Abstract
PULCON (Pulse Length Based Concentration Determination) is a powerful, versatile, non-invasive, and accurate technique for measuring solution concentrations during routine NMR spectroscopy. As solutes are quantified directly by their unique resonances, this technique avoids weight-based errors caused by contaminants (e.g. moisture), allows NMR samples to be directly employed in biological assays, and is particularly useful for quantifying small molecules, peptides, unstable molecules, and other materials that are difficult to weigh or handle. This article provides an introductory guide for biological and medicinal chemists, and highlights the diversity of applications.
Background
PULCON[1] (Pulse Length Based Concentration Determination) is a powerful, non-invasive, and accurate (> 98%) technique for measuring solute concentrations by quantitative NMR spectroscopy[2, 3] that is applicable to any NMR sensitive solute that can produce good quality NMR spectra. This article provides an introductory guide for biological and medicinal chemists, and highlights the diversity of applications.
PULCON can be conducted with standard NMR spectrometers without additional specialised equipment or software, and has many advantages, especially for preparing compound solutions for biological assays. Firstly, amounts of solute or solvent do not need to be measured, with individual solute concentrations directly quantified by their unique signals (resonances). This avoids weight-based errors caused by contaminants (e.g. moisture, salts) and counter-ions, and facilitates the solution preparation of viscous products or small amounts of material, which can be difficult to weigh. Secondly, internal standards are not needed, with concentrations determined by routine 1H NMR spectroscopy (typically in DMSO-d6 or 10 % D2O/H2O with water suppression). These allow the NMR sample to be directly employed in biological assays after spectral characterisation, particularly advantageous if the sample is scarce, unstable, or non-isolatable.
PULCON for most of the molecules encountered in biological and medicinal chemistry can be conducted as follows (Fig. 1). The 1H NMR spectra of the sample of interest and a reference sample of known concentration (e.g. 15 mM benzoic acid in DMSO-d6,[4] 10 mM sucrose in 10 % D2O/H2O,[1] or 40 mM cholesterol in CDCl3),[5] should be measured using the same spectrometer (probe) and pulse program. The solutions should fully fill the active coil volume. Viscous samples should be avoided due to line broadening and reduced signal to noise. After tuning, matching, and shimming, both spectra should be recorded using the accurately determined 90° pulse lengths unique to each sample (see Fig. 2), the same receiver gain, and a long relaxation time of 30 seconds (sufficient to fully relax most 1H nuclei[1, 6]). The temperature of the two samples can be different.
Figure 1.
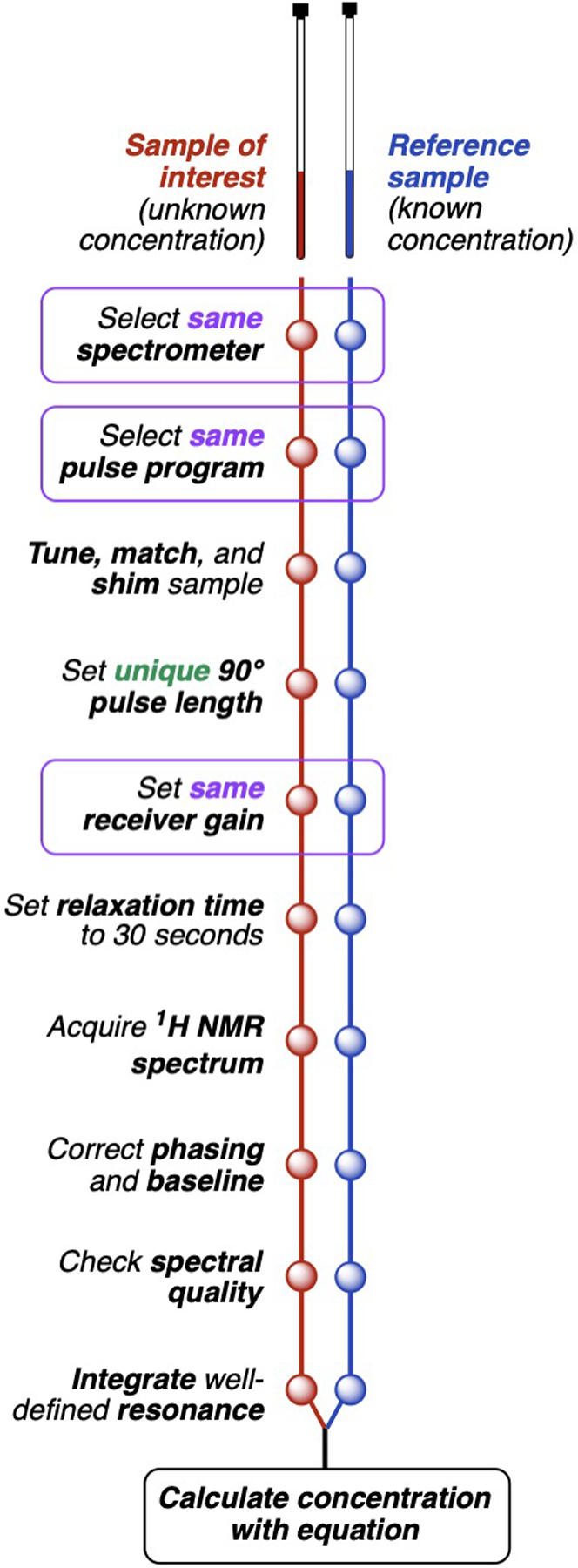
Flowchart of NMR sample concentration determination using PULCON.
Figure 2.
