Abstract
Increased reactive oxygen species (ROS) and hyperstabilized mutant p53 are common in cancer. Hyperstabilized mutant p53 contributes to its gain of function (GOF) which confers resistance to chemotherapy and radiotherapy. Targeting mutant p53 degradation is a promising cancer therapeutic strategy. We used a small-molecule NSC59984 to explore elimination of mutant p53 in cancer cells, and identified an inducible ROS-ERK2-MDM2 axis as a vulnerability for induction of mutant p53 degradation in cancer cells. NSC59984 treatment promotes a constitutive phosphorylation of ERK2 via ROS in cancer cells. The NSC59984-sustained ERK2 activation is required for MDM2 phosphorylation at serine-166. NSC59984 enhances phosphorylated-MDM2 binding to mutant p53, which leads to mutant p53 ubiquitination and degradation. High cellular ROS increases the efficacy of NSC59984 targeting mutant p53 degradation and antitumor effects. Our data suggest that mutant p53 stabilization has a vulnerability under high ROS cellular conditions, which can be exploited by compounds to target mutant p53 protein degradation through the activation of a ROS-ERK2-MDM2 axis in cancer cells.
Implications:
An inducible ROS-ERK2-MDM2 axis exposes a vulnerability in mutant p53 stabilization and can be exploited by small-molecule compounds to induce mutant p53 degradation for cancer therapy.
Introduction
The tumor suppressor p53 has mutations in over 50% of human cancers. Mutations not only result in loss of wild-type p53 function, but also endow mutant p53 with a gain of function (GOF). Mutant p53 GOF contributes to tumorigenesis, tumor proliferation, cell migration, and drug resistance via regulating gene transcription and chromatin modification (1–4). An emerging literature suggests that removal of mutant p53 suppresses tumor growth and sensitizes cancer cells to chemotherapy (5–7). These studies provide a rationale for depletion of mutant p53 protein by small molecules for cancer therapy. However, in contrast to wild-type p53, mutant p53 is hyperstabilized in tumors due to oncogenic signals and multiple cellular stresses (including oxidative stresses) caused by chemotherapy and radiotherapy (4, 8). The redox system is involved in the stabilization of mutant p53 in cancer cells (9), and mutant p53 further increases cellular ROS by regulating the redox system (10–12). These findings suggest a potential cross-talk between mutant p53 stabilization and reactive oxygen species (ROS) in cancer cells. There is a gap between in our current understanding of oxidative stresses (mostly caused by ROS) and the induction of mutant p53 protein degradation by small molecules in cancer therapy. It is urgently needed to determine whether increased ROS either imposes a barrier on the elimination of mutant p53 protein or renders upon mutant p53 a vulnerability that can be exploited by small molecules to abolish mutant p53 protein as a cancer therapeutic strategy.
Mutant p53 is stabilized in cancer cells due to its inability to induce expression of MDM2, an E3 ubiquitin ligase which forms a negative feedback loop with wild-type p53 protein for degradation. In addition, mutant p53 is protected by protein chaperones, such as HSPs, from protein degradation mediated by MDM2 or C-terminus of Hsc70-interacting protein (CHIP; refs. 13, 14). Few small molecules have been identified to induce mutant p53 degradation (15, 16). One of the strategies for depleting mutant p53 is to induce mutant p53 protein degradation via the ubiquitin-dependent proteasome pathway. For example, geldanamycin (GA), SAHA, and statins inhibit HSP chaperone binding to mutant p53, therefore mutant p53 is degraded by the proteasome through MDM2 (or CHIP)-mediated ubiquitination (5, 17). Exploitation of autophagy is another attractive strategy for eliminating mutant p53. A small-molecule spautin provides an example for depleting mutant p53 protein via lysosome ubiquitination (18). An increase in ROS level has been found in cancer cells treated with most of the mutant p53-destabilizing small molecules, such as GA, statins, and SAHA (19–21). Redox-status of p53 is well known for the stabilization and activation of wild-type p53 (22). The effect of increased ROS and its related signaling on mutant p53 degradation induced by small molecules remains unclear and needs to be investigated.
The extracellular signal-regulated kinases (ERK1/2) are at the center of signaling pathways that regulate cell proliferation and cell death in response to numerous stimuli (23). An altered ERK2 signaling pathway is common in cancer cells. Drugs that inhibit ERK2 pathway signaling have been approved by the FDA for cancer therapy (24). However, by contrast to the physiologic transient ERK2 activation, ERK2 is persistently activated by DNA damage agents (25, 26). Accumulating evidence shows that persistent ERK2 phosphorylation is associated with cell death (26, 27). ERK2 phosphorylates proteins and selectively induces protein degradation (28–31). ERK2 has been found to phosphorylate mutant p53 and MDM2 in cancer cells (32, 33). ERK2-mediated posttranslational modifications of proteins might therefore serve as a basis for designing more efficient strategies to eliminate mutant p53 via protein degradation in cancer cells.
Mutated p53 and accumulation ROS that occur commonly in cancer provide attractive therapeutic targets. NSC59984 is a first-in-class antitumor small molecule which induces restoration of the p53 pathway signaling and degradation of mutant p53 protein (depletion of GOF) in mutant p53-expressing colorectal cancer cells (34). We demonstrate here that NSC59984 exploits intracellular ROS to induce mutant p53 degradation and to restore p53 pathway signaling specifically via an ROS-ERK2-MDM2 axis. Furthermore, the mutant p53-degrading small-molecule NSC59984 synergizes with ROS-generating agents to suppress tumor growth. Thus, therapeutic approaches may take advantage of high ROS to destabilize mutant p53 to improve therapeutic efficacy against tumors.
Materials and Methods
Cell lines
SW480 (RRID:CVCL_0546) and HCT116 (RRID:CVCL_0291) cells which stably express a p53-regulated luciferase reporter were generated in our laboratory in 2003. HT29 (RRID:CVCL_0320), MRC5 (RRID:CVCL_0440), WI-38 (RRID:CVCL_0579), DLD-1 (RRID:CVCL_0248), KM12C (RRID:CVCL_9547), SW620 (RRID:CVCL_0547), BXPC3 (RRID:CVCL_01860), RXF 393 (RRID:CVCL_1673), SNB19 (RRID:CVCL_0535), U251 (RRID:CVCL_0021), T98G (RRID:CVCL_0556), BT549 (RRID:CVCL_1092), and Hop92 (RRID:CVCL_1286) were obtained from ATCC and cultured as recommended. Cells with less than 10 passages since thawing were used in the described experiments. Cells were regularly authenticated by bioluminescence, growth, and morphologic observation. Mycoplasma detection was routinely conducted with a PCR-based test using a Universal Mycoplasma Detection Kit (catalog no. 30-1012K, ATCC). All cell lines tested and used were Mycoplasma negative.
Reagents and antibodies
U0126 (catalog no. 9903) and PD98059 (catalog no. 9900) were purchased from Cell Signaling Technology. N-acetyl-cysteine (NAC, catalog no. A7250), L-buthionine-S, R-sulfoximine (BSO, catalog no. B2515), and EGF (catalog no. E9644) were purchased from Sigma. SB203580 (catalog no. S1076) was purchased from Selleck chemicals. SP600125 (catalog no. sc-200635) was purchased from Santa Cruz Biotechnology. VX-11e (catalog no. A3931) and SCH772984 (catalog no. A3805) were purchased from ApexBio, Z-VAD-FMK (catalog no. FMK001) was purchased from R&D Systems. NSC59984 was synthesized by Provid Pharmaceuticals Inc.
Anti-p53 (DO-1, catalog no. sc-126), anti-Ras (F234, catalog no. sc-30), and anti-MDM2 (SMP14, catalog no. sc-965) were purchased from Santa Cruz Biotechnology. Anti-p21(Ab-1, catalog no. OP64) and anti-Noxa (catalog no. OP180) were purchased from EMD Millipore. Anti-Ki67 (MIB-1, catalog no. IR626) was from DAKO. Anti-p73 (catalog no. A300-126A) was purchased from Bethyl Laboratories. The following antibodies were purchased from Cell Signaling Technology: anti-ERK1/2 (catalog no. 9102S), anti-phospho-ERK1/2 (thr202/tyr204, catalog no. 4370), anti-MEK (catalog no.9122), anti-Phospho-MEK1 (S217/211, catalog no. 9121), anti-Phospho-c-Jun (ser63, catalog no. 9261), anti-c-Jun (catalog no. 9165), anti-GFP (catalog no. 2555), anti-B-Raf (catalog no. 14814), anti-C-Raf (catalog no. 9422), anti-Phospho-MDM2 (ser166, catalog no. 3521), anti-cleaved PARP (catalog no. 9546), anti-RSK1-3 (catalog no. 9355), and anti-Phospho-P90RSK (ser380, catalog no. 12032). Anti-cleaved caspase 3 (catalog no. 559565) was purchased from BD Pharmingen.
siRNA#1 targeting ERK2 (s11138, catalog no. 4390824), siRNA#2 targeting ERK2 (s11139, catalog no. 4390824), siRNA#1 targeting MDM2 (s8628, no. 4390824), siRNA#2 targeting MDM2 (s8630, catalog no. 4390824), siRNA#2 targeting ERK1(s11140, catalog no. 4390824), and siRNA#2 targeting Ras (s7939, catalog no. 4390824) were purchased from Ambion. siRNA#1 targeting ERK1 (catalog no. 6436) were purchased from Cell Signaling Technology. siRNA#1 targeting Ras (catalog no. sc-35731), siRNA targeting C-Raf (catalog no. sc-29462), and siRNAs targeting B-Raf (catalog no. sc-36368) were purchased from Santa Cruz Biotechnology.
