Abstract
The global spread of critical-priority antimicrobial-resistant Enterobacterales by food is a public health problem. Wild-caught seafood are broadly consumed worldwide, but exposure to land-based pollution can favor their contamination by clinically relevant antimicrobial-resistant bacteria. As part of the Grand Challenges Explorations: New Approaches to Characterize the Global Burden of Antimicrobial Resistance Program, we performed genomic surveillance and cell culture-based virulence investigation of WHO critical priority Enterobacterales isolated from marine bivalves collected in the Atlantic Coast of South America. Broad-spectrum cephalosporin-resistant Klebsiella pneumoniae and Escherichia coli isolates were recovered from eight distinct geographical locations. These strains harbored blaCTX-M-type or blaCMY-type genes. Most of the surveyed genomes confirmed the convergence of wide virulome and resistome (i.e., antimicrobials, heavy metals, biocides, and pesticides resistance). We identified strains belonging to the international high-risk clones K. pneumoniae ST307 and E. coli ST131 carrying important virulence genes, whereas in vitro experiments confirmed the high virulence potential of these strains. Thermolabile and thermostable toxins were identified in some strains, and all of them were biofilm producers. These data point to an alarming presence of resistance and virulence genes in marine environments, which may favor horizontal gene transfer and the spread of these traits to other bacterial species.
Subject terms: Antimicrobial resistance, Food microbiology
Introduction
The rapid and global dissemination of critical-priority antimicrobial-resistant Enterobacterales is a public health problem that demands mitigation strategies and strengthening by genomic epidemiological surveillance investigations1. Given its epidemiological importance, there are increasing reports of critical-priority bacteria in aquatic environments2, including coastal waters3–5, reinforcing that polluted marine environments may act as potential reservoirs for waterborne pathogens.
Seafood consumption has increased significantly during the last decades6. Although domestic aquaculture farming has also presented a linear trend of increase, wild-caught seafood remains broadly consumed globally7–9. In this regard, marine bivalves have a wide geographic distribution, being considered an alternative food protein source to coastal populations worldwide since ancient times10. They have also been used as marine sentinels and investigated as potential ecological bioindicators to measure environmental impacts related to water pollution due to their activity in filtering particles suspended in the surrounding water11.
Recent studies have documented human gastroenteritis outbreaks associated with the consumption of contaminated bivalves12–14. In this regard, norovirus and Vibrio spp. infections have gathered scientific community attention due to the pathogenicity and virulence behavior of these etiologic agents13–15. Although some studies have speculated that the identification of multidrug-resistant (MDR) bacteria in marine bivalves could also represent a potential threat for seafood consumers12,16,17, there is scarce scientific evidence regarding this matter.
Here, we report the emergence of critical priority Enterobacterales producing CTX-M-type and CMY-type β-lactamases, and thermolabile and thermostable toxins in wild-caught marine bivalves in the Southeast coast of Brazil, South America's largest and most populated country. We performed a robust investigation using WGS and in vitro experiments to assay the virulence potential of these bacteria to infect human cells, highlighting a new and hidden threat for seafood consumers.
Results
Occurrence of multidrug-resistant clinical relevant bacteria in edible marine bivalves
Eight ceftriaxone-resistant Enterobacterales, including E. coli (n = 6) and K. pneumoniae (n = 2) isolates, were recovered from edible marine bivalves collected in different points on the southeast coast of Brazil (Fig. 1 and Table 1). Four E. coli and one K. pneumoniae were found in brown mussels, whereas two E. coli and one K. pneumoniae were obtained from oysters. All isolates displayed a MDR resistance profile, but still remained susceptible to carbapenems and colistin (Fig. 2). Seven isolates exhibited an ESBL phenotype, presenting cefotaxime MIC > 32 µg/mL. Additionally, one E. coli (EM1CRO) was cefoxitin-resistant (MIC > 32 µg/mL).
Figure 1.
Map showing 14 different collection sites on the coastal areas of São Vicente and Santos cities, Southeast Brazil. Critical priority Enterobacterales strains were isolated from bivalves at 6 of these sites (represented by black markers): (A)—E. coli TM2CRO [location, −23.972588 S, −46.391932 W]; (B)—K. pneumoniae JM2CRO [location, −23.972765 S, −46.384877 W]; (C)—E. coli MM and E. coli MO [location, −23.976040 S, −46.372580 W]; (D)—E. coli EM1CRO [location, −23.972388 S, −46.350241 W]; (E)—E. coli C6O and E. coli 6 M [location, −23.987125 S, −46.308609 W]; (F)—K. pneumoniae BO2 [location, −23.987841 S, −46.295134 W]). The map was generated using public domain shapefiles provided by the Brazilian Institute of Geography and Statistics (https://www.ibge.gov.br), and OpenStreetMap vector layers (©OpenStreetMap contributors), licensed under the Open Data Commons Open Database License (https://www.openstreetmap.org/copyright). OpenStreetMap vector layers were downloaded on QGIS v3.22.4 (https://qgis.org) with OSMDownloader plugin v1.0.3 (https://github.com/lcoandrade/OSMDownloader). Final layout was designed on ESRI ArcMap™ v10.7.
Table 1.
Epidemiological characteristics of critical priority WHO Enterobacterales strains isolated from edible marine bivalves.
| Strain | Bivalve species | Location | ST/CC* | Serotype | Accession number |
|---|---|---|---|---|---|
| E. coli EM1CRO | Brown mussels (Perna perna) |
−23.972388 S, −46.350241 W |
457/- | O11:H25 | NDBC00000000.1 |
| E. coli C6O | Oysters (Crassostrea spp.) |
−23.987125 S, −46.308609 W |
ND | O36:H5 | NDBB00000000.1 |
| E. coli 6 M |
Brown mussels (Perna perna) |
−23.987125 S, −46.308609 W |
38/38 | O86:H18 | NCWA00000000.1 |
| E. coli MM | Brown mussels (Perna perna) |
−23.976040 S, −46.372580 W |
4012/- | O8:H4 | NDYX00000000.1 |
| E. coli MO | Oysters (Crassostrea spp.) |
−23.976040 S, −46.372580 W |
131/131 | -:H4 | NCVZ00000000.1 |
| E. coli TM2CRO | Brown mussels (Perna perna) |
−23.972588 S, −46.391932 W |
ND | O9:H28 | NCVY00000000.1 |
| K. pneumoniae BO2 | Oysters (Crassostrea spp.) |
−23.987841 S, −46.295134 W |
2646/- | O2v2:K102 | NCVX00000000.1 |
| K. pneumoniae JM2CRO | Brown mussels (Perna perna) |
−23.972765 S, −46.384877 W |
307/- | O2v2:K102 | NCVW00000000.1 |
* CC, clonal complex; ST, sequence type; ND, not determined.
Figure 2.
Heatmap of virulome, resistome, plasmidome, MLST and serotype of critical priority WHO Enterobacterales isolated from wild edible marine bivalves recovered from anthropogenically-polluted area on the South America Atlantic coast.
