Abstract
Abnormal tau inclusions are hallmarks of Alzheimer's disease and predictors of clinical decline. Several tau PET tracers are available for neurodegenerative disease research, opening avenues for molecular diagnosis in vivo. However, few have been approved for clinical use. Understanding the neurobiological basis of PET signal validation remains problematic because it requires a large-scale, voxel-to-voxel correlation between PET and (immune)histological signals. Large dimensionality of whole human brains, tissue deformation impacting co-registration, and computing requirements to process terabytes of information preclude proper validation. We developed a computational pipeline to identify and segment particles of interest in billion-pixel digital pathology images to generate quantitative, 3D density maps. The proposed convolutional neural network for immunohistochemistry samples, IHCNet, is at the pipeline's core. We have successfully processed and immunostained over 500 slides from two whole human brains with three phospho-tau antibodies (AT100, AT8, and MC1), spanning several terabytes of images. Our artificial neural network estimated tau inclusion from brain images, which performs with ROC AUC of 0.87, 0.85, and 0.91 for AT100, AT8, and MC1, respectively. Introspection studies further assessed the ability of our trained model to learn tau-related features. We present an end-to-end pipeline to create terabytes-large 3D tau inclusion density maps co-registered to MRI as a means to facilitate validation of PET tracers.
Keywords: Machine learning, deep learning, convolutional neural networks, Alzheimer's disease, histopathology, digital pathology, big data, imaging
Introduction
An estimated 1 out of 9 people over the age of 65 has Alzheimer's (AD) (2021), and the cost of dementia surpassed the cost of heart conditions and cancer (Kelley et al., 2015). Neurodegenerative diseases feature abnormal protein deposits and neuronal loss that progressively overtake an expanding landscape of brain areas in stereotypical patterns, which provides the neuropathological basis of clinical staging systems (Seeley, 2017). AD shows a stereotypical spread of phospho-tau and Aβ deposits (Montine et al., 2012), which could only be detected in postmortem examination until recently.
Positron emission tomography (PET) radioligands allow spatially-resolved imaging of cerebral metabolism, neuroreceptors, and, more recently, pathological protein deposits (Dani et al., 2016; Tiepolt et al., 2019; Wang and Edison, 2019). PET-based labeling of tau and Aβ hold the promise to facilitate understanding of neurodegenerative diseases by enabling (differential) diagnosis, monitoring progression starting from preclinical stages, and evaluating therapeutic efficacy of new drugs in living patients (Rabinovici et al., 2019; Tiepolt et al., 2019).
From the early days of radiolabeling, rigorous criteria based on pharmacokinetics, binding properties in vitro, and match between autoradiography results with target protein burden measured by immunohistochemistry determine efficiency and suitability for novel tracers (Klunk, 2018).
However, the F.D.A requires postmortem correlation studies for testing clinical utility before approving PET tracers for clinical use. In vivo studies with humans testing "suitable" tau and Aβ tracer candidates soon started to show unexpected results. For instance, Aβ PET imaging shows white matter retention. Several phospho-tau tracers show PET signal in basal ganglia, areas that lack respective protein deposits in aging and most neurodegenerative diseases. Also, in vivo studies show signal retention in tauopathies to which the candidate tau tracer has a low affinity in vitro (Leuzy et al., 2019; Soleimani-Meigooni et al., 2020). Besides showing off-target binding, tau and Aβ tracers seem to lack sensibility to detect protein deposits at earlier disease stages (Leuzy et al., 2019; Salloway et al., 2017; Soleimani-Meigooni et al., 2020). Because of technical limitations, most postmortem correlation studies either compare PET signal with semi-quantitative pathological stagings, such as Braak stage, CERAD score, and Thal phases of β-amyloid deposition or compare regional PET signal with the density of targeted protein of approximately similar spatial areas instead of performing voxel-to-voxel comparison (Curtis et al., 2015; Leuzy et al., 2019; Pontecorvo et al., 2020; Sabri et al., 2015; Salloway et al., 2017). As a result, many candidate tracers have been stuck in experimental stages for years as their sensibility, accuracy, and specificity rates, the nature of the off-target binding, the influence of aging, and comorbid pathologies in their binding properties remain poorly understood (Barrio, 2018; Lemoine et al., 2018; Marquie et al., 2017; Smith et al., 2017). Lack of reliable validated means to directly assign molecular signals detected by PET imaging to the corresponding neural microstructures they originate from, persist as a pivotal barrier to widely incorporating PET tracers into clinical pipelines for neurodegenerative diseases.
We developed an end-to-end solution for performing large-scale, voxel-to-voxel correlations between PET and high-resolution histological signals using open-source resources and magnetic resonance imaging (MRI) as the common registration space. Our semi-automated computational pipeline introduces a wide range of computer vision techniques, modern deep learning (DL) algorithms, and high-performance computing (HPC) capabilities to process images of large-scale histological datasets to generate quantitative 3D maps of abnormal protein deposits of whole human brains. We provide a design rationale, a detailed description of the protocol, and results of validation and quality control steps supporting the robustness of our computational methods. The pipeline tacks two main limitations precluding histology to neuroimaging voxel-to-voxel correlations: inherited non-linear tissue deformation during processing that challenges co-registration and immense computational and wet lab capacity required to generate quantitative 3D protein maps of the whole human brain. To illustrate the utility of our pipeline for calculating anatomical priors to validate molecular neuroimaging methods accurately, we applied it to two postmortem human brains, harboring moderate and severe Alzheimer's disease (AD) pathology. We immunostained over 500 whole mount human brain slides and created 3D histology maps co-registered to the corresponding MRI volume of three different forms of abnormal tau protein, spanning several Gigabytes of data. Quality control steps showed excellent agreement between DL-based segmentation of tau inclusions against manual segmentation results based on the area under the receiver operating characteristic (ROC) curve and Dice coefficient metrics. This pipeline represents a way forward to decrease the time between PET tracer development and clinical approval in neurodegenerative diseases and possibly, other conditions.
METHODS
UCSF IRB approved this study under #13-12079. Algorithms used in this study can be found in (https://github.com/grinberglab/high-res-3D-tau). Figure 1 illustrates the workflow steps.
Figure 1:
Step-by-step pipeline for high-resolution 3D mapping of immunohistochemical findings in whole human brains. Blue shading represents unpublished steps, including IHCNET (darker blue shading).
Specimen Procurement, Histological Processing and brain slabbing
We tested the entire pipeline using two whole human brains. The first specimen, Case 1, belonged to a 88 years-old cognitively normal donor with moderate AD pathology (AD neuropathologic change A2B2C1 score (Montine et al., 2012)). The second specimen, Case 2, belonged to a 76 years-old donor diagnosed with dementia and severe AD pathology (AD neuropathologic changes A3B3C3 score). Case 1 underwent postmortem MPRAGE MRI acquisition, while Case 2 underwent postmortem SPGE acquisition within 10 hours of death (Fig. 1a). To minimize tissue deformation, upon autopsy, we stored the specimens upside down, hanging by the circle of Willis for three days. After that, we mounted them in plastic skulls, 3D-printed from the patient's computerized tomography (CT) images. Fixation continued for 18 days inside a bucket filled with buffered 4% paraformaldehyde (Alegro et al., 2016). Next, the specimens were embedded in celloidin. The brains were sectioned in serial sets of coronal slides, each set containing four 160 μm-thick sections. The section thickness was selected to balance the risk of tissue tear and the efficiency of antibody penetration. Representative 2 x 1” celloidin samples were cut for paraffin embedding for neuropathological diagnosis (Alegro et al., 2016). During sectioning, digital photographs were acquired directly from the blockface following each stroke using a high-definition, computer-controlled DSLR camera (EOS 5D Mark II, Canon, Tokyo, Japan) mounted on a copy stand arm (Kaiser Fototechnik, Germany) (Fig. 1b). During processing, brain tissue shrank about 30 to 40%. Thus, each coronal PET scan voxel would contain at least two complete histological sets, opening the opportunity to probe multiple antibodies per voxel. Details of tissue processing celloidin embedding and cutting have been described by us elsewhere (Alegro et al., 2016; Alegro et al., 2017; Theofilas et al., 2014).
CT and MRI acquisition
Postmortem structural T1-weighted MR imaging of Case 1 was performed on a 3T Siemens Skyra MRI system with a transmit and 32-channel receive coil using a 3D MPRAGE T1-weighted sequence with the following parameters: TR/TE/TI = 2300/2.98/900ms, 176 sagittal slices, within plane FOV = 256×240mm2, voxel size = 1×1×1mm3, flip angle = 9°, bandwidth = 240Hz/pix.
