-ROC |
|---|---|---|---|---|---|---|---|
| CUI-only | 0.750 | 0.688 | 0.638 | 0.392 | 0.576 | 0.689 | 0.911 |
| CUI + CCS | 0.778 | 0.723 | 0.677 | 0.414 | 0.597 | 0.706 | 0.913 |
| CCS-only | 0.728 | 0.679 | 0.640 | 0.390 | 0.561 | 0.668 | 0.905 |
Highest marks in boldface
Discussion on the Codes Prediction Results
These results directly state that the first-ranked probabilities predicted by our network agreed to the actual diseases observed in time t + 1 with a high precision. They indicate the feasible use of our method in a real-world scenario; the Precision@1 metric states that in 77.8% of the cases, the code with the highest probability was an actual CCS code to manifest in the next admission. Meanwhile, the Recall@30 metric indicates that among the 30-top ranked probabilities, the system was able to foresee 70.6% of all the CCS codes to appear in the next admission. A tool with such accuracy has the potential to make recommendations to the physician, who will be able to digest the clinical history of the patient in less time.
Table 5 presents the results of the McNemar test [29] to verify the null hypothesis that the probability of CUI+CCS being incorrect is the same as that of the CUI-only being incorrect. In other words, we test whether using the CCS data had an actual influence on the results. With a χ2 value of 47.44, the null hypothesis can be discarded with strong evidence (p-value of 0.000001). Numerically, one can see that CUI+CCS makes fewer mistakes than CUI-only; the McNemar test states that this difference is significant.
Table 5.
Statistical significance of the results presented in Table 4: McNemar test with Yates correction of 1.0 for diagnosis codes prediction
| Contingency matrix | p-value | χ2 | ||
|---|---|---|---|---|
| CUI+CCS (correct) | CUI+CCS (incorrect) | 0.000001 | 47.44 | |
| CUI-only (correct) | 51,898 | 45,646 | ||
| CUI-only (incorrect) | 47,752 | 220,044 | ||
Table 6 presents the results of comparing the use of inputs CUI-only and CCS-only for the three tasks. From the numbers, it becomes evident that the use of CUI codes results in a prediction performance more accurate than using CCS codes only. In addition, the conclusions obtained by experimenting over the task of diagnoses prediction also applied to the tasks of mortality and readmission.
Table 6.
Direct comparison between the use of CUI codes and the use of CCS codes for each of the three tasks
| Task | Metric | CUI-only input | CCS-only input |
|---|---|---|---|
| Diagnoses Prediction (FFN) | P@1 | 0.751 | 0.728 |
| P@2 | 0.688 | 0.679 | |
| P@3 | 0.638 | 0.64 | |
| R@10 | 0.392 | 0.39 | |
| R@20 | 0.576 | 0.561 | |
| R@30 | 0.689 | 0.668 | |
| AUC-ROC | 0.911 | 0.9046 | |
| Readmission Prediction (FFN) | 0.717 | 0.5961 | |
| Mortality Prediction (GRU) | 0.9223 | 0.7993 |
Experiments on Recurrent Neural Networks
In the next set of experiments, we proceeded to answer whether our diagnoses prediction problem could benefit from the long-term clinical history of the patients by using the memory capabilities of Recurrent Neural Networks. In the experiments using a Feed-Forward network, the prediction was based only on the last visit; by using RNNs, the network can exploit the whole sequence of hospital admissions. In case the admissions result from a series of chained events that tend to manifest throughout the clinical practice, the prediction is supposed to be more precise with RNNs. We test this hypothesis using RNNs based on two kinds of neuron units: Long Short-Term Memory (LSTM) and Gated Recurrent Units (GRUs).
Table 7 points out that the Feed-Forward Network had a remarkably superior performance if compared to LSTM and GRU. This is evidence that MIMIC-III has weak temporal dependencies in between admissions, presenting shorter clinical events — this is not a surprise for an intensive care unit. While this is not a definitive conclusion, these weak temporal dependencies are worthy investigating; this is because other works [14, 15, 28] have explored MIMIC-III by means of RNNs rather than by using FFNs, although Rodrigues-Jr et al. [15] has discussed the use of FFNs.
Table 7.