Immunoprecipitation
Briefly, 500 μg of cell lysate were incubated with 2 μg of antibody overnight at 4°C, then, mixed with 25 μL of protein A-Sepharose 6MB beads (GE healthcare, catalog no. 17-0469-01). The immunoprecipitated proteins were eluted from protein A-Sepharose beads by boiling with 2× sample buffer (Invitrogen, catalog no. NP0007) and subjected to SDS-PAGE.
Chromatin immunoprecipitation-PCR assay
Chromatin immunoprecipitation (ChIP) was performed according to the protocol of Upstate Biotechnology with slight modifications. Briefly, 2 × 106 cells were fixed with 1% formaldehyde in PBS for 10 minutes to cross-link chromatin, followed with sonication which sheared cross-linked DNA fragments to 200 and 1,000 bp in length. The sonicated chromatins were incubated with 3 μg of anti-p73 antibody (Bethyl Laboratories, catalog no. A300-126A) at 4°C overnight, and 50 μL of packed salmon sperm DNA/protein A-Sepharose beads at 4°C for 3 hours. The precipitated beads were washed with different washing buffers according to the protocol. ChIP-eluted DNA were reversed and extracted with phenol/chloroform. The eluted DNA was analyzed by quantitative Real-Time PCR with the following primers for ChIP (35): Noxa promoter Forward primer: 5′-CAG CGT TTG CAG ATG GTC AA-3′; Noxa promoter Reverse primer: 5′-CCC CGA AAT TAC TTC CTT ACA AAA-3′. P21 promoter Forward primer: 5′-GTG GCT CTG ATT GGC TTT CTG-3; P21 promoter Reverse primer: 5′-CTG AAA ACA GGC AGC CCA AG-3′.
Western blot analysis
Equal amount of cell lysates in RIPA buffer (Sigma-Aldrich, catalog no. R0278) were electrophoresed through 4%–12% SDS-PAGE (Thermo Fisher Scientific, catalog no. NP0323) then transferred to polyvinylidene difluoride (PVDF) membranes. The primary antibodies indicated in the figures were incubated with the transferred PVDF in blocking buffer at 4°C overnight. Antibody binding was detected on PVDF with appropriate IR Dye-secondary antibodies (LI-COR Biosciences) by the ODYSSEY infrared imaging system or with ECL Reagent (Thermo Fisher Scientific, catalog no. 32106) chemiluminescence reaction with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (Thermo Fisher Scientific, catalog no. no. 31460 for Goad anti-rabbit IgG and catalog no. 31430 for Goat anti-mouse IgG) by the Syngene imaging system.
Knockdown of gene expression by siRNA transfection
Cells were transfected with siRNA using lipofectamine 2000 (Life Technologies, catalog no. 11668-027) or lipofectamine RNAiMAX (Life Technologies, catalog no. 13778075) as described in the protocol. Forty-eight hours after transfection, cells were further treated as indicated in the figures.
FACS analysis
Briefly, cells were fixed with 70% ethanol and stained with propidium iodide, then subjected to analysis by an Epics Elite flow cytometer to measure the DNA content of the stained cells.
ROS assay
Cells seeded on 96-well plate were treated with the compounds as described in the figures for 16 hours. Cellular ROS was detected using 2′,7′-Dichlorofluorescin diacetate (Sigma-Aldrich, catalog no. D6883) on 96-well plate by an IVIS imager.
CellTiter-Glo luminescent cell viability assay
Cells were seeded at 4,000 cells/well on 96-well plates and treated as indicated. Cells were mixed with an equal volume of CellTiter-Glo reagents (Promega, catalog no. G7572) following the manufacturer's protocol, and bioluminescence imaging was performed using an IVIS imager.
Colony formation assay
A total of 250 cells/well were cultured in 12-well plates and treated with different compounds for 3 days, then cancer cells were cultured with drug-free complete medium for 2 weeks with fresh medium being changed every 3 days. Cells were fixed with 10% formalin and stained with 0.05% crystal violet at the end of the experiments.
In vivo experiments
All animal experiments were approved by the Institutional Animal Care and Use Committee of Brown University (Providence, RI). HT29 xenograft tumor were generated in CRL nude mice (female, 4–6 weeks old) as reported previously (34). Briefly, 2 × 106 cells were implanted with the same volume of Matrigel (Thermo Fisher Scientific, catalog no. 354234) subcutaneously in the flanks of nude mice. Three days later, mice were treated with NSC59984 (75 mg/kg) every 3 days with or without BSO (100 mg/kg) twice a day via intraperitoneally injection for 2 weeks.
IHC staining
Briefly, the tumor samples were fixed in formalin and embedded in paraffin, and 8 μm sections were sliced. The tumor sections were mounted on slides and hydrated. The sections were then incubated with antibodies mentioned in the figures, and stained with 3,3′-diaminobenzidine (Thermo Fisher Scientific, catalog no. NC9276270) after adding HRP-conjugated secondary antibodies (Vector Laboratories, catalog no. MP-7401-15 or MP-7402). At least 32,000 tumor cells were screened for calculating H-Score of cleaved-caspase 3 and Ki67 by VECTRA 2.0 Automated Quantitative Pathology Imaging system and Inform 2.0 software.
Statistical analysis
All results were obtained from triplicate treatments, unless other indicated. Statistical analyses were performed (Student t test). Statistical significances were determined by P < 0.05. Combination indices were calculated using the Chou-Talalay method with CompuSyn software.
The data generated in this study are available within the article and its Supplementary Data files.
Results
ROS is required for NSC59984 to induce sustained phosphorylation of ERK1/2 in cancer cells
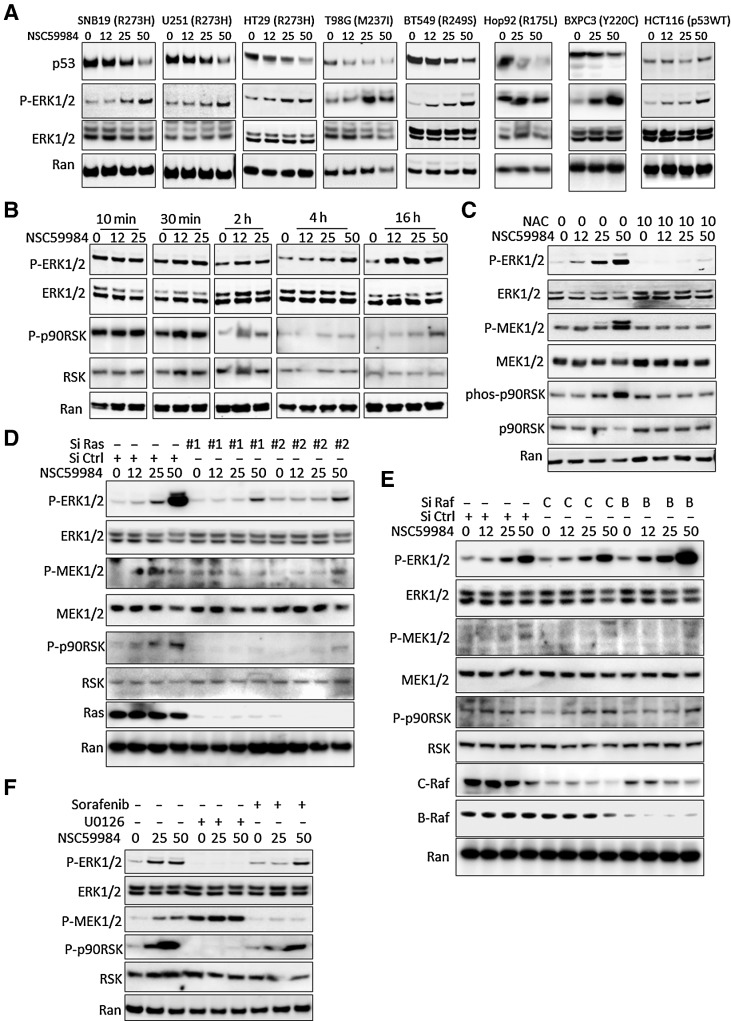
ERK1/2 are protein-serine/threonine kinases that play important roles in regulating cell proliferation and cell death in response to numerous stimuli including ROS (23). ERK1/2 have been found to selectively induce protein degradation (31). We sought to investigate whether ERK1/2 signaling may be involved in the mechanism of action of NSC59984. We detected a sustained ERK1/2 phosphorylation in different types of cancer cells regardless of p53 status in a dose-dependent manner at 16 hours of continuous NSC59984 treatment (Fig. 1A), but not in normal human fibroblast cells, MRC-5 and WI-38 (Supplementary Fig. S1). NSC59984 induced ERK1/2 phosphorylation as early as 30 minutes, and the phosphorylation gradually increased in a dose- and time-dependent manner in SW480 cancer cells (Fig. 1B). The NSC59984-sustained ERK1/2 phosphorylation was suppressed by N-acetyl-cysteine (NAC) in the cancer cells (Fig. 1C). NAC treatment also abrogated the effects of NSC59984 on the phosphorylation of MEK1/2, a component upstream of ERK1/2 in the cancer cells (Fig. 1C). NAC is a scavenger of ROS, and reduced ROS products in cancer cells (Supplementary Fig. S2A). Taken together, these results suggest that ROS is required for the NSC59884-sustained ERK1/2 phosphorylation.
Figure 1.
NSC59984 constitutively induces ERK1/2 phosphorylation. A, The phosphorylation of ERK1/2 in different cancer cells. Cancer cells were treated with NSC59984 (μmol/L) for 16 hours. B, The phosphorylation of ERK1/2 in SW480 cells treated with NSC59984 (μmol/L) in a dose and time course. C, The phosphorylation of ERK1/2 in SW480 cancer cells treated with NSC59984 (μmol/L) and NAC (mmol/L) for 16 hours. D, The phosphorylation of ERK1/2 in SW480 cancer cells with knockdown of Ras. SW480 cells were transfected with siRNAs (siRNA #1 and #2) to knockdown Ras, followed with NSC59984 (μmol/L) treatment for 16 hours. E, The phosphorylation of ERK1/2 in SW480 cells with knockdown of Raf expression. C-Raf or B-Raf expression was knocked down by siRNA in SW480 cells, followed with NSC59984 (μmol/L) treatment for 16 hours. F, The phosphorylation of ERK1/2 in SW480 cells treated with NSC59984 (μmol/L) and the different kinase inhibitors (μmol/L) as indicated for 16 hours.
