Summary
Background
Establishing objective and quantitative neuroimaging biomarkers at individual level can assist in early and accurate diagnosis of major depressive disorder (MDD). However, most previous studies using machine learning to identify MDD were based on small sample size and did not account for the brain connectome that is associated with the pathophysiology of MDD. Here, we addressed these limitations by applying graph convolutional network (GCN) in a large multi-site MDD dataset.
Methods
Resting-state functional MRI scans of 1586 participants (821 MDD vs. 765 controls) across 16 sites of Rest-meta-MDD consortium were collected. GCN model was trained with individual whole-brain functional network to identify MDD patients from controls, characterize the most salient regions contributing to classification, and explore the relationship between topological characteristics of salient regions and clinical measures.
Findings
GCN achieved an accuracy of 81·5% (95%CI: 80·5–82·5%, AUC: 0·865), which was higher than other common machine learning classifiers. The most salient regions contributing to classification were primarily identified within the default mode, fronto-parietal, and cingulo-opercular networks. Nodal topologies of the left inferior parietal lobule and left dorsolateral prefrontal cortex were associated with depressive severity and illness duration, respectively.
Interpretation
These findings based on a large, multi-site dataset support the feasibility and effectiveness of GCN in characterizing MDD, and also illustrate the potential utility of GCN for enhancing understanding of the neurobiology of MDD by detecting clinically-relevant disruption in functional network topology.
Funding
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81621003, 82027808, 81820108018).
Keywords: Deep learning, Graph theory, Magnetic resonance imaging, Graph neural network, Depression, Multi-site
Research in context.
Evidence before this study
We performed a comprehensive literature search in PubMed with the terms (depression” OR “depressive” OR “depressed” OR “unipolar” OR “MDD”) AND (“fMRI” OR “resting-state”) AND ("machine learning" OR "multivariate pattern recognition" OR "classification" OR “deep learning”) for machine learning studies using resting-state functional MRI to distinguish patients with MDD from controls. Substantial previous machine learning studies were performed using single-site dataset with small sample size, which leads to huge variability and poor generability in model performance. One multi-site study reported classification accuracy of 70% in identifying MDD patients. However, a relatively basic model (logistic regression) was used which may not fully capture the underlying complex pattern in brain data. Considering the neuropathophysiology of MDD is highly associated with brain network disruptions, applying machine learning models based on brain networks may reach a better performance.
Added value of this study
GCN is one of the most popular graph-based deep learning models, which can model graph data structure like networks. In this study, we attempted to examine reliable and objective performance of GCN on MDD characterization based on a multi-site dataset with over 1500 participants. The sample size of our study is larger than previous machine learning publications, enabling us to obtain robust and generalizable classification performance for MDD. Based on our findings, we found GCN could achieve better classification performance compared with other common classifiers, and this observed performance can remain stable in multiple validations. Moreover, our study identified the most salient network patterns contributing to classification, as well as network topological deficits related to depression severity and illness duration, further suggesting the neurobiological and clinical underpinnings of GCN modeling.
Implications of all the available evidence
GCN may serve as a powerful deep learning model, which is capable of achieving accurate characterization of MDD across multiple imaging sites, detecting reliable and generalizable regional topological deficits related to clinical measures at individual level, and thus enhancing the understanding of pathophysiology of MDD.
Alt-text: Unlabelled box
Introduction
Major depressive disorder (MDD) is one of the most common causes of disability worldwide.1 To date, the clinical diagnosis of MDD primarily relies on symptomatic and behavioral assessments. However, affected individuals with MDD manifest a wide range of heterogeneous symptoms, which typically lead to inaccurate and delayed diagnosis. In this context, establishing objective and quantitative biomarkers for the identification of MDD may not only provide insight into illness pathophysiology but also lead to the development of biologically-informed tests for clinical diagnosis and treatment planning.
One promising biomarker is derived from resting-state functional magnetic resonance imaging (rsfMRI) techniques that provide quantitative assessment of disrupted brain function in psychiatric disorders.2,54 Substantial rsfMRI-based evidence has demonstrated that patients with MDD exhibit abnormal brain function in multiple cortical and subcortical regions, such as prefrontal cortex, insula, amygdala, precuneus, and hippocampus.3, 4, 5, 6, 7 The implementation of machine learning further accelerates the transition of neuroimaging biomarker analysis from traditional population-level inferences to predictions about individual patients that may advance individualized clinical decisions.53 As shown in Table S1, we performed a comprehensive literature review of machine learning studies using rsfMRI measures to distinguish MDD patients and controls. We noted that while there has been an increasing number of publications, the results were inconsistent with their reported classification accuracies varied from 61·7% to 98·4%.
This dramatic divergence in classification accuracies can be primarily ascribed to the demographic and clinical heterogeneity across MDD studies that mostly trained models with small datasets.8 As the optimization of machine learning models typically requires adequate training data to mount generalizability across different samples, large sample size is critical to ensure population-representative model performance and provide reliable information on the biological underpinnings. Kambeitz et al. reported that rsfMRI can accurately identify depressed individuals with a robust and consistent accuracy of 84% based on meta-analytic integration.9 Nevertheless, the small sample size of each included study can still result in a high risk of systematic overestimation.10 In recent years, the rapid development of multi-site collaborations has increased the availability of large-scale datasets, which is critical for training a reliable and generalizable model. One multi-site machine learning study has demonstrated approximately 70% classification accuracy for MDD using rsfMRI metrics.11 This study, however, employed a simple model (i.e., logistic regression (LR)) which may not capture complex multivariate patterns within the brain data. Whether better performance in multi-site settings can be achieved with advanced models which capture these patterns remains to be explored.
