Abstract
Evernimicin (SCH 27899) is a new antibiotic with activity against a wide spectrum of gram-positive bacteria and activity against some gram-negative bacteria. Previous metabolic labeling studies indicated that evernimicin specifically inhibited protein synthesis in Staphylococcus aureus. Using a susceptible Escherichia coli strain, we demonstrated that evernimicin also inhibited protein synthesis in E. coli. In cell-free translation assays with extracts from either E. coli or S. aureus, evernimicin had a 50% inhibitory concentration of approximately 125 nM. In contrast, cell-free systems derived from wheat germ and rabbit reticulocytes were inhibited only by very high levels of evernimicin. Evernimicin did not promote transcript misreading. [14C]evernimicin specifically bound to the 50S subunit from E. coli. Nonlinear regression analysis of binding data generated with 70S ribosomes from E. coli and S. aureus and 50S subunits from E. coli returned dissociation constants of 84, 86, and 160 nM, respectively. In binding experiments, performed in the presence of excess quantities of a selection of antibiotics known to bind to the 50S subunit, only the structurally similar drug avilamycin blocked binding of [14C]evernimicin to ribosomes.
The emergence of vancomycin resistance among Enterococcus spp. and the demonstration that this resistance can be transferred to Staphylococcus aureus (14) have highlighted the need for alternative regimens to combat serious infections caused by gram-positive organisms. One potential new agent is evernimicin (SCH 27899), a member of the everninomicin class of antibiotics isolated from Micromonospora carbonaceae (5–7). Evernimicin is an oligosaccharide antibiotic with activity against a broad range of gram-positive pathogenic bacteria including glycopeptide-resistant enterococci, methicillin-resistant staphylococci, and penicillin-resistant streptococci (9). However, the mode of action of evernimicin is unclear. Two studies, a genetic characterization of a Streptococcus pneumoniae strain that displayed reduced susceptibility to evernimicin (3) and a metabolic labeling study with S. aureus (T. A. Black, W. Zhao, K. J. Shaw, and R. S. Hare, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. C-106, p. 99, 1998), suggested that evernimicin exerts its antibacterial effects by inhibiting protein synthesis. These data are consistent with the finding that avilamycin, which is structurally similar to evernimicin (Fig. 1), also inhibits translation in vivo and in vitro (24). An indication that avilamycin and evernimicin may share similar mechanisms of action came from a survey of avilamycin-resistant enterococci, which revealed a direct correlation between avilamycin resistance and a reduced susceptibility to evernimicin (1). In this report we document the biochemical effect of evernimicin on protein synthesis. Our data demonstrate that evernimicin binds ribosomes from S. aureus and Escherichia coli at what appears to be a unique site. In addition, evernimicin inhibits in vitro protein synthesis with ribosomes derived from both S. aureus and E. coli. However, in contrast to avilamycin, which was proposed to bind to the 30S subunit (24), evernimicin binds exclusively to the 50S subunit.
FIG. 1.
Structures of avilamycin and evernimicin.
MATERIALS AND METHODS
Antibiotics.
Evernimicin was isolated at Schering-Plough Research Institute. Linezolid was synthesized at Schering-Plough Research Institute. Quinupristin-dalfopristin and avilamycin were gifts from Rhone Poulenc-Rorer and Elanco Animal Health, respectively. Chloramphenicol, clindamycin, cycloheximide, erythromycin, lincomycin, thiostrepton, and puromycin were purchased from Sigma Chemical Co.
Bacterial strains.
E. coli A19 (rna-19) and HS227-5, a highly drug-susceptible strain, together with S. aureus RN450 were obtained from the Schering-Plough Research Institute culture collection.
Whole-cell labeling.
HS227-5 was grown in tryptic soy broth to an optical density at 600 nm (OD600) of 0.3. The culture was diluted (1:10) into 10 ml of fresh prewarmed tryptic soy broth containing 5 μCi of each of the following labeled substrates (all purchased from NEN): [14C]isoleucine (specific activity, 340 mCi/mmol), [14C]thymidine (specific activity, 60 mCi/mmol), and [14C]uracil (specific activity, 60 mCi/mmol). Test antibiotics were added when the culture reached an OD600 of 0.5. Duplicate 0.2-ml aliquots of culture were removed, both before and after addition of test antibiotic, and were subject to precipitation with trichloroacetic acid (TCA). Radioactivity was determined by liquid scintillation counting.