Postmortem structural T1-weighted MR imaging of Case 2 was performed on a GE Discovery 3T MR750 system with a transmit and 32-channel receive coil using a 3D SPGR T1-weighted sequence with the following parameters: TI = 400ms, 200 sagittal slices, within plane FOV = 256×256mm2, voxel size = 1×1×1mm3, flip angle = 11°, bandwidth = 31.25Hz/pix.
CT was acquired in life with a slice thickness of 1.25 mm, reconstruction diameter of 300 mm, and image matrix of 512x512 and only used as a model for the 3D printed skull (Fig. 1b)
Immunohistochemical labeling of abnormal tau protein inclusions
We created 3D histological maps for two widely used antibodies against phospho-tau (AT100 (pThr212+Ser214, Thermo), and AT8 (pSer202+Thr205, Thermo)) and one antibody that detects tau conformational changes (MC1, gift of Peter Davies). As a rule, 1st sections from each set were immunostained with AT8, 2nd sections from even-numbered sets were immunostained with AT100, and 3rd sections from even-numbered sets were immunostained with MC1. Free-floating immunohistochemistry reactions were done in batches of 50 (Fig. 1d). Each batch contained positive and negative controls for comparison. The controls were serial sections from a single subject not belonging to the study. Also, we excluded batches in which positive control showed low-quality staining, and in those, the staining process was repeated on the 2nd set of contiguous tissue sections. We performed single-label immunohistochemistry to avoid signal mixing and mounted each histological section on 6" x 4" glass slides.
Whole Slide Imaging
We built a cost-effective whole slide scanner (Fig. 1e, Fig. 2) to accommodate our histological slides (4" x 6"), which do not fit into regular microscope stages or cannot be fully imaged due to short stage travel range. The hardware comprises a high-precision, 6" travel range, industrial XY stage (Griffin Motion), an Olympus manual focusing box, color CCD camera (Qimaging Micro publisher 6), and 5.5X machine vision objective (Navitar Zoom 6000) mounted directly on the camera. Illumination is performed by a lightbox with diffuser, mounted on top of the XY stage. Sections were loaded directly to a 3D printed slide mount fixed on top of the lightbox. At 6.75x magnification, the objective field of view was 3.28 x 2.6 mm. The scanner was controlled by software developed in-house using Macro Manager 2.042, which has a user interface that allows defining the region-of-interest (ROI), performs white balance, and select lens magnification parameters. Macro Manager 2.0 computes the image tiles coordinates necessary to cover the selected ROI and synchronizes the XY stage movements with image capture. TeraStitcher (Bria and Iannello, 2012), which is capable of working with several Gigabytes of data while maintaining a small memory footprint were used to stitch full resolution tiles (1.22μm/ pixel resolution) and create histological images of each slide (Fig. 3). A 10% resolution version of each slide image was also created during the stitching process for visual quality control and aiding with histology pre-processing and registration steps (Fig. 1g). Our scanner software can be downloaded at (https://github.com/grinberglab/high-res-3D-tau).
Fig. 2-.
Scheme of the Slide Scanner set-up built for digitalizing and stitching 6 x 4” histological slides at 1.22μm/pixel resolution
Figure 3:
a) example of a stitched image (Case 1- AT8); b) zoom in of the area delimitated by the black square in 3a (scalebar is 10 μm) to illustrate tau inclusions our system is capable of detecting (green arrow: neurofibrillary tangle, red arrow: neuropil threads, and blue arrow: neuritic component of a plaque; c) example of training tile initially segmented using Weka (scalebar is 100 μm); d) result fo the tile segmentation after manual quality control (red: neurofibrillary tangle, green: threads and plaques, purple: background)
Creation of imaging datasets for IHCNet training and validation
We generated 1024x1024 pixels (1.25 x 1.25 mm) patches from randomly selected gray matter locations throughout the full-resolution brain image datasets to create training, testing, and validation datasets. Before patch extraction, we applied manually segmented white matter masks to the full-resolution brain image datasets. This step aimed to generate training and validation datasets enriched for gray matter patches (because tau pathology is predominantly located in gray matter in AD). We created 100 patches per antibody (AT100, AT8, and MC1) per case, totaling six datasets and 600 patches. The patch extraction routine was written in Python and completely automated, running on UCSFs' Wynton cluster (https://wynton.ucsf.edu), exploring computational parallelism while extracting patches, i.e., the different images are split into patches simultaneously (Fig. 1f). Each patch was manually masked for background and tau inclusion with the help of Fiji’s Trainable Weka Segmentation plugin (www.cs.waikato.ac.nz/ml/weka) (Fig. 3d-e) (Schindelin et al., 2012). Here, the user manually selected sample pixels belonging to tau and background classes. These pixels were used to compute Gaussian filters, Hessian, membrane projections, mean, maximum, anisotropic diffusion, Lipschitz, Gabor, Laplacian, entropy, Sobel, a difference of Gaussians, variance, minimum, median, bilateral filter, Kuwahara, derivatives, structure, and neighbor values. A linear SVM classifier (LibLinear) was then used to generate an initial tau segmentation (Fan et al., 2008). First, a user retrained and refined the initial segmentation using an image editor (Gimp) (https://www.gimp.org). Next, all final masks went through quality control by a highly experienced pathologist (LTG). Labeling took approximately 2 hours per patch (total 1200 h or 150 days of specialized work).
Finally, patches for each antibody (AT8, AT100, and MC1) were combined on bigger datasets of 100 or 200 patches each and were randomly split into 80% of patches for training, 10% for testing, and the remaining for validation.
Histology Images Pipeline
Pre-processing blockface images
Blockface images had their background segmented using a semi-automated graph-based algorithm (Fig. 1c). Briefly, the user selects brain and background sample pixels using a graphical user interface (GUI). Images are then converted to LAB color space (L* for perceptual lightness, and a* and b* for the four unique colors of human vision: red, green, blue, and yellow), and the algorithm computes mean color difference maps (ΔE). ΔE is defined as the distance in LAB space:
With Ldiff, Adiff, Bdiff being the difference values computed as:
Where Li, Ai and Bi are image LAB channels, Lr, Ar, Br are LAB channels of reference pixels selected by the user, and μ(.) is the mean. Pixels in ΔE, whose color is similar to the reference values, appear dark (smaller distance) while cell pixels are brighter (larger distance). We computed brain and background ΔE maps using the manually selected pixels as reference values and performed a global histogram threshold using the Otsu's method to obtain binary masks. We, in turn, combine both masks to obtain brain segmentation. It is expected that several undesired objects linger after the initial segmentation. The segmentation is further refined using a graph-based method to remove the undesired objects. In this method, image objects and their relationship are modeled as a weighted graph, where connected structures are considered the vertices. Edge weights are computed using a similarity function computed from color and distance values. The graph is partitioned using NCuts (Jianbo and Malik, 2000), leaving just the brain area. Commonly, the camera or brain must be repositioned several times during sectioning to adjust for changes in block size, causing the blockface images to be misaligned in relation to each other. We used Matlab's registration GUI to select landmarks for computing affine registrations (2018). Finally, the aligned blockface images were stacked together to form the blockface 3D volume using Insight Segmentation and Registration Toolkit (ITK) (Avants et al., 2014). This 3D volume was used as an intermediate space for creating the 3D tau-protein maps (Fig. 1h).
Low-resolution histology background segmentation
The 10% resolution histological image datasets were converted to LAB color space (Fig. 1g). In that space, background pixels are consistently darker than brain pixels, and segmentation is performed through histogram thresholding using the triangle algorithm (Zack et al., 1977). The resulting binary masks are used to erase all background pixels. Moreover, brain masks are combined with white matter masks to create gray matter masks that guided the entire segmentation process.
2D registration to blockface image
After background segmentation, the 10% resolution histological images are aligned to their respective blockface images using a combination of manual and automatic registration (Fig. 1g). Due to an excessive number of artifacts caused by histological processing, we initialized the registration manually using MIPAV spline-based registration (McAuliffe et al., 2001). Here, the user manually selects landmarks on both the histology and blockface images. MIPAV then generates a warped image and a registration warp file. After the initial registration, the image went through a diffeomorphic registration using the 2D SyN algorithm based on the large diffeomorphic deformation model metric mapping (LDDMM) method (Beg et al., 2005).