Genomic analyses reveal the convergence of broad virulome and resistome in seafood bacterial isolates
Resistome data of E. coli strains EM1CRO, MO, 6M were previously reported under an epidemiological context18,19. In the current study, we carried out an in-depth genomic analysis that also included new E. coli C60, MM and TM2CRO and K. pneumoniae BO2 and JM2CRO strains. The main results from WGS analysis are presented in Fig. 2. The convergence of wide virulome and resistome (antimicrobials, heavy metals, biocides, and pesticides resistance genes) was confirmed in most of the surveyed genomes. In this regard, all strains were positive to CTX-M variants, including blaCTX-M-8 (C6O), blaCTX-M-14 (MM), blaCTX-M-15 (BO2 and JM2CRO), blaCTX-M-27 (MO and 6M), and blaCTX-M-55 (TM2CRO), except for E. coli EM1CRO, which was positive for the plasmid-mediated AmpC (pAmpC) gene, blaCMY-2. Unfortunately, since we used a short read sequencing technology, we were not able to assemble the plasmids completely to verify all genes and their respective plasmid replicons. However, through mlplasmids tool (https://sarredondo.shinyapps.io/mlplasmids/) it was possible to predict the genes found in plasmids for E. coli strains (EM1CRO: mdfA; 6M: blaCTX-M-27; MO: aadA5, sul1, dfrA17, mphA; and TM2CRO: blaCTX-M-55).
A broad virulence repertoire was found in most strains, where lineages of phylogroups B2 (MO) and D (6M) exhibited a wider virulome than those of phylogroups A (MM), B1 (C6O and TM2CRO), and F (EM1CRO). MDR strains belonged to different STs, and worryingly an E. coli strain of ST131 (MO) and a K. pneumoniae strain of ST307 (JM2CRO) were detected. Overall, E. coli strains exhibited a large number of genes associated with adhesion to the host (i.e., iha, lpf, pap, ecp, fim, and afa/dr fimbriae), resistance to the host’s immune system (i.e., kps and neuC capsular polysaccharides; tra and iss complement and serum resistance), damage to the host (i.e., senB and astA enterotoxins) and iron/hemin uptake (i.e., chu, iuc, ent, irp, and ybt). The serotype and fimbriae type of E. coli strains were diverse and included O11:H25, O36:H5, O86:H18, O8:H4, and O9:H28; fimH145, fimH38, fimH30, and fimH32. K. pneumoniae strains also exhibited genes associated with adhesion to the host (i.e., fim operon), resistance to the host’s immune system (i.e., kfoC complement and serum resistance), damage to the host and iron/hemin uptake (i.e., iro and ent). We also identified the K-locus KL102/wzi173 in both BO2 and JM2CRO K. pneumoniae strains. Genome assembly revealed that IncF-type plasmids were the most prevalent among the strains. The limitations of short-read sequencing allowed only the type of plasmid (IncFII) associated with the blaCTX-M gene to be determined in the K. pneumoniae BO2 strain.
Virulent behavior of Enterobacterales isolated from edible marine bivalves
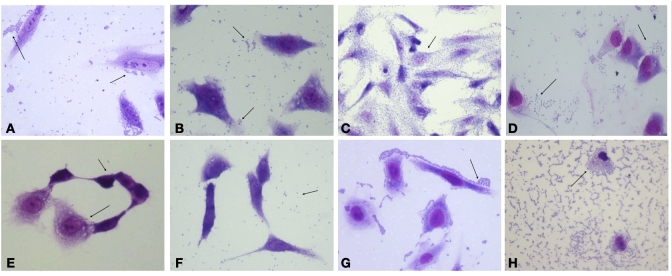
Bacterial isolates were tested to verify their ability to colonize and damage human epithelial cells. In this regard, all E. coli isolates were capable of adhering to HeLa cells after 6 h of incubation (Fig. 3), even though they displayed different adherence patterns with EM1CRO and C6O strains showing aggregative adherence, 6M and TM2CRO strains adhered diffusely to the cells, and MM and MO displaying an undefined adherence pattern. The two K. pneumoniae strains also exhibited aggregative adherence, both on the surface of the epithelial cells and on the glass slide. Additionally, E. coli strains 6M and C6O were capable of invading HeLa Cells after 6 h of incubation (Fig. 4), as observed through microscopy and confirmed by the gentamicin protection (invasion) assay.
Figure 3.
In vitro adhesion patterns displayed by the Enterobacterales strains isolated from edible marine bivalves, after 6 h incubation with HeLa cells. E.coli strain EM1CRO O11:H25 (A), E. coli strain C6O O36:H5 (B), E. coli 6 M O86:H18 (C), E. coli MM O8:H4 (D), E. coli MO O25:H4 (E), E. coli TM2CRO O9:H28 (F), K. pneumoniae BO2 (G) and K. pneumoniae JM2CRO (H). Arrows point to the adhered bacteria. Magnification 400x.
Figure 4.
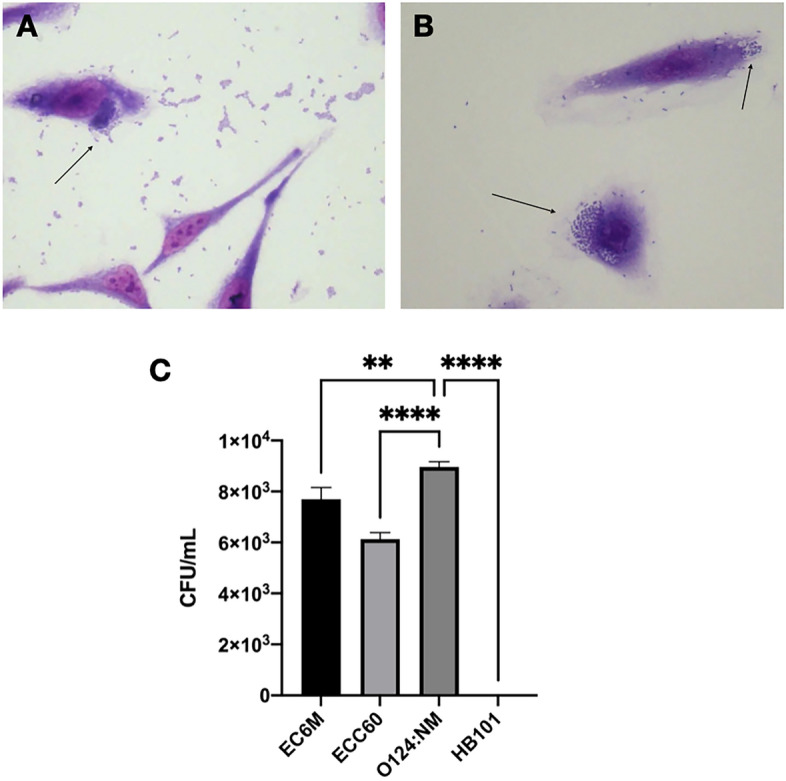
Invading phenotype exhibited in vitro by E. coli strains C6O O36:H5 (A) and 6 M O86:H18 (B). Arrows point to the invasion sites. Magnification 400x. The number of invading bacteria was determined by the invasion assay (C) and compared to 0124:NM (positive control) and HB101 (negative control) strains; ** < 0.01 **** < 0,0001.