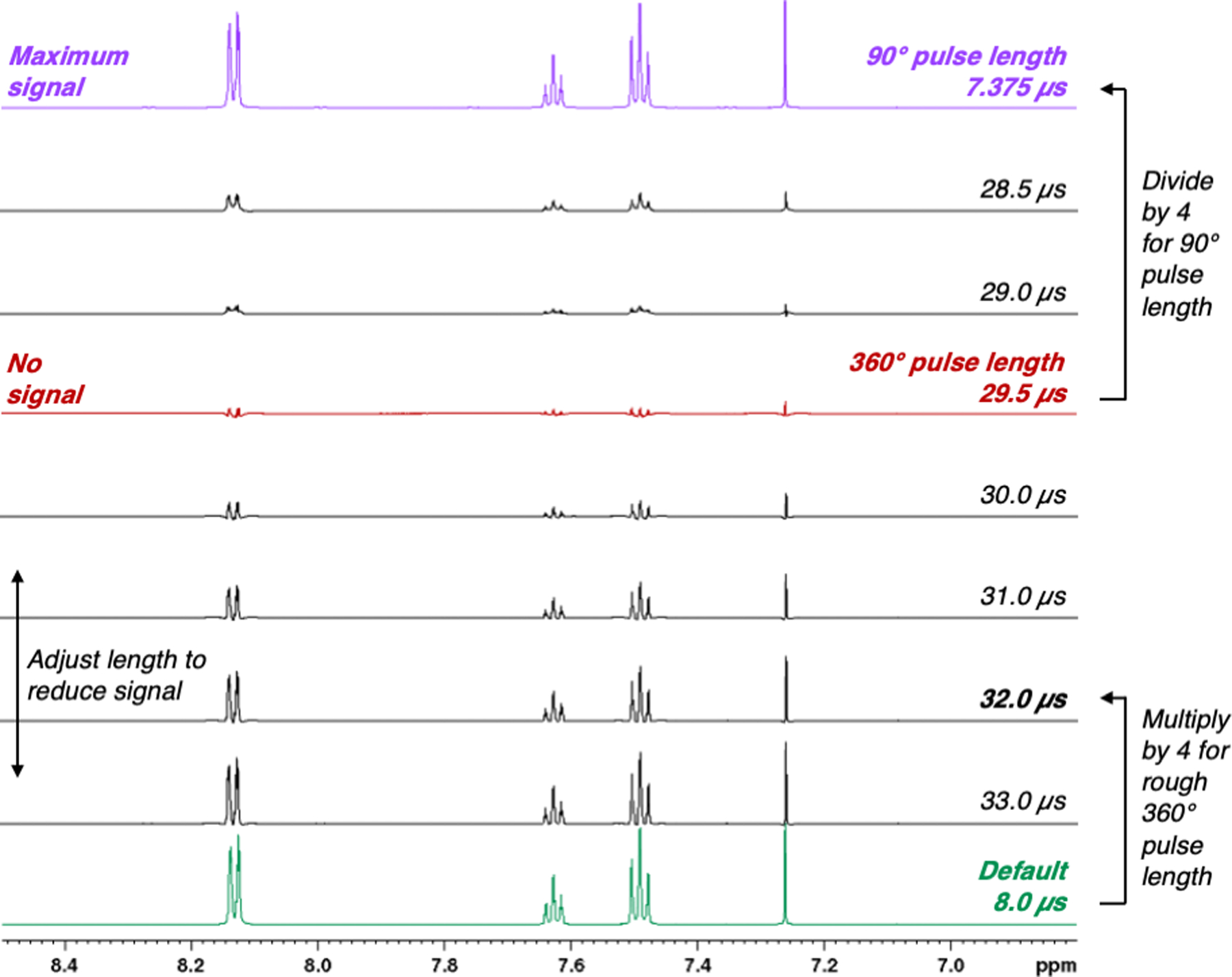
The 90° pulse length can be obtained by dividing the accurately measured 360° pulse length (no signal) by 4.
After careful phasing, baseline corrections, and checking for spectral quality (e.g. adequate signal to noise and solvent suppression [if required], no baseline distortions, see Fig. 3), a well-defined resonance should be integrated in each spectrum. Resonances that are broad (suggesting chemical or conformational exchange) or near solvent suppression should be avoided, as their integrations may be highly inaccurate (Fig. 3).
Figure 3.
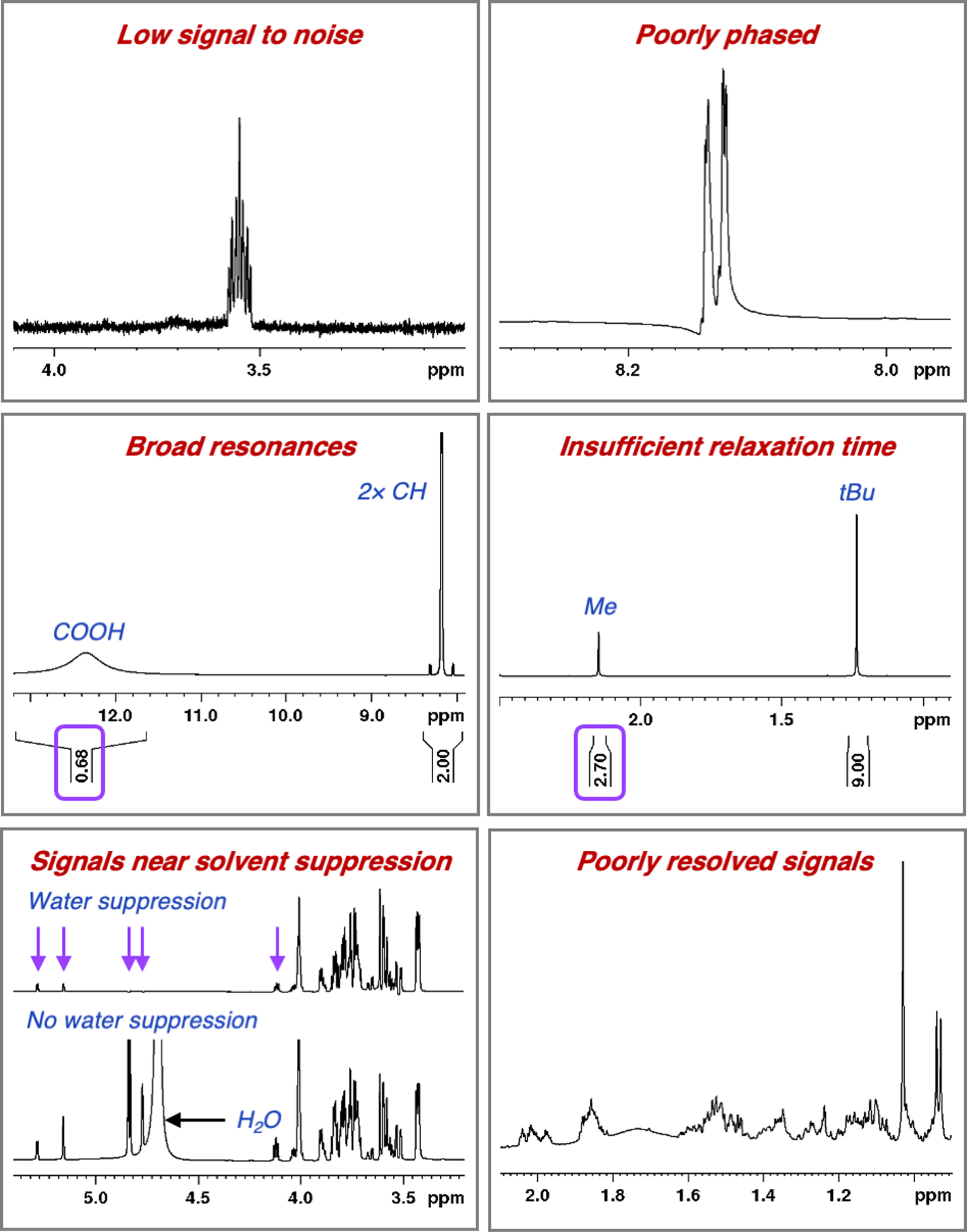
Instances where PULCON may be inaccurate.
