Abstract
Lake Sinai virus (LSV), an RNA virus, is suspected to be associated with poor health in honeybees (Apis mellifera). We examined LSV in 26 specimens of healthy honeybees and 44 specimens of wild arthropods in the Gifu Prefecture, Japan. LSV was found more frequently in honeybee specimens (11/26, 42.3%) than in wild arthropod specimens (1/44, 2.3%) (P<0.01). Phylogenetic and nucleotide sequence analysis revealed two lineages: LSV3 in honeybees, and LSV4 in both honeybees and wild arthropods. To our knowledge, this is the first report of LSV prevalence in honeybees and wild arthropods in Japan.
Keywords: arthropod, Apis mellifera, Gifu, Japan, Lake Sinai virus
Lake Sinai virus (LSV) is a positive-sense, single-stranded RNA virus that belongs to the Sinaivirus genus [2, 17]. LSV was first detected in reared western honeybees (Apis mellifera) during a survey following a massive loss of honeybee colonies between 2007 and 2008 in the United States of America (USA) [16]. Since its initial detection, LSV has been found in several additional countries including Belgium [13], Germany [4], Australia [15], and Slovenia [17]. According to phylogenetic clustering, LSV has been assigned to eight lineages to date: LSV1, LSV2, LSV3, LSV6, and LSV7 are distributed in the USA [3, 5, 16], LSV4 and LSV5 are distributed in Belgium [13], and LSV8 is distributed in Australia [15]. LSV has also been detected in hornets [18], bumble bees [6, 17], and ants [1]. In addition, the Varroa destructor mite, an ectoparasite of honeybees, is known as a vector for LSV [5, 14]. The pathogenicity of LSV in honeybees and wild arthropods is not well understood, although previous studies suggest that the prevalence of LSV in honeybees is related to the health of the colony [5, 7]. Additionally, although more than 30 species of honeybee viruses have been reported [2], there has been limited investigation of the species of viruses in honeybees in Japan [8,9,10]. In the present study, we conducted a field survey in the Gifu Prefecture of Japan to clarify the prevalence of LSV in honeybees and wild arthropods in the area.
All samples used in this study were collected for previous studies conducted in Gifu in 2018 and 2019 [8, 9, 12]. Twenty-six groups of honeybees (five pooled worker bees per group) were sampled from 26 clinically healthy apiaries [9]. Additionally, a total of 44 wild arthropods were collected and examined for LSV, including 28 specimens derived from three apiaries [8] and 16 specimens collected from non-apiary sources [12]. The wild arthropod species were identified at the order level using insect explorer (Japanese website available at https://insects.jp/) according to the methods used in our previous study [12] (Supplementary Table 1). This study was approved by the Ethics Committee for Animal Research and Welfare of Gifu University (approval numbers H30-009 and 17055). PCR detection of LSV was performed using the primer set (LSV1765-F and LSV2368-R), as previously described by Ravoet et al. [14]. In short, RNA was extracted using a NucleoSpin® RNA (Macherey Nagel, Düren, Germany) or QIAamp Viral RNA mini kit (QIAGEN, Venlo, Netherlands) according to the manufacturers’ protocol. Generation of cDNA from total RNA (5 μl) was performed with a random hexamer primer using a Prime Script™ RT-PCR Kit (TaKaRa Bio Inc., Kusatsu, Japan) or a Prime Script 1st strand cDNA Synthesis Kit (TaKaRa Bio Inc.) according to the manufacturers’ protocols (see Supplementary Materials 1 for the RT procedures). The PCR was conducted using TaKaRa Ex Taq (TaKaRa Bio Inc.) with 1 μl of the reverse transcription products and the primer set (see Supplementary Materials 1 for the PCR procedures).
LSV was detected in 11 of the 26 honeybee specimens (42.3%) and one of 44 wild arthropods (2.3%), the Japanese giant hornet (Vespa mandarinia) captured in apiaries (Table 1). In previous studies, LSV was observed among 41.9% and 27.0–45.0% of clinically healthy honeybees in Germany and Australia, respectively [4, 15]. The LSV prevalence of healthy honeybees in Gifu was similar to or higher than those in these countries. On the contrary, in Slovenia, a high prevalence of LSV (75.9%) was observed among honeybees in poor health [17]. As honeybee samples used in this study were collected from healthy colonies including reproductive performance in limited area in Gifu [9], further study using honeybee samples collected from honeybee colonies in varying states of health and with varying reproductive rates around Japan is needed. To our knowledge, this is the first report of LSV in honeybees and arthropods in Japan.
