Abstract
Purpose: As a promotor of tumor invasion and tumor microenvironment (TME) formation, the protein complex S100A8/S100A9 is associated with poor prognosis. Our aim was to further evaluate its origin and regulatory effects, and to establish an imaging biomarker for TME activity. Methods: S100A9−/−cells (ko) were created from syngeneic murine breast cancer 4T1 (high malignancy) and 67NR (low malignancy) wildtype (wt) cell lines and implanted into either female BALB/c wildtype or S100A9−/− mice (n = 10 each). Anti-S100A9-Cy5.5-targeted fluorescence reflectance imaging was performed at 0 h and 24 h after injection. Potential early changes of S100A9-presence under immune checkpoint inhibition (anti-PD-L1, n = 7 vs. rat IgG2b as isotype control, n = 3) were evaluated. Results: In S100A9−/−mice contrast-to-noise-ratios were significantly reduced for wt and S100A9−/−tumors. No significant differences were detected for 4T1 ko and 67NR ko cells as compared to wildtype cells. Under anti-PD-L1 treatment S100A9 presence significantly decreased compared with the control group. Conclusion: Our results confirm a secretion of S100A8/S100A9 by the TME, while tumor cells do not apparently release the protein. Under immune checkpoint inhibition S100A9-imaging reports an early decrease of TME activity. Therefore, S100A9-specific imaging may serve as an imaging biomarker for TME formation and activity.
Keywords: S100A8/S100A9, MRP8/MRP14, Tumor microenvironment, Target-specific imaging, Molecular imaging
Abbreviations: CNR, Contrast-to-noise-ratio; CRISPR/cas9, Clustered regularly interspaced short palindromic repeats/CRISPR associated genes; MDSC, Myeloid derived suppressor cells; TAM, Tumor associated macrophages; TME, Tumor microenvironment; TLR-4, Toll-like receptor 4
Introduction
Tumor growth, invasion and metastasis are strongly influenced and driven by tumor infiltrating immune cells, which form a characteristic inflammatory tumor microenvironment (TME) with considerable, intra- and intertumoral heterogeneity [1]. Within the TME, the protein heterodimer S100A8/S100A9 (synonym: myeloid related protein (MRP)8/MRP14) has been characterized as an important promoter of tumor invasiveness and metastasis [2]. As an important link between tumors and the protein heterodimer S100A8/S100A9, tumor cells express the Toll-like receptor 4 (TLR-4), the endogenous receptor of S100A8/S100A9 [3], mediating tumor growth [4]. Furthermore, the protein complex binds to tumor cells and, via activation of NF-kB, causes increased tumor cell proliferation [5]. S100A9 has protumoral properties by promoting immunosuppressive activity through polarization of myeloid derived suppressor cells (MDSC) and M2 macrophages [6]. Therefore, its presence has been associated with poor prognosis and is considered as an important pro-tumorigenic regulator [7].
S100A8/S100A9 is mainly present as an immune complex and as such biologically active. Thus, the lack of S100A9 leads to an inhibition of the overall function [5]. In this respect, the addressing of S100A9 reflects the presence of the protein complex.
Within the TME but also within the premetastatic niche the protein complex S100A8/S100A9 is predominantly secreted by infiltrating myeloid cells such as tumor associated macrophages (TAM), MDSC and neutrophils [8].
However, in the context of cancer, an expression also by tumor cells themselves has been reported: In proteomic and gene expression studies an overexpression of the protein complex has been found for various cancers such as gastric cancer [9, 10], lung cancer [11], hepatocellular [12] and colorectal carcinoma [13], but especially for breast cancer with an enhanced upregulation of S100A8 and S100A9 under estrogen stimulation in vitro [14]. Induced by monocytes, S100A8- and S100A9-mRNA expression has been described to be increased in cancer cells, promoting local invasiveness [15]. These studies have shown increased levels of the protein complex in tumor tissues an in the serum of cancer patients.
However, with respect to S100A8/S100A9, known to be highly expressed by monocytic cells in the tumor microenvironment, a TME derived origin of the protein [14] as opposed to an expression of S100A8/S100A9 by tumor cells [15] seems also probable.
In previous studies using a syngeneic murine breast cancer model we were not able to detect S100A8/S100A9 expression on the level of tumor cells [16]. However, we have shown its presence on tumor cell RNA, but without expression on protein level [17] as well as in MDSC derived exosomes [18].
Thus, overall, it has to be stated that the origin of S100A8/S100A9 within the tumor microenvironment as well as mechanisms of its secretion remain unclear.
In addition, in light of new therapy concepts in oncology such as immune checkpoint inhibition, there is a lack of imaging biomarkers for assessing therapy response and course. A purely morphological description of tumor lesions and the assessment of size dynamics are no longer sufficient in this context. Rather, an assessment based on functional tissue parameters for a prognostic assessment is highly desired [19].
Therefore, using the example of 4T1/67NR murine breast cancer, the purpose of this study was to analyze, whether the heterodimer is secreted by either primary tumor cells or tumor associated immune cells within the TME and thus reflects their local activity. S100A9-specific non-invasive in vivo imaging should allow for longitudinal examination of myeloid cell´s activity within the TME and provide further knowledge about the heterodimer´s regulatory influence on the TME. Furthermore, the potential of S100A8/S100A9 to serve as an imaging biomarker for the tumor microenvironment as well as possible changes within the TME under treatment with immune checkpoint inhibitors should be examined as a proof of principle.
Materials and methods
Cell lines and S100A9 knockout via CRISPR/cas9
As a syngeneic tumor model, murine breast cancer wildtype cell lines with the common genetic background of one maternal cell were used: 4T1 (CRL-2539, ATCC, Manassas, Virgina, USA) tumor cells with high (rapid local growth, lymphatic and hematogenous metastasis) and 67NR tumor cells (Barbara Ann Karmanos Cancer Intistute, Detroit, Michigan, USA) with low malignant potential (low-invasive and no metastasis) [16].
Additionally, in both cell lines a specific knockout of the S100A9 target gene was performed via the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/cas9 (CRISPR associated genes) system as described earlier [20]. Briefly, a S100A9-targeted guide RNA was used for the insertion into lentiCRISPRv2 plasmid (Addgene, Cambridge, USA, Nr. 52961). After virus propagation in HEK293T cells using lentiCRISPRv2-gRNA transfer plasmid, packaging plasmid (psPAX2) and envelope plasmid (pCMV-VSV-G) the 4T1 and 67NR cells were transduced with the virus-containing supernatants and selected with puromycin (InvivoGen, San Diego, USA) for two weeks. The successful knockout of S100A9 was analyzed by DNA sequencing of CRISPR/cas9 target region and again validated by western blotting.
Thus, four different tumor cell lines have been used in this study: 4T1 and 67NR wildtype tumor cells (4T1 wt, 67NR wt) as well as tumor cells with a specific knockout of the S100A9 target gene (4T1 ko, 67NR ko).
Lysate preparation
Tumor cells (1-5 × 106) were harvested, centrifuged (300 x g, 7 min) and washed with PBS. After re-centrifugation, cells were re-suspended in lysis buffer and incubated for 30 min on ice. The lysates were centrifuged (14000 x g, 20 min) and the supernatants were transferred to a fresh reaction tube. Protein content was analyzed using a standard microtiter plate assay with a BSA standard before storage of lysates at -80 °C.