Model configuration needs to take into account that the human brain is a highly interconnected network, and the emergence of psychiatric illness is generally thought to be underpinned by a disruption of normal functional integration amongst cortical and subcortical regions.12 Numerous studies have suggested that MDD cannot be explained in terms of localized dysfunction within specific brain areas and is better understood as a disruption of the brain connectome.13, 14, 15 However, most traditional models adopted previously captured information based on independent regional functional measures instead of modeling functional connectome. As the graph structure conveys the representation of brain connectomes and captures the brain network topology, the graph convolution network (GCN) modeling, which allows for direct convolution over graphs, may be the optimal model to collect more subtle network-level information. Unlike traditional convolutional neural networks limited to Euclidean inputs (e.g., 2D/3D images), GCN can work on non-Euclidean domains and implement convolutions on graphs by exploiting the input node features and neighborhood structure between nodes to generate new feature maps.16 While a few studies have validated the successful application of GCN in the identification of autism spectrum disorder,17 Alzheimer's disease,18 and attention deficit hyperactivity disorder,19 this approach has not been widely used in MDD. In addition, as a deep learning model, GCN can be expected to perform better with the large multi-center dataset, given the strong learning capability of deep learning under big data.20
In this study, we aimed to apply advanced graph-based deep learning techniques to characterize individuals with MDD in a large dataset of 1586 participants across 16 sites. Specifically, we trained a GCN model based on whole-brain functional connectivity networks to characterize MDD patients as well as MDD subtypes (i.e., first-episode drug-naïve (FEDN) and recurrent patients). The most salient brain regions contributing to the classification were identified, and the relationship between their network topological metrics and clinical measures was further explored. Our first hypothesis was that the application of GCN on brain functional networks would allow nominally higher classification performance than other common classifiers including support vector machine (SVM), multilayer perception (MLP), LR, random forests (RF), and BrainNetCNN. In addition, graph-based classifiers can learn discriminative patterns via neighborhood structures within the network, which enabled the identification of spatially segregated salient regions at the network level. Thus, our second hypothesis was that GCN could reveal regional saliency map showing network topological deficits that might be related to clinical variables.
Methods
Participants
Our study was performed based on 25 datasets from 17 hospitals in the Rest-meta-MDD consortium that included 1300 MDD patients and 1128 healthy controls (HC). Demographic and clinical information including age, sex, illness duration, medication status, episode status (i.e., first episode/recurrent), and 17-item Hamilton Depression Rating Scale (HAMD) were collected at each site. In this study, we excluded data with poor quality following standard quality control procedures in the Rest-meta-MDD consortium (Supplemental Information and Fig. S1).21 Finally, 1586 participants (821 MDD patients vs. 765 HC) were included in our analysis. According to available clinical data of included patients, 410 were first-episode patients, 208 were recurrent patients, 308 patients were drug-naïve, and 219 patients were treated before. Detailed demographic and clinical characteristics of the study population are shown in Table 1 (for information in each site, see Table S2).
Table 1.
Demographic and clinical characteristics of included multi-site participants.
| Variable | MDD | HC | p valuea |
|---|---|---|---|
| Sample size, N | 821 | 765 | - |
| Age, years, mean ± SD | 34·36 ± 11·55 | 34·50 ± 13·16 | 0·815 |
| Sex, N female (%) | 522 (63·6%) | 453 (59·2%) | 0·074 |
| Episode status | |||
| First-episode, N (%) | 410 (66·3%) | - | - |
| Recurrent, N (%) | 208 (25·3%) | - | - |
| Unknown, N (%) | 203 (24·7%) | - | - |
| Medication status | |||
| Medication-naïve, N (%) | 308 (37·5%) | - | - |
| Treated, N (%) | 219 (26·7%) | - | - |
| Unknown, N (%) | 294 (35·8%) | - | - |
| Duration of illnessb, months, mean ± SD | 38·81 ± 60·83 | - | - |
| HAMDc, mean ± SD | 21·26 ± 6·61 | ||
| HAMAd, mean ± SD | 18·99 ± 9·14 | - | - |
Abbreviations: MDD, major depressive disorder; HC, healthy controls; HAMD, Hamilton Depression scale; HAMA, Hamilton Anxiety scale.
P value were calculated using Two-sample t-test (age) and Pearson Chi-Square test (sex).
Data were available for 691 of 821 participants.
Data were available for 738 of 821 participants.
Data were available for 535 of 821 participants.
Image acquisition and processing
Resting-state functional MRI and three-dimensional structural T1-weighted MRI images were acquired from all participants at each local site. Acquisition parameters from all sites are presented in Table S3. A unified image preprocessing protocol was performed using the DPARSF toolbox.22 Preprocessing steps primarily included slice timing correction, head motion correction, normalization, and removal of confounds as described in detail previously.21 We parceled the whole brain into 160 regions of interests (ROI) according to Dosenbach's atlas.23 Time series of BOLD signals from voxels in each ROI were extracted and averaged. Functional connectivity between each pair of ROIs was evaluated using the Pearson correlation coefficient of the corresponding time series. Fisher's z-transformation was then applied to the correlation estimates yielding a 160 × 160 functional connectivity matrix for each participant.
Controlling for nuisance variables
A major challenge of training a neuroimaging-based classifier is the multiple nuisance variables that are unrelated to the diagnostic labels. In the multi-site settings, these nuisance variables are generally derived from inter-subject demographic heterogeneity that is known to affect imaging data, such as age and sex,23,24 as well as inter-site confounding effects including different scanners,25 acquisition parameters, and instructions to participants which are difficult to be removed even by a unified image preprocessing pipeline. The existence of these nuisance variables can make machine learning algorithms erroneously identify the pattern not related to neuropathological effects of interests and thus impair the model performance.26 To correct for unwanted nuisance variables, we firstly regressed out the inter-subject variance of age and sex effects by using a non-linear Gaussian process model as described in previous studies.27,28 For the site-varying effects, we applied ComBat harmonization. ComBat harmonization is essentially a model based on multivariate mixed linear regression, which was originally developed to adjust batch effects in genomic studies.29 This method is effective in removing site-related effects in different modalities from multi-site neuroimaging data.30, 31, 32 Compared with traditional harmonization methods, Combat uses Empirical Bayes to improve the parametric estimation of biological and site-varying effects, avoiding overcorrection on important biological variance during the correction on site-varying effects.33 To avoid information leakage which might inflate the classification performance, we estimated the parameters in above nuisance control procedures only using the training dataset and subsequently applied the model to the dataset.
Classification based on graph convolutional network
The pipeline of GCN model is shown in Figure 1. Individual whole-brain functional connectivity matrix was first represented as graph structure , where and are sets of nodes and edges, respectively, and is the weighted adjacency matrix. Nodes were defined as the 160 atlas-based brain regions, and node features reflected vector of nodal functional connectivity. To determine edges, we applied a k-nearest neighbors (KNN) algorithm to connect each node and its neighbors. Such graph modelling in GCN is consistent with previous publication reporting successful GCN application,34 which can fully capture information from connectivity and avoid introducing additional information beyond brain networks in classification.
