Abstract
This study aimed to determine whether causative pathogens in mastitic milk can be determined by Gram staining after the centrifugation of milk. Gram staining was performed using unconcentrated and concentrated milk cells. Using this method, we found that the background of microscopic image of unconcentrated milk cells was complex and bacteria were difficult to detect. In contrast, the background of the smears in the concentrated milk cells was translucent, and bacterial and somatic cells were clearly visible. The sensitivity and specificity of the Gram staining of concentrated milk cells were 84.4% and 86.0% and 50.0% and 94.5% for the detection of gram-positive and gram-negative bacteria, respectively. The presented method provides a simple and inexpensive means of determining mastitis-causing pathogens.
Keywords: dairy cow, diagnosis, Gram staining, mastitis
Mastitis in dairy cows is an inflammation of the mammary glands caused by invading pathogens, which can result in substantial economic losses for dairy farmers [9, 10] and has food safety implication for dairy products [11].
Effective treatment of mastitis requires the early detection and identification of causal pathogens. A diverse ranges of pathogens can cause mastitis in dairy cows [12, 17, 23], including bacteria; fungi, such as yeasts; and algae, such as Prototheca [4, 14]. Of these, bacteria, both gram-positive and gram-negative, are the principal pathogenic agents [18]. The findings of recent studies have indicated that the selection of appropriate mastitis treatment strategies is dependent on the causative pathogens. For example, whereas the use of antimicrobial agents can be effective for bacteria, it would be ineffective for other microorganisms such as fungi and algae. However, when the causative pathogen is a gram-negative bacterium or cannot be cultured on agar media, antimicrobial agents should not be administered, except in severe cases [5, 6]. In contrast, when the causative pathogen is gram-positive, antimicrobial treatments are recommended for treatment in clinical cases [24]. Accordingly, the strategy adopted for the treatment of mastitis should be determined based on the type of causative bacteria, particularly in cases where the causal agents are either gram-positive or gram-negative.
Culture-based methods are generally recommended for identifying the causative pathogens of mastitis [1, 18]. In the case of human infections, microbiological diagnosis is conducted based on a combination of direct microscopic examination of collected samples (using Gram staining) and culturing methods. For example, sputum [16] and cerebrospinal fluid [25] are used for Gram staining in cases of community-acquired pneumonia and bacterial meningitis, respectively. There are a large variety of bacterial pathogens that can cause community-acquired pneumonia, including Streptococcus pneumoniae, Staphylococcus aureus, Haemophilus influenzae, Klebsiella pneumoniae, and Pseudomonas aeruginosa [7]. The Gram staining sputum for bacterial diagnosis can provide information for pathogen-directed antimicrobial usage at the commencement of treatment [16]. With respect to dairy cows, several guidelines and review articles on the diagnosis of mastitis recommend culture-based methods to determine pathogens [1, 3, 18]. Although the Gram staining of milk is a rarely considered approach in this regard, it has been suggested that the direct Gram staining of milk can be used to detect gram-positive bacteria such as Staphylococcus and Streptococcus [20], although not other pathogens, such as gram-negative bacteria. This limitation of Gram staining in enabling the detection of mastitic pathogens in milk is one of the reasons why this technique is rarely applied, even though it produces results considerably more rapidly than culture-based methods and facilitates pathogen-targeted treatment decisions.
Suzuki et al. [22] centrifuged milk to remove milk proteins and fats to obtain somatic cell counts (SCCs). Bacteria in milk are also precipitated and harvested when the milk is centrifuged [8]. Thus, if cell pellets after centrifugation are used, milk fats and proteins that disturb microscopy can be removed and bacterial concentration can be elevated, which results in a successful distinction between gram-positive and gram-negative bacteria. Accordingly, in this study, we assessed the utility of a centrifugation-based method for detecting and discriminating between gram-positive and gram-negative bacteria in bovine milk in cases of clinical mastitis.