Image and Mask Tiling
Each full-resolution histological slide image was first tiled to reduce memory footprint during image segmentation, with tile size corresponding to approximately 5mm2 of tissue (Fig. 1f). Tile coordinates and dimensions are saved as XML metadata files. The histological images’ respective 10% resolution gray matter masks (whole image minus white matter masks) are rescaled to match their full-resolution dimension and tiles. Finally, histological tiles were masked using their respective gray matter tiles, leaving only the ROIs for image segmentation. Tiles with less than 5% of tissue pixels were ignored during segmentation to reduce the overall computational time. Image and mask tiling routines were developed in Python (Van Rossum and Drake Jr, 1995) and ran on UCSFs' Wynton cluster exploring computational parallelism, having one pipeline for each histological image.
Deep learning-based segmentation of particles of interest (Fig. 1f, Fig. 4)
Figure 4:
IHCNet architecture. The arrows indicate skip connections that are followed by cropped when the right-hand side layer is smaller than the left side layer
We designed IHCNet, a UNet (Ronneberger et al., 2015) based neural network capable of working with color information and outputting tau presence confidence maps that were later thresholded to create binary maps.
Model development and training
Our model has an 204x204x3 pixels input layer to accommodate RGB images – we chose this size to match 1mm2 of tissue in our whole slide imaging scanner resolution. Figure 4 shows the IHCNet architecture together with each layer tensor size. The image is pushed through three contractions blocks, the bottleneck, and upsampled by three expansion blocks in our model.
Each contraction block comprises two convolution layers that use 3x3 kernels, stride of 1 and ReLu activation, followed by a 2x2 max pooling layer and a 0.1 rate dropout. The bottleneck comprises two convolutional layers that use 1x1 kernels, stride of 1, and ReLu activation. Expansion blocks are composed of a 2x2 upsampling layer followed by a 0.1 rate dropout and two convolutional layers that use 3x3 kernels, stride of 1 and ReLu, except for the last expansion block that uses 3x3 upsampling. The last layer reshapes the data to a 20000x2 tensor and softmax activation.
Training is performed using standard backpropagation, with binary cross-entropy as the loss function and Adam algorithm for optimization. The learning rate is estimated using the cyclical learning rate method (Smith, 2015). The network was developed in Keras (https://keras.io) on top of TensorFlow (https://www.tensorflow.org). We trained the network using mini-batches of 32 images during 100 epochs or until the loss curve plateaued. We also used massive data augmentation, performing real-time random rotations, shear, horizontal and vertical flips. All training and inference were performed on an NVIDIA Titan V GPU with 12 GB of RAM. IHCNet outputs confidence maps of the existence of tau inclusions, which are thresholded to generate binary masks. The threshold value was set heuristically to 0.5 for AT100, AT8, and MC1 datasets, and pixel with confidence higher than this value was considered tau (Fig. 3d-e). IHCNet training took up to 2 days per antibody using a workstation with 2 GPUs. The networks were trained until we observed a plateau in the accuracy and loss function graphs.
Heatmap computation
The binary tiles were transferred back to the cluster for computing heatmaps (Fig. 1f). We computed the mean amount of tau on each tile, indicated by the mean number of pixels belonging to foreground, inside a 1 μm2 of tissue, an 8x8 pixels block. The heatmaps were generated as tiles having the same dimension of binary tiles where each 1 μm2 block is filed out with the mean number of pixels belonging to tau. Tiles are stitched together to create a heatmap having the same resolution as the original histological image and are later resized a 10% resolution.
Heatmap normalization
Each of the six heatmap datasets (AT100, AT8, and MC1 for each brain) was normalized to mitigate batch staining inhomogeneity (Fig. 1f). A trained neuropathologist (LTG) selected strong signal ROIs from slices whose staining is deemed optimal and weak signal ROIs from slices whose staining is suboptimal for each dataset. Outliers were removed from each ROI by thresholding the values between the first and third quartiles, and mean strong and weak signals were calculated. We then computed an increase factor as the absolute difference value between both means. All heatmaps from slices deemed suboptimal during quality control were adjusted by summing their non-zero values with this increase factor.
Heatmap to blockface alignment and stacking
The 2D registration maps computed during the 2D registration to blockface image step were applied to the 10% resolution normalized heatmaps, yielding heatmap registered to their respective blockface images. These images are then stacked together for a 3D volume (Fig. 1f).
3D Histological Reconstruction and Visualization and MRI-Histology registration
The 3D registration maps generated during the Histology to MRI 3D registration were inverted and used to warp the MRI to blockface space, allowing direct comparison between MRI signal and tau density. Freeview (Fischl, 2012) was used for visualizing 2D slices. Amira (ThermoFisher Scientific) was used for 3D visualization.
The accuracy of registration approaches for medical images relies on the presence and detection of homologous features in both target image (i.e., 3D blockface reconstruction) and the spatially warped image (i.e., structural T1-weighted MRI). To satisfy this prerequisite, we used the Advanced Normalization Tools (ANTs) (Avants et al., 2009), the N4 non-parametric non-uniform intensity normalization bias correction function (Tustison and Avants, 2013; Tustison et al., 2010) followed by skull-stripping (Fig. 1h). Briefly, structural T1-weighted MRIs were warped into the 3D image space of the corresponding blockface reconstructions using ANTs (Avants et al., 2008)). First, a linear rigid transformation was performed. Then a diffeomorphic transformation using the Symmetric Normalization (SyN) transformation model was performed. SyN uses a gradient-based iterative convergence using diffeomorphisms to converge on an optimal solution based on a similarity metric (e.g., cross-correlation) (Klein et al., 2009). To validate the registration, we calculated the Dice coefficient of ventricles (Dice, 1945).
RESULTS
Pipeline for creating High-Resolution 3D mapping of immunohistochemical findings in whole human brains
We engineered a comprehensive pipeline suitable for rendering quantitative 3D mapping of particles of interest (lesions, inclusions) detected by immunohistochemistry in whole postmortem human brains that can be co-registered with other histology- and neuroimaging-based 3D maps of the same brain. We broke down our pipeline into a series of histological and computation modules (Fig.1) that can be executed independently for scalability. The complete pipeline relies on three pillars: whole-brain histological processing and large slide immunostaining (Alegro et al., 2016; Alho et al., 2017), 3D and 2D registration algorithms (Alegro et al., 2016), and novel DL algorithms to segment and quantify particles of interest in terabytes-large imaging datasets.
Preliminary protocols for histological processing and registration have been published previously (Alegro et al., 2016; Alho et al., 2017; Theofilas et al., 2014). Here, we focused on pipeline development and quality control steps to enable scaling-up whole-brain slice immunostaining (including the hardware we assembled to accommodate oversized biological specimens), developing and validating DL algorithms for segmenting particles of interest, and integration of all steps. We processed two whole human brains in 1608 coronal, whole-brain sections, of which 524 underwent immunohistochemistry. Then, we used our computational pipeline to scan the histological sections at microscopic resolution, segment tau inclusions in each voxel, create 3D inclusion maps and register those maps to the corresponding MRI volume.
Briefly, the first module comprises acquiring structural MRI to create an MRI dataset (Neuroimaging Acquisition Module- Fig. 1a). We obtained postmortem MRI sequences in-cranio right before procurement (Alegro et al., 2016). Upon procurement, the brains were processed, embedded as a whole, and coronally cut (Alegro et al., 2016; Heinsen et al., 2000) (Histological Processing Module- Fig. 1b). Brain slicing generates one physical and one image datasets: (a) serial histological sections (about 800 per case, coronal axis), which are the input to the Immunostaining Module (Fig. 1d), and (b) blockface image dataset comprising digital images of each histological section generated by photographing the celloidin block before each microtome stroke (Theofilas et al., 2014). The blockface image dataset proceeds to the Blockface Imaging Processing Module (Fig. 1c), which delivers a digital blockface volume, an intermediate space for registering 2D histological maps, and the MRI volume to enable neuroimaging and histology voxel-to-voxel comparisons (Alegro et al., 2016). Further details about the Neuroimaging Acquisition, Histological Processing, and Blockface Imaging Processing modules are found in Methods and our previous publications (Alegro et al., 2016; Alho et al., 2017; Grinberg et al., 2009; Theofilas et al., 2014). Hereon, we will focus on newly developed Modules (blue/gray shaded in Fig. 1) and their integration into the pipeline.
Different types of abnormal tau protein accumulate along AD progression. Because autoradiography studies are only partially translatable to an in vivo condition (Okamura et al., 2018), it is often unclear which abnormal tau species correlates better to a tau PET signal. Therefore, a robust validation pipeline should enable probing different histological markers within the same PET voxel. In the Immunostaining Module (Fig. 1d), we used free-floating immunohistochemistry to independently detect three types of abnormal tau (detected by AT8, AT100, and MC1 antibodies) per case (6 whole-brain maps in total).
