Abstract
Background:
Blood-based biomarkers for gastric cancer risk stratification could facilitate targeting screening to people who will benefit from it most. The ABC Method, which stratifies individuals by their Helicobacter pylori (H. pylori) infection and serum-diagnosed chronic atrophic gastritis status, is currently used in Japan for this purpose. Most gastric cancers are caused by chronic H. pylori infection, but few studies have explored the capability of antibody response to H. pylori proteins to predict gastric cancer risk in addition to established predictors.
Methods:
We used the least absolute shrinkage and selection operator (Lasso) to build a predictive model of non-cardia gastric adenocarcinoma risk from serum data on pepsinogen and antibody response to 13 H. pylori antigens as well as demographic and lifestyle factors from a large international study in East Asia.
Results:
Our best model had a significantly (p < 0.001) higher area under the receiver operator curve of 73.79% (95% CI: 70.86%, 76.73%) than the ABC Method (68.75%; 95% CI: 65.91%, 71.58%). At 75% specificity, the new model had greater sensitivity than the ABC Method (58.67% vs. 52.68%) as well as NPV (68.24% vs. 66.29%).
Conclusion:
Along with serologically defined chronic atrophic gastritis, antibody response to the H. pylori proteins HP 0305, HP 1564 and UreA can improve the prediction of gastric cancer risk.
Impact:
The new risk stratification model could help target more invasive gastric screening resources to individuals at high risk.
Keywords: Epidemiology, Artificial intelligence & machine learning, stomach cancer, serum biomarkers, cancer risk biomarkers, early detection biomarkers, early detection, cancer risk assessment
Introduction
Gastric cancer is the fifth most common cancer in the world and the third leading cause of cancer death, with about one million incident cases and 750,000 deaths annually.1,2 Its incidence is particularly high in East Asia, especially in China, Japan, and Korea.1,2 More than 90% of diagnosed stomach cancers in East Asia are adenocarcinomas occurring in the gastric glands.3 Five-year survival from gastric cancer is low, but it can often be successfully treated if diagnosed early.4,5 To more accurately and non-invasively identify who would benefit from gastric cancer screening, Miki et al. created the ABC Method for risk stratification in Japan.6,7 The ABC Method combines two serum assays: one for infection with Helicobacter pylori (H. pylori), and another for pepsinogen-defined chronic atrophic gastritis (CAG). Helicobacter pylori (H. pylori) is a bacterium that lives in the gastric mucosa and has been implicated in gastric cancer carcinogenesis.8,9 Globally, H. pylori is estimated to infect 50% of the world’s population, and it is especially common in East Asia, particularly among older individuals.10,11 Chronic atrophic gastritis, which usually results from prolonged H. pylori infection, has been associated with gastric cancer incidence.8,12 CAG progression has also been associated with a stepwise reduction in serum pepsinogen levels.13 Pepsinogens are pepsin pro-enzymes that can be measured in the blood to detect changes to the gastric mucosa, including atrophic gastritis.14,15 The ABC Method defines chronic atrophic gastritis as an absolute serum pepsinogen I concentration ≤ 70 μg/L and a pepsinogen I : pepsinogen II ratio ≤ 3.
The ABC Method is limited, however, in that it does not account for differences in risk conferred by seroprevalence to different H. pylori proteins. For example, individuals seropositive for H. pylori who test positive for antibodies to the cytotoxin associated gene A (CagA) antigen in their serum generally have a higher risk of gastric cancer than individuals seropositive for H. pylori but not for CagA.16,17 Possibly for this reason, the ABC Method’s gastric cancer risk predictive capability is relatively poor – a 2019 study performed in a Chinese population at high risk of gastric cancer found that the ABC Method had an area under the curve of just 52.70% (95% CI: 47.60%, 57.90).18 Similarly, it does not account for the underlying continuous association between serum pepsinogen levels and gastric cancer risk, or for variety in pepsinogen thresholds for CAG dependent on H. pylori seropositive status.19,20
In this study, we used supervised machine learning techniques to build a predictive model of gastric cancer risk that incorporated antibody response to H. pylori-specific proteins and continuous coding of serum pepsinogen levels. We used data from a large East Asian nested case-control study of gastric cancer that collected individual-level data on demographic and lifestyle risk factors, serum pepsinogen, and host immune response to H. pylori-specific proteins.21 The model’s discrimination capability was assessed using receiver operator characteristic curves with special attention to sensitivity rather than specificity. Sensitivity was deemed more important because, at an early and minimally invasive stage of screening, it is more important to identify as many treatable precancers or early cancers as possible. Finally, we internally validated the model using leave-one-out cross-validation.
Materials and Methods
Study Population
This study uses data from three cohorts within the Helicobacter pylori Biomarker Cohort Consortium (HpBCC). The total HpBCC is a nested case-control study recruited from eight cohorts across Japan, China and the Republic of Korea. It was created to find potential biomarkers for H. pylori-related health outcomes including gastric cancer.22,23 The cohorts collected demographic and lifestyle information as well as blood samples from healthy individuals at baseline.23 The primary outcome of interest was non-cardia gastric cancer, defined by International Classification of Diseases for Oncology (including C16.1–C16.6, C16.8, C16.9). Three of these cohorts — the Japan Public Health Center Study (JPHC) I and JPHC II in Japan and the Linxian Nutrition Intervention Trial (NIT) in China — assessed pepsinogen from participants’ sera.24,25 Therefore, we used data from these three cohorts exclusively to build our predictive model.
The JPHC I and II used incidence density sampling to select controls. At the time of diagnosis of each index gastric cancer case, a control was selected randomly from among all currently living members of the cohort from which the case arose who had no history of gastric cancer or gastrectomy. JPHC I and II controls were individually matched to cases based on gender, birth date and blood collection date. NIT controls were frequency matched to cases based on gender and age.
The present study was approved by the institutional review boards of the University of North Carolina at Chapel Hill (Chapel Hill, NC, USA) and Duke University (Durham, NC, USA).
