Abstract
Coupled with the rise of data science and machine learning, the increasing availability of digitized health and wellness data has provided an exciting opportunity for complex analyses of problems throughout the healthcare domain. Whereas many early works focused on a particular aspect of patient care, often drawing on data from a specific clinical or administrative source, it has become clear such a single-source approach is insufficient to capture the complexity of the human condition. Instead, adequately modeling health and wellness problems requires the ability to draw upon data spanning multiple facets of an individual’s biology, their care, and the social aspects of their life. Although such an awareness has greatly expanded the breadth of health and wellness data collected, the diverse array of data sources and intended uses often leave researchers and practitioners with a scattered and fragmented view of any particular patient. As a result, there exists a clear need to catalogue and organize the range of healthcare data available for analysis. This work represents an effort at developing such an organization, presenting a patient-centric framework deemed the Healthcare Data Spectrum (HDS). Comprised of six layers, the HDS begins with the innermost micro-level omics and macro-level demographic data that directly characterize a patient, and extends at its outermost to aggregate population-level data derived from attributes of care for each individual patient. For each level of the HDS, this manuscript will examine the specific types of constituent data, provide examples of how the data aid in a broad set of research problems, and identify the primary terminology and standards used to describe the data.
Keywords: Healthcare analytics, Big data, Review, Standards
Introduction
Upon first consideration, it may be natural for one to view the United States (US) healthcare system as a single monolithic entity apportioning care across the country. In reality, care is provided by an intricate system of interconnected people, institutions, and resources working in concert to meet the health needs of the American people. While providing care remains the central function of this system, evolving economic, legislative, and social conditions have fostered the rise of numerous ancillary services. Ranging from finance to government reporting to evidence-based research programs, these services address the growing realization that healthcare is a cross-disciplinary endeavor requiring the integration of data from many fields.
As the breadth of available services continues to grow, healthcare is experiencing a dramatic shift in the amount and type of data needed to sustain their effective functioning. While most notably associated with the adoption and integration of electronic medical records (EMR) into clinical practice, this shift has occurred at a much broader scale. Together, industrial, academic, and government partners have worked to collectively transition the body of healthcare operations from paper-based documentation to electronic health and wellness data. This transition has elevated healthcare into a new, transformative era of “big data”. Fostering an explosion in the collection of health-related data predicted to grow by over 50 fold between 2012 and 2020 to a staggering 25,000 petabytes [40].
With the promise of facilitating new and increasingly complex analyses, the increasing scale, scope, and variety of digitized health data available holds great promise for the advancement of personalized healthcare. Yet as access to new tools and data sources become available, the variety of new data they generate will introduce challenges for managing the growing diversity of information. Without an organizational framework such diverse data will become fragmented. In turn making it increasingly difficult for researchers and practitioners to remain informed of the currently available health and wellness data and of the myriad purposes for which it may be collected and applied.
The work presented in this manuscript attempts to address exactly this task, organizing the diverse set of healthcare data from an interpretable, patient-centric view. While prior work has focused on the computational challenges presented by health data [70, 90, 99], little effort has been devoted to examining the challenges that arise from the fragmented provenance of the data itself. Our work seeks to integrate the diverse sources of healthcare data so that the ever-expanding silos can be catalogued, organized, and synthesized in ways practically useful for those who seek to consume it. For this purpose, we present an organizational framework that characterizes data as a hierarchy extending outward from the patient. We identify how different components of care fit into this framework and argue that the framework can be a powerful tool for addressing practical clinical questions and problems.
Organizational models have already proven a significant resource to researchers and practitioners alike. Among the most prominent examples of such can be found in the establishment of the social ecological model, which formulated how “social environmental and biological factors jointly influence health” of an individual [102]. However, the establishment of a framework is only one step. Rather, it is the continued development of these frameworks that drives the continued growth and success of informatics applications to problems in the healthcare domain.
The work presented in this manuscript aims to lay the groundwork for a new perspective on the interconnected nature of health data as it pertains to an individual, and is organized as follows. In Section 2, we define a patient-centric data model deemed the Healthcare Data Spectrum (HDS). Subsequent sections explore each level of the HDS in detail. Beginning with the inner-most level of data, Section 3 presents patient omics. Section 4 investigates patient demographics. Section 5 probes patient care. Section 6 considers patient wellness. Section 7 examines administrative attributes of care. Section 8 explores aggregate patient data. Within each section we examine the specific data elements that comprise the HDS level, offer a brief overview of how such data is being utilized in a broad set of research problems, and review the primary terminology and standards used to describe the data. Also highlighted are some relevant computational challenges associated with consuming and processing such data. Finally, in Sections 9 and 10, we provide a discussion of the most salient open problems in the organization and interoperability of healthcare data associated with the establishment of the HDS, and conclude with a summation of our work and contributions.
Healthcare Data Spectrum
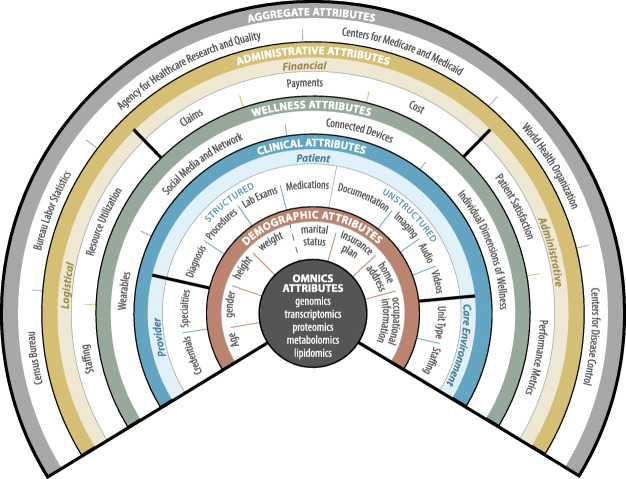
The HDS is an organizational framework designed to provide a comprehensive catalogue of the health and wellness data generated through all aspects of a patient’s care. The framework begins with micro-level omics and macro-level demographic data directly characterizing a patient. The next level extends to that of care. Moving further outwards, the attributes that comprise and quantity wellness. Beyond this are administrative attributes of care. Finally, the outermost level details aggregate population-level data derived from attributes of care across numerous patients. Through these levels, the HDS encapsulates and organizes all of the relevant data—both direct and indirect—that relate to patient health and wellness, from direct attributes of the patient all the way to the attributes of individuals and services from whom care is received. A visual representation of the HDS can be found in Fig. 1.
Fig. 1.
Healthcare data spectrum
Frameworks for organizing healthcare data are imperative for overcoming the negative impacts of data fragmentation and facilitating the continued growth and success of informatics applications to problems in the healthcare domain. Our formulation of the HDS distills the largely fragmented set of patient data available today into the essential components of health and wellness, providing an organizational foundation detailing the types of data available for analysis. Additionally, the HDS provides a detailed reference to the standards associated with each data type. Helping guide researchers to an accepted set of standards may promote consistency between future works, benefiting not only researchers but the field as a whole.
Omics Attributes
The inner-most level of a patient’s HDS is represented by omics data. Broadly defined, omics represents the study of information contained within an individual’s genome and the biological derivatives of these genes [49, 62]. Omics thus includes fields concerned with the study of genes (genomics), gene expression and RNA (transcriptomics), proteins (proteomics), and more recently metabolites (metabolomics) and lipids (lipidomics) [32, 107, 125]. The field of genomics is often broken down into additional subfields, which include genetic elements such as genes, single-nucleotide polymorphism (SNP), and short tandem repeat (STR) [123]. Although not a distinct omics discipline itself, the field of genome-wide association studies (GWAS) has been widely applied to omics data as a popular approach for assessing the association of SNPs with various phenotypic traits, as well as for assessing the genetic etiology of diseases [63]. Also, while we have focused primarily on fields that study groupings of molecules, a large body of work aims to define fields through their actions. Prior work by Greenbaum et al. discusses the identification and prevalence for a number of these emerging omics fields [51].
Research Applications
Often attributed to the shift from reactive to preventative care, the increasing availability of health data over the last decade has given rise to an even greater level of personalization in the emergence of a paradigm of care known as precision medicine [121]. Today, as massively parallel next-generation and high-throughput sequencing techniques become increasingly available, healthcare research finds itself awash in new information that many believe is posed to profoundly change the clinical landscape [25].
