The S100A9–ALDH1A1–retinoic acid signaling pathway mediates osimertinib resistance and brain relapse in models of EGFR-mutant lung cancer.
Abstract
The epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) osimertinib has significantly prolonged progression-free survival (PFS) in patients with EGFR-mutant lung cancer, including those with brain metastases. However, despite striking initial responses, osimertinib-treated patients eventually develop lethal metastatic relapse, often to the brain. Although osimertinib-refractory brain relapse is a major clinical challenge, its underlying mechanisms remain poorly understood. Using metastatic models of EGFR-mutant lung cancer, we show that cancer cells expressing high intracellular S100A9 escape osimertinib and initiate brain relapses. Mechanistically, S100A9 upregulates ALDH1A1 expression and activates the retinoic acid (RA) signaling pathway in osimertinib-refractory cancer cells. We demonstrate that the genetic repression of S100A9, ALDH1A1, or RA receptors (RAR) in cancer cells, or treatment with a pan-RAR antagonist, dramatically reduces brain metastasis. Importantly, S100A9 expression in cancer cells correlates with poor PFS in osimertinib-treated patients. Our study, therefore, identifies a novel, therapeutically targetable S100A9–ALDH1A1–RA axis that drives brain relapse.
Significance:
Treatment with the EGFR TKI osimertinib prolongs the survival of patients with EGFR-mutant lung cancer; however, patients develop metastatic relapses, often to the brain. We identified a novel intracellular S100A9–ALDH1A1–RA signaling pathway that drives lethal brain relapse and can be targeted by pan-RAR antagonists to prevent cancer progression and prolong patient survival.
This article is highlighted in the In This Issue feature, p. 873
Introduction
Lung cancer is the leading cause of cancer-related mortality (1). Somatic activating mutations in the epidermal growth factor receptor (EGFR) gene occur in up to 50% of patients with lung cancer worldwide (2–4). Targeted therapies with EGFR tyrosine kinase inhibitors (TKI) have transformed the treatment landscape for EGFR-mutant lung cancer (5–8). EGFR exon 19 (in-frame) deletions (del E746–A750) and exon 21 point mutations (L858R) comprise 90% of the EGFR mutations observed in lung cancer and lead to ligand-independent activation of the EGFR signaling pathway to promote proliferation, migration, and survival of cancer cells (7). Although the first- and second-generation EGFR TKIs (erlotinib and afatinib, respectively) showed promising initial responses in patients with EGFR-mutant lung cancer, patients eventually acquired therapy resistance (50% of cases with an acquired EGFRT790M mutation) within 9 to 14 months of treatment, and ultimately developed lethal metastatic relapse (9–11). The subsequent development of third-generation EGFR TKIs led to the discovery of osimertinib (AZD9291), which specifically targets EGFRT790M and the original sensitizing mutations (exon 19 deletion and L858R) while sparing wild-type EGFR and the toxicities associated with its inhibition (12). Remarkably, first-line treatment with osimertinib significantly extended the median progression-free survival (PFS) of patients with EGFR-mutant metastatic lung cancer from 10.2 months (involving treatment using earlier-generation EGFR TKIs) to 18.9 months (13, 14). Despite promising initial responses, osimertinib-treated patients eventually develop metastatic relapses and succumb to death (8, 15–17). Studies designed to explore osimertinib resistance mechanisms have found that on-target mutations in the EGFR gene that abrogate the binding of osimertinib to EGFR account for only 6% to 10% of osimertinib-resistant tumors with first-line treatment (17–19). Strikingly, 50% of osimertinib-refractory relapses instead arise from EGFR pathway–independent mechanisms, which remain poorly defined (17).
Metastasis to the central nervous system (CNS) is a frequent complication in patients with EGFR-mutant lung cancer (20–22). It is estimated that 25% of patients with EGFR-mutant lung cancer already present with CNS metastases at diagnosis, and the incidence of CNS metastasis increases to 45% at 3 years after diagnosis and TKI treatment (21, 23). Metastasis to the brain portends a poor prognosis, as it is associated with a significant decline in cognitive and motor function, impaired daily functioning, morbidity, and accelerated mortality (21–23). First- and second-generation EGFR TKIs showed poor blood–brain barrier permeability and consequently had minimal impact on brain metastases. In contrast, the superior blood–brain barrier permeability of osimertinib led to impressive clinical responses in patients with brain metastasis (23, 24). However, responses to osimertinib are not durable, and patients eventually relapse and die with osimertinib-refractory metastatic progression. In particular, CNS progression has been reported in 20% of patients with lung cancer treated with osimertinib (25), which adversely affects quality of life and shortens survival. Therefore, an understanding of post-osimertinib CNS relapse mechanisms is critical for improving the clinical management of patients with EGFR-mutant lung cancer.
The persistence of residual disease following osimertinib treatment likely contributes to metastatic relapse and presents a clinical challenge for patients with EGFR-mutant lung cancer. Tissue microenvironments can provide protective niches for the survival and expansion of residual cancer cells and enable the development of relapsed tumors (26, 27). Surprisingly, however, metastatic models are rarely used for studying osimertinib-refractory relapse mechanisms, which limits the potential for developing more effective therapies. Therefore, to identify mechanisms of osimertinib-refractory relapse in the context of metastatic progression, we utilized mouse models that closely resemble the metastatic progression and osimertinib response observed in human patients. We, therefore, generated long-term in vivo treatment models using osimertinib-sensitive EGFR-mutant human lung cancer cell lines (PC9 and H1650) that metastasize to distant organs, including the brain. These mice show remarkable initial responses to osimertinib that are analogous to human patients, with a long window of PFS followed by metastatic relapse. To identify the mechanisms underlying osimertinib-refractory relapse using these mouse models, we performed proteomic and transcriptomic profiling of relapsed brain metastatic cells and found that they express high levels of S100A9, a protein that is normally secreted by myeloid cells (28). Of note, clinical studies have found that S100A9 overexpression in lung cancer cells is correlated with poor prognosis in patients with lung cancer (29); however, the underlying molecular mechanisms remain unknown. Here, using mouse models and patient samples, we show that intracellular S100A9 expression in EGFR-mutant lung cancer cells drives brain relapse through a previously unknown S100A9–ALDH1A1–retinoic acid (RA) axis. We demonstrate that genetic inhibition of S100A9, ALDH1A1, or RA receptors (RAR), or pharmacologic inhibition of the RA pathway using pan-RAR antagonists, significantly reduces brain relapse from osimertinib-refractory cancer cells. Our study, therefore, reveals a novel S100A9–ALDH1A1–RA axis in EGFR-mutant lung cancer cells that drives osimertinib-refractory metastatic brain relapse and identifies a potential vulnerability in lung cancer cells that can be therapeutically targeted to prolong PFS in patients with EGFR-mutant lung cancer.
Results
Cancer Progression and Lethal Brain Relapse in Osimertinib-Treated EGFR-Mutant Metastatic Lung Cancer Models
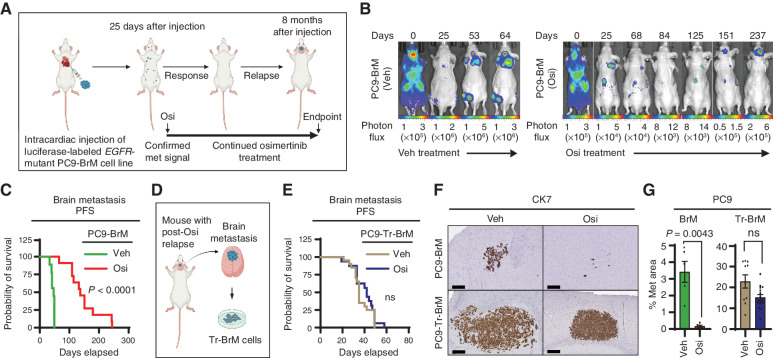
To model osimertinib response and relapse in mice, we used the human EGFR-mutant PC9-BrM3 lung cancer metastasis model (30, 31), which metastasizes to the brain, bone, and lymph nodes (31). The PC9-BrM3 cell line (referred to as “PC9-BrM” hereafter) was derived by in vivo selection for PC9 lung cancer cells (containing an EGFR exon 19 deletion) with a high incidence of brain metastasis. We engineered PC9-BrM cells to express luciferase for monitoring metastasis development by bioluminescence imaging and injected them into the arterial circulation of immunodeficient mice via intracardiac injection (Fig. 1A). After confirmation of metastatic signal at 25 days after injection by bioluminescence imaging, we initiated a long-term treatment study involving the regular administration of either vehicle or osimertinib (5 mg/kg body weight/day) 5 days per week. We monitored metastasis weekly by bioluminescence imaging (Fig. 1B). As in human patients (15, 16, 32, 33), osimertinib significantly prolonged brain metastasis PFS, from 47 days to 144 days (P < 0.0001), compared with vehicle-treated mice; however, all drug-treated mice eventually developed brain relapse and died (Fig. 1B and C). Interestingly, although cancer cells in the extracranial sites (bone and lymph nodes) in the body did not progress during continuous osimertinib treatment (Fig. 1B; Supplementary Fig. S1A), brain metastases gradually progressed (Fig. 1B and C; Supplementary Fig. S1A). To understand the underlying mechanisms of brain relapse using this model, we isolated brain metastatic cells from the relapsed brain of osimertinib-treated mice and designated them PC9-Tr-BrM (treated brain metastatic; Fig. 1D). We then injected either PC9-BrM or PC9-Tr-BrM cells into the arterial circulation of naïve immunodeficient mice and treated them with either vehicle or osimertinib. We found that the brain metastasis PFS of mice injected with PC9-Tr-BrM cells was no longer increased by osimertinib treatment (Fig. 1E). Instead, accelerated progression in the brain was observed in the PC9-Tr-BrM–injected mice compared with PC9-BrM–injected mice, as determined by histologic analysis of the brain metastasis surface area with cytokeratin 7 (CK7) immunostaining, despite continued osimertinib treatment (Fig. 1F and G). To validate these observations using a second, independent EGFR-mutant lung cancer model, we engineered H1650 lung cancer cells (harboring EGFR exon 19 and PTEN deletions; ref. 34) to express luciferase and injected them into immunodeficient mice to derive a new brain metastatic cell line (designated H1650-BrM) by the in vivo selection method (ref. 35; Supplementary Fig. S1B and S1C). We injected H1650-BrM cells into the arterial circulation of immunodeficient mice via intracardiac injection (Supplementary Fig. S1D), and after confirmation of metastasis development at 25 days after injection, we initiated a long-term treatment study involving the regular administration of either vehicle or osimertinib (5 mg/kg body weight/day) 5 days per week. Consistent with the PC9-derived model, osimertinib prolonged brain metastasis PFS in the H1650-BrM model (Supplementary Fig. S1E), albeit for a shorter duration than the PC9-BrM model (Fig. 1C). After a striking response period of 120 days, 100% of the osimertinib-treated mice developed brain relapse (45% of which also developed lung lesions) and died (Supplementary Fig. S1E). We then isolated brain metastatic cells from the osimertinib-treated mice (designated H1650-Tr-BrM), injected them into the arterial circulation of naïve immunodeficient mice, and treated the mice with either vehicle or osimertinib. Analogous to the PC9-Tr-BrM model (Fig. 1E), the brain metastasis PFS of mice injected with H1650-Tr-BrM cells was no longer prolonged by osimertinib treatment (Supplementary Fig. S1F), with rapid progression to the brain in 100% of the mice (Supplementary Fig. S1F–S1H). These results show that osimertinib initially delays metastatic progression, but eventually drug-tolerant cancer cells escape treatment and cause lethal brain relapse in two independent EGFR-mutant, metastatic lung cancer models.
Figure 1.
Fatal brain relapse in the osimertinib-treated, PC9-derived metastatic lung cancer mouse model. A, Schematic representation of the in vivo treatment model derived from the PC9-BrM cell line for metastatic EGFR-mutant lung cancer. Luciferase-labeled human EGFR-mutant PC9-BrM cells were injected into the arterial circulation of immunodeficient mice via intracardiac injection to generate metastases, which were detected by bioluminescence imaging. At 25 days after tumor cell injection, after confirmation of metastatic (met) signal, mice were administered long-term treatment with either vehicle or osimertinib (Osi) at 5 mg/kg body weight/day by oral gavage 5 days per week until the endpoint indicated in B and C, which averaged to 8 months after tumor cell injection. Periods of response to osimertinib and subsequent relapse were detected by bioluminescence imaging. B, Representative images for longitudinal monitoring of metastatic progression with vehicle (Veh) or osimertinib treatment by weekly bioluminescence imaging, with progressive development of osimertinib-refractory brain relapse in mice. Vehicle-treated mice developed bone, brain, and lymph node metastases and were euthanized when weight loss was >20% or when the body-conditioning score (BCS) reached 2. Osimertinib-treated mice were monitored for the emergence and progression of osimertinib-refractory metastasis in the brain and euthanized when either weight loss was >20%, the BCS reached 2, or mice developed paralysis or seizure-like symptoms due to brain metastasis. Days represent days after initial tumor cell injection. Photon-flux scales are indicated below the images. C, Kaplan–Meier plot for brain metastasis PFS of mice from the experiment described in A and B. Data were analyzed using the log-rank test: χ2 = 19.33, degrees of freedom (d.f.) = 1, P < 0.0001, n = 10 for vehicle-treated mice and 10 for osimertinib-treated mice. D, Schematic representation of the experimental design to derive osimertinib treatment–refractory Tr-BrM cells from relapsed brain metastases from the mice described in A–C that were injected with PC9-BrM cells and treated long-term with osimertinib. E, Kaplan–Meier plot for brain metastasis PFS of mice injected with PC9-Tr-BrM cells followed by treatment with either vehicle or osimertinib. Data were analyzed using the log-rank test: χ2 = 1.325, d.f. = 1, P value not significant (ns), n = 20 for vehicle-treated mice and 17 for osimertinib-treated mice. F, Representative images of human CK7 IHC on brain sections from mice injected with either PC9-BrM cells (top) or PC9-Tr-BrM cells (bottom) and treated with either vehicle (left) or osimertinib (right). Mice were euthanized at 7 weeks after tumor cell injection. Scale bars, 200 μm. G, Quantitative analysis of the percentage of CK7-immunostained brain sections covered by metastasis that are represented in F. Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test: n = 10 for vehicle-treated mice bearing PC9-BrM or PC9-Tr-BrM metastases and n = 11 for osimertinib-treated mice bearing PC9-BrM or PC9-Tr-BrM metastases.