Immunostained slides proceed to the Scanning and Stitching module (Fig. 1e). We designed a custom-built whole-slide scanner (Fig. 2) to digitalize large and thick tissue sections at the microscopic resolution. Each histological section generated about 1500 image tiles (20GBs of data) at a resolution of 1.22μm/pixel (Fig. 3). We scanned a total of 524 immunostained sections at one plane of focus. It took approximately 90 min to scan and two days to stitch each section. Thus we used an HPC system to perform stitching, exploring computational parallelism for each section since the tiles of individual sections can be stitched concurrently and independently from other sections. The stitching process generated two new imaging datasets: a high-resolution digital section dataset (Fig. 1f) with images of approximately 80000x50000 pixels per tissue section (another 20GBs of data per slide) and a low-resolution digital section dataset (Fig. 1g) in which the original digital sections were downsampled to a 10% resolution. Low-resolution images facilitate all computing steps that did not require microscopic resolution, including visual inspection of the final stitched images, masking background, or any brain area of interest. Low-resolution images also facilitated registering images of immunostained slides to the original blockface coordinates. We developed the 2D Histology Registration Module (Fig. 1g) to register each low-resolution histological digital image to the corresponding blockface image. First, we applied background masks to the low-resolution digital section dataset. Then, we used a combination of open-source registration algorithms to align these masked images to their corresponding pre-processed blockface images and create warping maps for each histological section. The warping results were visually inspected, and imperfections were corrected manually—most of the imperfections were located around the limbic structures because the region has delicate sulci and crests, making it more prone to tears and folding during processing.
Segmenting and quantifying of tau inclusions were performed in the high-resolution digital section dataset using the Inclusion Segmentation and Mapping Module (Fig. 1f). This module contains IHCNet for segmenting particles of interest in a setting of low signal-to-noise ratio. IHCNet is the core innovation of our pipeline, with higher segmentation accuracy and robustness to histological artifacts than thresholding algorithms. The architecture, rationale, and validation steps for IHCNet are described in Methods. We used the warping maps generated in the 2D Histology Registration Module to register the high-resolution heatmap of each histological section, created via IHCNet, to their corresponding blockface image. The 2D registered heatmaps were stalked and normalized to create 3D heatmaps of each tau type. The final 3D tau heatmaps were stored in standard nifti medical imaging file format
Finally, we used the 3D Histology Reconstruction Module (Fig. 1h) to register the histology to MRI 3D maps. Briefly, structural T1-weighted MRIs were warped into the 3D image space of the corresponding blockface reconstructions (Avants et al., 2008; Avants et al., 2014). The diffeomorphic transform matrices were then applied to all the tau inclusion heatmaps for mapping into the structural MR imaging space and resolution. The final product of our test was six 3D tau heatmaps registered to the corresponding MRI showing the distribution of tau detected by AT8, AT100, and MCI antibodies, from two different whole human brain samples (Fig. 5, Supplementary Videos 1-6).
Figure 5:
AT8, AT100 and MC1 quantitative heatmaps registered to blockface (1st column), MRI (2nd column) of Cases 1 and 2. The 3rd column depict the respective 3D renderings of each tau map. Heatmaps were normalized to the same scale to facilitate comparisions. As expected from neuropathological studies, AT8 detected higher amounts of tau than AT100 and MC1. Videos of the full 3D tau rendering are found in Supplementary Material.
IHCNet, a convolutional neural network (CNN) for semantic segmentation
Architecture and rationale
We assumed it would be possible to apply thresholding algorithms in our first attempts to segment tau deposits in histological images, assuming that the immunostaining label is brown (DAB −3,3′-Diaminobenzidine staining), and the background is transparent. However, the thresholding strategy failed despite several optimizing steps to ensure a high signal-to-noise ratio on immunohistochemistry. Conventional 5-um thick histological sections have a transparent background. However, we had to cut our brain blocks at a 160 μm minimum thickness to minimize tissue tearing. The background of those 32x thicker-than-conventional sections is opaque, especially in the white matter area because of the myelin. Second, hard-to-control factors such as submicrometric thickness variations and regional differences in hydration level influence the staining hue in large sections, making thresholding algorithms useless for the detection task. To address these challenges, we designed IHCNet, a CNN. IHCNet uses a UNet based architecture (Fig. 4), trained to compute a confidence map of the 2D distribution of particles of interest (i.e., tau inclusion here) instead of a single class confidence value. Furthermore, we extended IHCNet architecture to leverage color information and optimally work with microscopic imaging resolution (1.22 μm/pixel).
IHCNet has an input layer measuring 204×204×3 pixels corresponding to 1mm2 of tissue to accommodate RGB images. IHCNet features a validation hold-out scheme, where training and testing sets were used for model training, and a validation set was used for independent network performance statistics.
Validation of the network training
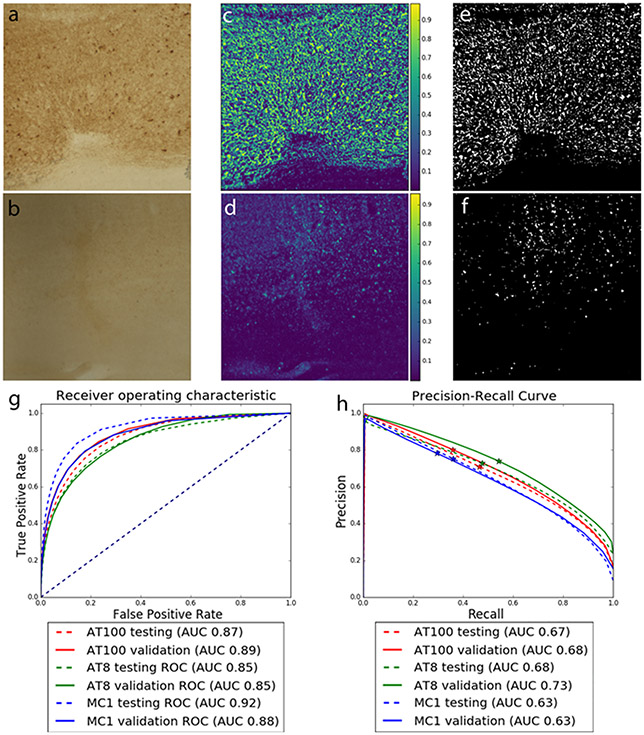
To validate the IHCNet model, we computed the ROC and precision-recall curves over the testing and validation sets for each antibody. For ROC, the AT8 model achieved an area under the curve (AUC) of 0.85 for both testing and validation datasets, while the AT100 model achieved an AUC of 0.87 on testing and 0.89 on the validation dataset. MC1 model achieved the best ROC results with an AUC of 0.91 on the testing and 0.88 on the validation dataset (Fig. 6). From thresholded masks, we calculated precision and recall values. We obtained precision values of 0.87, 0.87, 0.90 respectively for AT8, AT100, and MC1 in the testing datasets and 0.92, 0.92, 0.91 for AT8, AT100, and MC1, respectively, in the validation datasets. As for the recall, the thresholded masks obtained 0.16, 0.19, 0.12 respectively for AT8, AT100, and MC1 in the testing datasets and 0.18, 0.13, and 0.08 for AT8, AT100, and MC1, respectively, in the validation datasets.
Figure 6:
patches of digitalized images of an AD case (case #1) immunostained for AT8. Upper panel (a/c/e) - patch #1 and lower panel (b/d/f) – patch #2. a/b) ROIs of AT8 antibody-stained brain tissue with high amounts (a) and low amounts (b) of tau inclusions; c/d) their respective probability maps of tau signals generated by IHCNet. Green color symbolizes tau signal. The brighter the signal, higher the tau probability is and e/f) binary segmentation after thresholding c /d at 0.5. Graphs show ROC (g) and precision-recall curves (h) for models trained with all full datasets from all antibodies. Stars on the precision-recall graphs show where the 0.5 threshold is localized for each stain.
In terms of precision-recall, the AT8 model achieved an AUC of 0.68 for the testing and 0.72 for validation; AT100 had a 0.66 for testing and 0.68 for validation, and MC1 had a 0.63 for testing and 0.63 for validation.