We further investigated whether NSC59984 induces ERK1/2 phosphorylation through a ROS-Ras-Raf-MEK axis. We knocked down the gene expression of Ras or Raf using siRNA or inhibited the kinase activity of C-Raf and B-Raf using sorafenib, the inhibitor of tyrosine kinases including PDGFR and Raf. Ras expression was knocked down by two siRNA (siRNA #1 and siRNA#2) in cancer cells, Both siRNA#1 and siRNA#2 showed that the knockdown of Ras blocked the phosphorylation of ERK1/2 and MEK1/2 in the cells treated with NSC59984 (Fig. 1D). The knockdown of C-Raf or B-Raf did not block the NSC59984-induced phosphorylation of ERK1/2 in cancer cells (Fig. 1E). Similar results were observed in the cells treated with sorafenib (Fig. 1F). These results suggest that NSC59984 induces ERK1/2 phosphorylation independent of C-Raf and B-Raf through ROS-Ras. The treatment with U0126, a MEK inhibitor dramatically blocked the NSC59984-mediated ERK1/2 phosphorylation in SW480 cancer cells (Fig. 1F), suggesting that NSC59984-induced ERK1/2 phosphorylation depends on MEK1/2.
On the basis of these findings, we propose that NSC59984 takes advantage of high levels of ROS in cancer cells to strengthen ERK2 signaling in an MEK1-dependent manner partially through the ROS-Ras-MEK axis.
Cellular ROS is required for NSC59984-induced mutant p53 degradation through ERK2
Stabilization of mutant p53 is associated with oxidative stresses in cancer cells (8). We investigated the effects of ROS on NSC59984-induced mutant p53 degradation. The NAC treatment rescued mutant p53 protein from NSC59984-induced degradation (Fig. 2A), suggesting that ROS is required for NSC59984 to reduce mutant p53 expression.
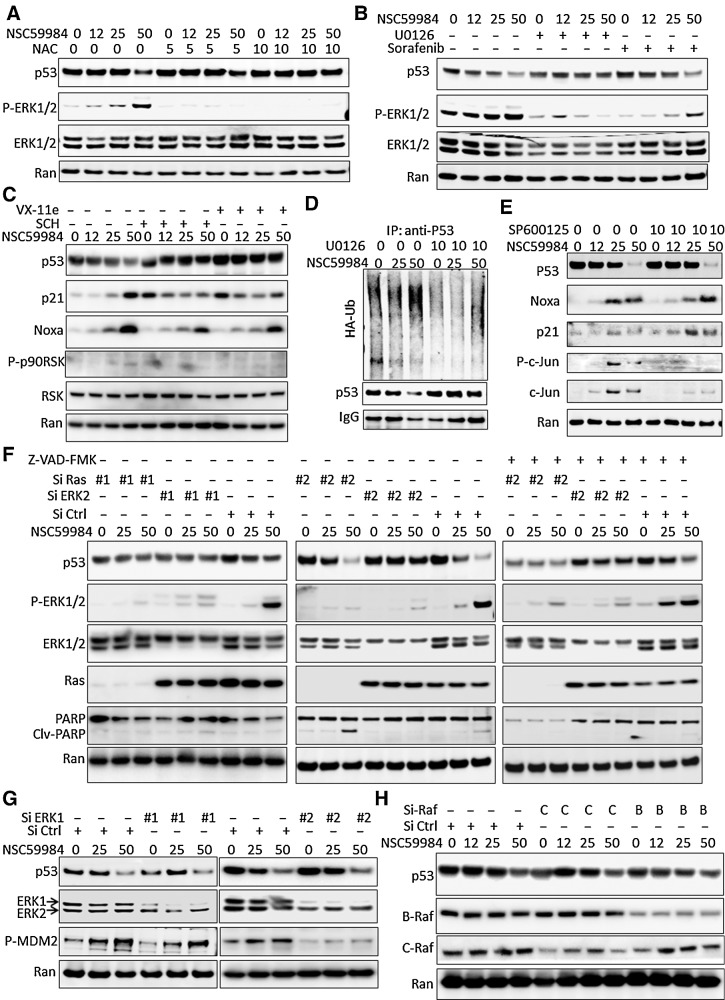
Figure 2.
ROS is required for NSC59984-induced mutant p53 degradation through ERK2. A, The expression of mutant p53 at the protein level in SW480 cells treated with NSC59984 (μmol/L) and NAC (mmol/L) for 16 hours. B, The expression of mutant p53 at the protein level in SW480 cancer cells treated with NSC59984 (μmol/L) and U0126 (10 μmol/L) or Sorafenib (32 μmol/L) for 16 hours. C, The expression of mutant p53, p21, and noxa at the protein levels in SW480 cancer cells treated with NSC59984 (μmol/L) and ERK1/2 inhibitor, SCH772984 (SCH, 1 μmol/L) or VX-11e (5 μmol/L) for 16 hours. D, The ubiquitination assay by IP in SW480 cells. SW480 cells were transfected with HA-Ub for 48 hours, followed with the treatment with NSC59984 (μmol/L) and U0126 (10 μmol/L) for an additional 8 hours. E, The protein level of mutant p53 and p53 targets in SW480 cells treated with NSC59984 (μmol/L) and SP600125 (10 μmol/L) for 16 hours. F, The expression of mutant p53 at the protein level in the cancer cells with knockdown of Ras and ERK2. Ras and ERK2 were knocked down in SW480 cells with siRNA (#1 and #2 for each target), followed by treatment with NSC59984 (μmol/L) and 20 μmol/L of Z-VAD-FMK for 16 hours. G, The expression of the mutant p53 at the protein level in the ERK1-knockdown SW480 cells treated with NSC59984 (μmol/L) for 16 hours. ERK1 was knocked down by two siRNAs. H, The expression of the mutant p53 at the protein level in the Raf-knockdown cancer cells. C-Raf and B-Raf were knocked down in SW480 cells by siRNA, followed with NSC59984 treatment (μmol/L) for 16 hours.
We further investigated whether the NSC59984-induced reduction of mutant p53 is mediated through the sustained ERK1/2 activation. The ERK1/2 signaling pathway was blocked by U0126 treatment, ERK1/2 inhibitors (VX-11e or SCH772984) or knockdown of ERK1/2 expression with siRNA. The treatment with U0126, a MEK1/2 inhibitor, rescued the mutant p53 protein in the cells treated with NSC59984 (Fig. 2B). Similar to MEK1/2 inhibitor, the ERK1/2 inhibitors, VX-11e and SCH772984 blocked NSC59984-induced reduction of mutant p53 in cells (Fig. 2C). Furthermore, U0126 treatment reduced the ubiquitination of mutant p53 from the NSC59984 treatment in the cancer cells (Fig. 2D). These results suggest that NSC59984 induces ERK1/2-dependent mutant p53 degradation in cancer cells. We further investigated whether JNK, one of the pathways regulated by ROS, is required for NSC59984 to induce mutant p53 protein degradation. The treatment with SP600125, a JNK inhibitor failed to block the NSC59984-induced reduction of mutant p53 protein (Fig. 2E), suggesting that JNK pathway is not required for the NSC59984-induced decrease of mutant p53 protein.
To clarify which one, ERK1 or ERK2 is required for NSC59984 to decrease mutant p53 protein, we knocked down ERK2 or ERK1 individually in the cancer cells by transient transfection with siRNA. ERK2 expression was knocked down by duplicate siRNA (siRNA#1 and siRNA#2). Both siRNA#1 and siRNA#2 specifically knocked down ERK2 expression in cells. The silencing of ERK2 expression by both siRNA #1 and siRNA #2 rescued mutant p53 from the NSC59984 treatment (Fig. 2F), but the knockdown of ERK1 did not. As shown in Fig. 2G, ERK1 expression was knocked down by two siRNAs (siRNA#1 and siRNA#2) specifically targeting ERK1. Both siRNA#1 and siRNA#2 showed that NSC59984 reduced mutant p53 protein in the ERK1-silenced cells (Fig. 2G). These results suggest that NSC59984-sustained phosphorylation of ERK2 is a major regulator required for NSC59984 to induce mutant p53 protein degradation in cancer cells.
Mutant p53 is stabilized because of tumor-associated stresses caused by mutant Ras in cancer cells (4, 8). Given the requirement of Ras for the NSC59984-sustained ERK2 phosphorylation in cancer cells (Fig. 1D), we further examined the mutant p53 protein level in SW480 cells by knocking down Ras. Ras expression was knocked down by two siRNAs (siRNA#1 and siRNA#2). As shown in Fig. 2F, silencing of Ras gene expression by both siRNA#1 and siRNA #2 partially blocked NSC59984 (25 μmol/L)-induced reduction of mutant p53 protein. The amount of mutant p53 rescued by the knockdown of Ras is less than that rescued by the knockdown of ERK2. The cleaved PARP was increased by 50 μmol/L of NSC59984 in the cells transfected with siRNA#2, suggesting cell death occurred. To avoid the possible effect of cell death on the mutant p53 protein degradation in these cells with knockdown of Ras by siRNA-#2, the cells were treated with Z-VAD-FMK, a pan-caspase inhibitor to block cellular apoptosis. NSC59984 reduced mutant p53 protein in the cells treated with Z-VAD-FMK, whereas knockdown of Ras partially rescued the mutant p53 protein from the NSC59984 treatment (Fig. 2F). Sorafenib, a Raf kinase inhibitor was not found to recover mutant p53 protein from the NSC59984-induced reduction (Fig. 2B). Consistent with the sorafenib experiment, the knockdown of C-Raf or B-Raf could not rescue mutant p53 from NSC59984-induced protein degradation in cancer cells (Fig. 2H).