For this study, we used milk samples collected from Holstein Friesian cows suffering from spontaneous clinical mastitis. Prior to collecting milk samples, the teats of cows were debrided and the initially obtained foremilk was discarded. Ten milliliters of each of the milk samples were collected by hand squeezing into sterilized tubes and immediately stored at 4°C until used for examination. All examinations were conducted within 24 hr of milk collection. This study was conducted in accordance with the guidelines for animal experiments issued by Hiroshima University (E19-3).
One of the 75 milk samples was used to compare Gram staining of milk with concentrated milk cells prepared by centrifugation with Gram staining of unconcentrated milk cells without centrifugation. To compare results of bacterial cultures with Gram staining of concentrated milk cells, all 75 milk samples were used.
Aliquots of the collected milk samples (10 μl each) were plated onto four different agar media. Chromogenic agar (CHROMagarTM Orientation, Kanto Chemical Co., Inc., Tokyo, Japan) was used for confirmation of colony morphology and to provide an indication of the type of bacteria; 5% sheep blood agar (Niisui Plate Sheep Blood Agar EX, Nissui Pharmaceutical Co., Ltd., Tokyo, Japan) was used to confirm colony morphology and hemolysis; deoxycholate-hydrogen sulfide-lactose agar (DHL) (Nissui Plate DHL Agar, Nissui Pharmaceutical Co., Ltd.) was used as a selective medium for Entarobacteriaceae; and Baird-Parker agar (Nissui Plate Baird-Parker agar, Nissui Pharmaceutical Co., Ltd.) was used as the selective medium for Staphylococcus spp., including S. aureus. Samples plated on all media were incubated under aerobic conditions at 37°C for 24 hr. Colonies with a single morphotype were observed on chromogenic agar, with Gram staining (0.2% Victoria blue solution for 1 min, 2% picric acid solution with methanol for 1 min, and 0.25% safranin solution for 1 min; Favor G, Nissui Pharmaceutical Co., Ltd.) being performed for a single colony collected from the agar. Catalase tests were performed on all isolates of gram-positive cocci. The latex slide agglutination test for S. aureus (DRY SPOT STAPHYTECT PLUS, Oxoid Ltd., Hampshire, UK) was used to detect catalase-positive isolates. Colonization on DHL and Baird-Parker agar was used to confirm gram-negative bacilli and catalase-positive gram-positive cocci, respectively. If colonies with differential morphotypes were observed on chromogenic agar, Gram staining was performed on a single colony of each type. Bacterial species were not identified in the present study.
To enhance the Gram staining of milk, we obtained concentrated preparations of the cells in mastic milk using the following method. Having plated samples onto agar, the remaining 10 ml of milk was centrifuged at 1,750 × g for 5 min, which separated the milk into three distinct layers, namely, a cell pellet, skimmed milk, and milk fat. The latter two fractions were discarded, and the remaining cell pellet was resuspended in 1 ml of sterilized saline solution (×10 concentrated). The concentrated milk cells were smeared on glass slides and fixed with 99.8% methanol for 2 min. Gram staining was performed as described previously. These slide preparation were examined by microscopy using a ×100 oil immersion objective lens. To compare Gram staining between milk cells concentrated by centrifugation and unconcentrated cells in milk that had not been centrifuged, we obtained a suspension of unconcentrated milk cells from a single sample of milk, which was smeared directly onto a glass slide and fixed with 99.8% methanol for 15 min. Gram staining for this sample was performed as described for the concentrated cell suspensions.
The sensitivity and specificity of Gram staining of concentrated milk cells for the detection of bacteria in mastitic milk were calculated. Achievement of bacterial detection was defined as the same bacterial color (gram-positive or gram-negative), and morphology was observed both in cell pellets and the colony from chromogenic agar. For gram-positive cocci, morphology (whether clustered or chained) was confirmed by a catalase test [26]. Sensitivity and specificity were calculated for the detection of gram-positive and gram-negative bacteria.