Preparation and fractionation of ribosomes.
Ribosomes were prepared from E. coli (21) and S. aureus (11) as described previously. Ribosomes and S100 extracts were resuspended and dialyzed against B3 (10 mM MgCl2, 20 mM Tris [pH 7.8], 30 mM NH4Cl, 0.1 mM EDTA, 6 mM β-mercaptoethanol), respectively. Ribosomes were washed with salt at high concentrations, prepared as described above, and then resuspended in B6 (which is the same as B3 except that NH4Cl is present at 1 M). After 12 h on ice the ribosomes were pelleted (40,000 rpm for 20 h) and resuspended in B3. Ribosomal subunits were prepared by resuspension of 70S ribosomes in B4 (which is the same as B3 except that NH4Cl is present at 100 mM and MgCl2 is present at 1 mM), followed by fractionation on 10 to 30% sucrose gradients. The subunits were pelleted and resuspended in B3. The purities of the preparations were assessed by centrifuging an aliquot on an analytical sucrose gradient.
In vitro translation reactions.
Duplicate reactions with the following labeled substrates (purchased from NEN) and templates were performed as described previously (22): poly(U) template with [14C]phenylalanine (specific activity, 500 mCi/mmol), poly(A) template with [14C]lysine (specific activity, 310 mCi/mmol), and MS2 with phenylalanine and isoleucine (specific activity, 340 mCi/mmol). Labeled proteins from reactions with poly(U) and MS2 templates were precipitated with 7% TCA and were then heated to 90°C for 15 min. Extracts were applied to 96-well filter plates (Millipore), and the radioactivity was determined by liquid scintillation counting. Proteins from reactions with polylysine were precipitated with 5% TCA containing 0.05% tungstic acid (16). Incorporation of radiolabel into TCA-precipitable material in both E. coli and S. aureus systems was template dependent and linear for at least 60 min (data not shown). In vitro translation reactions with rabbit reticulocyte and wheat germ cell extracts (Promega) were performed with luciferase and brome mosaic virus templates, respectively, as directed by the manufacturer. The level of incorporation of [35S]methionine into TCA-precipitable material was measured as directed by the manufacturer.
Binding assays.
[14C]evernimicin (specific activity, 8 mCi/mmol) was prepared as described previously (8), and [dichloroacetyl-1,2-14C]chloramphenicol (specific activity, 57 mCi/mmol) was purchased from NEN. Triplicate binding reactions, performed in B3, comprised 46 pmol of 70S ribosomes or ribosomal subunits (1A260 = 23 pmol of 70S ribosomes, 34.5 pmol of 50S, and 69 pmol of 30S) and labeled antibiotic. After 30 min at room temperature unbound antibiotic was removed by centrifuging the reaction through a 1-ml size-exclusion column (Bio-Gel P-30 rehydrated in B3 formed in a Micro Bio-Spin column; Bio-Rad). The radioactivity in the void volume was determined by liquid scintillation counting. By this method greater than 90% of the ribosomes were recovered, and in the absence of ribosomes there was no detectable flowthrough of radiolabeled material (data not shown). Dissociation constants (Kds) and binding maxima were calculated from nonlinear regression plots using EnzymeKinetics (Trinity software). Rates of dissociation of [14C]evernimicin-ribosome complexes were determined by first establishing a binding reaction with a fixed amount of [14C]evernimicin and ribosomes. At time zero a 500-fold excess of unlabeled evernimicin was added and the concentration of antibiotic-ribosome complexes [RD] was monitored over time. A plot of ln([RD]/[RD0]) versus time ([RD0] is the concentration of the antibiotic-ribosome complex at time zero) gave a straight line with a gradient equal to dissociation rate constant K−1. [14C]evernimicin-subunit complexes were generated by incubating the reaction mixtures in a buffer with magnesium at a low concentration (1 mM) and were then fractionated on 10 to 30% sucrose gradients.
Competition reactions.