With the cost of sequencing falling from well over $10 million per genome to within sight of the long-awaited $1,000 mark, several studies have already demonstrated the use of omics data to address a number of clinical problems [118].1 The most prevalent applications have focused on the identification of individuals’ diseases and disease risks, with over 2,000 genetic tests available to aid in the diagnosis and therapy for over 1,000 different diseases [7]. However, omics research extends much further. More recent work in pharmacogenomics has explored how omics data can be utilized to identify the treatment efficacy of various medications and medication dosages for a particular individual. In a field known as nutrigenomics, additional emerging work has investigated genome-wide influences on nutrition and the role of genetic polymorphisms in dietary-influenced disease [38, 89, 122].
Yet, despite these early successes, significant computational and biological challenges exist for researchers taking the final step from data to insights. As a consequence of sequencing advances, a considerable amount of research remains to understand the logistics of how to efficiently process and analyze this flood of information [6, 131]. Further, it is becoming increasingly clear that even at such a highly granular view of biological functioning, these systems do not exist in isolation. There exists a need to develop novel multi-omics techniques to capture and model the interrelated nature of these elements in an effort to develop a more complete picture of an individual [25].
Data Standards
In order to effectively use the wealth of information drawn from omics studies, significant effort has been put forth to develop standards for identifying the many variants of each molecule. A number of standards have also been developed to detail the minimal amount of information required to describe and reproduce experiments within the omics fields.
One of the most prolific standards for genomic data is the naming conventions maintained by the Gene Ontology Consortium. Additionally, organizations such as HUGO Gene Nomenclature Committee (HGNC) and the Human Genome Variation Society (HGVS) provide standardized nomenclature to human genes and their variations [27, 50, 61]. To account for accurate representations of genome sequences, the Genomic Standards Consortium has developed the Minimum Information about a Genome Sequence (MIGS) specification, which aims to align with data traditionally captured by the major nucleotide sequence repositories such as Genbank, EMBL, and DDBJ [45]. Other fields have developed their own standards initiatives, including the Metabolomics Standards Initiative (MSI) for metabolomics and the Proteomics Standards Initiative (PSI) for proteomics [44, 92].
To provide consistency in the annotation of the respective molecules, both the PSI and MSI maintain a set of controlled vocabularies (CV). These vocabularies primarily focus on PSI-mass spectrometry (MS), MSI-MS, and MSI-nuclear magnetic resonance (NMR). However, they may also include CV such as PSI-MI, which defines terminology for protein-protein interactions and MSI-GC for Gas chromatography. In a similar fashion, transcriptomics provides the MGED Ontology, which sets standards for the annotation of microarray experiments [23, 127]. To account for reproducibility, the PSI and MSI initiatives have developed minimal reporting sets, including the minimum information about a proteomics experiment (MIAPE) and the core information for metabolomics reporting (CIMR) respectively. Similarly, the transcriptomics field has developed the minimum information about a microarray experiment (MIAME) [17, 120].
Younger fields such as lipidomics do not, as of yet, have dedicated standard initiatives. In acknowledgment of the need to differentiate the increasing number of lipid molecules, the Lipid Metabolites and Pathways Strategy (Lipid MAPS) Classification System has been developed, funded by the National Institute of General Medical Sciences [39]. For more details on some of the more prominent reporting standards, including data format, transfer markup languages, and common analytic tools, see Section 3 of [26] and Table 1(iii) of [119].
Demographic Attributes
Moving outward a level on the HDS, we find demographic attributes. Demographics can be broadly defined as the epidemiologically objective characteristics of a population, which include age, marital status, income, and education level [81]. Drawing from this definition, demographic characteristics can be divided into two distinct categories: intrinsic and extrinsic attributes. Intrinsic attributes include physiological characteristics, such as age, gender, height, and to some extent weight. They may also include less obvious characteristics, such as allergies to foods or medications. By contrast, extrinsic attributes represent non-physiological characteristics derived from an individual’s environment and lifestyle, which may include his or her address, martial status, insurance plan, employment type, location, and salary [111].
Research Applications
Demographic attributes have been used extensively in the evaluation of clinical problems. Pol and Thomas define health demography as “the manner in which demographic attributes influence both the health status and health behavior of populations,” and argue that demographic techniques and perspectives provide a means of studying practically every aspect of health and wellness [97].
Broadly, the evaluation of both intrinsic and extrinsic demographic attributes, better known as an individual’s socio-demographic profile, has provided a significant body of work demonstrating the usefulness of demographic information in identifying various diagnoses and population-health risk factors. While detailing the effect of demography on each diagnosis and population subgroup is beyond the scope of this review, demographic attributes have been shown to hold a critical role in projections for public health programs, allocation of resources, planning for emergency services, and general estimations of health characteristics of a population [112].
Aside from the established importance in population health, researchers have investigated the influence of demographic attributes as it pertains to the health of an individual [96]. Establishing how demographic characteristics—particularly intrinsic attributes such as age and gender—can have a profound impact on an individual’s expression of various clinical attributes. As a result, these demographic attributes must be carefully controlled for to ensure proper analysis. Work by Skelly et al. has gone so far as to argue that a failure to consider demographic and clinical attributes as potential confounding factors can result in biased studies and incorrect conclusions [113, 114, 132]. Subsequent research has taken this idea one step further, highlighting the importance of understanding “the (biological) mechanisms through which demographic variables work” [29], noting how when properly controlled for, demographic attributes often represent underlying biological effects, such as the changes to an organism during aging.
Finally, earlier reflections by Eileen Crimmin’s on the 30-year state of demography note the shift from aggregate analysis of group-level data to an individualized approach was only the start. While demographic attributes are often modeled as static entities, in reality many evolve over time [28]. As such, there exists a need to capture the dynamic nature of these attributes, yet to do so will require a drastic change to the computational models in which these attributes are utilized. Today, the demographic attributes utilized to adjust or control for various subgroups are often considered as fixed effects, even during longitudinal evaluation of other independent variables. There remains an open and important problem as to how these effects can be dynamically adjusted for over repeated measurements of an individual—a problem that will become increasingly important as demographic data is collected more frequently and at a finer granularity.
Data Standards
Due to the large and diverse nature of the human population, demographic attributes have historically been difficult to standardize. One of the most effective demographic standards has been the Classification of Federal Data on Race and Ethnicity, created by the Office of Management and Budget (OMB) in 1995 and updated in 1997 [100]. These standards have since been built upon by the Department of Health and Human Services (HHS) to include standards for ethnicity, sex, primary language, and disability status [57]. Together, these standards provide a fairly comprehensive set of demographic attributes by which to characterize an individual. However, they still fail to capture many other important characteristics (e.g., marital status).
There are several standards that account for these missing health-related characteristics. One commonly used standard is the Logical Observation Identifiers Names and Codes (LOINC). LOINC provides a corpus of universal codes, created to unambiguously identify the set clinical and laboratory observations. It is maintained by the Regenstrief Institute, a non-profit medical research organization associated with Indiana University. The LOINC standard applies to data within many levels of the HDS [79]. Also, while not a demographic standard itself, the Health Level Seven International (HL7) standard can also apply to health-related patient characteristics. Currently at version 3, HL7 is a wide-ranging standard developed to provide “coherent, extensible standards that permit structured, encoded health care information of the type required to support patient care, to be exchanged between computer applications while preserving meaning.” It is one of several American National Standards Institute (ANSI) accredited Standards Developing Organizations (SDOs) and is supported by more than 1,600 members from over 50 countries, including over 500 corporate members. As with the LOINC codes, the standards developed by HL7 are used to provide standardized elements for a number of health-related attributes throughout the levels of the HDS.2
Clinical Attributes
The third level of the HDS represents the first in which the data are not a characteristic patient themselves, but rather generated as a result of care they receive. It is also the first level that intersects with entities beyond the patient, including the clinician(s) providing the care and the clinical environment in which the care is provided. At the patient level, clinical attributes are generated as a direct result of the care received through the individual’s interaction with a clinician. Clinical attributes are captured from a number of sources and may represent structured or unstructured data.
Structured clinical data represents those elements that can be discretely labeled or coded. These include the medications prescribed, diagnoses received, and procedures and lab tests performed. Unstructured data is associated with information that is not organized in a pre-defined manner and may be drawn from sources such as clinical documentation and medical imaging. Clinical documentation may include progress notes, consolations, procedural reports, and admission and discharge summaries. Medical imaging includes data recorded from techniques such as X-ray (projection radiography), X-ray computed tomography, nuclear medicine (SPECT/PET), ultrasound, and magnetic resonance imaging (MRI) [117]. Technological advances have further extended the purview of unstructured data to include digitally recorded audio and video.