H. pylori multiplex serology
Pepsinogen and host response to H. pylori proteins were assessed from 10 mL serum or plasma samples collected at the time of baseline questionnaire administration. Samples were collected during 1985 in the NIT, 1990–1992 for the JPHC I and 1993–1995 in the JPHC II. The median follow-up time between blood collection and cancer diagnosis was 7.0 years for the JPHC I, 4.2 years for the JPHC II, and 7.0 years for the NIT.23 In the NIT, samples were frozen and shipped from Linxian to Beijing where they were stored at −80°C.26 In the JPHC I and II, blood samples were centrifuged to obtain plasma and buffy coat layer within 12 hours of blood draw and stored at −80°C.27 Blood samples were assayed for the HpBCC in 2016. Antibody levels to 13 H. pylori proteins were assessed using multiplex serology. Samples were assayed at the German Cancer Research Center (DKFZ, Heidelberg). H. pylori multiplex serology is based on a glutathione S-transferase capture immunosorbent assay combined with fluorescent bead technology (Luminex) that detects human antibodies to 13 H. pylori recombinantly expressed fusion proteins (UreA, Catalase, GroEL, NapA, CagA, HP0231, VacA, HpaA, Cad, HyuA, HP1564, HcpC and HP0305).28,29 Antigen-specific median reporter fluorescence intensity (MFI) cutoff points were calculated using 17 H. pylori-negative sera that had previously been classified for H. pylori status. H. pylori seropositivity was defined as reactivity with at least four proteins, which has shown good agreement (kappa = 0.70) with commercial serological assay. This assay has 89% sensitivity and 82% specificity.28 A total of 24 quality control samples were included to test the assay’s reliability. The determination of seropositivity for all H. pylori proteins detected was highly consistent, with identical results for 98% (353/360) of the tests.23
Pepsinogen Assays
In the NIT, Serum pepsinogen I and pepsinogen II were measured using enzyme-linked immunosorbent assays (Biohit ELISA kit, Finland) which were performed by technicians who were blinded to the subjects’ case/control status.19 Two serum samples were taken per individual, and the average of these two was determined to be the final pepsinogen level.19 Among five participants, large differences were found between the two samples. For these, a third assay was conducted and the average of the two closer results was recorded as the final pepsinogen level. Correlation between duplicate samples was very high: the Pearson’s correlation coefficient was 0.995 for pepsinogen I and 0.997 for pepsinogen II.19 Each assayed plate also included two quality control (QC) samples.19 Using all QC samples together, the coefficients of variation for pepsinogen I and pepsinogen II assays were 6.5% and 2.7%, respectively.19 An additional 103 QC samples, aliquoted from a single large serum pool from the National Cancer Hospital in Beijing, were distributed among all assay plates. Considering all of these QC samples together, the coefficients of variation for pepsinogen I and pepsinogen II were 5.5% and 6.7%, respectively.19
In the JPHC I and II, Pepsinogen I and pepsinogen II levels were assessed from serum in a two-step enzyme immunoassay using commercial kits (E Plate “Eiken” Pepsinogen I, Eiken Kagaku) and (E Plate “Eiken” Pepsinogen II, Eiken Kagaku).27
Statistical Analysis
Person-time was calculated as the difference between the last date of follow-up and the start date of each cohort (March 1st 1985 for NIT, January 1st 1990 for JPHC I, and January 1st 1993 for JPHC II).30,31 For cases, the last date of follow-up was their date of diagnosis with gastric non-cardia adenocarcinoma. For JPHC I and II controls, the last follow-up date was their matched case’s date of diagnosis. NIT controls, who were not individually matched to cases, had for their last date of follow-up the maximum of the date of administrative censoring, date of death, or date of loss to follow-up. Dates of diagnosis for cases were accurate to the day, but many controls had only the year of loss to follow-up recorded. These controls’ last dates were therefore recorded as January 1st of their last year of follow-up. The Breslow method was used to manage ties.32 The survival package in R was used to estimate hazard ratios and 95% confidence intervals.33
We used the least absolute shrinkage and selection operator (Lasso) to build predictive models.34 Lasso is a parameter regularization technique that selects features by fitting a risk model under the constraint that the sum of the absolute values of the regression coefficients cannot exceed a pre-determined threshold: the tuning parameter, “λ.”34–36 When applied to the set of all covariates, this threshold excludes covariates that contribute least to predicting the outcome. The remaining covariates are thus selected as predictors of gastric cancer. In total, 37 variables were evaluated as potential features of the gastric cancer classification model. Continuous variables including age, pack-years of cigarette smoking, serum pepsinogen I, serum pepsinogen II, the ratio of serum pepsinogen I/II, and the 13 antibody response to H. pylori protein variables were evaluated by Lasso in straight line, monotonic functional form. Since the H. pylori MFI values are prone to statistical noise at the lower level, these continuous variables were recoded as 0 if they were below that antibody response variable’s predefined threshold for seroprevalence. Binary coded forms for the 13 H. pylori protein variables were also assessed as potential predictors. Gender, smoking status (current vs. former or never), H. pylori seropositivity, and family history of gastric cancer were coded as binary variables. Seropositivity to multiple H. pylori proteins (range: 0 to 13) was assessed as a 14-level categorical variable. A binary variable for serologically defined atrophic gastritis was also included (gastritis present = pepsinogen I ≤ 70 and pepsinogen I/II ratio ≤ 3; not present otherwise).
Ten-fold cross-validation was used to determine the tuning parameter: the HpBCC data set was split into ten subsets, then the classification model was fit to nine subsets (together comprising the training set) and the model’s classification capability was assessed by Harrell’s concordance index (c-index) in the omitted subset (the test set).37–39 This procedure was repeated ten times, omitting a different subset each time. Splitting the data into training and test sets safeguards Lasso from overfitting a model to the training data.36 We chose the λ value that gave the model with the fewest predictors whose c-index was within one standard error of that of the model with the largest c-index.40–43 We deemed this model more appropriate than the model with the largest c-index because (a) smaller models are less likely to be overfit to the training data and (b) a smaller model might shed more light on which risk factors are most strongly associated with gastric cancer.
Weighting
Incidence density sampling was used to select controls into the HpBCC from the underlying cohorts, except the NIT which was subsampled from a sex- and age-stratified case-cohort study.23,44 One control was sampled whenever a gastric cancer case was diagnosed. This means that controls can be selected into the nested case-control study more than once, provided that they are still at risk of the disease.45 From the hazard ratio, we can estimate the baseline hazard function and then the risk of gastric cancer in the cohort.45 However, this estimation is complicated by matching; controls were matched to cases by cohort, gender, and age. This will bias the estimate of the baseline hazard (and thus the risk), because certain individuals were more likely to be sampled as controls than others based on their matching characteristics. To account for this potential selection bias, we implemented time-varying inverse probability of sampling weights using the method from Salim et al.45 More detail on weight calculation is included in the Supplementary Materials and Methods.
Model Performance Analysis
We assessed the classifier’s ability to discriminate between gastric cancer cases and controls by calculating the area (AUC) under the receiver operator characteristic (ROC) curve at 10 years of follow-up within the training data set.41 In addition, we report sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). PPV and NPV were calculated using the prevalence of gastric cancer in the underlying cohorts and within relevant strata for stratified analyses. The timeROC package was used for constructing time-dependent ROC curves and their respective AUCs, sensitivity, and specificity.46
Assessing discrimination within the training data usually gives overly optimistic estimates of its ability to predict the case or non-case status of observations it has not seen before. Therefore, we internally validated the model using leave-one-out cross-validation (LOOCV).39,47 A detailed description of the strengths and weaknesses of LOOCV is included in the Supplementary Materials and Methods.
We also compared the Lasso model’s performance to that of the ABC Method, an established risk stratification model for gastric cancer currently in use in Japan and the Republic of Korea.7,48 The ABC Method classifies individuals via two binary variables: H. pylori infection status and serologically defined chronic atrophic gastritis. Its four levels, in ascending order of gastric cancer risk, are: A (H. pylori−, CAG−), B (H. pylori+, CAG−), C (H. pylori+, CAG+) and D (H. pylori−, CAG+). Owing to small numbers in the D category, and following previous research that found little difference in risk of gastric cancer between the two highest groups, we decided to code the ABC Method variable with only three categories (A, B, and C + D).49,50
All analyses were conducted in R statistical software, version 4.0.2.51
Data Availability
The data analyzed in this study are available from Duke University. Restrictions apply to the availability of these data, which were used under specific agreement for this study. Data are available from the authors upon reasonable request with the permission of both Duke University and the respective institutions that collected the specimens (the National Cancer Center Research Institute, Japan, the Cancer Institute, Chinese Academy of Medical Sciences, China, and the National Cancer Institute, USA).
Results
Descriptive Statistics
There were 708 cases and 714 controls in the analysis set (Table 1). Cases tended to be slightly older than controls (mean age: 57 vs. 55). About 45% of participants were recruited from the NIT, and 55% from the JPHC I and II. Cases were more likely than controls to test positive for H. pylori seropositivity (92% vs. 79%). The median serum pepsinogen I level was lower among cases than among controls (42 μg/L vs. 46 μg/L), as was the median ratio of serum pepsinogen I/II (2.9 vs. 4.5). Serologically diagnosed chronic atrophic gastritis was more common among cases than among controls (49% vs. 33%). The prevalence of current smoking and family history of gastric cancer was similar in both groups. The gender distribution was the same for cases and controls (about 66% men, 33% women) due to matching controls to cases by gender. As noted above, the age distribution, however, differed somewhat between groups. This is probably because controls from the NIT cohort were only matched to cases by 10-year age strata (i.e. 40–50 years, 50–60 years, and 60–70 years).