70S ribosomes (2 A260 units) were incubated with 40 μg of unlabeled competitor antibiotic in a volume of 48 μl for 25 min at room temperature. [14C]evernimicin (0.078 μg, 49 pmol) was added, and after a further 30 min the amount of [14C]evernimicin-ribosome complex formed was quantified as described above. A more accurate quantitation was performed by mixing various amounts of unlabeled competitor antibiotic with a fixed amount (0.156 μg, 98 pmol) of [14C]evernimicin. After addition of 70S ribosomes (2 A260 units), the amount of antibiotic-ribosome complex formed was measured as described above.
RESULTS
Effect of evernimicin on substrate incorporation.
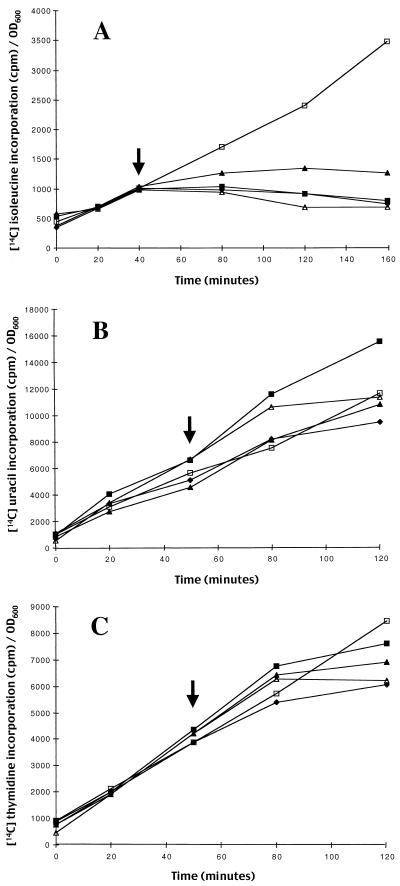
To determine if evernimicin inhibited protein synthesis in E. coli we monitored the effect of the antibiotic on incorporation of [14C]isoleucine, [14C]thymidine, and [14C]uracil in vivo. Since wild-type E. coli is not susceptible to growth inhibition by evernimicin, we used a modified E. coli strain, HS227-5, for which the evernimicin MIC is 4 μg/ml. Incorporation of [14C]isoleucine by HS227-5 was inhibited almost immediately after addition of evernimicin at 1× the MIC (Fig. 2A). In contrast, over the sampling period used in the present study inhibition of [14C]uracil and [14C]thymidine incorporation was largely unaffected at up to 4× the MIC (Figs. 2B and C).
FIG. 2.
Effect of evernimicin on incorporation of [14C]isoleucine (A) [14C]uracil (B), and [14C]thymidine (C) by E. coli HS227-5. Evernimicin was added to exponential-phase cultures at the time indicated by the arrow at the following multiples of the MIC (4 μg/ml): 0.5× (solid triangles), 1× (solid squares), and 2× (solid diamonds). Results for reactions with chloramphenicol at 20 μg/ml (open triangles) and no antibiotic (controls; open squares) were also included. The data were normalized, to correct for differences in growth rate resulting from addition of test antibiotics, by dividing the counts obtained at a given time point by the OD600.
Effect of evernimicin on translation in vitro.