These results taken together indicate that NSC59984 induces mutant p53 protein degradation mainly via ERK2 in a ROS-Ras-MEK-ERK2 axis in cancer cells.
NSC59984 induces mutant p53 protein degradation through ERK2-dependent MDM2 activation
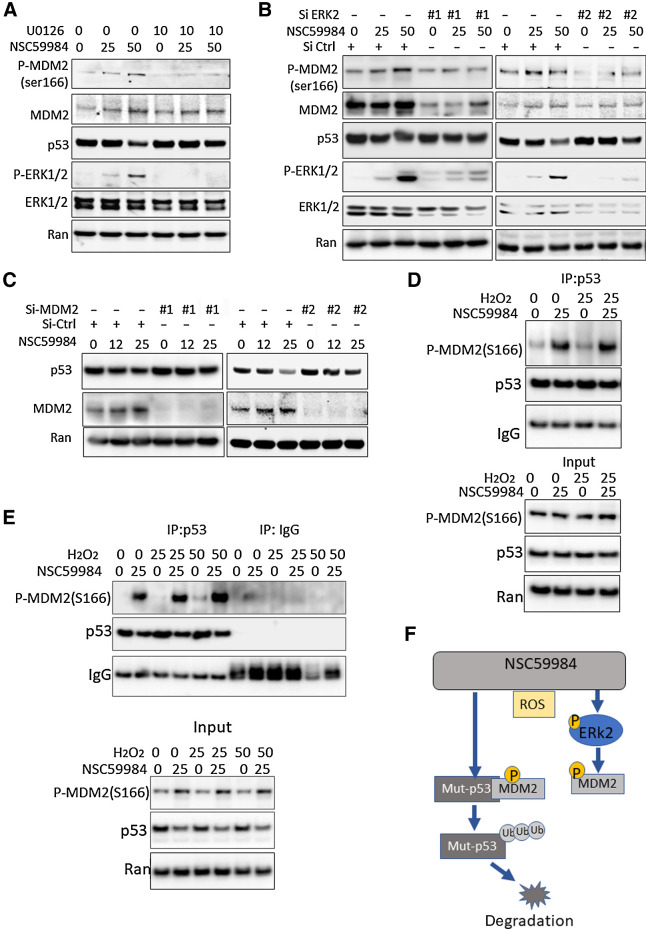
We previously reported that NSC59984 induced MDM2 phosphorylation at ser166 (34). Therefore, we investigated the effect of ERK2 on the phosphorylation of MDM2. U0126 treatment blocked NSC59984-mediated phosphorylation of MDM2 at ser166 (Fig. 3A). We further knocked down ERK2 expression using two siRNAs. Both siRNA#1 and siRNA#1 specifically silenced ERK2 expression in cells. Both siRNA# and siRNA#2 showed that the knockdown of ERK2 expression inhibited NSC59984-mediated phosphorylation of MDM2 at ser166 (Fig. 3B). We also knocked down ERK1 using two siRNAs. Both siRNA#1 and siRNA#2 showed that the knockdown of ERK1 did not block the NSC59984-mediated phosphorylation of MDM2 (Fig. 2G). These results suggest that NSC59984 induces ERK2-dependent phosphorylation of MDM2 at ser166 in cancer cells.
Figure 3.
NSC59984 induces MDM2-dependent mutant p53 degradation. A, Phosphorylation of MDM2 at ser166 in SW480 cells treated with NSC59984 (μmol/L) and U0126 (μmol/L) for 16 hours. B, Phosphorylation of MDM2 at ser166 in cells with knockdown of ERK2. The ERK2 was knocked down by two siRNAs (#1 and #2) in SW480 cells. The cells were treated with NSC59984 (μmol/L) for 16 hours. C, The expression of mutant p53 at the protein level in the cells with knockdown of MDM2. The MDM2 was knocked down with siRNA (#1 or #2) in SW480 cells. The cells were treated with NSC59984 (micromole/L) for 16 hours. D, The IP assay in SW480 cells. SW480 cells were treated with NCC59984 (μmol/L) and H2O2 (μmol/L) for 8 hours. E, The IP assay in HT29 cells. HT29 cells were treated with NCC59984 (μmol/L) and H2O2 (μmol/L) for 8 hours. F, The schematic of the mechanism of action of NSC59984 on mutant p53 degradation via an ROS-ERK2-MDM2 axis.
MDM2 has been found to be associated with mutant-p53 protein degradation (13, 14). We then examined the role of MDM2 in the effect of NSC59984 on mutant p53 degradation. The knockdown of MDM2 blocked NSC59984-mediated downregulation of mutant p53 in the cancer cells (Fig. 3C). This result is consistent with our previous report that Nutiln, an inhibitor of MDM2 abrogated NSC59984-mediated mutant p53 protein degradation (34). These results suggest that MDM2 is required for NSC59984 to decrease mutant p53 protein in cancer cells. We further performed an immunoprecipitation (IP) assay and found that NSC59984 treatment enhanced the phosphorylated MDM2 (ser166) binding to mutant p53 (Fig. 3D and E) in cancer cells. To investigate the effect of ROS levels on the MDM2 binding to mutant p53, the cells were treated with H2O2. We found that there was more phosphorylated-MDM2 bound to mutant p53 in the cells treated with H2O2 in combination with NSC59984 as compared with the NSC59984 treatment alone (Fig. 3D and E). These results taken together suggest that NSC59984 induces mutant p53 degradation via activation of MDM2 through a ROS-ERK2-MDM2 axis, and high levels of ROS enhance the efficacy of NSC59984-induced MDM2 binding to mutant p53 (Fig. 3F).
NSC5998 restores p53 pathway signaling through a ROS-ERK2 axis in p53-mutant cancer cells
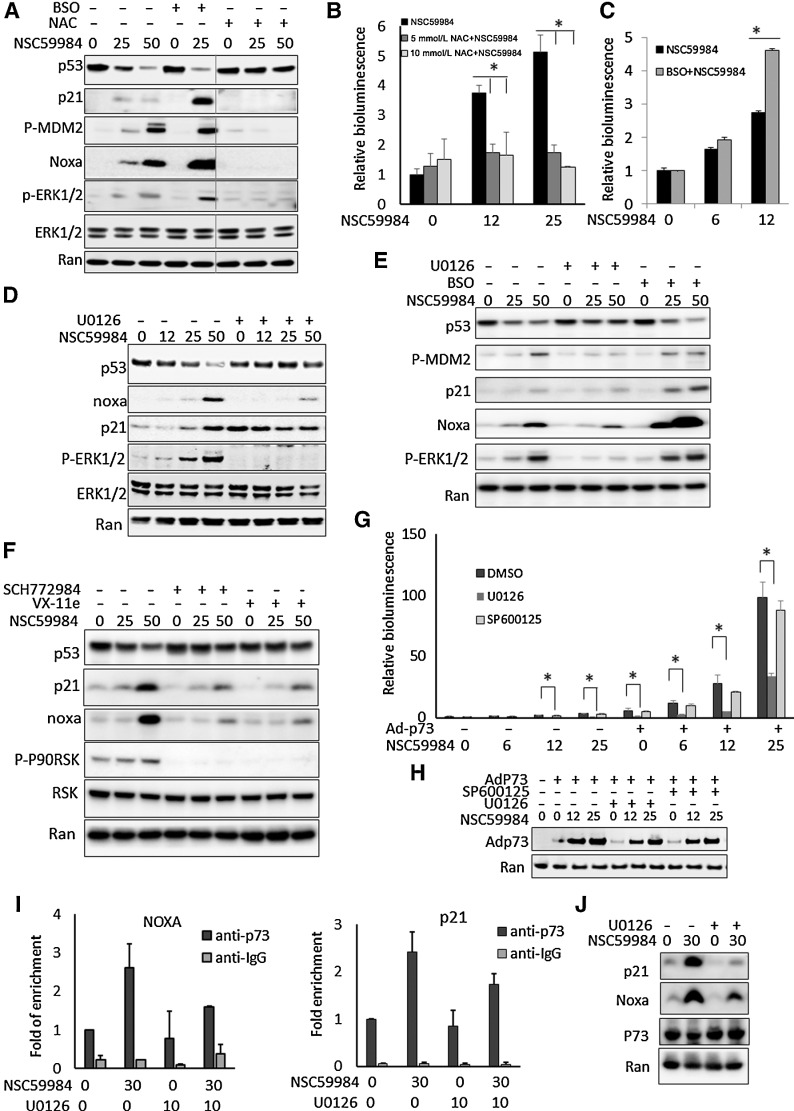
We previously reported that NSC59984-induced restoration of p53 pathway signaling correlates with the mutant p53 protein degradation in cancer cells (34). We further investigated the role of ROS in the effect of NSC59984 on p53 pathway signaling in mutant p53-expressing cancer cells. NAC treatment blocked NSC59984-mediated p53 pathway signaling based on the p53 targets (such as p21 and Noxa) at the protein level (Fig. 4A) and p53-responsive bioluminescence reporter assay (Fig. 4B). To increase the cellular ROS, cells were treated with BSO, a ROS-generating agent (Supplementary Fig. S2B). BSO treatment further elevated ERK1/2 phosphorylation, and substantially reduced mutant p53 protein expression in combination with NSC59984 in cancer cells carrying different p53 mutations (Fig. 4A and E). We also observed a significant increase in p53 targets and the p53-responsive reporter activity in the cancer cells in response to NSC59984 in combination with BSO (Fig. 4A, C, and E). These results suggest that ROS is required for NSC59984 to restore p53 pathway signaling correlated to the mutant p53 degradation. High levels of ROS enhance the efficacy of NS59984 in targeting mutant p53 degradation and in restoration of p53 pathway signaling.