Table 1:
Descriptive Statistics of the Analysis Set (n = 1422)
| Variable | Case | Control | |
|---|---|---|---|
| n | 708 | 714 | |
| Gender (%) | Women | 241 (34.0) | 240 (33.6) |
| Men | 467 (66.0) | 474 (66.4) | |
| Age Category, years (%) | ≤40 | 16 (2.3) | 53 (7.4) |
| 41–45 | 48 (6.8) | 78 (10.9) | |
| 46–50 | 87 (12.3) | 89 (12.5) | |
| 51–55 | 142 (20.1) | 135 (18.9) | |
| 56–60 | 181 (25.6) | 156 (21.8) | |
| >60 | 234 (33.1) | 203 (28.4) | |
| Study (%) | JPHC I | 205 (29.0) | 201 (28.2) |
| JPHC II | 192 (27.1) | 191 (26.8) | |
| NIT | 311 (43.9) | 322 (45.1) | |
| H. pylori Statusa (%) | H. pylori Positive | 651 (91.9) | 561 (78.6) |
| Serum Pepsinogen I (μg/L) Median (IQR) | 41.80 (21.78, 109.59) | 46.35 (24.83, 118.21) | |
| Median Pepsinogen I/II Ratio Median (IQR) | 2.93 (1.77, 6.04) | 4.49 (2.44, 8.42) | |
| Chronic Atrophic Gastritisb (%) | Chronic Atrophic Gastritis | 350 (49.4) | 235 (32.9) |
| Smoker (%) | Current Smoker | 77 (17.9) | 86 (19.2) |
| Family History (%) | Family History of Gastric Cancer | 58 (8.2) | 48 (6.7) |
Defined as seropositive to ≥ 4 H. pylori antigens from 13-plex.
Defined as pepsinogen I ≤ 70 and pepsinogen I/II ratio ≤ 3
We calculated crude hazard ratios for each potential predictor to describe their association with gastric cancer (Supplementary Table 1). Individuals seropositive for H. pylori were more likely to have gastric cancer than individuals without (HR = 3.34; 95% CI: 2.37, 4.71). All antibody response variables were associated with an increased hazard of gastric cancer. Stronger associations were observed for HP 1564 (HR = 3.02; 95% CI: 2.23, 4.07), CagA (HR = 2.96; 95% CI: 2.20, 3.97), and VacA (2.79; 95% CI: 2.10, 3.72) (Supplementary Table 1). Regarding the ABC Method, members of Groups B and C+D had a much higher hazard of gastric cancer than members of Group A (Supplementary Table 1). Median MFI values for host response to the 13 considered proteins are reported in Supplementary Table 2. Additionally, visualizations of univariate associations between log odds of gastric cancer and each continuous variable considered for inclusion in the Lasso model are reported in the Supplementary Materials and Methods.
Results from Lasso Model Building
Out of 37 potential features, Lasso selected six for classifying gastric cancer cases and controls in the new Lasso model at the pre-specified λ.1se threshold (Supplementary Table 1). Two features were demographic: gender (binary); age (linear; centered at 57 years). Three were host response to H. pylori biomarkers: UreA (linear); HP 0305 (binary); HP 1564 (binary). The remaining one was a pepsinogen variable: CAG (binary). The equation for the Lasso risk prediction model is included in the supplement (Supplementary Table 2).
Table 2 gives the Lasso-developed model’s coefficient values at ten years of follow-up. The strongest associations with gastric cancer were found with serologically defined CAG (HR=3.11), HP 1564 status (HR=1.85) and HP 0305 (HR=1.61).
Table 2:
Summary of the parameter estimates of the Lasso model in the whole data set (n = 1422)
| Parameter | Coefficient (SE) | Hazard Ratio (95% CI) |
|---|---|---|
| Gender (men; binary) | 0.428 (0.081) | 1.53 (1.18, 1.99) |
| Agea | 0.039 (0.005) | 1.04 (1.03, 1.05) |
| UreA (linear) | 0.00005 (0.00002) | 1.00 (1.00, 1.00) |
| HP 0305 (binary) | 0.473 (0.090) | 1.61 (1.21, 2.12) |
| HP 1564 (binary) | 0.616 (0.126) | 1.85 (1.28, 2.68) |
| CAGb (binary) | 1.31 (0.084) | 3.11 (2.35, 4.12) |
Centered at 57 years (median age in the data set)
Defined as pepsinogen I ≤ 70 μg/L and pepsinogen I/II ratio ≤ 3
We compared the Lasso model and ABC Method’s ability to discriminate between gastric cancer cases and controls within the whole data set by plotting ROC curves and reporting the area under the curve, sensitivity, specificity, positive predictive value and negative predictive value at ten years of follow-up (Table 3). The threshold for calculating sensitivity and specificity was set at a risk score > 1.0 which was the median risk score generated by the Lasso model (the median ABC Method risk score was 0.8). An individual’s risk score is the exponentiated sum of the β coefficients multiplied by their exposure status for each feature included in the predictive model. The ABC Method had an AUC of 68.75% (95% CI: 65.91, 71.58), a sensitivity of 52.68% (95% CI: 48.49%, 56.88%) and a specificity of 78.86% (95% CI: 75.37%, 82.35%). Compared to the ABC Method, the Lasso model had a higher AUC (73.79%; 95% CI: 70.86%, 76.73%; p < 0.001) and sensitivity (73.56%; 95% CI: 69.86%, 77.27%) at a lower specificity (60.76%; 95% CI: 56.59%, 64.94%). The ABC Method’s PPV (67.86%; 95% CI: 63.12%, 72.60%) was higher than that of the Lasso model (61.37%; 95% CI: 57.45%, 65.29%), although the 95% confidence intervals overlapped. The NPV of the Lasso model (73.06%; 95% CI: 69.22%, 76.90%) was higher than that of the ABC Method (66.29%; 95% CI: 62.90%, 69.68%), but with slight overlap of the 95% confidence intervals. We also compared the ABC Method and Lasso model at an equal specificity of 75% (Supplementary Table 3). At this level, the Lasso model had a greater sensitivity (58.67% vs. 52.68%) and NPV (68.24% vs. 66.29%) than the ABC level, and a similar but slightly smaller PPV (66.75% vs. 67.85%).
Table 3:
Lasso model and ABC Method Discrimination Capability (n = 1422, t = 10 years)
| Classification within the whole data set | ||||||
|---|---|---|---|---|---|---|
| Model Name | AUCa (%) | p d | Sensitivitye (%) | Specificity (%) | PPV (%) | NPV (%) |
| ABC Method b | 68.75 (65.91, 71.58) | < 0.001 | 52.68 (48.49, 56.88) | 78.86 (75.37, 82.35) | 67.86 (63.12, 72.60) | 66.29 (62.90, 69.68) |
| Lasso model c | 73.79 (70.86, 76.73) | 73.56 (69.86, 77.27) | 60.76 (56.59, 64.94) | 61.37 (57.45, 65.29) | 73.06 (69.22, 76.90) | |
| Prediction from Leave-One-Out Cross-Validation | ||||||
| ABC Method b | 56.60 (52.90, 60.29) | N/A | 52.68 (48.49, 56.88) | 78.86 (75.37, 82.35) | 67.86 (63.12, 72.60) | 66.29 (62.90, 69.68) |
| Lasso model c | 73.37 (70.42, 76.32) | 73.56 (69.86, 77.27) | 60.38 (56.19, 64.58) | 61.14 (57.22, 65.06) | 72.94 (69.10, 76.78) | |
Area under the receiver-operator characteristic curve. Time-dependent results reported at 10 years of follow-up.