Figure 1.
The overall pipeline of GCN classifier distinguishing between individuals with MDD and HC. (a) Constructing graph structure for each participant using whole-brain resting-state functional connectivity. (b) The architecture and implementation of GCN. Abbreviations: GCN, graph convolutional network; ReLu, Rectified Linear Unit; GAP, global average pooling; MDD, major depressive disorder; HC, healthy controls.
The core process of GCN model is the spectral graph convolution filter, which can implement the convolution operation on irregular graph data instead of typical Euclidean data (for detailed mathematical theory and formulation, see Supplemental Information). Similar to conventional CNN, GCN includes input layer, graph convolutional hidden layer, global average pooling layer, and fully connected layer. Each hidden layer is followed by Rectified Linear Unit (ReLU) activation function to introduce non-linearity. Fully connected output layer is activated by a Softmax function to encode output scalars into the predictive probability of each class. The parameters of GCN were optimized using grid search. Stratified ten-fold cross-validation and leave-one-site-out (LOSO) cross-validation were separately applied. For the 10-fold cross-validation, we split the samples into 10 non-overlapping parts. In each iteration of the scheme, one part was considered as the test set for model evaluation, and the remaining nine parts were defined as the training set. The ten-fold cross-validation was stratified to preserve the percentage of samples for each class in each fold consistent with that in the whole dataset. For the LOSO cross-validation, in each iteration, data from one site was used for model evaluation and data from the remaining sites was used as the training set. Model performance was evaluated in terms of accuracy, sensitivity, specificity, and area under receiver operating characteristic curve (AUC) value. Our GCN model was implemented by using Pytorch Geometric extension library based on Pytorch 1·7 (Python 3·7).35 Model parameter settings can be found in Supplemental Information.
In addition to the primary classification between patients with MDD and HC, we performed a series of subgroup analyses to distinguish between FEDN patients and HC, recurrent patients and HC, and FEDN patients and recurrent patients. For the three subgroup analyses, sites containing more than 10 patients were included. Specifically, 227 FEDN patients and 388 HC from five sites (sites 4, 5, 9, 13, 16), 187 recurrent patients and 423 HC from six sites (sites 3, 5, 7, 12, 13, 14), and 117 FEDN patients and 70 recurrent patients from two sites (sites 5, 13) were separately included (Table S2). The procedures of GCN model training kept consistent with the primary analysis.
Identifying the most salient regions contributing to classification
Class activation mapping (CAM) was used to estimate the contribution of each brain region to GCN classification. CAM was originally developed for traditional convolutional neural networks (CNN) in computer vision field, which can localize the discriminative image areas by estimating the attention of CNN classifier to each pixel when predicting a particular class.36 Recent introduction of CAM into GCN enables the identification of discriminative nodes in irregular graphs beyond traditional image data.34 CAM exploits information from the last graph convolutional layer and fully connected layer, providing class activation value for each node at the individual level. We subsequently calculated the average activation value across all the individuals to reflect the contribution of each region. Detailed calculation of CAM is shown in Supplemental Information and graphically depicted in Fig. S2.
Herein, we reported the top ten salient brain regions and the corresponding distribution in six functional subnetworks proposed by Dosenbach et al., including default mode network (DMN), fronto-parietal network (FPN), cingulo-opercular network (CON), sensorimotor network (SMN), occipital network (ON), and cerebellum network (CN).23
Post-hoc correlation analysis
To reveal the clinical relevance of GCN model, we explored the relationship between the identified salient regions and clinical measures. Considering the network structure utilized in GCN, we assumed that salient regions identified by GCN can reflect regional network topological deficits that may be clinically informative. Thus, we applied graph theoretical analysis to estimate the topological profiles of each salient region. Three nodal topological properties, including degree, betweenness, and efficiency, were calculated by using the GRETNA toolbox. Detailed calculation and definition of nodal topological properties were presented in Supplemental Information.
Secondary validation analyses
First, we examined the classification performance in each site to validate the variability of single-site performance based on small dataset. Second, we compared the performance of GCN with other classifiers, including SVM, MLP, LR, RF, and BrainNetCNN.37 Third, considering that there is no consensus in the field regarding optimal brain parcellation during network construction, we tested the stability of GCN performance across two atlases with different numbers of parcels, including the automatic anatomical labeling (AAL) atlas (116 parcels) and Craddock atlas (200 parcels).38 Fourth, to evaluate the influence of fMRI noise from head motion on the resulting classification performance, we trained classification model based on different head motion parameters to find whether only using noise features can reach significant accuracy. Fifth, given the imbalanced single-site performance and sample size across 16 sites, we separately excluded the site with the best single-site performance and the site with the largest sample size to validate whether the current classification performance was biased by these sites. The procedures of all the secondary validation analyses are described in Supplemental Information in detail.
Statistics
We used the chi-square test and independent two-sample t-test to examine significant between-group differences in categorical and continuous variables reported in Table 1, respectively. The performance of GCN model was examined using accuracy, sensitivity, specificity, and AUC value. The accuracy was determined as the percentage of correctly classified individuals among all subjects. The sensitivity and specificity were used to indicate the percentage of correct classifications in MDD patients and HC, respectively. For the AUC value, we plotted ROC curve showing the classification performance at all classification thresholds according to true positive rate (i.e., sensitivity) and false positive rate (i.e., 1 – specificity). The AUC value was thus calculated as the area under ROC curve to provide an aggregate measure of performance irrespective of classification thresholds selection. Pearson correlation or Kendall's correlation (non-normal data) was used to assess the significance level of post-hoc correlation between topological metrics and clinical measures. An FDR corrected p value < 0·05 indicated significant correlations.
Ethics
This machine learning study was approved by the Research Ethics Committee of West China Hospital of Sichuan University (ethical approval number: 2020(54)). All participants provided written informed consent prior to participation, and data collection at each site in Rest-meta-MDD was approved by the local Institutional Review Board. For more detailed information about the participants, see the previous consortium publications.21,49
Role of funding source
The funders had no role in study design, data collection, data analyses, interpretation, or writing of the paper.