Diagnoses prediction comparison of RNNs Long Short-Term Memory and Gated Recurrent Unit against a classic Feed-Forward Network (FFN)
| Architecture | P@1 | P@2 | P@3 | R@10 | R@20 | R@30 |
|---|---|---|---|---|---|---|
| LSTM | 0.5874 | 0.5483 | 0.5142 | 0.3117 | 0.4687 | 0.5764 |
| GRU | 0.5404 | 0.5085 | 0.4798 | 0.3001 | 0.4584 | 0.5712 |
| FFN | 0.750 | 0.688 | 0.638 | 0.392 | 0.576 | 0.689 |
Numbers computed using 5-fold cross validation over the CUI dataset produced with TUI list β and similarity threshold 0.9
Highest marks in boldface
Summary of Results
We present the best results for the three prediction tasks, considering the different architectures and data inputs — see Table 8. Notice that the diagnosis codes prediction simply reproduces the results presented in the previous sections.
Table 8.
Summary of results for tasks diagnosis codes prediction, mortality prediction, and readmission prediction. We report on FFN and GRU networks, and on data input CUI only and CUI+CCS, where applicable
| Task | Architecture | AUC-ROC |
|---|---|---|
| Diagnosis codes prediction | FFN | 0.911 (CUI only) |
| 0.913 (CUI+CCS) | ||
| GRU | 0.8721 (CUI+CCS) | |
| Mortality prediction | FFN | 0.8709 (CUI+CCS) |
| GRU | 0.9247 (CUI+CCS) | |
| 30-day readmission prediction | FFN | 0.717 (CUI only) |
| 0.719 (CUI+CCS) | ||
| GRU | 0.5335 (CUI+CCS) |
For task mortality prediction, we present the results obtained with data input CUI+CCS only, this is because previous experiments had already demonstrated that the use of CCS codes could improve results.
For task readmission prediction, we had results similar to what we obtained in the task of diagnosis codes prediction, in which the best results came with FFN architecture and data input CUI+CCS. This task had the smallest performance compared to the other tasks; possibly because it was modeled similar to the related works for precise comparison (refer to Section 4.1), with one single time frame of 30 days, which, if not satisfied, issued a negative result.
Statistical Validation
Following, we test the null hypothesis that our methodology, for the three tasks and considering metric AUC-ROC, is just as good as random guessing. This is a basic sanity check to provide confidence in our findings. We use a Two-sample Unpaired t-Test, whose results in Table 9 demonstrate that for all the cases, our marks largely refute the null hypothesis.
Table 9.
AUC-ROC statistical significance Two-sample Unpaired t-Test for tasks diagnosis codes prediction, mortality prediction, and readmission prediction
| Diagnosis codes | Mortality | Readmission | ||||
|---|---|---|---|---|---|---|
| Random | Our work | Random | Our work | Random | Our work | |
| AUC-ROC Fold 1 | 0.5850 | 0.9097 | 0.5753 | 0.9321 | 0.5363 | 0.7238 |
| AUC-ROC Fold 2 | 0.6068 | 0.9101 | 0.6184 | 0.8959 | 0.4826 | 0.7142 |
| AUC-ROC Fold 3 | 0.5939 | 0.9054 | 0.6874 | 0.9124 | 0.4601 | 0.7166 |
| AUC-ROC Fold 4 | 0.6066 | 0.9185 | 0.5184 | 0.9421 | 0.4844 | 0.7285 |
| AUC-ROC Fold 5 | 0.5930 | 0.9124 | 0.4038 | 0.9410 | 0.4478 | 0.7115 |
| Mean difference | 0.3142 | 0.3640 | 0.2367 | |||
| 95% confidence interval | [0.3032; 0.3251] | [0.2515; 0.4765] | [0.2009; 0.2724] | |||
| p-value | < 0.0001 | < 0.0001 | < 0.0001 | |||
Comparison to Related Works
Following, we compare our results to the related works discussed in Section 3 considering the tasks of diagnoses, mortality, and readmission prediction — Section 4.1.
The work of Sushil et al. [17] experiments on six different architectural configurations over MIMIC-III for the task of mortality prediction; and for the task of predicting the primary diagnostic category, which is similar to our Precision@1 metric. Their work considers variations of methods Bag-of-Words (BoW) [10], doc2vec [30], and CUI-based, similar to our work but using toolkit CLAMP [31]. They experiment using a regular Feed-Forward network and a stacked denoising autoencoder (SDAE) network [32]. For the task of mortality prediction, Sushil et al. achieved AUC-ROC of 0.95 for in-hospital death, 0.81 for death within up to 30 days, and 0.83 for death within up to 1 year — an average of 0.86 for the three cases. We achieved AUC-ROC of 0.9247 considering the three possible outcomes modeled as a 3-class problem; nearly 7.5% of improvement over the average of Sushil et al. but, yet, not as good as their in-hospital death prediction. As a last remark, dataset MIMIC-III is imbalanced as the number of deaths is much smaller than the number of survival cases. This fact is discussed in the work of Li and Liu [33] who explain how to improve the mortality prediction performance by taking the imbalance into account, that is, by augmenting the importance of the death cases during the learning process.