Three levels: A (H. pylori−, Chronic atrophic gastritis (CAG)−), B (H. pylori+, CAG−, C (H. pylori+, CAG+; or H. pylori−, CAG+)
Six predictors: gender (binary), age (continuous; centered at 57 years), UreA (continuous), HP 0305 (binary), HP 1564 (binary), serologically defined CAG (binary)
Testing the null hypothesis that |AUCLasso − AUCABC| = 0 at ten years of follow-up. No p value for prediction AUCs because they result from different data sets due to leave-one-out cross-validation
Threshold at gastric cancer risk score > 1
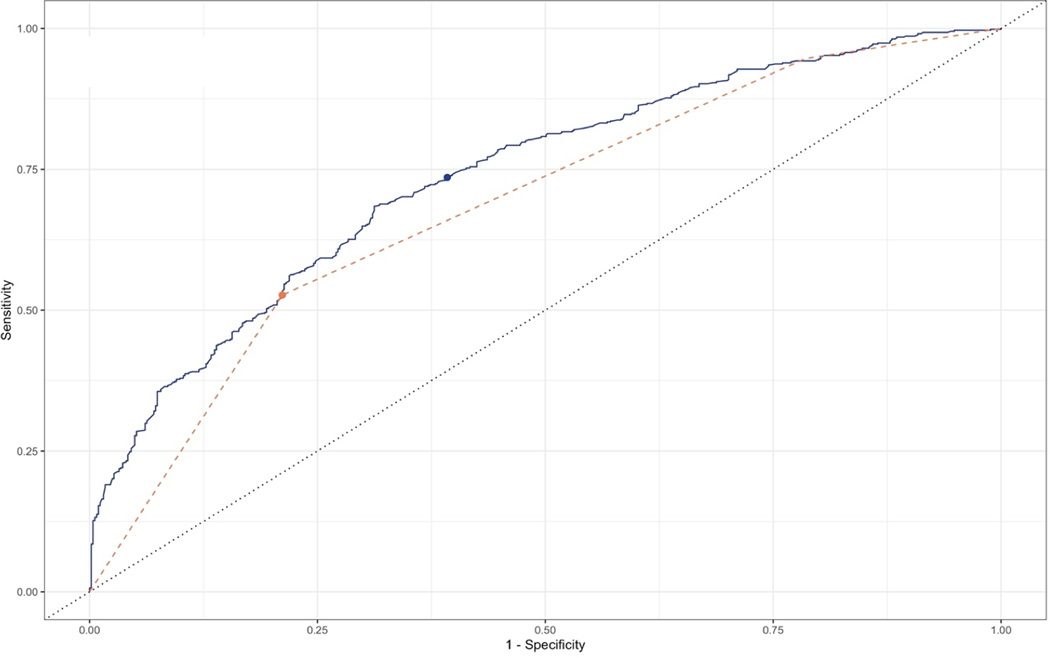
Figure 1 shows the two classification ROC curves for the Lasso and ABC Method models respectively, with points indicating the threshold of risk score > 1.0. The ABC Method ROC curve has a trapezoidal shape because it can only assign one of three predicted probabilities to each individual: one for those in Group A, one for those in Group B, and one for those in Group C+D. The same discrimination patterns were observed when comparing the two models at five years of follow-up (Supplementary Table 4). For further comparison, we assessed the discrimination capability of the ABC Method plus the variables selected into the Lasso model (Supplementary Table 3, Supplementary Table 4). This combined model performed almost identically to the Lasso model; the DeLong’s test of difference in AUC between these two models was not significant at 5 years of follow-up (p = 0.46) or at 10 years of follow-up (p = 0.37). Finally, the result of a likelihood ratio test of the Lasso model compared to the Lasso model plus the ABC Method variable was not statistically significant (p = 0.85).
Figure 1:
Receiver-operating characteristic curve for classification of gastric cancer in a subset of the Helicobacter pylori Biomarker Cohort Consortium (n=1422).
This figure compares the gastric cancer discrimination performance of the Lasso model (solid) to that of the ABC Method (dashed) within the training data set. Dots represent the threshold of gastric cancer risk score > 1 at which sensitivity and specificity were calculated for both models.
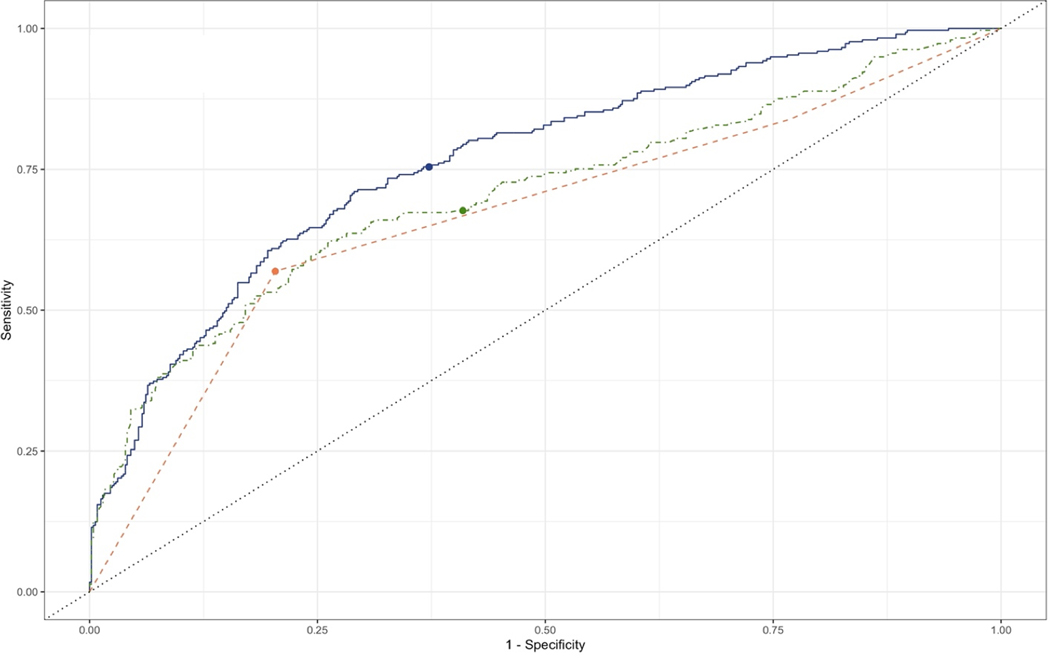
To assess how well the Lasso model may classify data it has not seen before, we used leave-one-out cross-validation (LOOCV). Under LOOCV, the ABC Method had an AUC of 56.60;( 95% CI: 52.90%, 60.29%), a sensitivity of 52.68% (95% CI: 48.49%, 56.88%) and a specificity of 78.86% (95% CI: 75.37%, 82.35%) (Table 3). The Lasso model’s AUC under LOOCV was 73.37% (95% CI: 70.42%, 76.32%), its sensitivity was 73.56% (95% CI: 69.86%, 77.27%) and its specificity was 60.38% (95% CI: 56.19%, 64.58%). The PPV and NPV for both models were very similar to their values in the training data. Figure 2 shows the two prediction ROC curves for the Lasso and ABC Method models respectively, with points indicating the risk score > 1.0 threshold. The ABC Method ROC curve in Supplementary Figure 2 has a peculiar shape due to LOOCV: the training data changed slightly every time a different individual was left out as the test set. Thus, individuals within the same level of the ABC Method received slightly different (usually in the fifth decimal place) predicted probabilities of gastric cancer. In reality, there are only three possible gastric cancer predicted probability values for the ABC Method, as in Figure 1.
Figure 2:
Sensitivity analysis. Receiver-operating characteristic curve for classification of gastric cancer in a subset of the Helicobacter pylori Biomarker Cohort Consortium (n=1422).