The solute concentration is then calculated using the adapted[1] equation below,[7] which relates concentrations (c) of the reference (R) and sample of interest (X) with the integration of the well-defined resonance (I), the number of protons corresponding to that resonance (A), the sample temperature in Kelvin (T), the 90° pulse length (θ), and the number of scans (n) (see Fig. 4), with f equal to 1 if receiver gain and the pulse program are the same for both samples.[8]
Figure 4.
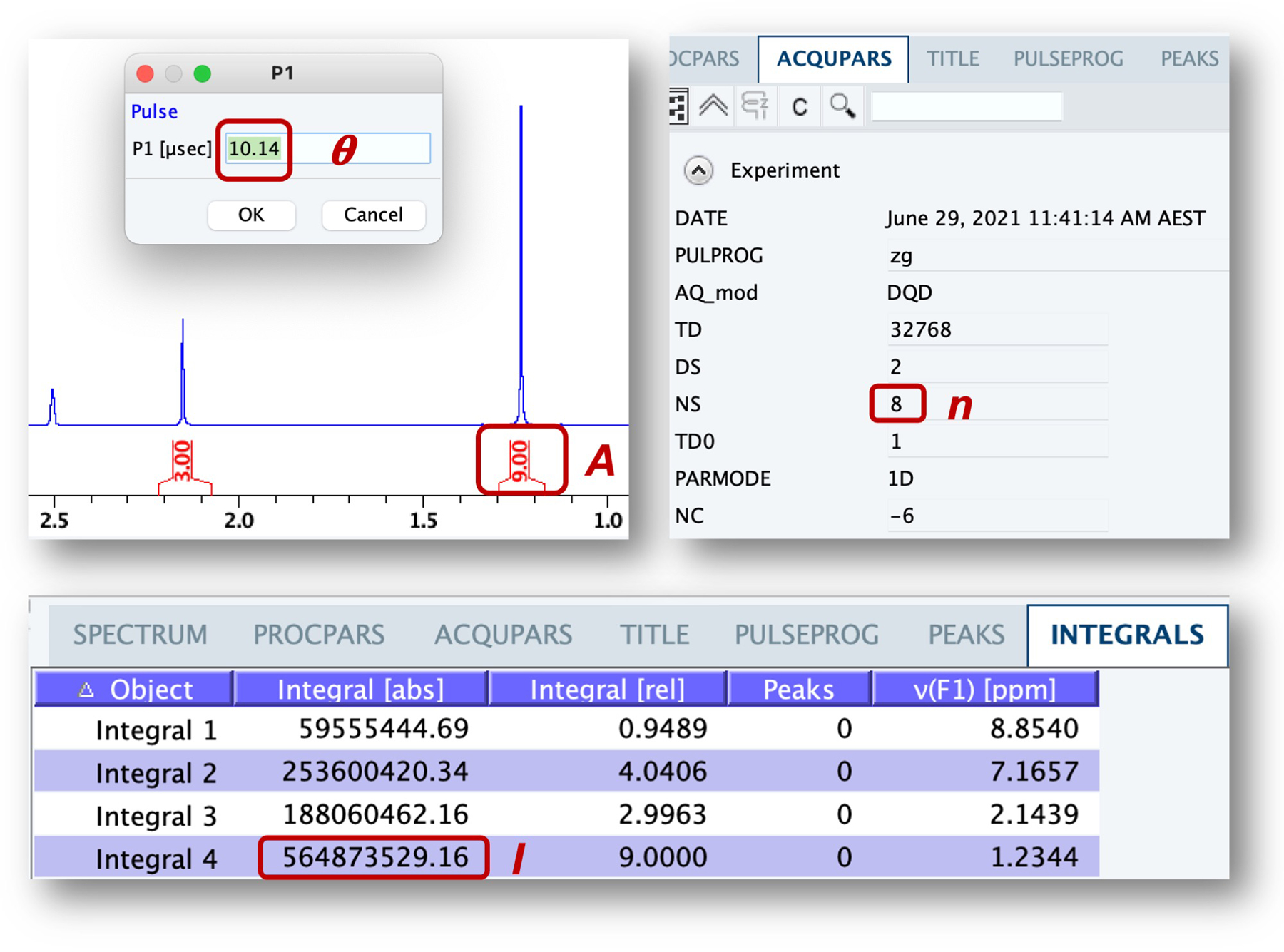
PULCON parameters and values are readily obtained from NMR software after spectroscopic analysis (TopSpin 4.02 shown).
Importantly, the values of the reference sample can then be used for subsequent samples on the same probe and pulse program,[1] even if the reference was in a different solvent[9]. The 2D NMR extension[10] enables quantification of molecules with significantly overlapped resonances.
Applications
Recent applications are wide-ranging. For small molecules in biological and medicinal chemistry (Fig. 5), PULCON was used to prepare solutions of physicochemical probes (1[11] and 2[12]) and radical scavengers (3)[13] for studying ligand-protein binding and oxidative stress, respectively. It was also used to analyse the DMSO solubility of 939 drug fragments,[14] and to support entire drug discovery campaigns (e.g. 4, 5 and 6),[15–17] demonstrating its capacity to facilitate rapid generation of structure-activity relationships.
Figure 5.