For the task of predicting the primary diagnostic category, Table 10 demonstrates that our methodology achieved results superior to all the competing configurations of Sushil et al. by nearly 10% (compared to BoCUI). The possible reasons for this superior results in both tasks are: (i) we explored different configurations for generating our CUI-based representation, while Sushil et al. used standard parameters; (ii) we used a TUI list for selecting the most relevant clinical concepts, while Sushil et al. used the whole set of retrieved concepts; (iii) we directly encoded the CUI codes as input to the network, while Sushil et al. used a Bag-of-Words over the CUI codes (BoCUI); (iv) we combined CCS codes to the input. Since we used a very similar network architecture, but for the cardinalities, the network architecture is not supposed to have influenced the results.
Table 10.
Direct comparison to the work of Sushil et al. [17] considering methods Bag-of-Words (BoW) [10], doc2vec [30], and CUI-based; using a regular Feed-Forward network and a stacked denoising autoencoder (SDAE) [32] network
| Architecture | P@1 |
|---|---|
| FFN (CUI + CCS) | 0.778 |
| BoW | 0.701 |
| SDAE-BoW | 0.650 |
| doc2vec | 0.681 |
| [doc2vec, SDAE-BoW] | 0.679 |
| BoCUI | 0.710 |
| SDAE-BoCUI | 0.665 |
Highest marks in bold face
The work of Grnarova et al. [6] achieved state-of-the-art results for mortality prediction. They achieved AUC-ROC of 0.963 for in-hospital death, 0.858 for death within up to 30 days, and 0.853 for death within up to 1 year — an average of 0.891 for the three cases. We achieved AUC-ROC of 0.9247 considering the three possible outcomes modeled as a classification problem, over 3.5% improvement. Furthermore, Grnarova et al. did not achieve the functionality of diagnoses prediction, a more demanding task in terms of preprocessing, modeling, representation, and neural network fine-tuning.
Considering the work of Dubois et al. [16], for task mortality prediction, their best performance refers to an AUC-ROC of 0.9 using the GloVe word embedding. This is the best performance of the related works but, still, 2.7% below our mark for the same task. We cannot directly compare to the other two tasks carried out by Dubois et al., as they diverge from our problem setting. Yet, for diagnosis codes prediction — a much more complex task, we achieved an AUC-ROC higher than what they achieved for simpler tasks patient admission, and emergency room visits. These comparisons suggest that our methodology is similar or superior to theirs since we are dealing with the same input, domain, and underlying principles. From a methodological perspective, our work learns patient representations from the full text of notes, which the very authors indicate as a promising approach.
In comparison to the work of Huang et al. [7], our prediction for 30-day readmission achieved an AUC-ROC of 0.719, which is superior to methods Logistic Regression based on Bag-of-Words with performance of 0.684, and method Bi-directional LSTM with 0.694; in comparison to method ClinicalBERT, whose mark was of 0.768, we stand 6% behind. This performance comes from a Transformer-based architecture much more complex than our FFN. Since our contribution focuses on the UMLS-based extraction of concepts, rather than on the network architecture, it is expected that ClinicalBERT could have an even superior performance by using our protocol, which, we indicate as a promising future work. Notwithstanding, the work of Huang et al. is not as versatile as our methodology, which reports on diagnosis, mortality, and readmission prediction; the last of those cannot perform over the BERT architecture.
Conclusions
We described a Machine Learning process based on Natural Language Processing and Artificial Neural Networks. The goal was to predict the clinical trajectory facts (diagnosis, mortality, and readmission) to occur in the future of a patient by inspecting the clinical notes in her/his Electronic Health Record. We started with data pre-processing and advanced until the stage of neural network fine-tuning to achieve a performance comparable to state-of-the-art works, which produced lessons that might guide future works in the field.