Tissue preparation and RNA isolation
Orthotopically grown tumors were explanted and shock frozen in liquid nitrogen for isolation of total RNA. Tissue homogenization for RNA isolation was performed using TRIZOL, the CK28 ceramic kit and the Precellys Evolution homogenizer (VWR, Darmstadt, Germany). After homogenization, tissue samples were centrifuged (5000 x g, 5 min) and supernatants were transferred to RNA isolation. RNA was isolated using the Qiagen RNeasy Kit (Qiagen, Hilden, Germany) in combination with the PeqGold DNAse kit (VWR, Darmstadt, Germany) according to the manufacturer's instruction. For analyses of RNA quality and quantity a Nanodrop spectrophotometer (VWR, Darmstadt, Germany) was used.
Reverse transcription and PCR
Reverse transcription of 1 µg of each RNA was performed using the Quantitect Reverse Transcription kit (Qiagen, Hilden, Germany) according to the manufacture`s protocol. PCR was done with Roboklon Taq Polymerase and dNTP`s (Roboklon, Berlin, Germany), Quantitect primer assays (Qiagen, Hilden, Germany; primer details see Supplementary Table S1) and the Real Plex Cycler (Eppendorf, Hamburg, Germany) using a standard PCR protocol. PCR products were analyzed by agarose gel electrophoresis.
Western blotting
Cells were lysed with a Mammalian Protein Extraction Reagent lysis buffer (ThermoFisher Scientific, Waltham, Massachusetts, USA) containing a protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Santa Cruz Biotechnology, Dallas, Texas, USA). An amount of 15-30 µg proteins was cooked in the SDS loading buffer at 95 °C for 10 min, separated by SDS-gel electrophoresis and transferred to a nitrocellulose membrane (ThermoFisher Scientific, Waltham, MA, USA). Membranes were blocked for 1 hour at room temperature in TBST containing 10 % milk powder (Carl Roth, Karlsruhe, Germany) followed by overnight incubation with primary antibodies diluted in TBST at 4°C. For detection, horseradish peroxidase-conjugated secondary antibody (Cell Signaling, Danvers, Massachusetts, USA) diluted in TBST containing 5 % milk powder were used in combination with ECL solution (Signal Fire ECL Reagent, Cell Signaling Technology, Frankfurt, Germany) to detect the bands via a Chemidoc XRS+ (BioRad, Hercules, California, USA).
Adhesion assays
Adhesion assays were customized according to Humphries et al. [21]. In brief, cells were washed, detached, resuspended in culture media containing 1 % BSA and counted using the TECAN Spark plate reader (Tecan, Maennedorf, Switzerland). A number of 3 × 104 cells were seeded in collagen-coated 96-well plates and incubated for the indicated times. Afterwards, cell suspension was removed and the remaining cells were washed, fixated and stained with crystal violet. After cell lysis using a citrate buffer, absorbance was measured at 570 nm using the TECAN Spark. The assays were repeated for at least three times.
Wound healing/Migration assay
For wound healing assays, 6-well plates were coated with collagen before 2-well culture inserts (ibidi, Graefelfing, Germany) were fitted to each well. Cells were washed, detached, resuspended in culture media and counted using the TECAN Spark plate reader (Tecan, Maennedorf, Switzerland). A number of 3 × 105 cells were seeded into each chamber of the culture insert and incubated overnight at 37 °C and 5 % CO2. Afterwards the culture inserts were removed and photographs of the cell layer wound using the Nikon Eclipse TE2000-U microscope (Nikon, Tokyo, Japan) were taken every hour for up to 24 h. Analysis of cell migration was done by measuring the closure of the wound over time. The migration rate was determined using linear regression.
In vivo experiments
All animal experiments described in this study were approved by the responsible authorities (“Landesamt für Natur, Umwelt und Verbraucherschutz NRW”, Germany, Protocol No. 84-02.04.2017.A011). Female BALB/c (Charles River Laboratories, Germany) mice (n = 56, age: 8–12 weeks) and female S100A9−/− mice on BALB/c background (n = 40, age: 8–12 weeks, Institute of Immunology, University of Muenster, Muenster, Germany) were implanted orthotopically (0.5 × 106 tumor cells in 50 μl of phosphate-buffered saline without additives) via injection into the lower left mammary fat pad (injection scheme in Table 1) using extra fine 29G syringes.
Table 1.
Injection scheme of tumor cell types and fluorescent probes.
| Injection of 0.5 × 106 cells | Fluorescent probe | Mouse strain | Number of mice |
|---|---|---|---|
| 4T1 (wildtype) | anti-S100A9-Cy5.5 | BALB/c | 10 |
| 4T1 (S100A9 ko) | anti-S100A9-Cy5.5 | BALB/c | 10 |
| 4T1 (wildtype) | anti-S100A9-Cy5.5 | S100A9−/− | 10 |
| 4T1 (S100A9 ko) | anti-S100A9-Cy5.5 | S100A9−/− | 10 |
| 67NR (wildtype) | anti-S100A9-Cy5.5 | BALB/c | 10 |
| 67NR (S100A9 ko) | anti-S100A9-Cy5.5 | BALB/c | 10 |
| 67NR (wildtype) | anti-S100A9-Cy5.5 | S100A9−/− | 10 |
| 67NR (S100A9 ko) | anti-S100A9-Cy5.5 | S100A9−/− | 10 |
Tumor sizes were measured daily using a digital caliper. To control for tumor size effects imaging experiments were initiated when tumors reached a maximum diameter of 4 mm (tumor volume approximately 268 mm3 (4/3 x π x r3)). The course of the tumor was given as relative tumor growth after calculating the difference to size at day 0.
After in vivo imaging, mice were sacrificed and tissue was harvested for flow cytometry and immunohistochemistry as described below.
Imaging probe and imaging procedure
A polyclonal antibody, binding the S100A9 subunit of the S100A8/S100A9 protein complex as representative for the biological functionality of the complex [5, 22] was labelled with the cyanine dye Cy5.5 (anti-S100A9-Cy5.5; λex/em 678/696 nm, GE Healthcare Bio-Sciences Corp.) as described in detail earlier [22].
According to the manufacturer´s protocol, 1.5 mg of purified antibody was labelled with Cy5.5 and separated from unbound dye by elution with PBS using a PD10 column. An labelling ratio of 3.0 dye molecules per antibody was confirmed via photometric analysis. The probes were injected normalized for the fluorescent dye with a tracer equivalent to 0.5 nmol Cy5.5 per mouse and imaging session. For evaluation of the probe's specificity and sensitivity in this study, polyclonal rabbit IgG was labelled with Cy5.5 accordingly for imaging of control mice (n = 6). In detail, the specificity of the probe has already been examined in vivo and in vitro in previous studies [16, 22].
The fluorescent conjugates were injected intravenously in a single dose once the individual tumor reached the aforementioned size threshold. For fluorescent dyes low toxicity has been reported for the similar IRDye800CW [23], which revealed no changes after coupling to an antibody for in vivo imaging [24]. Immediately before (0 h) and 24 h after probe injection and shaving of the abdominal region for reducing artefacts, fluorescence reflectance imaging was performed (MS-FX-Pro; Bruker BioSpin MRI GmbH, Ettlingen, Germany, excitation: 630 nm, emission: 700 nm) using a filter-equipped, high-sensitive charge-coupled device camera. Signal was recorded for a period of 5 s. Additionally, white light images (3 s acquisition time) were obtained for anatomical colocalization. During examination, mice were under inhalation anesthesia (isoflurane 2.0 % in 2 l/min air during in vivo imaging; 2.5 % for 2 min induction, immediately prior to imaging).