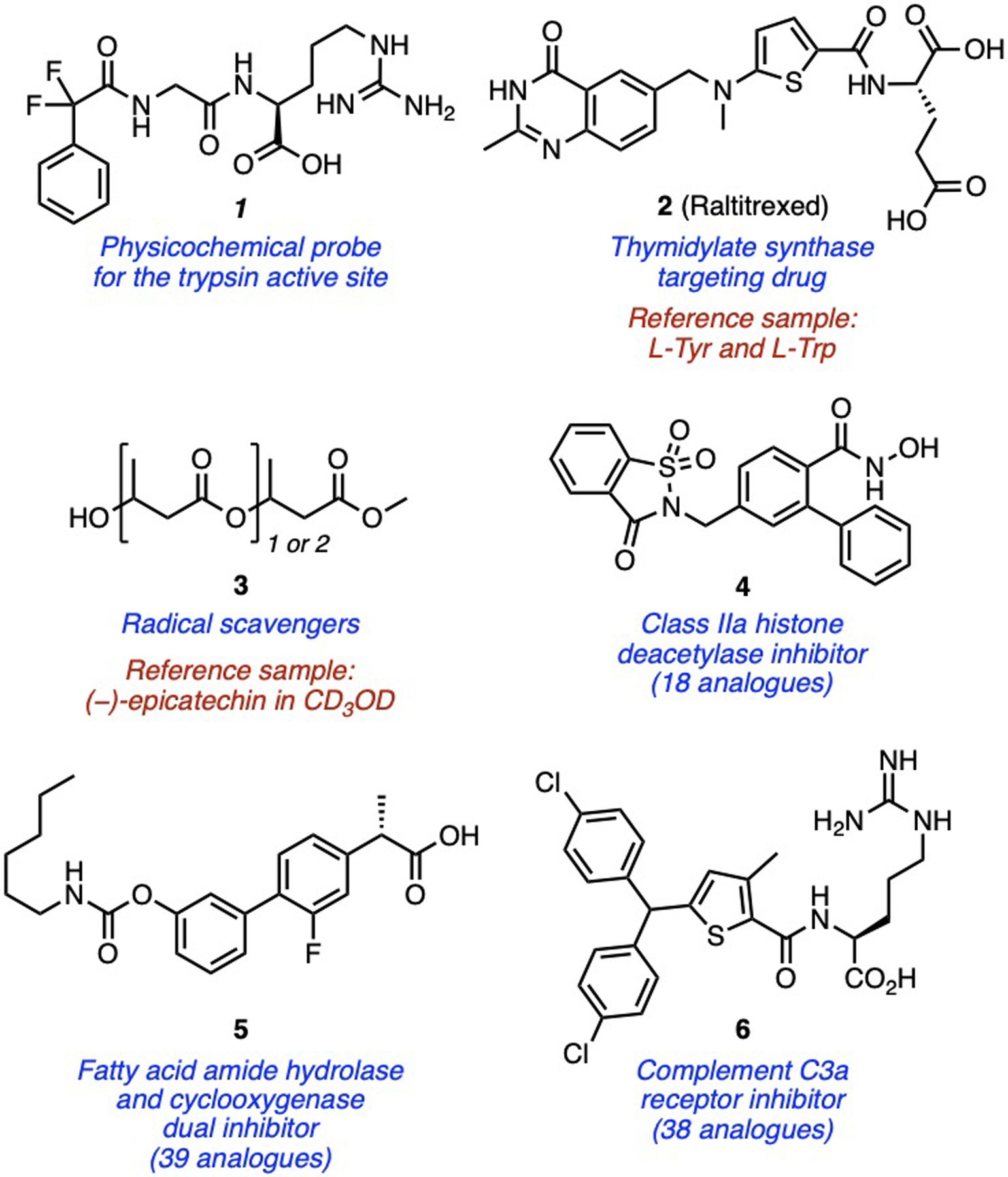
Recent examples of PULCON quantification of small molecules in biological and medicinal chemistry.
Metabolite mixtures from bacterial (7 – 11)[18] and cancer cells (7, 12 – 14)[19] could be spectroscopically quantified (Fig. 6). Coffee quality and authenticity were monitored via PULCON after CDCl3 extraction of natural products from coffee grounds (15 – 19, Fig. 7).[20]
Figure 6.
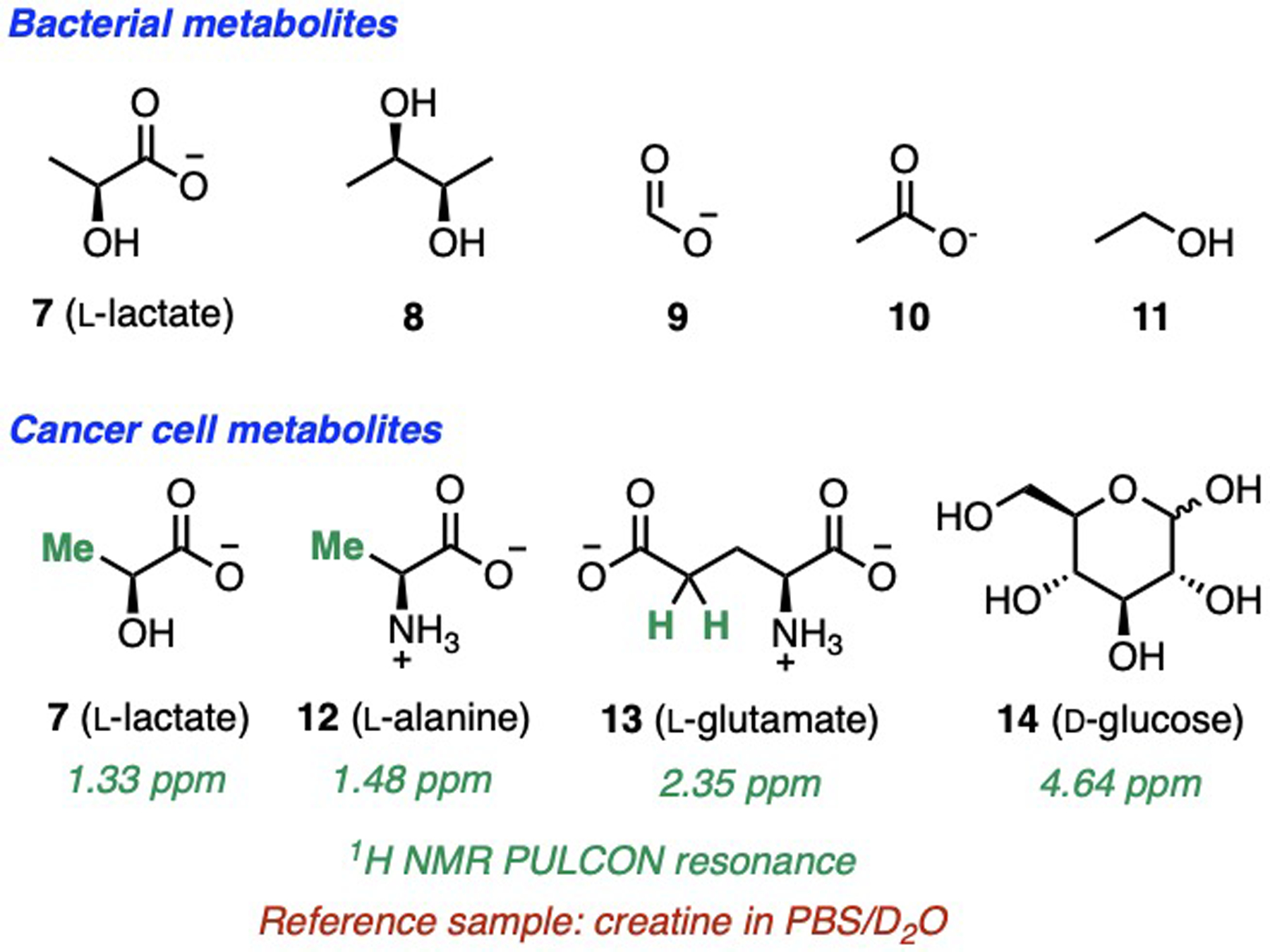
PULCON quantification of biological mixtures.