Results
Classification performance
Under stratified ten-fold cross-validation, GCN achieved an accuracy of 81·5% (95%CI: 80·5–82·5%) and AUC value of 0·865 for the classification between MDD patients and HC. FEDN patients were distinguished from HC with a classification accuracy of 74·1% (95%CI: 72·4–75·8%, AUC: 0·686), and recurrent patients were distinguished from HC with a classification accuracy of 78·1% (95%CI: 76·5–79·7%, AUC: 0·745). When differentiating between FEDN and recurrent patients, the classification accuracy was 70·9% (95%CI: 67·7–74·1%, AUC: 0·646). By using the LOSO cross-validation strategy, the accuracy was 83·1% (95%CI: 82·2–84·0%, AUC: 0·852) for the classification between the overall MDD patients and HC. In the subgroup analyses, FEDN patients were distinguished from HC with an accuracy of 68·3% (95%CI: 66·5–70·1%, AUC: 0·639), recurrent patients were distinguished from HC with an accuracy of 71·3% (95%CI: 69·5–73·1%, AUC: 0·586), and FEDN patients were distinguished from recurrent patients with an accuracy of 54·8% (95%CI: 51·3–58·3%, AUC: 0.547) (Table 2).
Table 2.
GCN model performance under different classification tasks.
| Model | Ten-fold stratified cross-validation |
Leave one site out cross-validation |
||||||
|---|---|---|---|---|---|---|---|---|
| ACC | SEN | SPE | AUC | ACC | SEN | SPE | AUC | |
| MDD vs. HC | 0·815 | 0·834 | 0·800 | 0·865 | 0·831 | 0·850 | 0·829 | 0·852 |
| FEDN vs. HC | 0·741 | 0·672 | 0·782 | 0·686 | 0·680 | 0·568 | 0·723 | 0·639 |
| Recurrent vs. HC | 0·781 | 0·718 | 0·822 | 0·745 | 0·713 | 0·601 | 0·739 | 0·586 |
| FEDN vs. recurrent | 0·709 | 0·743 | 0·681 | 0·646 | 0·548 | 0·573 | 0·648 | 0·547 |
Abbreviations: MDD, major depressive disorder; FEDN, first-episode drug-naïve; HC, healthy control; ACC, accuracy; SEN, specificity; SPE, specificity; AUC, area under receiver operating characteristic curve; GCN, graph convolutional network.
In the secondary validation analysis, we found that classification accuracy in each single site varied from 43·2% to 83·3%, confirming the variability of classification task with small sample size and the importance of using large dataset (Fig. S3). Additionally, GCN provided superior classification accuracy compared with other commonly used machine learning algorithms, including linear SVM, RF, MLP, and BrainNetCNN (Table S4). When using different atlases for parcellation, classification performance remained relatively stable, with accuracies of 81·1% for the AAL atlas and 78·8% for the Craddock atlas. The classification based on fMRI head motion noise obtained a poor random chance performance of 51·3% accuracy, 97·9% sensitivity, and 1·18% specificity (Fig. S4), suggesting that the classifier could not learn useful information from the noise and recognized the whole sample as MDD to minimize the training loss. We obtained an accuracy of 77·8% after removing the site with the largest sample size, whereas discarding the site with the best performance could still achieve an accuracy of 81·0%, consistent with the notion that the overall model performance was not biased by these individual sites.
Top salient regions contributing to classification
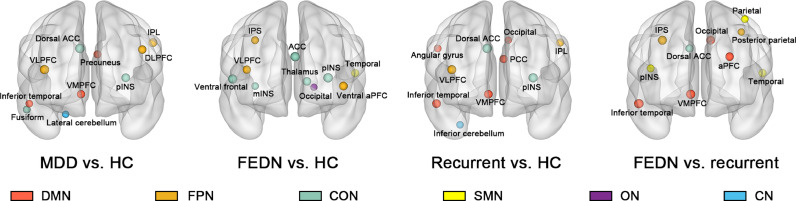
Based on the GCN model, the top ten salient regions contributing to group differentiation between MDD patients and HC were primarily located in the DMN, FPN, and CON, including the anterior cingulate cortex (ACC), prefrontal cortex, inferior parietal lobule (IPL), posterior insula, precuneus, fusiform, temporal cortex, and cerebellum. For the differentiation between FEDN patients and HC, the most salient regions were primarily in the FPN and CON. For the differentiation between recurrent patients and HC, saliency pattern was observed in DMN, FPN, and CON, which was similar to the main analysis. Finally, the DMN and SMN contributed to the classification between FEDN and recurrent patients (Table 3 and Figure 2).
Table 3.
Top salient regions contributing to classification based on graph convolutional network.
| Rank | MDD vs. HC |
FEDN vs. HC |
Recurrent vs. HC |
FEDN vs. recurrent |
||||
|---|---|---|---|---|---|---|---|---|
| Brain Region | Network | Brain Region | Network | Brain Region | Network | Brain region | Network | |
| 1 | R dorsal ACC | CON | L posterior insula | CON | R dorsal ACC | CON | L inferior temporal cortex | DMN |
| 2 | R VLPFC | FPN | L ACC | CON | L posterior insula | CON | R VMPFC | DMN |
| 3 | L IPL | FPN | R ventral frontal cortex | CON | R VLPFC | FPN | R IPS | FPN |
| 4 | L posterior insula | CON | L ventral anterior PFC | FPN | R inferior temporal cortex | DMN | L occipital cortex | DMN |
| 5 | L DLPFC | FPN | L thalamus | CON | R VMPFC | DMN | L anterior PFC | DMN |
| 6 | R VMPFC | DMN | R VLPFC | FPN | L PCC | DMN | L temporal cortex | SMN |
| 7 | L precuneus | DMN | L temporal cortex | SMN | L IPL | FPN | R dorsal ACC | CON |
| 8 | R fusiform | CON | R IPS | FPN | R angular gyrus | DMN | R posterior insula | SMN |
| 9 | R inferior temporal cortex | DMN | R middle insula | CON | R inferior cerebellum | CN | L parietal cortex | SMN |
| 10 | R lateral cerebellum | CN | L occipital cortex | ON | L occipital cortex | DMN | L posterior parietal cortex | FPN |
Abbreviations: L, left; R, right; MDD, major depressive disorder; HC, healthy control; FEDN, first-episode drug-naïve; ACC, anterior cingulate cortex; VLPFC, ventrolateral prefrontal cortex; IPL, inferior parietal lobule; DLPFC, dorsolateral prefrontal cortex; VMPFC, ventromedial prefrontal cortex; IPS, intraparietal sulcus; PCC, posterior cingulate cortex; DMN, default mode network; CON, cingulo-opercular network; FPN, fronto-parietal network; SMN, sensorimotor network; ON, occipital network; CN, cerebellum network.