Once MO, TM2CRO, and JM2CRO strains were visually confirmed as toxic to HeLa cells, cytotoxic assays on Vero cells were also performed (Fig. 5). Interestingly, when incubated with live bacterial cells, all strains were toxic to Vero cells. To verify if this was caused by cell–cell contact, Vero cells were incubated with dead bacteria and no effect was observed. To establish if secreted proteins were involved in toxicity, filtered supernatant from bacterial strains were incubated with Vero cells and only the TM2CRO strain resulted in cell damage. In order to determine if any secreted toxins were thermostable, boiled, filtered supernatant was added to the cells. This result suggested that E. coli TM2CRO expresses a thermolabile toxin since no effect on cells was observed after following this treatment. Interestingly, a vacuolating effect was observed in all MO treatments, which could indicate that this strain expresses a thermostable secreted toxin. Regarding the K. pneumoniae strains, JM2CRO and BO2 boiled supernatants caused nuclear damage to Vero cells, indicating that these strains probably secrete a thermostable toxin.
Figure 5.
Cytotoxic assay on Vero cells. BBC, boiled bacterial cells; SN, supernatant; BSN, boiled supernatant. Arrows point to vacuollation and nuclear damage induced by E. coli MO (O25:H4), and K. pneumoniae JM2CRO and BO2 strains, respectively. Magnification 400x.
All strains evaluated in this study were able to form biofilms, although they were considered, in their majority, to form weak or moderate biofilms under the tested conditions (Fig. 6a). However, the EM1CRO strain was an exception since it formed a biofilm as robust as the E. coli 4157, considered to be a highly biofilm-forming control strain (Fig. 6b).
Figure 6.
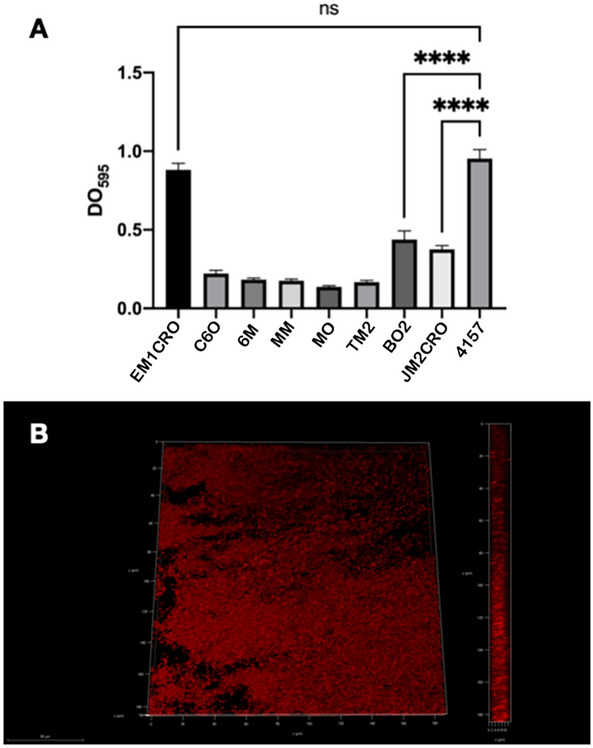
Biofilm formation by critical priority WHO Enterobacterales from isolated from wild edible marine bivalves. In A, crystal violet assay, after 24 h incubation at 37 °C in BHI medium. The values correspond to the average of three independent triplicates; p values < 0.001 were considered significant. Strains were considered to form weak, moderate, or strong biofilms, compared to a strong biofilm-forming E. coli strain (4157); In B, confocal laser scanning microscopy of EM1CRO O11:H25 biofilm on glass coverslips, after 24 h incubation at 37 °C in BHI medium and propidium iodide staining.
Discussion
The presence of ESBL and/or pAmpC-positive K. pneumoniae and E. coli strains contaminating bivalves in 6 of 14 investigated geographical points suggests heavy contamination of the marine environment of this geographical region, with critical priority bacteria. Indeed, nearby cities have a history of marine pollution due to lack of sewage treatment and sanitation facilities20, with the occurrence of critical priority bacteria in their coastal waters, and additional contamination of wild-caught fishes being recently documented21–25. Therefore, the presence of human fecal pollution in these coastal waters can support the origin of bioaccumulation of such pathogens in marine bivalves since they are filter-feeding organisms that remove a large amount of suspended material from the water.
While E. coli is a genetically heterogeneous species that can cause both diarrheal diseases and MDR extra-intestinal infections, K. pneumoniae has been a significant cause of antimicrobial-resistant healthcare-associated infections. In addition, worldwide dissemination of both species has been associated with well-established evolutionary pathogenic lineages, such as K. pneumoniae ST307 and E. coli ST131, which often display an MDR phenotype and a combination of virulence genes26,27.
Worryingly, in our study, we identified the presence of international CTX-M-27-producing E. coli ST131 and CTX-M-15-producing K. pneumoniae ST307 clones in oysters and mussel samples, respectively. This should be viewed as an epidemiological alert, since several studies have reported the rapid spread of these high-risk MDR clones at the human-animal interface, with several outbreaks worldwide28–30. The spread of virulence and MDR profiles is directly associated with the harboring of mobile genetic elements, and among them, plasmids are considered to play an essential role. A direct association between high-risk strains and the presence of IncF plasmids has been demonstrated, especially those containing FIA and FII replicon types31,32, which were detected in the majority of bacteria isolated in this study.
Currently, E. coli ST131 has been the most prevalent ExPEC clone circulating globally, being composed of multiple strains with distinct resistance profiles33, which might indicate that its emergence is involved in different events related to horizontal transfer of antimicrobial resistance genes. Indeed, each ST131 subclone is associated with a specific allele of the type 1 fimbrial adhesin gene (fimH), being the H30 the most prevalent. Of note, E. coli ST131-fimH30 has been responsible for urinary tract infections worldwide, being frequently associated to fluoroquinolone resistance34. On the other hand, the subclone H22 has been associated with foodborne infections35. Although the E. coli ST131 identified in study belongs to subclone H30, it could also represent a possible threat for foodborne infections since it has already been isolated from food-producing animals, including retail chicken36,37. Interestingly, most of the strains in subclone H30 belong to serotype O25:H4. In this study, the E. coli ST131-fimH30 (MO strain) was identified as H4 type flagellum, which has characteristics that aid in host colonization processes and is also capable of triggering enhanced induction of the cytokine IL-10, contributing to its fitness in the urinary tract38.