Neuronal network tau segmentation of the full resolution dataset
CNN-based segmentation is computationally intense. Thus, we first masked regions of interest to decrease computing time. Specifically, we masked out background and white matter tissue pixels to focus on the regions with tau deposition. Masks were created using the low-resolution digital section dataset and subsequently upsampled and applied to the high-resolution histological images. Moreover, to facilitate computation, we divided the high-resolution images into tiles. It took about four days for IHCNet to segment the largest dataset (AT8, case 2,160 tiles). The network output probability maps were thresholded to create binary masks. The exact cut-off values for thresholding were applied to all datasets. Figure 6 shows an example of the histology tiles, the probability maps created by the CNN, and the thresholded binary masks (top rows).
Transfer learning and additional validation experiments
Considering it is unfeasible to generate training data for each new antibody and case, we tested how our CNN behaves with different training datasets size and composition. We used the following training datasets for these experiments: #1: all labeled patches from cases 1 and 2, divided per antibody (AT8, AT100, and MC1), totalizing three databases with 200 patches each. #2 all labeled patches divided per antibody and case, totalizing six databases of 100 patches each. In summary, we created nine training models. The rationale for cross-testing is that morphology of tau inclusions detected by different tau antibodies is similar. Thus, in theory, networks trained for one tau antibody could segment histological images from a different tau antibody.
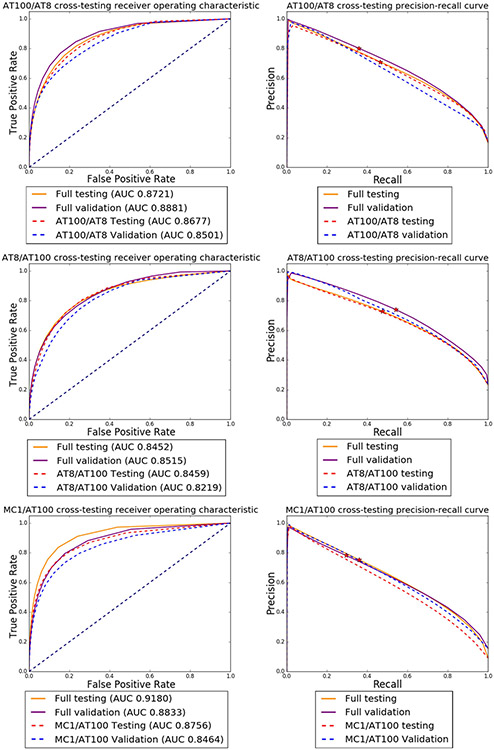
First, using these nine training modules, we tested how a CNN trained with labeled patches from a given tau antibody from a specific case(s) performs in segmenting images labeled with the same tau antibody in another case. For instance, the CNN trained using AT8 patches from case 1 was used to segment AT8 signal on case 2 and rendered an AUC of 0.7855 in the testing set and 0.7847 in the validation set. Inversely, when the network trained on AT8 patches from case 2 was applied to segment AT8 signal on case 1, we obtained an AUC of 0.8642 in the testing set and 0.7544 in the validation set (Fig. 7).
Figure 7:
Comparison of ROC (left) and precision-recall (right) curves for models trained with complete datasets (images of both cases/antibody) vs. models trained with a dataset comprised of images from one brain only/antibody. Top row: AT100; Middle row: AT8 and Botton row: MC1. The solid orange and solid purple lines show the results for the testing and validation using the full datasets. Dashed red and blue lines show testing and validation results for the model trained with images from case 2 alone and dashed green and brown lines show testing and validation results for the model trained with images from case 1 alone. Stars on the precision-recall graphs show where the 0.5 threshold is localized
Second, we used labeled patches of a given tau antibody to train a CNN and then applied the CNN to another tau antibody validation dataset (cross-testing). When a network trained on AT100 patches was applied to segment AT8 datasets, we achieved an AUC of 0.8459 in the AT8 testing dataset segmentation and 0.8219 in the AT8 validation dataset. Inversely, a network trained on AT8 patches when applied to segment AT100 datasets resulted in an AUC of 0.8677 in the AT100 testing dataset and 0.8501 in the AT100 validation dataset (Fig. 8).
Figure 8:
Comparison of ROC and precision-recall curves for cross-trained models, where we performed training using images from one antibody and tested the model on a dataset of a different antibody. Top row: dashed red and blue lines show testing and validation results for a model trained using AT100 data for segmenting AT8 images. For comparison, the orange and purple lines show the test and validation results for the model trained with AT8 data. Middle row: model trained on AT8 data to segment AT100 images. Bottom row: model trained on AT100 images to segment MC1 images. Stars on the precision-recall graphs show where the 0.5 threshold is localized.
The results show that the difference between models is negligible. The curves for the models trained with case 1 AT8-only and AT100-only images are slightly inferior to the dataset.
Network introspection experiments show the neural network correctly learns tau inclusions
Standard precision-recall and ROC curve evaluation cannot show whether the network effectively learns image features of particles of interest (tau inclusions) or is simply overfitting patterns. Thus, we used neural network inspection techniques to evaluate the network ability to learn. We first used gradient-guided class activation maps (Grad-CAM) (Selvaraju et al., 2017) on randomly selected images to evaluate the most critical features to drive the network to a particular decision. Then, we added perturbations for checking whether the network was segmenting tau inclusions solely based on spatial localization or using relevant features. Standard Grad-CAM works by computing the gradient between a user-defined class (artificial) neuron in the last network layer and an intermediary target layer and then multiplying its mean value with the target class activations. This technique was designed to work with networks with a fully connected or a global pooling layer (Cireşan et al., 2011), responsible for mapping the information spread across the convolutional layers to a single neuron at the last layer. As IHCNet outputs 2D probability maps of inclusions and lacks the property of fragmented mapping information to a single neuron, we adapted Grad-CAM by selecting multiple output neurons inside a tau inclusion, on randomly selected test images, i.e., neurons that should be activated for the class “tau,” with the help of a binary mask, and computing the mean of the Grad-CAM maps generated for each selected neuron. As seen in Figure 9, a visual inspection of the output CAM maps shows a good agreement between the features that drove network decision and tau signal. The network correctly responded to perturbations. We repeated the same experiment to interrogate if the network could correctly discriminate the background. We used a mask to select background pixels. The resulting Grad-CAM shows good localization of the background. Finally, we interrogated whether the network was learning to detect tau based on pixel spatial localization or using other relevant information. We created a perturbed image by partially covering a tangle with a background patch and repeated the background Grad-CAM experiment. The network correctly recognized the patch as background, suggesting that other relevant features inform the network.
Fig 9:
Network interpretability test using Grad-Cam and perturbation techniques. a. Example of original images with neuritic plaques (i.e. a1/f1 (lower magnification) and a2/f2 (higher magnification) and neurofibrillary tangles (i.e. a3/f3). Mask used for foreground pixels selection from (a), indicated by red arrow. c. Mask used to locate background pixels selection from (a), indicated by blue arrow. d. Grad-CAM result of original image using pixels from mask in (b) as references. e. Grad-CAM result of original image using pixels from mask in (c) as references. f. Perturbed image, where a background image patch (indicated by the yellow arrow) was artificially placed to partially cover the tau inclusion. g. Neural network model segmentation of the original image (area with in the green shapes). h. Neural network model segmentation of the perturbed image (area with in the green shapes). i. Grad-CAM result of perturbed image using pixels from mask in (b) as references. j. Grad-CAM result of perturbed image using pixels from mask in (c) as references. In Grad-CAM results (d, e, i, j), brighter/warmer heat-map values indicating stronger influence in the network decision.
Registration validation
We quantitatively evaluated registration quality by computing the Dice coefficient (Dice, 1945) between stacks of masks manually labeled on MRI and histological slices. The Dice coefficient measures how well two sets of images co-localize.
Histology to blockface 2D registration evaluation:
we manually labeled the lateral ventricles on histological images containing ventricles. All histology ventricle labels were registered to the blockface using the pre-computed registration maps. We computed the 2D Dice coefficient for all registered histology/block label pairs. The same procedure was performed for our two cases. Mean Dice coefficient values for case 1 were 0.9082±0.053 (AT8), 0.9112±0.0405 (AT100) and 0.88±0.0531 (MC1). Mean Dice coefficient values for case 2 were 0.8185±0.1070 (AT8), 0.8385±0.1444 (AT100) and 0.8605±0.1823 (MC1).
MRI to blockface 3D registration evaluation:
here, we used the lateral ventricle masks. The Dice coefficient values were then computed between each blockface ventricle label created to evaluate the 2D registration and its respective MRI ventricle label (after registration). The same procedure was performed for the two cases. We obtained a mean Dice coefficient of 0.9047 (±0.032) and 0.8570 (±0.095) for case 1 and case 2, respectively.