Figure 7.
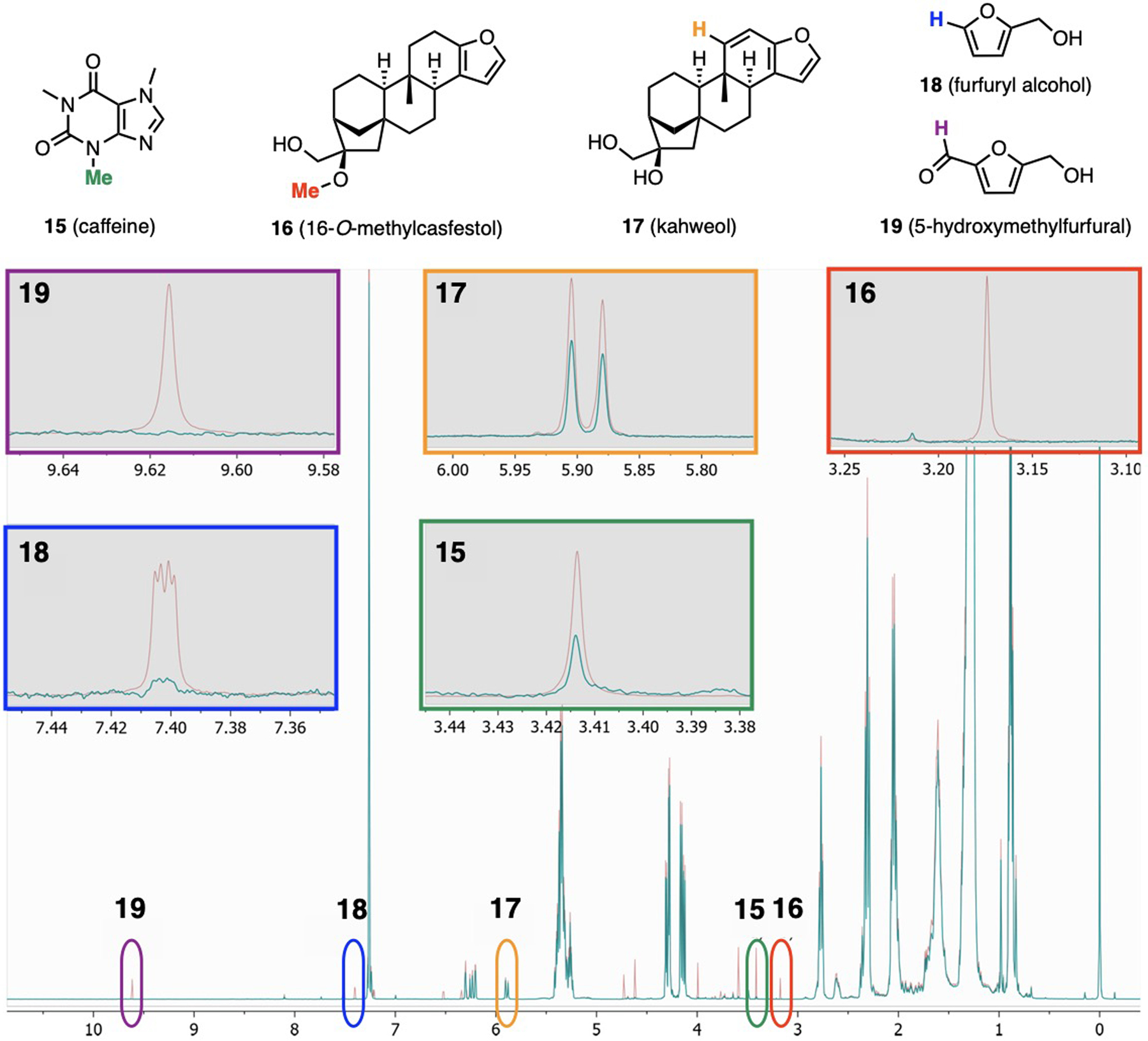
PULCON quantification of a coffee natural product mixture (teal trace) through marked resonances selected from spectra of pure 14 – 18 (maroon trace). Annotated 1H NMR spectrum adapted from reference [20].
Meanwhile, PULCON measurements of the structurally complex and scarce toxin okadaic acid (20) through multiple separate resonances (via ERETIC2)[7] were each consistent with an internal standard method (Fig. 8).[9]
Figure 8.
