Abstract
Inflammatory bowel disease is characterised by significant interindividual heterogeneity. With a wider selection of pharmacological and non-pharmacological interventions available and in advanced developmental stages, a priority for the coming decade is to determine accurate methods of predicting treatment response and disease course. Precision medicine strategies will allow tailoring of preventative and therapeutic decisions to individual patient needs. In this review, we consider the future of precision medicine in inflammatory bowel disease. We discuss the critical need to extend from research focussed on short-term symptomatic response, to integrative multi-omic systems biology strategies to identify and validate biomarkers that underpin precision approaches. Crucially, the international community has collective responsibility to provide well-phenotyped and curated, longitudinal datasets for scientific discovery and validation. Research must also study broader aspects of the immune response including components of the extracellular matrix to better understand biological pathways initiating and perpetuating tissue fibrosis and longer-term disease complications.
Keywords: Stratified medicine, Systems biology, Fibrosis, Biomarker, Natural history
Precision medicine
Why prediction of treatment response matters
As the global prevalence of inflammatory bowel disease (IBD) continues to rise, the need for effective tools to predict short-term or long-term outcomes becomes increasingly crucial. Established high rates of IBD in Western populations, now approaching 1%,1, 2 are coupled with a rising prevalence in developing countries.3 Subsequent impact on quality of life, psychological morbidity and financial costs to patients and healthcare services are substantial.4–13
In CD, whilst some patients follow a relatively indolent path that requires limited immunomodulation and results in few significant complications, others suffer from devastating penetrating and/or stricturing disease that necessitates sequential biologic therapy and surgery.14–16 Approximately 38–70% of patients with CD undergo surgery in the first 10 and 20 years after diagnosis, respectively17, 18 and the lifetime risk for need for surgical intervention in CD has been estimated to range from 50–80%.19 The use of biologics compared to no biologics has been shown to reduce the need for bowel resections in CD20–22 and an overall decline in surgeries for CD was noted,23 but their frequency remains significant.
Similarly, some UC patients require simple oral or topical mesalazine treatment, whilst others need biologics, frequent courses of corticosteroids, hospitalisation and surgery. Further, UC carries a 5- and 10-year cumulative risk for subtotal colectomy of 10–15%, and an up to 5% 30-year risk of colorectal cancer.24, 25 Additionally, the economic viability of continuing to treat all patients with ‘one-size-fits-all’ algorithms must be considered.
A predictive approach to clinical management is desirable to address the important balance of risk and benefit to IBD patients. This would allow those with a potentially complicated trajectory to be offered a tailored treatment plan, selecting the most likely medication, dietary or microbial intervention to be effective for the individual. A tailored approach should also avoid unnecessarily exposing those with a likely mild disease course to immune suppression or potential side-effects. Not only does this approach benefit the individual but it also minimises unnecessary spending on ineffective treatments, thus benefiting financially constrained healthcare systems.
Challenges to precision medicine
‘Predictive’, or ‘precision’, medicine involves electing specific treatments based on a patient’s various unique features.26 It is already a reality in other fields of medicine. For instance, patients with asthma who have high levels of eosinophils in the blood or respiratory tract are prescribed anti-interleukin (IL)-5 monoclonal antibodies27 and patients with HER2-positive breast cancer are treated with anti-HER2 antibody infusions.28 However, in IBD, a standardised approach remains, where treatment is instigated reactively and arbitrarily because of worsening clinical parameters. In some situations, such as the US healthcare system, these decisions may also be influenced by the payor. Without a stratified approach, data from landmark drug registration trials demonstrate that approximately 40% of patients experience primary non-response and almost 40% of the remainder suffer from secondary loss of response within one year.29–33 Real-world studies exploring drug persistence (the time between initiation and discontinuation of the drug) more closely reflect pragmatic clinical decision-making, including drug cessation due to intolerable side-effects, and are not bound by strict disease outcome parameters. These studies suggest that rates of drug withdrawal at 12-months are as high as 30–50% for anti-TNF medication and 20–30% for vedolizumab for IBD overall, and 10–20% for ustekinumab in Crohn’s, with higher rates in anti-TNF non-responders and those prescribed immunomodulator-free anti-TNF monotherapy.34–37 For these patients, there may be persisting symptoms, intestinal damage and psychological repercussions. Furthermore, even with seemingly effective anti-inflammatory treatment, disease may progress.38 A more nuanced approach to selecting pharmacological interventions is therefore required from the outset, particularly given that the overall chance of achieving remission reduces with successive post-anti-TNF treatment.39
The fundamental challenge to developing and validating precision medicine interventions in IBD is the wide heterogeneity in, and inability to conclusively predict, disease course. In addition, the time to complications varies greatly and may occur over decades. Any prediction model needs to account for environmental influences or treatment heterogeneity during this time.14–16, 24, 25 A further difficulty is the inconsistency in outcome measures for disease activity. Whilst mucosal healing appears the most significant in terms of predicting future relapse, complications and surgery in CD and UC,40, 41 the need for frequent endoscopic examination is resource-intense and may not be acceptable to patients, and may not be achievable in clinical practice as evidenced by numerous clinical trials where this endpoint was met only by subsets of patients.41 Instead, patient-reported outcomes (PROs) and non-invasive biomarkers, such as fecal calprotectin and C-reactive protein, are increasingly used to evaluate response to therapy in research studies and clinical practice, with non-standardised definitions of treatment response and remission. Interpretation of the performance and utility of the predictive biomarkers therefore becomes difficult. In addition, the STRIDE 2 consensus rightly argues that psychological and quality of life outcomes should inform our assessment of treatment response.41 The bidirectionality of the brain-gut axis seems increasingly evident, so effective candidate predictive biomarkers for IBD should consider improvement in both intestinal and psychological clinical parameters.42–44
Precise prevention of disease-related complications
Future precision approaches should aim to proactively escalate immune suppression based on pre-clinical biomarkers, to prevent an otherwise aggressive future disease course.40, 41 Approaches should identify features that would favour one immune targeted regimen over another in a specific individual. This is increasingly important as the number of drug classes effective in IBD rises.
Numerous associations have been found between clinical,45 genetic,46 microbial47, 48, transcriptional, metabolic and environmental factors on the one side and response to certain treatments or future disease course on the other.49–57 However, many studies have taken a hypothesis-free, rather than pathophysiological, approach to biomarker detection, potentially limiting applicability, and most have not been prospectively validated to ascertain their potential clinical relevance.
Notwithstanding the methodological limitations and discordance of biomarker prediction studies to-date,58, 59 the complex, multifaceted pathophysiology of IBD renders clinically relevant single biomarker detection difficult. At a basic level, IBD involves dysregulated interactions between the host immune system, intestinal epithelial barrier and luminal microbes in genetically susceptible individuals.60 Cytokines regulate the function of mucosal immune and non-immune compartments, including the epithelium and stroma, which leads to active and chronic inflammatory processes. These are driven by a variety of cells, including neutrophils, lymphocytes, monocytes, resident macrophages and fibroblasts, which contribute to intestinal damage, often-debilitating symptoms and reduced quality of life associated with the illness. Layers of complexity are added by the various environmental,50, 61, 62 microbiological,63 pharmacological,64 dietary,65 biochemical66 and psychological factors43, 67 that appear to be involved in disease inception, progression and relapse.
Prediction of short-term response to therapy
Current state of play
Examples of precision tools already widely used in IBD clinical practice to predict short term response or complications of therapy include clinical factors, blood, genetic or fecal biomarkers. They are used to assess immunogenicity, drug metabolism, treatment compliance or to predict flare or response to treatment.
Predictive clinical factors used to guide treatment decisions in routine practice include an understanding that early first relapse,68 particularly within the first 2 years69, is associated with greater likelihood of future active disease/relapses in UC. Age <40 years at diagnosis, female sex, and extraintestinal manifestations are also predictive of disease relapse in UC, whereas male sex and extensive disease predict colectomy.24 In contrast, mucosal healing in UC is predictive of reduced subsequent disease relapse and need for treatment intervention.70–72 In CD, predictors of worse disease course include age <40 years at diagnosis, perianal disease, stricturing or weight loss at diagnosis, and a need for steroids to treat the presenting inflammation.45, 73
Calprotectin is a cytosolic protein abundant in neutrophils with antimicrobial functions.74 Quantification of calprotectin in feces correlates with endoscopic disease activity,75, 76 histological inflammation77 and is thus predictive of relapse in asymptomatic IBD patients78, 79 or in the postoperative setting.80 Therefore, fecal calprotectin is a well-established non-invasive biomarker of mucosal inflammation used in screening for IBD in symptomatic patients pre-diagnosis and is widely used in longitudinal monitoring of IBD disease activity in clinical practice and research.81, 82
Up to 40% of patients have been shown to discontinue thiopurines due to adverse effects including leucopenia.83, 84 Testing for thiopurine methyltransferase (TPMT) genotype is cost effective.85 Assessment of TPMT activity is widely recommended prior to instituting thiopurine therapy in IBD.86, 87 In a prospective study of 207 patients, heterozygous TPMT genotype was a strong predictor of adverse effects when compared to those with wild type TPMT status (79% vs 35%).83 Dose reduction of thiopurines in patients pre-screened and identified as carrying TPMT variants led to a 10-fold reduction in haematological adverse effects without a negative impact on treatment efficacy.88 Following treatment commencement, measurement of the thiopurine metabolites thioguanine nucleotides (TGN) and methylmercaptopurine (MeMP) can support dose optimisation by identifying poor compliance, subtherapeutic dosing, thiopurine hypermethylation or supratherapeutic dosing.86
Multiple clinical studies have demonstrated a clear association between serum drug concentrations of biologics and outcome for IBD patients, including the association of low or undetectable drug levels and immunogenicity or non-response/loss of response.89–98 Therapeutic drug monitoring (TDM) describes the measurement of trough drug concentration and anti-drug antibodies in blood and can be used to assess adherence with treatment schedule, pharmacokinetics and immunogenicity, therefore guiding dose escalation or reduction decisions. TDM can guide when to switch therapies or add in immunomodulation. TDM is increasingly commonplace, particularly in reactive optimisation of anti-TNF therapy use in symptomatic patients to define and manage primary non response or secondary loss of response.86, 87 TDM may potentially be used more frequently in future proactive algorithms for patients with quiescent disease receiving anti-TNF treatment to support de-escalation99–102 and in optimisation of vedolizumab and ustekinumab depending on prospective data to support clinical utility, cost effectiveness and availability of TDM assays.103
On the verge of implementation
Identification of biomarkers to support precision medicine in clinical practice has been an intense area of research in recent years. Several biomarkers show promise and may enter common clinical practice subject to a variety of barriers including validation of clinical utility, cost effectiveness of testing, availability of assays, scalability and acceptance of testing.104
Exposome
The exposome refers to environmental factors that play a key role in IBD pathogenesis, flare or long-term complications. It is multifactorial including diet and dietary additives, exposure to medications, infections, air and water pollution, stress and sun exposure. The crucial importance of the exposome is evidenced by a rising prevalence of both UC and CD in newly industrialised countries in South America, Asia and the Middle East over recent decades.105, 106
Smoking, whilst protective against relapse in UC,68 is a major risk factor for disease progression in CD107 including need for hospitalisation and surgery.108 Smoking is also associated with poorer outcomes following colorectal surgery.109, 110 In a study of postoperative 6-mercaptopurine (6-MP) vs placebo to prevent recurrence of inflammation, 6-MP only demonstrated benefit in smokers, emphasizing the importance of considering postoperative prophylaxis in this high risk group.111
Evidence-based answers to diet as a precision tool in IBD are a current research priority.86 It has been observed that the ‘Western’ diet of fast or ultra-processed foods, high fat content and meat increase risk of disease development,112–114 and protective factors include higher intake of fibre, caffeine, fruit, vegetables, olive oil, fish, grain and nuts.112, 113, 115 Recently, it has been recognised that an association exists between dietary emulsifiers, detergent-like additives widely found in the food chain and gastrointestinal inflammation.116 In experimental colitis emulsifiers increase circulating inflammatory mediators, e.g. LPS, IL-1β, IL-6, and TNF, possibly mediated by gut microbial changes.117 Emulsifiers also induce thinning of the mucous layer of the gut epithelium with resultant encroachment of microbes at the epithelial surface, in theory altering intestinal permeability and risk of bacterial translocation.118–121
Formula-based exclusive enteral nutrition (EEN) diets can induce remission in CD but palatability and acceptability to patients limit its use, especially in adults. Exciting early data from the CD-TREAT programme that utilises a personalised food-based diet with similar composition to EEN has shown good tolerability, improved disease activity and changes to the gut microbiome and metabolome comparable to EEN.122
Further much needed studies are underway to determine the impact of environmental factors on the gut microbiome and intestinal inflammation in the context of IBD. These will also investigate whether precision approaches are possible to identify those most likely to respond, or to modify the gut microbiome or tailor dietary intake to enrich response.116, 122
Genetics and genomics
Genetic discovery has challenged the traditional view of IBD pathogenesis being a dichomotous condition comprised of UC and CD. Genetics stratifies subphenotypes of IBD, in particular a genetic distinction between ileal and colonic CD.123 Mechanistic pathways driving the inflammatory response in IBD have been discovered or confirmed, including microbial sensing (NOD2, CARD9, RIPK2), intestinal barrier function (C1orf106, HNF4A), innate and adaptive immune signalling (NLRP7, IL18RAP, CD28, IFNG, PTPN22, STAT4, IL6ST, IL23R, RORC, IL17RA), fibrosis (OSMR, SMAD3) and cellular homeostasis (ATG16L1, RNF186, ERGIC1).124
Despite the above advances, unfortunately few variants associate with prognosis or clinical outcomes or are ready for incorporation in clinical practice with a small number of notable exceptions. The ‘personalised anti-TNF therapy in CD study’ (PANTS) consortium longitudinally followed >1,000 biologic naïve CD patients before and after introduction of anti-TNF therapy. TNF failure was significantly associated with low drug levels and development of anti-drug antibodies.125 Immunogenicity to anti-TNF antibodies was associated with HLA-DQA1*05, with a hazard ratio approaching 2.126 Since 40% of Europeans carry the HLA-DQA1*05 allele it is a conceptually attractive biomarker candidate, and may be particularly useful in identifying patients prone to immunogenicity who may benefit from combination immunomodulator therapy to limit this risk.