Using the acquired white light images for anatomical orientation, the regions of interest (ROIs) were identified as the whole tumor region for for further image analysis. FRI data are presented as mean photon counts within the defined ROI in arbitrary units (AU). Reducing the background fluorescence the comparison of the fluorescence intensities is presented as the difference of contrast-to-noise-ratios with healthy tissue of the contralateral side as reference (CNR = (mean photon counts normalized for the ROI in the tumor – mean photon counts normalized for the ROI in healthy tissue of the contralateral side)/standard deviation of the background) between in vivo imaging 0 h and 24 h after probe injection ("CNR 24 h – 0 h").
Immediately after the second in vivo imaging, mice, still under anesthesia, were sacrificed by cervical dislocation.
Immune checkpoint inhibitor therapy
In a subsequent experiment 0.5 × 106 4T1 wildtype cells were implanted orthotopically into BALB/c wildtype mice (n = 10). Tumor volume was measured daily. Anti-S100A9-Cy5.5 was injected tumor size-dependent as described above followed by FRI at 0 h and after 24 h. Afterwards mice were randomly separated into two different treatment groups. The first group (n = 7) received an anti-PD-L1 immune checkpoint inhibitor (InVivo Mab rat anti-mouse PD-L1 (B7-H1, clone: 10F.9G2), Bio X Cell, Lebanon, New Hampshire, USA) via i.v. injection into the tail vein (10 mg/ kg body weight, dissolved in NaCl 0.9 %). Treatment was carried out immediately after the imaging procedure 24 h after probe injection (day 1) and subsequently on days 3 and 5 according to previously published protocols [25]. A second group received rat IgG2b as an isotype control (for antibody details see table S2) via i. v. injection 1(10 mg/ kg body weight, dissolved in NaCl 0.9 %) on days 1, 3 and 5.
Both groups were examined in a second in vivo imaging at 0 h and 24 h after injection of anti-S100A9-Cy5.5 on day 8. For both imaging time points “Delta fluorescence intensities” (mean fluorescence intensity 24 h – mean fluorescence intensity 0 h) were calculated.
Immunohistochemistry
Orthotopically grown tumors were explanted and tumor tissue was harvested for histology or flow cytometry. For immunohistochemistry, tumors were cryo-conserved and cut into 5 μm slices. Staining for S100A9 was performed, using the same antibody as for in vivo imaging, following established protocols [16].
Cryoblocks were sectioned at 10 µm thickness by a Leica CM1950 cryostat (Leica, Wetzlar, Germany) Cryosections were airdried and incubated in a glass jar with PBS in a 37 °C water bath for 10 min to remove OCT compounds. Sample sections were fixed by 4 % PFA at RT for 20 min and permeabilized with 0.2 % Triton/PBS for 25 min. Sections were then incubated with primary antibodies, biotin-anti-mouse/human CD11b (Biolegend 101204, Biolegend, San Diego, California, USA) at 1:500 dilution and the previously used rabbit anti-mouse S100A9 (see Supplementary Table S2) at 1:1000 dilution at RT for 30min. Followed by secondary antibodies, Streptavidin Alexa Fluor660 (Invitrogen S21377, Invitrogen, Carlsbad, California, USA) at 1:500 dilution, Alexa 594 Goat-anti-Rabbit (Invitrogen A-11012, Carlsbad, California, USA) at 1:500 dilution and together with other conjugated antibodies, anti-mouse Ly-6C-FITC (Biolegend 128006) at 1:200 dilution, anti-mouse F4/80-PE (Biolegend 123110) at 1:50 dilution, anti-mouse CD4-Alexa Fluor700 (Biolegend 100430) 1:100 dilution and anti-mouse CD8 (Biolegend 100802) conjugated with Dylight 633 (abcam ab201802, abcam, Cambridge, UK) 1:100 dilution incubated at RT for 1hr. Sections were cover-slipped after DAPI (Invitrogen D1306, Carlsbad, California, USA) staining at 1:1000 dilution at RT for 20 min.
Seven-color images were acquired on a Nikon A1R spectral deconvolution confocal microscope (Nikon, Toyko, Japan). Using a 32-channel A1-GaAsP-detector unit, fluorochrome emission can be split up in up to 32 bands from 400 to 750 nm with a spectral discrimination of 10 or 20 nm bandwidth when excited by a new laser-box with 4 solid state lasers 405, 488, 561 and 640 nm. The acquisition signals were then clearly and reliably distinguished by a process called “spectral unmixing” which is implemented in the NIS-Elements image acquisition and analysis software.
Flow cytometry
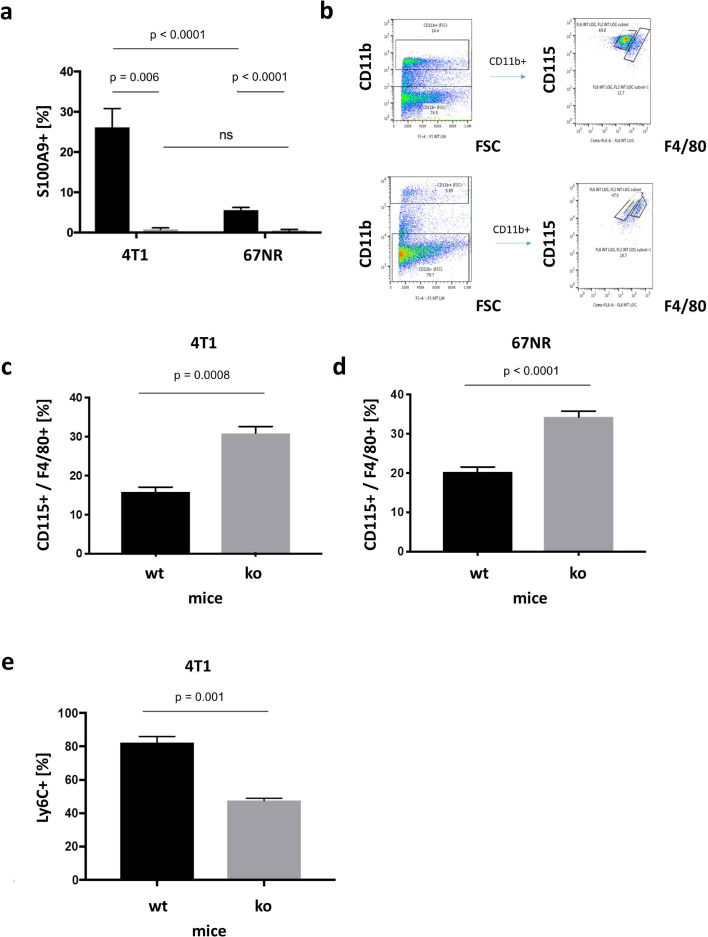
For preparation of flow cytometry, single-cell suspensions from tumor tissue were stained for S100A9, CD 115, CD 11b, Ly6C and F4/80 (for detailed information and corresponding isotype controls see Supplementary Table S3). Cell debris was excluded from analysis by filtering for size and granularity. Flow cytometry measurements were carried out using a FACSCalibur system (Becton Dickinson, Franklin Lakes, New Jersey) and analyzed via FlowJo software (FlowJo LLC, Ashland, Oregon). The data are presented as event frequencies. To minimize non-specific staining data were reduced by the individual isotype controls (for gating strategy see Fig. 3b).