To cause infection, microorganisms must be able to colonize their host and adherence is a crucial event to the colonization process. Bacterial pathogens usually express a variety of fimbrial and afimbrial adhesins that promote attachment to the host cells by interacting with surface receptors. For certain E. coli pathotypes, the presence of these structures may result in specific adhesion patterns on epithelial cells. EM1CRO and C6O strains exhibited an adhesion pattern known as aggregative. Enteroaggregative E. coli (EAEC), usually related to persistent diarrhea, often expresses AAF/I and AAF/II adhesins39, which were not detected by WGS. However, both strains harbor the lpfA gene that encodes the long polar fimbriae commonly found in EAEC40. Additionally, the presence of the fim operon, which encodes a type I fimbria, could explain the aggregative adhesion pattern observed in these two strains41. EM1CRO also harbors the air and eilA genes. Air is an adhesin that is under the control of eilA and promotes bacterial aggregation and colonization42,43, contributing to aggregative adherence. On the other hand, 6M and TM2CRO strains adhered diffusely to epithelial cells. Diffusely adherent E. coli (DAEC) comprise a heterogeneous group without specific genetic markers. Still, a large proportion of DAEC strains express Afa/Dr adhesins and are frequently involved in chronic diarrhea in children44. The genes encoding this adhesin were found only in the 6M strain.
MM and MO E. coli strains showed an undefined adhesion pattern. Both strains harbor the E. coli common pilus (ecp) operon, which mediates adherence to the host cell by pathogenic and commensal strains of E. coli45, and presented genes from the homologous pap operon, which encodes the P fimbriae. Interestingly, MM adhered preferably to the coverslips, while MO mainly adhered to epithelial cells. This difference could be due to the presence of the iha gene in the MO strain, which encodes a bacterial adherence-conferring protein that was described as sufficient to confer adherence phenotype in non-adherent E. coli strains46, and the presence of the fim operon.
Although not usually classified by adherence, K. pneumoniae BO2 and JM2CRO strains showed an aggregative adherence pattern. Genome sequencing detected the presence of genes that encode mannose-resistant Klebsiella-like hemagglutinins (Mrk) proteins in both strains. These proteins compose a type 3 fimbriae that participates in biofilm formation, and can result in the establishment of bacterial aggregates in biotic and abiotic surfaces47. In addition, these strains also have the fim operon, which encodes the type 1 fimbriae.
Bacterial attachment to host cells usually triggers host signal transduction cascades, which may induce the internalization of bacterial cells due to the consequent rearrangement of eukaryotic cytoskeleton components48. Bacterial pathogens capable of invading host cells and tissues can successfully evade host defenses and spread easily. Interestingly, 6M and C6O strains presented the cell invasion phenotype, although no gene encoding known E. coli invasins was detected. It is important to note that the 6M and MO strains met the requirements to be classified as ExPEC49, belonging to phylogroups D and B2, respectively, which are reported to show higher virulence in humans50. ExPEC lacks pathogenicity when it colonizes the host's intestine, but it can cause potentially fatal diseases when it is transferred to other sites in the body. These strains differ widely in their genetic content and have an extensive range of virulence factors that assist in colonization and invasion of extra-intestinal sites, causing it to be involved in urinary tract infections, meningitis, among other important diseases51. More worrying, in addition to the ecp operon, also known as fimbriae associated with meningitis and regulated by temperature (MAT), the MO strain carries the aslA gene, which has been shown to contribute to the invasion of the blood–brain barrier52.
The in vitro adherence assays showed that some bacterial strains (MO, TM2CRO, JM2CRO) were toxic to HeLa cells. In order to further analyze this phenotype, cytotoxicity assays on Vero cells were performed. Overall, the results indicated that all isolated strains are somewhat capable of damaging the host’s epithelia. Surprisingly, when Vero cells were cultivated with live bacteria, all strains showed a toxic effect, probably induced by a secreted substance rather than cell–cell contact, as pointed by the results obtained from incubation with dead bacterial cells. Therefore, we sought to test the effect of filtered supernatant. The TM2CRO strain resulted in cell damage, indicating that the observed cytotoxic effect was caused by a secreted toxin. Genome sequencing revealed that this strain harbors the astA gene that encodes the E. coli heat-stable toxin-1 (EAST-1). The presence of astA gene in E. coli strains has been reported worldwide, and it is commonly associated with diarrhea in humans53,54. However, when the boiled supernatant was tested, no cell damage was observed, leading to the conclusion that an unknown thermolabile toxin caused the cytotoxic effect. Interestingly, a vacuolating effect was observed in all ECMO treatments, indicating that this strain expresses a thermostable secreted toxin. Furthermore, genome sequencing revealed the presence of the sat gene, responsible for encoding the secreted autotransporter toxin (Sat). The cytopathic activity of Sat was reported on different epithelial cell lineages, such as HEp-2 and Vero cells55,56. This strain also carries the senB gene that encodes the secreted TieB enterotoxin. This toxin has been reported in different Shigella species and among other isolates of pathogenic and commensal E. coli, and plays a role in the development of severe diarrhea57,58.
Filtered supernatant of both K. pneumoniae strains also showed a cytotoxic effect. However, the damage was mainly on the cell nucleus. To determine if these secreted toxins were thermostable or thermolabile, boiled, filtered supernatants from JM2CRO and BO2 strains were tested, and nuclear damage to Vero cells was still observed, suggesting that these strains secrete a thermostable toxin, probably not yet identified. Interestingly, genes that code for a type 6 secretion system (T6SS) were detected in several strains evaluated in this study (C6O, 6M, TM2CRO, BO2, and JM2CRO). Although the primary role of T6SS is related to bacterial competition, the gene that encodes the effector protein Til1, detected in the BO2 strain, encodes a lipase that appears to have activity on eukaryotic cells59,60.
Evolutionary successful pathogenic bacterial lineages contain a plethora of virulence determinants that are essential for host colonization and pathogenesis. Among them, the iron/heme transport systems are crucial for colonization. All strains evaluated in this study carried at least one gene involved in iron acquisition, and those E. coli with ExPEC attributes (6M and MO) and both Klebsiella strains had more than 10 of these genes. The presence of different types of siderophores is an attribute of more virulent strains of Klebsiella61. This feature was also expected for ExPEC, since the concentration of iron in extra-intestinal sites is low, leading to the development of several strategies to sequester these molecules from the host62.
Another relevant aspect to be considered for the pathogenesis of bacterial strains isolated from seafood products investigated is the capacity to form biofilm, which was observed in all strains tested in this work. The ability to form biofilm can be considered an important virulence factor in gastrointestinal infections since it contributes to the colonization of the host by opportunistic pathogens and allows its persistence in the environment, where they can act as infectious agents for an extended period. In addition, life within the biofilm increases the resistance of the microorganism to the host's immune system and conventional antimicrobial drugs, making its eradication very difficult63. In fact, studies demonstrate that biofilm formation by pathogenic E. coli strains contributes to the persistence of the infection64,65, especially in strains that present an aggregative adhesion pattern, as observed in EM1CRO and C60 strains.
Although the most clinically significant K. pneumoniae biofilms are those formed on the internal surfaces of catheters and other internal devices, biofilm formation can also contribute to the colonization of the gastrointestinal tract, and the development of invasive infections, especially in immunocompromised patients65.