As noted prior, the third level of the HDS provides an intersection point between multiple entities. While data is generated as a result of patient care, this care is provided by clinicians who themselves exhibit a number of important attributes. These can be linked back to second-level demographic attributes (both intrinsic and extrinsic), such as age, gender, salary, and practice location. Further, clinician data as well as additional extrinsic attributes, including the medical school and residency programs they attended, their credentials (M.D., N.P., R.N., etc.), and their clinical specialty. Finally, the clinical environment in which the care is provided can offer its own set of attributes. Some of the most prevalent are the number of beds and the number of staff, though the complex nature of these environments may be represented by varying degrees of these attributes.
Research Applications
With electronic medical records emerging as a driving force in the digitalization of healthcare, research surrounding information collected at the clinical level of the HDS has thrived. While the primary goal of clinical data is to facilitate patient care, the persistence and accessibility of such data has facilitated its use in analyses across the entirety of the healthcare domain. Spanning from aiding in redefining clinical guidelines, to cost reduction, to population-level estimates of the Global Burden of Disease [15, 77], this repurposing of previously collected data is formally known as “secondary use,” and it has since become an established element of clinical research [105].
From a more general research perspective, structured and unstructured representations of clinical data are being employed in a range of applications to establish correlations between clinical variables, identify disease comorbidities, improve patient stratification, highlight novel drug interactions, and predict clinical outcomes [65]. Further, in an effort to continuously improve such analyses, longitudinal methods are being developed to understand the progression of these elements over time. Additionally, outside of attributes of the patient, analyses of hospital staffing and organization have been used beyond the standard evaluation on quality of care to examine aspects of job satisfaction, nurse burnout, and physician procedure selection [3, 41].
To date, extensive literature has been published around the influence of data mining and machine learning methodology on clinical data [64]. Yet despite—or perhaps because of—the progress and success of these models, the integration of new data sources has proceeded almost unchecked to include data collected from bedside monitors, laboratory tests, imaging procedures, and pharmacy prescriptions. Unfortunately, current computational modeling approaches are no longer sufficient to represent these data. Rather, significant work must be done to adapt these models to encompass the broad set of heterogeneous, noisy, high-dimensional, and irregularly sampled variables collected [42].
Data Standards
The expansive number of attributes at the clinical-level of the HDS yields a number of standards. One of the most pervasive standards is International Classification of Diseases (ICD). Published and maintained by the World Health Organization, the ICD standard is used to monitor the incidence and prevalence of diseases and other health problems. The standard has a notable extension, known as the clinical modification (ICD-CM), which provides standardized codes for the diagnoses and procedures associated with hospital utilization in the US. Unlike the ICD standard the ICD-CM standard is maintained by the National Center for Health Statistics (NCHS) and the Centers for Medicare and Medicaid Services (CMS). As an important distinction, within the newest revision of the ICD standard (ICD-10), the CM has been reserved solely for diagnosis coding, and a separate partition (ICD-10-PCS) has been created to encode procedures. However, prior revisions, such as ICD-9-CM, will continue to maintain codes for both diagnoses and procedures [33, 56, 84, 93].
Additionally, while the ICD standard holds the capacity to classify mental disorders, the American Psychiatric Association (APA) maintains what is often regarded as the standard for mental health professionals in the US, known as the Diagnostic and Statistical Manual of Mental Disorders (DSM) [9]. Though the ICD standard provides moderate support for the description of clinical procedures, additional standards have been created to more completely represent the breath of possible entities. One of the primary standards is drawn from the Current Procedural Terminology (CPT), a standard maintained by the American Medical Association (AMA) that provides unique codes to detail medical procedures and services under public and private health insurance programs. It should be noted that unlike most of the publicly available standards listed thus far, the CPT standard is privately owned and must be licensed from the AMA for use [8]. Another prominent procedural coding standard is the Healthcare Common Procedure Coding System (HCPCS), maintained by the CMS. The HCPCS standard is partitioned into two levels: level 1 represents analogous codes to the CPT standard, while level 2 is utilized primarily to identify products, supplies, and services that are not included in the CPT [84, 85].
To standardize the expansive set of pharmaceutical products, the Food and Drug Administration (FDA) maintains the National Drug Code (NDC). The NDC is intended to provide a “current list of all drugs manufactured, prepared, propagated, compounded, or processed for commercial distribution” [47]. Finally, the LOINC codes, which provided standards for a subset of demographic attributes, in fact provide the primary standard for laboratory tests. As of July 2002, the LOINC database carried records for more than 30,000 different observations, of which approximately 25,000 are categorized as laboratory test observations [79]. Although the majority of the coding standards reviewed thus far pertain to structured data, there has been a significant effort to provide standards encompassing unstructured data. In particular, HL7 working groups have provided standards for the structure and semantics of clinical documentation, known as the Clinical Document Architecture (CDA), and the American College of Radiology (ACR) have provided and maintained the Digital Imaging and Communications in Medicine (DICOM) for medical images and related information [34, 88]. Beyond the patient level, the CMS has created the National Provider Identifier (NPI) standard to effectively identify the link between patient care attributes and the clinicians who provide this care by uniquely identifying healthcare providers [86]. Moving beyond the clinician level to the clinical environment, to aid in the ever-increasing regulatory mandates, independent, not-for-profit organizations have taken to providing a standardized set of metrics for both care-units and hospital operations such as staffing. One of the most prominent is the Joint Commission, which as of 2011 accredits and certifies over 80% of the hospitals in the US [58].
Wellness Attributes
At the fourth level, we find an emerging aspect of the HDS, personal wellness data. While distinctly outside of the direct clinical care attributes, wellness data represents attributes that are typically collected outside of formal clinical environments and are often measured by the individual themselves.
Although recent media attention has emphasized the rise of wearable fitness tracking as a source of personal data, in actuality such data can be generated and collected through a number of mediums. These can include devices as simple as wireless scales and digital pill boxes or as complex as personal medical devices such as digital glucometers, personal blood pressure cuffs, and pulse oximeters. Further, advances in technology have allowed for increasingly pervasive monitoring tools including in-home sensors in beds, chairs, and fall detection within flooring [22].
In the evaluation of wellness data, it is important to highlight that the term “wellness” is itself ambiguous. In the context of health, the term wellness is most often associated with physical attributes, but formal research has established that it is more appropriate to consider wellness a multi-dimensional entity. In one of the earliest definitions, Hettler proposed a hexagon model, which included physical, emotional, social, intellectual, occupational, and spiritual wellness [59]. Since then, a number of works have revised and extended the definition of wellness to include social, physical, creative, and emotional factors [54]. As a result, wellness data extends far beyond physical sensors to the analysis of an individual’s social activity and dietary habits [133, 80].
Research Applications
Resulting from the relative infancy of formally collected wellness data, research into wellness data “lacks a clear theoretical basis, a set of data models, and empirically derived strategies for integrating tools and data into existing clinical applications and workflows” [104]. However, the applications of such data to understand an individual’s greater health condition has emerged as a key goal of the research community. In line with this goal, a working group of the American Medical Informatics Association (AMIA) investigated the policy, economic, and ethical implications of patient-generated data, highlighting “the potential to empower patients and support a transition from a role in which the patient is the passive recipient of care services to an active role in which the patient is informed, has choices, and is involved in the decision-making process” [31].
To this end, researchers have already begun to utilize a broad set of patient generated health data collected by sensors, direct data entry, and social media activities to aid in tasks ranging from determining clinical trials efficiency, evaluating novel therapeutics, and measuring functional recovery in patients [5, 130]. Parallel streams of research have focused on the utilization of mobile devices as a rich source of pervasive health and wellness information—a practice termed “m-health.” [66, 68]. Finally, outside of the applications, recent work has also begun to explore the implications of collecting this data in real-time, addressing multi-stream integration and discussing how the vast quantity of data can be presented in useful formats with new visualization techniques [108].
Such research brings to light a number of important caveats about wellness data. Open questions remain around the source, quality, and utility of such data for decision-making [109]. As wellness data is continuously integrated into medical contexts, it is vital to acknowledge the shift to data collection outside of traditional clinical settings. No longer collected and curated by trained medical professionals, moving forward it will become increasingly important for computational models to account for varying levels of data veracity [95]. Further compounding this difficulty we note, as opposed to largely standardized clinical tools, serious computational efforts must be made to capture and control for variability between the measurements captured by various consumer-grade devices.
Data Standards
Currently, there is no standard for the storage and collection of patient wellness data. Instead, many companies that produce products for measuring attributes of patient wellness have developed their own guidelines. To overcome these fragmented guidelines, the Consumer Electronics Association (CEA) has released the first set of standards related to wellness data, named the Guiding Principles on the Privacy and Security of Personal Wellness Data [10].
Administrative Attributes
Complex as it is with many interacting entities, the healthcare system generates a substantial amount of data on the peripheral attributes of care, including financial, logistical, and administrative data. The fifth level of the HDS moves beyond the characteristics of direct care to capture this broader set of data.