Table 1. Prevalence of Lake Sinai virus (LSV) in honeybees (Apis mellifera) and wild arthropods collected in the Gifu Prefecture, Japan.
| Sample | Sample site | n | LSV prevalence (%) |
||||
|---|---|---|---|---|---|---|---|
| Total | LSV1 | LSV2 | LSV3 | LSV4 | |||
| Honeybees (Apis mellifera) | 26 | 11 (42.3%) | 0 (0%) | 0 (0%) | 5 (19.2%) | 6 (23.1%) | |
| Total wild arthropods sampled | 44 | 1 (2.3%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (2.3%) | |
| Apiary | 28 | 1 (3.6%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (3.6%) | |
| Non-apiary | 16 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
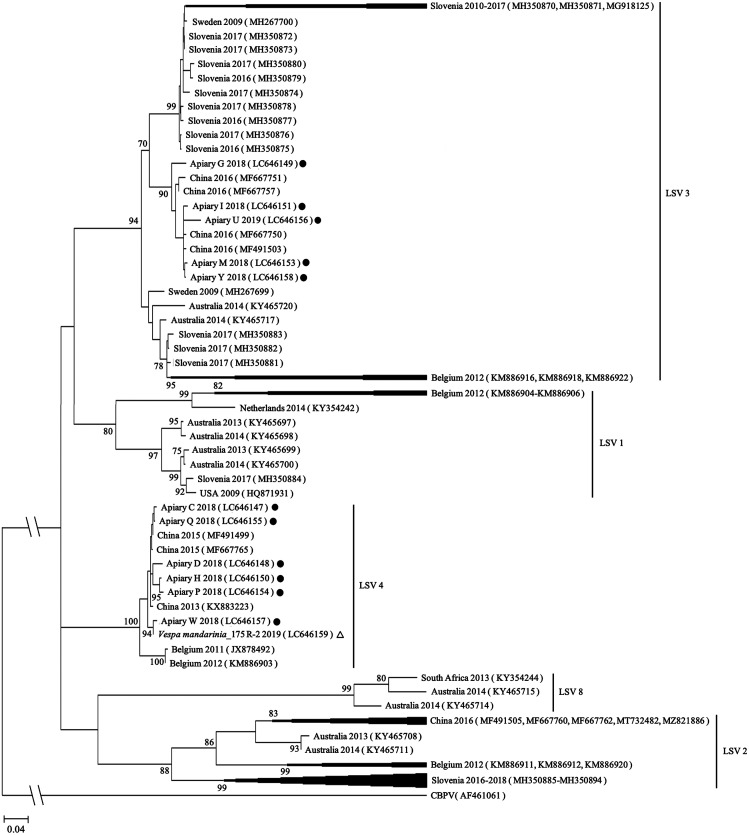
To elucidate the relationship between LSV isolates from Gifu and those from other countries, phylogenetic clustering of LSV was performed. The PCR products obtained from 12 samples were purified using Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) and then analyzed using Sanger sequencing at the Life Sciences Research Center, Gifu University. The datasets obtained in this study are available from the GenBank repository under the following accession numbers: LC646147–LC646151 and LC646153–LC646159. The nucleotide sequences obtained were subjected to searches for similar sequences using the Basic Local Alignment Search Tool (BLASTn) on the National Center for Biotechnology Information Search website (https://blast.ncbi.nlm.nih.gov/) with the following options: Database: standard databases with nucleotide collection (nr/nt); program selection: Megablast (accessed July 2021). The genome sequences were aligned with the Muscle tool using Molecular Evolutionary Genetics Analysis (MEGA)-X [11] (see Supplementary Materials 2 for the multiple alignment) and then the phylogenetic trees were constructed for the 540-bp partial RNA-dependent RNA polymerase (RdRp) gene using MEGA-X [11]. The phylogeny only included sequences from LSV1, 2, 3, 4, and 8 as sequences from other lineages were not available in this genetic region. The tree was constructed using maximum likelihood under the Kimura 2-parameter with a discrete gamma distribution model (K2+G). Bootstrap confidence limits were determined from 1,000 replications. Nodes supported by bootstrap values ≥70 are shown. The chronic bee paralysis virus (CBPV) (AF461061) sequence was used as an outgroup. The reference sequences are shown in Supplementary Table 2.