With regards to the chromogenic agar test, we observed colonies with a single morphotype on 38 (50.7%) plates and colonies of two morphotypes on seven (9.3%) plates (Table 1). For the remaining 30 milk samples (40.0%), no colonies grew on any of the four assessed media. Among the gram-positive bacteria detected, cocci, bacilli, and others (including fungal cells) were isolated from 25 (33.3%), 2 (3.0%), and 5 (7.0%) milk samples, respectively. All detected gram-negative bacteria, which were isolated from 20 (30.0%) milk samples, were confirmed to be bacilli.
Table 1. The results of bacterial culturing.
| Number | (%) | ||
|---|---|---|---|
| Gram positive | |||
| cocci | 25 | 33.3 | |
| bacilli | 2 | 3.0 | |
| others | 5 | 7.0 | |
| Gram negative | |||
| bacilli | 20 | 30.0 | |
| Not detected | 30 | 40.0 | |
| Total | 82 | ||
For seven milk samples, colonies with two morphotypes grew on the chromogenic agar.
Figure 1 shows microscopic images of gram-stained unconcentrated (Fig. 1a) and concentrated milk cells (Fig. 1b) from the same sample of mastitic milk. In the case of the unconcentrated cell preparation, the image background was complex and non-homogenous, and we were unable to detect the presence of bacteria (Fig. 1a). In contrast, the image background of the concentrated cell preparation was translucent, and bacteria and somatic cells were clearly detected (Fig. 1b). The morphology of these bacteria was found to be consistent with the Gram staining of isolates cultured from the same milk sample (Fig. 1c). Representative microscopic images of gram-stained concentrated milk cells are shown in Fig. 2. As shown in the figure, we observed clustered gram-positive cocci (Fig. 2a), chains of gram-positive cocci (Fig. 2b), gram-positive bacilli (Fig. 2c), gram-negative bacilli (Fig. 2d and 2e) and both clustered gram-positive cocci and gram-negative bacilli (Fig. 2f). We also attempted to detect pathogens directly from mastitic milk without intermediate culturing. Microscopic examination of bacteria in unconcentrated milk samples has long been reported [2]. Historically, methylene blue and Newman’s stain solutions have been used to stain bacteria and somatic cells in milk [15]. Shah et al. [20] reported Gram staining of heat-fixed smears of the unconcentrated milk of goats and sheep, although only gram-positive bacteria could be detected. In the present study, we found that the background of microscopic images of gram-stained unconcentrated milk cells from samples that had not been centrifuged was complex and non-homogeneous, thereby hampering the detection of bacteria. This unfavorable type of background could be attributed to the presence of certain milk constituents, including lipids and aggregated protein, which we found could be removed by centrifugation prior to Gram staining. Given that the method described herein, based on concentrated milk cells, can be used to detect both gram-positive and gram-negative pathogens, we considered that simple centrifugation was sufficient to clear milk lipids and proteins for Gram staining. Furthermore, we found that using a ten-fold concentration of bacteria enhanced our ability to identify bacteria, even if these had been phagocytosed by neutrophils.
Fig. 1.
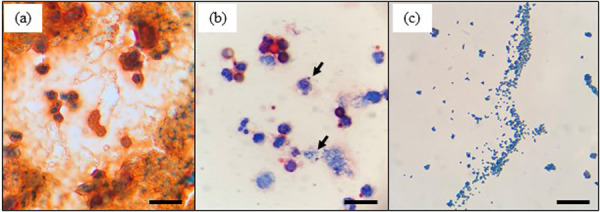
Microscopic images of gram-stained unconcentrated milk cells without centrifugation (a), concentrated milk cells with centrifugation (b), and isolates from the same mastitic milk sample (c). Arrows in (b) indicate the causal mastitis bacteria in milk. Scale bars=20 μm.
Fig. 2.