Fig. 3.
Ex vivo analyses via flow cytometry. (a) Flow cytometry of harvested tumor tissue of 4T1 and 67NR tumors in wildtype (black) and S100A9 knockout (grey) mice. The percentage of S100A9 positive cells was significantly higher in wildtype than in S100A9 ko mice. While in wildtype mice, the amount of S100A9 positive cells was significantly higher in 4T1 than compared with 67NR tumors, there were no significant differences in S100A9 knockout mice. (b) Gating strategy for two exemplary separations between CD115 (monocytic) from F4/80 (macrophages) positive cells out of the CD11b positive population. (c) In 4T1 tumors the percentage of CD115 and F4/80 positive cells was significantly lower in wildtype mice as compared with S100A9 ko mice; (d) a similar ratio also existed for 67NR tumors. (e) In 4T1 tumors the percentage of Ly6C positive cells was significantly higher in wildtype than compared with ko mice.
Statistical analysis
Data were analyzed using an unpaired student´s t test for normally distributed values (ex vivo analyses for tumor cell characterization) as well as Mann–Whitney–Wilcoxon test or a one-way ANOVA with Bonferroni post correction for in vivo experiments. Analyses were performed using the GraphPad Prism Software (version 8.4.3; GraphPad Software Inc., San Diego, California). A p-value of 0.05 or lower was considered significant.
Results
Source and characterization of S100A8/S100A9
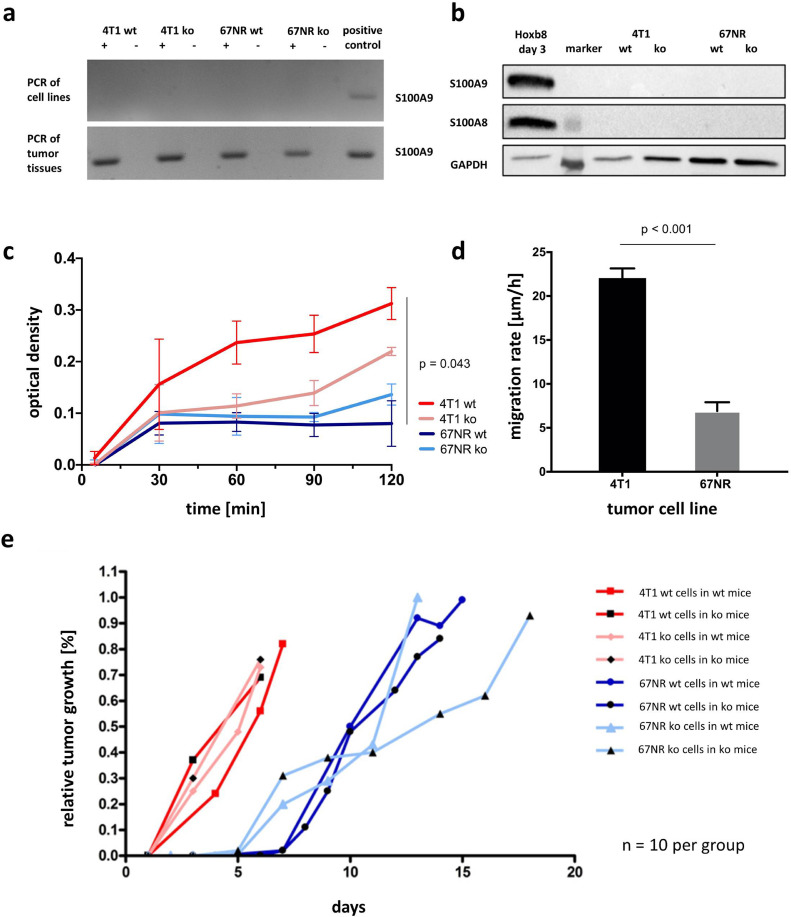
PCR of tumor cell lysates revealed the absence of S100A9 in all four tumor cell lines – wildtype and ko cells – for both tumor entities (Fig. 1a, upper row). However, PCR from orthotopical breast tumors showed presence of S100A9 in the respective tissue samples (Fig. 1a, lower row), suggesting a localization of S100A9 positive cells in the TME. Further analysis of the TME was carried out by flow cytometry (see below).
Fig. 1.
Characterization of wildtype and S100A9 knockout tumor cells. (a) PCR of tumor cell lysates (upper row) and harvested tumor tissue deriving from orthotopically grown breast tumors (lower row). (b) Western Blot of S100A8 and S100A9 regarding the four different tumor cell lines, Hoxb8 cells as positive control. (c) Adhesion/optical density over time as comparison of the four different cell lines. Optical density of 4T1 wt tumor cells is significantly higher as compared to the lower malignant 67NR wt tumor cells. (d) Migration rate comparing the wildtype tumor cells. 4T1 cells have a significantly higher migration rate as compared with 67NR. (e) Relative tumor growth over 20 days revealed no differences within the 4T1 or the 67NR system (for both wt and ko tumor cells in wt and ko mice each), but tumor growth of all 4T1 tumors was faster as compared with 67NR tumors.
Neither S100A8 nor S100A9 expression could be detected for any of the tested cell lines (4T1 wt, 4T1 ko, 67NR wt, 67NR ko) by western blotting (Fig. 1b).
In order to exclude a possible influence of the S100A9 knockout on tumor cell characteristis reflecting the different degrees of maligancy of 4T1 and 67NR tumors, tests on cell adhesion and migration were carried out. With regards to adhesion (Fig. 1c), there was no significant difference between wildtype and S100A9 knockout tumor cells within the same tumor cell line. 4T1 wt/ko and 67NR wt/ko cells did however differ regarding their adhesion properties as well as the (undirected) migration rate (Fig. 1d; 4T1: 22.05 µm/h vs. 67NR: 6.90 µm/h, p < 0.001; n = 3 each). However, compared to 67NR, the more invasive 4T1 cells showed stronger adhesion and a significantly higher migration rate as a correlate to the higher degree of malignancy.
Regarding the relative tumor growth over 20 days, no differences were observed between wildtype or S100A9 knockout for both tumor cells and mice within the same tumor cell system (Fig. 1e). However, 4T1 tumors grew faster as compared to 67NR tumors, reflecting their higher malignant potential.
S100A9-specific in vivo imaging
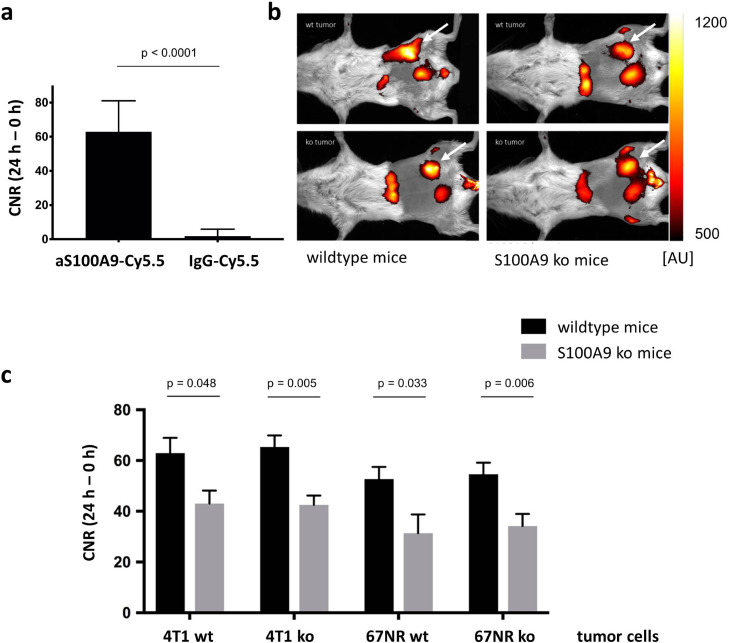
Anti-S100A9-Cy5.5 had significantly higher calculated contrast-to-noise-ratios as compared to rabIgG-Cy5.5 as a control for unspecific label distribution in 4T1 wildtype tumors in vivo (62.92, n = 9 vs. 1.83, n = 6, p < 0.0001, Fig. 2a). Therefore, the S100A9-driven in vivo imaging was assumed to be specific for the chosen target S100A9.