S100A9 Is a Key Mediator of Brain Relapse in Osimertinib-Refractory Lung Cancer Cells
To investigate the mechanisms of brain relapse from osimertinib treatment, we first explored whether Tr-BrM cells still showed EGFR pathway inhibition in response to osimertinib. We found that osimertinib treatment led to a similar dose-dependent inhibition of EGFR and ERK phosphorylation and similar cytotoxicity profiles in the BrM and Tr-BrM derivatives from both PC9 and H1650 cell lines (Fig. 2A; Supplementary Fig. S2A–S2C), thus confirming effective target inhibition of the EGFR pathway and cytotoxicity in vitro by osimertinib. To determine whether osimertinib effectively inhibits EGFR pathway activation in situ in the brain, we next performed immunostaining analysis of phospho-EGFR tyrosine 1068 (p-EGFR) on brain sections from mice injected with PC9-BrM and H1650-BrM cells and treated with either vehicle or osimertinib. Consistent with our in vitro findings (Fig. 2A; Supplementary Fig. S2A), we found that p-EGFR was significantly reduced in both micro- and relapsed metastatic lesions in osimertinib-treated mice compared with the vehicle-treated control (Fig. 2B and C; Supplementary Fig. S2D and S2E). However, in contrast to the in vitro findings, drug-tolerant cells were able to thrive in the brain by EGFR pathway–independent mechanisms. These results suggest that brain metastatic cells are able to resist the antiproliferative and cytotoxic effects of osimertinib-mediated EGFR inhibition and grow in the brain.
Figure 2.
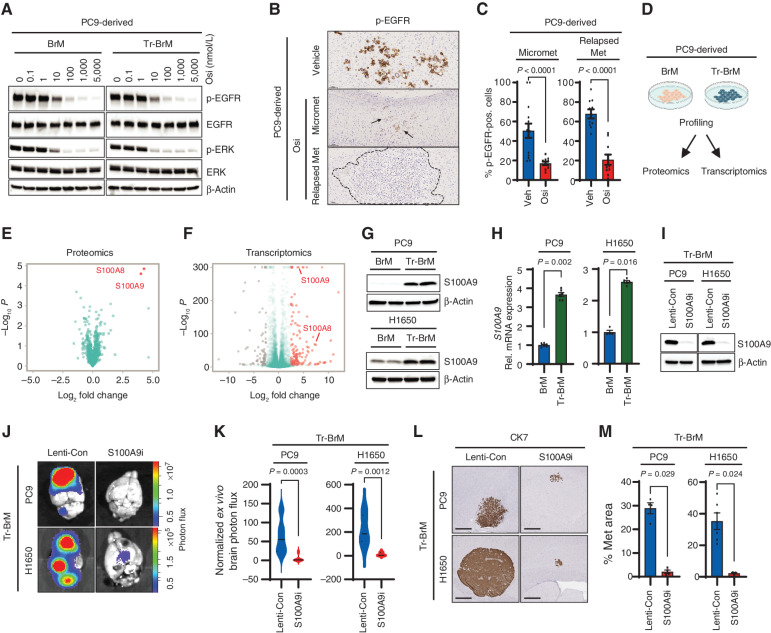
S100A9 is a key mediator of brain relapse in osimertinib-refractory lung cancer cells. A, Immunoblot analysis for inhibition of EGFR pathway activation in PC9-BrM and PC9-Tr-BrM cells treated with the indicated doses of osimertinib (Osi) and collected 6 hours after treatment. Antibodies against phospho-EGFR (Tyr1068), phospho-ERK (Thr202/Tyr204), total EGFR, total ERK, and β-actin (loading control) were used. Data are representative of three independent experiments. B, Brain sections from mice injected with PC9-BrM cells described in Fig. 1A and B were immunostained using an antibody against phospho-EGFR. PC9-BrM cells were injected into the arterial circulation of immunodeficient mice via intracardiac injection to generate metastases. Treatment was administered starting 25 days after tumor cell injection with either vehicle or osimertinib at 5 mg/kg body weight/day by oral gavage 5 days per week and continued until endpoint. The endpoint for vehicle-treated mice was 2 months after tumor cell injection (Vehicle). The endpoint for osimertinib-treated mice was 4 months after tumor cell injection for micrometastases (Micromet) and 8 months after tumor cell injection for relapsed metastatic lesion (Relapsed Met). For the osimertinib treatment group, mice were administered osimertinib by oral gavage. At 6 hours after treatment, mice were euthanized, and brain tissues were subsequently processed for histologic analysis. Representative images of IHC staining for phospho-EGFR in brain sections are shown. Arrows point to and dotted line surrounds the location of metastatic cells in the brain. Scale bars, 100 μm. C, The p-EGFR–immunostained brain sections described in B were quantitated using automated QuPath software to count p-EGFR–positive (pos.) cancer cells that were identified by setting a threshold for signal intensity (1+). Data are presented as mean values ± SEM. P values (indicated in the figures) were determined by a two-tailed, unpaired Mann–Whitney test: n = 10 for vehicle-treated mice, 14 for osimertinib-treated mice with micrometastases, and 10 for osimertinib-treated mice with relapsed metastases. Veh, vehicle. D, Schematic representation of the strategies used to compare PC9-BrM and PC9-Tr-BrM cells for differentially expressed proteins by quantitative label-free mass spectrometry and for differentially expressed genes by transcriptomics. E, Volcano plot shows the differentially expressed proteins between PC9-Tr-BrM and PC9-BrM cells identified by quantitative label-free mass spectrometry. Proteins with higher abundance in PC9-Tr-BrM cells compared with PC9-BrM cells have log2 fold changes with positive values and are labeled in red. N = 3 replicates per group. Data points referring to the top significantly differentially expressed proteins (S100A9 and S100A8) are labeled. F, Volcano plot of RNA-seq–based transcriptomic analysis shows the differentially expressed genes between PC9-Tr-BrM cells and PC9-BrM cells. Genes with significantly higher expression in PC9-Tr-BrM cells compared with PC9-BrM cells have log2 fold changes with positive values and are depicted in red. Genes with significantly lower expression in PC9-Tr-BrM cells compared with PC9-BrM cells have log2 fold changes with negative values and are depicted in gray. N = 3 replicates per group. Data points referring to the top significantly differentially expressed genes (S100A9 and S100A8) are labeled. G, Immunoblot analyses of lysates from BrM and Tr-BrM cells from both PC9- and H1650-derived models using antibodies against S100A9 and β-actin (loading control). The data are representative of three independent experiments. H, S100A9 expression was determined by qRT--PCR analysis of PC9- and H1650-derived BrM and Tr-BrM cells. GAPDH was measured as an internal control. Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test: n = 6 for PC9-BrM, n = 6 for PC9-Tr-BrM, n = 4 for H1650-BrM, and n = 5 for H1650-Tr-BrM. Rel., relative. I, Immunoblot analyses of lysates from PC9- and H1650-derived Tr-BrM cells infected with viruses expressing either a control gRNA (Lenti-Con) or an S100A9-specific gRNA (referred to as “S100A9i” throughout the figures). The indicated antibodies were used to confirm the loss of S100A9 protein expression following CRISPR/dCas9-mediated gene repression. β-Actin served as a protein loading control. Data are representative of three independent experiments. J,Ex vivo photon flux of brains from mice injected with PC9- or H1650-derived Tr-BrM cells expressing either Lenti-Con or S100A9i was determined by bioluminescence imaging. Mice were collected at 7 weeks after tumor cell injection. The photon-flux scale is indicated on the right side. K, Violin plots depicting normalized photon flux of brains imaged ex vivo from the mice described in J. The normalized photon flux for brain tissue was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (i.e., the day of injection) and multiplying that value by 100. Data are presented as mean values ± SEM. P values were determined by a two--tailed, unpaired Mann–Whitney test. For PC9 Tr-BrM, n = 10 for Lenti-Con and n = 9 for S100A9i. For H1650-Tr-BrM, n = 6 for Lenti-Con and n = 7 for S100A9i. L, Representative images of CK7 IHC on brain sections from mice injected with either PC9-derived Tr-BrM Lenti-Con–expressing (left) or S100A9i-expressing (right) cells in the top row, or H1650-derived Tr-BrM Lenti-Con–expressing (left) or S100A9i-expressing (right) cells in the bottom row. Brains were harvested from mice 7 weeks after tumor cell injection. Scale bars, 500 μm. M, Quantitative analysis of the percentage of brain sections covered by metastasis from the experiment described in L. Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test. For PC9-Tr-BrM, n = 4 for Lenti-Con and n = 4 for S100A9i. For H1650-Tr-BrM, n = 6 for Lenti-Con and n = 3 for S100A9i.
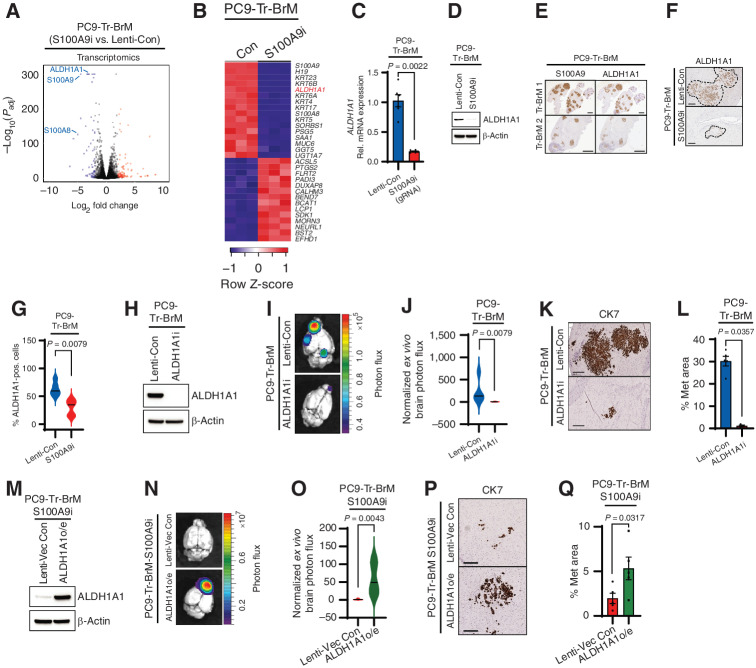
To identify pathways that promote the growth and survival of Tr-BrM cells in the brain, we performed quantitative label-free mass spectrometry and transcriptomics comparing PC9-BrM and PC9-Tr-BrM cells (Fig. 2D). S100A8 and S100A9, two calcium-binding proteins that form a heterodimer and are normally secreted by myeloid cells (36, 37), emerged as the top upregulated candidates in the PC9-Tr-BrM cells compared with PC9-BrM cells by proteomic profiling (Fig. 2E). Functional annotation analysis using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) showed S100A8 and S100A9 were enriched in 23 of 30 (77%) pathways (Supplementary Tables S1 and S2). Transcriptomic profiling by RNA sequencing (RNA-seq) and functional enrichment analysis by G:Profiler (38) identified broad gene ontology (GO) categories that were significantly enriched in the PC9- and H1650-derived BrM and Tr-BrM cells (Supplementary Fig. S2F–S2I). Consistent with the proteomics analysis (Fig. 2E), S100A8 and S100A9 were also among the significantly upregulated genes by RNA-seq in the Tr-BrM cells compared with BrM cells from both PC9 and H1650 models (Fig. 2F; Supplementary Fig. S2F and S2G; Supplementary Tables S3 and S4). Increased expression of S100A8 and S100A9 was validated by immunoblot and qRT-PCR analyses in both PC9- and H1650-derived Tr-BrM cells compared with their respective BrM controls (Fig. 2G and H; Supplementary Fig. S3A and S3B). Immunostaining analysis of brain sections showed prominent intracellular expression of S100A9 in metastatic cells derived from both PC9 and H1650 models (Supplementary Fig. S3C and S3D). Consistent with these observations, secreted S100A9 was not detected in the sera from mice bearing brain metastases in either the PC9- or H1650-derived models by ELISA analysis, and the majority of S100A9 was detected in the cell lysate rather than the culture supernatant (Supplementary Table S5).
To determine whether S100A8/S100A9 expression is causally linked with brain metastasis development, we performed loss-of-function studies. We suppressed the expression of S100A8 and S100A9 using CRISPR repression (CRISPRi) in both PC9- and H1650-derived Tr-BrM cells (Fig. 2I; Supplementary Fig. S3E–S3I). We found that individual repression of S100A8 and S100A9 in Tr-BrM cells derived from both PC9 and H1650 cell lines using independent guide RNAs (gRNA) led to a significant reduction in brain metastasis at endpoint (7 weeks after injection), as determined by quantitative bioluminescence imaging and by histologic analysis of the metastasis surface area with CK7 immunostaining (Fig. 2J–M; Supplementary Fig. S3J–S3O). S100A8 and S100A9 function together as a heterodimer, and S100A8 expression is downregulated in S100A9-deficient neutrophils (28, 36). We, therefore, examined whether S100A9 repression (S100A9i) leads to downregulation of S100A8 in EGFR-mutant lung cancer cell lines. Indeed, S100A8 expression was significantly reduced in the PC9- and H1650-derived Tr-BrM-S100A9i cells compared with their respective lenti-control Tr-BrM cells (Supplementary Fig. S3P and S3Q). We, therefore, used S100A9 repression as a surrogate for studying the functional loss of S100A8/9 in Tr-BrM cells in subsequent experiments. Our results demonstrate that elevated expression of S100A9 (and S100A8) promotes brain metastasis of EGFR-mutant lung cancer cells and becomes an alternative mechanism to thrive in the brain while under stress from EGFR pathway inhibition.
S100A9-Proficient Cancer Cells Promote Postcolonization Growth in the Brain
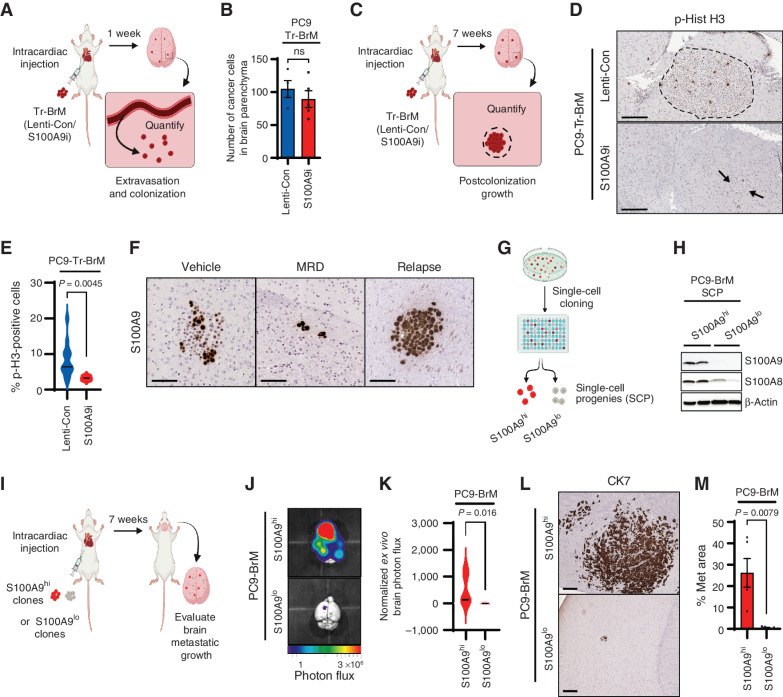
The ability of cancer cells to develop metastases in the brain depends on their ability to extravasate from blood vessels into the brain parenchyma (known as metastatic seeding) and subsequently adapt, survive, and grow in the brain microenvironment (known as postcolonization outgrowth; refs. 39–41). To determine how S100A9 drives brain metastasis, we asked which of these steps during brain metastasis require S100A9 expression. To test whether S100A9 is required for metastatic seeding in the brain, we injected PC9-derived Tr-BrM cells expressing lenti-control (Tr-BrM-Lenti-Con) or S100A9i (Tr-BrM-S100A9i) into the arterial circulation of immunodeficient mice via intracardiac injection (Fig. 3A). Seven days following injection, a time point when lung cancer cells extravasate and can be detected in the brain parenchyma (41), we harvested, sectioned, and immunostained brain tissues with an antibody against CK7 to quantitate the number of cancer cells that seeded in the brain. We found no difference in the number of extravasated cells in the brain parenchyma between the S100A9-proficient and S100A9-deficient groups (Fig. 3B), indicating that S100A9 is not required for metastatic seeding in the brain in the PC9-derived model. To test whether S100A9 is instead required for postcolonization growth in the brain, we immunostained brain sections from mice harvested seven weeks following tumor cell injection (Fig. 3C), using an antibody against phospho-histone H3 (Ser10; Fig. 3D). Consistent with larger metastatic lesions in the Tr-BrM-Lenti-Con group compared with the Tr-BrM-S100A9i group (Fig. 2L–M), a significantly higher number of mitotically active, phospho-histone H3–positive cells were observed in the Tr-BrM-Lenti-Con group (Fig. 3D and E). However, no differences in proliferation were observed in vitro between the BrM and Tr-BrM cells or the Tr-BrM-Lenti-Con and Tr-BrM-S100A9i cells that were derived from either PC9 or H1650 cell lines (Supplementary Fig. S3R–S3U). These results indicate that S100A9 is required for the postcolonization growth of metastatic lung cancer cells in situ in the brain.