Genetics may also be useful to predict complications of treatment. Variants in NUDT15 are associated with increased risk of thiopurine induced myelosuppression.127 The HLA-DQA1*02:01-HLA-DRB1*07:01 haplotype is predictive of thiopurine induced pancreatitis with a number needed to test of 76 to avoid one case.128 Current translation of these data into day to day practice is limited by cost, which will change once incorporated into routine clinical genotyping panels. Well curated, phenotyped, longitudinal cohorts such as IBD BioResource129 will be essential in the development of future precision genetics tools.
Molecular profiling
Deep molecular profiling of tissue and blood with multi-modal -omics technologies, including transcriptomics, metagenomics, metabolomics, proteomics and cytomics are bringing new insights into patient stratification. In particular, transcriptomics in either intestinal biopsies and/or peripheral blood shows considerable promise. Specific examples include single transcripts, such as OSM, OSMR or TREM1 separated responders from non-responders to anti-TNF induction.51, 53, 130–133 Colonic tissue gene expression of granzyme A (GZMA) and αE integrin (ITGAE) predicted outcome to etrolizumab therapy.134 The expression of 4 transcripts (RGS13, DCHS2, MAATS1, and PIWIL1) in colonic biopsies differentiated responders and non-responders to vedolizumab.52 A supervised list of individual transcripts, or pathway-specific transcripts can also be screened for enrichment in whole genome expression datasets. These approaches have identified TNF receptors (TNFR2) and TNF itself as predictors of response to anti-TNF therapy.135, 136 Similarly, overexpression of mucosal IL7R signalling gene transcripts distinguishes responders from non-responders to anti-TNF therapy.137
An excellent example of a precision medicine stratified trial translating insights from molecular profiling is the UK study ‘predicting outcome for CD using a molecular biomarker’ (PROFILE).138 Previous unsupervised transcriptional profiling of circulating CD8+ T cells was predictive of a more complicated disease course in CD and UC as defined by the need for future treatment escalation.139 Elevated expression of genes involved in antigen-dependent T cell responses, including signalling initiated by both IL-7 and TCR ligation, portended a poor prognosis with specificity for CD of 89% and UC of 84% and sensitivity for CD of 59% and UC of 77%. Based on this, the PROFILE trial is the first prospective randomized trial testing a biomarker strategy utilising these identified T cell signatures to stratify patients at diagnosis into an accelerated step-up standard of care arm and a top-down anti-TNF and immunomodulator arm.138 PROFILE exemplifies the scientific rigor, funding and time required to take academic discovery of -omic association through to randomized controlled trial, and hopefully later clinical implementation.
A different approach is to determine whether distribution of co-expressed genes with shared patterns of upregulation/downregulation, often termed “modules”, associate with clinical phenotypes. Transcriptional modules associated with OSM/OSMR expression effectively stratified clinical response to anti-TNF therapy.51 An advantage of identifying modules over individual transcripts, is provision of insights into biological mechanisms of treatment response/resistance. Cellular deconvolution methods, where cell-specific transcriptional programs are used to estimate cell proportions from genome-wide expression datasets, also promise to provide new insights into cellular mediators of treatment response or resistance. These methods are now being surpassed by single cell sequencing platforms, where transcriptional profiles are defined at single cell resolution. Together, these approaches are identifying mechanistically relevant transcriptional programs in specific intestinal cell types linked to clinical outcomes. Intriguingly, modules associated with inflammatory monocytes, neutrophils and their interactions with stromal cells are associated with treatment refractoriness to biological and conventional therapies.140, 141 These data also point to the possible notion of a “molecular severity”, or “molecular complexity” of disease, where induction of particular immune pathways has clinically meaningful relationships with tissue injury and patient trajectories, including responsiveness to therapeutic intervention. In the same way that activation of stromal cells, inflammatory subsets of mononuclear phagocytes neutrophils, IL-1 and IL-22 related pathways are linked to increased tissue injury and ulceration,140–142 activation of this axis is also implicated, potentially causally, in treatment resistance.
Microbiome
The microbiome is a key factor in initiation and perpetuation of the inflammatory response in IBD.143 Relative to non-IBD subjects the gut microbiome in IBD has a reduced diversity144 and lacks stability with greatest taxonomic and functional variability observed in the context of disease activity.145
In comparison to well powered human genetics studies with replication, to date microbiome studies have been relatively small though have yielded intriguing observations implicating the utility of microbiome data to predict outcome to therapeutic intervention. A prospective cohort of 85 patients with CD and UC commencing vedolizumab therapy, metagenomic sequencing of stool at baseline identified higher alpha diversity and greater abundance of Roseburia inulinivorans and a Burkholderiales species in CD patients who achieved remission at week 14.56 Enrichment of branched chain amino acid biosynthesis in those achieving remission was noted. Neural network modelling identified that microbial metadata alone outperformed clinical metadata alone in predicting remission. In 232 CD patients of the phase 2 CERTIFI study treated with ustekinumab146 Faecalibacterium and Bacteroides were enriched in baseline stool in CD patients who achieved remission after 6 weeks of treatment. Once again in predictive modelling algorithms, baseline microbiota profiles alone outperformed baseline clinical metadata alone in predicting remission at week 6.
Whilst these studies have been small, they support the concept that presence or absence of certain microbiota or species diversity may predict response to IBD therapy. What is not clear from these studies is mechanistically whether and how gut microbiota directly or indirectly influence treatment outcomes. This concept has been explored outside of IBD in studies of PD-1/PDL-1 targeted immune checkpoint therapy in the oncology field. Here, patients with metastatic melanoma responding to anti-PD-1 therapy had higher relative abundance of Faecalibacterium, whereas non-responders had higher relative abundance of Bacteroidales.147 In a separate study of metastatic melanoma patients responding to PD-1 directed therapy had higher abundance of Bifidobacterium longum, Collinsella aerofaciens and Enterococcus faecium, and non-response was associated with Ruminococcus obeum and Roseburia intestinalis.148 Similarly increased relative abundance of Akkermansia mucinophila was found in stool from responders to anti-PD-1 therapy in a study of non-small cell lung cancer and renal tract cancers.149 Fascinatingly in all three studies,147–149 researchers administered fecal microbial transplants (FMT) to mice that had implanted tumour cells. FMT from patients responding to PD-1 blockade into mice enhanced the anti-tumour effects in the animal model with an increase in anti-tumour CD8+ T cells within malignant tissue relative to FMT from non-responding patients. Supplementation of the microbiome from non-responding patients by oral administration of Akkermansia mucinophila improved the anti-tumour effect of PD-1 blockade by IL-12 dependent recruitment of CCR9+CXCR3+CD4+ T cells.149
These studies suggest enrichment of immunoregulatory microbes can both predict and influence responsiveness to systematically administered drugs. Whilst the mechanisms are unclear, it is possible these microbes reinforce epithelial barrier integrity and prevent bacterial translocation in the gut, and/or modify antigen presentation and subsequent anti-tumour T cell responses.147, 149, 150 Intriguingly these studies also raise the question of whether the microbiome can be manipulated to enhance systemic drug effects that perhaps in the future may benefit patients with IBD.
Prediction of future tissue damage and remodeling
The short-term treatment response is not clearly linked to prevention of future tissue damage and remodeling
Despite impressive progress in developing novel biologics and small molecules to treat IBD and improvement in the short-term treatment response, it becomes increasingly apparent that the prevention of future bowel damage and control of subsequent excessive tissue remodeling remains of high importance and is often underappreciated in short term cohort or intervention studies.38, 151 The rate of surgical resections is also of high importance and shows only modest decline over the past few decades.152 This observation is further supported by the pediatric RISK inception cohort.153 Pediatric CD patients were included close to diagnosis and patients with no complication were followed until the occurrence of complications or not.153 Out of 913 participants, within 3 years 9% developed either internal penetrating or stricturing disease. Interestingly, the early use of anti-TNF in this cohort prevented the occurrence of internal penetrating disease (HR 0.3), but not stricturing disease.153 In addition, observational data from organs other than the gut as well as from IBD suggest that despite the successful suppression of inflammation, intestinal damage and remodeling can take an independent course.38, 154 This indicates separate mechanisms driving inflammation and tissue damage.
In experimental animal models, successful anti-inflammatory therapy suppresses inflammatory mediators, but factors driving tissue remodeling, such as TGF-β1, remain high.155 In humans, a change in the intestinal immunophenotype is observed over time.156 The initially Th1 dominated T cell response close to diagnosis switches to a Th2 prone pro-repair response, which is characterized by the expression of pro-fibrotic factors.156 The same can be found in experimental colitis, where in the early phases cytokines such as IFNγ predominate, which undergoes a switch to a pro-repair TGF-β1 signature.157
Collectively, these data suggest that inflammation may initiate tissue damage and aberrant repair responses in IBD, but once tissue damage and subsequent excessive remodeling is established it can progress independently. This may explain the disconnect between the short-term response and the long-term outcome in IBD, which makes separate exploration of both a necessity.