Figure 2.
Top 10 salient regions contributing to GCN classification between different groups. Each salient brain region was shown as a sphere in glass brain from lateral and medial view. The size of the sphere reflects the rank of saliency. Region distributed in different networks were shown in different colors. Abbreviations: GCN, graph convolutional network; ACC, anterior cingulate cortex; DLPFC, dorsolateral prefrontal cortex; VLPFC, ventrolateral prefrontal cortex; VMPFC, ventromedial prefrontal cortex; mINS, middle insula; pINS, posterior insula; IPS, intraparietal sulcus; IPL, inferior parietal lobule; PCC, posterior cingulate cortex; FEDN, first-episode and drug-naïve; DMN, default mode network; FPN, fronto-parietal network; CON, cingulo-opercular network; SMN, sensorimotor network; ON, occipital network; CN, cerebellum network.
Correlation between topological characteristics of salient regions and clinical measures
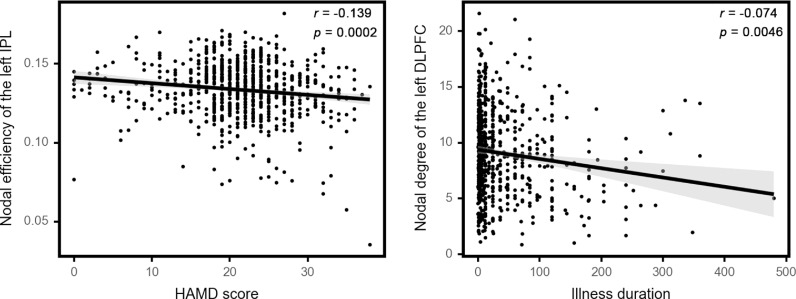
Among the most salient regions identified via GCN, the left dorsolateral prefrontal cortex (DLPFC) and left IPL exhibited significant associations with clinical measures. Specifically, the nodal efficiency of the left IPL was negatively associated with HAMD scores (r = -0·139, p = 0·0002 (Pearson correlation)). The nodal degree of the left DLPFC was negatively associated with illness duration (r = -0·074, p = 0·0046 (Kendall correlation)) (Figure 3). No significant associations between topological characteristics of the salient regions and HAMA scores were observed.
Figure 3.
Significant correlation between topological characteristics of the salient regions and clinical measures. The left panel shows the negative correlation between HAMD scores (n = 738) and nodal efficiency of the left IPL (p = 0·0002 (Pearson correlation)), and the right panel shows the negative correlation between illness duration (n = 691) and nodal degree of the left DLPFC (p = 0·0046 (Kendall correlation)). All these correlations survived FDR corrected p < 0·05. Abbreviations: IPL, inferior parietal lobule; HAMD, Hamilton Depression scale; DLPFC, dorsolateral prefrontal cortex; FDR, false discovery rate.
Discussion
In the current study, we applied GCN to characterize MDD patients using whole-brain functional networks based on a large multi-site dataset, which achieved classification performance with over 80% accuracy outperforming common machine learning methods used in previous studies. When identifying FEDN and recurrent patients, though model performance dropped, the accuracies remained at a level above 70%. Different patterns of salient regions contributing characterization of MDD, FEDN subtype, and recurrent subtype were mainly identified within the DMN, FPN, and CON. In the post-hoc correlation analysis, we found that topological profiles of the IPL and DLPFC were significantly correlated with depressive symptom severity and illness duration, respectively. These findings highlighted the relative advantages of using GCN to develop models for individualized case identification as well as brain dysfunction localization.
Although one previous study has demonstrated the capability of GCN for distinguishing individuals with MDD from HC,39 our work has significant advantages. First, the previous effort only included 29 individuals with MDD and 44 HC. Such small sample size can lead to a high risk of overfitting when training a deep learning algorithm which requires optimization of numerous parameters, and the variability of classification based on small dataset has also been suggested in previous publications and current investigation. Our study therefore utilized a multi-site MDD dataset containing over 1500 participants to evaluate GCN and included multiple validation strategies, which could provide more reliable and generalizable findings. Second, most previous GCN models constructed graphs at the group level based on inter-subject phenotypic association, while our GCN model established individual-level graphs with the similarity between node features. Compared with the group-level graphs, using image-based individual-level graphs may better fit the individualized clinical application and potentially reveal more accurate neurobiological underpinnings. Third, subgroup analysis regarding the characterization of FEDN and recurrent patients was performed in this study, providing evidence on the capability of GCN for specific subtypes. By exploring salient regional contribution in each subgroup, shared and distinct neurobiological mechanisms across subtypes were further illustrated.
The GCN exhibited superior classification performance compared with other common classifiers, paralleling previous observations in studies on Alzheimer's disease and autism.18 The pathophysiology of MDD is thought to include a disruption of the brain connectome,40 and the graphs are powerful tools for modeling brain connectome from neuroimaging data.41 Therefore, current promising performance of GCN may be ascribed to the consideration of graph structure to capture brain connectome information during model training. In contrast, most traditional classifiers typically extract and vectorize functional connectivity values as input, learning information from independent connections ignoring neighborhood relationships and complicated network structures. Notably, BrainNetCNN, another method that takes brain networks as input,37 also achieved better performance compared with the remaining methods that use connectivity values, suggesting better performance may be achieved using models based on brain networks not limited to GCN.