Data Sources
The primary source of financial data is public and private insurance claims. However, a variety of additional sources can be used to augment the scope and level of detail for financial data, including managed care plans, hospital discharge datasets, and revenue cycle management organizations [103]. Recent policy changes further promise to broaden the transparency of healthcare costs, providing new avenues by which to obtain financial data. Of note, the Department of Health and Human Services (HHS) announced on April 2nd, 2014 that the CMS would release a public dataset with information on the types of Medicare services, requested charges, and payments issued by providers across the country [18].
The complexities of the healthcare system provide opportunities to collect and analyze logistical data pertaining to several aspects of patient care, such as the care environment and resource utilization [74, 98]. Details of the care environment can include the care-team composition and staffing metrics, while observations of resource utilization can include an expansive set of service metrics (e.g., inpatient, outpatient, and emergency department visits), medication usage, and performed diagnostic tests and procedures [52]. Finally, further information is provided by assessments of the quality of patient care. These assessments may include performance analyses of the service providers (i.e., reviews of clinician performance) and reviews of customer satisfaction (patient satisfaction surveys) [72, 124].
Research Applications
Researchers have continuously explored how data can be used to improve the quality and efficiency of care provided. Such information can help researchers investigate how to enhance the patient experience and improve the efficiency of the healthcare systems providing care.
Continually rising healthcare costs have become a perennial concern associated with all levels of care for providers and patients alike. A prevalent use of administrative data has been to provide insight into the cost of patient care. Researchers have used financial data drawn from administrative claims to analyze the costs of care associated with the treatment of specific diagnoses [19, 73]. A tacit component of this cost is the resulting burden of fraud on the healthcare system. A large body of research has been dedicated to the application of patient claims data and statistical methods to detect healthcare fraud [76]. Research in this area continues to grow, with the burden of fraud so great that the European Healthcare Fraud and Corruption Network considers fighting it to be “the first and most effective step for...setting up cost cutting strategies in order to stop losses without reducing the access to and the quality of care [48].”
Researchers have demonstrated that predictive analytics techniques performed on data beyond the patient level can also be used to aid healthcare practices. For example, while financial data is collected primarily for the benefit of the hospitals and practices providing care, incorrectly coded and billed patient charges can be identified and recovered without the need for a manual review of all service claims, thereby improving the efficiency of these transactions for both the patient and provider [16]. Also, while data on care teams and unit staffing is collected primarily in an effort to ensure patient safety, it has been well-established that inadequate staffing is correlated to both an increased frequency of negative patient outcomes and increased rates of clinician burnout. Researchers have used staffing data not only to develop models to ensure adequate staffing, but to better understand the dynamics of the care environment from the provider’s perspective [53]. By targeting individuals, an intricate understanding of both the specific resources used by population sub-groups and how those resources are best utilized has the potential to greatly improve the quality of care [2, 115].
At an administrative level, patient satisfaction and clinical performance have always been an integral part of the evolving practice of medicine. Researchers have investigated the objectivity and utility of patient reviews, offering the increasingly accepted view that patients themselves can provide useful information on the delivery of care [35]. Researchers and hospital administrators have also suggested that patient satisfaction data can be applied to understand the role of medicine in satisfying patients’ physical and mental needs, as well as to improve the overall quality of care provided [91].
Beyond the patient and clinician quality and satisfaction, care environments such as hospitals have begun to analyze similar performance metrics. These metrics have shown impact on quality improvement, market share, and reputation [60]. Additionally, it should be noted that the data sources at this level are strongly interconnected. Recent work has begun to investigate the implicit relations between many of these factors, including satisfaction, utilization and outcomes [43].
Data Standards
The financial data practices established by hospitals and clinics are fundamentally similar to those employed by most businesses. A number of works have expressed how these standard financial techniques apply to the healthcare domain, while detailing the relation of financial regulations to managed care and to agencies such as Medicare and Medicaid [12, 82].
Insurance claims pose another interesting issue. Administrative claims data represent a critical source of healthcare financial data, but are often managed by external third parties. To help account for some of the inevitable variation, the Health Insurance Portability and Accountability Act (HIPAA) was introduced [24]. At the time that HIPAA legislation was enacted, more than 400 different “standard” claim forms were in use, and beyond the commonly associated aspect of patient privacy, HIPAA includes a number of provisions intended to provide standardization [36].
The collection of logistical data is somewhat more difficult, as it is often derived from care at an individual level. Similarly to omics data, a number of Minimum Data Set standards have been developed to ensure that data will be collected in a consistent manner between individuals and across units and institutions. Commonly, these can include standards such as the Nursing Minimum Data Set (NMDS), which provides standards for the documentation of “patient/client responses, interventions, patient-sensitive outcomes, and resource consumption [30].”
Data collected by various private and government agencies are typically structured based on internal guidelines. For larger datasets that may span many hospitals, these standards help to define a consistent reporting structure required for effective analysis. This can be seen in datasets such as the Healthcare Cost and Utilization Project (HCUP), which provides the basis for analyses on all aspects of inpatient hospital care [37, 101].
Finally, satisfaction and performance data is typically assessed through survey data. While there have been a mass of localized surveys, recent financial incentives have pushed for a more uniform metric between institutions. One emergent patient satisfaction standard has been created in the Hospital Consumer Assessment of Healthcare Providers and Systems (HCAHPS) survey [83], while a number of different approaches have been proposed to obtain, quantify, and standardize physician performance [11].
Aggregate Attributes
At the highest level of the HDS, we find aggregate data. Although upon first consideration, aggregate data may seem simply an extension of data encountered at the lower levels of the HDS, health-related data can be collected, inferred, and analyzed from a number of indirect sources.
Data Sources
A Bulletin of the World Health Organization has categorized data needs and sources into a hierarchy of community, facility, district, province, country, and global levels [1]. These sources include public health data typically curated by large national and government entities. Among the most prominent are the United States Census Bureau, Bureau of Labor Statistics, and the Agency for Healthcare Research and Quality (AHRQ). The Census Bureau conducts a number of surveys at varying intervals, intended to provide a comprehensive overview of the US population and government and industry, including the Decennial Census of Population and Housing, Economic Census, Census of Governments, American Community Survey (ACS) and Economic Indicators [20].
Traditionally, one of the fundamental uses of aggregate-level health data has been the review and analysis of population health [106]. Although the definition of “population health” often differs between sources, an overarching theme of population health is the idea that it “forces review of health outcomes in a population across determinants [71].” Determinants refer to the social, environmental and physical connections between structures, ideologies, policies, contexts, lifecourses or lifecycles, and their impact on health and well-being [128, 129].
While the data at this level primarily centers around population health and reporting, it can be drawn from many less-conventional channels, such as online social media [69]. In fact, the WHO maintains an extensive set of such indicators, which are often used for a variety of purposes including program management, allocation of resources, monitoring country progress, performance-based disbursement, global reporting, and so on [94]. It is the ability of aggregate data to draw from sources across these domains that positions it as such a powerful source of population health insights. As an example, work that investigated how income inequality may be related to health outcomes utilized data detailing investments in human capital and social resources. To address this question, data was derived from entities such as the United States Bureau of the Census, Bureau of Labor Statistics, Centers for Disease Control (CDC), Department of Agriculture, Education, and a number of additional aggregate level data sources [67].
Recent shifts in the regulatory landscape of the healthcare industry, including the passage of the Patient Protection and Affordable Care Act (ACA), have advanced the collection and analysis of aggregate-level health data though a number of initiatives. These include the establishment of the Patient-Centered Outcomes Research Institute, National Prevention, Health Promotion and Public Health Council, the promotion and implementation of accountable care organization (ACOs), and support for a new Prevention and Public Health Fund and community transformation grants [116].
Research Applications
As time passes and more extensive datasets become available for analysis, researchers have continued to find new ways in which to utilize aggregate health data. One of the most prominent examples for the utilization of such data has been the Institute for Healthcare Improvement (IHI) “Triple Aim” initiative. The Triple Aim, has been defined as an overarching goal of the US healthcare system to improve the experience of care, improve the health of populations, and reduce per capita costs of healthcare [13]. In their book “Big Data and Health Analytics,” Marconi and Lehmann state that “Health information is more readily available, and while walls have fallen to enable more sharing, there are needs beyond sharing of raw data to fully power a successful Triple Aim.” They note that “achieving better care, improved outcomes, and increased patient satisfaction requires analytics data to incorporate enterprise level aggregated information to ensure the best insights” [78].