In summary, this study shed light on the emergence of critical-priority Enterobacterales co-harboring a broad repertoire of virulence and resistance factors isolated from wild-caught marine bivalves. Our findings are particularly worrisome because they suggest that wild-caught bivalves have been contaminated with medically important bacteria in the Southeast coast of Brazil, which reinforce the urgent need to strengthen surveillance of seafood sold in countries with the highest level of antimicrobial resistance. Additionally, our virulence assays confirmed the high pathogenic potential of the isolated bacteria, revealing a red-alert threat to seafood consumers. Special attention should be paid for uncooked seafood (e.g., raw oysters), once the production and release of different toxins were documented in some strains. In this regard, the production of thermostable toxins is a worrisome prospect, as they would not be affected by cooking.
Conclusion
We report the occurrence of critical-priority Enterobacterales in edible bivalves from a polluted area on the South America Atlantic coast. Using in vitro experiments, we also demonstrated the convergence of MDR phenotype with broad virulence repertoire that enables these bacteria to infect and harm human hosts, revealing a high pathogenic potential that can jeopardize the health of seafood consumers and marine-related ecosystems.
Material and methods
Sample collection and MDR bacterial isolation
Between November 2016 and February 2017, oysters (Crassostrea spp.) and brown mussels (Perna perna) samples were collected at low tide from 14 different locations at the marine coast of Sao Vicente (23.963056 S 46.391944 W) and Santos (23.960833 S 46.333361 W), two densely populated cities located on the southeast coast of Brazil. Each sample consisted of approximately ten mussels and ten oysters of similar sizes. The oyster and mussel samples were processed separately. The bivalves’ collection and processing were authorized by the appropriate Brazilian authority (SISBIO licenses n. 58,570–1). Bivalve samples were kept refrigerated in sterile plastic bags until processing (no later than 3 h after collection). Subsequently, 25 g of bivalves were incubated at 37 °C for 24 h in new sterile plastic bags (Whirl–Pak, Nasco, WI, USA) filled with 225 mL of Brain–Heart Infusion (BHI) broth. Following incubation, 1 mL of the BHI culture was inoculated onto MacConkey agar plates containing colistin (2 μg/mL), ceftriaxone (2 μg/mL) or meropenem (2 μg/mL), and incubated for an additional 24 h at 37 °C.
Bacterial identification and antimicrobial susceptibility testing
All isolated Gram-negative bacterial colonies were identified by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analysis and tested for susceptibility to 15 different antimicrobials (amikacin, amoxicillin/clavulanic acid, aztreonam, cefepime, cefotaxime, cefoxitin, ceftazidime, ceftriaxone, ciprofloxacin, ertapenem, gentamicin, imipenem, meropenem, nalidixic acid, and sulfametoxazole-trimethoprim) by the Kirby-Bauer method, following the Clinical and Laboratory Standards Institute (CLSI) guidelines67. ESBL production was investigated using the double-disk synergy test (DDST)67 and minimum inhibitory concentrations (MICs) were determined using the E-test. The presence of genes encoding extended-spectrum β-lactamase (ESBL) (blaCTX-M-1, blaCTX-M-2, blaCTX-M-8 and blaCTX-M-9), cephamycin resistance (blaCMY-2), carbapenemase (blaKPC-2), and mobilized colistin resistance (mcr-1) was assessed by PCR. All PCR positive isolates were whole-genome sequenced.
Whole-genome sequence analysis
Extraction of total bacterial DNA was performed using PureLink™ Genomic DNA mini Kit, and library preparation followed the Nextera XT Illumina protocol. Whole bacterial genome sequencing was performed using the Illumina NextSeq platform with paired-end reads (150-bp). De novo assembly was carried out using Velvet v.1.2.1 pipeline and Geneious R9 software. Serotypes, MLST, virulence genes, antimicrobial resistance genes, plasmid replicons, fimbriae type, and pMLST of E. coli isolates were identified using the SerotypeFinder 2.0, MLST 2.0, Virulence Finder 2.0, ResFinder 4.1, PlasmidFinder 2.1, FimTyper 1.0. and pMLST 2.0 databases available at the Center for Genomic Epidemiology—CGE (http://genomicepidemiology.org/), respectively. Phylogenetic groups of E. coli were determined in silico according to Clermont v1.4.0 (https://github.com/A-BN/ClermonTyping). For K. pneumoniae, Kleborate (https://github.com/katholt/Kleborate) was used for capsule (K) and wzi prediction, species confirmation, and multilocus sequence type (ST). CGE database was used to determine antimicrobial resistance, plasmid type, and pMLST. Additionally, ABRicate v0.9.8 (https://github.com/tseemann/abricate) with VFDB database (https://github.com/haruosuz/vfdb) was used to assess the presence of virulence genes and Kleborate v2.0.4 (https://github.com/katholt/Kleborate) was used for typing and to screen for mutations in quinolone resistance determining regions in K. pneumoniae strains. The presence of genes associated with resistance to heavy metal (e.g., arsenic, lead, and mercury), disinfectants (e.g., quaternary ammonium compounds), and pesticide (e.g., glyphosate) was predicted by comparative in silico analysis against an in-house database. An identity threshold of > 98% was used to identify predicted genes. Genomic data were plotted on heatmaps for the presence/absence of virulence genes, resistance genes, and plasmids using iTOL v6 (https://itol.embl.de) with dummy trees.
In vitro infection assays of HeLa cells
Before infection, HeLa cells were cultured in a 5% CO2 atmosphere for 48 h in 24-well plates (Nest, USA) containing DMEM medium (Cultilab Ltd.), and washed three times with phosphate-buffered saline (PBS). To each well, 960 µL of supplemented DMEM (i.e. 2% fetal bovine serum and 1% D-mannose) and 40 µL of the bacterial pre-cultures in Luria Bertani broth (LB) were added. The plates were incubated for 3 h or 6 h. Adherence assays were performed as described previously68. Briefly, cells were washed four times with PBS, fixed with methanol, and stained with May-Grünwald and Giemsa staining solutions (Merck, Germany) before analysis by light microscopy. The enteropathogenic E. coli strains E2348⁄69 (localized adhesion) and BA320 (localized-like adhesion) were used as positive controls. The ability of bacteria to invade epithelial cells was accessed as described previously69, with some modifications: after 3 h incubation, the extracellular bacteria were killed by the addition of fresh DMEM containing 100 µg/mL gentamycin or meropenem, and plates were incubated for additional 3 h at 37ºC. Cells were washed with PBS and lysed with 1% TritonX-100. Serial dilutions of the cell suspensions were plated on MacConkey agar. An enteroinvasive E. coli strain (O124:NM) and the non-invasive E. coli strain HB101 were used as positive and negative controls, respectively. Cell invasion was considered positive when bacterial growth on MacConkey agar plates was observed after 18 h incubation at 37ºC.