The immune-stromal-epithelial interactome and the biology of treatment resistance
Traditional biologics and immunosuppressive therapies in IBD target immune cell function, with limited impact on non-immune cells.158 This is surprising considering the known contributions of non-immune cells to inflammation in other organs159, 160 as well as IBD,161, 162 and relevant given the fact that non-immune cells are major effector cell type in fibrosis, producing excessive amounts of extracellular matrix (ECM).38
A cell type that has been widely neglected in the considerations around treatment resistance are mesenchymal cells. Traditionally mesenchymal cell phenotypes have strongly contributed to understanding the field of remodeling across organs, but recent single cell genomics approaches indicate the role of mesenchymal cells in the anti-inflammatory therapy response. A recent study of the colonic mesenchyme revealed four previously unknown distinct subsets of dysregulated fibroblasts in intestinal inflammation,163 as indicated by fibroblast populations expressing interleukin (IL)-33, lysyl oxidases (LOX), TNFSF14 and fibroblastic reticular cell-associated genes. This highlights how intestinal fibroblasts may not only respond to, but drive inflammation and barrier dysfunction in IBD.163 In the context of ileal CD, cellular heterogeneity may contribute to resistance to treatment with anti-TNF therapy. Tissue in anti-TNF resistant CD patients contained two subsets of fibroblasts. One of those are CTHRC1+ fibroblasts, which express the highest concentration of collagen genes and exhibit pro-fibrotic function in vitro.164
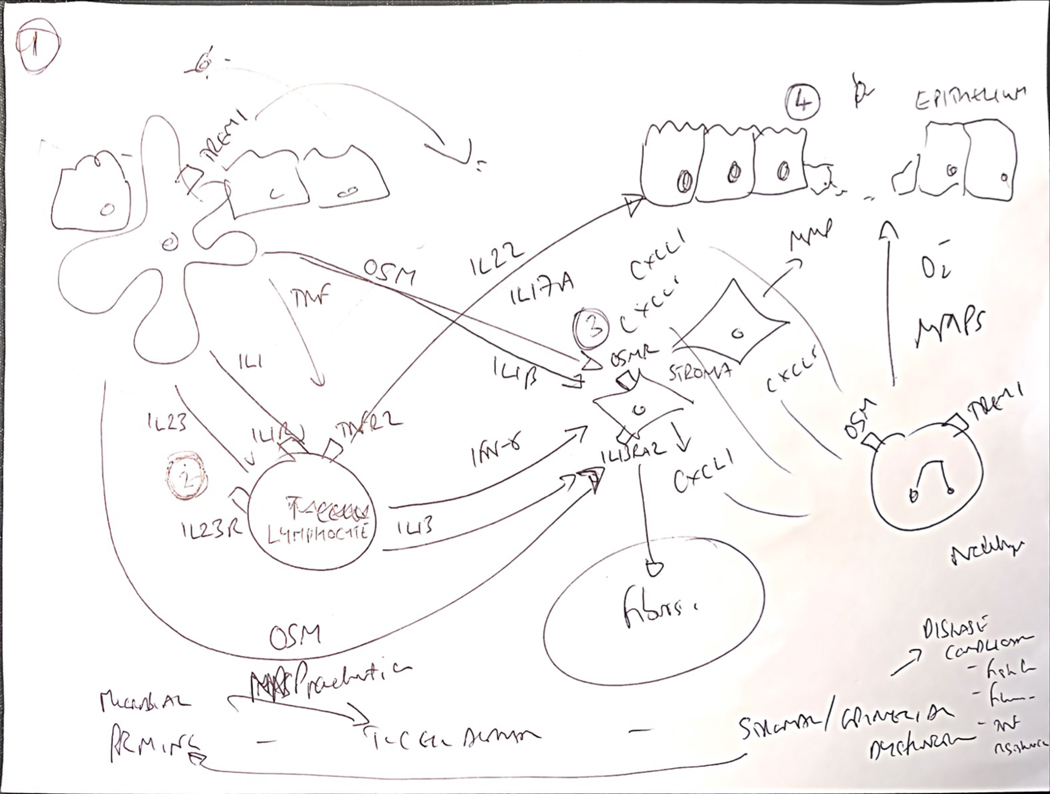
However, the exact role of these activated fibroblasts or other mesenchymal cell populations in the pathogenesis of treatment resistance (or development of complications) is not yet clear though a proposed model of cross-talk between immune, stromal and epithelial cells is presented in Figure 1. This will be one critical piece of the puzzle to not only understand short-term treatment response or lack thereof, but also long-term disease course. More recently, further mechanistic insights have been afforded by single cell transcriptomics and how the patterns of co-expressed genes detected in whole intestinal tissue specimens relate to morphological aspects of tissue injury, immune pathways and patient outcomes in intestinal ulceration. IL-1β responsive biological pathways were identified as key drivers of tissue damage, ulceration and severe inflammation. IL-1β activates fibroblasts to produce neutrophil-active chemokines, including the CXC family members, CXCL1, CXCL5 and CXCL8,141 which is associated with accumulation of tissue neutrophils, especially at ulcer bases. Modules linked to IL-1-activated stromal cells and neutrophil recruitment are strongly linked to poor patient outcomes, including lack of therapeutic response to multiple IBD therapies such as anti-TNF, vedolizumab and corticosteroids.141 IL-1β also triggers IL-23 production by activated intestinal inflammatory mononuclear phagocytes, and together both of these cytokines activate mucosal lymphocytes with induction of type 1 and type 3 immunity, including induction of IL-22, which in chronic inflammation is linked to induction of CXC family neutrophil-active chemokines by colonic epithelial cells and treatment refractoriness in IBD.140
Figure 1:
Towards a new conceptual framework of immunological mechanisms of treatment resistance integrating cross-talk between immune, stromal and epithelial interactions.
The immune mechanisms of treatment resistance are complex and likely multifactorial. Key observations from different independent experiments, including high resolution single cell transcriptomics and cytometry experiments, indicate suggests common immunological themes in treatment resistance, typically involving communication circuits between mucosal immune cells and non-immune compartments, including stromal cells and epithelial cells. It is possible to integrate these paradigms into a potentially unifying hypothetic framework to identify at least one of the biological pathways associated with poor patient outcomes, tissue injury and treatment refractoriness. 1. Key proximal steps responsible for initiating severe inflammation are likely to involve mononuclear phagocytes (MPs), and especially CD14+ CX3CR1+ subsets, which are enriched in diseased tissue of IBD patients and play an important role sampling and responding to luminal antigens, including dysbiotic bacterial communities and other components of the exposome. 2. These MPs are a key source of cytokines implicated in treatment resistance, such as IL-1β, IL-23, oncostatin M and TNF, and other immune molecules, such as TREM1. 3. Some of these cytokines act directly on key non-immune cells, including the stromal and epithelial compartments. The transcriptional response of different intestinal stromal cells to oncostatin M, and IL-1β in particular, is linked to resistance to steroids, anti-TNF and vedolizumab. Analysis of the biological pathway driven by these cytokines indicate activation of neutrophil attracting chemokines (CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL8), extracellular matrix disruption (including expression of metalloproteinases MMP1, MMP3, MMP13) and augmented expression of hallmark fibroblast genes, such as THY1, PDPN, IL1l, IL13RA2. 4. In agreement with increased expression of neutrophil-selective CXC family chemokines, treatment resistant patients with significant tissue injury, including ulcers have increased neutrophil accumulation in inflamed tissue. IL-1 and IL-23 are important drivers of lymphocyte activation, including conventional CD4+ T-cells, MAIT cells and innate lymphoid cells (ILCs), triggering induction of multiple effector cytokines, such as IFNγ and IL-22. Indeed, IL-22 is also a potent inducer of neutrophil-active cytokines, particularly in intestinal epithelial cells. Notably, IL-22-responsive genes are also enriched in IBD tissue, including ulcers. The mechanisms that underpin the association of enhanced neutrophil recruitment, tissue injury and poor patient outcomes are not known, but neutrophils themselves have potent inflammatory activity as producers of reactive oxygen species, tissue damaging metalloproteinases, extracellular traps and can produce cytokines.
Not only mesenchymal cells, but the ECM itself is considered an active participant in inflammation, tissue damage and remodeling.165, 166 Through relapsing and remitting cycles of inflammation and repair, the accumulation of ECM results in the change of its mechanical properties, mainly in increased stiffness. The stiffness of the inflamed IBD intestine is higher compared to non-inflamed tissue, irrespective of small bowel or colonic location.167, 168 This was also true for the stiffness of stricture areas from CD intestine that had up to 6-fold higher values compared to adjacent grossly normal tissue.167, 168 Increased stiffness activates intestinal mesenchymal cells through changing their morphology, cytoskeletal architecture, migratory properties, proliferation and importantly ECM deposition168–171 This may initiate a feed-forward mechanism that progresses independently of inflammation and hence is relevant for long-term outcomes.
The ECM also has important effects on the inflammatory process in IBD pathobiology by altering cellular processes and functions through sequestering cytokines, chemokines, and growth factors.165 The ECM can present bound mediators to immune and non-immune cells in the tissues to activate them. The ECM can promote adhesion and extravasation of leukocytes and therefore not only increase their number in the inflamed gut, but may also increase their retention. ECM components themselves, such as fibronectin, collagens, and laminin, interact with cell adhesion molecules expressed on the surface of essentially all immune and non-immune cells and manipulate their function through modulation of intracellular signaling pathways.165 A well-documented example is the ECM component hyaluronan, which plays a direct role in the recruitment of leukocytes in response to inflammatory stimuli.172
The increasingly recognized dynamic interactions within the ECM seem not only key drivers for stricture formation but also for penetrating complications in Crohn’s disease and disease progression in UC. The >1,000 molecules that contribute to and regulate the ECM, the ‘matrisome’,173 include a variety of mediators involved in tissue repair and remodeling, and dysregulation of this physiological function seems to be key in IBD pathobiology. Matrix metalloproteinases (MMPs) and elastases are upregulated in IBD,174, 175 with dysregulation preceding symptoms.176 In one study exploring matrisome dysregulation in colitis, neutrophil elastase was detected only in mice with severe DSS-induced disease, whilst those with mild disease demonstrated a balance between the activity of neutrophil elastase and its inhibitor, SerpinA3N. Extracellular secretion of SerpinA3N via an engineered bacterial strain was able to attenuate tissue damage, suggesting that the imbalance of proteolytic and anti-proteolytic activity is crucial in determining tissue damage.177 Similarly, tissue inhibitor of metalloproteinases-3 (TIMP3)-transgenic mice were resistant to 2,4,6-trinitrobenzene-sulfonic acid-induced colitis.178 Different types and degrees of dysregulation appear to affect the endothelium, facilitating intestinal entry of immune cells, and disrupt cell-cell adhesion and the integrity of the basement membrane, which increases epithelial permeability for luminal microbes.178 The matrisome may also directly influence response to biologic medication, and MMP3 and MMP12 have been shown to reduce the function of infliximab and adalimumab molecules through proteolytic cleavage, which is more pronounced in anti-TNF non-responders, suggesting a role in treatment resistance.179
Hence, one key to future prediction of long-term outcomes in IBD is to focus on the mesenchymal cell populations as well as their products, namely the ECM. While conceptually still in its infancy this notion has already found entry into the world of IBD.
Current predictors of long-term outcomes
Long-term complications in Crohn’s disease, such as strictures or fistulae, are linked to high morbidity, hospitalizations, need for surgery, and poor quality of life.38 The future potential for a patient without complications to develop a complicated phenotype is a major consideration that influences treatment decisions by clinicians. The striking variability in disease progression can also be seen in UC.180, 181
Currently the choice and timing of therapy are based exclusively upon clinical presentation of disease,182 which may be heterogenous and confounded by delays in diagnosis. Accurate biomarkers in this space would be a powerful patient management tool enabling clinicians to adjust the long-term therapy according to risk stratification. This may furthermore facilitate patient recruitment for natural history studies or allow the effective design for trials of medications preventing future complications and this should be a research priority.
This need is in contrast to the limited number of studies assessing long-term outcomes. The majority of the available publications examine genetics, serology, the microbiome, mucosal biopsy transcriptomic profiles or a combination of the above. Genetic variants are linked with the diagnosis of IBD, with disease location or drug metabolism.183 Initial studies assessing prognosis assessed gene variants that were identified with the diagnosis of IBD.184 Variants in NOD2 were found to be associated with bowel stenosis, bowel resection or complicated Crohn’s disease.184 Since genes linked with diagnosis may not be relevant for prognosis, Lee et al. performed a within-cases genome wide associated study in two large Crohn’s disease cohorts.185 Patients were grouped into aggressive (frequent flares, treatment refractory and at least 2 abdominal surgeries) and mild (no immunomodulators or surgery with at least 4 years of follow up) and consistent with prior investigations, genes linked with diagnosis were not linked with prognosis. Interestingly, and independent of disease location and follow-up, four significant gene loci had prognostic value, including XACT (X chromosome long non-coding RNA), MHC (antigen recognition), FOXO3 (TGF-β1 pathway) and IGFBP1-IGFBP3 (insulin like growth factor binding protein). Pathway analysis suggested innate and adaptive immune responses and responses to microorganisms to be involved in disease progression.
In fact, in IBD an intestinal immune response against intestinal luminal antigens can be found. This, together with a genetic predisposition, gives rise to the formation of anti-microbial antibodies, that can be detected in the circulation (serum) of patients with IBD, in particular Crohn’s disease. The presence or levels of several of those antibodies are linked with or predictive of complications and surgery in Crohn’s disease.186 The combination of clinical, serologic and genetic markers has a higher accuracy compared to any individual marker alone187 and, to allow use in clinical practice online, webtools have been developed to assist providers.187 Major limitations of those studies are the lack of prospective data and non-protocolized sampling schemes.
