Abstract
BACKGROUND:
Maternal diet during pregnancy can impact progeny health and disease by influencing the offspring’s gut microbiome and immune development. Gut microbial metabolism generates butyrate, a short-chain fatty acid which benefits intestinal health. Here, we assess the effects of antenatal butyrate on the offspring’s gastrointestinal health. We hypothesized that antenatal butyrate supplementation will induce protection against colitis in offspring.
METHODS:
C57BL/6 mice received butyrate during pregnancy and a series of experiments were performed on their offspring. RNA sequencing was performed on colonic tissue of 3-week-old offspring. Six to eight-week-old offspring were subjected to dextran sulfate sodium-induced colitis. Fecal microbiome analysis was performed on 6 to 8-week-old offspring.
RESULTS:
Antenatal butyrate supplementation dampened transcript enrichment of inflammation-associated colonic genes and prevented colonic injury in the offspring. Antenatal butyrate increased the offspring’s stool microbiome diversity and expanded the prevalence specific gut microbes.
CONCLUSIONS:
Antenatal butyrate supplementation resulted in down regulation of genes in the offspring’s colon that function in inflammatory signaling. In addition, antenatal butyrate supplementation was associated with protection against colitis and an expanded fecal microbiome taxonomic diversity in offspring.
INTRODUCTION
The gastrointestinal tract harbors a diverse ecosystem of several trillion microbial cells, which are paramount to human health and disease. The gut microbiome has an enormous functional capacity which influences health in a variety of ways, including, but not limited to pathogen exclusion, digestion of dietary components, and immunomodulatory activity1. In humans, the in utero environment was historically considered sterile. However, this dogma has recently been challenged as some studies have demonstrated bacterial signatures in the placenta2,3, amniotic fluid4, fetal tissue3,5 and meconium6,7, while other studies have reported contradicting results8,9. If the fetus does indeed harbor live microbes, then they may function in preparing the fetus for the larger inoculum that occurs at birth10.
The majority of microbes which colonize a newborn’s gastrointestinal (GI) tract originate from the mother at the time of birth11–13. Importantly, studies have shown that an individual’s dietary habits can strongly influence the gut microbiome composition14, including during pregnancy15. Furthermore, the maternal diet during pregnancy (the antenatal diet) has been reported to influence the diversity of a newborn’s microbiome15–17. Together, these observations support the notion that the maternal diet can not only influence the maternal microbiome, but also can influence the offspring’s microbiome composition, which is known to be a potent modulator of early life intestinal and immune development 18. Thus, an infant’s initial microbiome colonization may have enduring effects on their future health. For this reason, pregnancy represents a unique window of opportunity where modifications to the maternal diet could optimize health outcomes of the offspring.
One mechanism by which the gut microbiome influences host health is by the generation of bioactive metabolites such as short-chain fatty acids (SCFAs). SCFAs are the byproducts of fermentation of starch or dietary fiber by the gut microbiome19,20. The majority of SCFA generation occurs in the colon where the primary SCFAs produced include acetate, propionate, and butyrate21,22. The extent of SCFA production depends on the species and quantity of microbiome diversity in the colon, the substrate source and the gut transit time21,22.
Of the SCFAs produced by the colonic microbiome, butyrate has been the source of extensive research due to its protective effects on colonic health. Butyrate is the preferred energy source of colonocytes and through its actions as a histone deacetylase (HDAC) inhibitor, it modulates biological responses of gastrointestinal health by controlling gene transcription23. In addition, butyrate plays an important role in modulating immune responses and enhancing the intestinal barrier through the upregulation of genes that code for tight junction proteins such as zonula occludens-1 (ZO-1)24. Studies have also showed that butyrate acts as an anti-inflammatory agent by inhibiting pro-inflammatory cytokines, upregulating anti-inflammatory cytokines25, and inhibiting the activation of transcription factors involved in inflammatory signaling 26–28. In one study, butyrate provided protection against colonic injury in a rat model of dextran sulfate sodium (DSS)-induced colitis, through inhibition of the transcription factor, nuclear factor kappa B (NF-κB) 27. In addition, another study reported that butyrate diminished tumor necrosis factor alpha (TNFα) and nitric oxide (NO) production by LPS-stimulated neutrophils through its inhibitory effects on NF-κB 26. Thus, butyrate is a well-studied SCFA which attenuates host inflammatory responses through down regulation of NF-κB signaling 26–28.
Given the beneficial effects of butyrate on gut health and the influential developmental time period of pregnancy, we studied the effect of antenatal butyrate supplementation on the offspring’s gut health. Herein, we show that antenatal butyrate supplementation of pregnant murine dams leads to changes in colonic gene expression in their offspring, with the down regulation of colonic genes that function in inflammation and cholesterol synthesis. In addition, we report that antenatal butyrate supplementation confers powerful cytoprotective effects on the intestine of 6 to 8-week-old offspring when exposed to a model of DSS-induced colitis. Furthermore, antenatal butyrate was associated with changes in the offspring’s microbiome composition, shifting alpha and beta diversity, and leading to the expansion of Bifidobacteriaceae. Thus, butyrate supplementation during pregnancy has enduring effects on the offspring’s gastrointestinal health and may hold promise for enhanced cytoprotection in the offspring against conditions of gut inflammation and injury. Further investigations are warranted to understand the mechanisms of colonic protection resulting from antenatal butyrate supplementation.
METHODS
Animal studies
All experiments were performed using wild-type C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME). The mice were bred in an animal facility at Emory University. Animal procedures were approved by the Institutional Animal Care and Use of Committee of Emory University.
Mouse diets
Breeding pairs of mice (one male and one female) were randomly assigned to a diet which they received during pregnancy. The diets were given ad libitum and included either a diet supplemented with butyrate (butyrate group) or a regular diet with no butyrate supplementation (control group). The mice assigned to a butyrate diet had 1% sodium butyrate (Sodium Butyrate 98%, Sigma Aldrich, St. Louis, Missouri, Lot#MKCG7272) dissolved in their drinking water. The butyrate supplementation was discontinued when pups were born. Mice assigned to a regular diet received standard laboratory drinking water. Mice in both experimental groups (butyrate and control) were fed regular chow.