Figure 4.
ROS is required for NSC59984 to restore p53 pathway signaling through ERK2. A, The protein level of mutant p53 and p53 targets in HT29 cells following treatment with NSC59984 (μmol/L) in combination with NAC (10 mmol/L) or BSO (10 μmol/L) for 16 hours. B, p53-responsive reporter bioluminescence in SW480 cells treated with NSC59984 (μmol/L) and NAC (10 mmol/L) for 16 hours. C, p53-responsive reporter bioluminescence in SW480 cells treated with NSC59984 (μmol/L) and BSO (10 μmol/L) for 16 hours. D, The expression of mutant p53 and p53 targets at the protein level in SW480 cells treated with NSC59984 (μmol/L) and U0126 (10 μmol/L) for 16 hours. E, The protein level of mutant p53 and p53 targets in RXF393 cancer cells carrying mutant p53 (R175H). The cells were treated with NSC59984 (μmol/L) and U0126 (10 μmol/L) or BSO (20 μmol/L) for 16 hours. F, The expression of mutant p53 and p53 targets at the protein levels in HT29 cells treated with NSC59984 (μmol/L) and ERK1/2 inhibitor SCH772984 (1 μmol/L) or VX-11e (5 μmol/L) for 16 hours. G, The p53-responsive reporter bioluminescence assay in SW480 cells with the overexpression of Ad-p73. p73 was overexpressed in SW480 cells by adenovirus (Ad-p73) infection. The cells were treated with NSC59984 (μmol/L) and U0126 (10 μmol/L) or SP600125 (10 μmol/L) for 16 hours. Data represent mean ± SD. *, P < 0.05 compared with the NSC59984 treatment at each dosage. H, The Ad-p73 expressed protein levels in the cells with the adenovirus infection (G). I, ChIP-PCR assay. p73 was overexpressed in HT29 with adenovirus infection. Following treatment with NSC59984 (μmol/L) and U0126 (μmol/L) for 7 hours. ChIP assay was performed with anti-P73 and IgG as a control. The p21 or Noxa promoters were quantified by real-time PCR using the ChIP-eluted DNA. Data were normalized to the ChIP with anti-p73 in the cells treated with DMSO treatment as a control. Data represent mean ± SD, N = 2. ANOVA test, P < 0.05. J, P21 and Noxa at the protein level in HT29 cells by Western blot assay (I).
We further investigated the effect of the NSC59984-sustained ERK2 phosphorylation on the restoration of p53 signaling in cancer cells. The NSC59984-induced p53 targets (such as p21 and Noxa) and p53-responsive reporter bioluminescence were inhibited by the U0126 treatment in the cancer cells carrying different p53 mutations (Fig. 4D, E, and G). Consistent with these observations, the treatment with the ERK1/2 inhibitor, (SCH772984 or VX-11e) blocked the NSC59984-induced p21 and Noxa gene expression in different cancer cells (Figs. 4F and 2C). In contrast to the ERK1/2 blockade, the inhibition of JNK failed to abrogate the effect of NSC59984 on the restoration of the p53 (Figs. 2E and 4G).
We previously reported that NSC59984 restores p53 pathway signaling via p73 (34). We investigated the effect of ERK2 on p73-mediated p53 pathway restoration. p73 was overexpressed in cancer cells with adenovirus infection. The effect of NSC59984 on p53-responsive reporter bioluminescence was significantly blocked by U0126, but not by the JNK inhibitor, in the p73 overexpressing cancer cells (Fig. 4G and H). Consistently, the p53 targets such as p21 and Noxa were reduced at the protein level by U0126 treatment in the p73-overexpressing cancer cells (Fig. 4J). Furthermore, ChIP-PCR assay showed that NSC59984 increased the p73 binding to the p21 and Noxa promoters, and these effects were partially inhibited by U0126 treatment (Fig. 4I). These results taken together suggest that the sustained ERK2 phosphorylation is selectively required for NSC59984-induced restoration of p53 signal via p73.
NSC59984 induces ERK2-dependent cell death in cancer cells
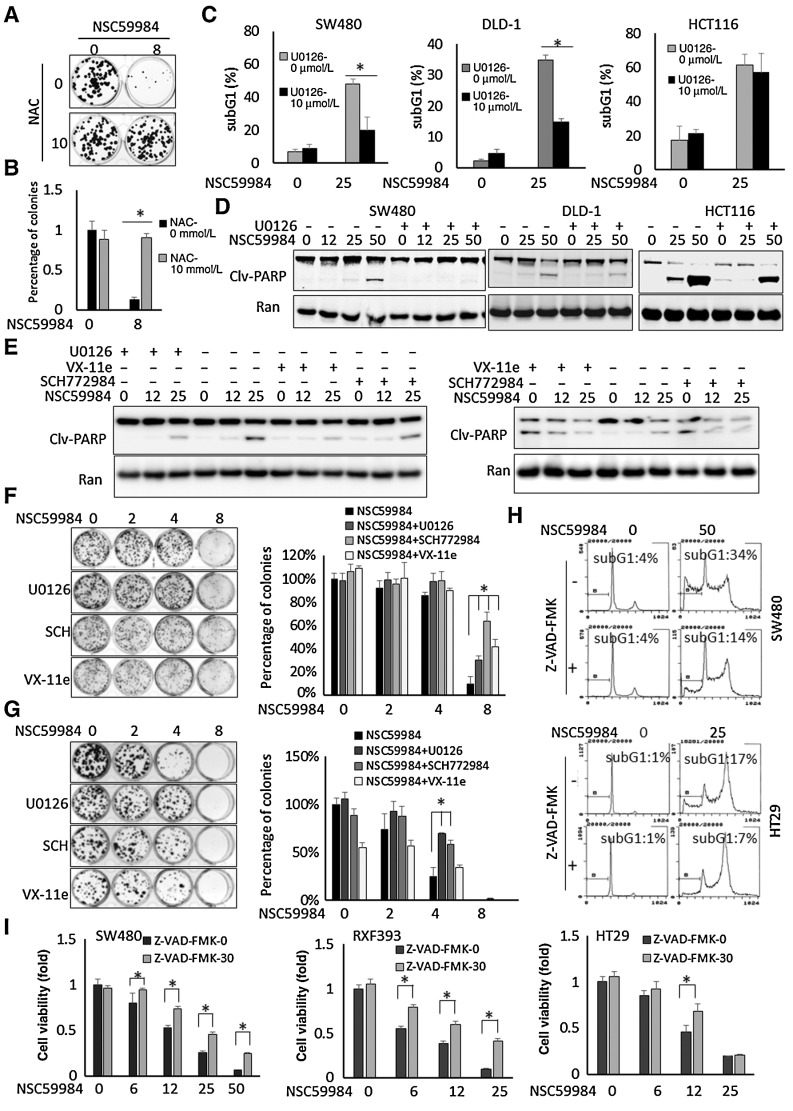
Given the requirement of ROS for NSC59984-sustained ERK2 phosphorylation and the mutant p53 degradation in cancer cells (Fig. 2), we further tested the effect of ROS on NSC59984-induced cell death. We observed that NAC treatment significantly blocked the suppressive effect of NSC59984 on colony formation in mutant p53-expressing colorectal cancer cells (Fig. 5A and B).These results suggest that cellular ROS is necessary for NSC59984 to induce cancer cell death.
Figure 5.
NSC59984 induces ERK2-dependent cell death in mutant p53-expressing cancer cells. A, Colony formation in HT29 cancer cells upon NSC59984 (μmol/L) treatment in combination with NAC (10 mmol/L) as described in the Materials and Methods. B, The relative number of the colonies (A). C, Sub-G1 flow cytometric analysis in different cancer cells treated with NSC59984 (μmol/L) and U0126 (10 μmol/L) for 72 hours. Data were obtained from two independent experiments. *, P < 0.05. D, Cleaved-PARP in SW480, DLD-1, and HCT116 cells treated with NSC59984 (μmol/L) and 10 μmol/L of U0126 for 36 hours. The cleaved-PARP was examined by Western blot analysis. E, Cleaved-PARP in Hop92 cells (left) and SW480 cells (right) treated with NSC59984 (μmol/L) and U0126 (10 μmol/L), SCH772984 (1 μmol/L) or VX-11e (5 μmol/L) for 36 hours. The cleaved-PARP was examined by Western blot analysis. F, The colony formation in HT29 cells. G, The colony formation in DLD-1 cells. The cells (F and G) were treated with NSC59984 (μmol/L) and U0126 (10 μmol/L), SCH772984 (SCH,1 μmol/L) or VX-11e (5 μmol/L). The percentages of colonies (B, F, and G) were obtained with DMSO treatment. Data represent mean ± SD from triplicate treatments. *, P < 0.05. H, The cell-cycle profiles of SW480 and HT29 cells treated with NSC59984 (μmol/L) in combination with Z-VAD-FMK (20 μmol/L) for 72 hours. I, Cell viability assay. Cancer cells were treated with NSC59984 (μmol/L) in the presence or absence of ZVAD-FMK (30 μmol/L) for 72 hours. Cell viability was determined by Cell Titer-Glo luminescence. Cell viability data were normalized to those of DMSO as a control. Data represent mean ± SD. *, P < 0.05 compared with the NSC59984 treatment at each dosage.