Figure 3.
S100A9-proficient cells promote postcolonization growth in the brain. A, Schematic representation of the experimental design to quantify seeding in the brain. PC9-Tr-BrM cells expressing either lenti-control (Lenti-Con) or S100A9i were injected into the arterial circulation of immunodeficient mice via intracardiac injection. Seven days later, brains were isolated, sectioned, and analyzed by IHC for human CK7. CK7-immunostained cancer cells were then counted to compare seeding of cancer cells in the brain parenchyma between experimental groups. B, Quantitative analysis of the experiment described in A. Tumor cells were counted in 10 sections of 20 μm each per brain. Data are presented as mean values ± SEM. The P value was determined by a two--tailed, unpaired Mann–Whitney test. N = 4 for Lenti-Con; n = 5 for S100A9i. ns, P value not significant. C, Schematic representation of the experimental design to analyze postcolonization growth in the brain. PC9-Tr-BrM cells expressing either Lenti-Con or S100A9i were injected into the arterial circulation of immunodeficient mice via intracardiac injection. At 7 weeks after injection, brain tissues were collected, and sections were analyzed by immunostaining for phospho-histone H3 (Ser10) to compare the number of mitotically active cancer cells between the experimental groups. D, Representative images of phospho-histone H3 (p-Hist H3) IHC on brain sections from the experiment described in C. Arrows point to and dotted line surrounds the location of metastatic cells in the brain. Scale bars, 100 μm. E, Quantitative analysis of the phospho-histone H3–positive cells within brain sections from the experiment described in C and represented in D. Immunostained sections were counted using the QuPath software, where positively stained cells are identified by setting a threshold for signal intensity (3+). Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test: n = 12 for Lenti-Con and n = 6 for S100A9i. F, Representative images of brain sections stained with an antibody against human S100A9. PC9-BrM cells were injected into the arterial circulation of immunodeficient mice via intracardiac injection. After metastatic signal was detected by bioluminescence imaging, treatment was started 25 days after tumor cell injection with either vehicle or osimertinib at 5 mg/kg body weight/day by oral gavage 5 days per week. Brain tissues were collected 2 months after tumor cell injection in the vehicle treatment group (Vehicle) 3 months after tumor cell injection in the osimertinib treatment group (minimal residual disease, or MRD) and 8 months after tumor cell injection in the osimertinib-treated relapse group (Relapse). Scale bars, 100 μm. Data are representative of 10 mice/group analyzed at each time point. G, Schematic representation of single-cell cloning from PC9-BrM cells. S100A9 high- and low-expressing single-cell progenies (SCP) are labeled as S100A9hi and S100A9lo, respectively. H, Immunoblot analysis of lysates from PC9-BrM–derived SCPs using antibodies against S100A8, S100A9, and β-actin (loading control). The data are representative of three independent experiments. I, Schematic representation of the brain metastasis assay to compare the ability of S100A9hi and S100A9lo SCPs to grow in the brain and generate metastases. J,Ex vivo photon flux of brains from mice injected with PC9-BrM–derived S100A9hi and S100A9lo SCPs was determined by bioluminescence imaging. Brains were collected from mice 7 weeks after tumor cell injection. Photon-flux scale is indicated below the images. K, Violin plots depicting normalized photon flux of brains imaged ex vivo from mice described in J. The normalized photon flux for brain tissue was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (i.e., the day of injection) and multiplying that value by 100. Data are presented as mean values ± SEM. P values were determined by a two--tailed, unpaired Mann–Whitney test. N = 5 for S100A9hi; n = 4 for S100A9lo. L, Representative images of CK7 IHC on brain sections from mice injected with PC9-BrM–derived S100A9hi and S100A9lo SCPs. Brains were harvested from mice 7 weeks after tumor cell injection. Scale bars, 200 μm. M, Quantitative analysis of the percentage of CK7-immunostained brain sections covered by metastasis (Met area) in the experiment described in L. Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test: n = 5 for S100A9hi and n = 5 for S100A9lo.
Based on these results, we reasoned that although both S100A9-proficient and S100A9-deficient cells can colonize the brain, the S100A9-proficient cells are likely to grow better in the brain, a biological trait that can potentially sustain their growth and survival after EGFR pathway inhibition by osimertinib. In line with this hypothesis, we observed the presence of both S100A9hi and S100A9lo cells in brain sections from PC9-BrM–injected mice treated with vehicle for 2 months (Fig. 3F, vehicle). However, this scenario changed dramatically after prolonged osimertinib treatment, where S100A9hi cells became prominent in the surviving metastatic cells following 3 months of osimertinib treatment (Fig. 3F, MRD), and predominated in relapsed brain metastases following 8 months of osimertinib treatment (Fig. 3F, relapse). Based on these findings, we reasoned that if osimertinib treatment selects for S100A9hi cells, then the PC9-BrM cell line should exhibit heterogeneity with respect to preexisting S100A9 expression levels that are present prior to drug treatment and brain metastasis. Indeed, single-cell cloning of the PC9-BrM line gave rise to distinct S100A9hi and S100A9lo single-cell progenies (SCP) in culture (Fig. 3G and H), which were then compared for their ability to grow in the brain. Bioluminescence imaging showed a striking increase in brain metastasis by S100A9hi SCPs compared with S100A9lo SCPs when an equal number of cells from each group were injected into the arterial circulation of immunodeficient mice (Fig. 3I–K). These results were further validated by histologic analysis of the metastasis surface area with CK7 immunostaining (Fig. 3L and M). These findings indicate that osimertinib treatment selects S100A9hi cells for growth and survival in the brain, from a preexisting pool of lung cancer cells that exhibit heterogeneity for S100A9 expression. S100A9hi cells thereby serve as seeds of future relapse from osimertinib treatment.
Association of S100A9 Expression with Brain Metastasis and Shorter PFS in Patients with Osimertinib-Treated Lung Cancer
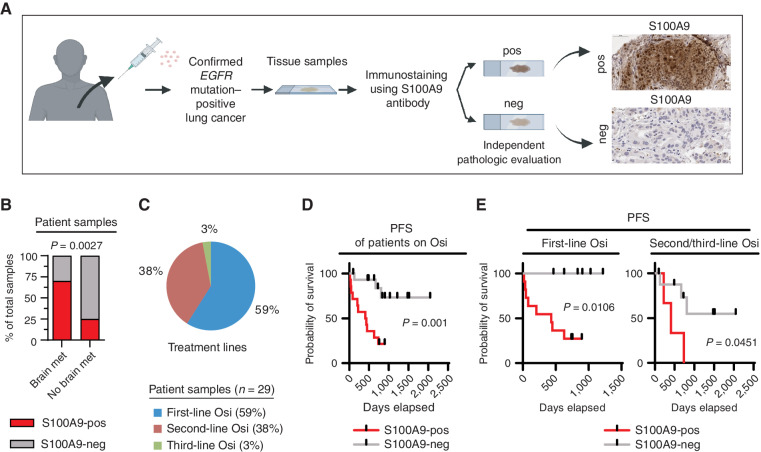
Our preclinical studies revealed two distinct functions of S100A9: to promote brain metastatic growth and to escape the growth-inhibitory effects of osimertinib. To clinically validate our experimental findings, we performed S100A9 immunostaining on tissue specimens that were obtained prior to osimertinib treatment from 29 patients with EGFR-mutant lung cancer (Fig. 4A; Supplementary Table S6). The immunostained samples were scored as either S100A9-positive (any percentage of clear, positive intracellular S100A9 staining in cancer cells) or S100A9-negative (no detectable S100A9 staining in cancer cells; Fig. 4A). Consistent with our preclinical observations (Fig. 2J–M and Fig. 3J–M), an independent blinded pathologic examination revealed a statistically significant association between S100A9 expression and the development of brain metastasis (P = 0.0027; Fig. 4B). We next asked whether S100A9 expression in pre–osimertinib treatment cancer cells correlated with osimertinib treatment response in a combined cohort of patients on first-, second-, and third-line osimertinib treatment (Fig. 4C and D). Indeed, high expression of S100A9 in cancer cells from pre–osimertinib treatment samples correlated significantly with worse PFS on osimertinib (n = 29, P = 0.0011), both in the combined cohort (Fig. 4D) and when stratified by treatment lines (n = 17, P = 0.0106 for first-line osimertinib patients; n = 12, P = 0.0451 for second- and third-line patients; Fig. 4E). Therefore, based on our preclinical studies and clinical validation, elevated S100A9 expression in cancer cells is significantly associated with brain metastasis and strongly correlates with progression in patients with osimertinib-treated lung cancer.
Figure 4.
High S100A9 expression is associated with brain metastasis and shorter PFS in patients with osimertinib-treated lung cancer. A, Schematic representation of the analysis of patient samples for S100A9 expression in cancer cells. S100A9 immunostaining was performed on tissue specimens (biopsies/resected material) from 29 patients with lung cancer with a validated EGFR mutation that were obtained prior to osimertinib treatment. The immunostained samples were scored by independent pathologists as either S100A9-positive (any percentage of clear, positive intracellular S100A9 staining in cancer cells) or S100A9-negative (no detectable S100A9 staining in cancer cells). neg, S100A9-negative; pos, S100A9-positive. B, Graphical representation of the association between S100A9 expression in the patient tissue specimens described in A and the development of brain metastasis (met) for 26 patients with a clinical annotation for the presence or absence of brain metastasis at diagnosis (three of 29 patients had unknown brain metastasis status at diagnosis). The P value was determined by a χ2 test: n = 10 samples from patients with brain metastasis and n = 16 samples from patients without brain metastasis. C, Distribution of patients on first-, second-, and third-line osimertinib (Osi) treatment from the 29-patient cohort described in A. D, Kaplan–Meier plot for the PFS of osimertinib-treated patients from the combined cohort described in A. Data were analyzed using the log-rank test: χ2 = 10.74, degrees of freedom (d.f.) = 1, P = 0.0001, n = 29 patients. Patients who had not progressed at the time of analysis were censored. E, Kaplan–Meier plot for PFS of the osimertinib-treated patients described in A and C. Data were analyzed using the log-rank test. For first-line osimertinib-treated patients: χ2 = 6.011, d.f. = 1, P = 0.0106, n = 17; for second- and third-line osimertinib-treated patients: χ2 = 4.015, d.f. = 1, P = 0.0451, n = 12. Patients who had not progressed at the time of analysis were censored.
S100A9 Promotes Brain Relapse through ALDH1A1
To further explore how S100A9 mediates the growth of brain metastatic lesions, we analyzed the transcriptome of S100A9-proficient (Tr-BrM-Lenti-Con) and S100A9-deficient (Tr-BrM-S100A9i) brain metastatic cells from the PC9- and H1650-derived models by RNA-seq (Fig. 5A and B; Supplementary Fig. S4A—S4C; Supplementary Tables S7 and S8). Consistent with our previous results (Supplementary Fig. S3P and S3Q), S100A8 was among the top downregulated genes in both PC9- and H1650-derived Tr-BrM-S100A9i cells (Fig. 5A and B; Supplementary Fig. S4A; Supplementary Tables S7 and S8). Interestingly, aldehyde dehydrogenase 1 family member A1 (ALDH1A1), which encodes an enzyme that catalyzes the conversion of retinaldehyde to RA, was among the top downregulated genes in both PC9- and H1650-derived Tr-BrM-S100A9i cells (Fig. 5B; Supplementary Fig. S4A). Gene set enrichment analysis further revealed a significant decrease in the expression of retinol metabolism genes in the Tr-BrM-S100A9i cells [GO Biological Process (GOBP) RA metabolic process, P = 0.024, and Kyoto Encyclopedia of Genes and Genomes (KEGG) retinol metabolism, P = 0.026; Supplementary Fig. S4D and S4E]. Moreover, the analysis of genes present at the leading edges of both the GOBP RA metabolic process and KEGG retinol metabolism gene sets confirmed enrichment for genes associated with RA metabolism in PC9-Tr-BrM compared with PC9-BrM cells (Supplementary Fig. S4F). The leading-edge genes from both GOBP RA metabolic process and KEGG retinol metabolism were also significantly downregulated upon S100A9 repression in PC9-Tr-BrM cells (Supplementary Fig. S4G). These results suggest that S100A9 activates the RA pathway in Tr-BrM cells.
Figure 5.