This figure compares the gastric cancer discrimination performance of the Lasso model including pepsinogen I (solid) to that of the ABC Method (dashed) and the Lasso model without pepsinogen I (dot-dashed) within the training data set. Points represent the threshold of gastric cancer risk score > 1 at which sensitivity and specificity were calculated for both models
We also evaluated the classification ability of the Lasso model determined from the whole HpBCC data set stratified by gender (men vs. women) and study site (JPHC I, JPHC II vs. NIT). The Lasso model had a significantly higher AUC than the ABC Method within all strata except the JPHC I under classification within the training data (Supplementary Table 5). Under LOOCV, the Lasso model also had a higher AUC than the ABC Method within all strata (Supplementary Table 6). The ABC Method had very high sensitivity within the NIT data (88.34%; 95% CI: 84.13%, 92.55%) compared to the Lasso model (81.17%; 95% CI: 76.03%, 86.30%). Within the JPHC I data, the Lasso model had a higher sensitivity (83.93%; 95% CI: 77.81%, 90.04%) than the ABC Method (76.23%; 95% CI: 69.06%, 83.41%). The Lasso model’s AUC was more than twice that of the ABC Method in the NIT and JPHC I strata, and 13% greater in the JPHC II stratum (Supplementary Table 5). In the JPHC II stratum, the ABC Method had a near-perfect sensitivity at 12.50% specificity, whereas that of the Lasso model was 86.85% (95% CI: 81.42%, 92.28%) at 25.00% specificity. However, the JPHC II statistics were imprecise due to the small sample size and the fact that all controls had been censored before 10 years of follow-up had been reached. Therefore, we reported discrimination statistics at 9 years of follow-up for the JPHC II. The ABC Method had a higher PPV than the Lasso model within gender strata but the relationship was different among cohort strata: in the NIT, the Lasso model had a higher PPV than the ABC Method, and the PPV estimates within the JPHC I and II were very similar for both models (Supplementary Table 5). The Lasso model’s NPV was higher than that of the ABC Method for gender strata and within the JPHC I. However, within the JPHC II, the ABC Method had a considerably higher NPV than the Lasso model (although the 95% confidence intervals were very wide). The NPV for both models was very similar within the NIT. When the specificity was held constant at 75%, the Lasso model outperformed the ABC Method in sensitivity and NPV within all gender and study site strata (Supplementary Table 7). These associations persisted under LOOCV (Supplementary Table 8).
The Lasso model was also assessed within strata of CAG status. It exhibited a higher AUC among CAG+ individuals (71.29%; 95% CI: 66.03%, 76.54%) than CAG− individuals (65.80%; 95% CI: 61.62%, 69.98%) (Supplementary Table 9). The same association was maintained under leave-one-out cross-validation: the AUC among CAG+ individuals was 70.65% (95% CI: 65.36%, 75.93%) whereas it was 64.83% (95% CI: 60.62%, 69.04%) among CAG− individuals (Supplementary Table 10).
Sensitivity Analyses
The association between continuous MFI level for H. pylori antibody response variables, such as UreA in our Lasso model, has been less consistently established than that of binary variables such as HP 0305 and HP 1564.52 Therefore, as a sensitivity analysis, we removed UreA from the Lasso model and reassessed its discrimination statistics. The model without UreA had a classification AUC of 73.41% (95% CI: 70.46%, 76.37%), which was higher than the AUC for the model with UreA (Table 3). At 60.00% specificity, the model without UreA had a sensitivity of 74.86% (95% CI: 71.21, 78.50). Under LOOCV, the model without UreA maintained the same AUC of 73.41% (70.46%, 76.37%) as it achieved within the whole data set but had a slightly lower sensitivity of 74.69% (95% CI: 71.05%, 78.34%) and lower specificity of 59.43% (95% CI: 55.23%, 63.62%) at the risk score threshold of 1.0.
In this data set, there were 17 individuals who were seronegative for H. pylori (meaning they were seropositive to < 4 H. pylori proteins) but seropositive for chronic atrophic gastritis. Such individuals have been found to have an extremely high risk of stomach cancer.7 It is possible that these people had a previous H. pylori infection which caused such severe gastric atrophy that it rendered the stomach inhospitable for the bacteria.7 Thus, labelling these people as H. pylori seronegative may have been a misclassification error in that they were previously infected with H. pylori, which highlights the limit of serology in capturing exposure history. Therefore, we conducted a sensitivity analysis in which we excluded these 17 observations from the data set before applying Lasso to build the predictive model. Additionally, previous research has suggested that using CagA seropositivity in addition to H. pylori seropositivity may classify H. pylori infection status more accurately than H. pylori seropositivity alone.27 We explored this by redefining H. pylori seropositivity as: positive antibody response to CagA and at least three additional H. pylori proteins. Individuals were classified as seronegative for H. pylori infection if they were neither seropositive for CagA nor seropositive for at least four H. pylori proteins. Those who were serodiscordant for H. pylori and CagA, i.e. seropositive for CagA but not for at least four H. pylori proteins, or seropositive for ≥ 4 H. pylori proteins but not CagA, were treated as missing and excluded from this sensitivity analysis set. Applying these exclusion criteria left 1,325 observations in the sensitivity analysis set (93% of observations used in the main analysis set). In this reduced data set, Lasso selected pepsinogen I in addition to the six variables chosen in the main analysis (Table 3). The AUC of the model including pepsinogen I was 76.56% (95% CI: 73.14%, 79.98%). This was significantly greater than that of the model without pepsinogen I in this sensitivity analysis data set, whose AUC was 70.60% (95% CI: 66.67%, 74.52%; p < 0.001, Figure 2). It was also significantly greater than the AUC of the ABC Method, which was 66.84% (95% CI: 63.01%, 70.67%; p < 0.001, Figure 2). The sensitivity of the model with pepsinogen I was 75.42% (95% CI: 70.52%, 80.32%) and its specificity was 62.76% (58.46%, 67.05%). By contrast, the sensitivity of the model without pepsinogen I was 67.68% (95% CI: 62.37%, 72.99%) and its specificity was 59.05% (95% CI: 54.68%, 63.42%). The sensitivity of the ABC Method in this data set was 56.90% (95% CI: 51.28%, 62.53%) and its specificity was 79.63% (95% CI: 76.04%, 83.22%). Through LOOCV, the model with pepsinogen I had a predictive AUC of 76.56% (73.14%, 79.99%), a sensitivity of 75.76% (95% CI: 70.89%, 80.64%) and a specificity of 62.35% (95% CI: 58.03%, 66.66%).
Discussion
Using data from a large East Asian nested case-control study, we built a predictive model of ten-year gastric cancer risk that incorporated detailed serum H. pylori protein and pepsinogen data as well as demographic and lifestyle risk factors. The new model had a greater classification AUC than the currently used ABC Method. In addition, leave-one-out cross-validation gave evidence that the Lasso model retained its predictive capability in independent data. Its discrimination metrics remained relatively stable, whereas the ABC Method’s AUC was considerably lower than it was within the training data set. The ABC Method did have greater specificity than the Lasso model at the threshold of risk score > 1.0. However, the importance of identifying high-risk individuals for early detection of gastric cancer makes sensitivity a more important measure of predictive models’ value at this early stage of screening. In a similar vein, it is noteworthy that the Lasso model had a higher NPV than the ABC Method – meaning the new model resulted in fewer false negatives (i.e. people with gastric cancer who would be classified as disease free: an immensely dangerous mistake). The Lasso model also had a greater sensitivity and NPV than the ABC Method, as well as comparable PPV, when both models were examined at 75% specificity. Furthermore, within strata of study cohort – which are more accurate approximations of real-world populations than the international HpBCC nested case-control study – the Lasso model exhibited a higher AUC than the ABC Method in addition to sensitivity and NPV when both models were compared at 75% specificity.