We further investigated the role of ERK2 activation in NSC59984-induced cell death. To address this issue, ERK1/2 signaling was blocked by MEK1/2 inhibitor U0126 or ERK1/2 inhibitor SCH772984 and VX-11e in different cancer cell lines. U0126 treatment reduced the percentage of sub-G1 DNA and the amount of cleaved-PARP in SW480 and DLD-1 cells treated with NSC59984 (Fig. 5C and D). The treatment with the ERK1/2 inhibitor, SCH772984 or VX-11e abrogated the NSC59984-induced PARP cleavage in different cancer cells (Fig. 5E). We further performed a colony formation assay in HT29 and DLD1 cancer cells. The results in these two cell lines are consistent. NSC59984 reduced cell colony formation in HT29 and DLD-1 in a dose-dependent manner. The inhibitory effect of NSC59984 on cell colony formation was blocked by U0126, an MEK1/2 inhibitor. The same results were observed in HT29 and DLD1 cells treated with ERK1/2 inhibitors SCH772984 and Vx-11e, except that VX-11e treatment alone reduced colony formation in DLD-1 cells (Fig. 5F and G). There were more colonies formed in HT29 and DLD-1 cells treated with NSC59984 and SCH772984 or VX-11e as compared with NSC59984 treatment alone (Fig. 5F and G). In contrast, the U0126 treatment could not significantly recover HCT116 cells from sub-G1, and only slightly blocked the PARP cleavage induced by NSC59984 at the tested doses (Fig. 5C and D).
To test whether the cell death induced by NSC59984 is due to cellular apoptosis, cells were treated Z-VAD-FMK, a pan-caspase inhibitor. Z-VAD-FMK treatment inhibited NSC59984-induced sub-G1 DNA content (Fig. 5H), suggesting that NSC59984 induces apoptosis. To determine whether NSC59984-induced apoptosis is important for the growth inhibitory effects of NSC59984, we performed a cell viability assay in cancer cells treated with NSC59984 in the presence or absence of Z-VAD-FMK. NSC59984 significantly reduced cell viability, with a diminished reduction in the presence of Z-VAD-FMK treatment in the cancer cells (Fig. 5I). These results using a pan-caspase inhibitor suggest that NSC59984-induced cell growth inhibition can be attenuated through caspase blockade and are consistent with other data with NSC59984 that apoptosis is one of the important mechanisms by which NSC59984 suppresses cancer growth. NSC59984-mediated G2–M arrest was still observed in the cells treated with Z-VAD-FMK (Fig. 5H). The G2–M arrest may partially contribute to the reduction of cell viability in the cells treated with NSC59984 in the presence of Z-VAD-FMK.
On the basis of these observations, we conclude that the ERK2 phosphorylation is required for NSC59984 to induce cell death in mutant p53-expressing colorectal cancer cells.
NSC59984 synergizes with ROS-generating agents to induce cell death in mutant p53-expressing colorectal cancer cells
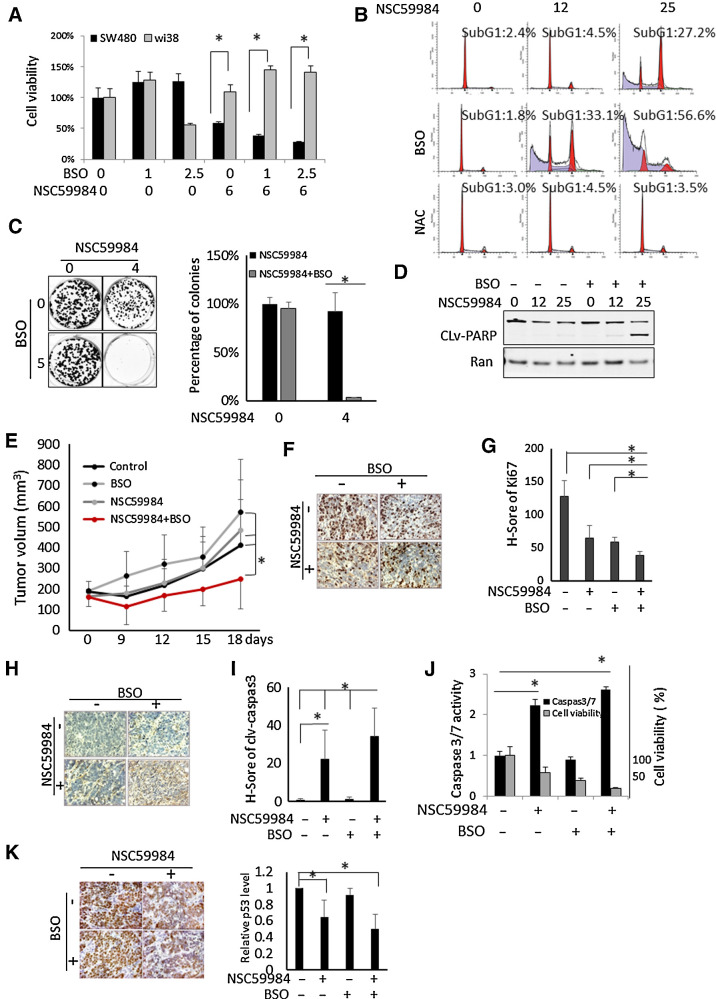
Targeting ROS by increasing its products is one of the promising strategies for cancer therapy owing to high intrinsic oxidative stresses in cancer cells (36). Given the requirement of ROS for NSC59984 to induce ERK2-dependent p53 restoration and mutant p53 degradation (Figs. 2 and 4), we further investigated whether combination of NSC59984 and ROS-generating agents synergistically suppresses tumor growth. A synergy between NSC59984 and BSO was observed in the colorectal cancer cells, but not in the normal cells at the tested doses (Fig. 6A; Supplementary Table S1). The combination of BSO and NSC59984 increased the sub-G1 DNA content and reduced colony formation (Fig. 6B and C). These results were further supported with the observation of PARP cleavage in the different cancer cells (Fig. 6D; Supplementary Fig. S4).
Figure 6.
NSC59984 in combination with BSO suppresses tumor growth. A, Cell viability of SW480 and normal WI-38 cells treated with NSC59984 (μmol/L) and BSO (μmol/L) for 72 hours. B, The cell-cycle profiles of SW480 cells treated with NSC59984 (μmol/L) in combination with NAC (10 mmol/L) or BSO (10 μmol/L) for 72 hours. C, The colony formation in HT29 cancer cells upon NSC59984 treatment in combination with BSO (5 μmol/L) as described in the Materials and Methods. D, Cleaved-PARP in SW480 cells treated with NSC59984 (μmol/L) and BSO (10 μmol/L) for 36 hours. E, Tumor volumes of HT29 xenografts in mice (n = 8). Tumor volumes were measured by a caliper every 3 days. Data are expressed as mean ± SD. *, P < 0.05. F, IHC staining for Ki67 in HT29 xenografted colorectal cancer tumors. G, The H-Score of Ki67 in HT29 xenograft tumors. H, IHC staining for cleaved-caspase 3 in HT29 xenograft tumors. I, The H-Score of cleaved caspase 3 in HT29 xenograft tumors. The H-Score (G and I) was calculated and analysis with VECTRA 2.0 as described in the Materials and Methods. J, Caspase 3/7 activity assays in SW480 cells. The cells were treated with NSC5994 (25 μmol/L) and BSO (10 μmol/L) for 30 hours for the caspase assay and 72 hours for the cell viability assay. The caspase 3/7 activity was normalized to the DMSO treatment as control. The cell viability was normalized to the cells treated with DMSO as control. Data are expressed as mean ± SD. *, P < 0.05. K, IHC staining for mutant p53 in HT29 xenografted tumors. The relative mutant p53 protein level was analyzed by image J and normalized to the nontreatment control. Data are expressed as mean ± SD. *, P < 0.05.
We further performed a combination of NSC59984 and BSO in vivo experiment using an aggressive HT29 CRC xenograft. As shown in Fig. 6E, NSC59984 in combination with BSO significantly suppressed tumor growth as compared with single-agent treatment, in agreement with our in vitro data (Fig. 6A–D; Supplementary Fig. S4). The combination of NSC59984 and BSO significantly increased the index of cleaved-caspase 3 and reduced Ki67 index in the xenograft as compared with NSC59984 treatment alone (Fig. 6F–I). We further detected a substantial increase in caspase 3 activity in the cancer cells treated with NSC59984 in combination with BSO (Fig. 6J), further validating the observations of cleaved caspase 3 in vivo. We further examined the mutant p53 expression level in the xenografted tumors. The IHC shows that the staining of mutant p53 is weaker in the tumors treated with NSC59984 or the combinational treatment of NSC59984 and BSO than in the tumors without NSC59984 treatment as a control (Fig. 6K), suggesting a decrease of mutant p53 at the protein level by NSC59984 in vivo. The reduction of mutant p53 is correlated to the high index of cleaved caspase 3 and low index of ki67 in the xenografted tumors treated with NSC59984 and the combinatorial treatment (Fig. 6G–I).
Discussion
Accumulating reports show an increase in ROS level in response to mutant p53-degrading small molecules in cancer cells (19–21). However, oxidative stress is associated with stabilization of mutant p53 in cancer cells (4, 8, 9). Little is known about the molecular mechanism of ROS related to the induction of mutant p53 degradation. This paradox promoted us to investigate the effect of ROS on small molecule–induced mutant p53 degradation in cancer cells. We for the first time demonstrate that a ROS-ERK2-MDM2 axis is inducible with small-molecule NSC59984 in cancer cells. NSC59984 takes advantage of high cellular ROS to increase ERK2 signaling and induce ERK2-dependent MDM2 phosphorylation. Small-molecule NSC59984 further enhances the phosphorylated MDM2 binding to mutant p53 under high cellular ROS levels, leading to mutant p53 protein degradation through MDM2-mediated ubiquitination. Our study about NSC59984 proposes that the activation of a ROS-ERK2-MDM2 axis exposes a vulnerability in mutant p53 stabilization that can be exploited to induce mutant p53 protein degradation.