In vitro cytotoxicity assay
VERO cells were cultivated in 24-well plates (Nest, USA) in DMEM medium (Cultilab Ltd.) containing 10% FBS, for 48 h at 37ºC, in a 5% CO2 atmosphere. The isolated strains were cultivated in LB broth for 18 h at 37ºC. An aliquot was centrifuged at 13,000 g for 2 min and the supernatant was filtered through 0.22 µm membrane filters (Millipore). Bacterial culture and supernatant filtrates aliquots were also boiled for 10 min. Fifty microliters of bacterial culture, supernatant, boiled bacterial culture or boiled supernatant were added to VERO cells, and after incubation for 18 h at 37ºC, the cells were washed three times with PBS, fixed with methanol and stained using May-Grünwald and Giemsa solutions. The results were compared with control cells. Cytotoxicity was considered positive when damaged cells were observed.
Biofilm formation in microtitre plates (crystal violet assay)
The quantitative analysis of biofilm formation was carried out in 96-well polystyrene plates following previously described methodologies70,71, with slight modifications. Overnight bacterial cultures were inoculated into BHI broth at a dilution of 1:100 in 96-well polystyrene plates (TPP, Switzerland) in a final volume of 100 µL. After 24 h of incubation at 37 °C, the plates were washed 3 times with PBS to remove planktonic bacteria. The formed biofilms were fixed for 10 min at room temperature with 125 µL of a 75% ethanol solution and stained for 5 min with 125 µL of a 0.5% crystal violet solution. After being washed four times with PBS, the plates were left at room temperature to dry. Then, 200 µL of 30% glacial acetic acid were added and the plates were left for 2 min at room temperature to solubilize the crystal violet. Absorbance was read at 595 nm on the Multiskan®EX ELISA reader (Thermo Fisher Scientific, USA). All assays were performed in triplicates. Strains were considered to form weak, moderate, or strong biofilms, compared to a strong biofilm-forming E.coli strain (4157).
Biofilm formation in glass slides and laser confocal microscopy
Microscopic analysis of biofilm formation was performed as described previously65. Briefly, overnight bacterial cultures were diluted 1:100 in fresh BHI broth and added to a 24-well cell culture plate with glass coverslips for a final volume of 1 mL. After 24 h of incubation at 37 °C and two PBS washes, the formed biofilms were fixed with 4% p-formaldehyde for 18 h at 4 °C, blocked for 30 min with a 0.2% bovine serum albumin solution, and permeabilized with 0.1% Triton X-100 for 5 min. After five PBS washes, the coverslips were removed from the plates, dried with filter paper, and fixed on glass slides with Mowiol (Calbiochem) added with Propidium Iodide (Molecular Probes) in a final concentration of 1:1000. After about 20 h at 4 °C, the slides were observed in a 630 × magnification confocal fluorescence microscope (Microscope LSM 510 Meta, Zeiss—Filters: BP500-530IR—488 nm; LP560—543 nm).
Statistical analyses
Unless otherwise stated, all experiments were carried out in three independent triplicates. Statistical analyses were performed by comparison of means between groups using One-Way Anova (GraphPad Prism 9.0). The Post Hoc analysis was performed using the Tukey or Dunnett methods, according to the test. p < 0.0001 values were considered statistically significant.
Acknowledgements
We thank Cefar Diagnóstica Ltda (Brazil) for kindly providing all antimicrobial discs used in the susceptibility tests. We also thank CEFAP-GENIAL for the DNA sequencing work.
Author contributions
V.B. and F.P.S. are responsible for the conceptualization and validation of the work, and wrote and edited the main manuscript text; V.B. and S.C.F.C. conducted the in vivo experiments (Figures 3, 4, 5 and 6); Q.M. and B.F. performed the sequencing and data curation; V.B. performed the formal data analysis; E.S. prepared Figures 1, 2; N.L. and M.P.N.C. provided resources, funding acquisition and text review. All authors reviewed the final manuscript.
Funding
This work was supported by the Bill & Melinda Gates Foundation (Grand Challenges Explorations Brazil OPP1193112), the Fundação de Amparo à Pesquisa do Estado de São Paulo (2020/08224-9), and Conselho Nacional de Desenvolvimento Científico e Tecnológico (AMR 443819/2018-1, 312249/2017-9 and 433128/2018-6). N.L. is a CNPq research fellow (312249/2017-9).
Data availability
All data generated or used during this work are presented in the paper. The whole-genome nucleotide sequence of the bacterial strains used in this work are available in the GenBank database under the following access numbers: E. coli EM1CRO (NDBC00000000.1), E. coli C6O (NDBB00000000.1), E. coli 6M (NCWA00000000.1), E. coli MM (NDYX00000000.1), E. coli MO (NCVZ00000000.1), E. coli TM2CRO (NCVY00000000.1), K. pneumoniae BO2 (NCVX00000000.1), and K. pneumoniae JM2CRO (NCVW00000000.1).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tacconelli E, et al. WHO Pathogens Priority List Working Group: Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Amarasiri M, Sano D, Suzuki S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: Current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020;50:2016–2059. doi: 10.1080/10643389.2019.1692611. [DOI] [Google Scholar]
- 3.Paschoal RP, et al. Concentration and variety of carbapenemase producers in recreational coastal waters showing distinct levels of pollution. Antimicrob. Agents Chemother. 2017;61:e01963–e2017. doi: 10.1128/AAC.01963-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen R, Paikin S, Rokney A, Rubin-Blum M, Astrahan P. Multidrug-resistant enterobacteriaceae in coastal water: An emerging threat. Antimicrob. Resist. Infect. Control. 2020;9:169. doi: 10.1186/s13756-020-00826-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewi DAPR, Götz B, Thomas T. Diversity and genetic basis for carbapenem resistance in a coastal marine environment. Appl. Environ. Microbiol. 2020;86:e02939–e3019. doi: 10.1128/AEM.02939-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guillen J, et al. Global seafood consumption footprint. Ambio. 2019;48:111–122. doi: 10.1007/s13280-018-1060-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tidwell JH, Allan GL. Fish as food: aquaculture's contribution. Ecological and economic impacts and contributions of fish farming and capture fisheries. EMBO Rep. 2001;2:958–963. doi: 10.1093/embo-reports/kve236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bostock J, et al. Aquaculture: global status and trends. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:2897–2912. doi: 10.1098/rstb.2010.0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reverter M, et al. Aquaculture at the crossroads of global warming and antimicrobial resistance. Nat. Commun. 2020;11:1870. doi: 10.1038/s41467-020-15735-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gormaz JG, Fry JP, Erazo M, Love DC. Public Health Perspectives on Aquaculture. Curr. Environ. Health Rep. 2014;1:227–238. doi: 10.1007/s40572-014-0018-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suárez-Ulloa V, et al. Bivalve omics: state of the art and potential applications for the biomonitoring of harmful marine compounds. Mar. Drugs. 2013;11:4370–4389. doi: 10.3390/md11114370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elbashir S, et al. Seafood pathogens and information on antimicrobial resistance: A review. Food Microbiol. 2018;70:85–93. doi: 10.1016/j.fm.2017.09.011. [DOI] [PubMed] [Google Scholar]