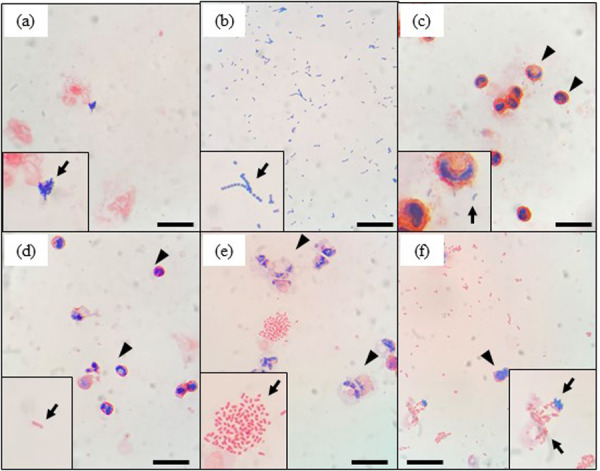
Microscopic images of gram-stained mastitic milk samples after centrifugation. For each image, insets show enlarged images of bacteria. The arrows in each image indicate a representative image of bacteria. Clustered gram-positive cocci (a and f), chains of gram-positive cocci (b), gram-positive bacilli (c), and gram-negative bacilli (d, e, and f) were observed. Arrowheads in d, e, and f indicate leukocytes in milk. Scale bars=20 μm.
In terms of the accuracy of Gram staining mastitic milk using the present method, we obtained sensitivity and specificity values of 84.4% and 86.0% for gram-positive bacteria and 50.0% and 94.5% for gram-negative bacteria, respectively. These values accordingly indicate that compared with gram-positive bacteria, the detection sensitivity for gram-negative bacteria was relatively low. This can be attributed to the fact that the background of slide preparations was pale red, which made it difficult to detect the red-stained gram-negative bacteria, whereas in contrast, the blue-stained gram-positive bacteria could be clearly distinguished. Another possible reason for this differential detection ability is that the numbers of gram-negative bacteria infecting our milk samples were relatively lower than those of gram-positive bacteria. Thus, future studies should examine the relationship between the amounts of different bacteria infecting mastitic milk and the accuracy of Gram staining. Comparatively, however, with respect to human medical care, a systematic review of the diagnostic accuracy of sputum Gram staining reported that the sensitivity and specificity for diagnosis of S. pneumoniae (gram-positive cocci) was 0.69 (95% credible interval, CrI; 0.56–0.80) and 0.91 (CrI, 0.83–0.96), respectively, whereas that for H. influenzae (gram-negative bacilli) was 0.76 (CrI, 0.60–0.87) and 0.97 (CrI, 0.91–0.99) [16]. Therefore, the accuracy of our Gram staining of concentrated milk cells in the present study is similar to that obtained for sputum Gram staining. In contrast, Fukuyama et al. [7] have reported that the sensitivity and specificity of sputum Gram staining were 9.1% and 100% for S. aureus (gram-positive cocci) and 39.5% and 98.2% for K. pneumoniae (gram-negative bacilli), respectively. Therefore, the sensitivity of Gram staining may differ even for bacteria of the same color and form (e.g., gram-positive cocci and gram-negative bacilli). Accordingly further studies are warranted to validate the efficacy of Gram staining for different species of mastitic pathogens in concentrated milk samples.
Our observations revealed the phagocytosis of gram-positive cocci (Fig. 3a) and gram-negative bacilli (Fig. 3b) by leukocytes present in milk samples. Consequently, Gram staining of concentrated milk cells may also have potential utility from the perspective immunopathological diagnosis, based on observations of leukocytes, particularly the proportions of polymorphonuclear neutrophils (PMNs). In this regard, a close relationship between the proportions of PMNs in milk and SCC have previously been reported [19]. Similarly, it has been found that milk with a low SCC and positive bacterial growth after culturing was characterized by a significantly higher population of PMNs compared with milk with a low SCC and negative bacterial growth after culturing [21]. These studies thus indicate that irrespective of SCC, there are potential differences in the immunological status of mammary glands. Consequently, if Gram staining of concentrated cells obtained from mastitic milk can be used to distinguish PMNs, this method could potentially facilitate both bacteriological and immunopathological diagnoses, thereby contributing to decisions on the use of both antimicrobial and anti-inflammatory drugs. Accordingly, the accuracy of PMN counts based on the Gram staining of concentrated milk cells warrants further examination.