To provide neurobiologically informative findings from our models, we investigated patterns of salient regions contributing to classification, and the most salient regions were mainly distributed in DMN, FPN, and CON areas. This is consistent with previous studies, which have highlighted the disruption of DMN as a neural correlate of MDD resulting in abnormal self-referential processing and rumination.42 In addition, greater connectivity within the DMN can predict the remission following 8-week antidepressant treatment.43 Altered DMN connectivity has also been reported to be associated with increased risk for MDD onset in adolescence.44 These findings suggest that DMN may play a critical role in various neurobiological mechanisms of MDD. The FPN and CON, known as top-down control system, play an important role in the processes of various cognitive functions which may be related to cognitive deficits in MDD patients.45 Mounting neuroimaging evidence indicates that abnormalities of these systems are associated with pathophysiology and potentially serve as treatment outcome predictors in MDD.43,46,47 Our study provides further evidence for the implication of DMN, FPN, and CON areas in MDD, verifying the plausibility of the current GCN model at the neurobiological aspect. Moreover, significant relationships between clinical measures and regional functional topology of the IPL and DLPFC were observed, supporting the ability of GCN to capture clinically-relevant topological deficits. As both IPL and DLPFC are key regions of the FPN, FPN may have deeper and specific involvement in the mechanism of depressive symptom severity and illness duration, which hold the potential to serve as a promising clinical indicator.
In the subgroup analysis, we observed higher classification performance for the characterization of recurrent patients compared with FEDN patients. Given the equivalent sample size in these two subgroups, such difference in performance may result from the long-term psychopathological development and medication from multiple depressive episodes in the recurrent depression group, leading to a pattern of more severe network disruption relative to controls that increased the inter-group discriminability.48 We noted that distinct saliency patterns were also identified for different classification tasks. Specifically, the most discriminative brain regions between recurrent patients and HC were distributed in the DMN, FPN, and CON which is similar to the main analysis, while the most salient regions for differentiation between FEDN patients and HC were only found in the FPN and CON. Our previous work has concluded that functional network topological deficits in the MDD population were primarily driven by recurrent patients rather than FEDN patients,49 which is in line with the current machine learning findings. Since the FPN and CON remained stable across different classification tasks, they may serve as generalizable biomarkers of MDD regardless of illness course and medication status. The DMN was only identified in the recurrent patients, suggesting a secondary salient biomarker besides FPN and CON potentially related to accumulating pathological and medication effects during multiple recurrent depressive episodes.
One previous multi-site machine learning study reported core abnormal functional connectivity profiles of prefrontal, limbic and striatal areas within DMN, FPN, and CON,50 which is in line with our findings. However, since alterations in the DMN, FPN, and CON have been associated with multiple psychiatric disorders51,52, the specificity of our findings for MDD may be limited. The pattern of these alterations across disorders and their relation to other networks needs to be evaluated in future research. We also noted subcortical areas that have been widely implicated in the pathophysiology of MDD, such as amygdala and hippocampus, were not among our salient regions. Since Dosenbach's brain functional atlas we used focus more on intrinsic functional networks instead of anatomical boundaries, it does not differentiate specific anatomical subdivisions of subcortical structures like most anatomical atlases did. Future studies are warranted to gain further insight into subcortical features and revisit this issue in a hypothesis driven fashion. Moreover, Drysdale et al., further investigated the heterogeneity of MDD using unsupervised machine learning techniques, clustering 4 connectivity-based biotypes of MDD. Although our findings on FEDN and recurrent MDD patients provide preliminary insights into specific clinical subtypes, novel neuroimaging-based subtypes may further illuminate the heterogeneity of MDD. Given the large MDD dataset we have, clustering novel connectome-based MDD subtypes with multi-site dataset will be the future work in our consortium.
Of note, there are several limitations in the current study. First, our multi-site dataset was exclusively collected from Chinese participants, so generalization to other racial/ethnic groups remains to be confirmed. Second, the current model requires replication on other independent datasets before any application in clinical decision-making. Third, the current study only investigated resting-state functional networks. Since other types of networks from different imaging modalities have been implicated in the pathophysiology of MDD, future studies can study gray matter covariance network, white matter connectivity network, or combination of different networks to find the optimal way for MDD identification. Fourth, given the age range of included participants in our current study, our findings may not apply to pediatric or geriatric depression. Fifth, although we considered FEDN and recurrent patients in the subgroup analysis, the ability to assess other confounds is limited. As MDD is highly heterogeneous, these factors, such as comorbidities, medication, illness duration and onset should be explored in the future to further enhance the classification accuracy.
In summary, this study explores the application value of GCN based on brain functional networks differentiating patients with MDD from HC. Based on a large multi-site dataset and various validation strategies, generalizable and reliable classification accuracy of over 80% can be achieved via GCN, indicating that GCN modeling is promising for the characterization of MDD. The investigation of saliency patterns contributing to GCN classification identified the most salient regions within the DMN, FPN, and CON, validating the plausibility of GCN at the neurobiological aspect. Moreover, topological deficits of partial top salient regions were associated with clinical measures such as symptom severity and illness duration. These findings provide promising direction towards the application of GCN to resting-state functional networks, with the ultimate aim of developing and validating biomarkers for clinical diagnosis and treatment planning in MDD and other psychiatric disorders.
Contributors
Q.G. joined the Rest-meta-MDD data sharing consortium as principal investigator and got full access to the multi-site dataset. K.Q., D.L., and Q.G. conceptualized, designed, and supervised the study. K.Q. and D.L. directly accessed and verified the underlying data. K.Q. and D.L. drafted and revised the manuscript. K.Q., D.L., Z.Z., W.L., and N.P. analyzed the data. W.H.L.P. and W.L. provided technical support. J.S. and A.M. reviewed the manuscript and provided constructive suggestions on data analysis and interpretation. All authors have read and approved the final version of the manuscript for submission.
Data sharing statement
The codes for the implementation of GCN are available at https://github.com/QKmeans0902/GCN_MDD_Classification. Multi-site MDD dataset is available from the corresponding author upon reasonable request or the Rest-meta-MDD consortium (http://rfmri.org/REST-meta-MDD).
Declaration of interests
Dr. Sweeney consults to VeraSci. Other authors report no conflicts of interest.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81621003, 82027808, and 81820108018). Drs. Sweeney and Gong received support from National Natural Science Foundation (No. 81820108018). We thank the other collaborative members of the REST-meta-MDD consortium for sharing the data.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103977.