Discussion
We developed a scalable pipeline for creating MR-registered 3D maps of particles-of-interest detected with immunohistochemistry in oversized (whole human brain) histological slides as a means to enable histology to neuroimaging voxel-to-voxel comparisons. This pipeline comprises histopathological and computational modules and incorporates DL algorithms and HPC capabilities into a previously developed framework to facilitate high precision 3D histology to neuroimaging registration. The pipeline is generalizable to a broad array of studies focusing on validating any neuroimaging modality that can be registered to MR space, such as PET, against stained-based histological data.
Molecular imaging is the most promising technique for quantifying proteins spatially-resolved in the living brain. Because neurodegenerative diseases spread stereotypically (Seeley, 2017), molecular imaging promises to render reliable preclinical diagnosis, monitor disease progression, and measure the efficacy of experimental treatments. Attempts to validate Aβ and tau tracers by comparing the histological properties of postmortem brains to the PET signal obtained closer to death proved valuable to facilitate clinical approval (Curtis et al., 2015; Leuzy et al., 2019; Pontecorvo et al., 2020; Sabri et al., 2015; Salloway et al., 2017; Soleimani-Meigooni et al., 2020). Still, methodological issues limiting large-scale voxel-to-voxel comparisons precluded these studies from answering several critical questions about the neurobiological basis of the observed PET signal, including the nature of off-target signal or each kind of posttranslational modification of the target protein better correlate with PET signal intensity. Thus, the diagnostic scope of clinically approved tracers remains limited, and the time between tracer development and clinical approval is still very long.
Immunohistochemistry and autoradiography are methods of choice to detect the anatomical distribution of a protein of interest in situ. The former is more broadly used because it takes less time, requires less equipment, avoids radioactivity, and is less prone to artifacts. Voxel-to-voxel comparison of PET signal to immunohistochemistry results can inform the neurobiological basis of PET signal. However, previous studies were limited by difficulties in locating precisely in PET imaging the areas probed with histology or performing histology at a large scale.
The resolution of PET scans is low. Anatomical parcellation, which is particularly relevant In neurodegenerative diseases because distribution and the regional load of abnormal protein deposition have implications for differential diagnosis and staging disease severity, is better achieved by co-registering PET to MRI space. Thus, co-registering histological data, the gold standard for detecting abnormal protein deposition, to MR space is crucial to facilitate voxel-to-voxel validation of PET signal. We and others have recently offered solutions to enable structural histology to MR co-registration of human whole-brain volumes (Alegro et al., 2016; Alkemade et al., 2020). Such solutions use modified brain tissue processing and computational algorithms to minimize and correct the significant non-linear tissue deformation inherited from tissue processing. Still, these solutions only use tissue stained with simple dyes, such as Nissl. Immunohistochemistry adds several processing steps, resulting in additional tissue deformation. Also, immunohistochemistry is optimized for thin tissues (3 to 10 um) to ensure good antibody penetration and signal-to-noise imaging results. Optimal histology to neuroimaging registration requires whole-brain histological 3D reconstructions. However, because brain tissue is only semi-rigid, cutting larger histological blocks thin results in tears and folds. Thus, the larger is the tissue area, the thicker it needs to be cut. Free-floating techniques facilitate antibody penetration on thicker histological slides, but the signal-to-noise ratio is always poor since thicker slides have many cell layers overlapped.
For incorporating immunohistochemistry 3D maps into our original pipeline (Alegro et al., 2016), we had to overcome significant challenges, including (1) how to digitalize images of hundreds of supersized and thick histological slides (that do not fit regular histological scanners, require stronger light sources, are prone to imaging inhomogeneity, and generate gigabytes of data each) and (2) how to perform large-scale image segmentation on several hundreds of terabytes of inhomogeneous images with accuracy and precision.
We addressed the first challenge by engineering a cost-effective whole slide imaging system with an extensive travel range capable of imaging the entire area of our slides. Options of open-hardware microscopy equipment expanded recently (Campbell et al., 2014; Nunez et al., 2017; Phillips et al., 2020; Rosenegger et al., 2014) with projects relying on a wide variety of materials, from inexpensive 3D printed parts to high-end optical kits. However, most designs accommodate small samples only. Hardware robustness and availability of customizable open-source software were imperative for our needs. Our configuration prioritizes cost-benefit to minimize production costs. Machine vision objectives offer a reasonable field of view while incorporating all the optical elements necessary for microscopic imaging in one set of lenses attached to the camera, thus improving scanning time. Our scanner took 50 to 90 minutes to image one plan of focus for each large section, plus loading and focusing time. The scanner never failed during the over 1000h of scanning time.
The second challenge was the most complex to solve. Microscopy lenses have a limited field of view. Thus large surfaces need to be imaged in several parts image inhomogeneity. Also, in histological thicker sections, several layers of cells overlap, resulting in an opaque background. These factors contributed to the failure of thresholding, or machine-learning-based segmentation algorithms developed to thin histological slides (Koga et al., 2021; Signaevsky et al., 2019) to segment tau inclusions in our preparations. Therefore, we took the challenge to create a CNN to segment tau signal with precision. We elected a DL algorithm for its ability to automatically discover unknown patterns that best characterize a raw data set by capturing semantic meaning (LeCun et al., 2015). DL models data in a bottom-up approach, using small low-level features such as edges, first in lower network layers, and increasing abstraction and complexity in the following layers. This approach makes DL more robust to identify immunostaining and tissue artifacts, signal inhomogeneity, and false positives.
IHCNet enabled segmenting morphologically diverse tau inclusions from blurred images. Moreover, its computational modules facilitate processing the large volume of data simultaneously by harnessing the power of HPC to run the different parts of the pipeline, using a paradigm known as embarrassingly parallel workload, where hundreds of copies of the same pipeline are run at the same time.
To test the robustness of IHCNet, we conducted thorough validation experiments. On ROC AUC, our classification results (mean 0.88 on the testing set and 0.87 on the validation set) were just below the reported values for cell culture (0.95) (Al-Kofahi et al., 2018). Noteworthy, cell culture images have a clearer background, fewer artifacts, and much smaller surfaces than our histological images. When compared to digital pathology methods for astrocyte detection (Suleymanova et al., 2018), which is a more challenging segmentation problem since digital pathology images usually have a cluttered background, our pipeline also yielded mean precision (0.88 for testing and 0.92 for validation) above the reported value of 0.86, although much worse recall performance (0.16 for testing, 0.13 validation) when compared to their reported value of 0.78. The same pattern of similar precision but worse recall rates were seen when comparing our results to other studies focusing on segmenting or classifying tau lesions (Koga et al., 2021; Signaevsky et al., 2019). This worse recall performance was expected because we dealt with the blurriness and hue variations inherited from thick and large histological slides.
In contrast, other studies using DL algorithms to segment tau from histological sections ~30x thinner and with higher spatial resolution (1.22 μm/pixel here vs. 0.5 μm/pixel), much smaller sampling areas (standard 2×1” vs. 4×6”). Our pipeline can accommodate increasing scanning spatial resolution (by replacing the lens) and improving image blurriness by compacting images acquired at different depths of focus to improve recall values. The choice of compromising spatial resolution and image sharpness vs. increasing computational capability needs in an already computing-demanding pipeline depends on the aim of each project. The other pipelines focused on classifying tau lesions to facilitate neuropathological staging and diagnosis, thus requiring better spatial resolution. The cost-benefit of increasing scanning resolution is limited for our goals because, at the current level of PET resolution, each PET voxel represents a summary signal from thousands of histology voxels, and histology-based maps need to be downsampled for MRI/PET co-registration. Although it is beyond the scope of this manuscript, assessing the distribution of signals from all these histology voxels relative to a single PET voxel could provide further insights into PET imaging mechanisms. Therefore, we opted to employ a tissue processing pipeline optimized for minimizing tissue deformation and enable staining of larger samples. A more recent investigation focused on segmenting tau at 50 μm thick immunostained slides from medial temporal lobes blocks that underwent MR ex-cranio (Yushkevich et al., 2021). This study aligned with ours and used similar elements of previous versions of our histology-to-imaging registration pipeline 20, reported AUC values similar to ours. However, precision and recall values are missing, precluding further comparisons. Here, we focused on 3D histological maps for PET tracer validation.
The capacity of transfer learning is another IHCNet strength as it allows us to scale up our pipeline. CNN training is time and resource-consuming because it requires a significant amount of labeled data for performing effectively. Cross-training results indicate that mixing images from different cases – as in the full model training where images from the two cases were combined, not only does not interfere with training, but it is beneficial. However, while the ROC AUC values do not show a considerable difference between models, the precision-recall curves show decreased model accuracy in some cases and should be computed before using transfer learning. Also, it may be necessary to add extra labels for applying IHCNet to other tauopathies beyond AD.