The best characterized data is derived from the North American pediatric RISK inception cohort153 A combination of clinical factors (age, race, disease location) and antimicrobial serologies predicted complicated disease with an accuracy of AUC 0.66. Presence of Ruminococcus at diagnosis was linked with future stricturing complications and Veillonella with penetrating complications. This study furthermore identified a gene set in ileal biopsies at diagnosis which was associated with future stricturing or internal penetrating disease, and when adding the gene signatures to the risk model the AUC increased to 0.72. Using deeper RNA sequencing and only focusing on future development of stricturing disease an ileal gene signature associated with ECM gene expression, including an oncostatin M co-expression signature were identified.188 An eight ECM gene set reached an accuracy for prediction of future strictures (with in 5 years) of AUC 0.82.188 When looking at development of future stricturing disease within 3 years, serum proteomics revealed ECM1 as a predictor of future strictures (HR 5.33).189 Plasma concentrations of collagen type III alpha 1 chain (COL3A1) and autoantibodies to colony-stimulating factor 2 (CSF2) at diagnosis identified patients with future B2 versus B1 disease with an AUC of 0.8.190
In totality, these available data make it apparent that current markers for long-term prediction are not accurate enough to be used in clinical practice. Further complicating the situation is the fact that the available markers predict a complicated disease course in general, rather than a specific complication, such as strictures or internal penetrating disease that could be targeted with a specific intervention.
Conclusions:
Practical approach to the future of precision medicine
As outlined in this review, the clear clinical unmet need is in stark contrast to a lack of validated biomarkers in essentially all major areas of IBD. Several research priorities must be addressed to develop evidence-based solutions to the challenges posed by effective precision medicine (Table 1). IBD is a highly complex heterogenous disease with gene and environment interactions. In traditional prediction studies, each of those components is examined separately from each other. This approach negates our increasing understanding of IBD pathogenesis, which suggests that they should be viewed in totality as opposed to single contributors. Each ‘ome’, such as the genome, microbiome or exposome, would then be individually analyzed, followed by their integration in a process termed ‘systems biology’ (Figure 2). Investigation of multi-omics data is believed to result in more than the sum of its individual components191 and may sharpen our approach to disease prediction in IBD.192 This is likely the case even more so when multi-omics data are examined longitudinally over time. While in other diseases systems biology is already commonly used,193–195 the journey in IBD has just begun.104, 196 To reach the ultimate goal several challenges need to be addressed.
Table 1:
Future research priorities in precision medicine
| Improved disease classification and understanding of natural history: |
| • Natural history: Patient stratification into complicated and indolent disease courses |
| • Exposome: Understand which environmental factors are linked to IBD pathogenesis, what influences susceptibility to these factors and at what time in an individual’s life these environmental factors have greatest impact. |
| • Patient heterogeneity: Improve IBD stratification by multi-omics approaches. Create more effective molecular sub classifications in IBD. Link stratifications to clinically meaningful outcomes. |
| Optimal design of cohort studies and clinical trials: |
| • Design of short-term studies: Factors governing treatment response and (primary and secondary) non-response. |
| • Design of long-term studies: Factors leading to long term consequences of IBD including malignancy, fibrosis and surgery risk. |
| • Definition of key clinical outcomes, surrogate endpoints and treatment targets: Short term (response and remission, mucosal healing, early drug side effects) and long term (hospitalisation, fibrosis, surgery, neoplasia, de-escalation of therapy, long term drug side effects). |
| • Phenotyping: Use of multi-omic technologies on relevant clinical material including blood, intestinal tissue, stool, saliva and urine. |
| • Inclusivity: Address inequalities in research methodologies including access across ethnic groups, ages and countries. |
| • Head to head studies: Better understand comparative efficacy of therapeutic interventions. |
| • Biomarker stratification: Discovery and validation of predictive or prognostic biomarkers embedded from outset of clinical development programmes. |
| • Tissue sampling: Defining mechanisms to support multicentre collection, shipping, cryopreservation and storage of samples. Development of governance to access, protocols and technologies to profile readily available clinical archival material (e.g. formalin fixed paraffin embedded (FFPE) tissue). |
| Computational integration: |
| • Digital health: Collection, interpretation and visualisation of healthcare data from electronic healthcare records for research. |
| • Collaboration: Encouragement of multicentre industry-academic-informatic partnerships. |
| • Bioinformatic training: Availability of training and open access sharing of bioinformatic pipelines including emerging artificial intelligence and machine learning approaches. |
| • Data integration/systems biology: Development of multi-omic integrative approaches to incorporate clinical, microbiome, metabolome, transcriptome, genome with environmental exposure. |
| Mechanistic biological validation: |
| • Functional experiments: Testing of biological implications of immune and microbial associations identified by -omic studies. |
| Incorporation of precision medicine strategies into clinical practice: |
| • Health economics: Delineation of models relevant to diverse healthcare settings, financial structures and populations globally. |
| • Accessibility: Availability of high throughput, accessible and cost-effective technology to introduce biomarkers in clinical practice. |
| • Clinical guidelines: Incorporation of validated biomarkers in treatment pathways. |
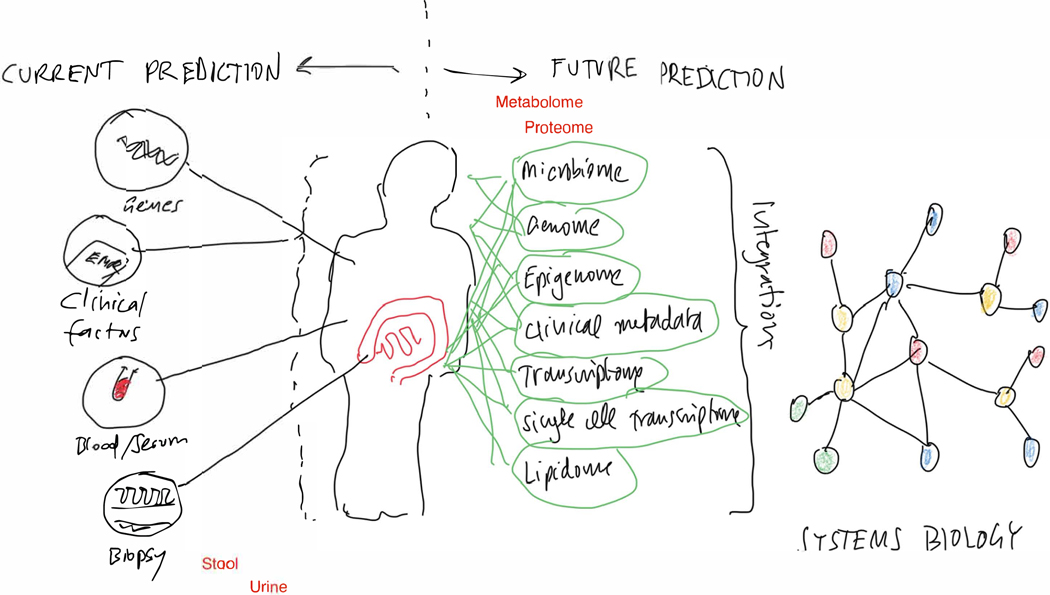
Figure 2:
Future of precision medicine discovery and validation: Intergation of multi-omics/systems biology.
The current approach of analysing individual omics separately needs to be changed to an integration of all omes’ using systems biology to identify central regulatory nodes allowing precision medicine.
Available systems biology tools have been largely developed for cancer investigations.197 In these diseases, often only a single specific gene mutation drives the pathology which may be, and is likely, different in IBD as a complex immune mediated disease. Hence, novel tools will need to be developed to account for this genetic heterogeneity. Current studies in IBD focus largely on individual -omics taken from larger populations and few to no datasets are available with multiple -omics from multiple tissues (biopsy, stool, blood, etc.) derived from only a few well characterized patients. It is however known that more -omics per patient with a smaller sample size reveal a more robust prediction compared to less omics in larger populations.198 Sample quality is of fundamental importance. This includes standardization and rigorous procurement protocols with documentation, when and how they were processed, storage conditions and quality controls. Finally, and perhaps of utmost importance is the clinical annotation of the samples. Examples include endoscopic evaluation only, without histologic confirmation or the lack of unified use of standardized and reliable scoring systems or patient reported outcome metrics. Only if the input material is solid can the output be trusted and be robust. What is not contributory is the fact that all published data is to be made available in publicly accessible datasets. These datasets all carry their own unique limitations, such as heterogeneity in patient cohorts, different procurement protocols, variable patient ages, missing data or different treatments, which then, even with the most sophisticated analysis pipeline, will yield incorrect results when combined with each other.
The solutions include a multi-omic analysis approach relying on the recruitment of significant numbers of patients in well planned, prospective, well characterized patient cohorts with standardized sampling schemes from multiple tissues (Figure 3). This is obtainable through multiple research designs, including registration randomized controlled interventional trials, cohort studies and increasingly prevalent IBD biorepositories.59 Sample collection should be ideally undertaken in a longitudinal fashion accounting for the environmental factors influencing IBD over time and to control for the fluctuations in disease activity. Once a biomarker signature is identified it should be validated in an independent dataset. What is often forgotten is the need to prospectively validate the relevance of the marker as otherwise any finding merely represents an association. Finally, even the perfect biomarker needs to show clinical impact. Prediction for the sake of prediction only is a weak sword against fighting IBD. For each biomarker, controlled trials needs to be performed to test if an intervention based on the biomarker ultimately improves outcome.
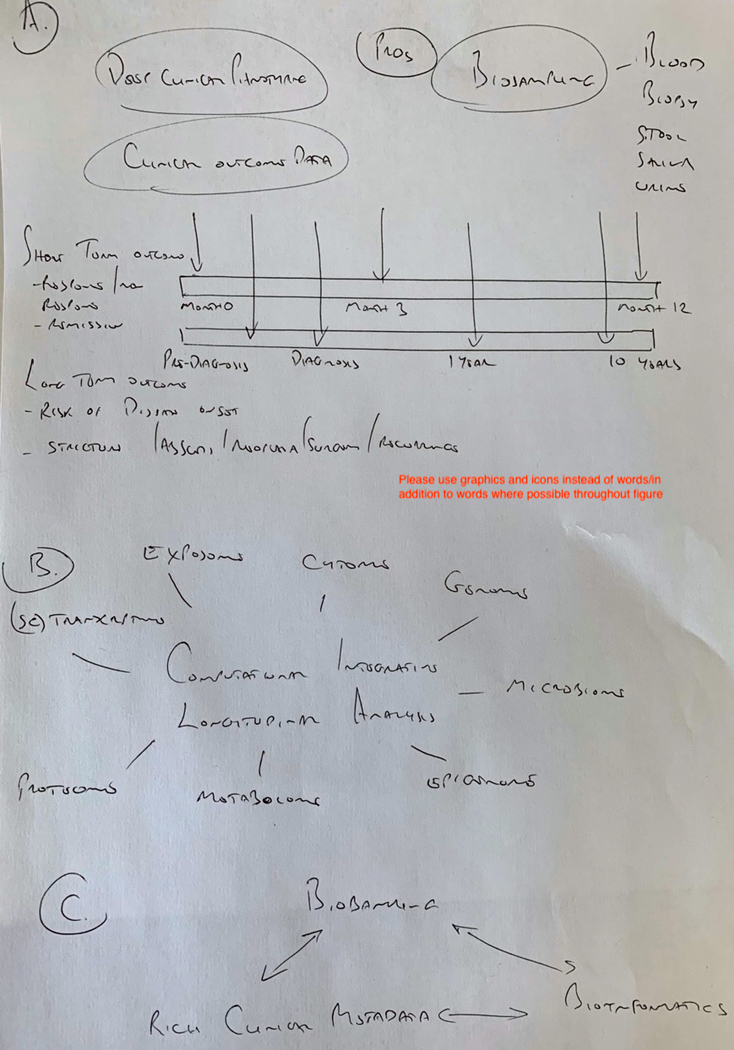
Figure 3:
Conceptual basis for optimal future precision medicine clinical trial and cohort design for short term and long-term outcomes in inflammatory bowel disease
A. Future observational cohort or interventional trials in precision medicine should be designed around relevant outcomes of interest. Short term outcomes such as treatment response/remission, non-response, loss of response and durable remission should have at least 1 year, ideally longer follow up. For Long term studies assessments should begin at diagnosis (or pre-diagnosis to assess disease risk or prevention) and continue for many years to study complications of disease such as stricture formation, abscess, neoplasia, surgery or postoperative recurrence. Crucial for both types of study is longitudinal collection of clinical outcome data, coupled with patient reported information, deep clinical phenotyping and appropriate biosampling to collect multiple tissue or sample types, at multiple timepoints. B. Biosamples should be analysed using multi-omic technologies or hypothesis-led targeted phenotypic assays and data integrated by computational means. This will provide multidimensional biological understanding, for example of disease progression, response to therapy, and biomarker discovery and validation. C. Remaining biosamples should be archived in biorepositories to facilitate targeted laboratory functional exploration or validation of biological insight informed by -omic approaches. Key to success is the association of these biorepositories with clinical metadata and bioinformatic pipelines to identify and refine big data discoveries and to formulate relevant hypotheses.