Appendix. Supplementary materials
References
- 1.Whiteford H.A., Degenhardt L., Rehm J., et al. Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet. 2013;382:1575–1586. doi: 10.1016/S0140-6736(13)61611-6. [DOI] [PubMed] [Google Scholar]
- 2.Lui S., Zhou X.J., Sweeney J.A., Gong Q. Psychoradiology: the frontier of neuroimaging in psychiatry. Radiology. 2016;281:357–372. doi: 10.1148/radiol.2016152149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lai C.H., Wu Y.T. Frontal-insula gray matter deficits in first-episode medication-naïve patients with major depressive disorder. J Affect Disord. 2014;160:74–79. doi: 10.1016/j.jad.2013.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Yao Z., Fu Y., Wu J., et al. Morphological changes in subregions of hippocampus and amygdala in major depressive disorder patients. Brain Imaging Behav. 2020;14:653–667. doi: 10.1007/s11682-018-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu X., Wang X., Xiao J., et al. Evidence of a dissociation pattern in resting-state default mode network connectivity in first-episode, treatment-naive major depression patients. Biol Psychiatry. 2012;71:611–617. doi: 10.1016/j.biopsych.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 6.Chen L., Wang Y., Niu C., et al. Common and distinct abnormal frontal-limbic system structural and functional patterns in patients with major depression and bipolar disorder. Neuroimage Clin. 2018;20:42–50. doi: 10.1016/j.nicl.2018.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang S., Li H., Lu L., et al. Anomalous functional connectivity of amygdala subregional networks in major depressive disorder. Depress Anxiety. 2019;36:712–722. doi: 10.1002/da.22901. [DOI] [PubMed] [Google Scholar]
- 8.Schnack H.G., Kahn R.S. Detecting neuroimaging biomarkers for psychiatric disorders: sample size matters. Front Psychiatry. 2016;7:50. doi: 10.3389/fpsyt.2016.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kambeitz J., Cabral C., Sacchet M.D., et al. Detecting neuroimaging biomarkers for depression: a meta-analysis of multivariate pattern recognition studies. Biol Psychiatry. 2017;82:330–338. doi: 10.1016/j.biopsych.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flint C., Cearns M., Opel N., et al. Systematic misestimation of machine learning performance in neuroimaging studies of depression. Neuropsychopharmacology. 2021;46:1510–1517. doi: 10.1038/s41386-021-01020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamashita A., Sakai Y., Yamada T., et al. Generalizable brain network markers of major depressive disorder across multiple imaging sites. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornito A., Zalesky A., Breakspear M. The connectomics of brain disorders. Nat Rev Neurosci. 2015;16:159–172. doi: 10.1038/nrn3901. [DOI] [PubMed] [Google Scholar]
- 13.He Z., Lu F., Sheng W., et al. Functional dysconnectivity within the emotion-regulating system is associated with affective symptoms in major depressive disorder: a resting-state fMRI study. Aust N Z J Psychiatry. 2019;53:528–539. doi: 10.1177/0004867419832106. [DOI] [PubMed] [Google Scholar]
- 14.Shi Y., Li J., Feng Z., et al. Abnormal functional connectivity strength in first-episode, drug-naïve adult patients with major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2020;97 doi: 10.1016/j.pnpbp.2019.109759. [DOI] [PubMed] [Google Scholar]
- 15.Zhang J., Wang J., Wu Q., et al. Disrupted brain connectivity networks in drug-naive, first-episode major depressive disorder. Biol Psychiatry. 2011;70:334–342. doi: 10.1016/j.biopsych.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 16.Bronstein M.M., Bruna J., LeCun Y., Szlam A., Vandergheynst P. Geometric deep learning: going beyond euclidean data. IEEE Signal Process Mag. 2017;34:18–42. [Google Scholar]
- 17.Ktena S.I., Parisot S., Ferrante E., et al. Metric learning with spectral graph convolutions on brain connectivity networks. Neuroimage. 2018;169:431–442. doi: 10.1016/j.neuroimage.2017.12.052. [DOI] [PubMed] [Google Scholar]
- 18.Parisot S., Ktena S.I., Ferrante E., et al. Disease prediction using graph convolutional networks: application to autism spectrum disorder and Alzheimer's disease. Med Image Anal. 2018;48:117–130. doi: 10.1016/j.media.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 19.Yao D., Sui J., Wang M., et al. A mutual multi-scale triplet graph convolutional network for classification of brain disorders using functional or structural connectivity. IEEE Trans Med Imaging. 2021;40:1279–1289. doi: 10.1109/TMI.2021.3051604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vieira S., Pinaya W.H., Mechelli A. Using deep learning to investigate the neuroimaging correlates of psychiatric and neurological disorders: methods and applications. Neurosci Biobehav Rev. 2017;74:58–75. doi: 10.1016/j.neubiorev.2017.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Yan C.G., Chen X., Li L., et al. Reduced default mode network functional connectivity in patients with recurrent major depressive disorder. Proc Natl Acad Sci U S A. 2019;116:9078–9083. doi: 10.1073/pnas.1900390116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chao-Gan Y., Yu-Feng Z. DPARSF: a MATLAB toolbox for "pipeline" data analysis of resting-state fMRI. Front Syst Neurosci. 2010;4:13. doi: 10.3389/fnsys.2010.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dosenbach N.U., Nardos B., Cohen A.L., et al. Prediction of individual brain maturity using fMRI. Science. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomasi D., Volkow N.D. Gender differences in brain functional connectivity density. Hum Brain Mapp. 2012;33:849–860. doi: 10.1002/hbm.21252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stonnington C.M., Tan G., Klöppel S., et al. Interpreting scan data acquired from multiple scanners: a study with Alzheimer's disease. Neuroimage. 2008;39:1180–1185. doi: 10.1016/j.neuroimage.2007.09.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yahata N., Morimoto J., Hashimoto R., et al. A small number of abnormal brain connections predicts adult autism spectrum disorder. Nat Commun. 2016;7:11254. doi: 10.1038/ncomms11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kostro D., Abdulkadir A., Durr A., et al. Correction of inter-scanner and within-subject variance in structural MRI based automated diagnosing. Neuroimage. 2014;98:405–415. doi: 10.1016/j.neuroimage.2014.04.057. [DOI] [PubMed] [Google Scholar]
- 28.Lei D., Pinaya W.H.L., Young J., et al. Integrating machining learning and multimodal neuroimaging to detect schizophrenia at the level of the individual. Hum Brain Mapp. 2020;41:1119–1135. doi: 10.1002/hbm.24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson W.E., Li C., Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 30.Fortin J.P., Cullen N., Sheline Y.I., et al. Harmonization of cortical thickness measurements across scanners and sites. Neuroimage. 2018;167:104–120. doi: 10.1016/j.neuroimage.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fortin J.P., Parker D., Tunç B., et al. Harmonization of multi-site diffusion tensor imaging data. Neuroimage. 2017;161:149–170. doi: 10.1016/j.neuroimage.2017.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu M., Linn K.A., Cook P.A., et al. Statistical harmonization corrects site effects in functional connectivity measurements from multi-site fMRI data. Hum Brain Mapp. 2018;39:4213–4227. doi: 10.1002/hbm.24241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radua J., Vieta E., Shinohara R., et al. Increased power by harmonizing structural MRI site differences with the ComBat batch adjustment method in ENIGMA. Neuroimage. 2020;218 doi: 10.1016/j.neuroimage.2020.116956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arslan S., Ktena S.I., Glocker B., Rueckert D. In: Graphs in Biomedical Image Analysis and Integrating Medical Imaging and Non-Imaging Modalities. Stoyanov D., Taylor Z., Ferrante E., Dalca A.V., editors. Springer; Granada, Spain. Berlin: 2018. Graph saliency maps through spectral convolutional networks: application to sex classification with brain connectivity; pp. 3–13. 2018 Sept 20. [Google Scholar]
- 35.Fey M., Lenssen J.E. Fast graph representation learning with PyTorch geometric [Internet]. arXiv [preprint]. 2019 [cited 2021 July 29]: arXiv:1903.02428. Available from: https://arxiv.org/abs/1903.02428.