ERK2 signaling is commonly altered or inappropriately activated in cancer cells due to different cellular stresses including high cellular ROS (24, 37). ERK2 is transiently phosphorylated via the balance between the Ras/Raf/MEK1/ERK2 kinase cascades and various negative feedback loops in response to the external stimuli (38). In contrast to the transiently phosphorylation form, NSC59984-induced phosphorylation of ERK2 is constitutively increased and sustained via MEK1. ROS can activate Ras-Raf-MEK-ERK1/2 pathway via oxidative modifications of the intracellular components upstream of ERK2 (such as MEK1 or/and Ras) or MKP1 (26, 39). The Ras and MEK1/2 play a major role in regulation of ERK1/2 signaling in response to ROS in cells treated with NSC59984 (Fig. 1). However, NSC59984 sustains ERK1/2 phosphorylation via MEK1/2 independent of B-Raf and C-Raf. These results suggest that other components downstream of ROS-Ras activate the MEK1/2-ERK1/2 cascades in response to NSC59984 treatment in cells. The molecular mechanism by which NSC59984 sustains ERK2 phosphorylation will be investigated in the future.
There are diverse signaling pathways related to Ras-MEK-ERK kinase cascades upon increased ROS (40). However, our results demonstrate that NSC59984 induces mutant p53 degradation and p53 pathway restoration specifically and mainly via ERK2 activation upon increased ROS in cancer cells (Fig. 2). Persistent ERK2 phosphorylation has been reported with multiple chemotherapies such as treatment with doxorubicin or cisplatin (26). Unlike NSC59984 treatment, treatment with doxorubicin or cisplatin cannot decrease mutant p53 protein in cancer cells in the context of persistent ERK2 phosphorylation (Supplementary Fig. S3A), in agreement with the previous reports (8). ROS-generating agents BSO and H2O2 could not reduce mutant p53 protein in cancer cells at the tested doses (Supplementary Fig. S3B and S3C), but enhanced the efficacy of NSC59984-induced mutant p53 degradation (Fig. 3). On the basis of these observations, we hypothesize that the sustained ERK2 phosphorylation may “prime” mutant p53 protein degradation (Fig. 3F). ERK2 activation has been reported to phosphorylate protein targets and induce selective protein degradation during cellular senescence (31). Protein posttranslational modifications mediated by ERK2 might “prime” mutant p53 destabilization. Indeed, we found ERK2-dependent MDM2 phosphorylation at ser166 (Fig. 3). MDM2 phosphorylation at ser166 has been reported to confer wild-type p53 protein degradation (33). Importantly, we found that NSC59984 enhanced the phosphorylated MDM2 binding to mutant p53 (Fig. 3). Although BSO and H2O2 increase cellular ROS, we could not detect either the interaction between phosphorylated MDM2 and mutant p53 or the reduction of mutant p53 protein under such a high cellular ROS. The activation of a ROS-ERK2-MDM2 axis may render the vulnerability of mutant p53 stabilization for NSC59984 to target degradation. As we observed, the higher the level of ROS, the more phosphorylated MDM2 was induced to bind to mutant p53 by NSC59984 (Fig. 3D and E), suggesting that this vulnerability is exploited by NSC59984 to induce mutant p53 degradation via activation of the ROS-ERK2-MDM2 axis. In addition, ROS may also be involved in mutant p53 protein oxidative modifications. ROS results in oxidative modifications of proteins which are degraded via multiple mechanisms (including via the proteasome) based on the strength and duration of ROS (41). Similar to wild-type p53, mutant p53 has thiol groups that can be modified by ROS (22). The requirement of ROS for NSC59984-induced mutant p53 degradation raises another possibility that NSC59984 may mediate mutant p53 structure modifications with ROS which might be susceptible to the mutant p53 binding to MDM2 for further ubiquitination. It also remains unclear whether NSC59984 induces mutant p53 binding to phosphorylated-MDM2 directly or indirectly. The chemical structure suggests that NSC59984 is a reactive covalent compound. Covalent drugs form covalent complexes with different proteins. Therefore, we suspect that NSC59984 may react with different proteins in addition to mutant p53, and cellular effects including effects on p53 protein expression are likely indirect and independent of any unproven interaction between NSC59984 and mutant p53 or any other cellular protein. Identification of cellular targets of NSC59984 as part of its further development may be warranted in future studies. Importantly, the screen that identified NSC59984 was a phenotypic cell-based screen looking for induction of p53 pathway–mediated reporter activity. Such functional screens do not typically yield p53-interacting compounds in the mediation of restored p53 pathway transcriptional activity. In fact, NSC59984 appears to require p73 for its functional activation of p53 pathway target genes and for its antitumor activity. Other compounds we have identified over the years through the functional cell-based phenotypic screen, such as PG3-Oc, or CB002 xanthine analogs, appear to induce an integrated stress response in tumor cells that upregulates pro-apoptotic target genes that are also regulated by p53 and which mediate cell death and antitumor effects (42, 43).
Mutant p53 GOF proteins can bind and inactivate p73 in cancer cells (44). Targeting p73 is one of the promising strategies bypassing restoration of wild type functions to mutant p53 in cancer therapy (45). Our laboratory has been working on small molecules targeting p73 and reported that p73 can be activated by small molecules via induction of p73 expression or interruption of p73 interaction with mutant p53 (46–48). In this study, we explored a new strategy for releasing p73 from the mutant p53 inhibitory complex by depleting mutant p53 using small-molecule NSC59984. The correlation between the mutant p53 degradation and the p73 activation supports the possibility that p73 most likely is released from the mutant p53 inhibitory complex due to the NSC59984-mediated mutant p53 degradation. It is also well known that p73 activity is regulated through a complex mechanism such as posttranslational modifications and protein–protein interactions (49). We observed that inhibition of ERK2 signaling partially blocked the p73 binding to the p21 and Noxa promoters and the gene expression in cells treated with NSC59984. These results raise a possibility that the released p73 may be further stimulated via the sustained ERK2 signaling, and functions as an activated transcription factor to bind to target promoters and regulate gene expression.
Cancer cells have higher baseline levels of ROS than normal cells (36). High cellular ROS may ensure appropriate cell status for drug treatment selectively in cancer cells (26, 36). NSC59984 targets cancer cells with high therapeutic index (34), probably due to high cellular ROS in cancer cells. We found the sustained phosphorylation of ERK2 in cancer cells, but not in normal cells (Fig. 1; Supplementary Fig. S1). High ROS sensitizes cancer cells to NSC59984 treatment including enhanced mutant p53 degradation, p53 pathway restoration and cell death, which is correlated with the persistent ERK2 phosphorylation. Our data support the hypothesis that high levels of ROS sustain ERK2 phosphorylation beyond a threshold, which induces cell death in cancer cells (26). The higher ROS levels render mutant p53-expressing cancer cells vulnerable to NSC59984 treatment to induce cell death via ERK2 activation in cells, probably due to mutant p53 degradation and consequent restoration of the p53 pathway through p73. Although the ERK2 blockade rescues most of the mutant p53 protein (Fig. 2), our data reveal that U0126 treatment cannot completely prevent cell death induced by NSC59984 (Fig. 5). Therefore, the possibility cannot be excluded that other signaling pathways related to ROS may, in part, be involved in NSC59984-induced cells death.
Mutant p53 is one of the major determinants of the tumorigenesis and tumor development, and cancer cells can be addicted to mutant p53. Our data show that the depletion of mutant p53 contributes to the antitumor effect of NSC59984 at least in part. How much contribution the reduction of p53 mutant has on the overall antitumor effect for NSC59984 will be further investigated in more detail in future studies.
Of note, NSC59984 induces ERK2 and acts through ROS to deplete mutant p53. While NSC59984 may appear to be a weak antitumor agent, and also may appear to not efficiently degrade mutant p53, it should be noted that we have not performed a MTD study to optimize or maximize its antitumor efficacy in preclinical models, or to maximize its pharmacokinetic or pharmacodynamic characteristics. Our insights into the role of ERK2 and ROS in the mechanism of mutant p53 degradation, synergistic interactions between NSC59984 and ROS-modifying agents, need for MTD/pharmacokinetic/pharmacodynamic studies, and our observations through knockdown studies suggesting that mutant p53 reduction is relevant to the antitumor effects of NSC59984, together provide a clear path for drug use optimization.
High intracellular ROS levels and mutant p53 are associated with poor prognosis of tumors, and contribute to drug resistance in cancer therapy. Toxicity of high ROS levels limits the application of ROS-generating agents in the clinic (36). The combination of NSC59984 and ROS-generating agents revealed a synergy to induce cell death in the mutant p53-expressing cancer cells with less toxicity (Fig. 6; Supplementary Table S1; Supplementary Fig. S5). Our data indicate that a high level of ROS and mutant p53 can be considered as a biomarker for NSC59984 administration in cancer therapy and warrant further evaluation of the combination of small molecules targeting mutant p53 degradation and ROS-generating agents. Our findings suggest that mutant p53 stabilization represents a vulnerability under high ROS cellular conditions, which can be exploited by compounds to target mutant p53 protein degradation through the activation of a ROS-ERK2-MDM2 axis in cancer cells. These effects on mutant p53 destabilization coupled with restoration of p53 target gene activation by p53 pathway restoring compounds such as NSC59984 provides a path forward to bring novel cancer therapeutic candidates for clinical testing.
Supplementary Material
Acknowledgments
W.S. El-Deiry is an American Cancer Society Research Professor. The work was supported, in part, by the ACS and NIH grant CA176289 to W.S. El-Deiry.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
Authors' Disclosures
W.S. El-Deiry reports a patent 10,744,133 licensed to p53-Therapeutics. No disclosures were reported by the other authors.