Provided these challenges clinical trials and cohort studies must change the current focus on only short term, symptomatic endpoints and consider predictive biomarkers and treatment targets within methodological designs. The wide spectrum of IBD phenotypes and severity of disease will need to be explored, if we are to understand treatment response and predict natural history. This includes older patients, those with mild-to-moderate disease, comorbidities as well as patients with prior response or non-response to anti-TNF or other biologic therapies.15, 24, 199 Such approaches could be facilitated by removing barriers to participation in research, using electronic patient records to identify potential participants, and promoting clinical and scientific principles in study design. This will become crucially important as predictive markers seek validation in randomized controlled trials. In these instances, whilst current registration trials often report efficacy data following a randomized responder study design, studies trialling potentially clinically tractable predictive biomarkers will need to be developed to explore intention-to-treat outcomes a priori. Furthermore, a holistic approach to the goals of a predictive approach in IBD will be required, focussing both on inflammation and quality of life.200
Future approaches to precision medicine through understanding the mechanistic basis of disease biology, short- and long-term treatment response, and natural history are promising (Figure 4), and will help to achieve a collective goal to control and reverse inflammation, tissue damage and control excessive remodeling, ultimately preventing long-term complications of IBD.
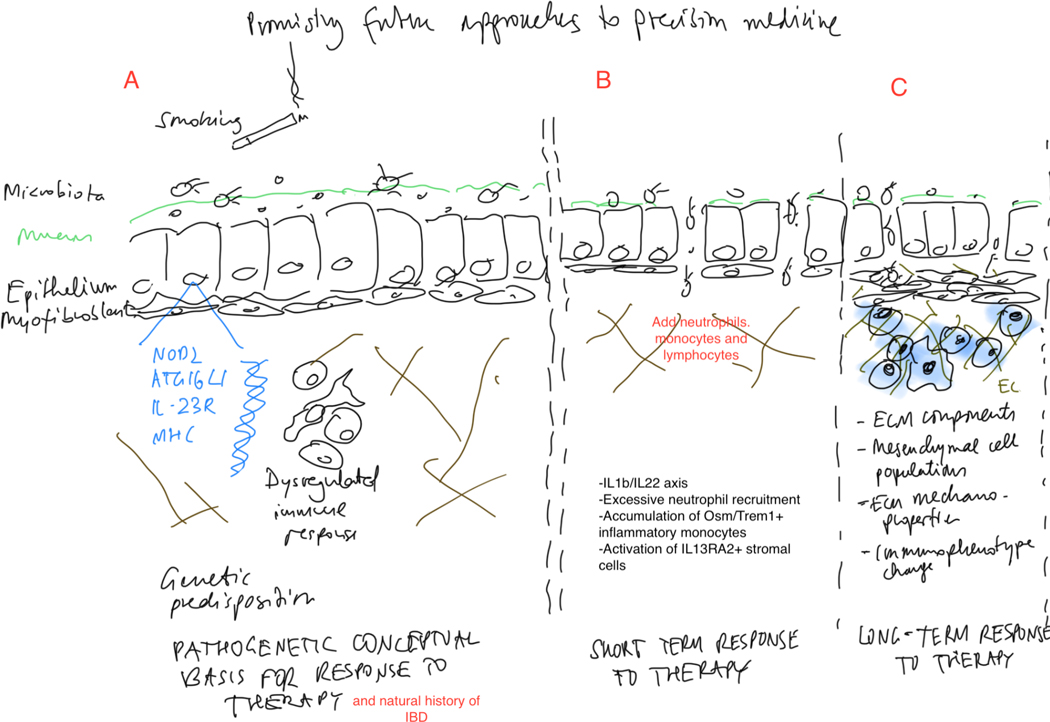
Figure 4:
Promising future approaches to precision medicine along the spectrum of the natural history of inflammatory bowel disease.
A. Pathogenetic conceptual basis for response to therapy and natural history of IBD: A complex interplay of environmental factors, genetic factors and a dysregulated inflammatory response including immune and non-immune cells. B. Potential markers for short term response to therapy. C. Potential markers for long term response to therapy/avoidance of complications of chronic inflammation and fibrogenesis. Abbreviations: ECM: Extracellular matrix; OSM: Oncostatin M; TREM: Triggering receptor expressed on myeloid cells 1; IL13RA2: Interleukin 13 receptor alpha 2; NOD: Nucleotide oligonerization domain; MHC: Major histocompatibility complex
Acknowledgments
Grant support:
This work was supported by the Helmsley Charitable Trust through the Stenosis Therapy and Anti-Fibrotic Research (STAR) Consortium (No. 3081 to F.R.) and the National Institute of Health (NIDDK K08DK110415 and R01DK123233 to F.R.). CAL acknowledges support from the NIHR Newcastle Biomedical Research Centre.
Dr. Christopher Lamb reports grants, consultancy and/or speaker fees from Genentech, Janssen, Takeda, AbbVie, Ferring, Eli Lilly, Pfizer, Roche, UCB Biopharma, Sanofi Aventis, Biogen IDEC, Orion OYJ, Dr Falk Pharma, and AstraZeneca.
Dr. Aamir Saifuddin has received conference travel grants from Dr Falk Pharma.
Dr. Nick Powell reports consultancy/advisory board fees, speaker fees and/or grants from Takeda, Janssen, Pfizer, Bristol-Myers Squibb, Abbvie, Roche, Lilly, Allergan, Celgene, Falk, Ferring,, Tillotts, and Vifor Pharma.
Dr. Florian Rieder is consultant to or on the advisory board of Agomab, Allergan, AbbVie, Boehringer-Ingelheim, Celgene, Cowen, Genentech, Gilead, Gossamer, Guidepoint, Helmsley, Index Pharma, Jannsen, Koutif, Metacrine, Morphic, Pfizer, Pliant, Prometheus Biosciences, Receptos, RedX, Roche, Samsung, Takeda, Techlab, Thetis, UCB and receives funding from the Crohn’s and Colitis Foundation of America, the Helmsley Charitable Trust, Kenneth Rainin Foundation and the National Institute of Health.
Footnotes
Conflicts of interest:
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jones GR, Lyons M, Plevris N, et al. IBD prevalence in Lothian, Scotland, derived by capture-recapture methodology. Gut 2019;68:1953–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamilton B, Green H, Heerasing N, et al. Incidence and prevalence of inflammatory bowel disease in Devon, UK. Frontline Gastroenterology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ananthakrishnan AN. Epidemiology and risk factors for IBD. Nat Rev Gastroenterol Hepatol 2015;12:205–17. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, MacIsaac D, Wong JJ, et al. Market share and costs of biologic therapies for inflammatory bowel disease in the USA. Aliment Pharmacol Ther 2018;47:364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burisch J, Vardi H, Schwartz D, et al. Health-care costs of inflammatory bowel disease in a pan-European, community-based, inception cohort during 5 years of follow-up: a population-based study. Lancet Gastroenterol Hepatol 2020;5:454–464. [DOI] [PubMed] [Google Scholar]
- 6.Irving P, Barrett K, Nijher M, et al. Prevalence of depression and anxiety in people with inflammatory bowel disease and associated healthcare use: population-based cohort study. Evidence Based Mental Health 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araki M, Shinzaki S, Yamada T, et al. Psychologic stress and disease activity in patients with inflammatory bowel disease: A multicenter cross-sectional study. PLoS One 2020;15:e0233365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keefer L. The Time Has Come to Integrate Behavioural Health Services Into IBD Centres. J Crohns Colitis 2019;13:817–818. [DOI] [PubMed] [Google Scholar]
- 9.Knowles SR, Graff LA, Wilding H, et al. Quality of Life in Inflammatory Bowel Disease: A Systematic Review and Meta-analyses-Part I. Inflamm Bowel Dis 2018;24:742–751. [DOI] [PubMed] [Google Scholar]
- 10.Topal F, Camyar H, Saritas Yuksel E, et al. Work Productivity Loss in Inflammatory Bowel Disease Patients in Turkey. Gastroenterol Res Pract 2020;2020:6979720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Valk ME, Mangen MJ, Leenders M, et al. Risk factors of work disability in patients with inflammatory bowel disease--a Dutch nationwide web-based survey: work disability in inflammatory bowel disease. J Crohns Colitis 2014;8:590–7. [DOI] [PubMed] [Google Scholar]
- 12.Feagan BG, Reinisch W, Rutgeerts P, et al. The effects of infliximab therapy on health-related quality of life in ulcerative colitis patients. Am J Gastroenterol 2007;102:794–802. [DOI] [PubMed] [Google Scholar]
- 13.Gray WN, Denson LA, Baldassano RN, et al. Disease activity, behavioral dysfunction, and health-related quality of life in adolescents with inflammatory bowel disease. Inflamm Bowel Dis 2011;17:1581–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosnes J, Cattan S, Blain A, et al. Long-term evolution of disease behavior of Crohn’s disease. Inflamm Bowel Dis 2002;8:244–50. [DOI] [PubMed] [Google Scholar]
- 15.Golovics PA, Mandel MD, Lovasz BD, et al. Inflammatory bowel disease course in Crohn’s disease: is the natural history changing? World J Gastroenterol 2014;20:3198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weimers P, Munkholm P. The Natural History of IBD: Lessons Learned. Curr Treat Options Gastroenterol 2018;16:101–111. [DOI] [PubMed] [Google Scholar]
- 17.Cosnes J, Gower-Rousseau C, Seksik P, et al. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011;140:1785–94. [DOI] [PubMed] [Google Scholar]
- 18.Solberg IC, Vatn MH, Hoie O, et al. Clinical course in Crohn’s disease: results of a Norwegian population-based ten-year follow-up study. Clin Gastroenterol Hepatol 2007;5:1430–8. [DOI] [PubMed] [Google Scholar]
- 19.Bernstein CN, Loftus EV Jr., Ng SC, et al. Hospitalisations and surgery in Crohn’s disease. Gut 2012;61:622–9. [DOI] [PubMed] [Google Scholar]
- 20.Khoudari G, Mansoor E, Click B, et al. Rates of Intestinal Resection and Colectomy in Inflammatory Bowel Disease Patients After Initiation of Biologics: A Cohort Study. Clin Gastroenterol Hepatol 2020. [DOI] [PubMed] [Google Scholar]
- 21.Holko P, Kawalec P, Pilc A. Impact of Biologic Treatment of Crohn’s Disease on the Rate of Surgeries and Other Healthcare Resources: An Analysis of a Nationwide Database From Poland. Front Pharmacol 2018;9:621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen GC, Nugent Z, Shaw S, et al. Outcomes of patients with Crohn’s disease improved from 1988 to 2008 and were associated with increased specialist care. Gastroenterology 2011;141:90–7. [DOI] [PubMed] [Google Scholar]
- 23.Rahman A, Jairath V, Feagan BG, et al. Declining hospitalisation and surgical intervention rates in patients with Crohn’s disease: a population-based cohort. Aliment Pharmacol Ther 2019;50:1086–1093. [DOI] [PubMed] [Google Scholar]
- 24.Fumery M, Singh S, Dulai PS, et al. Natural History of Adult Ulcerative Colitis in Population-based Cohorts: A Systematic Review. Clin Gastroenterol Hepatol 2018;16:343–356 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solberg IC, Lygren I, Jahnsen J, et al. Clinical course during the first 10 years of ulcerative colitis: results from a population-based inception cohort (IBSEN Study). Scand J Gastroenterol 2009;44:431–40. [DOI] [PubMed] [Google Scholar]
- 26.Konig IR, Fuchs O, Hansen G, et al. What is precision medicine? Eur Respir J 2017;50. [DOI] [PubMed] [Google Scholar]
- 27.FitzGerald JM, Bleecker ER, Nair P, et al. Benralizumab, an anti-interleukin-5 receptor alpha monoclonal antibody, as add-on treatment for patients with severe, uncontrolled, eosinophilic asthma (CALIMA): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet 2016;388:2128–2141. [DOI] [PubMed] [Google Scholar]
- 28.Vogel CL, Cobleigh MA, Tripathy D, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol 2002;20:719–26. [DOI] [PubMed] [Google Scholar]
- 29.Hanauer SB, Feagan BG, Lichtenstein GR, et al. Maintenance infliximab for Crohn’s disease: the ACCENT I randomised trial. Lancet 2002;359:1541–9. [DOI] [PubMed] [Google Scholar]
- 30.Feagan BG, Rutgeerts P, Sands BE, et al. Vedolizumab as induction and maintenance therapy for ulcerative colitis. N Engl J Med 2013;369:699–710. [DOI] [PubMed] [Google Scholar]
- 31.Sandborn WJ, Feagan BG, Rutgeerts P, et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N Engl J Med 2013;369:711–21. [DOI] [PubMed] [Google Scholar]
- 32.Sands BE, Marano C. Ustekinumab as Induction and Maintenance Therapy for Ulcerative Colitis. Reply. N Engl J Med 2020;382:91. [DOI] [PubMed] [Google Scholar]
- 33.Feagan BG, Sandborn WJ, Gasink C, et al. Ustekinumab as Induction and Maintenance Therapy for Crohn’s Disease. N Engl J Med 2016;375:1946–1960. [DOI] [PubMed] [Google Scholar]
- 34.Muhl L, Becker E, Muller TM, et al. Clinical experiences and predictors of success of treatment with vedolizumab in IBD patients: a cohort study. BMC Gastroenterol 2021;21:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blesl A, Binder L, Hogenauer C, et al. Limited long-term treatment persistence of first anti-TNF therapy in 538 patients with inflammatory bowel diseases: a 20-year real-world study. Aliment Pharmacol Ther 2021;54:667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ko Y, Paramsothy S, Yau Y, et al. Superior treatment persistence with ustekinumab in Crohn’s disease and vedolizumab in ulcerative colitis compared with anti-TNF biological agents: real-world registry data from the Persistence Australian National IBD Cohort (PANIC) study. Aliment Pharmacol Ther 2021;54:292–301. [DOI] [PubMed] [Google Scholar]