S100A9 promotes brain relapse through ALDH1A1. A, Volcano plot shows the significantly differentially expressed genes between PC9-Tr-BrM cells expressing either Lenti-Con or S100A9i gRNAs as identified by RNA-seq analysis. Genes with significantly higher expression in PC9-Tr-BrM-S100A9i cells compared with PC9-Tr-BrM-Lenti-Con cells have log2 fold changes with positive values and are depicted in red. Genes with significantly lower expression in PC9-Tr-BrM-S100A9i cells compared with PC9-Tr-BrM-Lenti-Con cells have log2 fold changes with negative values and are depicted in blue. N = 3 per group. Genes with an adjusted P value of less than 1.0 × 10−4 and an absolute value of the log2 fold change of greater than 2.4 were considered significant. B, Heat map of the top significantly upregulated and downregulated genes in PC9-Tr-BrM–derived S100A9i-expressing cells versus Lenti-Con–expressing cells (Con). Normalized gene expression above the row mean is indicated by progressively darker shades of red, and normalized gene expression below the row mean is indicated by progressively darker shades of blue. Genes with an adjusted P value of less than 1.0 × 10−4 and an absolute value of the log2 fold change of greater than 2.4 were considered significant. C,ALDH1A1 expression was determined by qRT--PCR analysis in PC9-derived Tr-BrM cells expressing either Lenti-Con or S100A9i. GAPDH was measured as an internal control. Data are presented as mean values ± SEM. The P value was determined by a two-tailed, unpaired Mann–Whitney test: n = 6 for Lenti-Con and n = 6 for S100A9i. D, Immunoblot analysis of lysates from PC9-Tr-BrM cells expressing either Lenti-Con or S100A9i using antibodies against ALDH1A1 and β-actin (loading control). The data are representative of three independent experiments. Rel., relative. E, Representative images of serial brain sections stained with an antibody against S100A9 (left) and ALDH1A1 (right) taken from two different mice (designated “Tr-BrM 1” and “Tr-BrM 2”) injected with PC9-Tr-BrM cells. Brains were collected at 7 weeks after tumor cell injection. Images are representative of eight mice analyzed per group. Scale bars, 2,000 μm. F, Representative images of brain sections stained with an antibody against ALDH1A1 taken from mice injected with PC9-Tr-BrM cells expressing either Lenti-Con or S100A9i and collected 7 weeks after tumor cell injection. Scale bars, 200 μm. G, Quantitative analysis of the ALDH1A1-positive cells (pos.) shown in F. Immunostained sections were counted using the QuPath software, where positively stained cells were identified by setting a threshold for signal intensity (1+). Data are presented as mean values ± SEM. The P value was determined by a two--tailed, unpaired Mann–Whitney test: n = 5 for Lenti-Con and n = 5 for S100A9i. H, Immunoblot analysis of lysates from PC9-Tr-BrM cells expressing either Lenti-Con or ALDH1A1i using antibodies against ALDH1A1 and β-actin (loading control). The data are representative of three independent experiments. I,Ex vivo photon flux of brains from mice injected with PC9-Tr-BrM–derived cells expressing either Lenti-Con or ALDH1A1i was determined by bioluminescence imaging. Brains were collected from mice 7 weeks after tumor cell injection. Photon-flux scale is indicated on the right side. J, Violin plots depicting normalized photon flux of brains imaged ex vivo from the mice described in I. The normalized photon flux for brain tissue was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (i.e., the day of injection) and multiplying that value by 100. Data are presented as mean values ± SEM. The P value was determined by a two--tailed, unpaired Mann–Whitney test: n = 5 for Lenti-Con and n = 5 for ALDH1A1i. K, Representative images from CK7 IHC on brain sections from mice injected with PC9-Tr-BrM-Lenti-Con cells (top) or PC9-Tr-BrM-ALDH1A1i cells (bottom). Brains were harvested from mice 7 weeks after tumor cell injection. Scale bars, 200 μm. L, Quantitative analysis of the percentage of CK7-immunostained brain sections covered by metastasis from the experiment described in K. Data are presented as mean values ± SEM. The P value was determined by a two-tailed, unpaired Mann–Whitney test: n = 5 for Lenti-Con and n = 3 for ALDH1A1i. M, Immunoblot analysis of lysates from PC9-Tr-BrM-S100A9i cells expressing either lenti-vector control (Lenti-Vec Con) or ALDH1A1 (“ALDH1A1o/e” denotes ALDH1A1 overexpression) using antibodies against ALDH1A1 and β-actin (loading control). Data are representative of three independent experiments. N,Ex vivo photon flux of brains from mice injected with PC9-Tr-BrM-S100A9i cells expressing either Lenti-Vec Con or ALDH1A1 was determined by bioluminescence imaging. Brains were collected from mice 7 weeks after tumor cell injection. The photon-flux scale is indicated on the right. O, Violin plots depicting normalized photon flux of brains imaged ex vivo from the mice described in N. The normalized photon flux for brain tissue was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (i.e., the day of injection) and multiplying that value by 100. Data are presented as mean values ± SEM. The P value was determined by a two--tailed, unpaired Mann–Whitney test: n = 6 for Lenti-Vec Con; n = 5 for ALDH1A1o/e. P, Representative images of CK7 IHC on brain sections from mice injected with PC9-Tr-BrM S100A9i cells expressing either Lenti-Vec Con (top) or ALDH1A1 (bottom). Brains were harvested from mice 7 weeks after tumor cell injection. Scale bars, 200 μm. Q, Quantitative analysis of the percentage of CK7-immunostained brain sections covered by metastasis (Met) from the experiment described in P. Data are presented as mean values ± SEM. The P value was determined by a two-tailed, unpaired Mann–Whitney test: n = 5 for Lenti-Vec Con and n = 6 for ALDH1A1o/e.
RA, an active metabolite of retinol (vitamin A), binds to nuclear hormone receptors to regulate diverse cellular processes, including proliferation, tissue remodeling, and differentiation (42, 43). For RA biosynthesis, retinol is first oxidized by alcohol dehydrogenase (ADH) enzymes to retinaldehyde. Retinaldehyde is further oxidized to RA by the aldehyde dehydrogenase (ALDH) family of enzymes, mainly ALDH1A1, ALDH1A2, and ALDH1A3. Among the ALDH1A family members, ALDH1A1 was only significantly downregulated by S100A9 repression in both PC9- and H1650-derived Tr-BrM cells (Supplementary Fig. S4H). We confirmed a reduction in RNA and protein expression of ALDH1A1 in PC9- and H1650-derived Tr-BrM-S100A9i cells compared with their respective controls (Fig. 5C and D; Supplementary Fig. S4I and S4J). Moreover, IHC analysis showed robust expression of ALDH1A1 and a striking overlap between S100A9 and ALDH1A1 in brain metastatic lesions from the PC9-Tr-BrM model (Fig. 5E). Importantly, ALDH1A1 was significantly downregulated by S100A9 repression in brain metastatic lesions from the PC9- and H1650-Tr-BrM models (Fig. 5F and G; Supplementary Fig. S4K). We, therefore, asked whether S100A9 promotes brain metastasis through upregulation of ALDH1A1. To test this possibility, we first analyzed whether repression of ALDH1A1 can phenocopy S100A9i in PC9- and H1650-derived Tr-BrM cells. We confirmed successful repression of ALDH1A1 in PC9- and H1650-derived Tr-BrM cells (Fig. 5H; Supplementary Fig. S4L) and found that ALDH1A1 repression in the Tr-BrM cells significantly reduced brain metastasis, as quantified by bioluminescence imaging and by histologic analysis of the metastasis surface area with CK7 immunostaining (Fig. 5I–L; Supplementary Fig. S4M–S4P). We also found that forced expression of ALDH1A1 (ALDH1A1o/e) was sufficient to rescue the S100A9i phenotype in both PC9- and H1650-derived models (Fig. 5M–Q; Supplementary Fig. S4Q–S4W), indicating that ALDH1A1 represents a key downstream effector of S100A9 that mediates brain metastasis. No differences in proliferation were observed in vitro among the PC9- and H1650-Tr-BrM cells transduced with lentivirus encoding either Lenti-Con, S100A9i, ALDH1A1i, or S100A9i–ALDH1A1o/e, suggesting that the S100A9–ALDH1A1–RA axis is not required for cell growth in vitro (Supplementary Fig. S4X and S4Y). These results therefore demonstrate that osimertinib-refractory lung cancer cells co-opt the S100A9–ALDH1A1 signaling axis to survive and grow in the brain despite inhibition of EGFR activity by osimertinib.
Site-Specific Effects of the S100A9–ALDH1A Axis
We next asked whether the S100A9–ALDH1A1 axis promotes the growth of cancer cells selectively in the brain or if it also promotes growth in the lung and bone, two additional sites of growth for H1650 and PC9 derivatives, respectively (Fig. 1; Supplementary Fig. S1). As H1650-derived BrM cells can grow in both the brain and lung (Supplementary Fig. S1C and S1E), we first asked whether the expression of S100A9 and ALDH1A1 is elevated in H1650-BrM–derived lung lesions post–osimertinib treatment (referred to as “Osi-relapse”) compared with lung lesions from vehicle-treated mice, similar to what we observe for brain lesions (Figs. 2 and 3; Supplementary Fig. S3). To address this question, we first injected H1650-BrM cells into the arterial circulation of immunodeficient mice, treated them with either vehicle or osimertinib, and then harvested lung and brain tissues for IHC analysis. Lung and brain tissues were collected at endpoint (2 months in the vehicle-treated group and 4 months after tumor cell injection in the Osi-relapse group). In contrast to the brain lesions, lung lesions showed no statistically significant increase in S100A9 and ALDH1A1 expression from the Osi-relapse group compared with the vehicle-treated group (Supplementary Fig. S5A–S5F). To evaluate the requirement of the S100A9–ALDH1A1 axis for cancer cell growth in the lung, we injected H1650-Tr-BrM cells (expressing high S100A9 and ALDH1A1 levels) that were transduced with lentivirus encoding either Lenti-Con, S100A9i (Fig. 2I), ALDH1A1i (Supplementary Fig. S4L), or S100A9i–ALDH1A1o/e (Supplementary Fig. S4R) into the arterial circulation of immunodeficient mice via intracardiac injection (Supplementary Fig. S5G). Compared with the robust brain metastasis phenotype (Fig. 2J–M; Supplementary Figs. S4M–S4P and S4T–S4W), we observed a modest but statistically significant reduction in the growth of lung lesions upon suppression of S100A9 or ALDH1A1, and a rescue by ALDH1A1 expression (in an S100A9i background), as determined by quantitative bioluminescence imaging (Supplementary Fig. S5H and S5I). To confirm these findings, we isolated H1650 lung derivatives from osimertinib-treated mice injected with H1650-BrM cells (abbreviated as “H1650-lung derivatives”) and transduced them with either Lenti-Con, S100A9i, ALDH1A1i, or S100A9i–ALDH1A1o/e (Supplementary Fig. S5J). We then directly implanted these cells in the lung of immunodeficient mice and evaluated the growth of these cells in the lung at endpoint (3 weeks after injection; Supplementary Fig. S5K). Consistent with our previous findings (Supplementary Fig. S5H and S5I), a modest but statistically significant reduction was observed in lung tumor growth upon S100A9 or ALDH1A1 repression, which was rescued by forced ALDH1A1 expression (in an S100A9i background; Supplementary Fig. S5L). Taken together, these data show that the S100A9–ALDH1A1 axis promotes the growth of EGFR-mutant lung cancer cells in the lung, albeit to a lesser extent than in the brain.
PC9-derived BrM cells can grow in the brain and bone after injection into the arterial circulation (Fig. 1B; Supplementary Fig. S1A). Therefore, we next tested whether the expression of S100A9 and ALDH1A1 is elevated in the PC9-BrM–derived bone lesions post–osimertinib treatment compared with vehicle-treated mice following tumor cell injection, analogous to what we observed for brain lesions. We analyzed the bone lesions at 5 months following tumor cell injection, which is an intermediate time point before osimertinib effectively eliminates bone metastatic lesions in this model. In contrast to the brain, IHC analysis showed no significant increases in either S100A9 or ALDH1A1 expression in the bone metastatic lesions from the osimertinib-treated group (referred to as “Osi-residual tumor”) compared with bone metastatic lesions from the vehicle-treated group (Supplementary Fig. S5M–S5R). To confirm these findings, we isolated PC9 bone derivatives from osimertinib-treated mice injected with PC9-BrM cells (referred to as “PC9-bone derivatives”) 5 months following tumor cell injection. We transduced the PC9-bone derivatives with lentivirus encoding either Lenti-Con, S100A9i, ALDH1A1i, or S100A9i–ALDH1A1o/e (Supplementary Fig. S5S). To evaluate the requirement of the S100A9–ALDH1A1 axis for growth in the bone, we directly implanted these transduced PC9-bone derivatives in the tibia bone of immunodeficient mice and evaluated the growth of these cells in the bone at endpoint (3 weeks after injection; Supplementary Fig. S5T). No significant differences were observed in tumor growth in the bone between these groups, as determined by quantitative bioluminescence imaging (Supplementary Fig. S5U). These results suggest site-specific functions of the S100A9–ALDH1A1 axis, which promotes metastatic growth in the brain, to a lesser extent in the lung but not in the bone.
Osimertinib-Refractory Tumor Cells from Brain Metastases Are Sensitive to Pan-RAR Inhibition
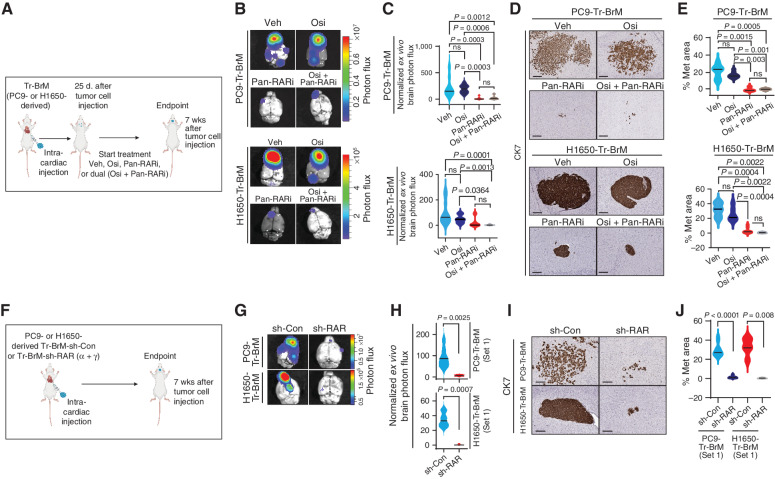
The physiologic functions of RA are mediated primarily through binding to two families of retinoid nuclear receptors, the RARs (RAR alpha, RAR beta, and RAR gamma) and RXRs (RXR alpha, RXR beta, and RXR gamma), that function as ligand-dependent transcription factors (43). RA-bound RAR/RXR heterodimers bind to target genes at RA-response elements (RARE) to regulate their transcriptional activation (43). As high expression of ALDH1A1 mediates S100A9-dependent brain metastasis, we hypothesized that activation of the RA-response pathway enables Tr-BrM cells to grow in the brain in the presence of osimertinib. To test the sensitivity of Tr-BrM cells to RAR pathway inhibition, we pharmacologically challenged PC9- and H1650-derived Tr-BrM cells with RAR pathway antagonists. Treatment of PC9- and H1650-Tr-BrM cells with retinol in serum-free media significantly induced expression of the RAR target gene STRA6 in vitro (Supplementary Fig. S6A and S6B), indicating functional RA biosynthetic and response pathways. Interestingly, STRA6 expression was only modestly inhibited when treated with either the RAR-alpha antagonist BMS195614 (abbreviated as RARαi) or the RAR-gamma antagonist MM11253 (abbreviated as RARγi), but was dramatically reduced with the pan-RAR antagonist AGN194310 (abbreviated as “pan-RARi”; Supplementary Fig. S6A and S6B). In line with these observations, retinol-treated PC9- and H1650-Tr-BrM cells showed a striking dose-dependent cytotoxicity with pan-RAR antagonism compared with PC9- and H1650-BrM cells (Supplementary Fig. S6C). Conversely, two other EGFR-mutant lung cancer cell lines, H1975 and HCC4006, that lack expression of S100A9 and ALDH1A1 did not show dose-dependent loss of viability with AGN194310 (Supplementary Fig. S6C–S6E).