Variables Included in the Lasso model
The Lasso-determined model included several H. pylori proteins that have been associated with greatly increased risk of gastric cancer in previous studies conducted in East Asia. HP 0305 and HP 1564 were both selected by Lasso as binary variables. These two proteins together have been associated with greater risk of gastric cancer in China.53 HP 0305 and HP 1564 may augment gastric cancer risk by increasing H. pylori-mediated inflammation through promoting key bacteria-gastric cell interactions that enable the delivery of oncogenic microbial cargo to vulnerable cells.54 In addition, HP 1564 has been found to translocate the oncoprotein CagA into gastric epithelial cells.54 UreA (urease alpha subunit), which is associated with H. pylori’s ability to neutralize gastric acid to facilitate life in the stomach, was also selected by Lasso.55 It is a curious choice, because UreA has not been found to have a strong association with gastric cancer in the past. It could be that UreA is strongly correlated with the presence of other H. pylori proteins that increase gastric cancer risk, such as HP 0305 and HP 1564. In a sensitivity analysis which excluded the continuous UreA variable from the Lasso model, the model without UreA exhibited a similar AUC to the model with UreA. Of note, our model did not include the CagA protein, which had the strongest univariate association with gastric cancer in our data set and has previously been associated with high risk of gastric cancer.17 This may be because CagA seropositivity is extremely common among individuals in East Asia living with H. pylori.8 In this data set, 87% of cases and 72% of controls were seropositive for CagA. VacA, another H. pylori protein that has been strongly associated with gastric cancer (including in this data set), was also not chosen by Lasso.23,28,56
Chronic atrophic gastritis was also selected as a predictor of gastric cancer in our model, likely because it represents a further step along the path from H. pylori infection to gastric adenocarcinoma.8,57 CAG in the absence of H. pylori infection has been found to have an even stronger positive association with gastric cancer than when infection is present, possibly because it implies that the atrophy has become so severe that it has rendered the stomach inhospitable for the bacterium.6,7
In a sensitivity analysis in which we excluded participants who were seronegative for H. pylori and seropositive for CAG or who were serodiscordant for H. pylori and CagA, Lasso selected pepsinogen I as a linear predictor in addition to the other six predictors. Including pepsinogen I substantially (and significantly) improved the predictive capability of the Lasso model when H. pylori seropositivity was defined this way. Pepsinogen I production only occurs in the acid-secreting glands of the gastric corpus; hence, as gastric atrophy spreads (usually from the pylorus, where H. pylori tends to colonize), the amount of pepsinogen I measurable in serum decreases.15,58,59 This is in line with our model finding an inverse association between serum pepsinogen I concentration and gastric cancer risk. However, absolute serum pepsinogen I has also been shown to increase due to mucosal inflammation caused by chronic H. pylori infection, which contradicts the association described previously.59,60 The fact that both pepsinogen I and CAG were selected by Lasso suggests that a lower pepsinogen I value within strata of serologically defined CAG may also be informative of gastric cancer risk.
Our Lasso-generated model components differed somewhat from a previous predictive model of ten-year gastric cancer risk designed using data from the JPHC II.49 Both models included age, gender and CAG. However, the JPHC II model included a binary variable for H. pylori infection status, and did not assess different H. pylori proteins. In addition, they included smoking status as a binary variable (current vs. former/never smokers). That model also included family history of gastric cancer, which was not selected by Lasso in our data set, and fish roe/fish gut consumption as a proxy measure for high salt intake. Our data set did not have a measure for salt consumption. The JPHC II model had a somewhat higher Harrell’s c-index (equivalent to AUC for a ROC curve) than our model: 76.8% vs. our model’s predictive AUC of 73.37% (95% CI: 70.42%, 76.32%).49 However, that model’s sensitivity (69.7%) was lower than ours (73.56% at 60.38% specificity). This may be because they used a different criterion to choose a threshold of sensitivity and specificity: 1.9% predicted probability of gastric cancer vs. our criterion of a risk score > 1.0. Comparison is also difficult because they did not report confidence intervals for their estimates. Upon external validation, the JPHC II model showed a higher c-index of 79.8% (95% CI: 72.5, 86.1) and a sensitivity of 74%.61 However, the precision of that model’s estimates is generally lower than ours, likely owing to the relatively small number of gastric cancer cases in their analysis sets (412 in the derivation sample and 33 in the validation sample, vs. 708 in our study sample).
Limitations and Strengths
We must acknowledge that measuring some of the variables included in the new Lasso model will be considerably more challenging than for the ABC Method. Currently, HP 0305, HP 1564 and UreA assays are only available in research laboratories. Technology transfer from such institutions to clinical laboratories will be necessary before any predictive model based on these H. pylori proteins could be implemented for widespread gastric cancer risk stratification. Commercial assays for pepsinogen are, however, readily available worldwide.62
There were two potential sources of measurement error in this study. First, the multiplex serological test for seropositivity to H. pylori, defined as reactivity to ≥ 4 H. pylori proteins, had 89% sensitivity and 82% specificity, meaning that about 11% of individuals recorded as seropositive for H. pylori may not have actually had an infection, and 18% of seronegative individuals may actually have had an infection.28 This may have affected which variables were chosen by Lasso for inclusion in the predictive model. However, the gold standard to which these discrimination measures were compared was an enzyme-linked immunosorbent assay (ELISA). Since ELISA is a technically different approach to multiplex serology and is itself imperfect at indicating current H. pylori infection, sensitivity and specificity less than 100% are not necessarily cause for alarm.63 Second, while all gastric cancer cases included in this analysis were non-cardia, we were not able to distinguish between histological subtypes of gastric adenocarcinoma in this analysis. Diffuse-type adenocarcinoma may have different H. pylori risk factors from intestinal-type. Seropositivity to CagA antibodies, for example, has been associated more strongly with diffuse-type than intestinal-type.64,65 Despite this, the majority of H. pylori proteins have been previously associated with both histological subtypes, which suggests that the findings of the Lasso model will be relevant to both.56 Additionally, the data included in this analysis were several decades old: the NIT blood samples were collected in 1985, and the JPHC I and II samples were collected in 1991–1993. The distribution of gastric cancer risk factors has surely changed in these populations since then, which may decrease these findings’ relevance to the present day. Finally, we chose to code the variables in the predictive model in simple linear or binary fashion despite the curvilinear univariate associations that some of them (including age and UreA) had with gastric cancer. Although this may not reflect the association between each of these variables and gastric cancer in the data set with high accuracy, we believe that coding the variables more simply reduces the likelihood that the model is overfit to the data in which it was constructed. Moreover, linear and binary coefficients are easier to interpret than more complex terms.
To our knowledge, this is the first study to use machine learning techniques to build a predictive model for gastric cancer using detailed H. pylori protein data. Our results suggest that adding host immune response to different H. pylori proteins to the model, rather than categorizing individuals simply by H. pylori infection status at large, improves the prediction of gastric cancer risk. Moreover, the large sample size of this data set, and large number of cases due to the nested case-control design, improved statistical precision and enabled us to explore stratified analyses of the model’s performance. Blood samples were collected at baseline, before any study participants developed gastric cancer, thus assuring that exposure preceded onset of disease.
Conclusion
Using machine learning techniques, we constructed a new predictive model for gastric cancer risk that incorporated host antibody response to the H. pylori proteins HP 0305, HP 1564 and UreA, serologically defined chronic atrophic gastritis, pepsinogen I, age and gender. The new model exhibited improved AUC and sensitivity over an existing risk stratification model, the ABC Method. Improved non-invasive gastric cancer risk stratification may streamline the allocation of more invasive screening modalities, such as radiography or endoscopy, to high-risk individuals who will benefit from them most and away from low-risk individuals who do not need them. This may not only save individuals and the healthcare system unnecessary expense, but could also promote patients’ peace-of-mind by encouraging high-risk individuals to seek further screening and reassuring low-risk individuals that they probably do not need such services. All in all, improved risk stratification could increase survival from this deadly disease in East Asia.