- 37.Ito T, Maemoto A, Katsurada T, et al. Long-Term Clinical Effectiveness of Ustekinumab in Patients With Crohn’s Disease: A Retrospective Cohort Study. Crohn’s & Colitis 360 2020;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rieder F, Fiocchi C, Rogler G. Mechanisms, Management, and Treatment of Fibrosis in Patients With Inflammatory Bowel Diseases. Gastroenterology 2017;152:340–350 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh S, George J, Boland BS, et al. Primary Non-Response to Tumor Necrosis Factor Antagonists is Associated with Inferior Response to Second-line Biologics in Patients with Inflammatory Bowel Diseases: A Systematic Review and Meta-analysis. J Crohns Colitis 2018;12:635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ungaro RC, Yzet C, Bossuyt P, et al. Deep Remission at 1 Year Prevents Progression of Early Crohn’s Disease. Gastroenterology 2020;159:139–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner D, Ricciuto A, Lewis A, et al. STRIDE-II: An Update on the Selecting Therapeutic Targets in Inflammatory Bowel Disease (STRIDE) Initiative of the International Organization for the Study of IBD (IOIBD): Determining Therapeutic Goals for Treat-to-Target strategies in IBD. Gastroenterology 2021;160:1570–1583. [DOI] [PubMed] [Google Scholar]
- 42.Gracie DJ, Guthrie EA, Hamlin PJ, et al. Bi-directionality of Brain-Gut Interactions in Patients With Inflammatory Bowel Disease. Gastroenterology 2018;154:1635–1646 e3. [DOI] [PubMed] [Google Scholar]
- 43.Kochar B, Barnes EL, Long MD, et al. Depression Is Associated With More Aggressive Inflammatory Bowel Disease. Am J Gastroenterol 2018;113:80–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moulton CD, Pavlidis P, Norton C, et al. Depressive symptoms in inflammatory bowel disease: an extraintestinal manifestation of inflammation? Clin Exp Immunol 2019;197:308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beaugerie L, Seksik P, Nion-Larmurier I, et al. Predictors of Crohn’s disease. Gastroenterology 2006;130:650–6. [DOI] [PubMed] [Google Scholar]
- 46.Adler J, Rangwalla SC, Dwamena BA, et al. The prognostic power of the NOD2 genotype for complicated Crohn’s disease: a meta-analysis. Am J Gastroenterol 2011;106:699–712. [DOI] [PubMed] [Google Scholar]
- 47.Bullock NR, Booth JC, Gibson GR. Comparative composition of bacteria in the human intestinal microflora during remission and active ulcerative colitis. Curr Issues Intest Microbiol 2004;5:59–64. [PubMed] [Google Scholar]
- 48.Rajca S, Grondin V, Louis E, et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm Bowel Dis 2014;20:978–86. [DOI] [PubMed] [Google Scholar]
- 49.Torres J, Caprioli F, Katsanos KH, et al. Predicting Outcomes to Optimize Disease Management in Inflammatory Bowel Diseases. J Crohns Colitis 2016;10:1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abegunde AT, Muhammad BH, Bhatti O, et al. Environmental risk factors for inflammatory bowel diseases: Evidence based literature review. World J Gastroenterol 2016;22:6296–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.West NR, Hegazy AN, Owens BMJ, et al. Oncostatin M drives intestinal inflammation and predicts response to tumor necrosis factor-neutralizing therapy in patients with inflammatory bowel disease. Nat Med 2017;23:579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verstockt B, Verstockt S, Veny M, et al. Expression Levels of 4 Genes in Colon Tissue Might Be Used to Predict Which Patients Will Enter Endoscopic Remission After Vedolizumab Therapy for Inflammatory Bowel Diseases. Clin Gastroenterol Hepatol 2020;18:1142–1151 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verstockt B, Verstockt S, Dehairs J, et al. Low TREM1 expression in whole blood predicts anti-TNF response in inflammatory bowel disease. EBioMedicine 2019;40:733–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Telesco SE, Brodmerkel C, Zhang H, et al. Gene Expression Signature for Prediction of Golimumab Response in a Phase 2a Open-Label Trial of Patients With Ulcerative Colitis. Gastroenterology 2018;155:1008–1011 e8. [DOI] [PubMed] [Google Scholar]
- 55.Aden K, Rehman A, Waschina S, et al. Metabolic Functions of Gut Microbes Associate With Efficacy of Tumor Necrosis Factor Antagonists in Patients With Inflammatory Bowel Diseases. Gastroenterology 2019;157:1279–1292 e11. [DOI] [PubMed] [Google Scholar]
- 56.Ananthakrishnan AN, Luo C, Yajnik V, et al. Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe 2017;21:603–610 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aldars-Garcia L, Chaparro M, Gisbert JP. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gisbert JP, Chaparro M. Clinical Usefulness of Proteomics in Inflammatory Bowel Disease: A Comprehensive Review. J Crohns Colitis 2019;13:374–384. [DOI] [PubMed] [Google Scholar]
- 59.Denson LA, Curran M, McGovern DPB, et al. Challenges in IBD Research: Precision Medicine. Inflamm Bowel Dis 2019;25:S31–S39. [DOI] [PubMed] [Google Scholar]
- 60.Fiocchi C. Current perspectives in inflammatory bowel disease: stress response and autophagy, host-microbe mutualism, immune duality and plasticity, and early versus late disease. Curr Opin Gastroenterol 2010;26:299–301. [DOI] [PubMed] [Google Scholar]
- 61.Johnson GJ, Cosnes J, Mansfield JC. Review article: smoking cessation as primary therapy to modify the course of Crohn’s disease. Aliment Pharmacol Ther 2005;21:921–31. [DOI] [PubMed] [Google Scholar]
- 62.Leong RW. Environmental risk factors in inflammatory bowel disease. J Gastroenterol Hepatol 2010;25:227–8. [DOI] [PubMed] [Google Scholar]
- 63.Scanlan PD, Shanahan F, O’Mahony C, et al. Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn’s disease. J Clin Microbiol 2006;44:3980–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ananthakrishnan AN. Environmental risk factors for inflammatory bowel disease. Gastroenterol Hepatol (N Y) 2013;9:367–74. [PMC free article] [PubMed] [Google Scholar]
- 65.Hou JK, Abraham B, El-Serag H. Dietary intake and risk of developing inflammatory bowel disease: a systematic review of the literature. Am J Gastroenterol 2011;106:563–73. [DOI] [PubMed] [Google Scholar]
- 66.Ananthakrishnan AN, Khalili H, Higuchi LM, et al. Higher predicted vitamin D status is associated with reduced risk of Crohn’s disease. Gastroenterology 2012;142:482–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ananthakrishnan AN, Long MD, Martin CF, et al. Sleep disturbance and risk of active disease in patients with Crohn’s disease and ulcerative colitis. Clin Gastroenterol Hepatol 2013;11:965–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoie O, Wolters F, Riis L, et al. Ulcerative colitis: patient characteristics may predict 10-yr disease recurrence in a European-wide population-based cohort. Am J Gastroenterol 2007;102:1692–701. [DOI] [PubMed] [Google Scholar]
- 69.Langholz E, Munkholm P, Davidsen M, et al. Course of ulcerative colitis: analysis of changes in disease activity over years. Gastroenterology 1994;107:3–11. [DOI] [PubMed] [Google Scholar]
- 70.Froslie KF, Jahnsen J, Moum BA, et al. Mucosal healing in inflammatory bowel disease: results from a Norwegian population-based cohort. Gastroenterology 2007;133:412–22. [DOI] [PubMed] [Google Scholar]
- 71.Meucci G, Fasoli R, Saibeni S, et al. Prognostic significance of endoscopic remission in patients with active ulcerative colitis treated with oral and topical mesalazine: a prospective, multicenter study. Inflamm Bowel Dis 2012;18:1006–10. [DOI] [PubMed] [Google Scholar]
- 72.Ardizzone S, Cassinotti A, Duca P, et al. Mucosal healing predicts late outcomes after the first course of corticosteroids for newly diagnosed ulcerative colitis. Clin Gastroenterol Hepatol 2011;9:483–489 e3. [DOI] [PubMed] [Google Scholar]
- 73.Loly C, Belaiche J, Louis E. Predictors of severe Crohn’s disease. Scand J Gastroenterol 2008;43:948–54. [DOI] [PubMed] [Google Scholar]
- 74.Steinbakk M, Naess-Andresen CF, Lingaas E, et al. Antimicrobial actions of calcium binding leucocyte L1 protein, calprotectin. Lancet 1990;336:763–5. [DOI] [PubMed] [Google Scholar]
- 75.D’Haens G, Ferrante M, Vermeire S, et al. Fecal calprotectin is a surrogate marker for endoscopic lesions in inflammatory bowel disease. Inflamm Bowel Dis 2012;18:2218–24. [DOI] [PubMed] [Google Scholar]
- 76.Schoepfer AM, Beglinger C, Straumann A, et al. Fecal calprotectin more accurately reflects endoscopic activity of ulcerative colitis than the Lichtiger Index, C-reactive protein, platelets, hemoglobin, and blood leukocytes. Inflamm Bowel Dis 2013;19:332–41. [DOI] [PubMed] [Google Scholar]
- 77.Zittan E, Kelly OB, Kirsch R, et al. Low Fecal Calprotectin Correlates with Histological Remission and Mucosal Healing in Ulcerative Colitis and Colonic Crohn’s Disease. Inflamm Bowel Dis 2016;22:623–30. [DOI] [PubMed] [Google Scholar]
- 78.Mao R, Xiao YL, Gao X, et al. Fecal calprotectin in predicting relapse of inflammatory bowel diseases: a meta-analysis of prospective studies. Inflamm Bowel Dis 2012;18:1894–9. [DOI] [PubMed] [Google Scholar]
- 79.De Vos M, Louis EJ, Jahnsen J, et al. Consecutive fecal calprotectin measurements to predict relapse in patients with ulcerative colitis receiving infliximab maintenance therapy. Inflamm Bowel Dis 2013;19:2111–7. [DOI] [PubMed] [Google Scholar]
- 80.Lamb CA, Mohiuddin MK, Gicquel J, et al. Faecal calprotectin or lactoferrin can identify postoperative recurrence in Crohn’s disease. Br J Surg 2009;96:663–74. [DOI] [PubMed] [Google Scholar]
- 81.Jukic A, Bakiri L, Wagner EF, et al. Calprotectin: from biomarker to biological function. Gut 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lamb CA, Mansfield JC. Measurement of faecal calprotectin and lactoferrin in inflammatory bowel disease. Frontline Gastroenterol 2011;2:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ansari A, Arenas M, Greenfield SM, et al. Prospective evaluation of the pharmacogenetics of azathioprine in the treatment of inflammatory bowel disease. Aliment Pharmacol Ther 2008;28:973–83. [DOI] [PubMed] [Google Scholar]
- 84.Jharap B, Seinen ML, de Boer NK, et al. Thiopurine therapy in inflammatory bowel disease patients: analyses of two 8-year intercept cohorts. Inflamm Bowel Dis 2010;16:1541–9. [DOI] [PubMed] [Google Scholar]
- 85.Winter J, Walker A, Shapiro D, et al. Cost-effectiveness of thiopurine methyltransferase genotype screening in patients about to commence azathioprine therapy for treatment of inflammatory bowel disease. Aliment Pharmacol Ther 2004;20:593–9. [DOI] [PubMed] [Google Scholar]
- 86.Lamb CA, Kennedy NA, Raine T, et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut 2019;68:s1–s106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feuerstein JD, Nguyen GC, Kupfer SS, et al. American Gastroenterological Association Institute Guideline on Therapeutic Drug Monitoring in Inflammatory Bowel Disease. Gastroenterology 2017;153:827–834. [DOI] [PubMed] [Google Scholar]
- 88.Coenen MJ, de Jong DJ, van Marrewijk CJ, et al. Identification of Patients With Variants in TPMT and Dose Reduction Reduces Hematologic Events During Thiopurine Treatment of Inflammatory Bowel Disease. Gastroenterology 2015;149:907–17 e7. [DOI] [PubMed] [Google Scholar]