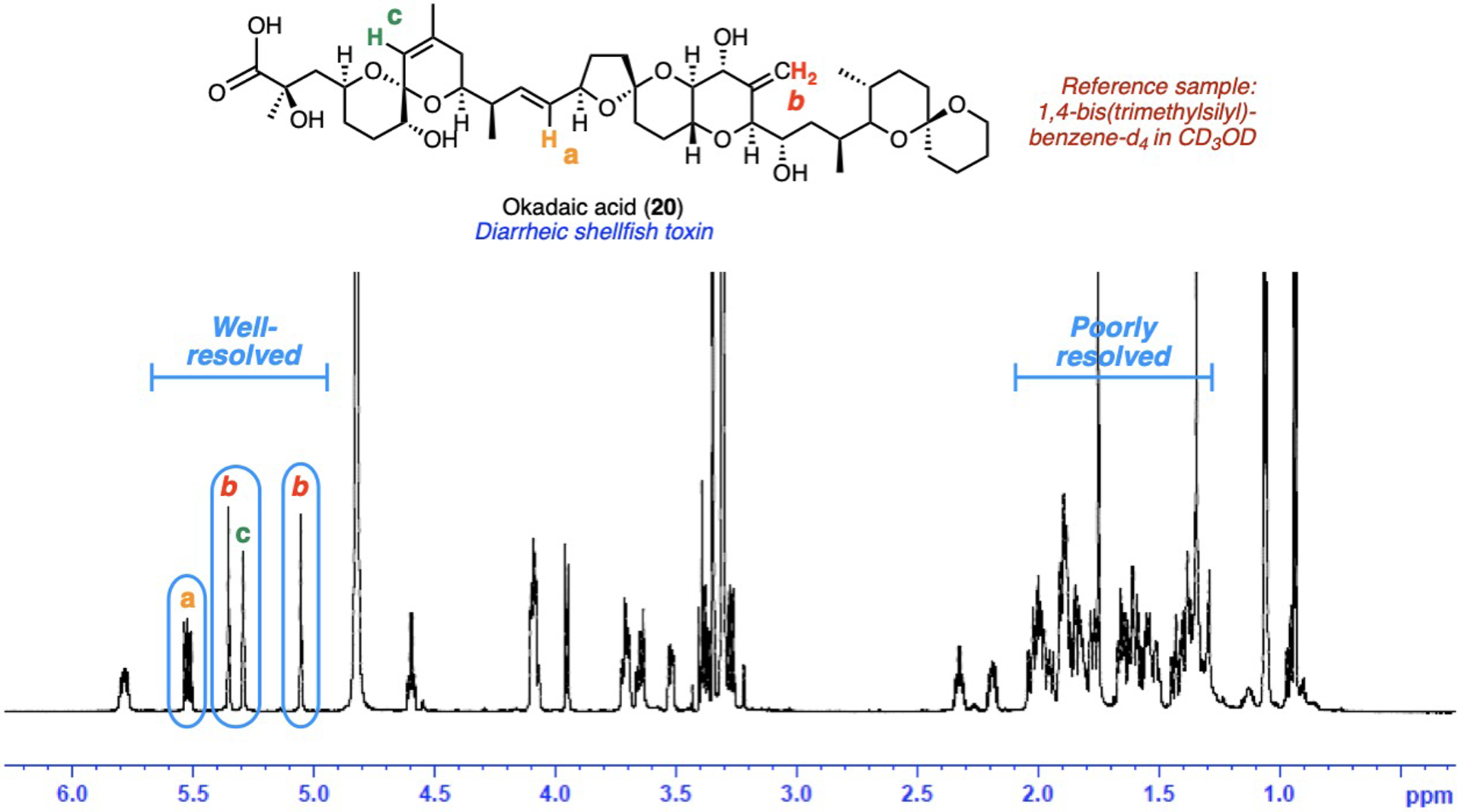
PULCON quantification of scarce toxin okadaic acid. Annotated 1H NMR spectrum adapted from reference [9].
For peptides, charged sidechains are often accompanied by unknown numbers of counter-ions (e.g. trifluoroacetates from HPLC eluants), which obfuscates their true molecular weights. PULCON bypasses this problem, and is typically performed in DMSO or 10 % D2O/H2O through integration of known numbers of amide protons, which are far away from the suppressed water resonance.[21] This has enabled the systematic quantification of diverse peptides, including long constrained peptides (e.g. 21),[22] cyclic lipopeptides (e.g. 22),[23] and cyclic pentapeptides (e.g. 23)[24] for biological assays and structural calculations via circular dichroism spectroscopy (Fig. 9). Hydroxyl groups in structurally complex lignin biopolymers (24) were quantified by in situ hydroxyl phosphitylation in pyridine/CDCl3 followed by PULCON on the 31P nuclei,[25] illustrating generality across NMR sensitive nuclei (Fig. 9).
Figure 9.
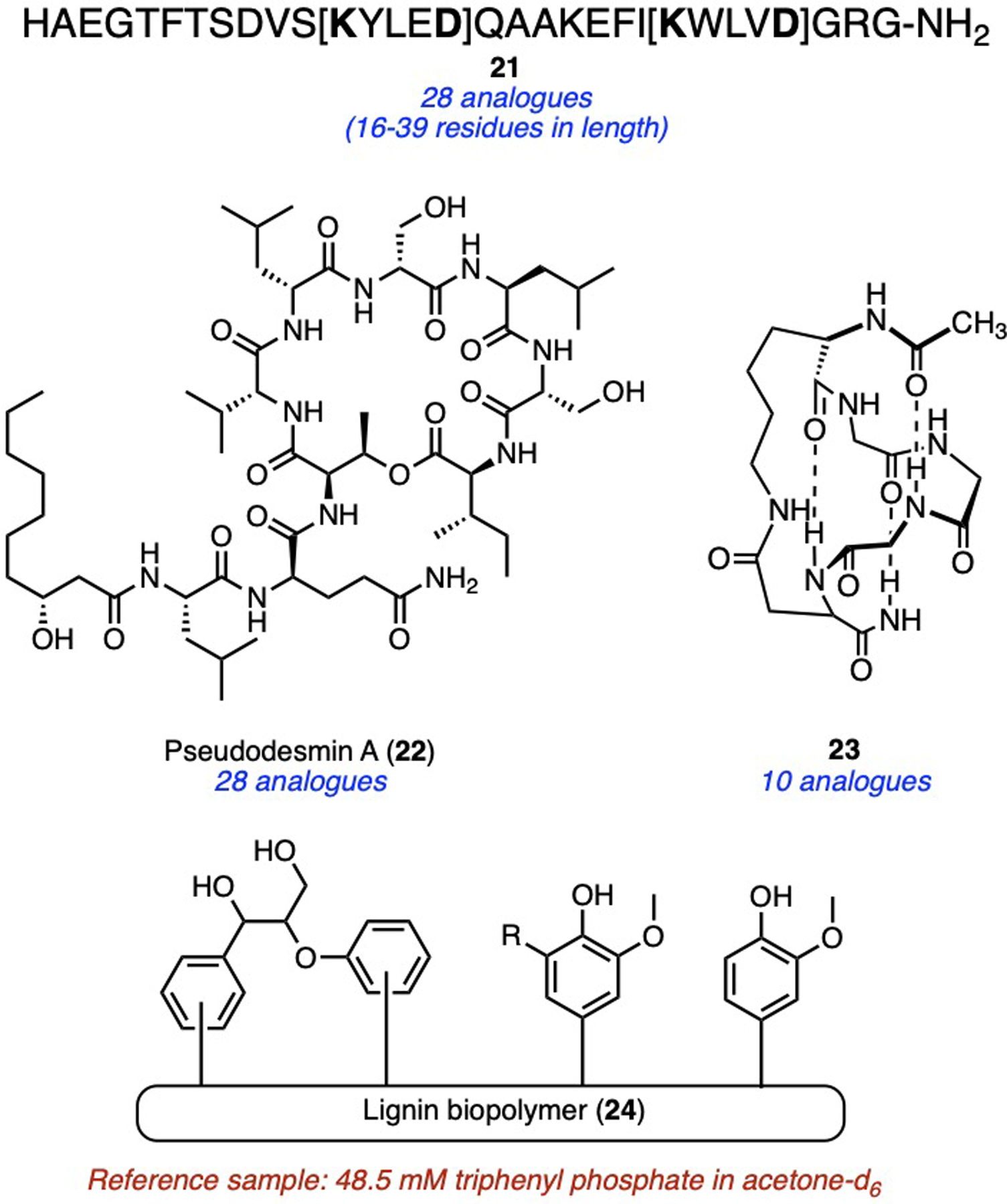
Recent examples of diverse peptides and other biopolymers quantified by PULCON.