Authors' Contributions
S. Zhang: Conceptualization, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. Zhou: Investigation, methodology, writing–review and editing. W.S. El-Deiry: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, project administration, writing–review and editing.
References
- 1. Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol 2013;15:2–8. [DOI] [PubMed] [Google Scholar]
- 2. Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119:847–60. [DOI] [PubMed] [Google Scholar]
- 3. Dittmer D, Pati S, Zambetti G, Chu S, Teresky AK, Moore M, et al. Gain of function mutations in p53. Nat Genet 1993;4:42–6. [DOI] [PubMed] [Google Scholar]
- 4. Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev 2008;22:1337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ 2011;18:1904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25:304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alexandrova EM, Yallowitz AR, Li D, Xu S, Schulz R, Proia DA, et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015;523:352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Suh YA, Post SM, Elizondo-Fraire AC, Maccio DR, Jackson JG, El-Naggar AK, et al. Multiple stress signals activate mutant p53 in vivo. Cancer Res 2011;71:7168–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Asher G, Lotem J, Tsvetkov P, Reiss V, Sachs L, Shaul Y. P53 hot-spot mutants are resistant to ubiquitin-independent degradation by increased binding to NAD(P)H:quinone oxidoreductase 1. Proc Natl Acad Sci U S A 2003;100:15065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lisek K, Walerych D, Del Sal G. Mutant p53-Nrf2 axis regulates the proteasome machinery in cancer. Mol Cell Oncol 2017;4:e1217967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu DS, Duong CP, Haupt S, Montgomery KG, House CM, Azar WJ, et al. Inhibiting the system xC-/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun 2017;8:14844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kalo E, Kogan-Sakin I, Solomon H, Bar-Nathan E, Shay M, Shetzer Y, et al. Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J Cell Sci 2012;125:5578–86. [DOI] [PubMed] [Google Scholar]
- 13. Li D, Marchenko ND, Schulz R, Fischer V, Velasco-Hernandez T, Talos F, et al. Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res 2011;9:577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Peng Y, Chen L, Li C, Lu W, Chen J. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem 2001;276:40583–90. [DOI] [PubMed] [Google Scholar]
- 15. Hong B, van den Heuvel AP, Prabhu VV, Zhang S, El-Deiry WS. Targeting tumor suppressor p53 for cancer therapy: strategies, challenges and opportunities. Curr Drug Targets 2014;15:80–9. [DOI] [PubMed] [Google Scholar]
- 16. Kastenhuber ER, Lowe SW. Putting p53 in context. Cell 2017;170:1062–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parrales A, Ranjan A, Iyer SV, Padhye S, Weir SJ, Roy A, et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol 2016;18:1233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vakifahmetoglu-Norberg H, Kim M, Xia HG, Iwanicki MP, Ofengeim D, Coloff JL, et al. Chaperone-mediated autophagy degrades mutant p53. Genes Dev 2013;27:1718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim JG, Lee SC, Kim OH, Kim KH, Song KY, Lee SK, et al. HSP90 inhibitor 17-DMAG exerts anticancer effects against gastric cancer cells principally by altering oxidant-antioxidant balance. Oncotarget 2017;8:56473–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wolf IM, Fan Z, Rauh M, Seufert S, Hore N, Buchfelder M, et al. Histone deacetylases inhibition by SAHA/Vorinostat normalizes the glioma microenvironment via xCT equilibration. Sci Rep 2014;4:6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qi XF, Zheng L, Lee KJ, Kim DH, Kim CS, Cai DQ, et al. HMG-CoA reductase inhibitors induce apoptosis of lymphoma cells by promoting ROS generation and regulating Akt, Erk and p38 signals via suppression of mevalonate pathway. Cell Death Dis 2013;4:e518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bykov VJ, Lambert JM, Hainaut P, Wiman KG. Mutant p53 rescue and modulation of p53 redox state. Cell Cycle 2009;8:2509–17. [DOI] [PubMed] [Google Scholar]
- 23. Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res 2010;44:479–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 2015;15:577–92. [DOI] [PubMed] [Google Scholar]
- 25. Navarro R, Martinez R, Busnadiego I, Ruiz-Larrea MB, Ruiz-Sanz JI. Doxorubicin-induced MAPK activation in hepatocyte cultures is independent of oxidant damage. Ann N Y Acad Sci 2006;1090:408–18. [DOI] [PubMed] [Google Scholar]
- 26. Cagnol S, Chambard JC. ERK and cell death: mechanisms of ERK-induced cell death–apoptosis, autophagy and senescence. FEBS J 2010;277:2–21. [DOI] [PubMed] [Google Scholar]
- 27. Yeh PY, Chuang SE, Yeh KH, Song YC, Chang LL, Cheng AL. Phosphorylation of p53 on Thr55 by ERK2 is necessary for doxorubicin-induced p53 activation and cell death. Oncogene 2004;23:3580–8. [DOI] [PubMed] [Google Scholar]
- 28. Ley R, Balmanno K, Hadfield K, Weston C, Cook SJ. Activation of the ERK1/2 signaling pathway promotes phosphorylation and proteasome-dependent degradation of the BH3-only protein, Bim. J Biol Chem 2003;278:18811–6. [DOI] [PubMed] [Google Scholar]
- 29. Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol 2008;10:138–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luciano F, Jacquel A, Colosetti P, Herrant M, Cagnol S, Pages G, et al. Phosphorylation of Bim-EL by Erk1/2 on serine 69 promotes its degradation via the proteasome pathway and regulates its proapoptotic function. Oncogene 2003;22:6785–93. [DOI] [PubMed] [Google Scholar]
- 31. Deschenes-Simard X, Gaumont-Leclerc MF, Bourdeau V, Lessard F, Moiseeva O, Forest V, et al. Tumor suppressor activity of the ERK/MAPK pathway by promoting selective protein degradation. Genes Dev 2013;27:900–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melnikova VO, Santamaria AB, Bolshakov SV, Ananthaswamy HN. Mutant p53 is constitutively phosphorylated at Serine 15 in UV-induced mouse skin tumors: involvement of ERK1/2 MAP kinase. Oncogene 2003;22:5958–66. [DOI] [PubMed] [Google Scholar]
- 33. Malmlof M, Roudier E, Hogberg J, Stenius U. MEK-ERK-mediated phosphorylation of Mdm2 at Ser-166 in hepatocytes. Mdm2 is activated in response to inhibited Akt signaling. J Biol Chem 2007;282:2288–96. [DOI] [PubMed] [Google Scholar]
- 34. Zhang S, Zhou L, Hong B, van den Heuvel AP, Prabhu VV, Warfel NA, et al. Small-molecule NSC59984 restores p53 pathway signaling and antitumor effects against colorectal cancer via p73 activation and degradation of mutant p53. Cancer Res 2015;75:3842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jackson JG, Pereira-Smith OM. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res 2006;66:8356–60. [DOI] [PubMed] [Google Scholar]
- 36. Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009;8:579–91. [DOI] [PubMed] [Google Scholar]
- 37. Yaeger R, Corcoran RB. Targeting alterations in the RAF-MEK pathway. Cancer Discov 2019;9:329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lake D, Correa SA, Muller J. Negative feedback regulation of the ERK1/2 MAPK pathway. Cell Mol Life Sci 2016;73:4397–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deschenes-Simard X, Kottakis F, Meloche S, Ferbeyre G. ERKs in cancer: friends or foes? Cancer Res 2014;74:412–9. [DOI] [PubMed] [Google Scholar]
- 40. Rezatabar S, Karimian A, Rameshknia V, Parsian H, Majidinia M, Kopi TA, et al. RAS/MAPK signaling functions in oxidative stress, DNA damage response and cancer progression. J Cell Physiol 2019 Feb 27. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 41. Pajares M, Jimenez-Moreno N, Dias IH, Debelec B, Vucetic M, Fladmark KE, et al. Redox control of protein degradation. Redox Biol 2015;6:409–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hernandez Borrero L, Dicker DT, Santiago J, Sanders J, Tian X, Ahsan N, et al. A subset of CB002 xanthine analogs bypass p53-signaling to restore a p53 transcriptome and target an S-phase cell cycle checkpoint in tumors with mutated-p53. Elife 2021;10:e70429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tian X, Ahsan N, Lulla A, Lev A, Abbosh P, Dicker DT, et al. P53-independent partial restoration of the p53 pathway in tumors with mutated p53 through ATF4 transcriptional modulation by ERK1/2 and CDK9. Neoplasia 2021;23:304–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Di Como CJ, Gaiddon C, Prives C. p73 function is inhibited by tumor-derived p53 mutants in mammalian cells. Mol Cell Biol 1999;19:1438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bisso A, Collavin L, Del Sal G. p73 as a pharmaceutical target for cancer therapy. Curr Pharm Des 2011;17:578–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang W, Kim SH, El-Deiry WS. Small-molecule modulators of p53 family signaling and antitumor effects in p53-deficient human colon tumor xenografts. Proc Natl Acad Sci U S A 2006;103:11003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hong B, Prabhu VV, Zhang S, van den Heuvel AP, Dicker DT, Kopelovich L, et al. Prodigiosin rescues deficient p53 signaling and antitumor effects via upregulating p73 and disrupting its interaction with mutant p53. Cancer Res 2014;74:1153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prabhu VV, Hong B, Allen JE, Zhang S, Lulla AR, Dicker DT, et al. Small-molecule prodigiosin restores p53 tumor suppressor activity in chemoresistant colorectal cancer stem cells via c-Jun-mediated DeltaNp73 inhibition and p73 activation. Cancer Res 2016;76:1989–99. [DOI] [PubMed] [Google Scholar]
- 49. Conforti F, Sayan AE, Sreekumar R, Sayan BS. Regulation of p73 activity by post-translational modifications. Cell Death Dis 2012;3:e285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.