LSV detected in 11 honeybee specimens was classified as LSV3 (5, 19.2%) and LSV4 (6, 23.1%) (Table 1). LSV4 has been reported in Belgium [13] and LSV3 has been detected in various European countries including Slovenia [17], from which queen bees are sourced to Japan every year. However, the LSV detected in Gifu clustered with lineages from China rather than those from European countries (Fig. 1), suggesting that the lineage in Gifu is most closely related to lineages from China. In our previous study [9], black queen cell virus (BQCV) and deformed wing virus (DWV) distributed in Gifu were also related to China. Therefore, LSV may have entered Japan via same route as BQCV and DWV. Japan imports apicultural materials, including pollen food, from China. It is important to clarify the route by which honeybee viruses, including LSV, entered Japan.
Fig. 1.
Phylogenetic tree of Lake Sinai virus (LSV) based on partial nucleotide sequences of the RNA-dependent RNA polymerase (RdRp) gene (540-bp). The current sequences are shown along with the apiary name and isolation year, with solid circles and open triangles showing sequences from honeybees and wild arthropods, respectively.
LSV4 was detected in a wild arthropod (Table 1). The LSV4 from the Japanese giant hornet had a high sequence similarity (96.3–99.1%) to that of the LSV4 isolated in honeybees. In addition, LSV was more often detected in honeybees (11/26, 42.3%) than in wild arthropods (1/44, 2.3%) (P<0.01, Fisher’s exact test) (Table 1). No arthropod specimens collected from non-apiary sources were positive for LSV. Although LSV was not found in honeybees in the apiary where LSV-positive hornet was captured (data not shown), the LSV detected in the hornet may have been associated with LSV derived from honeybees. Our results suggest that honeybees are more prominent than wild arthropods as LSV hosts in Japan.
The LSV prevalence in Japanese giant hornets was 9.1% (1/11) (Supplementary Table 1). In our previous study [8], Israeli acute paralysis virus (IAPV) and DWV were detected in 72.7% (8/11) and 27.3% (3/11) of Japanese giant hornets, respectively. In addition, in our previous study, Japanese yellow hornets (Vespa simillima xanthoptera) were positive for IAPV (12/12, 100%) and DWV (10/12, 83.3%) [8], but LSV was not observed in these wild arthropods in the present study. In the previous study conducted in China and France [18], IAPV and DWV were detected in adult hornets, whereas LSV was not found. In the USA, Dolezal et al. [6] reported that DWV (53.0%) and IAPV (20.0%) were detected more frequently than LSV (12.0%) in 148 wild non-honeybees including Andrena, Apidae, Colletidae, Halictidae, and Megachilidae. Therefore, these results suggest that wild bees may be less susceptible to LSV than to IAPV and DWV.
In conclusion, the present study constitutes the first detection of LSV in honeybees and wild arthropods in Japan. However, as this study was performed only on healthy honeybees, the impact of LSV on honeybees in Japan remains unclear. Therefore, to clarify the actual pathogenicity and clinical signs of LSV, further field surveys and laboratory experiments are both essential.
CONFLICT OF INTEREST
The authors declare no conflicts of interest associated with this manuscript.
Supplementary
Acknowledgments
We would like to thank the beekeepers in Gifu Prefecture for supplying the honeybee specimens. This work was supported by the Program for the Development of Young Researchers from the Discretionary Budget of the Dean of the United Graduate School of Veterinary Sciences, Gifu University.