- 36.Zhou B., Khosla A., Lapedriza A., Oliva A., Torralba A. Learning deep features for discriminative localization. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Las Vegas, United States; Seattle: IEEE; 2016. pp. 2921–2929. 2016 Jun 27-302016. [Google Scholar]
- 37.Kawahara J., Brown C.J., Miller S.P., et al. BrainNetCNN: Convolutional neural networks for brain networks; towards predicting neurodevelopment. Neuroimage. 2017;146:1038–1049. doi: 10.1016/j.neuroimage.2016.09.046. [DOI] [PubMed] [Google Scholar]
- 38.Craddock R.C., Holtzheimer P.E., Hu X.P., Mayberg H.S. Disease state prediction from resting state functional connectivity. Magn Reson Med. 2009;62:1619–1628. doi: 10.1002/mrm.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jun E., Na K.S., Kang W., Lee J., Suk H.I., Ham B.J. Identifying resting-state effective connectivity abnormalities in drug-naïve major depressive disorder diagnosis via graph convolutional networks. Hum Brain Mapp. 2020;41:4997–5014. doi: 10.1002/hbm.25175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong Q., He Y. Depression, neuroimaging and connectomics: a selective overview. Biol Psychiatry. 2015;77:223–235. doi: 10.1016/j.biopsych.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Bullmore E., Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 42.Whitfield-Gabrieli S., Ford J.M. Default mode network activity and connectivity in psychopathology. Annu Rev Clin Psychol. 2012;8:49–76. doi: 10.1146/annurev-clinpsy-032511-143049. [DOI] [PubMed] [Google Scholar]
- 43.Korgaonkar M.S., Goldstein-Piekarski A.N., Fornito A., Williams L.M. Intrinsic connectomes are a predictive biomarker of remission in major depressive disorder. Mol Psychiatry. 2020;25:1537–1549. doi: 10.1038/s41380-019-0574-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai Y., Elsayed N.M., Barch D.M. Contributions from resting state functional connectivity and familial risk to early adolescent-onset MDD: results from the adolescent brain cognitive development study. J Affect Disord. 2021;287:229–239. doi: 10.1016/j.jad.2021.03.031. [DOI] [PubMed] [Google Scholar]
- 45.Dosenbach N.U., Fair D.A., Cohen A.L., Schlaggar B.L., Petersen S.E. A dual-networks architecture of top-down control. Trends Cogn Sci. 2008;12:99–105. doi: 10.1016/j.tics.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaiser R.H., Andrews-Hanna J.R., Wager T.D., Pizzagalli D.A. Large-scale network dysfunction in major depressive disorder: a meta-analysis of resting-state functional connectivity. JAMA Psychiatry. 2015;72:603–611. doi: 10.1001/jamapsychiatry.2015.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X., Lin P., Yang J., Song H., Yang R., Yang J. Dysfunction of the cingulo-opercular network in first-episode medication-naive patients with major depressive disorder. J Affect Disord. 2016;200:275–283. doi: 10.1016/j.jad.2016.04.046. [DOI] [PubMed] [Google Scholar]
- 48.Dohm K., Redlich R., Zwitserlood P., Dannlowski U. Trajectories of major depression disorders: a systematic review of longitudinal neuroimaging findings. Aust N Z J Psychiatry. 2017;51:441–454. doi: 10.1177/0004867416661426. [DOI] [PubMed] [Google Scholar]
- 49.Yang H., Chen X., Chen Z.B., et al. Disrupted intrinsic functional brain topology in patients with major depressive disorder. Mol Psychiatry. 2021;26:7363–7371. doi: 10.1038/s41380-021-01247-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Drysdale A.T., Grosenick L., Downar J., et al. Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nat Med. 2017;23:28–38. doi: 10.1038/nm.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qin K., Lei D., Yang J., et al. Network-level functional topological changes after mindfulness-based cognitive therapy in mood dysregulated adolescents at familial risk for bipolar disorder: a pilot study. BMC Psychiatry. 2021;21:213. doi: 10.1186/s12888-021-03211-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godwin D., Ji A., Kandala S., Mamah D. Functional connectivity of cognitive brain networks in schizophrenia during a working memory task. Front Psychiatry. 2017;8:294. doi: 10.3389/fpsyt.2017.00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li F., Sun H., Biswal B.B., Sweeney J.A., Gong Q. Artificial intelligence applications in psychoradiology. Pyschoradiology. 2021;1:94–107. doi: 10.1093/psyrad/kkab009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gong Q. Psychoradiology, An Issue of Neuroimaging Clinics of North America. Vol. 30. Elsevier Inc; New York: 2020. pp. 1–123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.