Aggregated data has found application in a diverse range of research applications that seek to better understand how healthcare can and should be delivered. For instance, researchers have used aggregate data to model healthcare costs, using the data to model and estimate future medical and non-medical expenditures [87]. From a quality perspective, the Patient-Reported Outcomes Measurement Information System (PROMIS) program aimed to “create efficient measures that would be feasible to implement in busy office practices and that could provide a system of population health surveillance normed to the US general population” [55]. Finally from a population health perspective, macro-level interventions such as one provided by Kaiser Permanente “uses data about the population it serves, available through its system-wide electronic health record, to understand members’ health needs and the distribution of health outcomes. Using these data, Permanente offers a range of interventions tailored to the needs of different individuals and population groups to support people to remain healthy and to deliver the right treatments when they become ill” [4].
Yet, despite their careful curation, aggregate datasets present a number of inherent biases that must be accounted for as such data sources become an increasingly common component of larger computational models. First, in order to protect individual anonymity, data sources of low populous areas, infrequent diagnoses, or minimal response rates are often masked or removed from the final data. However, without correction, removal of this information can drastically shift interpretation of analyses that span multiple data elements. Further, it is important to note aggregate data reports are often collected across multiple years. As such, the utilization of even the most up to date information from multiple sources runs the risk of their collection occurring during different timespans, resulting in their capturing of potentially different latent factors creating an inherent and often undocumented bias in any analysis.
Data Standards
Aggregate level data provides an interesting challenge to the informatics community, as the format and collection standards of the data are governed by the individual agencies that manage the collection process. As might be expected, a number of practical issues have been identified with using data from large population level health databases [14]. As a result there has been a significant effort to design methods to improve the quality of data extracted from these databases, making it more suitable for analysis [46]
A number of works have investigated the implications of such fragmented collection techniques, including quantifying the validity and lifespan of clinical guidelines established using such data [110].
Future Research Directions
Moving forward, it is important to remember that the strength of organization frameworks lies beyond simply processing vast quantities of information into a cohesive structure. Their true value comes from the synergy this process creates as part of a larger body of research. Beyond organizing the diverse and interconnected collection of health data as it relates to an individual, the HDS framework can serve as a catalyst for researchers and practitioners to drive health informatics research forward. Such progress can take many forms, and in the following sections, we highlight a set of open research directions and their relation to the HDS.
Analytical Components
Perhaps the most direct application of the HDS can be found in the breadth of ongoing analytical research. The increasing scale, scope, and variety of available health care services permits a wealth of new and increasingly complex analyses. Although there exists a general understanding that these analyses will require data from multiple sources, these sources are still typically drawn from within a single level of the HDS. To unleash the full potential of health informatics, we must take a broader vision that employs data sources from several levels.
This idea of cross-level analysis has already exhibited early success, particularly within the fields of personalized and precision medicine. Identifying relations between clinical health and socio-demographic (clinical and demographic levels), clinical health and individual wellness (clinical and wellness attributes), and even clinical outcomes and omics data promise to offered tailored individual care and treatment plans. Utilization of the healthcare data spectrum offers a chance to push these types of analysis further. By offering a simple reference for those planning future research endeavors, the HDS allows researchers to identify a broader set of relevant data and to explore yet untapped relationships.
Interoperability
Identifying relevant data represents only one challenge of the advancing informatics environment. Once identified, there remain a number of open questions around how to obtain and prepare this data for analysis. Across the many levels of the HDS, it is clear that there are many sources of disparate data throughout the healthcare system, siloed away and confined for a narrow, particular use. Moreover, once acquired, considerable effort must be put forth to ensure consistency of the data elements across each of the sources from which they were collected.
The ability to connect data across these silos poses one of the primary obstacles for its broader use in translating research to practice. Current efforts in interoperability include the development of data standards for exchanging healthcare information electronically (such as the Fast Healthcare Interoperability Resource, FHIR), as well as the creation of reference infrastructure such as the Census Bureau Linkage Infrastructure (CBLI) to provide references to the methods, links, warehouses, and provisions of data [21, 126]. The goal of these standards has been to provide available and understandable electronic health data. However, much work remains to be done in the organization of these links. Such a need provides a number of possibilities through which researchers could utilize the HDS levels to understand which data elements are available for each source, the individual standards utilized by each, and the interoperability standards as they pertain to each individual element within and between levels.
Evolving Standards
Finally, the evolving nature of healthcare also provides an opportunity to extend the HDS itself. While the individual layers of the HDS are designed to represent a fixed portion of global health and wellness data, we are acutely aware that the standards through which the data are captured will continue to evolve over time. The introduction and deprecation of these standards presents its own set of challenges to the advancement of health analytics and interoperability. Where the lack of a clear connection between data captured at various time points may further serve to prohibit the use of historical data repositories, resulting in a significant loss of potentially useful information.
Once established, the HDS represents a promising structure through which to maintain references for such historical interoperability. As specified in this work, the HDS is two-dimensional, defining a set of data elements and the level in which they reside. However, this structure could be augmented to include a third and fourth dimension. The third dimension could capture the temporal evolution of standards for each data element over time. The fourth dimension could provide information pertaining to how a specific data entity maps to each standard. While this mapping is usually provided with each standard release, the centralization of these mappings—often referred to as data-crosswalks—would provide an incredibility valuable resource as health data continues to develop as a digital entity.
Conclusion
“Enormous amounts of new knowledge are barreling down the information highway, but they are not arriving at the doorsteps of our patients” [75]. This sentiment expressed by Dr. Claude Lenfant—the longest-serving Director of the National Heart, Lung, and Blood Institute—captures the state of informatics in healthcare today. Whether a result of government mandates, evidence-based practice guidelines, or perhaps even the generational shift in technology usage, access to digitized healthcare data continues to rise. Yet the enormity of the data alone cannot ensure its effective use, nor address the needs of clinicians and researchers. To achieve such a meaningful impact on the lives of individuals will require an intricate understanding of the strengths, limitations, and relations between the data available.
The work presented in this manuscript aims to take the first step in this direction, presenting a patient-centric organization of the variety of health data available today in the Healthcare Data Spectrum (HDS). The HDS provides a principal reference of the types of data available for analysis. It can aid experienced researchers as well as those who are just entering the health informatics field in building comprehensive multi-faceted models accounting for multiple aspects of an individual’s life and care. Further, it also offers a detailed listing of the organizations and standards that govern each data source, allowing those who utilize such data to do so in a more consistent manner, in turn supporting the push for accessible and reproducible research.
It is our hope that in this era of big data, such a framework can serve as a cornerstone in the organization of our vast supply of knowledge, helping guide it to its rightful destination in clinics, hospitals, and laboratories—connecting multiple sources of data, driving forward the field as a whole, and promoting health and wellness one data point, and one patient, at a time.
Funding Information
This work is supported in part by the National Science Foundation (NSF) Grant IIS-1447795.
Compliance with Ethical Standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Contributor Information
Keith Feldman, Email: kfeldman@nd.edu.