- 89.Reinisch W, Colombel JF, Sandborn WJ, et al. Factors associated with short- and long-term outcomes of therapy for Crohn’s disease. Clin Gastroenterol Hepatol 2015;13:539–547 e2. [DOI] [PubMed] [Google Scholar]
- 90.Vande Casteele N, Khanna R, Levesque BG, et al. The relationship between infliximab concentrations, antibodies to infliximab and disease activity in Crohn’s disease. Gut 2015;64:1539–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Papamichael K, Rakowsky S, Rivera C, et al. Association Between Serum Infliximab Trough Concentrations During Maintenance Therapy and Biochemical, Endoscopic, and Histologic Remission in Crohn’s Disease. Inflamm Bowel Dis 2018;24:2266–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gonczi L, Vegh Z, Golovics PA, et al. Prediction of Short- and Medium-term Efficacy of Biosimilar Infliximab Therapy. Do Trough Levels and Antidrug Antibody Levels or Clinical And Biochemical Markers Play the More Important Role? J Crohns Colitis 2017;11:697–705. [DOI] [PubMed] [Google Scholar]
- 93.Davidov Y, Ungar B, Bar-Yoseph H, et al. Association of Induction Infliximab Levels With Clinical Response in Perianal Crohn’s Disease. J Crohns Colitis 2017;11:549–555. [DOI] [PubMed] [Google Scholar]
- 94.Ungar B, Engel T, Yablecovitch D, et al. Prospective Observational Evaluation of Time-Dependency of Adalimumab Immunogenicity and Drug Concentrations: The POETIC Study. Am J Gastroenterol 2018;113:890–898. [DOI] [PubMed] [Google Scholar]
- 95.Baert F, Vande Casteele N, Tops S, et al. Prior response to infliximab and early serum drug concentrations predict effects of adalimumab in ulcerative colitis. Aliment Pharmacol Ther 2014;40:1324–32. [DOI] [PubMed] [Google Scholar]
- 96.Dreesen E, Verstockt B, Bian S, et al. Evidence to Support Monitoring of Vedolizumab Trough Concentrations in Patients With Inflammatory Bowel Diseases. Clin Gastroenterol Hepatol 2018;16:1937–1946 e8. [DOI] [PubMed] [Google Scholar]
- 97.Williet N, Boschetti G, Fovet M, et al. Association Between Low Trough Levels of Vedolizumab During Induction Therapy for Inflammatory Bowel Diseases and Need for Additional Doses Within 6 Months. Clin Gastroenterol Hepatol 2017;15:1750–1757 e3. [DOI] [PubMed] [Google Scholar]
- 98.Adedokun OJ, Xu Z, Gasink C, et al. Pharmacokinetics and Exposure Response Relationships of Ustekinumab in Patients With Crohn’s Disease. Gastroenterology 2018;154:1660–1671. [DOI] [PubMed] [Google Scholar]
- 99.Negoescu DM, Enns EA, Swanhorst B, et al. Proactive Vs Reactive Therapeutic Drug Monitoring of Infliximab in Crohn’s Disease: A Cost-Effectiveness Analysis in a Simulated Cohort. Inflamm Bowel Dis 2020;26:103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Papamichael K, Vajravelu RK, Vaughn BP, et al. Proactive Infliximab Monitoring Following Reactive Testing is Associated With Better Clinical Outcomes Than Reactive Testing Alone in Patients With Inflammatory Bowel Disease. J Crohns Colitis 2018;12:804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vaughn BP, Martinez-Vazquez M, Patwardhan VR, et al. Proactive therapeutic concentration monitoring of infliximab may improve outcomes for patients with inflammatory bowel disease: results from a pilot observational study. Inflamm Bowel Dis 2014;20:1996–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Amiot A, Hulin A, Belhassan M, et al. Therapeutic drug monitoring is predictive of loss of response after de-escalation of infliximab therapy in patients with inflammatory bowel disease in clinical remission. Clin Res Hepatol Gastroenterol 2016;40:90–8. [DOI] [PubMed] [Google Scholar]
- 103.Papamichael K, Cheifetz AS, Melmed GY, et al. Appropriate Therapeutic Drug Monitoring of Biologic Agents for Patients With Inflammatory Bowel Diseases. Clin Gastroenterol Hepatol 2019;17:1655–1668 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dart RJ, Ellul P, Scharl M, et al. Results of the Seventh Scientific Workshop of ECCO: Precision medicine in IBD - Challenges and future directions. J Crohns Colitis 2021. [DOI] [PubMed] [Google Scholar]
- 105.Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2017;390:2769–2778. [DOI] [PubMed] [Google Scholar]
- 106.Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol 2015;12:720–7. [DOI] [PubMed] [Google Scholar]
- 107.To N, Gracie DJ, Ford AC. Systematic review with meta-analysis: the adverse effects of tobacco smoking on the natural history of Crohn’s disease. Aliment Pharmacol Ther 2016;43:549–61. [DOI] [PubMed] [Google Scholar]
- 108.Lunney PC, Kariyawasam VC, Wang RR, et al. Smoking prevalence and its influence on disease course and surgery in Crohn’s disease and ulcerative colitis. Aliment Pharmacol Ther 2015;42:61–70. [DOI] [PubMed] [Google Scholar]
- 109.Sharma A, Deeb AP, Iannuzzi JC, et al. Tobacco smoking and postoperative outcomes after colorectal surgery. Ann Surg 2013;258:296–300. [DOI] [PubMed] [Google Scholar]
- 110.Baucom RB, Poulose BK, Herline AJ, et al. Smoking as dominant risk factor for anastomotic leak after left colon resection. Am J Surg 2015;210:1–5. [DOI] [PubMed] [Google Scholar]
- 111.Mowat C, Arnott I, Cahill A, et al. Mercaptopurine versus placebo to prevent recurrence of Crohn’s disease after surgical resection (TOPPIC): a multicentre, double-blind, randomised controlled trial. Lancet Gastroenterol Hepatol 2016;1:273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niewiadomski O, Studd C, Wilson J, et al. Influence of food and lifestyle on the risk of developing inflammatory bowel disease. Intern Med J 2016;46:669–76. [DOI] [PubMed] [Google Scholar]
- 113.D’Souza S, Levy E, Mack D, et al. Dietary patterns and risk for Crohn’s disease in children. Inflamm Bowel Dis 2008;14:367–73. [DOI] [PubMed] [Google Scholar]
- 114.Narula N, Wong ECL, Dehghan M, et al. Association of ultra-processed food intake with risk of inflammatory bowel disease: prospective cohort study. BMJ 2021;374:n1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ananthakrishnan AN, Khalili H, Konijeti GG, et al. A prospective study of long-term intake of dietary fiber and risk of Crohn’s disease and ulcerative colitis. Gastroenterology 2013;145:970–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bancil AS, Sandall AM, Rossi M, et al. Food Additive Emulsifiers and Their Impact on Gut Microbiome, Permeability, and Inflammation: Mechanistic Insights in Inflammatory Bowel Disease. J Crohns Colitis 2021;15:1068–1079. [DOI] [PubMed] [Google Scholar]
- 117.Jiang Z, Zhao M, Zhang H, et al. Antimicrobial Emulsifier-Glycerol Monolaurate Induces Metabolic Syndrome, Gut Microbiota Dysbiosis, and Systemic Low-Grade Inflammation in Low-Fat Diet Fed Mice. Mol Nutr Food Res 2018;62. [DOI] [PubMed] [Google Scholar]
- 118.Chassaing B, Koren O, Goodrich JK, et al. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015;519:92–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lock JY, Carlson TL, Wang CM, et al. Acute Exposure to Commonly Ingested Emulsifiers Alters Intestinal Mucus Structure and Transport Properties. Sci Rep 2018;8:10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Delahunty T, Recher L, Hollander D. Intestinal permeability changes in rodents: a possible mechanism for degraded carrageenan-induced colitis. Food Chem Toxicol 1987;25:113–8. [DOI] [PubMed] [Google Scholar]
- 121.Roberts CL, Keita AV, Duncan SH, et al. Translocation of Crohn’s disease Escherichia coli across M-cells: contrasting effects of soluble plant fibres and emulsifiers. Gut 2010;59:1331–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Svolos V, Hansen R, Nichols B, et al. Treatment of Active Crohn’s Disease With an Ordinary Food-based Diet That Replicates Exclusive Enteral Nutrition. Gastroenterology 2019;156:1354–1367 e6. [DOI] [PubMed] [Google Scholar]
- 123.Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Graham DB, Xavier RJ. Pathway paradigms revealed from the genetics of inflammatory bowel disease. Nature 2020;578:527–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kennedy NA, Heap GA, Green HD, et al. Predictors of anti-TNF treatment failure in anti-TNF-naive patients with active luminal Crohn’s disease: a prospective, multicentre, cohort study. Lancet Gastroenterol Hepatol 2019;4:341–353. [DOI] [PubMed] [Google Scholar]
- 126.Sazonovs A, Kennedy NA, Moutsianas L, et al. HLA-DQA1*05 Carriage Associated With Development of Anti-Drug Antibodies to Infliximab and Adalimumab in Patients With Crohn’s Disease. Gastroenterology 2020;158:189–199. [DOI] [PubMed] [Google Scholar]
- 127.Walker GJ, Harrison JW, Heap GA, et al. Association of Genetic Variants in NUDT15 With Thiopurine-Induced Myelosuppression in Patients With Inflammatory Bowel Disease. JAMA 2019;321:773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heap GA, Weedon MN, Bewshea CM, et al. HLA-DQA1-HLA-DRB1 variants confer susceptibility to pancreatitis induced by thiopurine immunosuppressants. Nat Genet 2014;46:1131–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Parkes M, Investigators IBDB. IBD BioResource: an open-access platform of 25 000 patients to accelerate research in Crohn’s and Colitis. Gut 2019;68:1537–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bertani L, Fornai M, Fornili M, et al. Serum oncostatin M at baseline predicts mucosal healing in Crohn’s disease patients treated with infliximab. Aliment Pharmacol Ther 2020;52:284–291. [DOI] [PubMed] [Google Scholar]
- 131.Minar P, Lehn C, Tsai YT, et al. Elevated Pretreatment Plasma Oncostatin M Is Associated With Poor Biochemical Response to Infliximab. Crohns Colitis 360 2019;1:otz026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Verstockt B, Verstockt S, Blevi H, et al. TREM-1, the ideal predictive biomarker for endoscopic healing in anti-TNF-treated Crohn’s disease patients? Gut 2019;68:1531–1533. [DOI] [PubMed] [Google Scholar]
- 133.Prins MM, Verstockt B, Ferrante M, et al. Monocyte TREM-1 Levels Associate With Anti-TNF Responsiveness in IBD Through Autophagy and Fcgamma-Receptor Signaling Pathways. Front Immunol 2021;12:627535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tew GW, Hackney JA, Gibbons D, et al. Association Between Response to Etrolizumab and Expression of Integrin alphaE and Granzyme A in Colon Biopsies of Patients With Ulcerative Colitis. Gastroenterology 2016;150:477–87 e9. [DOI] [PubMed] [Google Scholar]
- 135.Mascheretti S, Hampe J, Kuhbacher T, et al. Pharmacogenetic investigation of the TNF/TNF-receptor system in patients with chronic active Crohn’s disease treated with infliximab. Pharmacogenomics J 2002;2:127–36. [DOI] [PubMed] [Google Scholar]
- 136.Pierik M, Vermeire S, Steen KV, et al. Tumour necrosis factor-alpha receptor 1 and 2 polymorphisms in inflammatory bowel disease and their association with response to infliximab. Aliment Pharmacol Ther 2004;20:303–10. [DOI] [PubMed] [Google Scholar]
- 137.Belarif L, Danger R, Kermarrec L, et al. IL-7 receptor influences anti-TNF responsiveness and T cell gut homing in inflammatory bowel disease. J Clin Invest 2019;129:1910–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Parkes M, Noor NM, Dowling F, et al. PRedicting Outcomes For Crohn’s dIsease using a moLecular biomarkEr (PROFILE): protocol for a multicentre, randomised, biomarker-stratified trial. BMJ Open 2018;8:e026767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lee JC, Lyons PA, McKinney EF, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Aschenbrenner D, Quaranta M, Banerjee S, et al. Deconvolution of monocyte responses in inflammatory bowel disease reveals an IL-1 cytokine network that regulates IL-23 in genetic and acquired IL-10 resistance. Gut 2021;70:1023–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Friedrich M, Pohin M, Jackson MA, et al. IL-1-driven stromal-neutrophil interaction in deep ulcers defines a pathotype of therapy non-responsive inflammatory bowel disease. bioRxiv 2021. [Google Scholar]