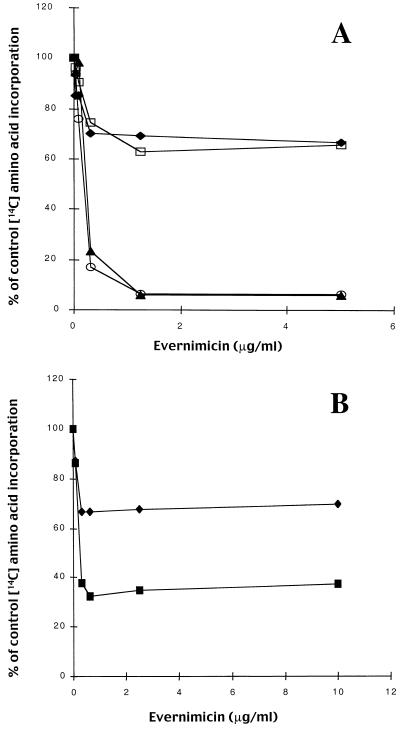
In vitro translation reactions were performed with ribosomes and S100 extracts isolated from E. coli A19 and S. aureus RN450. In the absence of antibiotic there were no significant differences in the amount of radiolabeled protein synthesized in either the E. coli- or the S. aureus-based reactions, indicating that the overall rates of protein synthesis were similar (data not shown). Evernimicin was equally effective at inhibiting translation in both reactions based on both gram-positive and gram-negative ribosomes (Fig. 3A). For both the poly(U)- and poly(A)-based reactions, the 50% inhibitory concentrations (IC50s) were 125 nM. However, the maximal level of inhibition was template dependent. With a poly(U) template the maximal level of inhibition was 35% and occurred at 0.3 μg/ml (180 nM). Under the same conditions, kirromycin (3 μg/ml) and tetracycline (100 μg/ml) inhibited expression by over 90%, while erythromycin, lincomycin, and clindamycin at 800 μg/ml did not inhibit expression (data not shown). In a poly(A)-based reaction, evernimicin at 1.2 μg/ml (720 nM) caused 90% inhibition. Under the same conditions we observed 85% inhibition with kirromycin and erythromycin at 5 μg/ml and 70% inhibition with lincomycin and clindamycin at 50 μg/ml (data not shown). Using a native template, phage MS2, in an E. coli-based reaction, we observed approximately 70% inhibition (Fig. 3B).
FIG. 3.
Effect of evernimicin on in vitro translation reactions with various mRNA templates, ribosomes washed with salt at low concentrations, and S100 extracts from E. coli A19 and S. aureus RN450. Incorporation of the various 14C-labeled amino acids into TCA-precipitable material is expressed as a percentage of that in the control (no antibiotic) reaction. (A) Poly(U)-based reaction with components from E. coli (solid diamonds) and S. aureus (open squares) and a poly(A)-based reaction with components from E. coli (solid triangles) and S. aureus (open circles). (B) E. coli components with poly(U) (solid diamonds) and phage MS2 (solid squares) templates.
To determine if inhibition is specific to prokaryotic ribosomes, we repeated these experiments using cell extracts from rabbit reticulocytes and wheat germ cells (data not shown). In both systems there was no inhibition by evernimicin at concentrations up to 40 μg/ml (25 μM). At concentrations above 40 μg/ml protein synthesis in the rabbit reticulocyte system was progressively inhibited, reaching a maximum of 90% at 160 μg/ml (100 μM). In contrast, protein synthesis in the wheat germ system showed no inhibition at 640 μg/ml (400 μM). Addition of puromycin (800 μg/ml) reduced translation in both systems by 99%.
Effect of evernimicin on misreading.
To determine if evernimicin promotes misreading we separately measured incorporation of [14C]isoleucine and [14C]phenylalanine in a poly(U)-based in vitro translation assay. In the absence of either antibiotic isoleucine incorporation was below detectable levels (data not shown). In separate experiments streptomycin, added over a range of 0.78 to 800 μg/ml, progressively inhibited incorporation of phenylalanine (maximal level of inhibition, 50% at 800 μg/ml) and increased the level of incorporation of isoleucine (i.e., promoted misreading; maximal increase, 40-fold at 800 μg/ml). Addition of evernimicin (over a range of 0.08 to 640 μg/ml) progressively inhibited incorporation of phenylalanine (maximal level of inhibition, 65% at 640 μg/ml) but did not promote isoleucine incorporation.
Binding of [14C]evernimicin to 70S ribosomes.
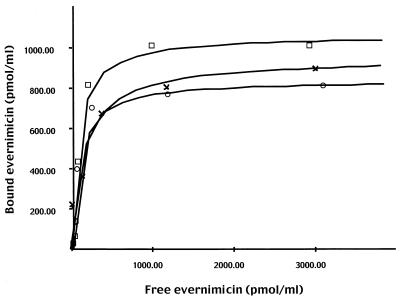
[14C]evernimicin bound to ribosomes from E. coli and S. aureus with equal affinity. Nonlinear regression plots returned Kds of 84 and 86 nM for ribosomes from E. coli and S. aureus, respectively (Fig. 4). At saturation, the stoichiometry of [14C]evernimicin to ribosome was approximately 1:1 (960 pmol of ribosomes from E. coli and S. aureus bound 1,060 and 830 pmol of [14C]evernimicin, respectively). Under the same conditions the Kd for [14C]chloramphenicol binding to ribosomes from E. coli was 1 μM (data not shown), which is in agreement with previous data (4). Washing of E. coli ribosomes with salt at a high concentration (1 M NH4Cl) had no effect on binding of [14C]evernimicin (data not shown). The rates of dissociation (K−1) of [14C]evernimicin from both sets of ribosomes were similar; ribosomes from E. coli and S. aureus returned K−1 values of 0.034 min−1 and 0.035 min−1, respectively (data not shown). These values equate to antibiotic-ribosome complex half-lives of approximately 20 min.