Reid A. Johnson, Email: rjohns15@nd.edu
Nitesh V. Chawla, Email: nchawla@nd.edu
References
- 1.AbouZahr C, Boerma T. Health information systems: the foundations of public health. Bull World Health Organ. 2005;83(8):578–583. [PMC free article] [PubMed] [Google Scholar]
- 2.Adashi EY, Geiger HJ, Fine MD. Health care reform and primary care—the growing importance of the community health center. England J Med. 2010;362(22):2047–2050. doi: 10.1056/NEJMp1003729. [DOI] [PubMed] [Google Scholar]
- 3.Aiken LH, Clarke SP, Sloane DM. Hospital staffing, organization, and quality of care: cross-national findings. Nurs Outlook. 2002;50(5):187–194. doi: 10.1067/mno.2002.126696. [DOI] [PubMed] [Google Scholar]
- 4.Alderwick H, Ham C, Buck D (2015) Population health systems. Going beyond integrated care. The King’s Fund
- 5.Appelboom G, Yang AH, Christophe BR, Bruce EM, Slomian J, Bruyère O, Bruce SS, Zacharia BE, Reginster JY, Connolly ES. The promise of wearable activity sensors to define patient recovery. J Clin Neurosci. 2014;21(7):1089–1093. doi: 10.1016/j.jocn.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Ashley EA. Towards precision medicine. Nat Rev Genet. 2016;17(9):507. doi: 10.1038/nrg.2016.86. [DOI] [PubMed] [Google Scholar]
- 7.Association AM Genetic testing. http://www.ama-assn.org/ama/pub/physician-resources/medical-science/genetics-molecular-medicine/related-policy-topics/genetic-testing.page. Accessed 31 May (2016)
- 8.Association AM (2007) Current procedural terminology: CPT. American Medical Association
- 9.Association AP et al. (2013) Diagnostic and Statistical Manual of Mental Disorders (DSM-5). American Psychiatric Pub
- 10.Association CE Guiding principles on the privacy and security of personal wellness data. Online (2015). https://fpf.org/wp-content/uploads/2015/10/CEA-Guiding-Principles-on-the-Privacy-and-Security-of-Personal-Wellness-Data-102215.pdf. Accessed 31 May (2016)
- 11.Barro AR. Survey and evaluation of approaches to physician performance measurement. Acad Med. 1973;48(11):1047–93. doi: 10.1097/00001888-197311000-00017. [DOI] [PubMed] [Google Scholar]
- 12.Berger S (2008) Fundamentals of health care financial management: a practical guide to fiscal issues and activities. Wiley
- 13.Berwick DM, Nolan TW, Whittington J. The triple aim: care, health, and cost. Health Aff. 2008;27(3):759–769. doi: 10.1377/hlthaff.27.3.759. [DOI] [PubMed] [Google Scholar]
- 14.Bibb SCG. Issues associated with secondary analysis of population health data. Appl Nurs Res. 2007;20(2):94–99. doi: 10.1016/j.apnr.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 15.Bloomrosen M, Detmer DE. Informatics, evidence-based care, and research; implications for national policy: a report of an american medical informatics association health policy conference. J Am Med Inform Assoc. 2010;17(2):115–123. doi: 10.1136/jamia.2009.001370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradley P, Kaplan J. Turning hospital data into dollars: healthcare financial executives can use predictive analytics to enhance their ability to capture charges and identify underpayments. Healthc Financ Manage. 2010;64(2):64–69. [PubMed] [Google Scholar]
- 17.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (miame)—toward standards for microarray data. Nat Genet. 2001;29(4):365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 18.Brennan N, Conway PH, Tavenner M. The medicare physician-data release—context and rationale. England J Med. 2014;371(2):99–101. doi: 10.1056/NEJMp1405026. [DOI] [PubMed] [Google Scholar]
- 19.Brown ML, Riley GF, Potosky AL, Etzioni RD. Obtaining long-term disease specific costs of care: application to medicare enrollees diagnosed with colorectal cancer. Med Care. 1999;37(12):1249–1259. doi: 10.1097/00005650-199912000-00008. [DOI] [PubMed] [Google Scholar]
- 20.Bureau UC Census product catalog (2012). http://www.census.gov/mp/www/cat/index.html. Accessed 31 May (2016)
- 21.Bureau UC Census bureau linkage infrastructure (cbli) (2016). https://www.census.gov/about/adrm/data-linkage/what.html. Accessed 31 May (2016)
- 22.Carroll R, Cnossen R, Schnell M, Simons D. Continua: an interoperable personal healthcare ecosystem. Pervas. Comput. IEEE. 2007;6(4):90–94. doi: 10.1109/MPRV.2007.72. [DOI] [Google Scholar]
- 23.Castle AL, Fiehn O, Kaddurah-Daouk R, Lindon JC. Metabolomics standards workshop and the development of international standards for reporting metabolomics experimental results. Brief Bioinform. 2006;7(2):159–165. doi: 10.1093/bib/bbl008. [DOI] [PubMed] [Google Scholar]
- 24.Centers for Medicare & Medicaid Services (1996) The Health Insurance Portability and Accountability Act of 1996 (HIPAA). Online at http://www.cms.hhs.gov/hipaa/
- 25.Chen R, Snyder M. Promise of personalized omics to precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2013;5(1):73–82. doi: 10.1002/wsbm.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chervitz Stephen A., Deutsch Eric W., Field Dawn, Parkinson Helen, Quackenbush John, Rocca-Serra Phillipe, Sansone Susanna-Assunta, Stoeckert Christian J., Taylor Chris F., Taylor Ronald, Ball Catherine A. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2011. Data Standards for Omics Data: The Basis of Data Sharing and Reuse; pp. 31–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium GO. The gene ontology (go) database and informatics resource. Nucl Acids Res. 2004;32(suppl 1):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crimmins EM. Demography: the past 30 years, the present, and the future. Demography. 1993;30(4):579–591. doi: 10.2307/2061807. [DOI] [PubMed] [Google Scholar]
- 29.Crimmins EM, Seeman T (2001) Integrating biology into demographic research on health and aging (with a focus on the macarthur study of successful aging). In: Cells and surveys: should biological measures be included in social science research? National Academies Press (US) [PubMed]
- 30.Delaney C, Moorhead S. The nursing minimum data set, standarized language, and health care quality. J Nurs Care Q. 1995;10(1):16–30. doi: 10.1097/00001786-199510010-00005. [DOI] [PubMed] [Google Scholar]
- 31.Demiris G, Afrin LB, Speedie S, Courtney KL, Sondhi M, Vimarlund V, Lovis C, Goossen W, Lynch C. Patient-centered applications: use of information technology to promote disease management and wellness. A white paper by the amia knowledge in motion working group. J Am Med Inform Assoc. 2008;15(1):8–13. doi: 10.1197/jamia.M2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dettmer K, Hammock BD. Metabolomics—a new exciting field within the “omics” sciences. Environ Health Perspect. 2004;112(7):A396. doi: 10.1289/ehp.112-1241997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Centers for Disease Control and Prevention (2014) Classification of diseases, functioning, and disability. International classification of diseases, tenth revision, clinical modification (ICD-10-CM) CDC web site
- 34.Dolin RH, Alschuler L, Boyer S, Beebe C, Behlen FM, Biron PV, Shabo A. Hl7 clinical document architecture, release 2. J Am Med Inform Assoc. 2006;13(1):30–39. doi: 10.1197/jamia.M1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Draper M, Cohen P, Buchan H. Seeking consumer views: what use are results of hospital patient satisfaction surveys? Int J Qual Health Care. 2001;13(6):463–468. doi: 10.1093/intqhc/13.6.463. [DOI] [PubMed] [Google Scholar]
- 36.Dwyer SJIII, Weaver AC, Hughes KK. Health insurance portability and accountability act. Secur Issues Digit Med Enterp. 2004;72(2):9–18. [Google Scholar]
- 37.Eisenberg JM (2000) Quality research for quality healthcare: the data connection. Health services research 35(2) xii
- 38.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286(5439):487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 39.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CR, Shimizu T, Spener F, van Meer G, Wakelam MJ, Dennis EA. Update of the lipid maps comprehensive classification system for lipids. J Lipid Res. 2009;50(Supplement):S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feldman B, Martin EM, Skotnes T (2012) Big data in healthcare hype and hope. October 2012. Dr Bonnie, 360
- 41.Feldman K, Chawla NV. Does medical school training relate to practice? Evidence from big data. Big Data. 2015;3(2):103–113. doi: 10.1089/big.2014.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feldman K, Faust L, Wu X, Huang C, Chawla NV (2017) Beyond volume: the impact of complex healthcare data on the machine learning pipeline. In: Towards Integrative machine learning and knowledge extraction. Springer, pp 150–169