- 142.Powell N, Pantazi E, Pavlidis P, et al. Interleukin-22 orchestrates a pathological endoplasmic reticulum stress response transcriptional programme in colonic epithelial cells. Gut 2020;69:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Frank DN, St Amand AL, Feldman RA, et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 2007;104:13780–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Doherty MK, Ding T, Koumpouras C, et al. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn’s Disease Patients. mBio 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gopalakrishnan V, Spencer CN, Nezi L, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Matson V, Fessler J, Bao R, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018;359:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Routy B, Le Chatelier E, Derosa L, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018;359:91–97. [DOI] [PubMed] [Google Scholar]
- 150.Jobin C. Precision medicine using microbiota. Science 2018;359:32–34. [DOI] [PubMed] [Google Scholar]
- 151.Rieder F, Zimmermann EM, Remzi FH, et al. Crohn’s disease complicated by strictures: a systematic review. Gut 2013;62:1072–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cosnes J, Nion-Larmurier I, Beaugerie L, et al. Impact of the increasing use of immunosuppressants in Crohn’s disease on the need for intestinal surgery. Gut 2005;54:237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389:1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.McQuitty CE, Williams R, Chokshi S, et al. Immunomodulatory Role of the Extracellular Matrix Within the Liver Disease Microenvironment. Front Immunol 2020;11:574276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wu F, Chakravarti S. Differential expression of inflammatory and fibrogenic genes and their regulation by NF-kappaB inhibition in a mouse model of chronic colitis. J Immunol 2007;179:6988–7000. [DOI] [PubMed] [Google Scholar]
- 156.Kugathasan S, Saubermann LJ, Smith L, et al. Mucosal T-cell immunoregulation varies in early and late inflammatory bowel disease. Gut 2007;56:1696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fichtner-Feigl S, Strober W, Geissler EK, et al. Cytokines mediating the induction of chronic colitis and colitis-associated fibrosis. Mucosal Immunol 2008;1 Suppl 1:S24–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Duijvestein M, Battat R, Vande Casteele N, et al. Novel Therapies and Treatment Strategies for Patients with Inflammatory Bowel Disease. Curr Treat Options Gastroenterol 2018;16:129–146. [DOI] [PubMed] [Google Scholar]
- 159.Chang SK, Noss EH, Chen M, et al. Cadherin-11 regulates fibroblast inflammation. Proc Natl Acad Sci U S A 2011;108:8402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Croft AP, Campos J, Jansen K, et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 2019;570:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Vogel JD, West GA, Danese S, et al. CD40-mediated immune-nonimmune cell interactions induce mucosal fibroblast chemokines leading to T-cell transmigration. Gastroenterology 2004;126:63–80. [DOI] [PubMed] [Google Scholar]
- 162.Danese S, Sans M, Spencer DM, et al. Angiogenesis blockade as a new therapeutic approach to experimental colitis. Gut 2007;56:855–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kinchen J, Chen HH, Parikh K, et al. Structural Remodeling of the Human Colonic Mesenchyme in Inflammatory Bowel Disease. Cell 2018;175:372–386 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Tsukui T, Sun KH, Wetter JB, et al. Collagen-producing lung cell atlas identifies multiple subsets with distinct localization and relevance to fibrosis. Nat Commun 2020;11:1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Petrey AC, de la Motte CA. The extracellular matrix in IBD: a dynamic mediator of inflammation. Curr Opin Gastroenterol 2017;33:234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Henderson NC, Rieder F, Wynn TA. Fibrosis: from mechanisms to medicines. Nature 2020;587:555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Stidham RW, Xu J, Johnson LA, et al. Ultrasound elasticity imaging for detecting intestinal fibrosis and inflammation in rats and humans with Crohn’s disease. Gastroenterology 2011;141:819–826 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Johnson LA, Rodansky ES, Sauder KL, et al. Matrix stiffness corresponding to strictured bowel induces a fibrogenic response in human colonic fibroblasts. Inflamm Bowel Dis 2013;19:891–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hinz B. Tissue stiffness, latent TGF-beta1 activation, and mechanical signal transduction: implications for the pathogenesis and treatment of fibrosis. Curr Rheumatol Rep 2009;11:120–6. [DOI] [PubMed] [Google Scholar]
- 170.Johnson LA, Rodansky ES, Haak AJ, et al. Novel Rho/MRTF/SRF inhibitors block matrix-stiffness and TGF-beta-induced fibrogenesis in human colonic myofibroblasts. Inflamm Bowel Dis 2014;20:154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Li J, Mao R, Kurada S, et al. Pathogenesis of fibrostenosing Crohn’s disease. Transl Res 2019;209:39–54. [DOI] [PubMed] [Google Scholar]
- 172.Kessler SP, Obery DR, de la Motte C. Hyaluronan Synthase 3 Null Mice Exhibit Decreased Intestinal Inflammation and Tissue Damage in the DSS-Induced Colitis Model. Int J Cell Biol 2015;2015:745237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Naba A, Clauser KR, Ding H, et al. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol 2016;49:10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Gao Q, Meijer MJ, Kubben FJ, et al. Expression of matrix metalloproteinases-2 and - 9 in intestinal tissue of patients with inflammatory bowel diseases. Dig Liver Dis 2005;37:584–92. [DOI] [PubMed] [Google Scholar]
- 175.Meijer MJ, Mieremet-Ooms MA, van der Zon AM, et al. Increased mucosal matrix metalloproteinase-1, −2, −3 and −9 activity in patients with inflammatory bowel disease and the relation with Crohn’s disease phenotype. Dig Liver Dis 2007;39:733–9. [DOI] [PubMed] [Google Scholar]
- 176.Shimshoni E, Adir I, Afik R, et al. Distinct extracellular-matrix remodeling events precede symptoms of inflammation. Matrix Biol 2021;96:47–68. [DOI] [PubMed] [Google Scholar]
- 177.Barry R, Ruano-Gallego D, Radhakrishnan ST, et al. Faecal neutrophil elastase-antiprotease balance reflects colitis severity. Mucosal Immunol 2020;13:322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Monteleone I, Federici M, Sarra M, et al. Tissue inhibitor of metalloproteinase-3 regulates inflammation in human and mouse intestine. Gastroenterology 2012;143:1277–1287 e4. [DOI] [PubMed] [Google Scholar]
- 179.Biancheri P, Brezski RJ, Di Sabatino A, et al. Proteolytic cleavage and loss of function of biologic agents that neutralize tumor necrosis factor in the mucosa of patients with inflammatory bowel disease. Gastroenterology 2015;149:1564–1574 e3. [DOI] [PubMed] [Google Scholar]
- 180.Farmer RG, Easley KA, Rankin GB. Clinical patterns, natural history, and progression of ulcerative colitis. A long-term follow-up of 1116 patients. Dig Dis Sci 1993;38:1137–46. [DOI] [PubMed] [Google Scholar]
- 181.Filippi J, Allen PB, Hebuterne X, et al. Does anti-TNF therapy reduce the requirement for surgery in ulcerative colitis? A systematic review. Curr Drug Targets 2011;12:1440–7. [DOI] [PubMed] [Google Scholar]
- 182.Gomollon F, Dignass A, Annese V 3rd, et al. European Evidence-based Consensus on the Diagnosis and Management of Crohn’s Disease 2016: Part 1: Diagnosis and Medical Management. J Crohns Colitis 2017;11:3–25. [DOI] [PubMed] [Google Scholar]
- 183.Ye BD, McGovern DP. Genetic variation in IBD: progress, clues to pathogenesis and possible clinical utility. Expert Rev Clin Immunol 2016;12:1091–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet 2008;40:955–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lee JC, Biasci D, Roberts R, et al. Genome-wide association study identifies distinct genetic contributions to prognosis and susceptibility in Crohn’s disease. Nat Genet 2017;49:262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Rieder F, Kugathasan S. Circulating antibodies against bacterial wall products: are there arguments for early immunosuppression? Dig Dis 2012;30 Suppl 3:55–66. [DOI] [PubMed] [Google Scholar]
- 187.Siegel CA, Horton H, Siegel LS, et al. A validated web-based tool to display individualised Crohn’s disease predicted outcomes based on clinical, serologic and genetic variables. Aliment Pharmacol Ther 2016;43:262–71. [DOI] [PubMed] [Google Scholar]
- 188.Haberman Y, Minar P, Karns R, et al. Mucosal Inflammatory and Wound Healing Gene Programs Reveal Targets for Stricturing Behavior in Pediatric Crohn’s Disease. J Crohns Colitis 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Stidham RW, Wu J, Shi J, et al. Serum Glycoproteome Profiles for Distinguishing Intestinal Fibrosis from Inflammation in Crohn’s Disease. PLoS One 2017;12:e0170506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ballengee CR, Stidham RW, Liu C, et al. Association Between Plasma Level of Collagen Type III Alpha 1 Chain and Development of Strictures in Pediatric Patients With Crohn’s Disease. Clin Gastroenterol Hepatol 2019;17:1799–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Buescher JM, Driggers EM. Integration of omics: more than the sum of its parts. Cancer Metab 2016;4:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fiocchi C, Iliopoulos D. IBD Systems Biology Is Here to Stay. Inflamm Bowel Dis 2021;27:760–770. [DOI] [PubMed] [Google Scholar]
- 193.Phipps AI, Limburg PJ, Baron JA, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015;148:77–87 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sharma A, Menche J, Huang CC, et al. A disease module in the interactome explains disease heterogeneity, drug response and captures novel pathways and genes in asthma. Hum Mol Genet 2015;24:3005–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Leopold JA, Maron BA, Loscalzo J. The application of big data to cardiovascular disease: paths to precision medicine. J Clin Invest 2020;130:29–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Fiocchi C, Dragoni G, Iliopoulos D, et al. Results of the Seventh Scientific Workshop of ECCO: Precision medicine in IBD - what, why, and how. J Crohns Colitis 2021. [DOI] [PubMed] [Google Scholar]
- 197.Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Huang S, Chaudhary K, Garmire LX. More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front Genet 2017;8:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Dubinsky MC, Collins R, Abreu MT, et al. Challenges and Opportunities in IBD Clinical Trial Design. Gastroenterology 2021. [DOI] [PubMed] [Google Scholar]
- 200.Dulai PS, Jairath V, Khanna R, et al. Development of the symptoms and impacts questionnaire for Crohn’s disease and ulcerative colitis. Aliment Pharmacol Ther 2020;51:1047–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]