The strong point of our research is that we evaluated different strategies on using UMLS-based concepts extraction to represent the clinical notes. Our results demonstrated that the choice of a list of concept types and of a similarity threshold can narrow the scope of CUI codes to a more dense representation. By comparison to previous works, we verified that this course of action is more effective than using the entire set of retrieved concepts, and also more effective than using word-embedding techniques without first extracting concepts. The drawback of our approach was the more intricate preprocessing stage, and the size of our input, which demanded days of processing due to its order of magnitude, around dozens of thousand elements.
Still, in comparison to previous works, we did not follow a strict methodology from the beginning. As we experimented on different models and neural network architectures, we verified that by using a Feed-Forward architecture, fed by the last admission only, worked better than using the entire history of admissions over Recurrent Neural Networks. We also learned that by combining the diagnosis codes with the clinical notes, it was possible to significantly improve the results, achieving a performance suitable for real-world applications. Our Precision@1 metric indicated that the code with the highest probability was an actual CCS code to manifest in the next admission in 77.8% of the cases. In turn, the Recall@30 metric indicated that among the 30-top ranked probabilities, the system was able to foresee 70.6% of all the CCS codes to appear in the next admission.
Although we experimented with the real-world dataset MIMIC-III, it is desirable to test our methodology on a private dataset. This is because the culture of different hospitals leads to clinical notes with particular structures, which shall demand adaptations on our preprocessing and concepts extraction steps. Furthermore, despite achieving results comparable to the related works, we did not use negation detection, available in concept recognizers such as cTAKES (based on algorithm Negex [34]). Nevertheless, negation has potential to improve the data representation even more by counting on a finer semantic analysis — we suggest the use of negation as a straight prominent future work. Either, we did not explore the imbalance of the classes in MIMIC-III; just as discussed, the work of Li and Liu [33] has benefited from this characteristic to reach even better results. This is an open issue here that, together with negation, could produce a more advanced investigation. It is also desirable to test word-embedding techniques over our extracted concepts; a more concise representation has potential for shorter fine-tuning cycles and, consequently, for improved network configurations. Finally, we focused on the process of concepts extraction more than on the neural network techniques; there is room for experimenting with the more advanced neural network techniques that appear every day in the fast-paced field of Machine Learning.
Funding
This research was financed by French agency Multidisciplinary Institute in Artificial Intelligence (Grenoble Alpes, ANR-19-P3IA-0003); and by Brazilian agencies Fundacao de Amparo a Pesquisa do Estado de Sao Paulo (2018/17620-5, and 2016/17078-0); and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (406550/2018-2, and 305580/2017-5).
Declarations
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jamil Zaghir, Email: jamil.zaghir@grenoble-inp.org.
Jose F Rodrigues-Jr, Email: junio@usp.br.
Lorraine Goeuriot, Email: lorraine.goeuriot@univ-grenoble-alpes.fr.
Sihem Amer-Yahia, Email: Sihem.Amer-Yahia@univ-grenoble-alpes.fr.
References
- 1.Seymour T, Frantsvog D, Graeber T, et al. Electronic health records (EHR) Am J Health Sci (AJHS) 2012;3(3):201. doi: 10.19030/ajhs.v3i3.7139. [DOI] [Google Scholar]
- 2.Bellot A, Schaar MVD. Flexible modelling of longitudinal medical data: A Bayesian nonparametric approach. ACM Trans Comput Healthcare. 2020;1(1):1. doi: 10.1145/3377164. [DOI] [Google Scholar]
- 3.Goodfellow I, Bengio Y, Courville A. Deep Learning, vol 1. Cambridge: MIT Press; 2016. [Google Scholar]
- 4.Wu S, Roberts K, Datta S, Du J, Ji Z, Si Y, Soni S, Wang Q, Wei Q, Xiang Y, et al. Deep learning in clinical natural language processing: a methodical review. J Am Med Inform Assoc. 2020;27(3):457. doi: 10.1093/jamia/ocz200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenbloom ST, Denny JC, Xu H, Lorenzi N, Stead WW, Johnson KB. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grnarova P, Schmidt F, Hyland SL, Eickhoff C (2016) Neural document embeddings for intensive care patient mortality prediction. arXiv:1612.00467