FIG. 4.
Nonlinear regression analysis of [14C]evernimicin binding to 70S ribosomes from E. coli A19 (open squares) and S. aureus RN450 (open circles) washed with salt at low concentrations and purified 50S subunits from E. coli A19 (crosses).
Binding of [14C]evernimicin to the 50S subunit.
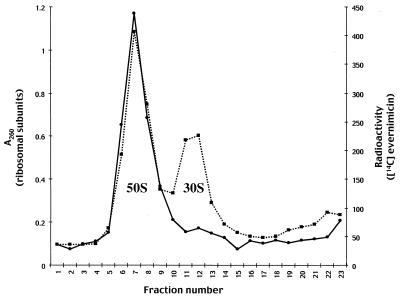
The binding reactions were repeated with a buffer with magnesium at a low concentration to promote dissociation of 70S ribosomes. Fractionation of the resulting subunit-[14C]evernimicin complexes on sucrose gradients demonstrated that evernimicin bound to only the 50S subunit (Fig. 5). A repeat assay with a 100-fold excess of labeled antibiotic gave the same result (data not shown). We confirmed these data by purifying 30S and 50S subunits from E. coli. Analytical sucrose gradients demonstrated that the 30S and 50S preparations were contaminated with small amounts (less than 5%) of the corresponding subunit (data not shown). [14C]evernimicin bound to the purified 50S subunits with a 1:1 stoichiometry and a Kd of 160 nM (Fig. 4). We also observed a small amount (less than 10% of that seen with the 50S subunits) of binding to the 30S subunit which we attribute to the contaminating 50S subunits (data not shown).
FIG. 5.
Binding of [14C]evernimicin to ribosomal subunits from E. coli A19. 70S ribosomes (115 pmol) washed with salt at low concentrations and [14C]evernimicin (98 pmol) were incubated in buffer with a low concentration of magnesium, and the resultant antibiotic-subunit complexes were resolved on a 10 to 30% sucrose gradient. The gradient was fractionated from the bottom and radioactivity (solid line) and the A260 (dashed line) were determined for each fraction. The peaks corresponding to the 30S and 50S subunits are labeled.
Competition binding assays.
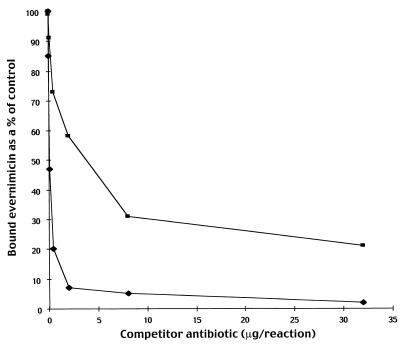
The 50S-binding antibiotics, avilamycin, chloramphenicol, clindamycin, erythromycin, lincomycin, linezolid, puromycin, thiostrepton, and a combination of quinupristin-dalfopristin were tested for their abilities to block binding of [14C]evernimicin to E. coli ribosomes. Only avilamycin and unlabeled evernimicin inhibited binding; compared to control reactions both compounds reduced the level of binding of [14C]evernimicin by 85 to 90% (data not shown). To rule out the possibility that avilamycin binds to the 30S subunit, as suggested previously (24), in a manner that precludes binding of evernimicin to the 50S subunit, we repeated the competition assay using purified 50S subunits. Avilamycin again blocked the binding of evernimicin (data not shown). To more accurately quantify the relative affinities of avilamycin and evernimicin for the ribosome we measured the amount of unlabeled antibiotic required to inhibit binding of a fixed amount of [14C] evernimicin by 50% (IB50). The IB50 of avilamycin was 32-fold higher than that measured for evernimicin (Fig. 6).