Although we and others have previously developed and successfully tested algorithms for registering images of whole brain-histological sections dyed with Nissl staining on blockface images (Alegro et al., 2016; Alkemade et al., 2020), co-registering the digital images from immunostained slices to the blockface images represented an additional challenge in our pipeline. Immunohistochemistry processing requires harsher chemicals than Nissl staining, causing more tissue deformation and tears. These artifacts were prominent on the severely affected case 2, in which the brain tissue was extremely atrophic and friable. We had to add a manual affine registration step during the 2D histology to blockface alignment. Although this solution is robust to resolve histology artifacts, it impairs the scalability of our pipeline.
In conclusion, we present an unprecedented solution to improve PET tracer validation against histopathological changes measured on postmortem tissue. A crucial component of this pipeline is IHCNet, a CNN capable of processing large image datasets to locate and segment particles of interest. IHCNet has robust transfer learning capability. Our pipeline innovates for its ability to process massive datasets and perform well in blurred images with broad hue variations and obtain excellent co-registration results. Our 3D mapping at microscopic resolution coupled with our previously developed 3D registration algorithms for combining histological and imaging volumes can potentially open avenues for thorough and systematic validation of new neuroimaging tracers and expediting their approval for clinical use.
Supplementary Material
ACKNOWLEDGEMENTS and FUNDING
We thank the brain donors for contributing tissue to this study. This work was supported by AVID Radiopharmaceuticals, NIH R01AG056573, R01AG056573, U54 NS100717 and BrightFocus Foundation. MA and DU were partially funded by the Berkeley Institute of Data Science and the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Abbreviations
- AD
Alzheimer's disease
- AUC
area under the curve
- CNN
convolutional neural network
- DL
deep learning
- Grad-CAM
gradient-guided class activation maps
- HPC
high-performance computing
- MRI
magnetic resonance imaging
- PET
positron emission tomography
- ROC
receiver operating characteristic
Footnotes
COMPETING INTERESTS
The authors declare no competing interests.
ETHICS DECLARATIONS
This project was approved the UCSF IRB (protocol 13-12079).
REFERENCES
- 2018. MATLAB The MathWorks Inc, Natick, Massachusetts. [Google Scholar]
- 2021. 2021 Alzheimer's disease facts and figures. Alzheimers Dement 17, 327–406. [DOI] [PubMed] [Google Scholar]
- Al-Kofahi Y, Zaltsman A, Graves R, Marshall W, Rusu M, 2018. A deep learning-based algorithm for 2-D cell segmentation in microscopy images. BMC Bioinformatics 19, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alegro M, Júnior EA, Loring B, Heinsen H, Alho EJL, Zöllei L, Ushizima D, Grinberg LT, 2016. Multimodal Whole Brain Registration: MRI and High Resolution Histology. 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 634–642. [Google Scholar]
- Alegro M, Theofilas P, Nguy A, Castruita PA, Seeley W, Heinsen H, Ushizima DM, Grinberg LT, 2017. Automating cell detection and classification in human brain fluorescent microscopy images using dictionary learning and sparse coding. Journal of Neuroscience Methods 282, 20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alho A, Hamani C, Alho EJL, da Silva RE, Santos GAB, Neves RC, Carreira LL, Araujo CMM, Magalhaes G, Coelho DB, Alegro MC, Martin MGM, Grinberg LT, Pasqualucci CA, Heinsen H, Fonoff ET, Amaro E Jr., 2017. Magnetic resonance diffusion tensor imaging for the pedunculopontine nucleus: proof of concept and histological correlation. Brain Struct Funct 222, 2547–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkemade A, Pine K, Kirilina E, Keuken MC, Mulder MJ, Balesar R, Groot JM, Bleys R, Trampel R, Weiskopf N, Herrler A, Moller HE, Bazin PL, Forstmann BU, 2020. 7 Tesla MRI Followed by Histological 3D Reconstructions in Whole-Brain Specimens. Front Neuroanat 14, 536838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison N, Song G, 2009. Advanced normalization tools (ANTS). Insight 2. [Google Scholar]
- Avants BB, Tustison NJ, Stauffer M, Song G, Wu B, Gee JC, 2014. The Insight ToolKit image registration framework. Front Neuroinform 8, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrio JR, 2018. The Irony of PET Tau Probe Specificity. J Nucl Med 59, 115–116. [DOI] [PubMed] [Google Scholar]
- Beg MF, Miller MI, Trouvé A, Younes L, 2005. Computing Large Deformation Metric Mappings via Geodesic Flows of Diffeomorphisms. International Journal of Computer Vision 61, 139–157. [Google Scholar]
- Bria A, Iannello G, 2012. TeraStitcher - a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell RA, Eifert RW, Turner GC, 2014. OpenStage: a low-cost motorized microscope stage with sub-micron positioning accuracy. PLoS One 9, e88977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cireşan DC, Meier U, Masci J, Gambardella LM, Schmidhuber J, 2011. Flexible, High Performance Convolutional Neural Networks for Image Classification. Proceedings of the Twenty-Second International Joint Conference on Artificial Intelligence 2, 1237–1242. [Google Scholar]
- Curtis C, Gamez JE, Singh U, Sadowsky CH, Villena T, Sabbagh MN, Beach TG, Duara R, Fleisher AS, Frey KA, Walker Z, Hunjan A, Holmes C, Escovar YM, Vera CX, Agronin ME, Ross J, Bozoki A, Akinola M, Shi J, Vandenberghe R, Ikonomovic MD, Sherwin PF, Grachev ID, Farrar G, Smith AP, Buckley CJ, McLain R, Salloway S, 2015. Phase 3 trial of flutemetamol labeled with radioactive fluorine 18 imaging and neuritic plaque density. JAMA Neurol 72, 287–294. [DOI] [PubMed] [Google Scholar]
- Dani M, Brooks DJ, Edison P, 2016. Tau imaging in neurodegenerative diseases. Eur J Nucl Med Mol Imaging 43, 1139–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dice LR, 1945. Measures of the Amount of Ecologic Association Between Species. Ecology 26, 297–302. [Google Scholar]
- Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ, 2008. LIBLINEAR: A Library for Large Linear Classification J. Mach. Learn. Res 9, 1871–1874. [Google Scholar]
- Fischl B, 2012. FreeSurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinberg LT, Amaro Junior E, da Silva AV, da Silva RE, Sato JR, dos Santos DD, de Paula Pacheco S, de Lucena Ferretti RE, Paraizo Leite RE, Pasqualucci CA, Teipel SJ, Flatz WH, Brazilian Aging Brain Study, G., Heinsen H, 2009. Improved detection of incipient vascular changes by a biotechnological platform combining post mortem MRI in situ with neuropathology. J Neurol Sci 283, 2–8. [DOI] [PubMed] [Google Scholar]
- Heinsen H, Arzberger T, Schmitz C, 2000. Celloidin mounting (embedding without infiltration) - a new, simple and reliable method for producing serial sections of high thickness through complete human brains and its application to stereological and immunohistochemical investigations. J Chem Neuroanat 20, 49–59. [DOI] [PubMed] [Google Scholar]
- Jianbo S, Malik J, 2000. Normalized cuts and image segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 22, 888–905. [Google Scholar]
- Kelley AS, McGarry K, Gorges R, Skinner JS, 2015. The burden of health care costs for patients with dementia in the last 5 years of life. Ann Intern Med 163, 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV, 2009. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage 46, 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klunk WE, 2018. Molecular imaging: What is right and what is an illusion? Alzheimers Dement (Amst) 10, 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga S, Ghayal NB, Dickson DW, 2021. Deep Learning-Based Image Classification in Differentiating Tufted Astrocytes, Astrocytic Plaques, and Neuritic Plaques. J Neuropathol Exp Neurol 80, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G, 2015. Deep learning. Nature 521, 436–444. [DOI] [PubMed] [Google Scholar]