Supplementary Material
Acknowledgments
Financial support
John D. Murphy was supported by grant T32 CA057726/CA/NCI NIH HHS/United States.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest for this research project.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 3.Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO Classification of Tumours of the Digestive System. Lyon, France: IARC; 2010. World Heal Organ Classif Tumours. 3. [Google Scholar]
- 4.Allemani C, Weir HK, Carreira H, et al. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet (London, England). 2015;385(9972):977–1010. doi: 10.1016/S0140-6736(14)62038-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colquhoun A, Arnold M, Ferlay J, Goodman KJ, Forman D, Soerjomataram I. Global patterns of cardia and non-cardia gastric cancer incidence in 2012. Gut. 2015;64(12):1881–1888. doi: 10.1136/gutjnl-2014-308915 [DOI] [PubMed] [Google Scholar]
- 6.Ohata H, Kitauchi S, Yoshimura N, et al. Progression of chronic atrophic gastritis associated withHelicobacter pylori infection increases risk of gastric cancer. Int J Cancer. 2004;109(1):138–143. doi: 10.1002/ijc.11680 [DOI] [PubMed] [Google Scholar]
- 7.Miki K. Gastric cancer screening by combined assay for serum anti-Helicobacter pylori IgG antibody and serum pepsinogen levels - “ABC method”. Proc Jpn Acad Ser B Phys Biol Sci. 2011;87(7):405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correa P, Piazuelo MB. Natural history of Helicobacter pylori infection. Dig Liver Dis. 2008;40(7):490–496. doi: 10.1016/j.dld.2008.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Warren JR, Marshall B. Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet. 1983;321(8336):1273–1275. [PubMed] [Google Scholar]
- 10.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet (London, England). 2016;388(10060):2606. doi: 10.1016/S0140-6736(16)32226-7 [DOI] [PubMed] [Google Scholar]
- 11.Shiota S, Mahachai V, Vilaichone R, et al. Seroprevalence of Helicobacter pylori infection and gastric mucosal atrophy in Bhutan, a country with a high prevalence of gastric cancer. J Med Microbiol. 2013;62(Pt 10):1571–1578. doi: 10.1099/jmm.0.060905-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pimentel-Nunes P, Libânio D, Marcos-Pinto R, et al. Management of epithelial precancerous conditions and lesions in the stomach (MAPS II): European Society of Gastrointestinal Endoscopy (ESGE), European Helicobacter and Microbiota Study Group (EHMSG), European Society of Pathology (ESP), and Sociedade Portuguesa de Endoscopia Digestiva (SPED) guideline update 2019. Endoscopy. 2019;51(04):365–388. doi: 10.1055/a-0859-1883 [DOI] [PubMed] [Google Scholar]
- 13.Miki K, Ichinose M, Shimizu A, et al. Serum pepsinogens as a screening test of extensive chronic gastritis. Gastroenterol Jpn. 1987;22(2):133–141. http://www.ncbi.nlm.nih.gov/pubmed/3596151. Accessed August 29, 2019. [DOI] [PubMed] [Google Scholar]
- 14.Dinis-Ribeiro M, Yamaki G, Miki K, Costa-Pereira A, Matsukawa M, Kurihara M. Meta-analysis on the validity of pepsinogen test for gastric carcinoma, dysplasia or chronic atrophic gastritis screening. J Med Screen. 2004;11(3):141–147. doi: 10.1258/0969141041732184 [DOI] [PubMed] [Google Scholar]
- 15.Samloff IM. Pepsinogens, pepsins, and pepsin inhibitors. Gastroenterology. 1971;60(4):586–604. http://www.ncbi.nlm.nih.gov/pubmed/4324336. Accessed August 1, 2019. [PubMed] [Google Scholar]
- 16.Blaser MJ, Perez-Perez GI, Kleanthous H, et al. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 1995;55(10):2111–2115. http://www.ncbi.nlm.nih.gov/pubmed/7743510. Accessed August 29, 2019. [PubMed] [Google Scholar]
- 17.Shiota S, Matsunari O, Watada M, Yamaoka Y. Serum Helicobacter pylori CagA antibody as a biomarker for gastric cancer in east-Asian countries. Future Microbiol. 2010;5(12):1885–1893. doi: 10.2217/fmb.10.135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cai Q, Zhu C, Yuan Y, et al. Development and validation of a prediction rule for estimating gastric cancer risk in the Chinese high-risk population: A nationwide multicentre study. Gut. 2019;68(9):1576–1587. doi: 10.1136/gutjnl-2018-317556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren J-S, Kamangar F, Qiao Y-L, et al. Serum pepsinogens and risk of gastric and oesophageal cancers in the General Population Nutrition Intervention Trial cohort. Gut. 2009;58(5):636–642. doi: 10.1136/gut.2008.168641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park CH, Kim EH, Jung DH, et al. The new modified ABCD method for gastric neoplasm screening. Gastric Cancer. 2016;19(1):128–135. 10.1007/s10120-015-0473-4. [DOI] [PubMed] [Google Scholar]
- 21.You W, Blot WJ, Li J, et al. Precancerous gastric lesions in a population at high risk of stomach cancer. Cancer Res. 1993;53(6):1317–1321. [PubMed] [Google Scholar]
- 22.Tsugane S, Sawada N. The JPHC Study: Design and Some Findings on the Typical Japanese Diet. Jpn J Clin Oncol. 2014;44(9):777–782. doi: 10.1093/jjco/hyu096 [DOI] [PubMed] [Google Scholar]
- 23.Cai H, Ye F, Michel A, et al. Helicobacter pylori blood biomarker for gastric cancer risk in East Asia. Int J Epidemiol. 2016;45(3):774–781. doi: 10.1093/ije/dyw078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsugane S, Sobue T. Baseline Survey of JPHC Study Design and Participation Rate. J Epidemiol. 2001;11(6sup):24–29. doi: 10.2188/jea.11.6sup_24 [DOI] [PubMed] [Google Scholar]
- 25.Li B, Taylor PR, Li JY, et al. Linxian nutrition intervention trials. Design, methods, participant characteristics, and compliance. Ann Epidemiol. 1993;3(6):577–585. doi: 10.1016/1047-2797(93)90078-i [DOI] [PubMed] [Google Scholar]
- 26.Blot WJ, Hsing AW, Fraumeni JF. Correlations of Dietary Intake and Blood Nutrient Levels With Esophageal Cancer Mortality in China. Nutr Cancer. 1990;13(3):121–127. doi: 10.1080/01635589009514053 [DOI] [PubMed] [Google Scholar]
- 27.Sasazuki S, Inoue M, Iwasaki M, et al. Effect of Helicobacter pylori infection combined with CagA and pepsinogen status on gastric cancer development among Japanese men and women: A nested case-control study. Cancer Epidemiol Biomarkers Prev. 2006;15(7):1341–1347. doi: 10.1158/1055-9965.EPI-05-0901 [DOI] [PubMed] [Google Scholar]
- 28.Michel A, Waterboer T, Kist M, Pawlita M. Helicobacter pylori Multiplex Serology. Helicobacter. 2009;14(6):525–535. doi: 10.1111/j.1523-5378.2009.00723.x [DOI] [PubMed] [Google Scholar]
- 29.Waterboer T, Sehr P, Michael KM, et al. Multiplex Human Papillomavirus Serology Based on In Situ-Purified Glutathione S-Transferase Fusion Proteins. Clin Chem. 2005;51(10):1845–1853. doi: 10.1373/clinchem.2005.052381 [DOI] [PubMed] [Google Scholar]
- 30.Li JY, Taylor PR, Li GY, et al. Intervention studies in Linxian, China--an update. J Nutr growth cancer. 1986. [Google Scholar]
- 31.Watanabe S, Tsugane S, Sobue T, Konishi M, Baba S. Study design and organization of the JPHC study. J Epidemiol. 2001;11(6sup):3–7. [DOI] [PubMed] [Google Scholar]
- 32.Breslow N. Covariance analysis of censored survival data. Biometrics. 1974:89–99. [PubMed] [Google Scholar]
- 33.Therneau T, Lumley T. R survival package. 2013. [Google Scholar]
- 34.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B. 1996;58(1):267–288. [Google Scholar]