FIG. 6.
Competition between avilamycin and evernimicin for binding to 70S ribosomes from E. coli A19 washed with salt at low concentrations. Various amounts of unlabeled avilamycin (squares) and evernimicin (diamonds) were mixed with a fixed amount (0.156 μg, 98 pmol) of [14C]evernimicin and incubated with 70S ribosomes from E. coli washed with salt at low concentrations. Following binding the amount of bound antibiotic was determined.
DISCUSSION
Prior studies (3; Black et al., 38th ICAAC) suggested that protein synthesis is the likely target of evernimicin. In the study described in this paper we examined the effect of evernimicin on translation in vitro using synthetic and native templates. Synthetic templates lack initiation signals, ribosome-binding sites, and initiation codons, as well as termination codons. Consequently, translational initiation with a synthetic template does not require N-formylmethionine, and the assay can be regarded as a specific test of elongation. By these criteria, evernimicin is a potent inhibitor of elongation. In a poly(A)-based system the inhibition caused by evernimicin approached 100%. In contrast, the maximal level of inhibition in a poly(U)-based reaction never exceeded 35%. Erythromycin exhibited the same template dependency, and this effect has been noted previously (15). These differences have been attributed to the conformation of the nascent polypeptides; polyphenylalanine aggregates in an insoluble hydrophobic mass in the vicinity of the peptidyltransferase center, whereas polylysine peptides extend away from the peptidyltransferase center into the aqueous environment (17). Erythromycin is proposed to bind in the vicinity of the peptidyltransferase center and to block the exit of the nascent polypeptide (21). Therefore, the atypical conformation adopted by polyphenylalanine is thought to allow it to escape inhibition by erythromycin. Whether the fact that evernimicin exhibits a pattern of inhibition similar to that of erythromycin implies that it inhibits protein synthesis by the same mechanism remains to be determined. Further evidence that evernimicin acts at the level of elongation came from assays with native templates. It was shown that agents that specifically blocked initiation of translation are sensitive to the amount of template in the reaction mixture (20). A 2.5-fold reduction in the amount of MS2 template had no effect on inhibition by evernimicin (data not shown), suggesting that it does not inhibit translation initiation.
The marked in vivo differences in the susceptibility of E. coli and S. aureus to inhibition by evernimicin were not mirrored by the respective cell-free translation reactions. This suggests that the evernimicin binding site is conserved between the two bacteria and that the susceptibility differences are due to penetration issues. In vivo labeling studies with an evernimicin-sensitive E. coli strain and measurement of labeled evernimicin binding to both 70S ribosomes and 50S subunits confirmed this supposition. Evernimicin bound to a single high-affinity site on the 50S subunit. Exposure of ribosomes to a wash with salt at a high concentration did not change the level of binding of evernimicin, implying that the antibiotic does not bind to a loosely associated protein.
Competition assays with either 70S ribosomes or 50S subunits demonstrated that only the structurally similar drug, avilamycin, blocked evernimicin binding. These data strongly suggest that avilamycin also binds to the 50S subunit, which contradicts a previous study in which avilamycin was shown to bind to the 30S subunit (24). However, in the earlier study the investigator determined which subunit avilamycin bound to by reconstituting functional 70S ribosomes from either 30S or 50S subunits that had been preincubated with excess avilamycin and the appropriate untreated subunit. The differences between treated 30S and 50S subunits were relatively minor and thus may explain the contradictory conclusions obtained. We note that in the competition reactions an excess of unlabeled evernimicin effectively blocks binding of labeled evernimicin. The ability of a given labeled antibiotic to bind to the ribosome in the presence of a large excess of the same unlabeled compound was previously used to measure nonspecific binding (10). Therefore, by these criteria, evernimicin binds to the ribosome with a high degree of specificity.
Killing curve data suggested that evernimicin is not uniformly bactericidal (9). We used a misincorporation assay to determine if evernimicin, like streptomycin, exerts its limited bactericidal effects through promotion of misreading. We found no evidence that SCH 27899 promotes misreading.