PULCON is especially useful for unstable molecules. After suspending a mixture of oxidation prone amine 5-A-RU (25) and inorganic stabilisers in DMSO-d6, followed by filtration of stabilisers, PULCON of 25 enabled the precise addition of methylglyoxal (1.1 eq) to yield bacterial metabolite 5-OP-RU (26, Scheme 1).[27] This compound is highly prone to hydrolysis and cyclisation in water (to 27) and cannot be readily purified,[27] but PULCON critically enabled the NMR sample to be used as a potent immunostimulant[28] for protecting mice against bacterial infections[29–31] and tumours[32].
Scheme 1.
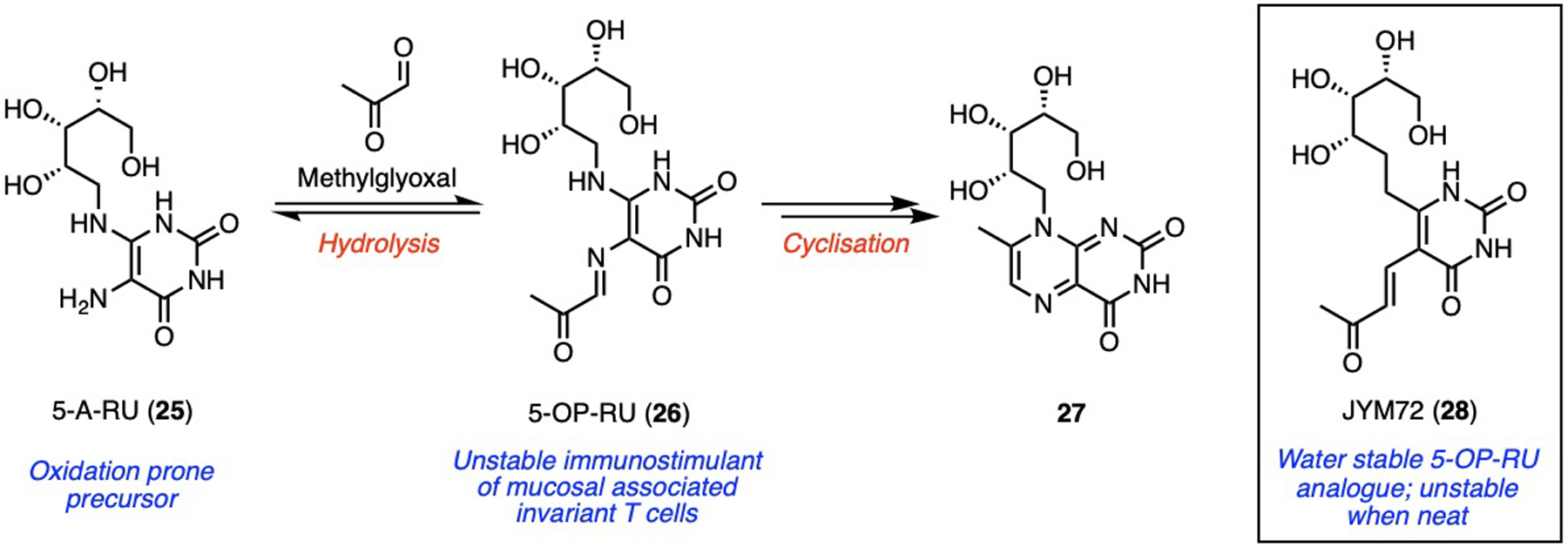
PULCON enabled in situ quantification of unstable precursor 5-A-RU (25), and unstable immunostimulants 5-OP-RU (26) and JYM72 (28).
Similarly, its water stable analogue JYM72 (28) is unstable when neat (Scheme 1).[27] However, partial concentration of its aqueous HPLC eluants,[27] accurate dilution into 10 % D2O/H2O, and then PULCON, enabled its preparation as a cancer immunotherapy tool[32]. Together, these exemplify PULCON as a powerful solution quantification technique for biological and medicinal chemistry.
Acknowledgements
This work was funded by the US National Institutes of Health grant RO1 AI148407-01A1. The author thanks Dr Huy Hoang and Dr Martin Stoermer for insightful discussions.
Biography

Jeffrey Mak was awarded a University Medal (University of Queensland) before undertaking his PhD in total synthesis with Prof. Craig Williams. He joined the Fairlie group at the Institute for Molecular Bioscience, where his interests include biological chemistry, chemical biology, and medicinal chemistry. In 2014, he was part of a multidisciplinary Australian team that discovered the unstable microbial metabolites (e.g. 5-OP-RU) that potently activate mucosal associated invariant T cells. Dr Mak was selected as a CAS SciFinder Future Leader (2017), lectures undergraduate synthetic chemistry at UQ, and was recently promoted to Research Fellow.
Footnotes
Conflicts of Interest
JYWM is a named inventor on a patent describing 5-OP-RU and 5-OP-RU analogues as immunological reagents.
References
- [1].Wider G, Dreier L, Journal of the American Chemical Society 2006, 128, 2571. [DOI] [PubMed] [Google Scholar]
- [2].Singh S, Roy R, Expert Opinion in Drug Discovery 2016, 11, 695. [DOI] [PubMed] [Google Scholar]
- [3].Pierens GK, Carroll AR, Davis RA, Palframan ME, Quinn RJ, Journal of Natural Products 2008, 71, 810. [DOI] [PubMed] [Google Scholar]
- [4].JEOL, Application of PULCON to purity assay of Acetaminophen, https://www.jeol.co.jp/en/applications/detail/1229.html (verified 13-Aug-2021).
- [5].Any pure, non-hygroscopic, and fully soluble compounds that can produce well-resolved spectra could also be used as references.