- 7.Huang K, Altosaar J, Ranganath R (2019) Clinicalbert: Modeling clinical notes and predicting hospital readmission. arXiv:1904.05342
- 8.Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl_1):D267. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Official ICD reference. https://www.who.int/standards/classiffications/classiffication-of-diseases. Accessed March, 2021
- 10.Zhang Y, Jin R, Zhou ZH. Understanding bag-of-words model: a statistical framework. Int J Mach Learn Cybern. 2010;1(1-4):43. doi: 10.1007/s13042-010-0001-0. [DOI] [Google Scholar]
- 11.Unified medical language system (UMLS). https://www.nlm.nih.gov/research/umls/index.html, accessed March, 2021
- 12.Metathesaurus’s unique identifiers. https://www.nlm.nih.gov/research/umls/new_users/online_learning/Meta_005.html, accessed March, 2021
- 13.Clinical classifications software databases. https://www.hcup-us.ahrq.gov/toolssoftware/ccs/ccs.jsp, accessed March, 2021
- 14.Choi E, Bahadori MT, Schuetz A, Stewart WF, Sun J (2016) Doctor ai: Predicting clinical events via recurrent neural networks [PMC free article] [PubMed]
- 15.Rodrigues-Jr JF, Spadon G, Brandoli B, Amer-Yahia S (2019) Patient trajectory prediction in the Mimic-III dataset, challenges and pitfalls. arXiv:1909.04605
- 16.Dubois S, Romano N, Kale DC, Shah N, Jung K. Learning effective representations from clinical notes. Stat. 2017;1050:15. [Google Scholar]
- 17.Sushil M, Šuster S, Luyckx K, Daelemans W. Patient representation learning and interpretable evaluation using clinical notes. J. Biomed. Informatics. 2018;84:103. doi: 10.1016/j.jbi.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 18.MIT’s MIMIC-III (). https://mimic.physionet.org/, accessed March, 2021
- 19.Clinical language annotation, modeling, and processing toolkit. https://clamp.uth.edu/, accessed March, 2021
- 20.Pennington J, Socher R, Manning CD (2014) Glove: Global vectors for word representation. In: Proceedings of the 2014 conference on empirical methods in natural language processing (EMNLP), pp 1532–1543
- 21.Devlin J, Chang MW, Lee K, Toutanova K (2018) Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv:1810.04805
- 22.Johnson AE, Pollard TJ, Shen L, Li-Wei HL, Feng M, Ghassemi M, Moody B, Szolovits P, Celi LA, Mark RG. MIMIC-III, a freely accessible critical care database. Scientific Data. 2016;3(1):1. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soldaini L, Goharian N (2016) Quickumls: a fast, unsupervised approach for medical concept extraction. MedIR workshop, sigir 1–4
- 24.Quickumls github link. https://github.com/Georgetown-IR-Lab/QuickUMLS, Accessed March, 2021
- 25.Kingma DP, Ba J (2014) Adam: A method for stochastic optimization. arXiv:1412.6980
- 26.Sra S, Nowozin S, Wright SJ. Optimization for Machine Learning. Cambridge: MIT Press; 2012. [Google Scholar]
- 27.Kumar SK (2017) On weight initialization in deep neural networks. arXiv:1704.08863
- 28.Pham T, Tran T, Phung D, Venkatesh S. Predicting healthcare trajectories from medical records: A deep learning approach. J Biomed Informa. 2017;69:218. doi: 10.1016/j.jbi.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Hawass N. Comparing the sensitivities and specificities of two diagnostic procedures performed on the same group of patients. British J Radiology. 1997;70(832):360. doi: 10.1259/bjr.70.832.9166071. [DOI] [PubMed] [Google Scholar]
- 30.Le Q, Mikolov T (2014) Distributed representations of sentences and documents. In: International conference on machine learning (PMLR), pp 1188–1196
- 31.Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, Xu H. CLAMP–a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc. 2018;25(3):331. doi: 10.1093/jamia/ocx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent P, Larochelle H, Lajoie I, Bengio Y, Manzagol PA, Bottou L (2010) Stacked denoising autoencoders: Learning useful representations in a deep network with a local denoising criterion. J Mach Learn Res 11(12)
- 33.Li L, Liu G (2020) In-hospital mortality prediction for ICU patients on large healthcare MIMIC datasets using class imbalance learning, IEEE
- 34.Chapman WW, Hilert D, Velupillai S, Kvist M, Skeppstedt M, Chapman BE, Conway M, Tharp M, Mowery DL, Deleger L. Extending the NegEx lexicon for multiple languages. Studies in Health Technology and Informatics. 2013;192:677. [PMC free article] [PubMed] [Google Scholar]