- 35.Tibshirani R. The lasso method for variable selection in the cox model. Stat Med. 1997;16(4):385–395. doi: [DOI] [PubMed] [Google Scholar]
- 36.Liu Y, Chen P-HC, Krause J, Peng L. How to read articles that use machine learning: users’ guides to the medical literature. Jama. 2019;322(18):1806–1816. [DOI] [PubMed] [Google Scholar]
- 37.Pavlou M, Ambler G, Seaman SR, et al. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015;351. doi: 10.1136/bmj.h3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon RM, Subramanian J, Li MC, Menezes S. Using cross-validation to evaluate predictive accuracy of survival risk classifiers based on high-dimensional data. Brief Bioinform. 2011;12(3):203–214. doi: 10.1093/bib/bbr001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian J, Simon R. An evaluation of resampling methods for assessment of survival risk prediction in high-dimensional settings. Stat Med. 2011;30(6):642–653. doi: 10.1002/sim.4106 [DOI] [PubMed] [Google Scholar]
- 40.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1. [PMC free article] [PubMed] [Google Scholar]
- 41.Harrell FE Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15(4):361–387. [DOI] [PubMed] [Google Scholar]
- 42.Pencina MJ, D’Agostino RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2004;23(13):2109–2123. [DOI] [PubMed] [Google Scholar]
- 43.Antolini L, Boracchi P, Biganzoli E. A time-dependent discrimination index for survival data. Stat Med. 2005;24(24):3927–3944. [DOI] [PubMed] [Google Scholar]
- 44.F K YL Q MJB, et al. Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. Br J Cancer. 2007;96(1):172–176. doi: 10.1038/SJ.BJC.6603517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salim A, Delcoigne B, Villaflores K, et al. Comparisons of risk prediction methods using nested case-control data. Stat Med. 2017;36(3):455–465. doi: 10.1002/sim.7143 [DOI] [PubMed] [Google Scholar]
- 46.Blanche P, Blanche MP. Package ‘timeROC.’ 2019. [Google Scholar]
- 47.Molinaro AM, Simon R, Pfeiffer RM. Prediction error estimation: A comparison of resampling methods. Bioinformatics. 2005;21(15):3301–3307. doi: 10.1093/bioinformatics/bti499 [DOI] [PubMed] [Google Scholar]
- 48.Heo J, Jeon SW, Kim SK, Yang HM. Application of ABC system into clinical practice in Korea: Is it worthwhile? United Eur Gastroenterol J. 2015;3(5):A310. doi: 10.1177/2050640615601623 LK - http://vb3lk7eb4t.search.serialssolutions.com?sid=EMBASE&issn=20506406&id=doi:10.1177%2F2050640615601623&atitle=Application+of+ABC+system+into+clinical+practice+in+Korea%3A+Is+it+worthwhile%3F&stitle=United+Eur.+Gastroenterol.+J.&title=United+European+Gastroenterology+Journal&volume=3&issue=5&spage=A310&epage=&aulast=Heo&aufirst=J.&auinit=J.&aufull=Heo+J.&coden=&isbn=&pages=A310-&date=2015&auinit1=J&auinitm= [DOI] [Google Scholar]
- 49.Charvat H, Sasazuki S, Inoue M, et al. Prediction of the 10-year probability of gastric cancer occurrence in the Japanese population: the JPHC study cohort II. Int J Cancer. 2016;138(2):320–331. doi: 10.1002/ijc.29705 [DOI] [PubMed] [Google Scholar]
- 50.Tatemichi M, Sasazuki S, Inoue M, Tsugane S, Grp JS. Clinical Significance of IgG Antibody Titer against Helicobacter pylori. Helicobacter. 2009;14(3):231–236. doi: 10.1111/j.1523-5378.2009.00681.x [DOI] [PubMed] [Google Scholar]
- 51.R Core Team. R: A Language and Environment for Statistical Computing. 2020. https://www.r-project.org/.
- 52.Michel A, Waterboer T, Kist M, Pawlita M. Helicobacter pylori multiplex serology. Helicobacter. 2009;14(6):525–535. doi: 10.1111/j.1523-5378.2009.00723.x [DOI] [PubMed] [Google Scholar]
- 53.Varga MG, Wang T, Cai H, et al. Helicobacter pylori blood biomarkers and gastric cancer survival in China. Cancer Epidemiol Biomarkers Prev. 2018;27(3):342–344. doi: 10.1158/1055-9965.EPI-17-1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varga MG, Wood CR, Butt J, et al. Immunostimulatory membrane proteins potentiate H. pylori-induced carcinogenesis by enabling CagA translocation. Gut Microbes. 2021;13(1):1–13. doi: 10.1080/19490976.2020.1862613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and gastric cancer: Factors that modulate disease risk. Clin Microbiol Rev. 2010;23(4):713–739. doi: 10.1128/CMR.00011-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song H, Michel A, Nyrén O, Ekström AM, Pawlita M, Ye W. A CagA-independent cluster of antigens related to the risk of noncardia gastric cancer: Associations between Helicobacter pylori antibodies and gastric adenocarcinoma explored by multiplex serology. Int J Cancer. 2014;134(12):2942–2950. doi: 10.1002/ijc.28621 [DOI] [PubMed] [Google Scholar]
- 57.Piazuelo MB, Correa P. Gastric cáncer: Overview. Colomb medica (Cali, Colomb. 2013;44(3):192–201. http://www.ncbi.nlm.nih.gov/pubmed/24892619. Accessed October 2, 2019. [PMC free article] [PubMed]
- 58.Agréus L, Kuipers EJ, Kupcinskas L, et al. Rationale in diagnosis and screening of atrophic gastritis with stomach-specific plasma biomarkers. Scand J Gastroenterol. 2012;47(2):136–147. doi: 10.3109/00365521.2011.645501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Mario F, Cavallaro LG, Moussa AM, et al. Usefulness of Serum Pepsinogens in Helicobacter pylori Chronic Gastritis: Relationship With Inflammation, Activity, and Density of the Bacterium. Dig Dis Sci. 2006;51(10):1791–1795. doi: 10.1007/s10620-006-9206-1 [DOI] [PubMed] [Google Scholar]
- 60.Iijima K, Sekine H, Koike T, Imatani A, Ohara S, Shimosegawa T. Serum pepsinogen concentrations as a measure of gastric acid secretion in Helicobacter pylori-negative and -positive Japanese subjects. J Gastroenterol. 2005;40(10):938–944. doi: 10.1007/s00535-005-1677-x [DOI] [PubMed] [Google Scholar]
- 61.Charvat H, Shimazu T, Inoue M, et al. Estimation of the performance of a risk prediction model for gastric cancer occurrence in Japan: Evidence from a small external population. Cancer Epidemiol. 2020;67:101766. [DOI] [PubMed] [Google Scholar]
- 62.Leja M, Camargo MC, Polaka I, et al. Detection of gastric atrophy by circulating pepsinogens: A comparison of three assays. Helicobacter. 2017;22(4):e12393. doi: 10.1111/hel.12393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salama SM, Wefuan JN, Shiro-Koulla S, et al. Value of whole-cell antigen extracts for serologic detection of Helicobacter pylori. J Clin Microbiol. 1993;31(12):3331–3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ekström AM, Held M, Hansson L-E, Engstrand L, Nyrén O. Helicobacter pylori in gastric cancer established by CagA immunoblot as a marker of past infection. Gastroenterology. 2001;121(4):784–791. [DOI] [PubMed] [Google Scholar]
- 65.Parsonnet J, Friedman GD, Orentreich N, Vogelman H. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut. 1997;40(3):297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data analyzed in this study are available from Duke University. Restrictions apply to the availability of these data, which were used under specific agreement for this study. Data are available from the authors upon reasonable request with the permission of both Duke University and the respective institutions that collected the specimens (the National Cancer Center Research Institute, Japan, the Cancer Institute, Chinese Academy of Medical Sciences, China, and the National Cancer Institute, USA).