To determine whether treatment with AGN194310 inhibits brain metastasis in vivo, we injected PC9- and H1650-Tr-BrM cells into the arterial circulation of immunodeficient mice via intracardiac injection (Fig. 6A). We confirmed metastatic signal by bioluminescence imaging at 25 days and initiated a treatment study involving the administration 5 days per week of either (i) vehicle, (ii) AGN194310 (0.5 mg/kg body weight/day), (iii) osimertinib (5 mg/kg body weight/day), or (iv) AGN194310 (0.5 mg/kg body weight/day) plus osimertinib (5 mg/kg body weight/day). A striking reduction in brain metastasis was observed in mice treated with AGN194310, compared with vehicle control and osimertinib-alone groups, by bioluminescence imaging and histologic analysis of the metastasis surface area with CK7 immunostaining (Fig. 6B–E; Supplementary Fig. S6F and S6G). To complement our pharmacologic inhibition studies, we generated PC9- and H1650-Tr-BrM cells expressing two independent sets of short hairpins (shRNA) targeting both RARα and RARγ (Supplementary Fig. S6H–S6K). Consistent with the pharmacologic studies (Fig. 6B–E), a striking reduction in brain metastasis was observed with both sets of shRNAs targeting RARα and RARγ in both PC9- and H1650-derived cells, as determined by quantitative bioluminescence imaging and by histologic analysis of the metastasis surface area with CK7 immunostaining (Fig. 6F–J; Supplementary Fig. S6L–S6O). We next asked whether AGN194310 also affects tumor growth in the bone or lung. Similar to our genetic suppression experiments (Supplementary Fig. S5), AGN194310 treatment modestly reduced lung (but not bone) lesions in the PC9- and H1650-derived models when directly implanted in the lung and bone, respectively (Supplementary Fig. S6P–S6S). Our study therefore reveals a therapeutic vulnerability in osimertinib-refractory, brain metastatic lung cancer cells that can be targeted by pan-RAR antagonism.
Figure 6.
Osimertinib-refractory cancer cells are sensitive to pan-RAR inhibition. A, Schematic representation of the treatment of PC9- and H1650-derived Tr-BrM cells with vehicle, osimertinib alone, a pan-RAR antagonist (AGN194310) alone, or AGN194310 in combination with osimertinib. At 25 days after tumor cell injection, after confirmation of metastatic signal, mice were administered long-term treatment with either (i) vehicle (Veh), (ii) AGN194310 (Pan-RARi; 0.5 mg/kg body weight/day), (iii) osimertinib (Osi; 5 mg/kg body weight/day), or (iv) AGN194310 (0.5 mg/kg body weight/day) plus osimertinib (5 mg/kg body weight/day) by oral gavage 5 days per week until the endpoint (7 weeks after tumor cell injection). B,Ex vivo photon flux of posttreatment brains from the experiment described in A was determined at endpoint by bioluminescence imaging. The photon-flux scale is indicated on the right. C, Violin plots depicting normalized photon flux of brains imaged ex vivo from the mice described in B. The normalized photon flux for brain tissue was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (i.e., the day of injection) and multiplying that value by 100. Data are presented as mean values ± SEM. P values were determined by a two--tailed, unpaired Mann–Whitney test. For PC9-Tr-BrM mice: n = 8 for vehicle; n = 8 for osimertinib; n = 7 for pan-RARi; and n = 7 for osimertinib plus pan-RARi. For H1650-Tr-BrM mice: n = 12 for vehicle; n = 8 for osimertinib; n = 10 for pan-RARi; and n = 6 for osimertinib plus pan-RARi. ns, P value not significant. D, Representative images of CK7 IHC on posttreatment brain sections collected at endpoint from the experiment described in A. Scale bars, 200 μm for top (PC9-Tr-BrM) and 100 μm for bottom (H1650-Tr-BrM). E, Quantitative analysis of the percentage of CK7-immunostained brain sections covered by metastasis (Met) shown in D. Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test. For PC9-Tr-BrM mice: n = 11 for vehicle; n = 9 for osimertinib; n = 4 for pan-RARi; and n = 5 for osimertinib plus pan-RARi. For H1650-Tr-BrM mice: n = 6 for vehicle; n = 6 for osimertinib; n = 9 for pan-RARi; and n = 6 for osimertinib plus pan-RARi. F, Schematic representation of the experiment testing the effect of RAR gene knockdown on brain metastasis development. Mice were injected with either PC9- or H1650-derived Tr-BrM cells with one of two sets of shRNA-mediated stable dual knockdown of RARα and RARγ (sh-RARα + γ), or control shRNA (sh-Con), via intracardiac injection. Experiments involving shRNA set #1 are shown in G–J, whereas experiments involving shRNA set #2 are shown in Supplementary Fig. S6L–S6O. Mice were euthanized 7 weeks after tumor cell injection, and brains were collected for analysis. G,Ex vivo photon flux of posttreatment brains from the experiment described in F was determined at endpoint by bioluminescence imaging. The photon-flux scale is indicated on the right. H, Violin plots depicting normalized photon flux of brains imaged ex vivo from the mice represented in G. The normalized photon flux for brain tissue was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (i.e., the day of injection) and multiplying that value by 100. Data are presented as mean values ± SEM. P values were determined by a two--tailed, unpaired Mann–Whitney test. For PC9-Tr-BrM: n = 7 for sh-Con; n = 5 for sh-RAR. For H1650-Tr-BrM: n = 8 for sh-Con; n = 6 for sh-RAR. I, Representative images of CK7 IHC on posttreatment brain sections collected at endpoint from the experiment described in F. Scale bars, 100 μm for all images. J, Quantitative analysis of the percentage of CK7-immunostained brain sections covered by metastasis shown in I. Data are presented as mean values ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test. For PC9-Tr-BrM: n = 10 for sh-Con; n = 10 for sh-RAR. For H1650-Tr-BrM: n = 5 for sh-Con; n = 5 for sh-RAR.
The Combination of Osimertinib with Pan-RAR Antagonism Reduces Residual Cancer Cells in the Brain
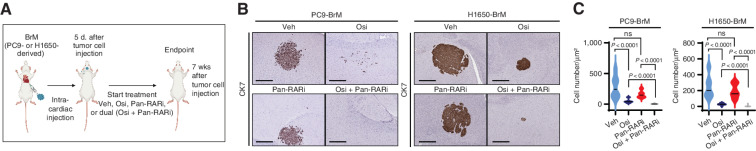
Based on the existence of clonal heterogeneity in S100A9 expression in PC9-BrM cells prior to osimertinib treatment in the brain (Fig. 3), we asked whether treatment with the combination of osimertinib and AGN194310 could reduce residual cancer cells in the brain. We injected unselected, treatment-naïve PC9- and H1650-BrM cells into the arterial circulation of immunodeficient mice via intracardiac injection (Fig. 7A). We initiated a prevention study 5 days after tumor cell injection involving the administration 5 days per week of either (i) vehicle, (ii) AGN194310 (0.5 mg/kg body weight/day), (iii) osimertinib (5 mg/kg body weight/day), or (iv) AGN194310 (0.5 mg/kg body weight/day) plus osimertinib (5 mg/kg body weight/day). At endpoint (7 weeks after tumor cell injection), we harvested, sectioned, and immunostained brain tissues with a CK7 antibody to identify cancer cells that were below the detection limit of bioluminescence imaging (Fig. 7B and C). Compared with the vehicle-treated mice, there was a significant reduction in the number of residual cancer cells in the brain in all treatment groups (Fig. 7B and C). Importantly, mice in the AGN194310 plus osimertinib group showed a significantly greater reduction in the residual disease burden in the brain compared with either osimertinib or AGN194310 alone (Fig. 7B and C). These preclinical studies suggest that the combination of therapies (AGN194310 plus osimertinib) could prevent or delay the emergence of osimertinib-refractory disease in the brain of patients with EGFR-mutant lung cancer.
Figure 7.
The combination of osimertinib and pan-RAR antagonism reduces residual cancer cells in the brain. A, Schematic representation of the experimental treatment protocol for the prevention trial. Osimertinib-sensitive PC9- or H1650-BrM cells were injected into mice via intracardiac injections. At 5 days after tumor cell injection, mice were administered treatment with either (i) vehicle (Veh), (ii) AGN194310 (Pan-RARi; 0.5 mg/kg body weight/day), (iii) osimertinib (Osi; 5 mg/kg body weight/day), or (iv) AGN194310 (0.5 mg/kg body weight/day) plus osimertinib (5 mg/kg body weight/day) by oral gavage 5 days per week until the endpoint (7 weeks after tumor cell injection). B, Representative images of CK7 IHC on posttreatment brain sections at endpoint from the experiment described in A. Scale bars, 500 μm for PC9-BrM and 100 μm for H1650-BrM. C, Quantitative analysis of the CK7-immunostained brain metastatic cancer cell number per μm2 represented in B. Data are presented as the mean number of cancer cells per μm2 of the brain tissue section ± SEM. P values were determined by a two-tailed, unpaired Mann–Whitney test. For PC9-BrM: n = 12 for vehicle; n = 16 for osimertinib; n = 10 for pan-RARi; and n = 10 for osimertinib plus pan-RARi. For H1650-BrM: n = 17 for vehicle; n = 12 for osimertinib; n = 10 for pan-RARi; and n = 14 for osimertinib plus pan-RARi. ns, P value not significant.
Discussion
Metastatic relapse resulting from treatment failure has been a formidable challenge to finding a cure for EGFR-mutant lung cancer. Metastasis to the brain is a critical complication for 45% of patients with EGFR-mutant lung cancer that drastically reduces their quality of life and survival (20, 21). The brain is also a frequent site of relapse following the administration of early-generation EGFR TKIs, as most of these drugs have limited penetration of the blood–brain barrier. This limitation was overcome with osimertinib, which can permeate the blood–brain barrier (24, 44) and elicit striking responses in patients with brain metastases. However, the challenge with osimertinib treatment is that the responses are transient, and many patients eventually progress, often in the brain (25). The gradual evolution of drug resistance generally observed in human patients has not been previously modeled in the physiologic context of brain metastasis, and as such, mechanisms that drive brain relapses have remained poorly understood. Here, we generated osimertinib treatment-response-and-relapse mouse models using human lung cancer cells harboring osimertinib-sensitive EGFR-activating mutations to study brain relapse mechanisms. Our studies show that the S100A9–ALDH1A1–RA signaling axis endows cancer cells with the ability to thrive in the brain despite on-target inhibition of EGFR activity by osimertinib. Arising from clonal heterogeneity, S100A9-high expressors acquire tolerance to osimertinib and are selected to grow in the brain. Osimertinib-refractory cells therefore co-opt an EGFR mutation–independent bypass pathway that preexists in brain metastatic cells in order to escape osimertinib treatment and initiate brain relapse.
Our studies reveal site-specific requirements for the S100A9–ALDH1A1–RA axis to promote the growth and survival of metastatic lung cancer cells. Following osimertinib treatment, the expression of S100A9 and ALDH1A1 was highly enriched in metastatic cells located in the brain, but not in the lung and bone, which could be due to clonal heterogeneity or suppressive signals by the local microenvironment. Moreover, it remains to be investigated how S100A9 regulates the expression of ALDH1A1 in cancer cells. In this context, nuclear S100A9 has been reported in inflamed keratinocytes (45) and in models of cellular transformation (46), suggesting a role for S100A9 in transcriptional regulation. It will be interesting to further explore how S100A9 regulates the transcription of ALDH1A1 and potentially other genes in brain metastatic lesions.
Our genetic studies in both PC9- and H1650-derived models showed that although brain metastatic cells are highly dependent on the S100A9–ALDH1A1–RA axis for growth, bone metastatic cells are not. Such tissue-specific dependencies of metastatic cells on the RA signaling axis might be determined by the expression and activity of the receptors that transport retinol, enzymes that convert retinol to RA, and catabolizing enzymes that degrade RA (47, 48). Interestingly, in contrast to the brain, bone metastatic cells from the PC9-derived model, which lack the S100A9–ALDH1A1–RA signaling axis, are still dependent on the EGFR pathway and are effectively eliminated by osimertinib treatment.
We envision that lung cancer cells with distinct EGFR mutations might have different pathway dependencies and routes to EGFR treatment resistance and that the S100A9–ALDH1A1–RA axis represents one such escape mechanism for EGFR-mutant lung cancer cells. Future studies are therefore needed to develop metastatic lung cancer models with distinct EGFR mutations to study osimertinib drug resistance in vivo. Furthermore, the development of immunocompetent mouse models of EGFR-mutant lung cancers that metastasize to multiple organ sites will enable the study of drug resistance and clonal heterogeneity in the context of the adaptive immune system, which is a limitation of the models used in our studies.
We highlight the following key implications from our findings that could improve the clinical management of the disease. First, using osimertinib-response-and-relapse mouse models, we have elucidated an underlying mechanism for progression of EGFR-mutant lung cancer cells on osimertinib that does not involve on-target EGFR resistance mutations and could not have been identified through mutational analysis of tumors from either mouse models or patients. Our model system with long-term drug treatment enabled us to elucidate a novel relapse mechanism that might be especially relevant to patients with lung cancer because most relapses occur late (1 to 2 years) into osimertinib treatment. Indeed, we found that the presence of S100A9-positive cancer cells in pretreatment tumor tissue from patients with EGFR-mutant lung cancer is associated with brain metastasis and correlated with poor PFS following osimertinib treatment. These findings suggest that the assessment of intracellular S100A9 expression in pretreatment samples might be useful to determine which patients are at high risk for brain metastasis and progression following osimertinib treatment. It is also important to note that routinely performed plasma genotyping for mutations will not detect intracellular S100A9 expression. Therefore, it is necessary to perform IHC analysis of S100A9 in patient biopsies and/or resected tissues to predict the risk of progression in patients. Second, S100A8/9 is normally expressed and secreted by myeloid cells and can also be packaged as cargo in their exosomes (28, 49–53). We and others have demonstrated how S100A9 that has been secreted from myeloid cells promotes breast (36) and colon cancer progression (54). In contrast, we show here that intracellular S100A9 in cancer cells activates the ALDH1A1–RA pathway in EGFR-mutant lung cancer cells to promote relapse. Because the S100A9 detected in serum samples might be derived from the secretome of myeloid cells, our studies underscore the importance of analyzing the intracellular levels of S100A9 specifically in cancer cells from patients by IHC or after isolation of cancer cells by intracellular flow cytometry. Third, the clinical management of EGFR-mutant lung cancer will benefit from new strategies that circumvent osimertinib-resistance mechanisms and enhance the effectiveness of osimertinib in combination treatments. Our preclinical prevention studies showed a significant reduction in the number of residual brain metastatic cells when a pan-RAR antagonist was combined with osimertinib in both PC9- and H1650-derived mouse models of EGFR-mutant lung cancer. Our findings therefore offer a new therapeutic strategy to target the S100A9–ALDH1A1–RA signaling axis using pan-RAR antagonists that, in combination with osimertinib, can be tested in clinical trials to prolong PFS and to prevent brain relapses in patients with EGFR-mutant lung cancer. Future studies are needed to investigate whether treatment with the combination of RAR antagonists and osimertinib promotes the regression of advanced brain metastatic lesions in patients with EGFR-mutant lung cancer. Of relevance, multiple high-affinity RAR antagonists were developed in the late 1990s (55) and are currently the subject of renewed interest for their ability to inhibit prostate cancer cell growth (56, 57). With the increasing prevalence of brain metastasis in patients with EGFR-mutant lung cancer and its associated poor prognosis (39), therapeutic strategies that can prevent or treat brain relapses in these patients will be of high translational relevance and direct clinical benefit.