- 43.Fenton JJ, Jerant AF, Bertakis KD, Franks P. The cost of satisfaction: a national study of patient satisfaction, health care utilization, expenditures, and mortality. Arch Intern Med. 2012;172(5):405–411. doi: 10.1001/archinternmed.2011.1662. [DOI] [PubMed] [Google Scholar]
- 44.Fiehn O, Robertson D, Griffin J, van der Werf M, Nikolau B, Morrison N, Sumner LW, Goodacre R, Hardy NW, Taylor C, et al. The metabolomics standards initiative (msi) Metabolomics. 2007;3(3):175–178. doi: 10.1007/s11306-007-0070-6. [DOI] [Google Scholar]
- 45.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (migs) specification. Nat Biotechnol. 2008;26(5):541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher ES, Baron JA, Malenka DJ, Barrett J, Bubolz TA. Overcoming potential pitfalls in the use of medicare data for epidemiologic research. Am J Public Health. 1990;80(12):1487–1490. doi: 10.2105/AJPH.80.12.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Food U, Administration D et al. National drug code directory. Internet address: http://www.fda.gov/cder/ndc/ (2011)
- 48.Gee J, Button M, Brooks G (2010) The financial cost of healthcare fraud: what data from around the world shows. Tech. rep., MacIntyre Hudson
- 49.Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res. 2009;154(6):277–287. doi: 10.1016/j.trsl.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Gray Kristian A., Yates Bethan, Seal Ruth L., Wright Mathew W., Bruford Elspeth A. Genenames.org: the HGNC resources in 2015. Nucleic Acids Research. 2014;43(D1):D1079–D1085. doi: 10.1093/nar/gku1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greenbaum D, Luscombe NM, Jansen R, Qian J, Gerstein M. Interrelating different types of genomic data, from proteome to secretome:’oming in on function. Genome Res. 2001;11(9):1463–1468. doi: 10.1101/gr.207401. [DOI] [PubMed] [Google Scholar]
- 52.Greenfield S, Nelson EC, Zubkoff M, Manning W, Rogers W, Kravitz RL, Keller A, Tarlov AR, Ware JE. Variations in resource utilization among medical specialties and systems of care: results from the medical outcomes study. Jama. 1992;267(12):1624–1630. doi: 10.1001/jama.1992.03480120062034. [DOI] [PubMed] [Google Scholar]
- 53.Hall LM, Doran D, Pink GH. Nurse staffing models, nursing hours, and patient safety outcomes. J Nurs Admin. 2004;34(1):41–45. doi: 10.1097/00005110-200401000-00009. [DOI] [PubMed] [Google Scholar]
- 54.Hattie JA, Myers JE, Sweeney TJ. A factor structure of wellness: theory, assessment, analysis, and practice. J Counsel Develop. 2004;82(3):354–364. doi: 10.1002/j.1556-6678.2004.tb00321.x. [DOI] [Google Scholar]
- 55.Hays RD, Spritzer KL, Thompson WW, Cella D. Us general population estimate for ”excellent” to ”poor” self-rated health item. J Gen Intern Med. 2015;30(10):1511–1516. doi: 10.1007/s11606-015-3290-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.of Health UD, Services H et al. (1980) ICD 9 CM. The International Classification of Diseases. 9. Rev: Clinical Modification.; Vol. 1: Diseases: Tabular List. ; Vol. 2: Diseases: Alphabetic Index; Vol. 3: Procedures: Tabular List and Alphabetic Index. US Government Printing Office
- 57.of Health UD, Services H et al. (2011) Us department of health and human services implementation guidance on data collection standards for race, ethnicity, sex, primary language and disability status
- 58.on Accreditation of Healthcare Organizations JC (1991) Accreditation manual for hospitals, vol. 1 Joint Commission on Accreditation of Healthcare Organizations
- 59.Hettler B. Wellness: encouraging a lifetime pursuit of excellence. Health Values. 1984;8(4):13. [PubMed] [Google Scholar]
- 60.Hibbard JH, Stockard J, Tusler M. Hospital performance reports: impact on quality, market share, and reputation. Health Aff. 2005;24(4):1150–1160. doi: 10.1377/hlthaff.24.4.1150. [DOI] [PubMed] [Google Scholar]
- 61.Horaitis O, Cotton RG. The challenge of documenting mutation across the genome: the human genome variation society approach. Human Mutation. 2004;23(5):447–452. doi: 10.1002/humu.20038. [DOI] [PubMed] [Google Scholar]
- 62.Horgan RP, Kenny LC. ’omic’technologies: genomics, transcriptomics, proteomics and metabolomics. Obstetr Gynaecol. 2011;13(3):189–195. [Google Scholar]
- 63.Huang YT. Integrative modeling of multiple genomic data from different types of genetic association studies. Biostatistics. 2014;15(4):587–602. doi: 10.1093/biostatistics/kxu014. [DOI] [PubMed] [Google Scholar]
- 64.Jacob SG, Ramani RG (2012) Data mining in clinical data sets: a review. IJAIS-ISSN: 2249-0868 Foundation of Computer Science FCS, New York USA 4(6)
- 65.Jensen PB, Jensen LJ, Brunak S. Mining electronic health records: towards better research applications and clinical care. Nat Rev Gen. 2012;13(6):395. doi: 10.1038/nrg3208. [DOI] [PubMed] [Google Scholar]
- 66.Kailas A, Chong CC, Watanabe F. From mobile phones to personal wellness dashboards. Pulse, IEEE. 2010;1(1):57–63. doi: 10.1109/MPUL.2010.937244. [DOI] [PubMed] [Google Scholar]
- 67.Kaplan GA, Pamuk ER, Lynch JW, Cohen RD, Balfour JL. Inequality in income and mortality in the united states: analysis of mortality and potential pathways. Bmj. 1996;312(7037):999–1003. doi: 10.1136/bmj.312.7037.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaplan WA. Can the ubiquitous power of mobile phones be used to improve health outcomes in developing countries? Global Health. 2006;2(1):1. doi: 10.1186/1744-8603-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kass-Hout TA, Alhinnawi H. Social media in public health. British Med Bull. 2013;108(1):5–24. doi: 10.1093/bmb/ldt028. [DOI] [PubMed] [Google Scholar]
- 70.Kayyali B, Knott D, Van Kuiken S (2013) The big-data revolution in us health care: accelerating value and innovation. Mc Kinsey & Company, pp 1–13
- 71.Kindig D, Stoddart G. What is population health? Am J Public Health. 2003;93(3):380–383. doi: 10.2105/AJPH.93.3.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landon BE, Normand SLT, Blumenthal D, Daley J. Physician clinical performance assessment: prospects and barriers. Jama. 2003;290(9):1183–1189. doi: 10.1001/jama.290.9.1183. [DOI] [PubMed] [Google Scholar]
- 73.LAVE JUDITH R., PASHOS CHRIS L., ANDERSON GERARDF., BRAILER DAVID, BUBOLZ THOMAS, CONRAD DOUGLAS, FREUND DEBORAH A., FOX STEVEN H., KEELER EMMETT, LIPSCOMB JOSEPH, LUFT HAROLD S., PROVENZANO GEORGE. Costing Medical Care. Medical Care. 1994;32(Supplement):JS90. doi: 10.1097/00005650-199407001-00007. [DOI] [PubMed] [Google Scholar]
- 74.Lemieux-Charles L, McGuire WL. What do we know about health care team effectiveness? a review of the literature. Med Care Res Rev. 2006;63(3):263–300. doi: 10.1177/1077558706287003. [DOI] [PubMed] [Google Scholar]
- 75.Lenfant C. Clinical research to clinical practice—lost in translation? England J Med. 2003;349(9):868–874. doi: 10.1056/NEJMsa035507. [DOI] [PubMed] [Google Scholar]
- 76.Li J, Huang KY, Jin J, Shi J. A survey on statistical methods for health care fraud detection. Health Care Manag Sci. 2008;11(3):275–287. doi: 10.1007/s10729-007-9045-4. [DOI] [PubMed] [Google Scholar]
- 77.Lopez AD, Mathers CD, Ezzati M, Jamison DT, Murray CJ. Global and regional burden of disease and risk factors, 2001: systematic analysis of population health data. Lancet. 2006;367(9524):1747–1757. doi: 10.1016/S0140-6736(06)68770-9. [DOI] [PubMed] [Google Scholar]
- 78.Marconi K, Lehmann H (2014) Big data and health analytics. CRC Press
- 79.McDonald CJ, Huff SM, Suico JG, Hill G, Leavelle D, Aller R, Forrey A, Mercer K, DeMoor G, Hook J, et al. Loinc, a universal standard for identifying laboratory observations: a 5-year update. Clin Chem. 2003;49(4):624–633. doi: 10.1373/49.4.624. [DOI] [PubMed] [Google Scholar]
- 80.McGrath MJ, Scanaill CN (2013) Wellness, fitness, and lifestyle sensing applications. In: Sensor technologies. Springer, pp 217–248
- 81.McGraw-Hill Concise dictionary of modern medicine. Online (2002). Accessed 31 May (2016)
- 82.McLean R (2002) Financial management in health care organizations. Cengage Learning
- 83.for Medicare & Medicaid Services C Hospital consumer assessment of healthcare providers and systems. Online. http://www.hcahpsonline.org/home.aspx. Accessed 31 May (2016)
- 84.for Medicare & Medicaid Services C ICD-9-CM, ICD-10-CM, ICD-10-PCS, CPT, and HCPCS code sets. Online (2015). Accessed 31 May 2016. ICN: 900943
- 85.for Medicare & Medicaid Services C et al. (2003) Healthcare Common Procedure Coding System (HCPCS) Centers for Medicare & Medicaid Services
- 86.Centers for Medicare & Medicaid Services H et al. (2004) Hipaa administrative simplification: standard unique health identifier for health care providers. Final rule. Fed Register 69(15):3433 [PubMed]