Fig. 2.
S100A9-specific in vivo imaging. (a) Evaluation of the probe´s specificity. The signal for anti-S100A9-Cy5.5 was significantly higher as compared with the unspecific binding IgG-Cy5.5. (b) Exemplary overlay images of fluorescence signals and white light images for anatomic orientation in 4T1 tumors 24 h after probe injection (tumor region highlighted with white arrows): As the scaling indicates, in wt mice both the wt and ko tumors had higher fluorescence intensities as in S100A9 ko mice. Tumor signals can be identified within the images lateral to the urinary bladder. In addition, a non-specific uptake of the probe could be detected in the liver. (c) Contrast-to-noise ratios of S100A9-specific imaging comparing the 8 different imaging groups. In S100A9 ko mice CNR were significantly reduced for wt and S100A9 ko tumors. No significant differences were detected for 4T1 ko and 67NR ko cells as compared to wildtype cells. In all subgroups, CNR were higher in 4T1 as compared to 67NR tumors.
In all 8 imaging groups (as indicated in Table 1) knockout of S100A9 in tumor cells did not result in reduced calculated CNRs for anti-S100A9-Cy5.5 in vivo (62.9 (4T1 wt) vs. 65.33 (4T1 ko), p = 0.759, and 52.73 (67NR wt) vs. 54.61 (67NR ko), p = 0.817). Thus, the absence of S100A9 in the tumor cells had no effect on the presence of the protein complex within the tumor microenvironment. However, the specific fluorescence signals were significantly higher in wildtype as compared to S100A9−/− mice (4T1 wt tumor cells: 62.92 vs. 43.02, p = 0.048; 4T1 ko tumor cells: 65.33 vs. 42.51, p = 0.005; 67NR wt tumor cells: 52.73 vs. 27.26, p = 0.033; 67NR ko tumor cells: 54.61 vs. 34.19, p = 0.006). Thus, in wildtype mice, a significantly increased presence of S100A9 within the TME was found, which in this respect presumably resulted from immigrated immune cells. In contrast to this, no S100A9 was present in the TME of S100A9−/− mice, which was reflected by the lower CNRs.
In principle, a relatively high background signal has been detected in S100A9−/− mice as described in previous studies due to vascularity and increased Fc receptor affinity [16]. Overall, CNRs were higher in 4T1 as compared to 67NR tumors (Fig. 2b,c), corresponding with their higher malignant potential.
Effects of S100A8/S100A9 on the TME
Flow cytometry of harvested tumor tissue confirmed the results of the in vivo imaging (Fig. 3a). The percentage of S100A9 positive monocytic cells of 4T1 tumors was significantly higher in wildtype mice as compared to S100A9 ko mice (26.1 %, n = 3 vs. 0.77 %, n = 3, p = 0.006). Similar conditions were seen in 67NR tumors (wt mice: 5.58 %, n = 7 vs. ko mice: 0.49 %, n= 7, p < 0.0001). While the number of S100A9 positive cells in wildtype mice was significantly higher for 4T1 tumors than for 67NR (p < 0.0001), there were no significant differences between both tumors in S100A9 ko mice (p = 0.61). Thus, these ex vivo measurements reflect the data from S100A9-specific in vivo imaging. With regard to the further differentiation of monocytic cells, the percentage of CD115 [26] and F4/80 positive cells from the group of CD11b positive cells (for exemplary gating strategy see Fig. 3b) in 4T1 tumors was significantly lower in wild-type than in knockout mice (15.8% vs. 30.9% n = 3 (ko), n = 4 (wt), p = 0.0008, Fig. 3c). Similarly, in 67NR tumors, implanted in S100A9 knockout mice, CD115 and F4/80 positive cells were significantly higher in knockout than in wildtype mice (34.26 % vs. 20.3 %, n = 5 each, p < 0.0001, Fig. 3d). Supporting this observation, the number of Ly6C positive cells from the group of CD11b positive monocytes, indicating CD115(pos)Gr-1(pos) monocyte precursor of resident tissue macrophages [26], which is the immunosuppressive population, was significantly higher in wildtype mice as compared to S100A9 knockout mice after implantation of 4T1 tumors (82.2 % vs. 47.53 %, n = 3 each, p = 0.0011, Fig. 3e).
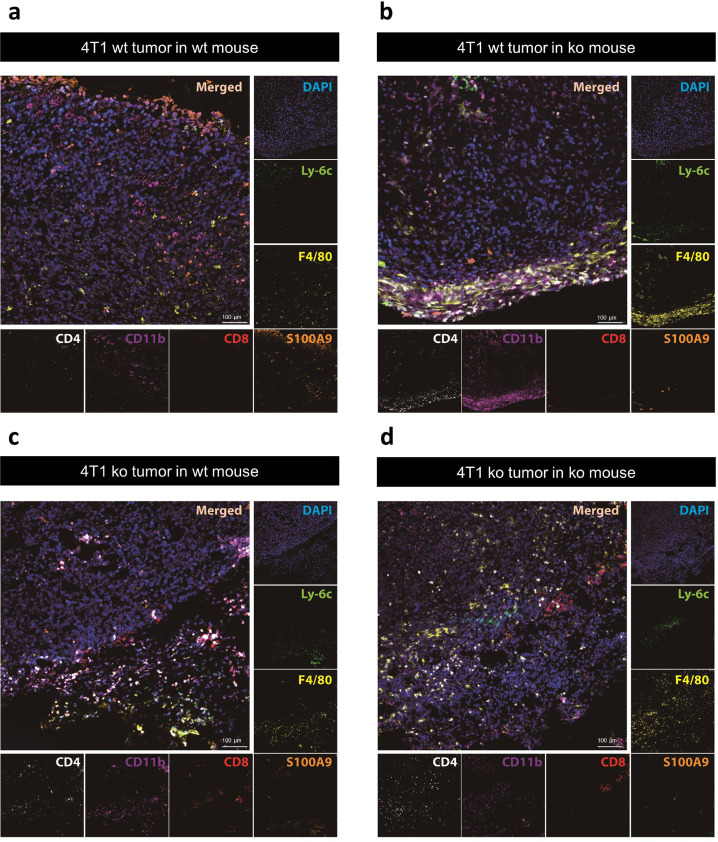
Using 7-channel staining a broad spectrum of immune cells within the TME could be visualized (Fig. 4). Representative tumor sections, immunohistochemistry revealed different compositions and distributions of immune cells within the TME, specific for the respective combinations of implanted wt and ko tumors in wt or S100A9-ko mice. Analogous to the number of F4/80 positive cells already identified via flow cytometry, F4/80 positive cells (yellow) could also be detected via immunohistochemistry especially in S100A9 ko mice (Fig. 4b,d). In addition to a significant overall number of monocytic cells (CD11b positive, purple, Fig. 4a-d), CD4 positive T helper cells (white) also seem to be more abundant within the TME of the exemplary tumor preparations than the CD8 positive cytotoxic T cells (red).