Animal models
A series of experiments were performed on the offspring of the breeding pairs. Breeding pairs were randomly selected from cages of males and female mice. In each experiment, breeding pairs of male and female mice originated from the same cage, thereby ensuring that the microbiome composition was homogenous between control and experimental groups at the beginning of the experiment. When appropriate, the offspring were weaned at 3 weeks of age according to their sex and experimental group and received a regular diet with standard laboratory drinking water thereafter.
RNA sequencing
When male offspring from the antenatal butyrate group and the control group reached 3 weeks of age, they were sacrificed, and their colons were harvested for RNA sequencing (RNAseq). There were 9 male offspring from the butyrate group and 8 male offspring from the control group. The sacrifice was performed prior to weaning. Since sex is known to have an effect in biological responses29–31 and the majority of the pups from both samples were males, we only included male offspring in this analysis to prevent bias in our results. Samples were sequenced on an Illumina NovaSeq6000 system by the Emory Integrated Genomics Core (EIGC). Sequences were quality-checked using FastQC for completeness, depth and read quality. Sequences were aligned to the HG38 reference genome using STAR aligner32. Gene quantification was done using HTSeq-count33. Analysis of RNA sequencing data was performed by the Emory Integrated Computational Core (EICC). Further analysis of the RNA sequencing data was done by performing a gene set enrichment analysis34,35 using Molecular Signatures Database Hallmark gene set collection. RNA data sets generated in this study are deposited in gsea-msigdb.org.
DSS-induced colitis
Acute or chronic colitis was induced in offspring from antenatal butyrate group and control group at 6–8 weeks of age through administration of enteral DSS (Thermo Scientific, Fair Lawn, New Jersey, Lot # 187742), established previously as a murine model of colitis36,37. To induce colitis, DSS was dissolved in the drinking water of the mice. The mice in the acute colitis model received 3% DSS for 5 days. Mice in the chronic colitis model received 2% DSS for 7 days, followed by 7 days of recovery with no DSS and then another 7 days of 2% DSS. The degree of colitis was evaluated by using a disease activity index (DAI) scoring, colon length measurement, histomorphological injury scoring and immunofluorescence staining. The DAI scoring is an established method for measuring degree of colitis, taking into account the stool consistency, presence of blood in the stool and percent weight loss of each mouse38. DAI scores range from 0 to 6 and the score is proportional to the degree of intestinal injury with a score of ‘0’ being no injury, and a score of ‘6’ indicating severe colitis. DAI scoring began at day 3 of the DSS colitis model. Colon length is another measure of colonic injury, as colitis causes contraction and shortening of the colon39. Thus, colon length is inversely related to the severity of colitis and was measured at sacrifice.
Hematoxylin-eosin staining and scoring of tissue for intestinal inflammation
The colon of each mouse was fixed in 4% formalin, dehydrated, paraffin embedded and then sectioned. The sections underwent deparaffinization, hydration in alcohol gradient, and staining with Hematoxylin and Eosin (H&E). Images were scored for the degree of colitis using a 2-tiered scoring system, adapted from an established scoring guideline 40. The histologic activity index (HAI) score is based on the quality and dimension of inflammatory cell infiltrates and mucosal architecture. HAI score ranges from 0–6 and the score is directly proportional to the degree of injury. The inflammatory cell infiltrates are considered mild, moderate or severe based on the degree of inflammatory cells in the colonic tissue. Inflammatory cell infiltrates in the mucosa are considered mild (score of 1), in the mucosa and submucosa are considered moderate (score of 2) and transmural inflammatory cell infiltrates are considered severe (score of 3). Next, the extent of epithelial injury is determined based on architectural epithelial changes such as focal erosions (score of 1), erosions with focal ulcerations (score of 2) or extended ulcerations with or without granulation tissue or pseudo polyps (score of 3). The HAI score is then calculated as a combination of the inflammatory cell infiltrate score (0–3) and epithelial architecture score (0–3). Sections were scored in blinded fashion.
Antibodies and immunofluorescence
Sections underwent deparaffinization, hydration and blockage in 5% goat serum albumin (GSA) at 4° Celsius. Paraffin sections were then incubated with the primary antibody β-Catenin mouse antibody cat# 610154, (Brand BD Transduction Laboratories, San Jose, CA) to evaluate the health of the epithelium and ZO-1 rabbit anti-human antibody cat #108952 (GeneTex, Irvine, CA) to evaluate the colonic tight junctions. On the second day, sections were washed three times with phosphate-buffered saline (PBS) for 5 minutes and then incubated with secondary antibodies for 1 hour at room temperature. The nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). ZO-1 immunofluorescence was measured using ImageJ software, measuring the fluorescence and calculating the corrected total cell fluorescence (CTCF).
Microbiome analysis
Stool samples were collected from 6 to 8-week-old offspring prior to colitis experiments. The stool pellets were analyzed by 16S ribosomal RNA (rRNA) gene microbiome analysis by ZymoBIOMICS (Zymo Research, Irvine, CA). DNA was extracted from stool using ZymoBIOMICS-96 MagBead DNA kit (Zymo Research, Irvine, CA). The ZymoBIOMICS Microbial Community Standard was used as a positive control for each DNA extraction. The ZymoBIOMICS Microbial Community DNA Standard was used as a positive control for each targeted library preparation. Negative controls were included to assess the level of bioburden carried by the wet-lab process. The DNA samples were then prepared for targeted sequencing with Quick-16S Primer Set V3-V4 Library (Zymo Research, Irvine, CA). The sequencing library was prepared using innovative library preparation process in which PCR reactions were performed in real-time PCR machines to control cycles and limit PCR chimera formation. The final PCR products were quantified with qPCR fluorescence readings and pooled together based on equal molarity. The final pooled library was cleaned up with the Select-a-Size DNA Clean & Concentrator (Zymo Research, Irvine, CA), then quantified with TapeStation (Agilent Technologies, Santa Clara, CA) and Qubit (Thermo Fisher Scientific, Waltham, WA). The final library was sequenced on Illumina MiSeq with a v3 reagent kit (600 cycles). The sequencing was performed with 10% PhiX spike-in.