The data above presented suggest that evernimicin binds to the 50S subunit and blocks some aspect of the elongation phase of translation. Both these findings are consistent with the observation that substitutions in L16, a component of the 50S subunit, result in reduced susceptibility to evernimicin in both S. pneumoniae (3) and S. aureus (unpublished data). L16 has been implicated in both aminoacyl-tRNA binding (2, 13, 25) and peptidyltransferase activity (12, 19), although it remains to be determined if L16 is absolutely required for the latter activity (23). The question of whether evernimicin binds directly to L16 remains to be determined. One observation that argues against this is that L16 has been implicated in binding to chloramphenicol (18). Our studies strongly suggest that the chloramphenicol and evernimicin binding sites do not overlap. Further indirect evidence that evernimicin acts at the level of elongation comes from studies on the mode of action of avilamycin. Avilamycin partially blocked nonenzymatic aminoacyl-tRNA binding and completely blocked enzymatic aminoacyl-tRNA binding (24). The structural similarities between evernimicin and avilamycin and the finding that they compete for a common binding site suggest that evernimicin may also inhibit translation by interfering with aminoacyl-tRNA binding.
In conclusion, evernimicin is an inhibitor of translation both in vivo and in vitro, and this inhibition is specific to prokaryotic ribosomes. Furthermore, the evernimicin binding site on the 50S subunit does not overlap with the binding site of other drugs currently in clinical use and is likely the reason why cross-resistance is not prevalent in recent clinical isolates (R. S. Hare and F. J. Sabatelli, Abstr. 38th Intersci. Conf. Antimicrob. Agents Chemother., abstr. E-119, p. 188, 1998).
ACKNOWLEDGMENTS
We thank Alexander Mankin and Keith Marotti for advice on ribosome preparation.
REFERENCES
- 1.Aarestrup F M. Association between decreased susceptibility to a new antibiotic for treatment of human diseases, everninomicin (SCH 27899), and resistance to an antibiotic used for growth promotion in animals, avilamycin. Microb Drug Resist. 1998;4:137–141. doi: 10.1089/mdr.1998.4.137. [DOI] [PubMed] [Google Scholar]
- 2.Abdurashidova G G, Turchinsky M F, Aslanov K A, Budowsky E I. Polynucleotide-protein interactions in the translation system. Identification of proteins interacting with tRNA in the A- and P-sites of E. coli ribosomes. Nucleic Acids Res. 1979;6:3891–3909. doi: 10.1093/nar/6.12.3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adrian P V, Zhao W, Black T A, Shaw K J, Hare R S, Klugman K P. Characterization of mutations in ribosomal protein L16 conferring reduced susceptibility to everninomicin SCH27899: implications for mechanism of action. Antimicrob Agents Chemother. 2000;44:732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fernandez-Munoz R, Monro R E, Vazquez D. Ribosomal peptidyltransferase: binding of inhibitors. Methods Enzymol. 1971;20:481–490. [Google Scholar]
- 5.Ganguly A K, Girijavallabhan V, Miller G H, Sarre O Z. Chemical modification of everninomicin. J Antibiot. 1982;35:561–570. doi: 10.7164/antibiotics.35.561. [DOI] [PubMed] [Google Scholar]
- 6.Ganguly A K, Pramanik B, Chan T M, Sarre O Z, Liu Y-T, Morton J, Girijavallabhan V. The structure of new oligosaccharide antiobiotics, 13-384 components 1 and 5. Heterocycles. 1989;28:83–88. [Google Scholar]
- 7.Ganguly A K, McCormick J P, Chan T M, Saksena A K, Das P R. Determination of the absolute stereochemistry at the C16 orthoester of everninomicin antiobiotics; a novel acid-catalysed isomerization of orthoesters. Tetrahedron Lett. 1997;38:7989–7992. [Google Scholar]
- 8.Hesk D, Gunnarsson I, Koharski D, McNamara P, Schwartz J L, Thonoor M, Wirth M. Synthesis of 3H and 14C-SCH 27899 by fermentation and evaluation of in vivo label stability. J Labeled Compd Radiopharm. 1999;42:159–167. [Google Scholar]