Fig. 4.
Immunohistochemistry of exemplary 4T1 tumors. Representative tumor sections after 7-channel staining of DAPI (blue), Ly-6C (green), F4/80 (yellow), S100A9 (orange), CD8 (red), CD11b (purple) and CD4 (white) with correlating merged images. Sections of implanted 4T1 wt tumors in wt (a) and S100A9 ko (b) mice as well as 4T1 ko tumors in wt (c) and S100A9 ko (d) mice.
Alterations of S100A9-driven TME activity after immunomodulating therapy
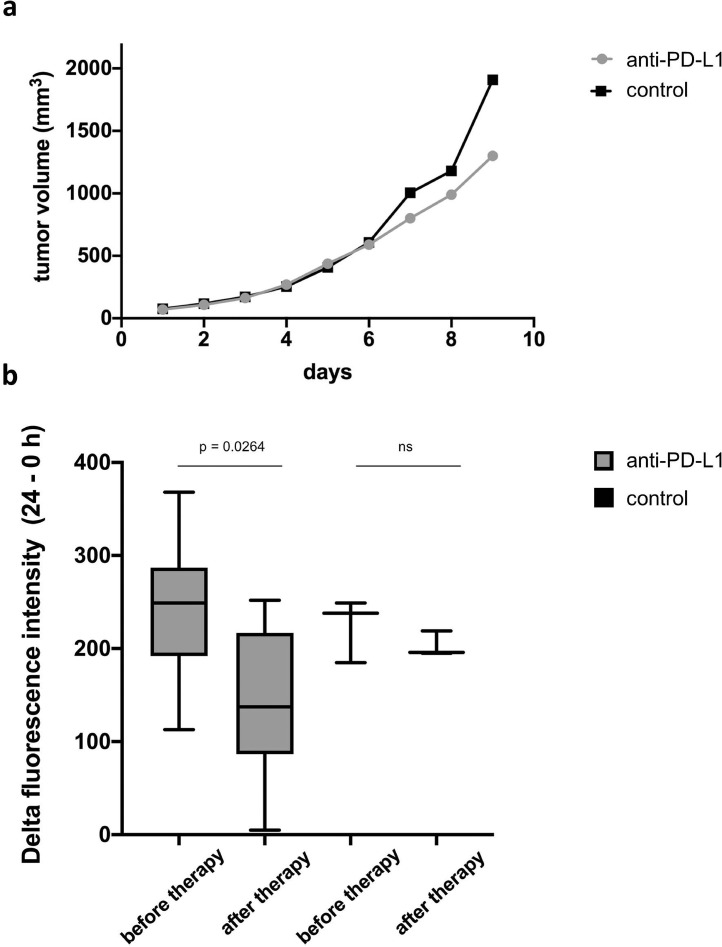
The tumor growth was not inhibited by anti-PD-L1 treatment as compared to IgG isotype control over the study period of 9 days (Fig. 5a). S100A9-specific in vivo imaging, however revealed a significant reduction of the fluorescence intensity under anti-PD-L1 treatment as early as one week after the treatment when compared to the first imaging prior to therapy (224.29 AU vs. 124.57 AU, p = 0.0264, n= 7). No significant changes of the fluorescence intensities were detected for the control group receiving an IgG isotype control (224.0 AU vs. 203.33 AU, p = 0.386, n = 3, Fig. 5b).
Fig. 5.
In vivo imaging of 4T1 tumors under anti-PD-L1 therapy. (a) Measured tumor volumes over a period of 9 days. In the early course of treatment, no significant differences between the treatment groups receiving anti-PD-L1 (grey) and the control group (IgG-isotype control, black) were measurable. (b) S100A9-specific in vivo imaging revealed a significant decrease of fluorescence intensities under anti-PD-L1 treatment compared to the first imaging before therapy start, whereas no significant changes were detected in the control group.
It was therefore possible to detect significant changes in the S100A8 / S100A9 presence within the TME even before macroscopic changes with regard to the tumor size were visible as a possible therapy response. Since the presence of the protein complex in the TME is associated with poor prognosis, in vivo imaging may therefore show a possible therapeutic response at an early time point during tumor growth by changes of the TME activity.
Thus, as in vivo imaging reflects the local S100A9 activity in the tumor due to its short half-life [27], it is not only suitable as an imaging biomarker for assessing tumor aggressiveness (in this study using the example of 4T1 and 67NR tumors), but also for detection of early changes under immune checkpoint therapy.
Discussion
The protein complex S100A8/S100A9 is a major regulator of innate immunity and in this context crucially involved not just in regular inflammatory conditions but in tumor-immune interaction as well [28]. S100A9 regulates the inhibition of dendritic cells as well as the accumulation of MDSC and therefore promotes a pro-tumorigenic shift of the TME [29]. Furthermore, the protein complex is involved in reactivation of dormant tumor cells and tumor recurrence after successful treatment of primary tumors [30]. Thus, S100A9-dependent immune-suppressive activity of macrophages results in a negative clinical outcome in cancer patients [6].
We have shown that elevated S100A8/S100A9-levels in tumor lesions reflect local invasiveness and precede accelerated growth [16] while local S100A8/S100A9 expression in distant tissue such as the lungs indicates premetastatic niche establishment [17].
Being representative for changes of the immune cell environment in malignant lesions as well as in target tissues of metastasis, S100A8/S100A9 could provide a long sought-after possibility to monitor tumor-immune interaction using either blood tests or target-specific imaging.
To facilitate this however, the so far elusive, specific role of S100A8/S100A9 and the source within the TME needs to be elucidated. While our own results suggest that tumor cells, although in some cases expressing S100A8/S100A9 on RNA level, do not produce the protein heterodimer [17], other groups report differently for gastric cancer [9, 10], lung cancer [11], hepatocellular [12] and colorectal carcinoma [13] as well as for breast cancer [14]. However, most studies do not further differentiate an increased expression in the TME with regard to the exact cellular origin of S100A9.
Using CRISPR/cas9 technology, we created knock-out variants of well-established breast cancer cell lines that could be used in syngeneic model systems, again available as a control with a respective knock-out. The cross-over implantation studies described in this paper enabled the evaluation of S100A8/S100A9 effects with respect to the protein source.
The cross-over implantation studies presented here with S100A9-knock out tumors in wild type and S100A9-knock out mice as well as wild type tumors in both murine systems found little influence of the primary tumor's genetic equipment on the S100A8/S100A9 levels in tumor tissue. In knock-out mice, even the implantation of wild type tumors, equipped with S100A8/S100A9 DNA and RNA, no protein expression could be detected. It can be concluded that S100A9 in the TME originates from cells, infiltrating the tumor microenvironment, not from the tumor cells themselves. Thus, since S100A8 / S100A9 reflects the TME activity, it can serve as an activity marker regardless of the tumor model.
The knock-out tumor cells were not otherwise compromised, as far as the testing we conducted showed. S100A9-blockade or knock-out in monocytes or other migratory cells could be shown to lead to a loss in directional migration [31] as the heterodimer is involved in the necessary tubulin turnover. For the local functionality of tumor cells however, no apparent diversion could be detected.