- 13.Spaur M, et al. A systematic review of post-harvest interventions for Vibrio parahaemolyticus in raw oysters. Sci. Tot. Environ. 2020;745:140795. doi: 10.1016/j.scitotenv.2020.140795. [DOI] [PubMed] [Google Scholar]
- 14.Razafimahefa RM, Ludwig-Begall LF, Thiry E. Cockles and mussels, alive, alive, oh-The role of bivalve molluscs as transmission vehicles for human norovirus infections. Transbound. Emerg. Dis. 2020;67:9–25. doi: 10.1111/tbed.13165. [DOI] [PubMed] [Google Scholar]
- 15.Campos CJ, Lees DN. Environmental transmission of human noroviruses in shellfish waters. Appl. Environ. Microbiol. 2014;80:3552–3561. doi: 10.1128/AEM.04188-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vignaroli C, et al. Multidrug-resistant and epidemic clones of Escherichia coli from natural beds of Venus clam. Food Microbiol. 2016;59:1–6. doi: 10.1016/j.fm.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Grevskott DH, Svanevik CS, Sunde M, Wester AL, Lunestad BT. Marine bivalve mollusks as possible indicators of multidrug-resistant Escherichia coli and other species of the Enterobacteriaceae Family. Front. Microbiol. 2017;8:24. doi: 10.3389/fmicb.2017.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sellera FP, et al. Draft genome sequence of a blaCMY-2/IncI1-harbouring Escherichia coli D:ST457 isolated from coastal benthic organisms. J. Glob. Antimicrob. Resist. 2018;14:83–84. doi: 10.1016/j.jgar.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 19.Fernandes MR, et al. Emergence of CTX-M-27-producing Escherichia coli of ST131 and clade C1–M27 in an impacted ecosystem with international maritime traffic in South America. J. Antimicrob. Chemother. 2020;75:1647–1649. doi: 10.1093/jac/dkaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamparelli CC, et al. Are fecal indicator bacteria appropriate measures of recreational water risks in the tropics: A cohort study of beach goers in Brazil? Water Res. 2015;87:59–68. doi: 10.1016/j.watres.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Fernandes MR, et al. Colistin-resistant mcr-1-positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob. Agents Chemother. 2017;61:e00234–e317. doi: 10.1128/AAC.00234-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sellera FP, et al. Draft genome sequence of Enterobacter cloacae ST520 harbouringblaKPC-2, blaCTX-M-15 and blaOXA-17 isolated from coastal waters of the South Atlantic Ocean. J. Glob. Antimicrob. Resist. 2017;10:279–280. doi: 10.1016/j.jgar.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 23.Andrade VC, Caetano T, Mendo S, Oliveira AJFC. Carbapenem resistant Enterobacteriaceae from port areas in São Paulo State (Brazil): Isolation and molecular characterization. Mar. Pollut. Bull. 2020;159:111329. doi: 10.1016/j.marpolbul.2020.111329. [DOI] [PubMed] [Google Scholar]
- 24.Fernandes MR, et al. Identification and genomic features of halotolerant extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in urban-impacted coastal waters, Southeast Brazil. Mar. Pollut. Bull. 2020;150:110689. doi: 10.1016/j.marpolbul.2019.110689. [DOI] [PubMed] [Google Scholar]
- 25.Sellera FP, Fernandes MR, Moura Q, Carvalho MPN, Lincopan N. Extended-spectrum-β-lactamase (CTX-M)-producing Escherichia coli in wild fishes from a polluted area in the Atlantic Coast of South America. Mar. Pollut. Bull. 2018;135:183–186. doi: 10.1016/j.marpolbul.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Dahbi G, et al. Emergence of new variants of ST131 clonal group among extraintestinal pathogenic Escherichia coli producing extended-spectrum β-lactamases. Int. J. Antimicrob. Agents. 2013;42:347–351. doi: 10.1016/j.ijantimicag.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 27.Mathers AJ, Peirano G, Pitout JD. Escherichia coli ST131: The quintessential example of an international multiresistant high-risk clone. Adv. Appl. Microbiol. 2015;90:109–154. doi: 10.1016/bs.aambs.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Bonura C, et al. An update of the evolving epidemic of blaKPC carrying Klebsiella pneumoniae in Sicily, Italy, 2014: emergence of multiple non-ST258 clones. PLoS ONE. 2015;10:e0132936. doi: 10.1371/journal.pone.0132936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Long, S.W. et al. Population genomic analysis of 1,777 extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates, Houston, Texas: Unexpected abundance of clonal group 307. mBio8, e00489–17 (2017). 10.1128/mBio.00489-17 [DOI] [PMC free article] [PubMed]
- 30.Castanheira M, et al. Rapid expansion of KPC-2-producing Klebsiella pneumoniae isolates in two Texas hospitals due to clonal spread of ST258 and ST307 lineages. Microb. Drug Resist. 2013;19:295–297. doi: 10.1089/mdr.2012.0238. [DOI] [PubMed] [Google Scholar]
- 31.Carattoli A. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. 2009;53:2227–2238. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calhau V, Ribeiro G, Mendonça N, Da Silva GJ. Prevalent combination of virulence and plasmidic-encoded resistance in ST 131 Escherichia coli strains. Virulence. 2013;4:726–729. doi: 10.4161/viru.26552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee R, Johnson JR. A New Clone Sweeps Clean: the Enigmatic Emergence of Escherichia coli Sequence Type 131. Antimicrob. Agents Chemother. 2014;58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson JR, et al. Abrupt emergence of a single dominant multidrug-resistant strain of Escherichia coli. J. Infect. Dis. 2013;207:919–928. doi: 10.1093/infdis/jis933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, C.M. et al. Escherichia coli ST131-H22 as a foodborne uropathogen. mBio9, e00470–18 (2018). 10.1128/mBio.00470-18 [DOI] [PMC free article] [PubMed]
- 36.Vincent C, et al. Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 2010;16:88–95. doi: 10.3201/eid1601.091118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cortes P, et al. Isolation and characterization of potentially pathogenic antimicrobial-resistant Escherichia coli strains from chicken and pig farms in Spain. Appl. Environ. Microbiol. 2010;76:2799–2805. doi: 10.1128/AEM.02421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kakkanat, A. et al. The role of H4 flagella in Escherichia coli ST131 virulence Sci. Rep.5, 16149 (2015). 10.1038/srep16149 [DOI] [PMC free article] [PubMed]
- 39.Czeczulin JR, et al. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 1997;65:4135–4145. doi: 10.1128/IAI.65.10.4135-4145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valat C, et al. Phylogenetic grouping and virulence potential of extended-spectrum-β-lactamase-producing Escherichia coli strains in cattle. Appl. Environ. Microbiol. 2012;78:4677–4682. doi: 10.1128/AEM.00351-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreira CG, Carneiro SM, Nataro JP, Trabulsi LR, Elias WP. Role of type I fimbriae in the aggregative adhesion pattern of enteroaggregative Escherichia coli. FEMS Microbiol. Lett. 2003;226:79–85. doi: 10.1016/S0378-1097(03)00561-5. [DOI] [PubMed] [Google Scholar]