- 9.Jones R N, Barrett M S. Antimicrobial activity of SCH 27899, oligosaccharide member of the everninomicin class with a wide gram-positive spectrum. J Clin Microb Infect. 1995;1:35–43. doi: 10.1111/j.1469-0691.1995.tb00022.x. [DOI] [PubMed] [Google Scholar]
- 10.Lin A H, Murray R W, Vidmar T J, Marotti K R. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob Agents Chemother. 1997;41:2127–2131. doi: 10.1128/aac.41.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mahmood R, Compagnone-Post P, Khan S A. An in vitro coupled transcription-translation system from Staphylococcus aureus. Gene. 1991;106:29–34. doi: 10.1016/0378-1119(91)90562-p. [DOI] [PubMed] [Google Scholar]
- 12.Maimets T, Ustav M, Villems R. The role of protein L16 and its fragments in the peptidyltransferase activity of 50S ribosomal subunits. Eur J Biochem. 1983;135:127–130. doi: 10.1111/j.1432-1033.1983.tb07627.x. [DOI] [PubMed] [Google Scholar]
- 13.Maimets T, Remme J, Villems R. Ribosomal protein L16 binds to the 3′-end of transfer RNA. FEBS Lett. 1984;166:53–56. doi: 10.1016/0014-5793(84)80043-5. [DOI] [PubMed] [Google Scholar]
- 14.Nobel W C, Virani Z, Cree R G A. Co-transfer of vancomycin and other resistance genes from Enterococcus faecalis NCTC 12201 to Staphylococcus aureus. FEMS Microbiol Lett. 1992;72:195–198. doi: 10.1016/0378-1097(92)90528-v. [DOI] [PubMed] [Google Scholar]
- 15.Otaka T, Kaji A. Release of (oligo) peptidyl-tRNA from ribosomes by erythromycin A. Proc Natl Acad Sci USA. 1975;72:2649–2652. doi: 10.1073/pnas.72.7.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pezzuto J M, Hecht S M. Amino acid substitutions in protein biosynthesis. Poly(A)-directed polyphenylalanine synthesis. J Biol Chem. 1980;255:865–869. [PubMed] [Google Scholar]
- 17.Picking W D, Odom O W, Tsalkova T, Serdyuk I, Hardesty B. The conformation of nascent polylysine and polyphenylalanine peptides on ribosomes. J Biol Chem. 1991;266:1534–1542. [PubMed] [Google Scholar]
- 18.Pongs O, Bald R, Erdmann V A. Identification of chloramphenicol-binding protein in Escherichia coli ribosomes by affinity labeling. Proc Natl Acad Sci USA. 1973;70:2229–2233. doi: 10.1073/pnas.70.8.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulze H, Nierhaus K H. Minimal set of ribosomal components for reconstitution of the peptidyltransferase activity. EMBO J. 1982;1:609–613. doi: 10.1002/j.1460-2075.1982.tb01216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shinabarger D L, Marotti K R, Murray R W, Lin A H, Melchior E P, Swaney S M, Dunyak D S, Demyan W F, Buysee J M. Mechanism of action of oxazolidinones: effects of linezolid and eperezolid on translation reactions. Antimicrob Agents Chemother. 1997;41:2132–2136. doi: 10.1128/aac.41.10.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Spahn C M, Prescott C D. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Med. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 22.Staehlin T, Marglott D R. Preparation of Escherichia coli ribosomal subunits active in polypeptide synthesis. Methods Enzymol. 1971;20:449–455. [Google Scholar]
- 23.Tate W P, Schulze H, Nierhaus K H. The importance of the Escherichia coli ribosomal protein L16 for the reconstitution of the peptidyl-tRNA hydrolysis activity of peptide chain termination. J Biol Chem. 1983;258:12810–12815. [PubMed] [Google Scholar]
- 24.Wolf F. Avilamycin, an inhibitor of the 30S ribosomal subunit function. FEBS Lett. 1973;36:181–186. doi: 10.1016/0014-5793(73)80364-3. [DOI] [PubMed] [Google Scholar]
- 25.Wower J, Hixson S S, Zimmermann R A. Labeling the peptidyltransferase center of the Escherichia coli ribosome with photoreactive tRNA(Phe) derivatives containing azidoadenosine at the 3′ end of the acceptor arm: a model of the tRNA-ribosome complex. Proc Natl Acad Sci USA. 1989;86:5232–5236. doi: 10.1073/pnas.86.14.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]