In the TME, S100A8/S100A9 promotes alternatively activated M2-leaning macrophages and immature monocytes as MDSC dominate over mature, differentiated macrophages [6]. The dominant, immature monocyte population under regulation of MDSC support tumor development by provision of growth factors and promotion of angiogenesis [32]. In parallel, MDSC drive the accumulation of regulatory CD8 positive T cells which suppress anti-tumor immunity manifested in CD4 positive cells and NK cells [33].
S100A8/S100A9 plays a crucial role in MDSC activation and accumulation and serves as chemokine for immature monocytes [34, 35].
The differences in TME composition as observed in the cross-over implantation studies, mainly with regards to the maturity and differentiation of the monocytic population and the differentiation of the T cell population further support S100A8/S100A9 as promoter of a tumor-supportive TME as it maintains the dominance of undifferentiated, tumor-supportive immune cells. In knock-out mice, lacking S100A8/S100A9 in the TME, the number of pro-tumorigenic monocytes and regulatory T cells decreases (Fig. 3), the number of anti-tumor T cells and fully differentiated macrophages increases as the regulating, differentiation-inhibiting effect of S100A8/S100A9 is missing.
In the last decade treatment with T cell targeted immunomodulatory drugs blocking the immune checkpoints CTLA-4, PD1 and PD-L1 represented a great achievement in cancer therapy and are now well esthablished and safely applied in the clinical context. As single agents or in combination with chemotherapy they have been approved as first or second line therapies for about 50 human cancer types [36] such as melanoma, renal cell carcinoma, non-small-cell lung cancer and urothel carcinoma and aim at reversal of the pro-tumorigenic shifts in immunity. In patients with metastatic melanoma, the presence of S100A9-positive monocytes in tumor tissue has been associated with poor response to anti-PD-1 antibody treatment [6]. Established diagnostics fail to provide early measures for response to this kind of treatment [19]. The measurement of tumor size, indicative of response to conventional chemotherapy proved inadequate as the effect of immune-modulatory tumor therapy is often delayed and might not result in a measurable change of tumor size at all. Diagnostic tools for dynamic, early assessment of TME activity are urgently needed [37].
Especially in advanced tumor stages, regularly being referred to immunotherapy, interlesional heterogeneity might lead to different degrees of response to therapy in different tumor lesions. While a blood test, screening for specific serum markers or cellular components might be indicative of therapy response or failure, a site-specific estimation of therapy response, to guide further therapy or biopsy could only be achieved through whole body imaging of features or markers, reflective of tumor-immune interaction. The local level of S100A8/S100A9 is highly sensitive to changes in the TME as induced by anti-PD-L1 treatment. Here, we present non-invasive S100A9-driven imaging that allows to display changes of myeloid activity in the TME at a very early time point after immunomodulatory treatment, even before changes in tumor growth are detectable. Therefore, it might represent an imaging biomarker for early treatment response as well as the evaluation of the consecutive tumor development, above meeting the requirements for imaging in modern oncology and could therefore overcome the limitations of the clinically established modalities described.
Regarding a clinical translation of S100A9-specific imaging, however, the full-length antibodies used to address the target structures are very large at 10-12 kDa [38] and are therefore eliminated hepatically. In the available preclinical work, a measurement time of around 24 hours after intravenous tracer injection turned out to be ideal for specific imaging [16]. Therefore, for a possible translation, either further developments regarding the use of radioisotopes with a shorter half-life such as zirconium-89-based methods or the use of smaller particles such as antibody fragments or nanoparticles are to be discussed.
Conclusions
Our results confirm a secretion of S100A8/S100A9 by components of the TME in the 4T1/67NR breast cancer model and not apparently by tumor cells themselves. In vivo imaging of S100A9 is therefore reflective of myeloid activity within the TME. Moreover, we show here, that immunomodulatory therapy effects on the TME activity and composition can be measured and visualized as early as several days after treatment. Using S100A9 as an imaging biomarker might therefore meet the requirements of immunomodulatory therapeutic approaches in modern oncology. With regard to clinical translation, further studies evaluating the use of smaller probes and PET / SPECT-based applications are desirable.
Credit authorship contribution statement
Anne Helfen: Conceptualization, Funding acquisition, Investigation, Visualization, Writing – original draft; Jan Rieß: Investigation, Data curation, Software; Olesja Fehler: Funding acquisition, Methodology; Miriam Stölting: Formal analysis, Methodology; Zhengwen An: Investigation; Vanessa Kocman: Investigation; Annika Schnepel: Data curation; Christiane Geyer: Validation; Mirjam Gerwing: Investigation; Max Masthoff: Validation; Thomas Vogl: Resources, Formal analysis; Carsten Höltke: Validation; Johannes Roth: Conceptualization; Resources; Tony Ng: Supervision; Moritz Wildgruber: Project administration, Resources; Michel Eisenblätter: Project administration, Supervision, Writing – original draft.
Financial support
This research was funded by Cells in Motion Cluster of Excellence/Deutsche Forschungsgemeinschaft (grant number PP-2017-01).
Informed consent statement
Not applicable.
Data availability statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Declaration of Competing Interests
The authors have no relevant financial or non-financial interests to disclose.
Acknowledgments
We thank Klaudia Niepagenkemper, Irina Arnhold, Heike Berheide and Claudia Terwesten-Solé for excellent technical support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100792.