- 42.Harrington SM, et al. The Pic protease of enteroaggregative Escherichia coli promotes intestinal colonization and growth in the presence of mucin. Infect. Immun. 2009;77:2465–2473. doi: 10.1128/IAI.01494-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheikh J, et al. EilA, a HilA-like regulator in enteroaggregative Escherichia coli. Mol. Microbiol. 2006;61:338–350. doi: 10.1111/j.1365-2958.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- 44.Servin AL. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dradhesins (Afa/Dr DAEC): current insights and future challenges. Clin. Microbiol. Rev. 2014;27:823–869. doi: 10.1128/CMR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rendón, M.A. et al. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. USA104, 10637–42 (2007). 10.1073/pnas.0704104104 [DOI] [PMC free article] [PubMed]
- 46.Tarr PI, et al. Iha: A novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 2000;68:1400–1407. doi: 10.1128/iai.68.3.1400-1407.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilksch JJ, et al. MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 2011;7:e1002204. doi: 10.1371/journal.ppat.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finlay BB, Cossart P. Exploitation of mammalian host cell functions by bacterial pathogens. Science. 1997;276:718–725. doi: 10.1126/science.276.5313.718. [DOI] [PubMed] [Google Scholar]
- 49.Johnson JR, Gajewski A, Lesse AJ, Russo TA. Extraintestinal pathogenic Escherichia coli as a cause of invasive nonurinary infections. J. Clin. Microbiol. 2003;41:5798–5802. doi: 10.1128/jcm.41.12.5798-5802.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iranpour D, et al. Phylogenetic groups of Escherichia coli strains from patients with urinary tract infection in Iran based on the new Clermont phylotyping method. Biomed. Res. Int. 2015;2015:846219. doi: 10.1155/2015/846219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Johnson JR, Russo TA. Extraintestinal pathogenic Escherichia coli: "the other bad E coli". J. Lab. Clin. Med. 2002;139:155–162. doi: 10.1067/mlc.2002.121550. [DOI] [PubMed] [Google Scholar]
- 52.Hoffman JA, Badger JL, Zhang Y, Huang SH, Kim KS. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect. Immun. 2000;68:5062–5067. doi: 10.1128/iai.68.9.5062-5067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ménard LP, Dubreuil JD. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): A new toxin with an old twist. Crit. Rev. Microbiol. 2002;28:43–60. doi: 10.1080/1040-840291046687. [DOI] [PubMed] [Google Scholar]
- 54.Zhou, Z. et al. An outbreak of gastroenteritis in Osaka, Japan due to Escherichia coli serogroup O166:H15 that had a coding gene for enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1). Epidemiol. Infect.128, 363–71 (2002). 10.1017/s0950268802006994 [DOI] [PMC free article] [PubMed]
- 55.Guyer DM, Henderson IR, Nataro JP, Mobley HL. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 2000;38:53–66. doi: 10.1046/j.1365-2958.2000.02110.x. [DOI] [PubMed] [Google Scholar]
- 56.Guyer DM, Radulovic S, Jones FE, Mobley HL. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect. Immun. 2002;70:4539–4546. doi: 10.1128/iai.70.8.4539-4546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yavzori M, Cohen D, Orr N. Prevalence of the genes for Shigella enterotoxins 1 and 2 among clinical isolates of Shigella in Israel. Epidemiol. Infect. 2002;128:533–535. doi: 10.1017/s0950268802006866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farfán MJ, Toro CS, Barry EM, Nataro JP. Shigella enterotoxin-2 is a type III effector that participates in Shigella-induced interleukin 8 secretion by epithelial cells. FEMS Immunol. Med. Microbiol. 2011;61:332–339. doi: 10.1111/j.1574-695X.2011.00778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Basler, M., Mekalanos, J.J. Type 6 secretion dynamics within and between bacterial cells. Science337, 815 (2012).10.1126/science.1222901 [DOI] [PMC free article] [PubMed]
- 60.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. USA. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Marr CM, Russo TA. Hypervirulent Klebsiella pneumoniae: a new public health threat. Expert. Rev. Anti Infect. Ther. 2019;17:71–73. doi: 10.1080/14787210.2019.1555470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao Q, et al. Roles of iron acquisition systems in virulence of extraintestinal pathogenic Escherichia coli: salmochelin and aerobactin contribute more to virulence than heme in a chicken infection model. BMC Microbiol. 2012;12:143. doi: 10.1186/1471-2180-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ito A, Taniuchi A, May T, Kawata K, Okabe S. Increased antibiotic resistance of Escherichia coli in mature biofilms. Appl. Environ. Microbiol. 2009;75:4093–4100. doi: 10.1128/AEM.02949-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaur P, Chakraborti A, Asea A. Enteroaggregative Escherichia coli: an emerging enteric food borne pathogen. Interdiscip. Perspect. Infect. Dis. 2010;2010:254159. doi: 10.1155/2010/254159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Culler HF, et al. Atypical enteropathogenic Escherichia coli strains form biofilm on abiotic surfaces regardless of their adherence pattern on cultured epithelial cells. Biomed. Res. Int. 2014;2014:845147. doi: 10.1155/2014/845147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vuotto, C. et al. 2017. Biofilm formation and antibiotic resistance in Klebsiella pneumoniae urinary strains. J. Appl. Microbiol.123, 1003–1018 (2010). 10.1111/jam.13533 [DOI] [PubMed]
- 67.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2020).
- 68.Scaletsky IC, Silva ML, Trabulsi LR. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect. Immun. 1984;45:534–536. doi: 10.1128/IAI.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Luck SN, Bennett-Wood V, Poon R, Robins-Browne RM, Hartland EL. Invasion of epithelial cells by locus of enterocyte effacement-negative enterohemorrhagic Escherichia coli. Infect. Immun. 2005;73:3063–3071. doi: 10.1128/IAI.73.5.3063-3071.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.hristensen, G. D. et al. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol.22, 996–1006 (2005). 10.1128/JCM.22.6.996-1006.1985 [DOI] [PMC free article] [PubMed]
- 71.Sheikh, J., Hicks, S., Dall’Agnol, M., Phillips, A.D., Nataro, J.P. Roles for Fis and YafK in biofilm formation by enteroaggregative Escherichia coli. Mol. Microbiol.41, 983–97 (2001). 10.1046/j.1365-2958.2001.02512.x [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or used during this work are presented in the paper. The whole-genome nucleotide sequence of the bacterial strains used in this work are available in the GenBank database under the following access numbers: E. coli EM1CRO (NDBC00000000.1), E. coli C6O (NDBB00000000.1), E. coli 6M (NCWA00000000.1), E. coli MM (NDYX00000000.1), E. coli MO (NCVZ00000000.1), E. coli TM2CRO (NCVY00000000.1), K. pneumoniae BO2 (NCVX00000000.1), and K. pneumoniae JM2CRO (NCVW00000000.1).