Fig. 3.
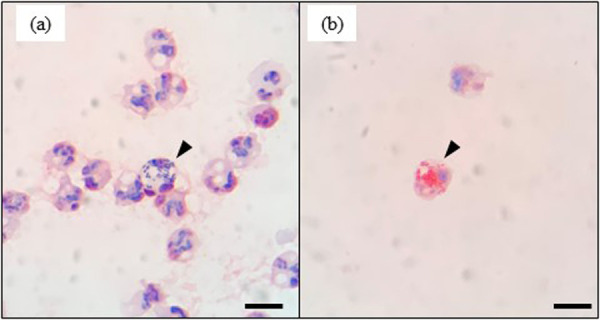
Microscopic images showing leukocyte phagocytosis of gram-positive cocci (a, arrowhead) and gram-negative bacilli (b, arrowhead). Scale bars=10 μm.
Although culture-based methods are widely recommended for detection of the bacteria causing mastitis [1, 18], it is typically necessary to culture samples for more than 24 hr to enable determinations. Consequently, more rapid means of detections would make a valuable contribution to the timely and effective management of mastitis [5, 13, 24]. In this regard, culture-independent methods, such as PCR-based methods, loop-mediated isothermal amplification [1], and fluorescent in situ hybridization [8], can facilitate rapid detection and identification of causative agents. In contrast, although Gram staining of concentrated milk cells can be used to detect bacteria, it does not enable an identification of species. However, this method can be used to discriminate gram-positive or gram-negative bacteria and enables examinations of bacterial morphology, which can provide valuable information for determining initial treatment strategies, as has been demonstrated in human clinical care.
In terms of veterinary clinical care for dairy cows, although the method presented herein can be readily performed in the laboratory, it would be impracticable in the case of farm visits. Nevertheless, results can be obtained within 1 hr from the start of the procedure, and could thereby provide veterinarians with the necessary information regarding initial treatment decisions in non-urgent cases. Moreover, the pre-treatment detection of fungal or algal agents using this method could contribute to the more prudent administration of antibacterial drugs, by reducing the use of antibacterial drugs in cases of fungal or algal mastitis. Furthermore, given its simplicity and low cost, this method can be used worldwide. However, to confirm the efficacy of this method, further studies are desirable to determine the accuracy and limits of detection for different bacterial species.
POTENTIAL CONFLICTS OF INTEREST
The authors declare that they have no conflicts of interest associated with this study.