Methods
Animal Studies
All animal protocols and treatment of mice were approved by the Columbia University Institutional Animal Care and Use Committee (IACUC), and all animal experiments were conducted according to the ethical regulations described in the institutional guidelines of the Columbia University Medical Center (CUMC) Institute of Comparative Medicine, in compliance with the U.S. National Research Council's Guide for the Care and Use of Laboratory Animals, the U.S. Public Health Service's Policy on Humane Care and Use of Laboratory Animals, and the Guide for the Care and Use of Laboratory Animals. Mice were maintained in the CUMC barrier facility under conventional conditions with constant temperature and humidity and fed a standard diet (Labdiet 5053). For animal experiments performed at the Memorial Sloan Kettering Cancer Center (MSKCC) by the Antitumor Assessment Core Facility at Sloan Kettering Institute, all animal protocols and treatment of mice were approved by the MSKCC IACUC, and all animal experiments were conducted according to the ethical regulations described in the institutional guidelines of MSKCC, in compliance with the U.S. National Research Council's Guide for the Care and Use of Laboratory Animals, the U.S. Public Health Service's Policy on Humane Care and Use of Laboratory Animals, and the Guide for the Care and Use of Laboratory Animals. Male athymic mice ages 8 to 9 weeks obtained from Envigo were intracardially injected with 1 × 105 PC9-BrM cells, H1650-BrM cells, or their derivative sublines. Metastasis was monitored by bioluminescence imaging using the In Vivo Imaging System (IVIS, PerkinElmer) weekly. Briefly, mice were anesthetized with 3% to 4% isoflurane and injected with 150 ng of D-Luciferin (Fisher Scientific) via intraperitoneal injections. The mice were then placed inside the PerkinElmer IVIS Spectrum Optical Imaging System for measuring bioluminescence. Total photon flux was calculated using Living Image 4.7.3.software (PerkinElmer). Mice were weighed weekly and were monitored twice a week. Following IACUC guidelines from Columbia University, mice with weight loss of more than 20% or a body-conditioning score (BCS) of 2 or less or mice exhibiting signs of hunched posture, impaired locomotion, or respiratory distress were criteria followed for prompt euthanasia. Additionally, mice that develop brain metastases may develop paralysis or seizure-like activity, so mice with these indicators were also euthanized promptly. Mice with a BCS of less than 3 were monitored by the investigative staff at least daily and provided with additional supportive care or as directed by an Institute of Comparative Medicine veterinarian. Euthanasia with carbon dioxide inhalation with secondary method of decapitation/cervical dislocation was followed. Timed collection was performed in matching cohorts, which are indicated in figure legends. Bone and lungs were collected from mice as indicated in the figures.
For intratibial injections, we followed the procedure outlined in ref. 58. Briefly, mice were anesthetized with isoflurane. Mice were injected with buprenorphine XR 3.25 mg/kg subcutaneously. The hind limb was prepared with a 70% ethanol swab. The lateral malleolus, medial malleolus, and lower half of the tibia were grasped and the leg bent in a combination of flexion and lateral rotation to expose the knee. Using a 28-G 1/2-inch needle and a drilling motion, the needle was inserted through the patellar ligament percutaneously and into the anterior intercondylar area of the tibia. The cell suspension was slowly injected into the previously drilled tibia tract. Ten microliters of luciferase-labeled 5 × 104 cells were injected. Mice were kept on a heating pad until they recovered from anesthesia. Mice were monitored daily for 48 hours and then twice a week. The mice were imaged weekly by bioluminescence imaging, and bones were collected by ex vivo imaging.
For intrathoracic injections or lung orthotopic injections, we followed the procedure outlined in ref. 59. The mouse was anesthetized with isoflurane administered with a precision vaporizer. Depth of anesthesia was monitored at least every 15 minutes throughout the procedure by observing that there was no change in the respiratory rate associated with surgical manipulation and/or toe pinch. The mouse was placed on a snuggle safe pad with a barrier between mouse and pad. Following confirmation that a suitable anesthetic plane (no response to stimulation) had been attained, sterile eye lubricant was applied to both eyes to prevent corneal drying. The area around the injection site was shaved and sterility prepared with betadine and 70% alcohol consisting of three alternate swabs. A 5-mm incision was made on the side of the left thoracic cavity. A 5-mm incision was made through the fat and muscle surrounding the rib cage. It was ensured that the incision was only through the skin and fat/muscle layer and did not enter the thoracic cavity. If the thoracic cavity was opened, the mouse was humanely euthanized. One-milliliter tuberculin syringes were used to inject the cell inoculum into the left lateral thorax, at the lateral dorsal axillary line, approximately 1.5 cm above the lower rib line just below the inferior border of the scapula once the lungs were visualized through the ribs. The needle was quickly advanced approximately 6 mm into the thorax and injected into the lungs while they were extended. Thirty microliters of 1 × 105 luciferase-labeled cancer cells in HBSS were injected. The needle was quickly removed after the injection cell suspension. The muscles were sutured together using Vicryl sutures. Wound clips were used to close the skin. The procedure took 10 to 15 minutes per mouse. The mice were thereafter imaged weekly by bioluminescence imaging, and lungs were collected by ex vivo imaging.
For ex vivo imaging at endpoint, mice were injected with luciferin intraperitoneally, and after 3 minutes, mice were euthanized and tissues (brain and any other extracranial site with luminescent signal) were harvested for ex vivo imaging. The normalized photon flux for brain tissue ex vivo was calculated by dividing the photon flux from brain collected ex vivo by the total photon flux at day 0 (day of injection), and that value was multiplied by 100. Brain metastasis PFS was analyzed using the log-rank (Mantel–Cox) test. Survival curves and data analyses were performed with Prism 6 (GraphPad Software).
For drug treatments, mice were randomized into treatment groups. Osimertinib was administered by oral gavage in mice with a dose of 5 mg/kg/day 5 days per week. Osimertinib treatment was started 25 days after tumor cell injection for metastasis trials for both PC9- and H1650-Tr-BrM models, and 5 days after tumor cell injection for prevention trials for both PC9- and H1650-BrM models. For intratibial and lung orthotopic injections, mice were treated starting 7 days after tumor cell injection for another 2 weeks (endpoint at 3 weeks after tumor cell injection). For experiments involving testing of p-EGFR activation in brain metastatic cells in tissues, mice were gavaged with osimertinib (dose of 5 mg/kg body weight), and 6 hours later, mice were euthanized and brains were collected and fixed in 4% paraformaldehyde in PBS for 24 hours at 4°C and subsequently processed for histologic analysis. AGN194310 was prepared in DMSO and reconstituted in 1% polysorbate/PBS and was administered to the mice by oral gavage with a dose of 0.5 mg/kg/day 5 days per week. After euthanasia, mouse brain was immediately dissected out and cut in half along the middle line. The two hemispheres were fixed with 4% paraformaldehyde in PBS for 24 hours at 4°C. Paraffin-embedded brain tissue was sectioned sagittally at 5-μm thickness and processed for histologic analysis.
PC9 and H1650 Derivatives and Generation of Stable Cell Lines Expressing Luciferase
PC9-BrM3 cells stably expressing GFP luciferase were obtained from the Massague Laboratory. Cancer cells were isolated from the lung after osimertinib treatment after injecting H1650-BrM cells in arterial circulation at endpoint (4 months after tumor cell injection) are labeled as H1650-lung derivatives. Cancer cells were isolated from the bone after osimertinib treatment after injecting PC9-BrM cells in arterial circulation at endpoint (5 months after tumor cell injection) and are labeled as PC9-BrM bone derivatives. All cancer cell lines (PC9-BrM and H1650 derivatives) used in this study were stably infected with lentivirus expressing luciferase enzyme and a hygromycin resistance marker as described previously (35). Briefly, cells were plated at 30% confluency (adherent cells), and lentivirus was produced using the pLVX-Hygro vector expressing the luciferase gene. Target cells were transduced with the viral supernatant for 3 hours. At 48 hours following infection, stable cells integrated with the vector were selected using 100 μg/mL of hygromycin B (Invitrogen) for 1 week and were tested by gene expression and immunoblot analysis.
Immunoblot Analysis
Cells were washed with cold PBS, collected in lysis buffer consisting of 25 mmol/L Tris, pH 7.5, 150 mmol/L NaCl, 1% Triton X-100, 0.5% SDS, and supplemented with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktail (Thermo Scientific). After sonication, cell lysates were centrifuged at 18,000 × g for 5 minutes at 4°C. Protein concentration in the supernatant was determined by the BCA protein assay (Pierce), and a known amount of protein samples was mixed with an appropriate volume of 4× SDS-PAGE sample buffer and incubated at 95°C for 5 minutes. After separation of proteins by SDS-PAGE, protein bands were transferred to nitrocellulose membranes and immunoblotted with rabbit antibodies to phospho-EGFR (Y1068), phospho-ERK1/2 (T202/Y204), ALDH1A1, S100A9, S100A8, and a mouse mAb to β-actin, followed by the corresponding horseradish peroxidase (HRP)–conjugated secondary antibodies (Sigma). The membranes were developed using an ECL substrate (Bio-Rad), and immunoblotted specific protein bands were visualized on a Bio-Rad ChemiDoc Imaging System (Bio-Rad). RAR-alpha and -gamma antibodies were from Cell Signaling Technology (#62294 and #8965). For other antibodies, ALDH1A1 (D9Q8E) XP rabbit mAb #54135 from Cell Signaling Technology was used at 1:800, S100A9 (D5O6O) rabbit mAb #72590 from Cell Signaling Technology was used at 1:500, anti-MRP8/S100A8 rabbit antibody #Ab92331 from Abcam was used at 1:500, anti–β-actin mouse mAb (A1978) from Sigma was used at 1:2,000, phospho-EGF receptor (Tyr1068; D7A5) XP rabbit mAb #3777 was used at 1:1,000, EGF receptor (D38B1) XP rabbit mAb #4267 was used at 1:1,000, and phospho-p44/42 MAPK (Erk1/2; Thr202/Tyr204) rabbit antibody #9101 and p44/42 MAPK (Erk1/2; 137F5) rabbit mAb #4695 (all Cell Signaling Technology) were used at 1:1,000. Anti-mouse IgG (whole molecule)–peroxidase antibody produced in rabbit (A9044) from Sigma was used at 1:2,000, anti-rabbit IgG (whole molecule)–peroxidase antibody produced in goat (A0545) from Sigma was used at 1:5,000, and peroxidase AffiniPure donkey anti-goat IgG (H + L) #705035147 tertiary antibody from Jackson ImmunoResearch was used for S100A8 at a dilution of 1:2,000.
RNA Isolation and qRT-PCR
Total RNA (500 ng) was isolated using TRIzol and an RNeasy Mini Kit. RNA was then reverse-transcribed to cDNA using a cDNA Synthesis Kit (Applied Biosystems; Thermo Fisher Scientific). qRT-PCR was performed with 10 ng of cDNA per sample using gene-specific primers and the SYBR Green PCR master mix (Applied Biosystems; Thermo Fisher Scientific). GAPDH primers were used as an internal control. An Applied Biosystems 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher Scientific) was used to run all the samples, and data were exported to Excel (Microsoft) for gene expression analysis using the 2−ΔΔCt method. The qRT-PCR primer sequences used in this study are shown below:
hALDH1A1
forward primer: 5-CTGCTGGCGACAATGGAGT-3
reverse primer: 5-CGCAATGTTTTGATGCAGCCT-3
hS100A8
forward primer: 5-ATGCCGTCTACAGGGATGAC-3
reverse primer: 5-ACGCCCATCTTTATCACCAG-3
hS100A9
forward primer: 5-TCATCAACACCTTCCACCAA-3
reverse primer: 5-GTGTCCAGGTCCTCCATGAT-3
hSTRA6
forward primer: 5-CCACAGAGGACTACTCCTATGG-3
reverse primer: 5-CAGCACAAGGATTGACAGCG-3
hRAR alpha
forward primer: 5-GGGCAAATACACTACGAACAACA-3
reverse primer: 5-CTCCACAGTCTTAATGATGCACT-3
hRAR gamma
forward primer: 5-ATGCTGCGTATCTGCACAAG-3
reverse primer: 5-AGGCAAAGACAAGGTCTGTGA-3
hGAPDH
forward primer: 5-AATCCCATCACCATCTTCCA-3
reverse primer: 5-TGGACTCCACGACGTACTCA-3
Proteomic Profiling
PC9-BrM and PC9-Tr-BrM cells were cultured at 80% confluency. Cells were washed three times with cold PBS and collected in TBS buffer (25 mmol/L Tris-HCl, 150 mmol/L NaCl, pH 7.2) supplemented with protease inhibitor (Roche) and phosphatase inhibitor (Thermo Scientific) on ice. After ultrasound sonication, cell lysates were centrifuged at 18,000 × g for 10 minutes, and supernatants were collected for proteomic analysis, which was performed at the Whitehead Proteomics Core Facility (WPCF). At the WPCF, the fractionated samples were further purified by TCA precipitation, resuspended in a Tris/urea buffer, reduced, alkylated, and digested with trypsin at 37°C overnight. This solution was subjected to solid-phase extraction to concentrate the peptides and remove unwanted reagents, followed by injection onto a Waters NanoAcquity HPLC equipped with a self-packed Aeris 3-mm C18 analytical column, 0.075 mm by 20 cm (Phenomenex). Peptides were eluted using standard reverse-phase gradients. The effluent from the column was analyzed using a Thermo Orbitrap Elite mass spectrometer (nanospray configuration) that was operated in a data-dependent manner for 120 minutes. The resulting fragmentation spectra were correlated against custom databases using PEAKS Studio X (Bioinformatics Solutions).
Sample Preparation for RNA-seq Analysis
PC9-BrM and H1650-BrM cells and their derivatives were cultured at 80% confluency. Cells were washed with cold PBS and collected in TRIzol reagent (Thermo Fisher) on ice. Total RNA was isolated using the RNeasy Mini Kit following the manufacturer's instructions (Qiagen). RNA was quantified using Nanodrop (Thermo Scientific), and RNA quality was assessed by capillary gel electrophoresis (Agilent 2100 Bioanalyzer; Agilent Technologies, Inc.).
RNA-seq
Illumina-compatible, poly-A–enriched RNA-seq libraries were generated from total RNA and sequenced to a total depth of approximately 30M paired-end, 150-base-pair-long reads at Genewiz. Raw RNA-seq reads were mapped to GRCh38 together with the Gencode comprehensive gene annotation version 38 (https://www.gencodegenes.org), and read counts were summarized to the gene level using STAR version 2.9.7 in quantitative mode. Gene counts were normalized to the individual library size, and all statistical tests (including Wald test for significance) were performed using DESeq2 version 1.28.1 in R version 4.0.1. RNA-seq data have been deposited in the NCBI Gene Expression Omnibus (GEO) with the accession number GSE19074. G:Profiler and DAVID were used to determine GO term enrichment and functional annotation of RNA-seq and proteomics data.