- [6].While a delay of 30 seconds is more than sufficient for the majority of samples, the delay should be increased if integrations do not scale with number of scans. For information on the importance of relaxation time constants in quantitative NMR, how they can be measured, and example values for various nuclei, see; Wei R, Dickson CL, Uhrín D, Lloyd-Jones GC, Journal of Organic Chemistry 2021, 86, 9023. [DOI] [PubMed] [Google Scholar]
- [7].Users could also use ERETIC2, which is a potentially convenient PULCON tool built into Bruker TopSpin NMR processing software. Refer to: Tyburn J-M, Coutant J. Electronic to Access In-Vivo Concentration User Manual 2016. (Bruker; ). [Google Scholar]
- [8].The factor f accounts for signal intensity differences between the reference and the sample of interest if the two spectra were obtained using different pulse programs and receiver gains.
- [9].Watanabe R, Sugai C, Yamazaki T, Matsushima R, Uchida H, Matsumiya M, Takatsu A, Suzuki T, Toxins 2016, 8, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Dreier L, Wider G, Magnetic Resonance in Chemistry 2006, 44, S206. [DOI] [PubMed] [Google Scholar]
- [11].Buratto R, Mammoli D, Canet E, Bodenhausen G, Journal of Medicinal Chemistry 2016, 59, 1960. [DOI] [PubMed] [Google Scholar]
- [12].Sapienza PJ, Lee AL, Biochemistry 2016, 55, 5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Koskimäki JJ, Kajula M, Hokkanen J, Ihantola E-L, Kim JH, Hautajärvi H, Hankala E, Suokas M, Pohjanen J, Podolich O, Kozyrovska N, Turpeinen A, Pääkkönen M, Mattila S, Campbell BC, Pirttilä AM, Nature Chemical Biology 2016, 12, 332. [DOI] [PubMed] [Google Scholar]
- [14].Baybekov S, Marcou G, Ramos P, Saurel O, Galzi J-L, Varnek A, Molecules 2021, 26, 3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mak JYW, Wu K-C, Gupta PK, Barbero S, McLaughlin MG, Lucke AJ, Tng J, Lim J, Loh Z, Sweet MJ, Reid RC, Liu L, Fairlie DP, Journal of Medicinal Chemistry 2021, doi: 10.1021/acs.jmedchem.0c01967. [DOI] [PubMed] [Google Scholar]
- [16].Migliore M, Habrant D, Sasso O, Albani C, Bertozzi SM, Armirotti A, Piomelli D, Scarpelli R, European Journal of Medicinal Chemistry 2016, 109, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rowley JA, Reid RC, Poon EKY, Wu K-C, Lim J, Lohman R-J, Hamidon JK, Yau M-K, Halili MA, Durek T, Iyer A, Fairlie DP, Journal of Medicinal Chemistry 2020, 63, 529. [DOI] [PubMed] [Google Scholar]
- [18].Troitzsch A, Loi VV, Methling K, Zühlke D, Lalk M, Riedel K, Bernhardt J, Elsayed EM, Bange G, Antelmann H, Pané-Farré J, J Bacteriol 2021, 203, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Itkonen HM, Gorad SS, Duveau DY, Martin SES, Barkovskaya A, Bathen TF, Moestue SA, Mills IG, Oncotarget 2016, 7, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Okaru AO, Scharinger A, Rajcic de Rezende T, Teipel J, Kuballa T, Walch SG, Lachenmeier DW, Foods 2020, 9, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].2D NMR can be used to verify the number of amide protons.
- [22].Plisson F, Hill TA, Mitchell JM, Hoang HN, de Araujo AD, Xu W, Cotterell A, Edmonds DJ, Stanton RV, Derksen DR, Loria PM, Griffith DA, Price DA, Liras S, Fairlie DP, European Journal of Medicinal Chemistry 2017, 127, 703. [DOI] [PubMed] [Google Scholar]
- [23].De Vleeschouwer M, Van Kersavond T, Verleysen Y, Sinnaeve D, Coenye T, Martins JC, Madder A, Frontiers in Microbiology 2020, 11, 646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].de Araujo AD, Hoang HN, Kok WM, Diness F, Gupta P, Hill TA, Driver RW, Price DA, Liras S, Fairlie DP, Angewandte Chemie International Edition 2014, 53, 6965. [DOI] [PubMed] [Google Scholar]
- [25].Lagerquist L, Rahkila J, Eklund P, ACS Sustainable Chemistry & Engineering 2019, 7, 9002. [Google Scholar]
- [26]. Trace amounts of salts may remain, but PULCON is not affected by salt content nevertheless. See reference [1].
- [27].Mak JYW, Xu W, Reid RC, Corbett AJ, Meehan BS, Wang H, Chen Z, Rossjohn J, McCluskey J, Liu L, Fairlie DP, Nature Communications 2017, 8, 14599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mak JYW, Liu L, Fairlie DP, Accounts of Chemical Research 2021, 54, 3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang H, D’Souza C, Lim XY, Kostenko L, Pediongco TJ, Eckle SBG, Meehan BS, Shi M, Wang N, Li S, Liu L, Mak JYW, Fairlie DP, Iwakura Y, Gunnersen JM, Stent AW, Godfrey DI, Rossjohn J, Westall GP, Kjer-Nielsen L, Strugnell RA, McCluskey J, Corbett AJ, Hinks TSC, Chen Z, Nature Communications 2018, 9, 3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang H, Kjer-Nielsen L, Shi M, D’Souza C, Pediongco T, Cao H, Kostenko L, Lim X, Eckle S, Meehan B, Zhu T, Wang B, Zhao Z, Mak J, Fairlie D, Teng M, Rossjohn J, Yu D, Fazekas de St Groth B, Lovrecz G, Lu L, McCluskey J, Strugnell R, Corbett A, Chen Z, Science Immunology 2019, 4, [DOI] [PubMed] [Google Scholar]
- [31].Zhao Z, Wang H, Shi M, Zhu T, Pediongco T, Lim XY, Meehan BS, Nelson AG, Fairlie DP, Mak JYW, Eckle SBG, de Lima Moreira M, Tumpach C, Bramhall M, Williams CG, Lee HJ, Haque A, Evrard M, Rossjohn J, McCluskey J, Corbett AJ, Chen Z, Nature Communications 2021, 12, 4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Petley EV, Koay H-F, Henderson MA, Sek K, Todd KL, Keam SP, Lai J, House IG, Zethoven M, Chen AXY, Oliver AJ, Michie J, Freeman AJ, Giuffrida L, Chan JD, Pizzolla A, Mak JYW, R. MT, Souza-Fonseca-Guimaraes F, Kearney CJ, Millen R, Ramsay RG, Huntington ND, McCluskey J, Oliaro J, Fairlie DP, Neeson PJ, Godfrey DI, Beavis PA, Darcy PK, Nature Communications 2021, 12, 4746. [DOI] [PMC free article] [PubMed] [Google Scholar]