REFERENCES
- 1.Bigot D., Dalmon A., Roy B., Hou C., Germain M., Romary M., Deng S., Diao Q., Weinert L. A., Cook J. M., Herniou E. A., Gayral P.2017. The discovery of Halictivirus resolves the Sinaivirus phylogeny. J. Gen. Virol. 98: 2864–2875. doi: 10.1099/jgv.0.000957 [DOI] [PubMed] [Google Scholar]
- 2.Beaurepaire A., Piot N., Doublet V., Antuñez K., Campbell E., Chantawannakul P., Chejanovsky N., Gajda A., Heerman M., Panziera D., Smagghe G., Yañez O., de Miranda J. R., Dalmon A.2020. Diversity and global distribution of viruses of the western honey bee, Apis mellifera. Insects 11: 239. doi: 10.3390/insects11040239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cornman R. S., Tarpy D. R., Chen Y., Jeffreys L., Lopez D., Pettis J. S., vanEngelsdorp D., Evans J. D.2012. Pathogen webs in collapsing honey bee colonies. PLoS One 7: e43562. doi: 10.1371/journal.pone.0043562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D’Alvise P., Seeburger V., Gihring K., Kieboom M., Hasselmann M.2019. Seasonal dynamics and co-occurrence patterns of honey bee pathogens revealed by high-throughput RT-qPCR analysis. Ecol. Evol. 9: 10241–10252. doi: 10.1002/ece3.5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daughenbaugh K. F., Martin M., Brutscher L. M., Cavigli I., Garcia E., Lavin M., Flenniken M. L.2015. Honey bee infecting Lake Sinai viruses. Viruses 7: 3285–3309. doi: 10.3390/v7062772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dolezal A. G., Hendrix S. D., Scavo N. A., Carrillo-Tripp J., Harris M. A., Wheelock M. J., O’Neal M. E., Toth A. L.2016. Honey bee viruses in wild bees: viral prevalence, loads, and experimental inoculation. PLoS One 11: e0166190. doi: 10.1371/journal.pone.0166190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faurot-Daniels C., Glenny W., Daughenbaugh K. F., McMenamin A. J., Burkle L. A., Flenniken M. L.2020. Longitudinal monitoring of honey bee colonies reveals dynamic nature of virus abundance and indicates a negative impact of Lake Sinai virus 2 on colony health. PLoS One 15: e0237544. doi: 10.1371/journal.pone.0237544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kitamura Y., Asai T.2021. Detection of bee virus species from wild arthropods in apiaries in Gifu Prefecture. Nippon Juishikai Zasshi 74: 427–431. [Google Scholar]
- 9.Kitamura Y., Odoi J. O., Nagai M., Asai T.2021. Prevalence of honeybee viruses in Apis mellifera in Gifu prefecture of Japan. J. Vet. Med. Sci. 83: 1948–1951. doi: 10.1292/jvms.21-0379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kojima Y., Toki T., Morimoto T., Yoshiyama M., Kimura K., Kadowaki T.2011. Infestation of Japanese native honey bees by tracheal mite and virus from non-native European honey bees in Japan. Microb. Ecol. 62: 895–906. doi: 10.1007/s00248-011-9947-z [DOI] [PubMed] [Google Scholar]
- 11.Kumar S., Stecher G., Li M., Knyaz C., Tamura K.2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. doi: 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Odoi J. O., Yamamoto M., Sugiyama M., Asai T.2021. Antimicrobial resistance in Enterobacteriaceae isolated from arthropods in Gifu City, Japan. Microbiol. Immunol. 65: 136–141. doi: 10.1111/1348-0421.12878 [DOI] [PubMed] [Google Scholar]
- 13.Ravoet J., Maharramov J., Meeus I., De Smet L., Wenseleers T., Smagghe G., de Graaf D. C.2013. Comprehensive bee pathogen screening in Belgium reveals Crithidia mellificae as a new contributory factor to winter mortality. PLoS One 8: e72443. doi: 10.1371/journal.pone.0072443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ravoet J., De Smet L., Wenseleers T., de Graaf D. C.2015. Genome sequence heterogeneity of Lake Sinai Virus found in honey bees and Orf1/RdRP-based polymorphisms in a single host. Virus Res. 201: 67–72. doi: 10.1016/j.virusres.2015.02.019 [DOI] [PubMed] [Google Scholar]
- 15.Roberts J. M. K., Anderson D. L., Durr P. A.2017. Absence of deformed wing virus and Varroa destructor in Australia provides unique perspectives on honeybee viral landscapes and colony losses. Sci. Rep. 7: 6925. doi: 10.1038/s41598-017-07290-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Runckel C., Flenniken M. L., Engel J. C., Ruby J. G., Ganem D., Andino R., DeRisi J. L.2011. Temporal analysis of the honey bee microbiome reveals four novel viruses and seasonal prevalence of known viruses, Nosema, and Crithidia. PLoS One 6: e20656. doi: 10.1371/journal.pone.0020656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Šimenc L., Kuhar U., Jamnikar-Ciglenečki U., Toplak I.2020. First complete genome of Lake Sinai Virus lineage 3 and genetic diversity of Lake Sinai Virus strains from honey bees and bumble bees. J. Econ. Entomol. 113: 1055–1061. doi: 10.1093/jee/toaa049 [DOI] [PubMed] [Google Scholar]
- 18.Yang S., Gayral P., Zhao H., Wu Y., Jiang X., Wu Y., Bigot D., Wang X., Yang D., Herniou E. A., Deng S., Li F., Diao Q., Darrouzet E., Hou C.2019. Occurrence and molecular phylogeny of honey bee viruses in Vespids. Viruses 12: 6. doi: 10.3390/v12010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.