Cell Culture
The human lung cancer cell lines (PC9 and H1650 derivatives) used in this study were cultured in RPMI 1640 media supplemented with 10% FBS and grown at 37°C in a humidified CO2 incubator (5% CO2). The human lung cancer cell lines (H1975 and HCC4006) were purchased from ATCC and authenticated by PCR analysis and checked for Mycoplasma contamination (Lonza) periodically every 3 months in the lab (last checked in October 2021). Both cells were cultured in RPMI 1640 media supplemented with 10% FBS and grown at 37°C in a humidified CO2 incubator (5% CO2). All media were supplemented with 100 IU/mL penicillin and 100 μg/mL streptomycin (Life Technologies). We generated single-cell clones by the limited dilution method using a standard protocol. In brief, we dissociated PC9-BrM cells (grown at 30%–50% confluency) with trypsin and prepared a single-cell suspension by passage through a 40-μm cell strainer. Subsequently, we counted the cells using the Bio-Rad TC20 Automated Cell Counter and diluted the cells with growth media (RPMI 1640 with 10% FBS and penicillin–streptomycin) following a serial dilution method to obtain 5 cells/mL. After homogeneously mixing the cell suspension, we plated 100 μL (one cell in every two wells) to each well of a 96-well plate. We changed the media every 3 days. After a week, when proliferating clones were visible, we trypsinized and transferred those clones individually from each well of a 96-well plate to individual wells of 24-well plates. After 4 weeks, we tested the clones for S100A9 expression by Western blotting, which detected clones expressing varying levels of S100A9. We collected and preserved the clones expressing the lowest and highest levels of S100A9 and designated those as S100A9hi and S100A9lo clones.
The viability of PC9- and H1650-Tr-BrM cells with drug treatment was determined by the MTS assay using Promega CellTiter 96 AQueous One Solution Cell Proliferation Assay kit containing tetrazolium compound following manufacturer's instructions. Briefly, 10,000 cells in 0.1 mL of growth medium were plated in a 96-well plate and allowed to grow for 16 to 24 hours. Cells were treated in 0.2 mL of serum-free and phenol red–free colorless medium for 3 days containing either DMSO (vehicle) or the pan-RAR antagonist AGN194310 (Sigma), all in the presence of 5 μmol/L retinol. For cytotoxicity assay using osimertinib, cells were treated with either vehicle (DMSO) or various concentrations of osimertinib, as mentioned in the figures. Cell viability was measured by CellTiter 96 AQueous One Solution Reagent (Promega) following the manufacturer's protocol.
After 1 to 2 hours of incubation at 37°C in a CO2 incubator, the amount of soluble formazan was determined by absorbance at 450 nm. The viability was calculated as a percentage of viable cells in vehicle-treated controls (designated as 100% viability).
Cell proliferation assay was performed by plating a fixed predetermined number of cells in culture media and counting the cell numbers after every 24 hours for 3 days. Briefly, 2.5 × 105 cells were plated in 60-mm tissue culture dishes, and cell numbers were determined by counting the cells using Bio-Rad TC20 cell counter. For all cell lines, each data point represents the mean values ± SEM of triplicate plates from three independent experiments. All data points were normalized with the cell number (3 × 105) at 24 hours after cell plating.
For determining STRA6 expression in PC9- and H1650-derived Tr-BrM cells, cells were cultured for 2 days in serum-free RPMI 1640 medium with DMSO alone, 5 μmol/L retinol alone, or cell medium with both 5 μmol/L retinol and 5 μmol/L antagonists (either the RARα antagonist BMS-195614, the RARγ inhibitor antagonist MM-11253, or the pan-RAR antagonist AGN194310). After 2 days, cells were washed and collected for RNA isolation as described above.
ELISA Assay
The S100A8 and S100A9 ELISA assay kits were used from R&D Systems. The ELISA plate used for coating was purchased from Nunc (Nunc MaxiSorp 96-well plate). For collecting culture supernatant, cancer cells (PC9- and H1650-Tr-BrM) were plated in a 60-mm tissue culture dishes with culture media containing 10% FBS with 50% to 60% confluence and allowed to grow for 24 hours in a CO2 incubator. Subsequently, the culture media were removed by aspiration from the culture plates and washed twice with 5 mL of serum-free media. The cells were then incubated in 5 mL serum-free media for 48 hours in a CO2 incubator. After 48 hours, the culture supernatant was collected and centrifuged at 10,000 × g for 5 minutes at 4°C to remove any floating dead cells and debris. The supernatant was then directly used for detecting S100A9 by ELISA assay other than for positive control, which was diluted 1:50. For positive control, H1650-Tr-BrM-S100A9i cells were engineered to ectopically express S100A9 cDNA carrying Ig-kappa chain-leader secretory sequence. For preparing cell lysates, cells were collected by gentle scraping in cold 1× TBS and centrifuged to harvest cell pellets, which were lysed in 1× TBS buffer containing protease inhibitors cocktail (Roche) by sonication. The cell lysates were subsequently centrifuged at 12,000 × g for 10 minutes at 4°C to remove cell debris, lysates were collected, and protein concentrations were determined by the BCA method. Cell lysates containing 10 μg protein were used for the ELISA. Each sample was determined in triplicate, and sample details are provided in Supplementary Table S5.
Gene Repression by CRISPR
We used the CRISPR/dCas9–KRAB-mediated gene repression strategy to knock down specific target genes following a previously described method (60) with some modifications. To generate a single viral vector plasmid that expresses both the gRNA and KRAB–dCas9 fusion protein, we removed the EF1a-Cas9-NLS fragment from LentiCRISPRv2 (Addgene plasmid #52961; ref. 61) by EcoRI/BamHI digestion and replaced it with SFFV-KRAB-dCas9 from the pHR-SFFV-KRAB-dCas9-P2A-mCherry plasmid (Addgene plasmid #60954; ref. 60) using the InFusion cloning method (Takara). The resultant chimeric plasmid was named LentiCRISPRv2-SFFV-KRAB-dCas9. We designed the oligos for the gRNAs using the CRISPR-ERA online tool (http://crispr-era.stanford.edu) and cloned them into the BsmBI-digested LentiCRISPRv2-SFFV-KRAB-dCas9 following the procedure outlined by Feng Zhang's group (61). We detected positive clones by PCR using the human H6 forward primer and the reverse oligo for the corresponding gRNA.
gRNA oligos for gene repression by the CRISPRi method are listed below.
S100A9-gRNA1-Oligo1: 5′-CACCGAGTGAGCTGCCAGCTTCCCC-3′
S100A9-gRNA1-Oligo2: 5′-AAACGGGGAAGCTGGCAGCTCACTC-3′
S100A9-gRNA2-Oligo1: 5′-CACCGTCCCTTGTCAACCCAAACTG-3′
S100A9-gRNA2-Oligo2: 5′-AAACCAGTTTGGGTTGACAAGGGAC-3′
S100A8-gRNA-Oligo1: 5′-CACCGAGACTGTAGCAACTCTGGCA-3′
S100A8-gRNA-Oligo2: 5′-AAACTGCCAGAGTTGCTACAGTCTC-3′
ALDH1A1-gRNA-Oligo1: 5′-CACCGTTTGCATACTCGGATACGAT-3′
ALDH1A1-gRNA-Oligo2: 5′-AAACATCGTATCCGAGTATGCAAAC-3′
shRNA-Mediated Knockdown of RAR Alpha and RAR Gamma
Following lentiviral clones with specific shRNAs against human RAR alpha and RAR gamma were purchased from Sigma, and two sets with highest level of knockdowns were selected for subsequent downstream analysis. The two sets are: RARα (TRCN0000020370 and TRCN0000020373) and RARγ (TRCN0000236363 and TRCN0000236364). We generated lentiviral particles for each of these shRNAs and subsequently transduced the target cells following standard procedures. Dual knockdown of both RARα and RARγ was achieved by first transducing with one lentivirus, selected with puromycin, and followed by transduction with second lentivirus. The efficiency of knockdown was determined by measuring the mRNA expression by qRT-PCR and protein level by Western blotting analysis.
Forced ALDH1A1 Expression
To express the ALHD1A1 gene, we cloned the cDNA encoding human ALDH1A1 into the pLV-EF1a-IRES-Blast plasmid (Addgene #85133; ref. 62) within BamHI/EcoRI sites. We produced lentiviral particles following standard procedure using a third-generation packaging system. To stably express ALDH1A1 in S100A9-repressed cells, we infected S100A9-repressed cells with lentivirus carrying ALDH1A1 cDNA and selected with blasticidin following 48 hours of infection. After 1 week of antibiotic selection, we tested ALDH1A1 expression by real-time PCR as well as by Western blotting.
Human Tissues
Human tissue samples (n = 29) were obtained from the Columbia University Irving Medical Center (CUIMC) after deidentification with institutional review board (IRB)–approved protocols. Lines of treatment for the 29 patients are included in Supplementary Table S6. All research was conducted in compliance with ethical regulations outlined by the IRB. Sections from paraffin-embedded tissues (biopsies or resected samples) were immunostained and are described below.
IHC Staining
Paraffin-embedded brain tissues from mice were sectioned at 5- or 20-μm thickness, as described in the figure legends. Slides were baked at 60°C for 1 hour and deparaffinized, rehydrated, and treated with 1% hydrogen peroxide for 10 minutes. Antigen retrieval was performed using either pH 6.0 citrate buffer (Vector Laboratories) or pH 9.0 Tris-based buffer (Vector Laboratories) in a steamer apparatus for 30 minutes, and endogenous avidin and biotin were blocked using avidin- and biotin-blocking reagents (Vector Laboratories), respectively. The slides were further blocked with BSA and goat serum, and tissue sections were incubated with primary antibodies, including rabbit antibodies against human CK7 (1:25, Roche), human S100A9 (1:500, Cell Signaling Technology), human ALDH1A1 (1:500, Cell Signaling Technology), human phospho-histone H3 (Ser10; 1:100, Cell Signaling Technology), GFP (1:1,000, Aves Labs), and collagen IV (1:500, Millipore Sigma), followed by incubation with the corresponding biotinylated secondary antibodies (1:250, Vector Laboratories). The ABC kit and DAB kit (Vector Laboratories) were used for detection following the manufacturer's instructions. Sections were subsequently counterstained with hematoxylin, dehydrated, and mounted using Cytoseal XYL (Richard-Allan Scientific) for subsequent histologic analysis. Automated immunostaining for phospho-EGFR was performed at the Molecular Cytology Core Facility at MSKCC. Paraffin-embedded tissue sections were cut at 5 μm and heated at 58°C for 1 hour. Samples were loaded into Leica Bond RX and pretreated with EDTA-based epitope retrieval ER2 solution (Leica, AR9640) for 20 minutes at 95°C. The rabbit mAb p-EGFR (Cell Signaling Technologies; #3777, 2 μg/mL) was applied for 60 minutes and processed with the Polymer Refine Detection Kit (Leica, DS9800). Antibody Leica Bond Polymer anti-rabbit HRP was used, followed by the Refine Detection Kit Mixed DAB for 10 minutes, and the Refine Detection Kit Hematoxylin counterstaining for 20 minutes. After staining, sample slides were washed in water, dehydrated using an ethanol gradient (70%, 90%, 100%), washed three times in HistoClear II (National Diagnostics, HS-202), and mounted in Permount (Fisher Scientific, SP15).
For counting cells in the brain parenchyma (metastatic seeding), mice were euthanized 7 days after the intracardiac injection of 1 × 105 PC9 derivative (Tr-BrM lenti-control or S100A9i) cells (Fig. 3) or 7 weeks after tumor cell injection of both PC9- and H1650-BrM cell lines (Fig. 7) after vehicle, RAR antagonist, osimertinib, or RAR antagonist with osimertinib treatment. Mouse brains were dissected and cut in half along the mid-sagittal plane and put into 4% paraformaldehyde solution in PBS for a 24-hour fixation at 4°C. After washing with PBS, the brain was processed and embedded in paraffin, and blocks were sectioned throughout the brain at a 20-μm thickness/section. We performed immunostaining analysis for counting the number of cancer cells in the brain parenchyma. To quantitate the number of cancer cells in the brain parenchyma, sections were immunostained using an antibody against human CK7 (cancer cells), and 10 sections of 20-μm thickness/section were counted per brain sections and were quantitated using a brightfield microscope (Leica; Fig. 3) or using ImageJ described below (Fig. 7).
For the brain metastasis area, the immunostained slide images with CK7 staining marking the metastatic cells in the brain were analyzed with ImageJ (NIH). The scale bar was set using the Set Scale function under the Analyze tool. Using the region-of-interest (ROI), measure, and analyze tools, we calculated the area of the metastatic lesion and the total brain area under the field of view for each section. We calculated the percentage of the area of brain sections covered by metastasis.
For calculating staining intensity or number of positively stained cells, QuPath 0.2.3 (https://qupath.github.io/) was used. Image type was set as Brightfield (H-DAB). For phospho-EGFR–stained IHC slides, metastatic areas were selected with the polygon tool as ROIs to be analyzed. To carry out positive cell detection, hematoxylin optical density (OD) was set for image detection. Default parameters were used for nucleus and intensity parameters. Single threshold was selected, and the threshold 1+ (weak staining) parameter was set to 0.0634 to capture positive staining cells above threshold of 1+. For phospho-histone H3 (Ser10) slide quantitation, strongly positive cells were quantitated based on their optical intensity and cell:DAB OD mean, with the signal-intensity setting threshold of 3+ (strong staining) equating to 0.6. For ALDH1A1 slide quantitation, single threshold+1 was set to 0.2 and used as the cutoff for capturing total positive cells above 1+ based on their OD and nucleus:DAB OD mean. For all staining intensity quantitation, percentage of positive cells was determined by calculating the percentage of cells that showed positive staining with a specific antibody within the marked ROI of metastatic lesions.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 8. Power analysis was performed for sample size calculation for animal experiments. All statistical tests are described in the figure legends. All values were determined as the mean ± SEM. P values < 0.05 were considered statistically significant.
Data Availability Statement
RNA-seq data have been deposited in the NCBI GEO with the accession number GSE19074. Additional data, reagents, and materials generated in this study can be obtained by written request to the corresponding authors.