Statistics
Statistical significance was determined by Student’s T test using Prism 8 (GraphPad, San Diego, CA) for paired samples, unless stated otherwise. A p-value of <0.05 was defined as statistically significant. All data are presented as mean ± standard error of the mean (SEM), unless stated otherwise. For RNA sequencing, DESeq2 was used to determine differentially expressed genes between the two experimental groups41–43. Genes with low counts were filtered by mean normalized counts in DESeq2. Raw p-values generated were transformed using the Benjamini-Hochberg correction to account for multiple hypothesis testing. Genes considered significantly differentially expressed were those with high posterior likelihoods and FDR ≤ 0.01.
RESULTS
Antenatal butyrate supplementation down regulates the expression of inflammation-associated genes in the colon of 3-week-old offspring
Male and female C57BL/6 mice originating from the same cages were randomly sorted into breeding pairs and received regular chow with either butyrate supplementation (butyrate) or no butyrate supplementation (control) during pregnancy. Three-week-old male offspring from each experimental group were sacrificed, their colons were harvested, and their RNA was isolated for sequencing (Fig. 1a). A total of 9 male offspring from the butyrate group and 8 male offspring from the control group were sacrificed. RNA sequencing analysis detected that 11 genes were upregulated, and 26 genes were down regulated in the colon of 3-week-old offspring as a result of antenatal butyrate supplementation (Fig. 1b). Of the 37 genes that were differentially expressed between the antenatal butyrate group and the control group offspring, 17 genes have ascribed functions (Fig. 1c). Of those 17 genes, 5 were associated with cholesterol synthesis and/or lipid metabolism, all of which were downregulated in offspring from the antenatal butyrate group and 3 genes were involved in inflammatory signaling and were downregulated in offspring from the antenatal butyrate group (Fig. 1d). Furthermore, analysis of RNA sequencing data set using Molecular Signatures Database Hallmark gene set collection revealed 11 signaling pathways that were negatively enriched with antenatal butyrate supplementation and were statistically significant based on nominal p-value (Fig 1e). One of these pathways, TNFα signaling via NFκB was negatively enriched with antenatal butyrate supplementation, with a false discovery rate (FDR) <25% and a nominal p-value <0.01 (Fig.1f). Of note, 43 genes were identified in the leading-edge subset of the TNFα via NF-κB signaling pathway (Fig. 1g).
Figure 1: Antenatal butyrate supplementation down regulates the expression of inflammation-associated genes in the gut of 3-week-old offspring.

a Experimental design. Pregnant dams were randomly assigned to either a diet with 1% butyrate supplementation or no butyrate supplementation during pregnancy. Three-week-old male offspring from antenatal butyrate group and no antenatal butyrate group (control group) were sacrificed and their colons were harvested for RNA sequencing analysis (n>5 per group).
b Volcano plot of RNA sequencing data, FDR ≤ 0.01 with Log (2) fold change cut off set at 1.0. 11 genes were uprated while 26 genes were down regulated with antenatal butyrate exposure.
c Heatmap of RNA sequencing data. 37 genes were differentially expressed between control group and butyrate group, with 17 of these genes present on heatmap, have a known function. Color and intensity of boxes represent counts transformed using a variance stabilizing transformation, similar to a log2 transformation but accounting for sampling variability of genes with low counts. Samples show some separation by group in differentially expressed genes.
d Normalized counts plots of three genes, Nos2, Pla2g2a and Slpi which were down regulated with antenatal butyrate exposure and associated with inflammation.
e Analysis of RNA sequencing data from Gene Set Enrichment Analysis showed 11 statistically significant pathways associated with negative enrichment with antenatal butyrate exposure.
f Enrichment plot from Gene Set Enrichment Analysis of Hallmark revealed antenatal butyrate was negative associated with TNFα via NF-κβ signaling pathway.
g Heatmap from Gene Set Enrichment Analysis of 43 genes involved in TNFα via NF-κβ signaling pathway. Upregulated genes are colored red (positive z-scores) and down regulated genes are colored blue (negative z-scores).
*p<0.05, **p<0.01 by Student’s T test using Prism 8 (GraphPad, San Diego, CA) for paired samples.
Antenatal butyrate supplementation elicits cytoprotection in offspring subjected to a model of acute colitis
In order to examine if the differential gene expression detected by RNA sequencing had physiological consequences, colitis was induced in the six to eight-week-old offspring from each experimental group. Breeding pairs were randomly selected to receive butyrate supplementation during pregnancy (butyrate) or no butyrate supplementation during pregnancy (control). For breeding pairs in butyrate group, butyrate supplemetation was discontinued on the day offspring were born. The offspring of both groups were weaned at 3-weeks of age, according to sex and experimental group. After weaning, the offspring were housed in cages with standard laboratory drinking water and regular chow up until 6 to 8-weeks of age whereupon they were subjected to a model of acute colitis through DSS-induced injury (Fig. 2a). In this approach, 3% DSS was included in the drinking water of offspring from both groups. There were 15 mice in the butyrate group and 11 mice in the control group. The severity of colitis was measured by DAI score of each mouse starting at day 3 following the introduction of DSS into the drinking water. The mice were sacrificed at day 5 following the commencement of DSS treatments and colonic tissue was harvested. Offspring whose mothers received butyrate supplementation during pregnancy had significantly lower DAI scores compared to the control groups on days 3, 4 and 5 of the acute colitis experiments (p-value <0.01 on each day of DAI measurement) (Fig. 2b). In addition, at sacrifice, mice from the antenatal butyrate group had significantly longer colon lengths, compared to controls (p-value <0.05) (Fig. 2c). Furthermore, HAI scoring revealed that the offspring exposed to butyrate during fetal development had significantly lower colonic injury compared to controls (Fig.2d–e). Immunofluorescence staining using antibodies against the tight junction protein ZO-1 detected higher abundance of this protein in the butyrate group (Fig. 2f). Consistent with H&E staining, immunofluorescence staining using antibodies against β-catenin (Fig. 2g) revealed markedly preserved colonic morphology and physiological architecture in mice whose mothers received butyrate supplementation during pregnancy. By contrast, mice from the control group exhibited considerable epithelial erosion and marked tissue damage.
Figure 2: Antenatal butyrate supplementation elicits cytoprotection in offspring subjected to a model of acute colitis.