Appendix. Supplementary materials
References
- 1.Ganesh K., Massague J. Targeting metastatic cancer. Nat Med. 2021;27:34–44. doi: 10.1038/s41591-020-01195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gebhardt C., Nemeth J., Angel P., Hess J. S100A8 and S100A9 in inflammation and cancer. Biochem Pharmacol. 2006;72:1622–1631. doi: 10.1016/j.bcp.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Vogl T., Tenbrock K., Ludwig S., Leukert N., Ehrhardt C., van Zoelen M.A., et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13:1042–1049. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 4.Kallberg E., Vogl T., Liberg D., Olsson A., Bjork P., Wikstrom P., et al. S100A9 interaction with TLR4 promotes tumor growth. PLoS One. 2012;7:e34207. doi: 10.1371/journal.pone.0034207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrchen J.M., Sunderkotter C., Foell D., Vogl T., Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 6.Kwak T., Wang F., Deng H., Condamine T., Kumar V., Perego M., et al. Distinct Populations of Immune-Suppressive Macrophages Differentiate from Monocytic Myeloid-Derived Suppressor Cells in. Cancer. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roth J., Vogl T., Sorg C., Sunderkotter C. Phagocyte-specific S100 proteins: a novel group of proinflammatory molecules. Trends in immunology. 2003;24:155–158. doi: 10.1016/s1471-4906(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 8.Arai K., Takano S., Teratani T., Ito Y., Yamada T., Nozawa R. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Current cancer drug targets. 2008;8:243–252. doi: 10.2174/156800908784533445. [DOI] [PubMed] [Google Scholar]
- 9.El-Rifai W., Moskaluk C.A., Abdrabbo M.K., Harper J., Yoshida C., Riggins G.J., et al. Gastric cancers overexpress S100A calcium-binding proteins. Cancer Res. 2002;62:6823–6826. [PubMed] [Google Scholar]
- 10.Zhao Z., Zhang C., Zhao Q. S100A9 as a novel diagnostic and prognostic biomarker in human gastric cancer. Scand J Gastroenterol. 2020;55:338–346. doi: 10.1080/00365521.2020.1737883. [DOI] [PubMed] [Google Scholar]
- 11.Arai K., Teratani T., Nozawa R., Yamada T. Immunohistochemical investigation of S100A9 expression in pulmonary adenocarcinoma: S100A9 expression is associated with tumor differentiation. Oncol Rep. 2001;8:591–596. doi: 10.3892/or.8.3.591. [DOI] [PubMed] [Google Scholar]
- 12.Meng J., Gu F., Fang H., Qu B. Elevated Serum S100A9 Indicated Poor Prognosis in Hepatocellular Carcinoma after Curative Resection. J Cancer. 2019;10:408–415. doi: 10.7150/jca.28409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou M., Li M., Liang X., Zhang Y., Huang H., Feng Y., et al. The Significance of Serum S100A9 and TNC Levels as Biomarkers in Colorectal Cancer. J Cancer. 2019;10:5315–5323. doi: 10.7150/jca.31267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cross S.S., Hamdy F.C., Deloulme J.C., Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 15.Lim S.Y., Yuzhalin A.E., Gordon-Weeks A.N., Muschel RJ. Tumor-infiltrating monocytes/macrophages promote tumor invasion and migration by upregulating S100A8 and S100A9 expression in cancer cells. Oncogene. 2016;35:5735–5745. doi: 10.1038/onc.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker A., Große Hokamp N., Zenker S., Flores-Borja F., Barzcyk K., Varga G., et al. Optical in vivo imaging of the alarmin S100A9 in tumor lesions allows for estimation of the individual malignant potential by evaluation of tumor-host cell interaction. J Nucl Med. 2015;56:450–456. doi: 10.2967/jnumed.114.146688. [DOI] [PubMed] [Google Scholar]
- 17.Eisenblaetter M., Flores-Borja F., Lee J.J., Wefers C., Smith H., Hueting R., et al. Visualization of Tumor-Immune Interaction - Target-Specific Imaging of S100A8/A9 Reveals Pre-Metastatic Niche Establishment. Theranostics. 2017;7:2392–2401. doi: 10.7150/thno.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke M., Choksawangkarn W., Edwards N., Ostrand-Rosenberg S., Fenselau C. Exosomes from myeloid-derived suppressor cells carry biologically active proteins. J Proteome Res. 2014;13:836–843. doi: 10.1021/pr400879c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerwing M., Herrmann K., Helfen A., Schliemann C., Berdel W.E., Eisenblatter M., et al. The beginning of the end for conventional RECIST - novel therapies require novel imaging approaches. Nat Rev Clin Oncol. 2019;16:442–458. doi: 10.1038/s41571-019-0169-5. [DOI] [PubMed] [Google Scholar]
- 20.Gran S., Honold L., Fehler O., Zenker S., Eligehausen S., Kuhlmann M.T., et al. Imaging, myeloid precursor immortalization, and genome editing for defining mechanisms of leukocyte recruitment in vivo. Theranostics. 2018;8:2407–2423. doi: 10.7150/thno.23632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Humphries MJ. Cell adhesion assays. Mol Biotechnol. 2001;18:57–61. doi: 10.1385/MB:18:1:57. [DOI] [PubMed] [Google Scholar]
- 22.Vogl T., Eisenblätter M., Völler T., Zenker S., Hermann S., van Lent P., et al. Alarmin S100A8/S100A9 as a biomarker for molecular imaging of local inflammatory activity. Nat Commun. 2014;5:4593. doi: 10.1038/ncomms5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marshall M.V., Draney D., Sevick-Muraca E.M., Olive DM. Single-dose intravenous toxicity study of IRDye 800CW in Sprague-Dawley rats. Mol Imaging Biol. 2010;12:583–594. doi: 10.1007/s11307-010-0317-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pei J., Juniper G., van den Berg N.S., Nisho N., Broadt T., Welch A.R., et al. Safety and Stability of Antibody-Dye Conjugate in Optical Molecular Imaging. Mol Imaging Biol. 2021;23:109–116. doi: 10.1007/s11307-020-01536-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim J., Jeong Ryu S., Oh K., Ju J.M., Yeong Jeon J., Nam G., et al. Memory programming in CD8(+) T-cell differentiation is intrinsic and is not determined by CD4 help. Nat Commun. 2015;6:7994. doi: 10.1038/ncomms8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacDonald K.P., Palmer J.S., Cronau S., Seppanen E., Olver S., Raffelt N.C., et al. An antibody against the colony-stimulating factor 1 receptor depletes the resident subset of monocytes and tissue- and tumor-associated macrophages but does not inhibit inflammation. Blood. 2010;116:3955–3963. doi: 10.1182/blood-2010-02-266296. [DOI] [PubMed] [Google Scholar]
- 27.Foell D., Roth J. Proinflammatory S100 proteins in arthritis and autoimmune disease. Arthritis Rheum. 2004;50:3762–3771. doi: 10.1002/art.20631. [DOI] [PubMed] [Google Scholar]
- 28.Veglia F., Perego M., Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol. 2018;19:108–119. doi: 10.1038/s41590-017-0022-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng P., Corzo C.A., Luetteke N., Yu B., Nagaraj S., Bui M.M., et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perego M., Tyurin V.A., Tyurina Y.Y., Yellets J., Nacarelli T., Lin C., et al. Reactivation of dormant tumor cells by modified lipids derived from stress-activated neutrophils. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abb5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raquil M.A., Anceriz N., Rouleau P., Tessier PA. Blockade of antimicrobial proteins S100A8 and S100A9 inhibits phagocyte migration to the alveoli in streptococcal pneumonia. J Immunol. 2008;180:3366–3374. doi: 10.4049/jimmunol.180.5.3366. [DOI] [PubMed] [Google Scholar]
- 32.Srikrishna G. S100A8 and S100A9: new insights into their roles in malignancy. J Innate Immun. 2012;4:31–40. doi: 10.1159/000330095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He Y.M., Li X., Perego M., Nefedova Y., Kossenkov A.V., Jensen E.A., et al. Transitory presence of myeloid-derived suppressor cells in neonates is critical for control of inflammation. Nat Med. 2018;24:224–231. doi: 10.1038/nm.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Austermann J., Spiekermann C., Roth J. S100 proteins in rheumatic diseases. Nat Rev Rheumatol. 2018;14:528–541. doi: 10.1038/s41584-018-0058-9. [DOI] [PubMed] [Google Scholar]
- 35.Scalea J.R., Lee Y.S., Davila E., Bromberg JS. Myeloid-Derived Suppressor Cells and Their Potential Application in Transplantation. Transplantation. 2018;102:359–367. doi: 10.1097/TP.0000000000002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robert C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat Commun. 2020;11:3801. doi: 10.1038/s41467-020-17670-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Helfen A., Roth J., Ng T., Eisenblaetter M. In Vivo Imaging of Pro- and Antitumoral Cellular Components of the Tumor Microenvironment. J Nucl Med. 2018;59:183–188. doi: 10.2967/jnumed.117.198952. [DOI] [PubMed] [Google Scholar]
- 38.Ghavami S., Chitayat S., Hashemi M., Eshraghi M., Chazin W.J., Halayko A.J., et al. S100A8/A9: a Janus-faced molecule in cancer therapy and tumorgenesis. Eur J Pharmacol. 2009;625:73–83. doi: 10.1016/j.ejphar.2009.08.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.