REFERENCES
- 1.Ashraf A., Imran M.2018. Diagnosis of bovine mastitis: from laboratory to farm. Trop. Anim. Health Prod. 50: 1193–1202. doi: 10.1007/s11250-018-1629-0 [DOI] [PubMed] [Google Scholar]
- 2.Breed R. S., Brew J. D.1917. The control of public milk supplies by the use of the microscopic method. J. Dairy Sci. 1: 259–271. doi: 10.3168/jds.S0022-0302(17)94379-6 [DOI] [Google Scholar]
- 3.Britten A. M.2012. The role of diagnostic microbiology in mastitis control programs. Vet. Clin. North Am. Food Anim. Pract. 28: 187–202. doi: 10.1016/j.cvfa.2012.03.006 [DOI] [PubMed] [Google Scholar]
- 4.Farnsworth R. J., Sorensen D. K.1972. Prevalence and species distribution of yeast in mammary glands of dairy cows in Minnesota. Can. J. Comp. Med. 36: 329–332. [PMC free article] [PubMed] [Google Scholar]
- 5.Fuenzalida M. J., Ruegg P. L.2019. Negatively controlled, randomized clinical trial to evaluate intramammary treatment of nonsevere, gram-negative clinical mastitis. J. Dairy Sci. 102: 5438–5457. doi: 10.3168/jds.2018-16156 [DOI] [PubMed] [Google Scholar]
- 6.Fuenzalida M. J., Ruegg P. L.2019. Negatively controlled, randomized clinical trial to evaluate use of intramammary ceftiofur for treatment of nonsevere culture-negative clinical mastitis. J. Dairy Sci. 102: 3321–3338. doi: 10.3168/jds.2018-15497 [DOI] [PubMed] [Google Scholar]
- 7.Fukuyama H., Yamashiro S., Kinjo K., Tamaki H., Kishaba T.2014. Validation of sputum Gram stain for treatment of community-acquired pneumonia and healthcare-associated pneumonia: a prospective observational study. BMC Infect. Dis. 14: 534. doi: 10.1186/1471-2334-14-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gey A., Werckenthin C., Poppert S., Straubinger R. K.2013. Identification of pathogens in mastitis milk samples with fluorescent in situ hybridization. J. Vet. Diagn. Invest. 25: 386–394. doi: 10.1177/1040638713486113 [DOI] [PubMed] [Google Scholar]
- 9.Heikkilä A. M., Liski E., Pyörälä S., Taponen S.2018. Pathogen-specific production losses in bovine mastitis. J. Dairy Sci. 101: 9493–9504. doi: 10.3168/jds.2018-14824 [DOI] [PubMed] [Google Scholar]
- 10.Hogeveen H., Huijps K., Lam T. J.2011. Economic aspects of mastitis: new developments. N. Z. Vet. J. 59: 16–23. doi: 10.1080/00480169.2011.547165 [DOI] [PubMed] [Google Scholar]
- 11.Kümmel J., Stessl B., Gonano M., Walcher G., Bereuter O., Fricker M., Grunert T., Wagner M., Ehling-Schulz M.2016. Staphylococcus aureus entrance into the dairy chain: tracking S. aureus from dairy cow to cheese. Front. Microbiol. 7: 1603. doi: 10.3389/fmicb.2016.01603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makovec J. A., Ruegg P. L.2003. Results of milk samples submitted for microbiological examination in Wisconsin from 1994 to 2001. J. Dairy Sci. 86: 3466–3472. doi: 10.3168/jds.S0022-0302(03)73951-4 [DOI] [PubMed] [Google Scholar]
- 13.Milner P., Page K. L., Hillerton J. E.1997. The effects of early antibiotic treatment following diagnosis of mastitis detected by a change in the electrical conductivity of milk. J. Dairy Sci. 80: 859–863. doi: 10.3168/jds.S0022-0302(97)76008-9 [DOI] [PubMed] [Google Scholar]
- 14.Möller A., Truyen U., Roesler U.2007. Prototheca zopfii genotype 2: the causative agent of bovine protothecal mastitis? Vet. Microbiol. 120: 370–374. doi: 10.1016/j.vetmic.2006.10.039 [DOI] [PubMed] [Google Scholar]
- 15.Newman R. W.1927. A one-solution technique for the direct microscopic method of counting bacteria in milk. Proc. Soc. Exp. Biol. Med. 24: 323–325. doi: 10.3181/00379727-24-3351 [DOI] [Google Scholar]