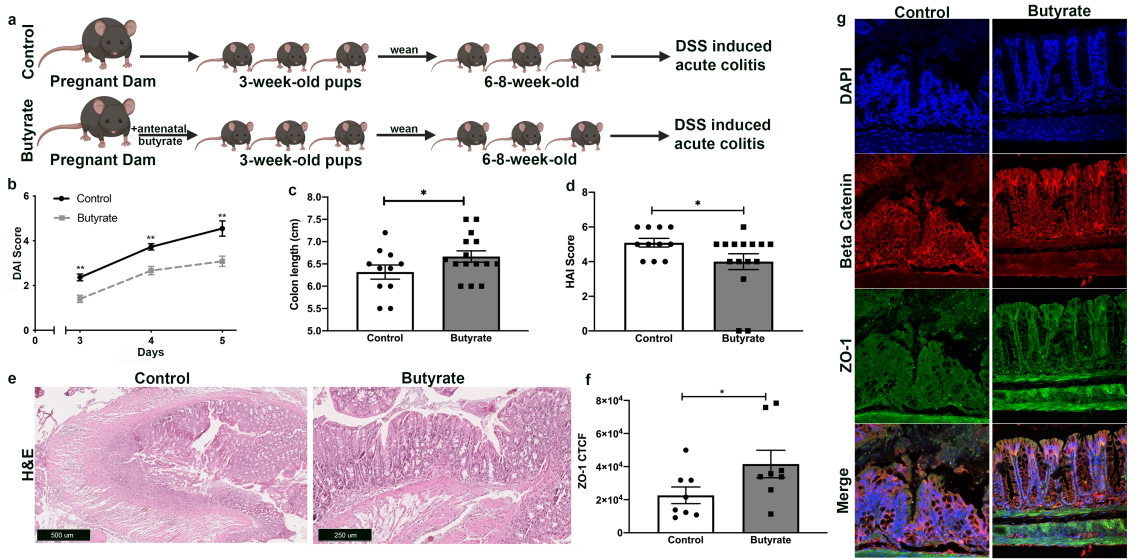
a Experimental design. 6–8-week-old offspring from mating pairs who received butyrate supplementation during pregnancy (butyrate) or no butyrate supplementation during pregnancy (control) were exposed to 3% DSS for 5 days, inducing acute colitis.
b Quantification of intestinal injury by measuring the DAI score on days 3–5 of colitis model for each mouse (n >10 per group; **p <0.01). DAI score ranges from 0 to 6 and is proportional to degree of intestinal injury.
c The colon length of each mouse was measured at the time of sacrifice, (n>10 per group, *p<0.05).
d H&E of proximal colonic tissue obtained at end of acute colitis experiment. HAI score quantifies colonic injury by a validated scoring tool which revealed a decreased injury score in antenatal butyrate offspring. HAI score ranges from 0 to 6 and is proportional to degree of intestinal injury, (n>10 per group; *p<0.05).
e Representative H&E images of proximal colonic tissue at end of acute colitis experiment. Scale bar for control group is 500 𝜇m, scale bar for butyrate group is 250 𝜇m
f Quantification of fluorescence intensity, corrected total cell fluorescence (CTCF), using ImageJ (*p<0.05).
g Photomicrographs of colonic cells, at end of acute colitis experiment, immunostained for ZO-1(green), beta catenin (red) and DAPI (blue).
*p<0.05, **p<0.01 by Student’s T test using Prism 8 (GraphPad, San Diego, CA) for paired samples.
Antenatal butyrate supplementation elicits cytoprotection in offspring subjected to a model of chronic colitis
Next, we induced a chronic colitis injury model in 6 to 8-week-old offspring from each experimental group (Fig. 3a). In this model, 2% DSS was included in the drinking for 7 days and DAI was measured. After the first 7 days of 2% DSS, DSS was removed whereupon the mice had a recovery period of 7 days. They then received 2% DSS in the drinking for 7 more days. There were 8 mice in the butyrate group and 5 mice in the control group. Similar to the DSS acute colitis model, the offspring whose mothers received antenatal butyrate supplementation had significantly lower DAI scores compared to the control group between days 3 and 7 days (first round of 2% DSS), and between days 18 and 22 (second round of 2% DSS) (Fig. 3b). In addition, the offspring from the antenatal butyrate group exhibited faster recovery from the first round of DSS with lower DAI scores on the first two days of recovery at days 8 and 9 (Fig. 3b). The mice were sacrificed at day 22 of the chronic DSS treatments and colonic tissue was harvested. The colon lengths of mice whose mothers were supplemented with butyrate were markedly longer than controls but did not reach statistical significance (p-value: 0.06) (Fig. 3c). Interestingly, upon evaluation of HAI, there was no statistically significant differences detected (Fig. 3d), and no significant differences in the level of ZO-1 immunofluorescence were detected (Fig. 3f). Nevertheless, histological analysis by H&E staining (Fig. 3e) and by immunofluorescence staining (Fig. 3g), revealed markedly preserved colonic morphology and physiological architecture of mice whose mothers received butyrate supplementation during pregnancy, whereas mice from the control group exhibited marked inflammatory cell infiltration, epithelial erosion and tissue damage.
Figure 3. Antenatal butyrate supplementation elicits cytoprotection in offspring subjected to a model of chronic colitis.