- Lemoine L, Leuzy A, Chiotis K, Rodriguez-Vieitez E, Nordberg A, 2018. Tau positron emission tomography imaging in tauopathies: The added hurdle of off-target binding. Alzheimers Dement (Amst) 10, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzy A, Chiotis K, Lemoine L, Gillberg PG, Almkvist O, Rodriguez-Vieitez E, Nordberg A, 2019. Tau PET imaging in neurodegenerative tauopathies-still a challenge. Mol Psychiatry 24, 1112–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquie M, Normandin MD, Meltzer AC, Siao Tick Chong M, Andrea NV, Anton-Fernandez A, Klunk WE, Mathis CA, Ikonomovic MD, Debnath M, Bien EA, Vanderburg CR, Costantino I, Makaretz S, DeVos SL, Oakley DH, Gomperts SN, Growdon JH, Domoto-Reilly K, Lucente D, Dickerson BC, Frosch MP, Hyman BT, Johnson KA, Gomez-Isla T, 2017. Pathological correlations of [F-18]-AV-1451 imaging in non-alzheimer tauopathies. Ann Neurol 81, 117–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuliffe MJ, Lalonde FM, McGarry D, Gandler W, Csaky K, Trus BL, 2001. Medical Image Processing, Analysis and Visualization in clinical research. Proceedings 14th IEEE Symposium on Computer-Based Medical Systems. CBMS 2001, pp. 381–386. [Google Scholar]
- Montine TJ, Phelps CH, Beach TG, Bigio EH, Cairns NJ, Dickson DW, Duyckaerts C, Frosch MP, Masliah E, Mirra SS, Nelson PT, Schneider JA, Thal DR, Trojanowski JQ, Vinters HV, Hyman BT, 2012. National Institute on Aging-Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: a practical approach. Acta Neuropathologica 123, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez I, Matute T, Herrera R, Keymer J, Marzullo T, Rudge T, Federici F, 2017. Low cost and open source multi-fluorescence imaging system for teaching and research in biology and bioengineering. PLoS One 12, e0187163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura N, Harada R, Ishiki A, Kikuchi A, Nakamura T, Kudo Y, 2018. The development and validation of tau PET tracers: current status and future directions. Clinical and Translational Imaging 6, 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ZF, Dean S, Recht B, Waller L, 2020. High-throughput fluorescence microscopy using multi-frame motion deblurring. Biomedical Optics Express 11, 281–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontecorvo MJ, Keene CD, Beach TG, Montine TJ, Arora AK, Devous MD Sr., Navitsky M, Kennedy I, Joshi AD, Lu M, Serrano GE, Sue LI, Intorcia AJ, Rose SE, Wilson A, Hellstern L, Coleman N, Flitter M, Aldea P, Fleisher AS, Mintun MA, Siderowf A, 2020. Comparison of regional flortaucipir PET with quantitative tau immunohistochemistry in three subjects with Alzheimer's disease pathology: a clinicopathological study. EJNMMI Res 10, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovici GD, Gatsonis C, Apgar C, Chaudhary K, Gareen I, Hanna L, Hendrix J, Hillner BE, Olson C, Lesman-Segev OH, Romanoff J, Siegel BA, Whitmer RA, Carrillo MC, 2019. Association of Amyloid Positron Emission Tomography With Subsequent Change in Clinical Management Among Medicare Beneficiaries With Mild Cognitive Impairment or Dementia. JAMA 321, 1286–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF (Eds.), Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Springer International Publishing, Cham, pp. 234–241. [Google Scholar]
- Rosenegger DG, Tran CH, LeDue J, Zhou N, Gordon GR, 2014. A high performance, cost-effective, open-source microscope for scanning two-photon microscopy that is modular and readily adaptable. PLoS One 9, e110475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabri O, Sabbagh MN, Seibyl J, Barthel H, Akatsu H, Ouchi Y, Senda K, Murayama S, Ishii K, Takao M, Beach TG, Rowe CC, Leverenz JB, Ghetti B, Ironside JW, Catafau AM, Stephens AW, Mueller A, Koglin N, Hoffmann A, Roth K, Reininger C, Schulz-Schaeffer WJ, Florbetaben Phase 3 Study, G., 2015. Florbetaben PET imaging to detect amyloid beta plaques in Alzheimer's disease: phase 3 study. Alzheimers Dement 11, 964–974. [DOI] [PubMed] [Google Scholar]
- Salloway S, Gamez JE, Singh U, Sadowsky CH, Villena T, Sabbagh MN, Beach TG, Duara R, Fleisher AS, Frey KA, Walker Z, Hunjan A, Escovar YM, Agronin ME, Ross J, Bozoki A, Akinola M, Shi J, Vandenberghe R, Ikonomovic MD, Sherwin PF, Farrar G, Smith APL, Buckley CJ, Thal DR, Zanette M, Curtis C, 2017. Performance of [(18)F]flutemetamol amyloid imaging against the neuritic plaque component of CERAD and the current (2012) NIA-AA recommendations for the neuropathologic diagnosis of Alzheimer's disease. Alzheimers Dement (Amst) 9, 25–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A, 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, 2017. Mapping Neurodegenerative Disease Onset and Progression. Cold Spring Harb Perspect Biol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D, 2017. Grad-CAM: Visual Explanations from Deep Networks via Gradient-Based Localization. 2017 IEEE International Conference on Computer Vision (ICCV), pp. 618–626. [Google Scholar]
- Signaevsky M, Prastawa M, Farrell K, Tabish N, Baldwin E, Han N, Iida MA, Koll J, Bryce C, Purohit D, Haroutunian V, McKee AC, Stein TD, White CL 3rd, Walker J, Richardson TE, Hanson R, Donovan MJ, Cordon-Cardo C, Zeineh J, Fernandez G, Crary JF, 2019. Artificial intelligence in neuropathology: deep learning-based assessment of tauopathy. Lab Invest 99, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LN, 2015. cyclical learning Rates for Training Neural Networks. ArXiv150601186 Cs. [Google Scholar]
- Smith R, Schain M, Nilsson C, Strandberg O, Olsson T, Hagerstrom D, Jogi J, Borroni E, Scholl M, Honer M, Hansson O, 2017. Increased basal ganglia binding of (18) F-AV-1451 in patients with progressive supranuclear palsy. Mov Disord 32, 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soleimani-Meigooni DN, Iaccarino L, La Joie R, Baker S, Bourakova V, Boxer AL, Edwards L, Eser R, Gorno-Tempini ML, Jagust WJ, Janabi M, Kramer JH, Lesman-Segev OH, Mellinger T, Miller BL, Pham J, Rosen HJ, Spina S, Seeley WW, Strom A, Grinberg LT, Rabinovici GD, 2020. 18F-flortaucipir PET to autopsy comparisons in Alzheimer's disease and other neurodegenerative diseases. Brain 143, 3477–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suleymanova I, Balassa T, Tripathi S, Molnar C, Saarma M, Sidorova Y, Horvath P, 2018. A deep convolutional neural network approach for astrocyte detection. Sci Rep 8, 12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theofilas P, Polichiso L, Wang X, Lima LC, Alho AT, Leite RE, Suemoto CK, Pasqualucci CA, Jacob-Filho W, Heinsen H, Brazilian Aging Brain Study, G., Grinberg LT, 2014. A novel approach for integrative studies on neurodegenerative diseases in human brains. J Neurosci Methods 226, 171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiepolt S, Patt M, Aghakhanyan G, Meyer PM, Hesse S, Barthel H, Sabri O, 2019. Current radiotracers to image neurodegenerative diseases. EJNMMI Radiopharm Chem 4, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, 2013. Explicit B-spline regularization in diffeomorphic image registration. Front Neuroinform 7, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29, 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Rossum G, Drake FL Jr, 1995. Python tutorial. . Centrum voor Wiskunde en Informatica, Amsterdam, The Netherlands. [Google Scholar]
- Wang YT, Edison P, 2019. Tau Imaging in Neurodegenerative Diseases Using Positron Emission Tomography. Curr Neurol Neurosci Rep 19, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Lopez MM, Martin M, Ittyerah R, Lim S, Ravikumar S, Bedard ML, Pickup S, Liu W, Wang J, Hung LY, Lasserve J, Vergnet N, Xie L, Dong M, Cui S, McCollum L, Robinson JL, Schuck T, de Flores R, Grossman M, Tisdall MD, Prabhakaran K, Mizsei G, Das SR, Artacho-Perula E, Jimenez M, Rabal M, Romero FJM, Sanchez SC, Gonzalez JCD, de la Rosa-Prieto C, Parada MC, Lee EB, Trojanowski JQ, Ohm DT, Wisse LEM, Wolk DA, Irwin DJ, Insausti R, 2021. Three-dimensional mapping of neurofibrillary tangle burden in the human medial temporal lobe. Brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack GW, Rogers WE, Latt SA, 1977. Automatic measurement of sister chromatid exchange frequency. J Histochem Cytochem 25, 741–753. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.