Supplementary Material
Acknowledgments
We thank Dajiang (Kevin) Sun, Hanina Hibshoosh, Vanessa Chin, and Christina Zahonero, Matthias Szabolcs (CUIMC), Lorraine Gudas (Weill Cornell Medicine), Dan Macalinao, Afsar Barlas, Radoslaw A. Junka, and Christina Falcon (MSKCC) for their assistance in our studies. We thank Eric Spooner (MIT) for his assistance with mass spectrometry analysis. This work was supported by K99/R00 CA172697 from the NCI (S. Acharyya), the Department of Defense Lung Cancer Research Program career development program (S. Acharyya), the Irma T. Hirschl Monique Weill-Caulier Trust Award (S. Acharyya), Schaefer Scholar Program (S. Acharyya), start-up funds from CUIMC (S. Acharyya), P01-CA129243 (J. Massague), P30 CA008748 (MSKCC), and the Alan and Sandra Gerry Metastasis and Tumor Ecosystems Center (MSKCC). S. Acharyya was supported by the Pershing Square Sohn Award and the Columbia University Irving Scholars Program. These studies used the resources of the Herbert Irving Comprehensive Cancer Center Core facilities, which are funded in part through Center Grant P30CA013696. Schematic models were created using BioRender.com with an institutional license.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
A.K. Biswas reports grants from the NIH and the Department of Defense Lung Cancer Research Program during the conduct of the study. H.A. Yu reports personal fees and other support from AstraZeneca, Cullinan, Janssen, and Daiichi, other support from Novartis, Pfizer, and Lilly, and personal fees from C4 Therapeutics and Blueprint Medicines outside the submitted work. P.K. Paik reports personal fees from EMD Serono, other support from Boehringer Ingelheim, and personal fees and other support from Bicara outside the submitted work. A. Saqi reports other support from Genentech and personal fees from Genentech, Bristol Myers Squibb, Dedham Group, Roche, Fieldworks/Inspirata, Huron, Trinity Partners, and Medscape outside the submitted work. C.A. Shu reports personal fees from AstraZeneca during the conduct of the study, as well as personal fees from Genentech, Mirati, and Janssen outside the submitted work. M.G. Kris reports grants from the NCI/NIH, personal fees from AstraZeneca, Janssen, Sanofi, Pfizer, and Daiichi Sankyo, and nonfinancial support from Hoffmann-La Roche outside the submitted work. J. Massague reports grants from the NIH during the conduct of the study, as well as a patent for PCT/US2013/032617 issued, a patent for PCT/US2014/056379 issued, a patent for PCT/US2015/051057 issued, and a patent for PCT/US2017/045145 pending. S. Acharyya reports grants from the NIH, the Department of Defense Lung Cancer Research Program, and a Columbia University Irma T. Hirschl Monique Weill-Caulier Trust Award during the conduct of the study. No disclosures were reported by the other authors.
Authors’ Contributions
A.K. Biswas: Conceptualization, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. S. Han: Data curation, software, formal analysis, validation, investigation, visualization, methodology. Y. Tai: Formal analysis, validation, investigation, visualization, methodology, writing–review and editing. W. Ma: Data curation, software, formal analysis, investigation, visualization, methodology, writing–review and editing. C. Coker: Formal analysis, validation, investigation, visualization, methodology. S.A. Quinn: Data curation, software, formal analysis, validation, investigation, writing–review and editing. A.R. Shakri: Validation, investigation, methodology. T.J. Zhong: Investigation, methodology, writing–review and editing. H. Scholze: Investigation, methodology. G.G. Lagos: Data curation, validation, investigation, methodology, writing–review and editing. A. Mela: Resources, investigation. K. Manova-Todorova: Resources, investigation, visualization, methodology, writing–review and editing. E. de Stanchina: Resources, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. A.A. Ferrando: Resources, formal analysis, investigation, writing–review and editing. C. Mendelsohn: Resources, formal analysis, investigation, methodology, writing–review and editing. P. Canoll: Resources, validation, investigation, visualization, methodology. H.A. Yu: Investigation, writing–review and editing. P.K. Paik: Formal analysis, validation, investigation, visualization, methodology. A. Saqi: Resources, data curation, formal analysis, investigation, visualization, methodology. C.A. Shu: Resources, data curation, validation, investigation, visualization, methodology, writing–review and editing. M.G. Kris: Formal analysis, validation, investigation, visualization, methodology, writing–review and editing. J. Massague: Resources, formal analysis, validation, investigation, writing–review and editing. S. Acharyya: Conceptualization, resources, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal Aet al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Shi Y, Au JS, Thongprasert S, Srinivasan S, Tsai CM, Khoa MTet al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non-small-cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014;9:154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Midha A, Dearden S, McCormack R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: a systematic review and global map by ethnicity (mutMapII). Am J Cancer Res 2015;5:2892–911. [PMC free article] [PubMed] [Google Scholar]
- 4. Du X, Yang B, An Q, Assaraf YG, Cao X, Xia J. Acquired resistance to third-generation EGFR-TKIs and emerging next-generation EGFR inhibitors. Innovation 2021;2:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rosell R, Moran T, Queralt C, Porta R, Cardenal F, Camps Cet al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009;361:958–67. [DOI] [PubMed] [Google Scholar]
- 6. Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BWet al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39. [DOI] [PubMed] [Google Scholar]
- 7. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169–81. [DOI] [PubMed] [Google Scholar]
- 8. Peters S, Mok T, Passaro A, Janne PA. The promising evolution of targeted therapeutic strategies in cancer. Cancer Discov 2021;11:810–4. [DOI] [PubMed] [Google Scholar]
- 9. Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer 2017;17:637–58. [DOI] [PubMed] [Google Scholar]
- 10. Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel Set al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004;304:1497–500. [DOI] [PubMed] [Google Scholar]
- 11. Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MFet al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005;2:e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cross DA, Ashton SE, Ghiorghiu S, Eberlein C, Nebhan CA, Spitzler PJet al. AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated resistance to EGFR inhibitors in lung cancer. Cancer Discov 2014;4:1046–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KHet al. Osimertinib in untreated EGFR-mutated advanced non-small-cell lung cancer. N Engl J Med 2018;378:113–25. [DOI] [PubMed] [Google Scholar]
- 14. Ramalingam SS, Vansteenkiste J, Planchard D, Cho BC, Gray JE, Ohe Yet al. Overall survival with osimertinib in untreated, EGFR-mutated advanced NSCLC. N Engl J Med 2020;382:41–50. [DOI] [PubMed] [Google Scholar]
- 15. Piotrowska Z, Isozaki H, Lennerz JK, Gainor JF, Lennes IT, Zhu VWet al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov 2018;8:1529–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gregorc V, Lazzari C, Karachaliou N, Rosell R, Santarpia M. Osimertinib in untreated epidermal growth factor receptor (EGFR)-mutated advanced non-small cell lung cancer. Transl Lung Cancer Res 2018;7:S165–S70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer 2019;121:725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ricordel C, Friboulet L, Facchinetti F, Soria JC. Molecular mechanisms of acquired resistance to third-generation EGFR-TKIs in EGFR T790M-mutant lung cancer. Ann Oncol 2018;29:i28–37. [DOI] [PubMed] [Google Scholar]
- 19. Yu HA, Tian SK, Drilon AE, Borsu L, Riely GJ, Arcila MEet al. Acquired resistance of EGFR-mutant lung cancer to a T790M-specific EGFR inhibitor: emergence of a third mutation (C797S) in the EGFR tyrosine kinase domain. JAMA Oncol 2015;1:982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shin DY, Na II, Kim CH, Park S, Baek H, Yang SH. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J Thorac Oncol 2014;9:195–9. [DOI] [PubMed] [Google Scholar]
- 21. Rangachari D, Yamaguchi N, VanderLaan PA, Folch E, Mahadevan A, Floyd SRet al. Brain metastases in patients with EGFR-mutated or ALK-rearranged non-small-cell lung cancers. Lung Cancer 2015;88:108–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ge M, Zhuang Y, Zhou X, Huang R, Liang X, Zhan Q. High probability and frequency of EGFR mutations in non-small cell lung cancer with brain metastases. J Neurooncol 2017;135:413–8. [DOI] [PubMed] [Google Scholar]
- 23. Heon S, Yeap BY, Britt GJ, Costa DB, Rabin MS, Jackman DMet al. Development of central nervous system metastases in patients with advanced non-small cell lung cancer and somatic EGFR mutations treated with gefitinib or erlotinib. Clin Cancer Res 2010;16:5873–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ballard P, Yates JW, Yang Z, Kim DW, Yang JC, Cantarini Met al. Preclinical comparison of osimertinib with other EGFR-TKIs in EGFR-mutant NSCLC brain metastases models, and early evidence of clinical brain metastases activity. Clin Cancer Res 2016;22:5130–40. [DOI] [PubMed] [Google Scholar]
- 25. Reungwetwattana T, Nakagawa K, Cho BC, Cobo M, Cho EK, Bertolini Aet al. CNS response to osimertinib versus standard epidermal growth factor receptor tyrosine kinase inhibitors in patients with untreated EGFR-mutated advanced non-small-cell lung cancer. J Clin Oncol 2018. Aug 28 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 26. Gilbert LA, Hemann MT. DNA damage-mediated induction of a chemoresistant niche. Cell 2010;143:355–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carlson P, Dasgupta A, Grzelak CA, Kim J, Barrett A, Coleman IMet al. Targeting the perivascular niche sensitizes disseminated tumour cells to chemotherapy. Nat Cell Biol 2019;21:238–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hobbs JA, May R, Tanousis K, McNeill E, Mathies M, Gebhardt Cet al. Myeloid cell function in MRP-14 (S100A9) null mice. Mol Cell Biol 2003;23:2564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kawai H, Minamiya Y, Takahashi N. Prognostic impact of S100A9 overexpression in non-small cell lung cancer. Tumour Biol 2011;32:641–6. [DOI] [PubMed] [Google Scholar]
- 30. Bos PD, Nguyen DX, Massague J. Modeling metastasis in the mouse. Curr Opin Pharmacol 2010;10:571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi Met al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell 2009;138:51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SSet al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med 2017;376:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Planchard D, Boyer MJ, Lee JS, Dechaphunkul A, Cheema PK, Takahashi Tet al. Postprogression outcomes for osimertinib versus standard-of-care EGFR-TKI in patients with previously untreated EGFR-mutated advanced non-small cell lung cancer. Clin Cancer Res 2019;25:2058–63. [DOI] [PubMed] [Google Scholar]
- 34. Sos ML, Koker M, Weir BA, Heynck S, Rabinovsky R, Zander Tet al. PTEN loss contributes to erlotinib resistance in EGFR-mutant lung cancer by activation of Akt and EGFR. Cancer Res 2009;69:3256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang G, Biswas AK, Ma W, Kandpal M, Coker C, Grandgenett PMet al. Metastatic cancers promote cachexia through ZIP14 upregulation in skeletal muscle. Nat Med 2018;24:770–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris PGet al. A CXCL1 paracrine network links cancer chemoresistance and metastasis. Cell 2012;150:165–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bresnick AR, Weber DJ, Zimmer DB. S100 proteins in cancer. Nat Rev Cancer 2015;15:96–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson Het al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 2019;47:W191–W8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak Let al. Brain metastases. Nat Rev Dis Primers 2019;5:5. [DOI] [PubMed] [Google Scholar]
- 40. Winkler F. The brain metastatic niche. J Mol Med 2015;93:1213–20. [DOI] [PubMed] [Google Scholar]
- 41. Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH, Lee DJet al. Serpins promote cancer cell survival and vascular co-option in brain metastasis. Cell 2014;156:1002–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J 1996;10:940–54. [PubMed] [Google Scholar]
- 43. Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol 2011;6:345–64. [DOI] [PubMed] [Google Scholar]
- 44. Colclough N, Chen K, Johnstrom P, Strittmatter N, Yan Y, Wrigley GLet al. Preclinical comparison of the blood-brain barrier permeability of osimertinib with other EGFR TKIs. Clin Cancer Res 2021;27:189–201. [DOI] [PubMed] [Google Scholar]
- 45. Schonthaler HB, Guinea-Viniegra J, Wculek SK, Ruppen I, Ximenez-Embun P, Guio-Carrion Aet al. S100A8-S100A9 protein complex mediates psoriasis by regulating the expression of complement factor C3. Immunity 2013;39:1171–81. [DOI] [PubMed] [Google Scholar]
- 46. Song R, Struhl K. S100A8/S100A9 cytokine acts as a transcriptional coactivator during breast cellular transformation. Sci Adv 2021;7:eabe5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Li Y, Wongsiriroj N, Blaner WS. The multifaceted nature of retinoid transport and metabolism. Hepatobiliary Surg Nutr 2014;3:126–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kelly M, von Lintig J. STRA6: role in cellular retinol uptake and efflux. Hepatobiliary Surg Nutr 2015;4:229–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hiratsuka S, Watanabe A, Aburatani H, Maru Y. Tumour-mediated upregulation of chemoattractants and recruitment of myeloid cells predetermines lung metastasis. Nat Cell Biol 2006;8:1369–75. [DOI] [PubMed] [Google Scholar]
- 50. Rafii S, Lyden D. S100 chemokines mediate bookmarking of premetastatic niches. Nat Cell Biol 2006;8:1321–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cheng P, Corzo CA, Luetteke N, Yu B, Nagaraj S, Bui MMet al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med 2008;205:2235–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Burke M, Choksawangkarn W, Edwards N, Ostrand-Rosenberg S, Fenselau C. Exosomes from myeloid-derived suppressor cells carry biologically active proteins. J Proteome Res 2014;13:836–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wortzel I, Dror S, Kenific CM, Lyden D. Exosome-mediated metastasis: communication from a distance. Dev Cell 2019;49:347–60. [DOI] [PubMed] [Google Scholar]
- 54. Ichikawa M, Williams R, Wang L, Vogl T, Srikrishna G. S100A8/A9 activate key genes and pathways in colon tumor progression. Mol Cancer Res 2011;9:133–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Johnson AT, Wang L, Standeven AM, Escobar M, Chandraratna RA. Synthesis and biological activity of high-affinity retinoic acid receptor antagonists. Bioorg Med Chem 1999;7:1321–38. [DOI] [PubMed] [Google Scholar]
- 56. Hammond LA, Van Krinks CH, Durham J, Tomkins SE, Burnett RD, Jones ELet al. Antagonists of retinoic acid receptors (RARs) are potent growth inhibitors of prostate carcinoma cells. Br J Cancer 2001;85:453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Petrie K, Urban-Wojciuk Z, Sbirkov Y, Graham A, Hamann A, Brown G. Retinoic acid receptor gamma is a therapeutically targetable driver of growth and survival in prostate cancer. Cancer Rep 2020;3:e1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Maloney C, Edelman MC, Kallis MP, Soffer SZ, Symons M, Steinberg BM. Intratibial injection causes direct pulmonary seeding of osteosarcoma cells and is not a spontaneous model of metastasis: a mouse osteosarcoma model. Clin Orthop Relat Res 2018;476:1514–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Safari R, Meuwissen R. Practical use of advanced mouse models for lung cancer. Methods Mol Biol 2015;1267:93–124. [DOI] [PubMed] [Google Scholar]
- 60. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EHet al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell 2014;159:647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 2014;11:783–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hayer A, Shao L, Chung M, Joubert LM, Yang HW, Tsai FCet al. Engulfed cadherin fingers are polarized junctional structures between collectively migrating endothelial cells. Nat Cell Biol 2016;18:1311–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the NCBI GEO with the accession number GSE19074. Additional data, reagents, and materials generated in this study can be obtained by written request to the corresponding authors.