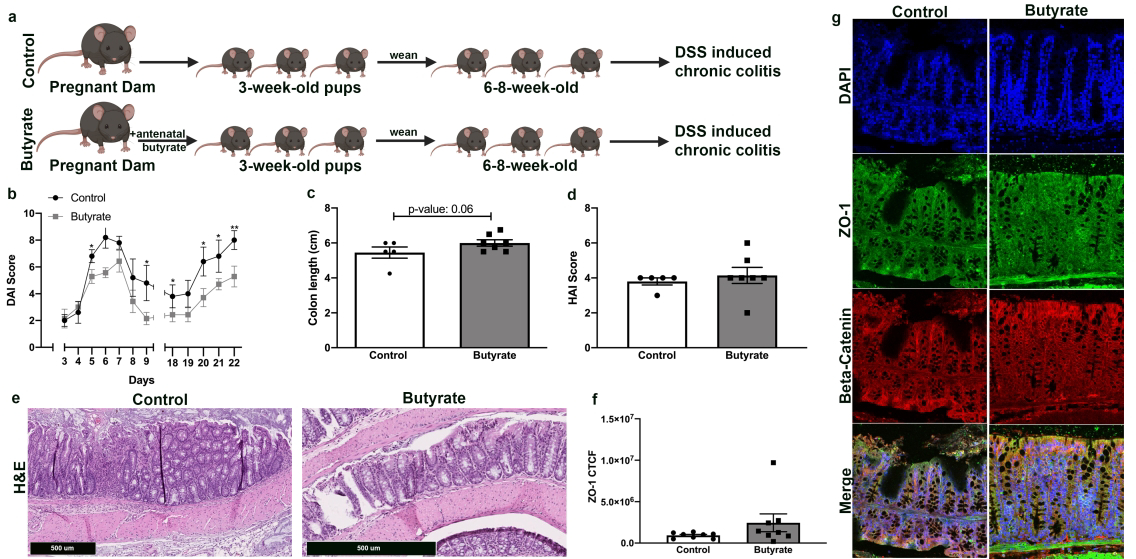
a Experimental design. 6–8-week-old offspring from mating pairs who received butyrate supplementation during pregnancy (butyrate) or no butyrate supplementation during pregnancy (control) were exposed to 2% DSS for 7 days, followed by 7–10 days of recovery with no DSS, then another 7 days of 2% DSS, inducing chronic colitis.
b Quantification of intestinal injury by measuring the DAI score starting on day 3 of DSS exposure for the first and second round of DSS administration and on the first two days of recovery, days 8 and 9 on the graph (n ≥ 5 per group; *p<0.05, **p<0.001). DAI score ranges from 0 to 6 and is proportional to degree of intestinal injury.
c The colon length of each mouse was measured at the time of sacrifice (n ≥ 5 per group).
d H&E of proximal colonic tissue obtained at end of chronic colitis experiment. HAI score quantifies colonic injury by a validated scoring tool did not detect a difference in colonic injury, (n>5 per group). HAI score ranges from 0 to 6 and is proportional to degree of intestinal injury.
e Representative H&E images of colonic tissue proximal colonic tissue at end of chronic colitis experiment (scale bar = 500 𝜇m).
f Quantification of fluorescence intensity, corrected total cell fluorescence (CTCF), using ImageJ.
g Photomicrographs of colonic cells, at end of chronic colitis experiment, immunostained for ZO-1(green), beta catenin (red) and DAPI (blue).
*p<0.05, **p<0.01 by Student’s T test using Prism 8 (GraphPad, San Diego, CA) for paired samples
Antenatal butyrate supplementation is associated with greater fecal microbiome diversity among 6 to 8-week-old offspring
Since dietary modifications can change the gut microbiome, 16S rRNA gene sequencing was conducted on stool samples collected from 6 to 8-week-old male offspring from each experimental group (n=5 per group). Firmicutes was the most abundant phyla in both groups, followed by Bacteroidetes. The offspring from the antenatal butyrate group had higher alpha diversity compared to controls, as evidenced by higher phylogenetic diversity (PD) index (p-value <0.05), Chao 1 diversity index (p-value 0.059) and observed diversity index (p-value 0.063) (Fig. 4a). In addition, based on analysis of Bray-Curtis distance metric, the antenatal butyrate supplemented group had higher beta diversity (p-value <0.01) (Fig. 4b). The community structure of individual samples was compared by principal coordinate analysis (PCoA) of beta diversity. The PCoA plot revealed a clear separation between samples based on antenatal dietary supplementation (Fig. 4c). Furthermore, at the family taxonomic level, the antenatal butyrate supplemented group had higher levels of Clostridiales defluviitaleaceae, and Mollicutes (Fig. 5a). In the antenatal butyrate group, enrichments in Bifidobacteriaceae and Actinobacteriacea were noted while the control group had enrichments in Coriobacteriaceae (Fig. 5b). Finally, the presence of gut microbes from the order Bifidobacteriales was identified in the butyrate group, but undetected in the control group. Specifically, analysis to the species level detected Bifidobacterium choreinum pseudolongum in the butyrate group, but not in the control group. Indeed, at the species level, the antenatal butyrate supplemented group had anywhere from 0.3%−3.7% Bifidobacterium choreinum pseudolongum within the microbiome while the offspring from the control group had no detectable Bifidobacterium (Fig. 5c).
Figure 4: Antenatal butyrate supplementation is associated with greater microbiome diversity among 6–8-week-old offspring.

a Box and whisker plots showing fecal microbial diversity based on Shannon, Observed, Chao 1 and phylogenetic diversity (PD) indices. Horizontal line in middle of each box represents the median and the top and bottom borders of each box represent the 75th and 25th percentiles, respectively. P value for Shannon index 0.37, P value for Observed index 0.063, P value for Chao1 index 0.059, P value for PD index <0.05; (n = 5 per group; *p<0.05).
b Box plot of Bray-Curtis distance as a measure of beta diversity; (n = 5 per group; **p<0.01).
c Three-dimensional principal coordinates analysis (PCoA) based on Bray-Curtis distance between bacterial communities.
Figure 5: Antenatal butyrate supplementation is associated with greater fecal microbiome diversity among 6 to 8-week-old offspring.

a Composition bar chart comparing the relative abundance at bacterial family level between offspring from antenatal butyrate and control groups (n=5 per group; *p<0.05). P value for Coriobacteriaceae is 0.039, P value for Defluviitaleaceae is 0.025, P value for Mollicuteso family unknown is 0.03, P value for Verrucomicrobiaecea is 0.049.
b A cladogram representation of relative abundance of bacteria between offspring from antenatal butyrate and control groups. Red, taxa enriched in offspring from antenatal butyrate group; green, taxa enriched in offspring from control group (n=5 per group).
c Relative abundance of Bifidobacterium in stool from offspring from antenatal butyrate and control groups (n=5 per group).