- 16.Ogawa H., Kitsios G. D., Iwata M., Terasawa T.2020. Sputum Gram stain for bacterial pathogen diagnosis in community-acquired pneumonia: a systematic review and Bayesian meta-analysis of diagnostic accuracy and yield. Clin. Infect. Dis. 71: 499–513. doi: 10.1093/cid/ciz876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliveira L., Hulland C., Ruegg P. L.2013. Characterization of clinical mastitis occurring in cows on 50 large dairy herds in Wisconsin. J. Dairy Sci. 96: 7538–7549. doi: 10.3168/jds.2012-6078 [DOI] [PubMed] [Google Scholar]
- 18.Ruegg P. L.2017. A 100-year review: mastitis detection, management, and prevention. J. Dairy Sci. 100: 10381–10397. doi: 10.3168/jds.2017-13023 [DOI] [PubMed] [Google Scholar]
- 19.Schwarz D., Diesterbeck U. S., König S., Brügemann K., Schlez K., Zschöck M., Wolter W., Czerny C. P.2011. Flow cytometric differential cell counts in milk for the evaluation of inflammatory reactions in clinically healthy and subclinically infected bovine mammary glands. J. Dairy Sci. 94: 5033–5044. doi: 10.3168/jds.2011-4348 [DOI] [PubMed] [Google Scholar]
- 20.Shah A., Darzi M. M., Kamil S. A., Mir M. S., Maqbool R., Ali R., Kashani B., Wani H., Bashir A., Dar A. A., Qureshi S.2017. Somatic cell alteration in healthy and mastitic milk of sheep and goats. J. Entomol. Zool. Stud. 5: 27–33. [Google Scholar]
- 21.Souza F. N., Blagitz M. G., Batista C. F., Takano P. V., Gargano R. G., Diniz S. A., Silva M. X., Ferronatto J. A., Santos K. R., Heinemann M. B., De Vliegher S., Della Libera A. M. M. P.2020. Immune response in nonspecific mastitis: What can it tell us? J. Dairy Sci. 103: 5376–5386. doi: 10.3168/jds.2019-17022 [DOI] [PubMed] [Google Scholar]
- 22.Suzuki N., Yuliza Purba F., Hayashi Y., Nii T., Yoshimura Y., Isobe N.2020. Seasonal variations in the concentration of antimicrobial components in milk of dairy cows. Anim. Sci. J. 91: e13427. doi: 10.1111/asj.13427 [DOI] [PubMed] [Google Scholar]
- 23.Thomas V., de Jong A., Moyaert H., Simjee S., El Garch F., Morrissey I., Marion H., Vallé M.2015. Antimicrobial susceptibility monitoring of mastitis pathogens isolated from acute cases of clinical mastitis in dairy cows across Europe: VetPath results. Int. J. Antimicrob. Agents 46: 13–20. doi: 10.1016/j.ijantimicag.2015.03.013 [DOI] [PubMed] [Google Scholar]
- 24.Tomazi T., Sumnicht M., Tomazi A. C. C. H., Silva J. C. C., Bringhenti L., Duarte L. M., Silva M. M. M., Rodrigues M. X., Bicalho R. C.2021. Negatively controlled, randomized clinical trial comparing different antimicrobial interventions for treatment of clinical mastitis caused by gram-positive pathogens. J. Dairy Sci. 104: 3364–3385. doi: 10.3168/jds.2020-18830 [DOI] [PubMed] [Google Scholar]
- 25.Tunkel A. R., Hartman B. J., Kaplan S. L., Kaufman B. A., Roos K. L., Scheld W. M., Whitley R. J.2004. Practice guidelines for the management of bacterial meningitis. Clin. Infect. Dis. 39: 1267–1284. doi: 10.1086/425368 [DOI] [PubMed] [Google Scholar]
- 26.Wald R., Baumgartner M., Urbantke V., Stessl B., Wittek T.2017. Diagnostic accuracy of a standardized scheme for identification of Streptococcus uberis in quarter milk samples: a comparison between conventional bacteriological examination, modified Rambach agar medium culturing, and 16S rRNA gene sequencing. J. Dairy Sci. 100: 1459–1466. doi: 10.3168/jds.2016-11786 [DOI] [PubMed] [Google Scholar]