- 87.Meltzer D. Accounting for future costs in medical cost-effectiveness analysis. J Health Econ. 1997;16(1):33–64. doi: 10.1016/S0167-6296(96)00507-3. [DOI] [PubMed] [Google Scholar]
- 88.Mildenberger P, Eichelberg M, Martin E. Introduction to the dicom standard. Europ Radiol. 2002;12(4):920–927. doi: 10.1007/s003300101100. [DOI] [PubMed] [Google Scholar]
- 89.Müller M, Kersten S. Nutrigenomics: goals and strategies. Nat Rev Gen. 2003;4(4):315–322. doi: 10.1038/nrg1047. [DOI] [PubMed] [Google Scholar]
- 90.Murdoch TB, Detsky AS. The inevitable application of big data to health care. Jama. 2013;309(13):1351–1352. doi: 10.1001/jama.2013.393. [DOI] [PubMed] [Google Scholar]
- 91.Nelson CW, Niederberger J. Patient satisfaction surveys: an opportunity for total quality improvement. Hosp Health Serv Admin. 1990;35(3):409–428. [PubMed] [Google Scholar]
- 92.Orchard S, Hermjakob H, Apweiler R. The proteomics standards initiative. Proteomics. 2003;3(7):1374–1376. doi: 10.1002/pmic.200300496. [DOI] [PubMed] [Google Scholar]
- 93.Organization WH et al. International classification of diseases (ICD) (2012)
- 94.Organization WH et al. Global reference list of 100 core health indicators (2015)
- 95.Ostherr K, Borodina S, Bracken RC, Lotterman C, Storer E, Williams B. Trust and privacy in the context of user-generated health data. Big Data Soc. 2017;4(1):2053951717704,673. doi: 10.1177/2053951717704673. [DOI] [Google Scholar]
- 96.Pol LG, Thomas RK (2000) The demography of health and health care. Springer Science & Business Media
- 97.Pol L G, Thomas RK (2013) Health demography: an evolving discipline. In: The demography of health and healthcare. Springer, pp 1–12
- 98.Poulton BC, West MA. The determinants of effectiveness in primary health care teams. J Interprof Care. 1999;13(1):7–18. doi: 10.3109/13561829909025531. [DOI] [Google Scholar]
- 99.Raghupathi W, Raghupathi V. Big data analytics in healthcare: promise and potential. Health Inf Sci Syst. 2014;2(1):3. doi: 10.1186/2047-2501-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Registrar F. Revisions to the standards for the classification of federal data on race and ethnicity. Fed Registr. 1997;62:58,781–58,790. [Google Scholar]
- 101.Retchin SM, Ballard D. Commentary: establishing standards for the utility of administrative claims data. Health Serv Res. 1998;32(6):861. [PMC free article] [PubMed] [Google Scholar]
- 102.Richard L, Gauvin L, Raine K. Ecological models revisited: their uses and evolution in health promotion over two decades. Ann Rev Public Health. 2011;32:307–326. doi: 10.1146/annurev-publhealth-031210-101141. [DOI] [PubMed] [Google Scholar]
- 103.Riley GF. Administrative and claims records as sources of health care cost data. Med Care. 2009;47(7_Supplement_1):S51–S55. doi: 10.1097/MLR.0b013e31819c95aa. [DOI] [PubMed] [Google Scholar]
- 104.Rosenbloom S Trent. Person-generated health and wellness data for health care. Journal of the American Medical Informatics Association. 2016;23(3):438–439. doi: 10.1093/jamia/ocw059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Safran C, Bloomrosen M, Hammond WE, Labkoff S, Markel-Fox S, Tang PC, Detmer DE. Toward a national framework for the secondary use of health data: an american medical informatics association white paper. J Am Med Inform Assoc. 2007;14(1):1–9. doi: 10.1197/jamia.M2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schiller JS, Adams PF, Nelson ZC. Summary health statistics for the us population: national health interview survey, 2003. Vital and health statistics. Series 10. Data Nat Health Surv. 2005;2005(224):1–104. [PubMed] [Google Scholar]
- 107.Schneider MV, Orchard S (2011) Omics technologies, data and bioinformatics principles. Bioinforma Omics Data: Methods Protocols, 3–30 [DOI] [PubMed]
- 108.Shameer K, Badgeley MA, Miotto R, Glicksberg BS, Morgan JW, Dudley JT (2016) Translational bioinformatics in the era of real-time biomedical, health care and wellness data streams. Briefings in bioinformatics p bbv118 [DOI] [PMC free article] [PubMed]
- 109.Shapiro M, Johnston D, Wald J, Mon D (2012) Patient-generated health data. RTI International
- 110.Shekelle PG, Ortiz E, Rhodes S, Morton SC, Eccles MP, Grimshaw JM, Woolf SH. Validity of the agency for healthcare research and quality clinical practice guidelines: how quickly do guidelines become outdated? Jama. 2001;286(12):1461–1467. doi: 10.1001/jama.286.12.1461. [DOI] [PubMed] [Google Scholar]
- 111.Shryock HS, Siegel JS, Larmon EA (1973) The methods and materials of demography. US Bureau of the Census
- 112.Siegel JS (2011) The demography and epidemiology of human health and aging. Springer Science & Business Media
- 113.Skelly AC, Dettori JR, Brodt ED. Assessing bias: the importance of considering confounding. Evidence-based Spine-care J. 2012;3(1):9. doi: 10.1055/s-0031-1298595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smith HL. Some thoughts on causation as it relates to demography and population studies. Popul Dev Rev. 2003;29(3):459–469. doi: 10.1111/j.1728-4457.2003.00459.x. [DOI] [Google Scholar]
- 115.Stanhope M, Lancaster J (2015) Public health nursing: population-centered health care in the community. Elsevier Health Sciences
- 116.Stoto MA. Population health in the Affordable Care Act era, vol 1. Washington, DC: AcademyHealth; 2013. [Google Scholar]
- 117.Suetens P (2009) Fundamentals of medical imaging. Cambridge University Press
- 118.Taber KAJ, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Int Med. 2014;174(2):275–280. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 119.Taylor CF. Standards for reporting bioscience data: a forward look. Drug Discov Today. 2007;12(13):527–533. doi: 10.1016/j.drudis.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 120.Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, et al. The minimum information about a proteomics experiment (miape) Nature Biotechnol. 2007;25(8):887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 121.Tebani A, Afonso C, Marret S, Bekri S. Omics-based strategies in precision medicine: toward a paradigm shift in inborn errors of metabolism investigations. Int J Molec Sci. 2016;17(9):1555. doi: 10.3390/ijms17091555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Ommen B, Stierum R. Nutrigenomics: exploiting systems biology in the nutrition and health arena. Curr Opin Biotechnol. 2002;13(5):517–521. doi: 10.1016/S0958-1669(02)00349-X. [DOI] [PubMed] [Google Scholar]
- 123.Veeramah KR, Hammer MF. The impact of whole-genome sequencing on the reconstruction of human population history. Nat Rev Gen. 2014;15(3):149–162. doi: 10.1038/nrg3625. [DOI] [PubMed] [Google Scholar]
- 124.Ware JE, Snyder MK, Wright WR, Davies AR. Defining and measuring patient satisfaction with medical care. Eval Program Plan. 1983;6(3):247–263. doi: 10.1016/0149-7189(83)90005-8. [DOI] [PubMed] [Google Scholar]
- 125.Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4(7):594–610. doi: 10.1038/nrd1776. [DOI] [PubMed] [Google Scholar]
- 126.West M, Ginsburg GS, Huang AT, Nevins JR. Embracing the complexity of genomic data for personalized medicine. Genome Res. 2006;16(5):559–566. doi: 10.1101/gr.3851306. [DOI] [PubMed] [Google Scholar]
- 127.Whetzel PL, Parkinson H, Causton HC, Fan L, Fostel J, Fragoso G, Game L, Heiskanen M, Morrison N, Rocca-Serra P, et al. The mged ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22(7):866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 128.Wilkinson RG, Marmot MG (2003) Social determinants of health: the solid facts. World Health Organization
- 129.Williams GH. The determinants of health: structure, context and agency. Sociol Health Illness. 2003;25(3):131–154. doi: 10.1111/1467-9566.00344. [DOI] [PubMed] [Google Scholar]
- 130.Wood WA, Bennett AV, Basch E. Emerging uses of patient generated health data in clinical research. Molec Oncol. 2015;9(5):1018–1024. doi: 10.1016/j.molonc.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wu PY, Cheng CW, Kaddi CD, Venugopalan J, Hoffman R, Wang MD. –omic and electronic health record big data analytics for precision medicine. IEEE Trans Biomed Eng. 2017;64(2):263–273. doi: 10.1109/TBME.2016.2573285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wunsch G. Confounding and control. Demograph Res. 2007;16(4):97–120. doi: 10.4054/DemRes.2007.16.4. [DOI] [Google Scholar]
- 133.Yumak Z, Pu P. Survey of sensor-based personal wellness management systems. BioNanoScience. 2013;3(3):254–269. doi: 10.1007/s12668-013-0099-0. [DOI] [Google Scholar]