DISCUSSION
There is a growing body evidence which highlight the influence of the antenatal diet on the offspring’s microbiome and immune function44. In this study, we show that butyrate supplementation to pregnant mice leads to enduring changes in the offspring’s colonic gene expression, microbiome composition and importantly, protection against colonic injury in adolescent aged offspring. To evaluate the effect of antenatal butyrate supplementation on fetal gut development, we first evaluated the offspring’s colonic gene expression. Antenatal butyrate supplementation led to changes in colonic gene expression in the offspring, with 37 differentially expressed genes, of which 17 have established function. Notably, antenatal butyrate supplementation was associated with a down regulation of genes involved in inflammatory signaling and cholesterol synthesis in 3-week-old offspring. The inflammatory genes downregulated by antenatal butyrate include nitric oxide synthase II (NOS2), phospholipase A2 group IIA (PLA2G2A) and secretory leukocyte protease inhibitor (SLPI). NOS2 is an enzyme responsible for the production of NO. When NO is released in an inflammatory site it leads to the recruitment and activation of leukocytes, enhancing the inflammatory process26. PLA2G2A encodes a protein which is a secreted enzyme involved in host antimicrobial defense, inflammatory response and tissue regeneration45. SLPI inhibits several leukocyte serine proteases46 and plays important roles in normal neutrophil formation and the recruitment of innate and adaptive immune cells to sites of inflammation26,47. Thus, antenatal butyrate led to down regulation of multiple genes that are integral to proinflammatory signaling.
Next, we challenged the offspring with models of DSS-induced acute or chronic colitis. DSS induces colonic injury by causing epithelial damage, resulting in bacterial translocation and a massive inflammatory response in colonocytes48. Based on butyrate’s effects on inflammatory pathways detected by RNA sequencing, we expected offspring from the antenatal butyrate group to have less colonic injury when exposed to DSS. We evaluated the severity of colonic injury by measuring the DAI, colon length and histologically by HAI scoring. The colitis model was induced when the offspring reached 6 to 8-weeks of age, which is equivalent to adolescence-early adulthood in humans. Through the DSS-induced acute colitis model, we found that mice exposed to antenatal butyrate supplementation had significantly lower colonic injury, as evidenced by lower DAI scores, longer colon lengths and lower HAI scores. Thus, butyrate supplementation during pregnancy resulted in protection against acute colitis in adolescent aged offspring. Antenatal butyrate supplementation also resulted in lower DAI scores among offspring in the butyrate group during the chronic colitis model. The colon length of the mice in the chronic colitis model did not reach statistical significance (p-value: 0.06), which may be due to lower power intrinsic in the experimental design. Despite the DAI scores, we did not find a significant difference in the HAI scoring which may be due to limitations of the scoring matrix itself since we detected that mice in the control group had preserved architecture (as detected by beta-catenin staining) compared to the butyrate group.
Finally, we evaluated the microbiome of 6 to 8-week-old offspring prior to starting the DSS-induced colitis model to determine whether the antenatal butyrate supplementation had induced changes in the offspring’s microbiome composition. Through 16S rRNA analysis, we found that there is broader diversity, both alpha (PD) and beta diversity, in the gut microbiome of mice whose mothers received butyrate supplementation during pregnancy. Additionally, we found that butyrate induced the expansion of the abundance of Bifidobacteria while the control mice had no detectable level of Bifidobacteria. Intriguingly, a systematic review of patients with inflammatory bowel disease (IBD) found that compared to patients in remission, patients with active IBD had lower abundance of Bifidobacterium in their stool (mean difference: −0.37, confidence interval: −0.56 to −0.17)49. In additon, in a study of infant gut dysbiosis, absence of Bifidobacterium infantis was associated with increased intestinal inflammation50. Thus, the expansion of Bifidobacterium among mice exposed to butyrate during fetal development may partly explain their protection against colitis.
The fetal-maternal dyad is complex and there are many possible explanations to account for our findings. First, we postulate that antenatal butyrate supplementation may cause an increase in maternal serum butyrate levels which then may be transferred vertically, through the placenta into the fetus and potentiate local effects on the colonic gene expression in the fetus. A second possibility is that antenatal butyrate supplementation may also result in increased butyrate levels in the amniotic fluid, leading to local changes in the fetus’ colonic gene expression through butyrate’s properties as an HDAC inhibitor. In support of this notion is a report of human amniotic fluid containing SCFAs4. Third, antenatal butyrate supplementation may cause changes in the maternal microbiome composition, which is then passed on to the offspring. Butyrate-mediated modification to the microbiome may lead to protection against colitis or it may lead to higher levels of butyrate production and hence changes to colonic gene expression in the offspring. Fourth, if the fetus is indeed exposed to microbes in utero, this may have an effect on local butyrate levels in the colon, resulting in gene expression changes in utero. Identifying which of these mechanisms drive the antenatal effects of butyrate on the offspring is the focus of intense scrutiny in our research group.
In conclusion, antenatal butyrate supplementation mitigates intestinal injury in adolescent/early adulthood aged murine offspring in the setting of colitis. Our experiments support other studies which have shown that maternal dietary intervention during pregnancy can have enduring effects on the offspring’s gastrointestinal health. For example, it has been reported that in mice, a high fat diet during pregnancy changes the offspring’s microbiome and increases the offspring’s susceptibility to gut inflammatory challenges51. In addition, other animal models have reported that maternal high-fat diet and maternal obesity increase the offspring’s susceptibility to developing inflammatory bowel disease (IBD)52–57. Furthermore, it has been reported that administration of the microbially-derived aryl hydrocarbon receptor (AHR) ligand indole-3-carbinol (I3C) to pregnant mice prevents necrotizing enterocolitis in offspring58. Although there are a limited number of human studies that have focused on the role of the antenatal diet, these pre-clinical studies in mice serve to support the notion that the maternal diet can shape the microbiome composition of both the mother and the offspring17. Together, these examples comprise a growing body of literature that highlight the influence of antenatal diet on the offspring’s health.
In our study, the protective effects of antenatal butyrate in the setting of colonic injury are likely from the butyrate-induced down regulation of genes involved in inflammatory signaling. In addition, antenatal butyrate supplementation has an effect on the offspring’s microbiome, increasing its diversity and expanding the presence of bacteria that may be protective in the setting of colitis. Therefore, in addition to directly influencing gene expression, antenatal butyrate can exert influence on the microbiome diversity, which is firmly established as a key modulator of host immunity and overall health59. Together, our data may support future clinical studies focused on assessing the extent to which butyrate supplementation during pregnancy protects offspring from colonic inflammation and injury. This would be relevant to women at risk of preterm delivery as preterm infants are at higher risk of intestinal inflammation60, or in obese women as their offspring are at risk of developing IBD57. At this time, further studies are needed to determine the precise mechanisms and functional elements that mediate antenatal butyrate’s protection against colitis in offspring. Nevertheless, our data highlights the important potential impact of the antenatal diet on offspring health and disease and demonstrates that antenatal supplementation with bioactive gut microbe-generated metabolites could imprint lasting effects on the offspring’s responses to gut injury later in life.
IMPACT.
Dietary butyrate supplementation to pregnant mice led to down regulation of colonic genes involved in inflammatory signaling and cholesterol synthesis, changes in the fecal microbiome composition of the offspring, and protection against experimentally induced colitis in offspring.
These data support the mounting evidence that the maternal diet during pregnancy has enduring effects on the offspring’s long-term health and disease risk.
Although further investigations are needed to identify the mechanism of butyrate’s effects on fetal gut development, the current study substantiates the approach of dietary intervention during pregnancy to optimize the long-term gastrointestinal health of the offspring.
ACKNOWLEDGEMENTS
The Emory Integrated Core Facilities are supported by the Emory Neuroscience NINDS Core Facilities, the Georgia Clinical & Translational Science Alliance, and the Emory University School of Medicine.
Financial support: MEB is supported in part by American Academy of Pediatrics Marshall Klaus Neonatal-Perinatal Research Award and Emory University Department of Pediatrics and Children’s Healthcare of Atlanta Warshaw Fellow’s Research Award. JAO is supported by the NIH through F31CA247415. CRN is supported by an American Heart Association fellowship 19POST34370006. PD is supported by NIH through 5P01HL086773. RMP is supported by NIH through K23HL128942. RMJ is supported, in part, by NIH Grant R01DK098391 and R01CA179424
Footnotes
Disclosures: The authors have no conflicts of interest.
Figures 1a, 2a and 3a were created with Biorender.com.
REFERENCES
- 1.Jones RM & Neish AS Gut Microbiota in Intestinal and Liver Disease. Annu Rev Pathol (2020). [DOI] [PubMed] [Google Scholar]
- 2.Aagaard K et al. The Placenta Harbors a Unique Microbiome. Sci Transl Med 6, 237ra265 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra A et al. Microbial Exposure During Early Human Development Primes Fetal Immune Cells. Cell 184, 3394–3409 e3320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stinson LF, Boyce MC, Payne MS & Keelan JA The Not-So-Sterile Womb: Evidence That the Human Fetus Is Exposed to Bacteria Prior to Birth. Front Microbiol 10, 1124 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rackaityte E et al. Viable Bacterial Colonization Is Highly Limited in the Human Intestine in Utero. Nat Med 26, 599–607 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jimenez E et al. Is Meconium from Healthy Newborns Actually Sterile? Res Microbiol 159, 187–193 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Hu J et al. Diversified Microbiota of Meconium Is Affected by Maternal Diabetes Status. PLoS One 8, e78257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Goffau MC et al. Human Placenta Has No Microbiome but Can Contain Potential Pathogens. Nature 572, 329–334 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perez-Munoz ME, Arrieta MC, Ramer-Tait AE & Walter J A Critical Assessment of the “Sterile Womb” and “in Utero Colonization” Hypotheses: Implications for Research on the Pioneer Infant Microbiome. Microbiome 5, 48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez JM et al. The Composition of the Gut Microbiota Throughout Life, with an Emphasis on Early Life. Microb Ecol Health Dis 26, 26050 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benson AK et al. Individuality in Gut Microbiota Composition Is a Complex Polygenic Trait Shaped by Multiple Environmental and Host Genetic Factors. Proc Natl Acad Sci U S A 107, 18933–18938 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKnite AM et al. Murine Gut Microbiota Is Defined by Host Genetics and Modulates Variation of Metabolic Traits. PLoS One 7, e39191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez-Bello MG et al. Delivery Mode Shapes the Acquisition and Structure of the Initial Microbiota across Multiple Body Habitats in Newborns. Proc Natl Acad Sci U S A 107, 11971–11975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yatsunenko T et al. Human Gut Microbiome Viewed across Age and Geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maher SE et al. The Association between the Maternal Diet and the Maternal and Infant Gut Microbiome: A Systematic Review. Br J Nutr, 1–29 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Chu DM et al. The Early Infant Gut Microbiome Varies in Association with a Maternal High-Fat Diet. Genome Med 8, 77 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mirpuri J Evidence for Maternal Diet-Mediated Effects on the Offspring Microbiome and Immunity: Implications for Public Health Initiatives. Pediatr Res (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belkaid Y & Hand TW Role of the Microbiota in Immunity and Inflammation. Cell 157, 121–141 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macfarlane GT & Macfarlane S Bacteria, Colonic Fermentation, and Gastrointestinal Health. Journal of AOAC International 95, 50–60 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Bach Knudsen KE Microbial Degradation of Whole-Grain Complex Carbohydrates and Impact on Short-Chain Fatty Acids and Health. Advances in nutrition 6, 206–213 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong JM, de Souza R, Kendall CW, Emam A & Jenkins DJ Colonic Health: Fermentation and Short Chain Fatty Acids. Journal of clinical gastroenterology 40, 235–243 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Fernandes J, Su W, Rahat-Rozenbloom S, Wolever TM & Comelli EM Adiposity, Gut Microbiota and Faecal Short Chain Fatty Acids Are Linked in Adult Humans. Nutrition & diabetes 4, e121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamer HM et al. Review Article: The Role of Butyrate on Colonic Function. Aliment Pharmacol Ther 27, 104–119 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Goncalves P, Araujo JR & Di Santo JP A Cross-Talk between Microbiota-Derived Short-Chain Fatty Acids and the Host Mucosal Immune System Regulates Intestinal Homeostasis and Inflammatory Bowel Disease. Inflamm Bowel Dis 24, 558–572 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Mowat AM & Agace WW Regional Specialization within the Intestinal Immune System. Nat Rev Immunol 14, 667–685 (2014). [DOI] [PubMed] [Google Scholar]
- 26.Vinolo MA et al. Suppressive Effect of Short-Chain Fatty Acids on Production of Proinflammatory Mediators by Neutrophils. J Nutr Biochem 22, 849–855 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Venkatraman A et al. Amelioration of Dextran Sulfate Colitis by Butyrate: Role of Heat Shock Protein 70 and Nf-Kappab. Am J Physiol Gastrointest Liver Physiol 285, G177–184 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Aguilar EC et al. Butyrate Impairs Atherogenesis by Reducing Plaque Inflammation and Vulnerability and Decreasing Nfkappab Activation. Nutr Metab Cardiovasc Dis 24, 606–613 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Markle JG et al. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science 339, 1084–1088 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Cross TL, Kasahara K & Rey FE Sexual Dimorphism of Cardiometabolic Dysfunction: Gut Microbiome in the Play? Mol Metab 15, 70–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elderman M, de Vos P & Faas M Role of Microbiota in Sexually Dimorphic Immunity. Front Immunol 9, 1018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dobin A et al. Star: Ultrafast Universal Rna-Seq Aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anders S, Pyl PT & Huber W Htseq--a Python Framework to Work with High-Throughput Sequencing Data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mootha VK et al. Pgc-1alpha-Responsive Genes Involved in Oxidative Phosphorylation Are Coordinately Downregulated in Human Diabetes. Nat Genet 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Wirtz S et al. Chemically Induced Mouse Models of Acute and Chronic Intestinal Inflammation. Nat Protoc 12, 1295–1309 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Darby TM et al. Lactococcus Lactis Subsp. Cremoris Is an Efficacious Beneficial Bacterium That Limits Tissue Injury in the Intestine. iScience 12, 356–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang L et al. Heme Oxygenase-1 Ameliorates Dextran Sulfate Sodium-Induced Acute Murine Colitis by Regulating Th17/Treg Cell Balance. J Biol Chem 289, 26847–26858 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao H et al. Protective Effect of Naringin on Dss-Induced Ulcerative Colitis in Mice. J Agric Food Chem 66, 13133–13140 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Erben U et al. A Guide to Histomorphological Evaluation of Intestinal Inflammation in Mouse Models. Int J Clin Exp Pathol 7, 4557–4576 (2014). [PMC free article] [PubMed] [Google Scholar]
- 41.Hardcastle TJ & Kelly KA Bayseq: Empirical Bayesian Methods for Identifying Differential Expression in Sequence Count Data. BMC Bioinformatics 11, 422 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson MD, McCarthy DJ & Smyth GK Edger: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W & Anders S Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mirpuri J Evidence for Maternal Diet-Mediated Effects on the Offspring Microbiome and Immunity: Implications for Public Health Initiatives. Pediatr Res 89, 301–306 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishizaki J et al. Cloning and Characterization of Novel Mouse and Human Secretory Phospholipase a(2)S. J Biol Chem 274, 24973–24979 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Barrios VE, Jarosinski MA & Wright CD Proteinase-Activated Receptor-2 Mediates Hyperresponsiveness in Isolated Guinea Pig Bronchi. Biochem Pharmacol 66, 519–525 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Williams SE, Brown TI, Roghanian A & Sallenave JM Slpi and Elafin: One Glove, Many Fingers. Clin Sci (Lond) 110, 21–35 (2006). [DOI] [PubMed] [Google Scholar]
- 48.Okayasu I et al. A Novel Method in the Induction of Reliable Experimental Acute and Chronic Ulcerative Colitis in Mice. Gastroenterology 98, 694–702 (1990). [DOI] [PubMed] [Google Scholar]
- 49.Prosberg M, Bendtsen F, Vind I, Petersen AM & Gluud LL The Association between the Gut Microbiota and the Inflammatory Bowel Disease Activity: A Systematic Review and Meta-Analysis. Scand J Gastroenterol 51, 1407–1415 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Henrick BM et al. Colonization by B. Infantis Evc001 Modulates Enteric Inflammation in Exclusively Breastfed Infants. Pediatr Res 86, 749–757 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Babu ST et al. Maternal High-Fat Diet Results in Microbiota-Dependent Expansion of Ilc3s in Mice Offspring. JCI Insight 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobson K, Mundra H & Innis SM Intestinal Responsiveness to Experimental Colitis in Young Rats Is Altered by Maternal Diet. Am J Physiol Gastrointest Liver Physiol 289, G13–20 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Yan X et al. Maternal Obesity Induces Sustained Inflammation in Both Fetal and Offspring Large Intestine of Sheep. Inflamm Bowel Dis 17, 1513–1522 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xue Y, Wang H, Du M & Zhu MJ Maternal Obesity Induces Gut Inflammation and Impairs Gut Epithelial Barrier Function in Nonobese Diabetic Mice. J Nutr Biochem 25, 758–764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gruber L et al. Maternal High-Fat Diet Accelerates Development of Crohn’s Disease-Like Ileitis in Tnfdeltaare/Wt Offspring. Inflamm Bowel Dis 21, 2016–2025 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Bibi S, Kang Y, Du M & Zhu MJ Maternal High-Fat Diet Consumption Enhances Offspring Susceptibility to Dss-Induced Colitis in Mice. Obesity (Silver Spring) 25, 901–908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Al Nabhani Z et al. Excess Calorie Intake Early in Life Increases Susceptibility to Colitis in Adulthood. Nat Metab 1, 1101–1109 (2019). [DOI] [PubMed] [Google Scholar]
- 58.Lu P et al. Maternal Aryl Hydrocarbon Receptor Activation Protects Newborns against Necrotizing Enterocolitis. Nat Commun 12, 1042 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones RM & Neish AS Gut Microbiota in Intestinal and Liver Disease. Annu Rev Pathol 16, 251–275 (2021). [DOI] [PubMed] [Google Scholar]
- 60.Lin PW & Stoll BJ Necrotising Enterocolitis. Lancet 368, 1271–1283 (2006). [DOI] [PubMed] [Google Scholar]


